Characterization of open chromatin reveals wide spread occurrence of cis-regulatory DNA sequences derived from transposable elements in the maize genome.
Abstract
Genomic regions free of nucleosomes, which are hypersensitive to DNase I digestion, are known as DNase I hypersensitive sites (DHSs) and frequently contain cis-regulatory DNA elements. To investigate their prevalence and characteristics in maize (Zea mays), we developed high-resolution genome-wide DHS maps using a modified DNase-seq technique. Maize DHSs exhibit depletion of nucleosomes and low levels of DNA methylation and are enriched with conserved noncoding sequences (CNSs). We developed a protoplast-based transient transformation assay to assess the potential gene expression enhancer and/or promoter functions associated with DHSs, which showed that more than 80% of DHSs overlapping with CNSs showed an enhancer function. Strikingly, nearly 25% of maize DHSs were derived from transposable elements (TEs), including both class I and class II transposons. Interestingly, TE-derived DHSs (teDHSs) homologous to retrotransposons were enriched with sequences related to the intrinsic cis-regulatory elements within the long terminal repeats of retrotransposons. We demonstrate that more than 80% of teDHSs can drive transcription of a reporter gene in protoplast assays. These results reveal the widespread occurrence of TE-derived cis-regulatory sequences and suggest that teDHSs play a major role in transcriptional regulation in maize.
Plant growth and development rely on precise spatiotemporal transcription of genes. Transcription is orchestrated by interactions between regulatory proteins and cis-regulatory elements (CREs; Kaufmann et al., 2010; Sparks et al., 2013). Although identification of CREs is of great importance for understanding the regulation of gene expression, this task is impaired by inherent short sequence signatures and the lack of a locational and orientational restriction to CREs, such as enhancers (Marand et al., 2017). In eukaryotic species, nuclear DNA is organized as chromatin, in which DNA is packaged into a string of nucleosomes. Chromatin containing active CREs is required to be accessible for the binding of regulatory proteins, which is achieved by depletion or eviction of nucleosomes (Henikoff et al., 2009; Tsompana and Buck, 2014). Thus, “open chromatin” is prone to cleavage by enzymes compared to immediately neighboring chromatin that is tightly bound by nucleosomes. Several technologies were developed to capture signatures of open chromatin based on their biochemical characteristics, such as FAIRE-seq (Giresi et al., 2007), sono-seq (Auerbach et al., 2009), DNase-seq (Song and Crawford, 2010), and ATAC-seq (Buenrostro et al., 2013). These technologies were successfully applied to identify functional CREs in several model animal species, including humans (The ENCODE Project Consortium, 2004), mouse (Stamatoyannopoulos et al., 2012), Caenorhabditis elegans, and Drosophila melanogaster (Hesselberth et al., 2009; Gerstein et al., 2010). Similar efforts have recently been reported in plants (Zhang et al., 2012a; Zhang et al., 2012b; Pajoro et al., 2014; Sullivan et al., 2014; Cumbie et al., 2015; Qiu et al., 2016), which showed that open chromatin possesses unique epigenetic modifications (Zhang et al., 2012a) and is associated with DNA sequences bound to transcription factors (TFs; Zhang et al., 2012b) and with enhancer function (Zhu et al., 2015). These studies provided insights on the regulatory landscape and transcription factor networks in plant species.
Transposable elements (TEs) are a major DNA component of most plant genomes. TEs are classified into two classes based on their replicative mechanisms. Class I or retrotransposons transpose via an RNA intermediate, whereas class II or DNA transposons transpose via a DNA-mediated mechanism (Wicker et al., 2007). TEs were traditionally considered as “selfish” or “junk DNA” because of their parasitic nature and evolutional neutrality in hosts (Ohno, 1972; Orgel and Crick, 1980). However, recent studies have shown that TEs have a strong influence on the host genomes (Feschotte, 2008; Bennetzen and Wang, 2014). Although TE insertions within genes are frequently detrimental, TEs can play important roles in the evolution of gene regulation (Slotkin and Martienssen, 2007; Feschotte, 2008; Chuong et al., 2017; Hirsch and Springer, 2017). Comparative genomic analysis in animals showed that TE insertions contributed to a large portion of conserved noncoding elements and were under positive selection (Nishihara et al., 2006; Lowe et al., 2007; Mikkelsen et al., 2007). Thus, there has been an increasing amount of evidence suggesting that TEs are a rich source of cis-regulatory elements, either from de novo evolution or from preexisting binding sites within TEs (Rebollo et al., 2012). The participation of TEs in gene regulation has also been reported in plant species (Lisch, 2013; Hirsch and Springer, 2017). For example, in rice (Oryza sativa), the DNA transposon mPing preferentially inserts in the 5′ regions of genes and up-regulates the expression of adjacent genes under stress conditions (Naito et al., 2009). A recent genome-wide study in maize (Zea mays) showed that TEs confer a regulatory role to nearby genes under stresses (Makarevitch et al., 2015). These studies suggest that TEs have a global impact on gene regulation in plant species, although the underlying mechanism is not well understood.
Maize is a classical model plant species for various biological phenomena and is also an important crop. Major research efforts have been devoted to discovering the temporal and spatial transcriptional landscapes in maize (Soderlund et al., 2009; Sekhon et al., 2011, 2013; Chen et al., 2014), which is crucial for the identification and functional annotation of genes. However, scarce information is available describing the underlying regulatory networks of maize transcriptional landscapes. With the sequencing of the maize genome, characterizing the functional elements of the remaining noncoding sequence will be essential to fully understand gene expression and regulation during growth and development. The binding sites of several maize TFs have been identified using chromatin immunoprecipitation (ChIP) coupled with sequencing (ChIP-seq; Bolduc et al., 2012; Morohashi et al., 2012; Eveland et al., 2014; Li et al., 2015; Pautler et al., 2015). In addition, open chromatin associated with micrococcal nuclease (MNase) sensitivity has recently been mapped in maize (Rodgers-Melnick et al., 2016). However, the daunting task of identifying the binding sites of thousands of TFs in the maize genome remains. We developed a modified DNase-seq technique to identify DNase I hypersensitivity sites (DHSs) in the maize genome. We also developed a protoplast-based transient transformation assay to validate the promoter and enhancer functions of randomly selected maize DHSs. We discovered that a significant portion of maize DHSs were derived from TEs, supporting the notion that TEs contribute to gene expression regulation in the maize genome.
RESULTS
Development of the Modified DNase-Seq Protocol in Maize
We initially developed and sequenced a DNase-seq library from maize leaf chromatin using our original DNase-seq protocol that generates ∼20-bp sequence reads (Zhang et al., 2012a). Only 16.1% of these ∼20-bp DNase-seq reads could be mapped to unique positions in the maize genome. By contrast, nearly 70% of the DNase-seq reads can be mapped to unique positions in the Arabidopsis (Arabidopsis thaliana) genome (Zhang et al., 2012b). Thus, the ∼20-bp reads from the original DNase-seq protocol are not ideal for highly repetitive and complex plant genomes such as maize. To overcome this limitation, we modified our DNase-seq library construction method (modified DNase-seq [mDNase-seq]) to generate longer sequence reads (Fig. 1; see details in “Materials and Methods”). The mDNase-seq method generated reads with an average length of 100 bp. We developed mDNase-seq libraries from both leaf and root tissues and generated 176 and 279 million 100-bp single-end reads, respectively. The increased read length effectively doubled the mappability of sequencing data. We mapped 59 million (33.3%) and 98 million (35.2%) mDNase-seq reads, from leaf and root tissues, respectively, to unique locations in the maize genome. These reads were used for further DHS peak calling.
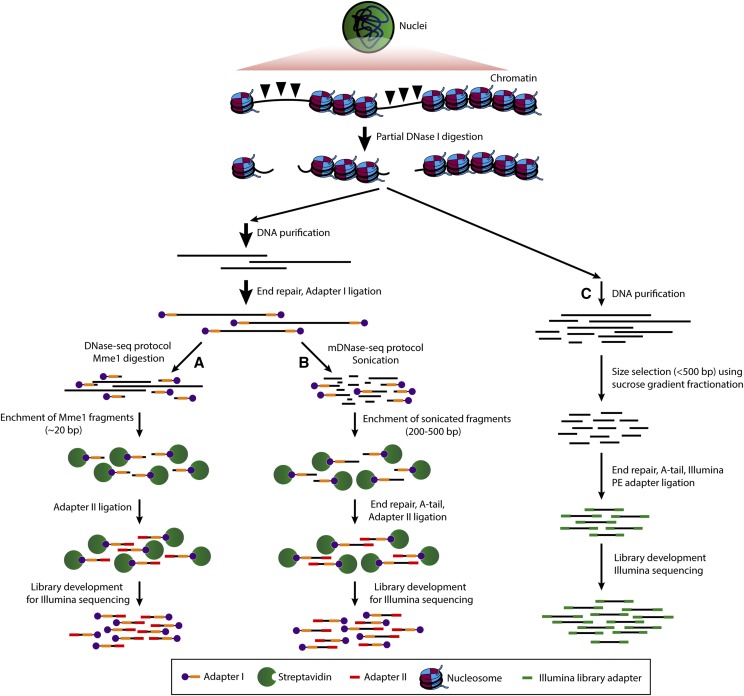
Figure 1.
Principles of the DNase-seq and mDNase-seq used in this study. Chromatin was partially digested by DNase I. A and B, High molecular weight DNA was extracted, end repaired, and ligated with adaptor I. In the traditional DNase-seq protocol (A), the DNA sample is digested by MmeI, resulting in ∼20-bp fragments. In mDNase-seq (B), the DNA was sheared into 200- to 500-bp fragments by sonication. DNA fragments associated with adaptor I were enriched, end repaired, and ligated with adaptor II. DNA fragments with both adaptors were amplified for library development. C, The DNase-seq technique developed by Hesselberth et al. (2009). Chromatin was digested with an amount of DNase I to release short (<300 bp) DNA fragments, which are isolated and sequenced (Hesselberth et al., 2009).
To examine the saturation and reproducibility of our data, we sampled one-quarter, one-half, and three-quarters of the total mDNase-seq data for DHS identification using F-seq (Boyle et al., 2008) and Popera, an in-house developed software (https://github.com/forrestzhang/Popera). The number of peaks identified was linear to the data size (Supplemental Fig. S1A) and did not plateau, suggesting that DHSs were not saturated in our data set. However, most DHSs (86–93%) from the downsampled data sets overlapped with the top 50% of DHSs from the whole data set (Supplemental Fig. S1B), suggesting that the strongest DHSs were consistently identified.
Genomic and Epigenomic Features and Functional Validation of Maize DHSs
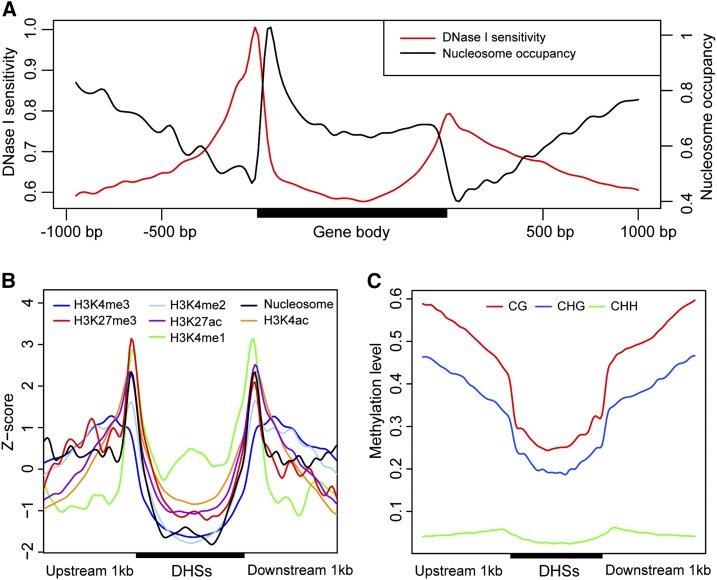
We identified 35,822 and 36,467 DHSs using F-seq (Boyle et al., 2008) and Popera (false discovery rate [FDR] < 0.0064) in leaf and root tissues, respectively. The distribution of DHSs along the maize chromosomes followed a similar pattern as genes but was antagonistic to the density of TEs along the chromosomes (Supplemental Fig. S2A). Indeed, nearly half (47–56%) of DHSs were mapped to genes and their surrounding 1-kb regions (Supplemental Fig. S2B). Similarly, maize genomic regions differentially sensitive to MNase digestion localized mainly around active genes (Rodgers-Melnick et al., 2016). We investigated nucleosome occupancy at genic regions by analyzing the distribution of DNA fragments associated with mononucleosomes (Struhl and Segal, 2013). Briefly, the chromatin was digested into single nucleosomes and the resulted DNA fragments were isolated and sequenced (Zhao et al., 2016). The length of these fragments showed a peak at 147 bp, suggesting that these fragments represent the DNA sequences that are protected by mononucleosomes. DHS peaks around genes coincided with a low level of nucleosome occupancy (Fig. 2A). Maize DHSs showed depletion of nucleosomes, very low levels of histone modifications (Fig. 2B), and DNA methylation (Fig. 2C). Analysis of the differential nuclease sensitivity based on MNase digestion (Rodgers-Melnick et al., 2016) of the DHS regions showed that DHSs are associated with a high levels of differential nuclease sensitivity (Supplemental Fig. S3). Thus, the genomic features of maize DHSs are consistent with the known characteristics of open chromatin observed in both maize (Rodgers-Melnick et al., 2016) and other plant species (Zhang et al., 2012a, 2012b).
Figure 2.
Chromatin characteristics of maize DHSs. A, Distribution of DNase I sensitivity and nucleosome occupancy at genic regions. Note the high levels of DNase I sensitivity and depletion of nucleosomes at the beginning and the end of the gene body. B, Distribution of histone modifications at DHSs. C, Distribution of DNA methylation at DHSs.
Conserved noncoding sequences (CNSs) are considered as regions harboring potential regulatory elements (Freeling and Subramaniam, 2009). We analyzed the profile of mDNase-seq reads that overlapped with CNSs, which were identified based on sequence conservation between maize and rice (Turco et al., 2013). Aggregated mDNase-seq reads showed a clear peak at the center of CNSs (Supplemental Fig. S4). We found that 8,888 of 63,237 (14%) of the CNSs overlap DHSs (Supplemental Resource 1).
Tissue specificity analysis revealed that 22,413 DHSs (62.6%) are specific to leaves and 23,058 DHSs (63.2%) are specific to roots. To analyze the relationship between DNase sensitivity and gene expression in these two tissues, we developed replicated RNA-seq data sets using the same leaf and root tissues that were used for mDNase-seq (see “Materials and Methods”). We identified 28,383 and 28,716 genes differentially expressed in leaves and roots, respectively. DNase sensitivity was positively correlated with gene expression levels (Supplemental Fig. S5A). We then divided the genes into three groups according to the expression patterns in the two tissues: (1) preferentially expressed in leaves, (2) preferentially expressed in roots, (3) and not preferentially expressed in either tissue. DNase I sensitivity at transcriptional start sites (TSSs) was positively correlated with gene expression levels in different tissues (Supplemental Fig. S5B). The genes differentially expressed between leaf and root tissues were commensurate with changes to DNase I sensitivity at TSSs.
Chromatin Accessibility in Centromeric Regions
Centromeric nucleosomes contain CENH3, a centromere-specific H3 variant (Henikoff et al., 2001). Centromeric chromatin often exhibits distinct genomic and epigenomic features (Fukagawa and Earnshaw, 2014). Several maize centromeres have been well sequenced and provide a rare opportunity to analyze chromatin accessibility of maize centromeres. Analysis of the chromosome-wide distribution of DHSs showed that centromeric/pericentromeric regions contain low number of DHSs (Supplemental Fig. S2A). Only 51 DHSs were found within 12.6 Mb of sequenced centromeric regions (4 DHSs/Mb). The density of DHSs in centromeric regions was significantly lower than that in chromosome arms (17 DHS/Mb, empirical P value < 1 × 10−5). We identified 12 centromeric genes that were expressed in leaf and root tissues (Zhao et al., 2016). However, only four genes were associated with DHSs within the gene body or 1-kb flanking regions. This could be a result of the low levels of saturation of our data in addition to the repetitiveness of centromeric sequences. Nevertheless, the centromeric regions showed a low level of chromatin accessibility, which is consistent with a lack of transcriptional activity observed in maize centromeres (Zhao et al., 2016).
Functional Validation of DHSs Using a Protoplast-Based Transient Transformation Assay
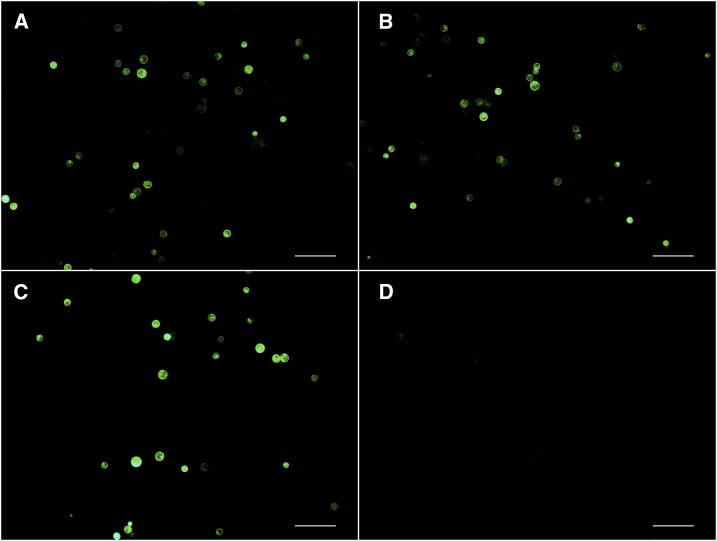
We developed a protoplast-based transient transformation assay (see “Materials and Methods”) to validate the potential regulatory function (enhancer and/or promoter) of putative DHSs. We first selected 11 CNS-associated DHSs (DHSs that overlapped CNSs with at least one base pair) for the assay. DNase sensitivity of these 11 CNS-associated DHSs was within 10 to 90% of all DHSs, representing DNase sensitivity levels of most DHSs. Briefly, target DNA sequences were amplified from genomic DNA of the maize inbred B73 and inserted into a modified pJIT163-hGFP vector that contains GFP driven by a mini35S promoter. The constructs were used to transform maize protoplasts prepared from young leaf tissue. The mini35S promoter alone cannot drive the transcription of GFP in the assay (Figure 3D). Of the 11 DHSs examined (Supplemental Fig. S6), nine (82%) constructs resulted in consistent expression of the GFP marker when the DHS was placed in front of the mini35S in the forward direction, or in both forward and reverse directions (Fig. A and B; Supplemental Table S1). We also examined three DHSs (DHS-a, -b, and -c in Supplemental Fig. S7) that do not overlap with CNSs. These three DHSs were associated with ribulose bisphosphate carboxylase small subunit1 (SSU1; GRMZM2G098520). DHS-c showed enhancer function, whereas both DHS-b and DHS-c showed promoter activity (Supplemental Fig. S7).
Figure 3.
Functional validation of a CNS-cognate DHS using the protoplast-based transient transformation assay. A, Protoplast transformation using a construct with GFP expression driven by the DHS in forward orientation. B, Protoplast transformation using a construct with GFP expression driven by the DHS in reverse orientation. C, Positive control of protoplast transformation using a construct with GFP expression driven by the 35S promoter. D, Negative control of protoplast transformation using a construct with GFP expression driven by a mini35S promoter. Note: Nonambiguous GFP signal was not observed in protoplasts transformed with the negative control. Bars = 10 μm.
DHSs Are Associated with Regulation of Retained Duplicated Genes
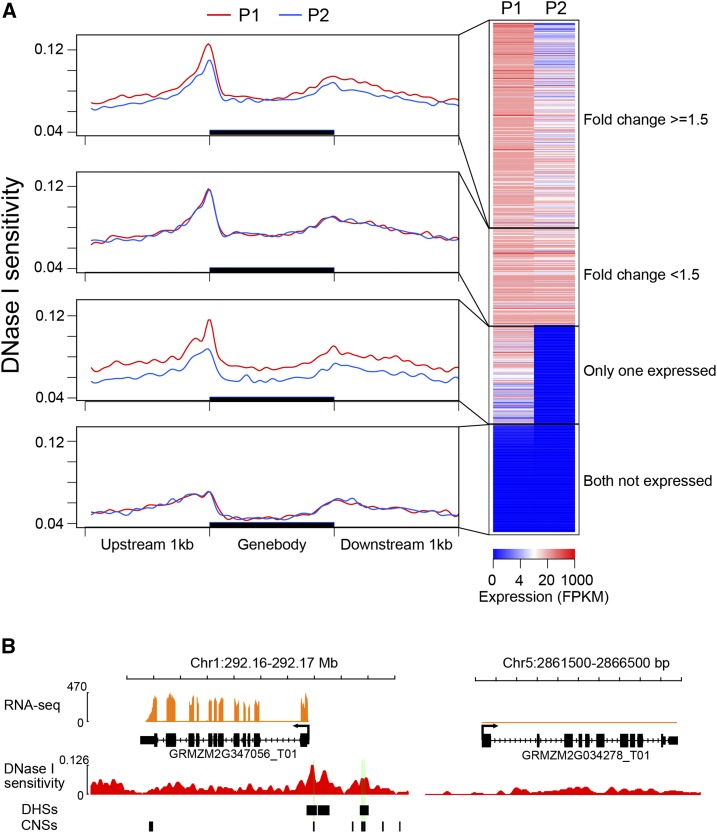
The maize genome underwent a whole-genome duplication between 5 to 12 million years ago (Swigonová et al., 2004). The majority of duplicated genes tend to be functionally lost after a duplication event (Sémon and Wolfe, 2007; Schnable et al., 2009). The retained duplicated genes often develop new functions, a process known as subfunctionalization (Moore and Purugganan, 2005). Subfunctionalization could be due to missense mutation since the fidelity of duplicated genes is typically under less selective constraint (Ohno, 1970; Kondrashov et al., 2002). Alternatively, changes to regulatory elements can lead to differential expression patterns (Lockton and Gaut, 2005; Freeling and Subramaniam, 2009). To investigate the potential contribution of regulatory elements to transcriptional divergence of duplicate genes, we examined the expression patterns of 3,984 pairs of duplicated genes in leaf tissue using RNA-seq data. The expression patterns of these duplicated pairs can be divided into four categories: pairs with at least 1.5-fold change (40%), pairs with only one copy expressed (20%), pairs with <1.5-fold change (21%), and pairs with both copies silenced (19%; Fig. 4A). Duplicated pairs with differential expression levels (fold change ≥ 1.5) showed distinct changes in DNase I sensitivity around TSSs and throughout gene bodies (Fig. 4A). In contrast, duplicated pairs with no differences in expression (fold change < 1.5) showed the same level of DNase I sensitivity.
Figure 4.
DNase I sensitivity of duplicated genes. A, Correlation between DNase I sensitivity and expression level among duplicated genes. P1 represents genes with higher expression level of the pairs, and P2 represents genes with lower expression level of the pairs. B, An example of a duplicated gene pair. Gene GRMZM2G347056 is expressed in leaves and contains three DHSs and six CNSs located in its promoter region. GRMZM2G034278, a duplicated copy of GRMZM2G347056, is not expressed and does not contain DHS or CNS in its promoter region.
We then compared the numbers of DHSs associated with duplicated genes. We focused on 2,452 duplicated gene pairs of which at least one copy of a duplicated pair contains DHSs within genic and/or the 1-kb flanking regions. We found that the duplicated gene copy with a higher expression level contained a larger count of DHSs (P value < 2.2 × 10−16, Wilcoxon rank-sum test; Supplemental Fig. S8). We also analyzed the number of CNSs from the entire set of 3,983 duplicated pairs of which at least one copy of a duplicated pair was associated with CNSs. Consistent with the results associated with DHSs, the duplicated gene copy with a higher expression level contained more counts of CNSs (P value = 5.3 × 10−7, Wilcoxon rank-sum test; Supplemental Fig. S8). For example, gene GRMZM2G347056 is highly expressed in leaves and contains six CNSs located in the upstream promoter region (Fig. 4B). Three of the CNSs overlapped with leaf DHSs, suggesting that these CNSs play regulatory roles in leaves. GRMZM2G034278, which is not expressed in leaves, is the duplicated copy of GRMZM2G347056. Consistently, DHSs and CNSs are absent in GRMZM2G034278. Thus, the orthologous regulatory elements of GRMZM2G034278 may have decayed following the duplication event.
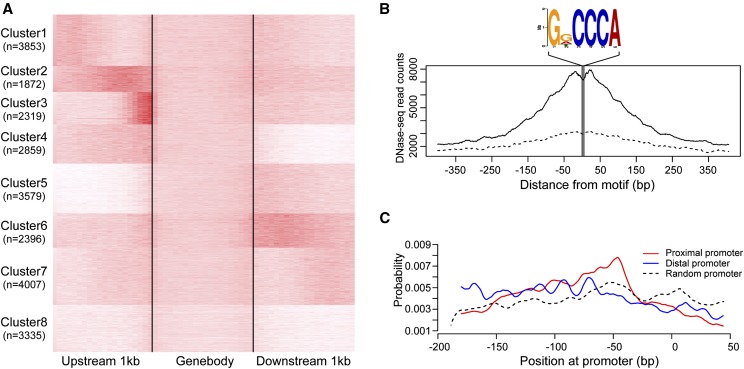
DHSs and DNA Sequence Motifs Associated with Proximal and Distal Promoters
We partitioned genes according to the distribution of their associated DHSs by k-means clustering (Fig. 5A). We found two distinct classes of DHSs based on their locations within promoters (clusters 2 and 3) that displayed higher levels of DNase I sensitivity compared to other clusters (Fig. 5A). DHSs in cluster 3 had the highest overall DNase I sensitivity and were restricted to within 200 bp upstream of TSSs (proximal DHSs). The mDNase-seq reads of DHSs in cluster 2 were distributed more broadly across the 500 bp upstream of TSSs (distal DHSs; Fig. 5A). For convenience, we refer to proximal/distal promoters for promoters containing proximal/distal DHSs, respectively. Gene Ontology (GO) analysis revealed that genes with proximal promoters are enriched in basic biological activities, including translation and cellular protein metabolic processes. By contrast, genes with distal promoters are enriched with functions related to biological regulation, including posttranslational protein modifications and regulation of gene transcription (Supplemental Table S2). The average expression level of genes with proximal DHSs was higher than that of genes with distal DHSs (61.0 versus 40.2 fragments per kilobase of transcript per million mapped reads [FPKM], P value < 7.0 × 10−5, Wilcoxon rank-sum test).
Figure 5.
Distinct patterns of DNase I sensitivity associated with promoters. A, Grouping of genes by the pattern of DNase I sensitivity at promoters. B, A footprint of the GDCCCA DNA motif (D represents G or A or T) located at proximal promoters. C, Positional preference of the GDCCCA motif in proximal and distal promoters.
We next conducted DNA motif enrichment analysis for +50 to −200 bp and −200 to −500 bp regions corresponding to proximal and distal promoters, respectively, using MEME suite programs (Bailey and Elkan, 1994). Interestingly, a DNase I footprint containing the GDCCCA motif was discovered in the proximal promoters (Fig. 5B). To analyze the organization of this motif in promoters, we conducted positional preference analysis of this motif in the proximal and distal promoters (Bailey and Machanick, 2012). We observed that 41% of proximal promoters contain the GDCCCA motif (D represents G or A or T), which exhibited a positional preference of between −155 to −35 bp and peaked at −55 bp, consistent with the distribution of DNase I sensitivity at proximal promoters (Fig. 5C). Although the GDCCCA motif was also detected in 31% of distal promoters, it showed less sensitivity and a broader positional preference compared to those located in proximal promoters (Fig. 5, B and C).
The GDCCCA motif matches the consensus binding sites (GGNCCCAC) of type I TCP proteins, which are plant-specific transcription factors (Kosugi and Ohashi, 2002; Martín-Trillo and Cubas, 2010) that are known to promote plant growth morphogenesis (Hervé et al., 2009) and to regulate genes involved in cell division and growth (Li et al., 2005; Welchen and Gonzalez, 2006). Thus, the overrepresented GDCCCA motifs in proximal promoters are likely involved in functional regulation of the corresponding genes mediated by type I TCP proteins, which mainly function in cellular housekeeping, including translation and biosynthetic processes. In contrast with proximal promoter regions, the lack of positional preference motifs at distal promoters suggests that genes with distal promoters are possibly regulated by a wider range of transcription factors (Fig. 5C).
Maize Genes Expressed at Different Levels Are Associated with Distinct Patterns of Chromatin Accessibility and Histone Modifications
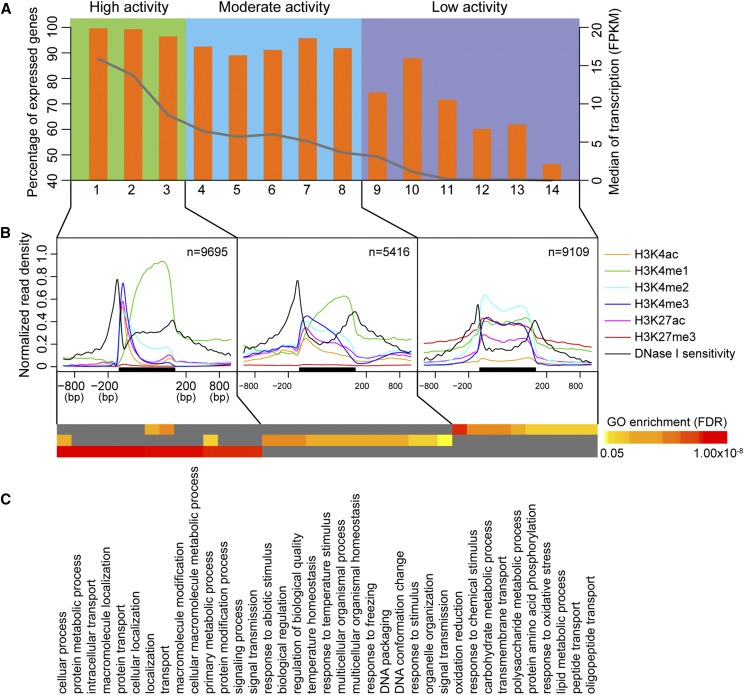
To assess the combinatorial influence of histone modifications and DHSs on transcriptional regulation, we conducted ChIP-seq using antibodies against five active histone modifications (H3K4me1, H3K4me2, H3K4me3, H3K4ac, and H3K27ac) and one repressive modification (H3K27me3) using maize leaf tissue. We specifically focused on genes that contain DHSs (n = 24,220), because the cis-regulatory sequences of these genes are putatively accessible to regulatory transfactors. Histone marks, together with DNase I sensitivity of these genes, were analyzed by k-means clustering, which resulted in 14 distinct clusters (Supplemental Fig. S9). To analyze how the chromatin state relates to gene regulation, we assessed the expression of genes in each cluster. Transcriptional activity varied among clusters. For example, more than 99.5% of genes in cluster 1 were expressed, whereas only 46.2% of genes in cluster 14 were expressed. Additionally, the expression levels (measured by median FPKM of genes in each cluster) varied among clusters (Fig. 6A; Supplemental Fig. S9).
Figure 6.
Chromatin accessibility and histone modifications orchestrate gene expression. A, k-means clustering of maize genes based on their levels of expression. Orange bar, percentage of expressed genes; gray line, median of expression level in each cluster. B, Distribution of histone modifications and DNase I sensitivity at genic regions. The black bar represents the gene body. C, Functional enrichments of genes in each group.
Several clusters shared similar transcriptional activity. For example, genes from clusters 1, 2, and 3 were highly expressed, whereas genes from clusters 12, 13, and 14 were expressed at low levels (Fig. 6A). Hence, we merged clusters that shared similar transcriptional activity and generated three groups: high activity, moderate activity, and low activity (Fig. 6A; Supplemental Fig. S9). We observed distinct profiles of DNase I sensitivity and histone modifications, specifically at TSSs and promoter regions. In the high activity group, DNase I sensitivity and four active histone modification (H3K4me2, H3K4me3, H3K4ac, and H3K27ac) form sharp peaks and aggregated around TSSs (Fig. 6B). However, the aggregation of DNase I sensitivity and histone modifications around TSSs was noticeably reduced in the moderate group and was accompanied by a broader distribution for each signal (Fig. 6B). These results suggest that the regulation of moderately expressed genes is mediated by distal promoter regions, which is likely due to the involvement of distal cis-regulatory elements (Fig. 5A). Consistent with this notion, genes in the moderate activity group exhibited tissue specificity and were enriched for functions related to “response to stimulus” (see below). In the low group, DNase I sensitivity and the four active histone marks were no longer aggregated around TSSs. The repressive mark H3K27me3 was enriched throughout the gene body and the flanking regions, which is consistent with the low transcriptional activity of this group. However, there is still substantial DNase I sensitivity upstream of TSSs (Fig. 6B; Supplemental Fig. S9).
To investigate whether there are any functional differences in the corresponding genes for each group, we analyzed gene ontology enrichment and compared expression patterns between the leaf and root. We found that the three groups were enriched for genes associated with distinct biological processes. The high activity group was enriched in basic cellular processes, including “protein metabolism,” “macromolecule modification,” and “signaling” (Fig. 6C). The moderate activity group was enriched with genes associated with “response to abiotic stimuli” and “biological regulation” (Fig. 6C). The low activity group mainly functioned in polysaccharide, carbohydrate, and lipid metabolic processes, as well as “peptide transport.” Interestingly, genes in the high activity group were consistently expressed in both leaf and root tissues, whereas genes in the moderate group, especially in clusters 4 and 5, exhibited preferential expression in leaf tissue (Supplemental Figs. S9 and S10). In contrast, the low activity group showed preference for expression in roots (clusters 11 and 14; Supplemental Figs. S9 and S10).
Identification and Functional Assays of DHSs Derived from TEs
The maize genome is dominated by TEs (Schnable et al., 2009). Recent studies have revealed that TEs are a potential source of regulatory elements and may act as a driving force in the evolution of regulatory networks, a process known as TE exaptation (Feschotte, 2008; Rebollo et al., 2012). We scanned the maize genome for TE-derived DHSs (teDHSs). Briefly, we used RepeatMasker (http://www.repeatmasker.org/) and Censor (Jurka et al., 1996) in sensitive mode to annotate all DHSs against transposable elements. A teDHS is defined if more than 50% of the DHS sequence is annotated as a TE. Strikingly, a total of 8,950 teDHSs were identified, which account for 25% of the 35,822 DHSs identified in leaf tissue. Retrotransposons and DNA transposons account for 6,558 (73.3%) and 2,389 (26.7%) of the teDHSs, respectively. DNA transposons account for only 11.5% of all transposons in the maize gnome. Thus, the percentage of teDHSs derived from DNA transposons was greater than by chance, using the whole-genome background (26.7% versus 11.5%, P value < 2.2 × 10−16, Fisher’s exact test). This may reflect the preferential insertion of DNA transposons into genic regions, whereas retrotransposons tend to be present in gene-poor regions (Bennetzen, 2000).
To exclude the possibility that teDHSs are a result of mapping bias at repetitive DNA sequences, we generated 100-bp artificial reads from the sequences of teDHSs and regular DHSs that are not related to TEs. We then mapped these artificial reads to the B73 genome. Nearly all of the artificial reads (99% for teDHSs and 99% for regular DHSs) were uniquely mapped to the correct locations, confirming that these teDHS, although related to TEs, are unique in the B73 genome and not the result of mapping bias. Indeed, average similarity between teDHSs and their related TEs (the closest homologous TEs in the genome) was 78%, indicating that the TE sequences related to DHSs were highly decayed. The average similarity between the artificial reads from teDHSs and the related TEs was 90%. TE families that are highly repetitive were excluded because we only retained uniquely mapped reads for further analysis. Therefore, our analysis likely underestimates the number of teDHSs. Nevertheless, these results suggest that a significant proportion of the regulatory elements are derived from decayed TEs in the maize genome.
To validate the functional role of teDHSs, we randomly selected 10 teDHSs (Supplemental Fig. S11) to assess in the protoplast-based transient transformation assay. We designed two different types of constructs for each teDHS. The first type of construct consisted of the teDHS, an enhancer derived from the 35S promoter, and GFP (Supplemental Fig. S12). This construct lacks a functional promoter and was used to test the potential core promoter function of each teDHS (Hernandez-Garcia and Finer, 2014). The second type of construct, which consisted of the teDHS, the mini35S promoter, and GFP (Supplemental Fig. S12), was used to examine the potential enhancer function of each teDHS (Zhu et al., 2015). Each construct was designed to include the teDHS in either forward or reverse orientation to further examine the directionality of DHS function. We found that seven teDHSs exhibited promoter function and three teDHSs showed both promoter and enhancer functions (Supplemental Table S3). The proportion of teDHSs with enhancer function was lower than that of CNS-associated DHSs (30% versus 82%), suggesting that teDHSs mostly function in local regulation.
Contribution of Retrotransposon LTRs to Open Chromatin in the Maize Genome
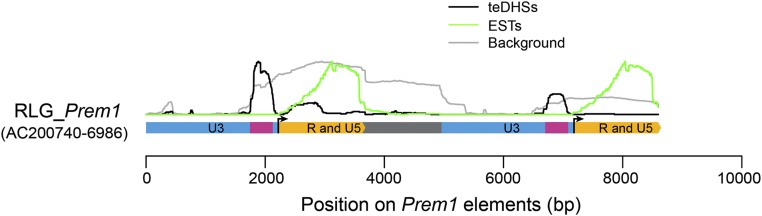
teDHSs could be derived from intrinsic regulatory elements associated with TEs or formed de novo. Strikingly, more than 57% of teDHSs were derived from the LTR (long terminal repeat) of Gypsy and Copia retrotransposons. The Gypsy and Copia retrotransposons contain two identical LTRs at their 5′ and 3′ ends with lengths ranging from hundreds of base pairs to several kilobases (Kumar and Bennetzen, 1999). LTRs can be divided into U3, R, and U5 regions, which contain the promoters and enhancers required for transcription of the retrotransposon. In order to determine whether LTRs are a potential source of novel cis-regulatory elements, we analyzed the distribution of DHSs along the full length of the Prem1 retrotransposon, which contains a 3-kb LTR (SanMiguel and Vitte, 2009). Ten subfamilies of Prem1 were identified (http://maizetedb.org/). We found that most of the teDHSs localize to the LTR regions of all 10 subfamilies (Fig. 7; Supplemental Fig. S13A). To rule out the influence of overrepresented LTR sequences in the genome, we mapped simulated artificial reads from the maize B73 genome to the full-length Prem1 elements. We found that the distribution of teDHSs on Prem1 is distinct from the background based on the simulated reads, suggesting that the enrichment of DHSs in LTR regions is not due to overrepresentation of LTR sequences in the genome. We then mapped maize ESTs to Prem1 elements to identify the TSSs within LTRs (Du et al., 2010a). A total of 2,013 maize ESTs can be mapped to Prem1 elements. Strikingly, teDHSs were located upstream of TSSs, as defined by the distribution of ESTs (Fig. 7). The location of teDHSs on LTRs is consistent with the regulatory roles of these teDHSs. Interestingly, some subfamilies of Prem1 were associated with additional teDHSs that were located distantly upstream of TSSs, rather than immediately upstream of the TSS (Supplemental Fig. S13A). We speculate that these distal teDHSs may act as enhancers for the transcription of cognate TEs. We conducted the same analysis on DNA transposons, but did not observe a similar pattern as for LTRs (Supplemental Fig. S13B). These results suggest that LTR sequences can donate their intrinsic regulatory sequences (Rebollo et al., 2012), whereas DNA transposons likely gain de novo binding sites during evolution.
Figure 7.
Distribution of DHSs and ESTs on Prem1 retrotransposon. Purple bars represent positions of teDHSs. Light-blue bars represent U3 regions, whereas yellow bars represent R and U5 regions. Positions of U3, R, and U5 of the LTR were determined by EST alignment. The arrows show transcriptional start sites within LTRs, which were determined by mapping ESTs to Prem1 retrotransposon sequence. The gray bar represents internal sequences of the Prem1 element. The black line represents the distribution of teDHSs. The gray line represents the distribution of artificial reads that were simulated from B73 reference genome sequences. The green line represents the distribution of ESTs.
We hypothesize that teDHSs may contribute to the distinct classes of transcriptional expression of their cognate genes, compared to non-TE DHS-associated genes. The expression levels of genes with teDHSs located in their promoters were higher than those of genes without DHSs (P value < 2.2 × 10−16, Wilcoxon rank-sum test; Supplemental Fig. S14). This is consistent with the protoplast result (Supplemental Table S3) and suggests that teDHSs can positively regulate expression of proximal genes. However, genes associated with teDHSs showed lower expression levels than did genes associated with non-TE derived DHSs (P value = 9.7 × 10−9, Wilcoxon rank-sum test). We found that the majority of teDHS-associated genes (821/1,071 genes, 76.7%) contain only teDHSs (that is, an absence of non-TE DHSs), suggesting that teDHSs are the major source of regulatory elements within the proximal promoter regions (as marked by mDNase-seq) of these genes in the tissues that we examined. This may be a consequence of cis-regulatory destruction by insertion of the TE and subsequent establishment of novel cis-regulatory sequences derived from the newly inserted element. Approximately 79% (7,052/8,950) of teDHSs were specific to leaves. The locations of teDHSs tended to show a broader distribution at promoters compared to non-TE derived DHSs (Supplemental Fig. S15). The remaining genes (250 genes, 23.3%) contain both teDHSs and non-TE DHSs. Non-TE DHSs of these genes were primarily located near TSSs, similar to the genome-wide pattern of DHSs, and teDHSs tended to locate further upstream of promoters (Supplemental Fig. S15). These patterns suggest that teDHSs participate in the regulation of gene expression, either by acting as the lone source of regulatory sequences or by contributing additional sequences in addition to the standing cis-regulatory landscape.
DISCUSSION
DNase-seq has been used in both animal and plant species to capture open chromatin in genomes. In our original DNase-seq method, small fragments (∼20 bp) were collected and sequenced for identification of open chromatin. While the short length of such sequence reads is not a problem for genomes with less repetitive sequences, such as Arabidopsis and rice, it hinders the application of this technology in species with highly repetitive genomes because of the poor mappability of the 20-bp reads. Therefore, we developed a modified DNase-seq method (mDNase-seq) and collected ∼100-bp sequences around DNase I cutting sites (Fig. 1). We demonstrated that the longer reads generated from mDNase-seq doubled the mappability. This method can potentially be further optimized to conduct paired-end sequencing of libraries with longer insert size, which will resolve more repetitive sequences and further increase the power of DHS detection. Differential MNase sensitivity analysis (Rodgers-Melnick et al., 2016) can also identify open chromatin in the maize genome. Data from MNase- and DNase I-based methodologies should be complementary to recover the open chromatin regions in the maize genome. Maize DHSs exhibit depletion of nucleosomes, low levels of histone modifications, and low level of DNA methylation, which is consistent with the characteristics of DHSs in rice and Arabidopsis (Zhang et al., 2012a, 2012b). We compared the width of DHS peaks in Arabidopsis, rice, maize, and human HeLa cells and found a similar overall distribution of width of DHSs in the four species.
DHSs are typically associated with cis-regulatory elements such as enhancers and promoters (Jiang, 2015). However, computationally identified DHSs represent putative cis-regulatory regions and therefore require validation through experimental assays. Traditional validation methods based on plant transformation are often technically demanding and time consuming. Thus, we developed a protoplast-based transient transformation assay. Using this methodology, we demonstrated that 82% (9/11) of CNS-cognate DHSs enable transcriptional activity of GFP linked to a null promoter. Thus, combining DHS and CNS data sets provides a high success rate for enhancer identification. We speculate that the DHSs that did not activate GFP transcription may function in specific cell types or developmental stages or otherwise act as insulators or repressors. Although our protoplast-based assay is faster than plant transformation-based assays (Zhu et al., 2015), it is cost-prohibitive and time-consuming to validate a large number of DHSs. Thus, further development of functional assays will be required to validate development- and environment-specific DHS catalogs and their associated genes. Similarly, we detected promoter and/or enhancer function of 80% (8/10) of teDHSs based on the protoplast-based transient transformation assay.
Transcriptional regulation is a complex process that involves both regulatory factors and epigenetic modifications. We found that maize genes are associated with different patterns of DNase I sensitivity at promoter regions. Genes involved in basic biological functions possess DHSs close to the TSS, presumably to maintain a high level of expression. However, genes involved in biological regulation have DHSs comparatively distal to TSSs, which may facilitate the regulation of these genes by a wide range of regulatory factors. The correlation between gene expression and histone modifications is evidenced by genome-wide analysis in both plant and animal species. Previous studies showed that transcription factors interact with histone modifications during transcription regulation. Histone methyltransferase Set1 is associated with Pol II and generates H3K4me3 at the beginning of transcription elongation (Ng et al., 2003). In humans, basal transcription factor TFIID directly binds to H3K4me3 via a PHD domain of TAF3 and promotes gene transcription (Vermeulen et al., 2007). We showed that maize genes exhibit different patterns of DNase I sensitivity and histone modifications around TSSs. Genes associated with different patterns of these chromatin signatures were enriched in different functional catalogs and showed distinct expression levels and expression patterns between leaf and root tissues. Thus, the combinatorial specificity of chromatin accessibility and histone modifications at genic regions may allow for the precise regulation of gene expression levels.
TEs are a major component of genomes, especially in complex eukaryotes. There is an increasing amount of evidence that TEs play crucial roles in genome evolution via various mechanisms, including gene mutagenesis, sequence rearrangement, epigenetic regulation, and exaptation of TE sequences (Feschotte, 2008; Lisch, 2013; Bennetzen and Wang, 2014; Friedli and Trono, 2015; Hirsch and Springer, 2017). The host genome can co-opt TEs into de novo coding sequences or regulatory elements (Feschotte, 2008). Some TEs can be activated by various abiotic stresses (Beguiristain et al., 2001; Naito et al., 2009; Pecinka et al., 2010). Genome-wide studies revealed a correlation between TE insertions and expression of nearby genes under stress conditions (Naito et al., 2009; Ito et al., 2011; Makarevitch et al., 2015). It is likely that TEs may benefit the host genome by promoting environmental adaptation via exaptation of their regulatory elements (Casacuberta and González, 2013). Another interesting question is whether TEs contribute to speciation. In animals, expansion of TEs has been shown to promote species-specific regulatory networks (Mariño-Ramírez et al., 2005; Wang et al., 2007; Jacques et al., 2013; Sundaram et al., 2014).
We demonstrated that thousands of regulatory elements in the maize genome are derived from decayed TEs, either by exaptation of intrinsic regulatory elements located within LTRs or by de novo co-opt of TE sequences. Remarkably, eight of the 10 randomly selected teDHSs can drive the transcription of a reporter gene in protoplast-based functional assays (Supplemental Table S3). The TE-derived regulatory elements are likely to be fixed in the maize genome, as these elements are highly decayed and remain functional in normal conditions. It is important to note that we may significantly underestimate the percentage of teDHSs in our current data set. Recently evolved teDHSs will be difficult to identify because of their sequence similarity to TEs and stress-related teDHSs cannot be detected under normal growth conditions. Thus, further investigations are required to obtain insights into the influence of teDHSs on environmental adaption and diversity of the transcriptional regulatory landscape among maize populations.
MATERIALS AND METHODS
mDNase-Seq, ChIP-Seq, and RNA-Seq
Leaf and root tissues were collected from 10-d-old B73 maize (Zea mays) plants grown in a greenhouse and were divided for mDNase-seq, ChIP-seq, and RNA-seq. Briefly, three pots with 10 seeds in each pot were planted in the greenhouse at the same time; all leaf tissues except for the first leaf were collected from the 30 plants and were pooled for the downstream process. All pooled leaf and root tissues were ground into a fine powder using liquid nitrogen. mDNase-seq library construction, including nuclei isolation, was performed according to published DNase-seq protocols (Zhang and Jiang, 2015) with modifications. Specific modifications (Fig. 1) of the procedures included adaptor I-ligated HMW DNA fragmentation into 200- to 500-bp segments using sonication in place of MmeI cleavage. After purification using a Qiagen PCR purification column, sonicated DNA was incubated with dynal M-280 beads (Invitrogen) for the enrichment of adaptor I ligation products. Adaptor I ligated DNA fragments were treated following the standard protocol for preparing ChIP-seq libraries from Illumina, including end repair, adding “dA” to the 3′ ends of blunt-ended DNA fragments, and ligation with PE adaptors. mDNase-seq libraries were sequenced on the Illumina Hi-Seq 2000 platform after final amplification by PCR using linker-specific primers and agarose-based purification.
ChIP experiments were performed following published protocols (Zhang et al., 2012a) using commercial antibodies against H3K4me2 (07-030; Millipore), H3K27me3 (07-449; Millipore), H3K4ac (07-539; Millipore), H3K4me1 (ab8895; abcam), H3K4me3 (ab8580; abcam), and H3K27ac (ab4729; abcam). ChIP-seq libraries were prepared and sequenced on the Illumina Hi-Seq 2000 platform following published protocols (Zhang et al., 2012a). Total RNA from two biological replicates were extracted from 10-d-old leaf tissue and treated with DNase I. Approximately 10 µg of total RNA was reverse-transcribed into cDNA subjected to library construction following the manufactures protocol (Illumina). Bar-coded mRNA-seq libraries were sequenced on the Illumina Hi-Seq 2000 platform.
Protoplast Transformation
Maize protoplasts were isolated from leaf tissue harvested from 10-d-old seedlings and transfected with the GFP fusion constructs using the polyethylene glycol-mediated method as described previously (Zhang et al., 2011). Briefly, maize leaf tissue was collected and cut into strips followed by digestion for 5 to 6 h in the dark using 10 mL of enzyme solution (1.5% Cellulase “Onozuka” R-10 [Yakult Pharmaceuticals], 0.75% MacerozymeR-10 [Yakult Pharmaceuticals], 0.6 m mannitol, 10 mm MES, pH 5.7, 10 mm CaCl2, and 0.1% BSA). The protoplasts were released and purified by washing several times. The purified protoplasts were resuspended using MMG solution (0.6 m mannitol, 15 mm MgCl2, and 4 mm MES, pH 5.7) and were ready for polyethylene glycol-mediated transfection according to the published protocol (Yoo et al., 2007). The protoplasts transfected with vectors containing DHSs were cultured in the dark at room temperature for 16 to 20 h. GFP signals were observed and recorded with a confocal microscope. Each construct was triplicated for transformation. The DHSs for validation were randomly selected, except that (1) the DNase sensitivity was 10 to 90% percentile of all DHSs (extreme values removed), (2) it was possible to design primers at the very ends of the DHS interval so that no flanking sequences of DHSs will be included in the constructs, and (3) a single band was amplified using the primers.
Read Mapping and DHS Identification
The reads from mDNase-seq, ChIP-seq, and MNase-seq (SRR2000648; Zhao et al., 2016) were aligned to the Maize B73 genome (Schnable et al., 2009) version 3 (AGRv3.22, http://plants.ensembl.org/Zea_mays) using BWA-MEM (Li et al., 2009) with default parameters. Reads with mapping quality greater than 20 were extracted using SAMtools (Li et al., 2009) for further analysis. RNA-seq data from leaf and root tissue were analyzed using TopHat (Trapnell et al., 2009) and Cufflinks (Trapnell et al., 2010) with default parameters. Bisulfite sequencing reads (SRR850328; Li et al., 2014) of B73 were analyzed using bismark (-q –N 1). DHSs were identified using F-seq (Boyle et al., 2008) with parameter “-l 300 -t 9 -of bed -f 0” and Popera (https://github.com/forrestzhang/Popera) with parameter “-b 300.” DHSs identified by both tools were retained and DHS scores generated from F-seq were assigned to each DHS. To calculate the FDR of DHSs, we used random reads from the B73 genome to identify DHSs. FDR was calculated as the ratio of DHSs identified from random data sets to DHSs from mDNase-seq. We set the cutoff of DHS score to 0.07, corresponding to FDR < 0.0064, based on manual inspection of DHS peaks. To determine the saturation of mDNase-seq data, we sampled 25, 50, and 75% of reads from our leaf and root data. DHSs were identified for each subset of data, and the number of DHSs was then plotted against the total reads used.
DNase I Sensitivity Profile and Epigenomic Features of DHSs
The distribution of DHSs was analyzed by dividing the maize genome into 1-Mb nonoverlapping windows and calculating the number of DHSs in each window. To analyze DNase I sensitivity, histone modification, and nucleosome occupancy, we divided genic regions, gene bodies, and 1-kb up- and downstream of genes into 20 bins. DNase sensitivity was calculated as the number of 5′ ends of mDNase-seq reads in each bin averaged across all genes. Histone modification densities and nucleosome occupancy were calculated as the numbers of ChIP-seq and MNase-seq reads in each bin averaged across all genes. DNase sensitivity and nucleosome occupancy were then scaled from 0 to 1. To analyze the cutting frequency of DNase I around CNSs, CNSs were centered at their midpoints and the number of 5′ end of mDNase-seq reads was counted at each base pair from the midpoint. To analyze the epigenomic features of DHSs, DHSs and the 1-kb flanking regions were divided into 20 bins. The profile of histone modifications and nucleosome occupancy over DHSs were generated by calculating the average number ChIP-seq and MNase-seq reads at each bin across all DHSs. The average number of reads obtained from these data sets was transformed to Z-scores to allow comparison. The DNA methylation level of each bin was calculated and averaged across all DHSs.
k-Means Clustering, Motif Identification, and Transposable Element Derived DHSs
The DNase I sensitivity and histone densities of genes (see above) were formatted as a matrix. DNase I sensitivity and histone modification density were scaled from 0 to 1 and analyzed using k-means clustering in R. We conducted k-means clustering using K from 5 to 20. The best K was chosen to minimize the sum of squares of the K clusters. K was determined as 8 for DNase I sensitivity of genes (Fig. 4A) and 15 for DNase I sensitivity and histone modifications (Supplemental Fig. S5). Motif identification, local motif enrichment, and motif annotation were conducted using DREME (Bailey, 2011), CentriMo (Bailey and Machanick, 2012), and Tomtom (Gupta et al., 2007) from the MEME Suite with default parameters. Transposable elements were annotated using RepeatMasker (http://www.repeatmasker.org/) and Censor (Jurka et al., 1996). To remove potential genic sequences, DHSs with more than 70% sequence similarity to CDSs from rice (Oryza sativa) or sorghum (Sorghum bicolor) were removed. RepeatMasker was run against maize transposable element database (maizetedb.org) with parameter “-norna –s –no_is –gff” and censor was run against transposable element from Repbase with parameter “-mode sens.” The output from RepeatMasker and censor were combined. We defined teDHSs as DHSs that overlap with annotated TEs by at least 50 bp and the annotated TEs cover at least 50% of the DHS. GO annotation was conducted using agriGO (Du et al., 2010b). Duplicated genes were identified using SynMap (Lyons et al., 2008) with parameters “-D 20 –A 6” and “-Dm 25” for Quota Align to merge syntenic blocks.
Accession Numbers
All Illumina sequence data from this study were submitted to the NCBI Sequence Read Archive under project number PRJNA382414.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Saturation and rediscovery rate of DHSs.
Supplemental Figure S2. Distribution of DHSs in the maize genome.
Supplemental Figure S3. Differential nuclease sensitivity over DHSs.
Supplemental Figure S4. Cutting frequency of DNase I centered at CNSs.
Supplemental Figure S5. DNase I sensitivity and differentially expressed genes in maize.
Supplemental Figure S6. Annotations of the 11 CNS-cognate DHSs used for protoplast-based transient transformation.
Supplemental Figure S7. Protoplast-based transformation assay of three DHSs associated with the SSU1 gene.
Supplemental Figure S8. DHSs and CNSs associated with duplicated genes.
Supplemental Figure S9. Heat map of histone modifications, DNase sensitivity, and expression levels of genes associated with DHSs.
Supplemental Figure S10. Tissue-specific expression of genes in specific clusters.
Supplemental Figure S11. Annotations of the 10 teDHSs used for protoplast-based transformation.
Supplemental Figure S12. Diagrams of constructs designed to examine the potential promoter and/or enhancer function of teDHSs.
Supplemental Figure S13. Distribution of DHSs and ESTs on transposable elements.
Supplemental Figure S14. Expression levels of genes associated with different types of DHSs.
Supplemental Figure S15. Distribution of different types of DHSs at promoter regions.
Supplemental Table S1. Summary of functional validation of DHSs using protoplast-based transient transformation assay.
Supplemental Table S2. GO enrichment of genes with distal and proximal promoters.
Supplemental Table S3. Summary of functional validation of teDHSs using protoplast-based transient transformation assay.
Supplemental Resource 1. List of DHSs associated with CNSs.
Footnotes
Articles can be viewed without a subscription.
This research was supported by National Natural Science Foundation of China Grants 31571579 and 31371239 and an “Innovation and Enterprise Scholar” of Jiangsu Province to W.L.Z., and by National Science Foundation Grant IOS-1444514 to J.J.
References
- Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrançois P, Struhl K, Gerstein M, Snyder M (2009) Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci USA 106: 14926–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguiristain T, Grandbastien M-A, Puigdomènech P, Casacuberta JM (2001) Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol 127: 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42: 251–269 [PubMed] [Google Scholar]
- Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65: 505–530 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS (2008) F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J (2013) The impact of transposable elements in environmental adaptation. Mol Ecol 22: 1503–1517 [DOI] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J (2014) Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol 166: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C (2017) Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumbie JS, Filichkin SA, Megraw M (2015) Improved DNase-seq protocol facilitates high resolution mapping of DNase I hypersensitive sites in roots in Arabidopsis thaliana. Plant Methods 11: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Bowen NJ, Schmutz J, Shoemaker RC, Ma J (2010a) Bifurcation and enhancement of autonomous-nonautonomous retrotransposon partnership through LTR Swapping in soybean. Plant Cell 22: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010b) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Goldshmidt A, Pautler M, Morohashi K, Liseron-Monfils C, Lewis MW, Kumari S, Hiraga S, Yang F, Unger-Wallace E, et al. (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res 24: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Subramaniam S (2009) Conserved noncoding sequences (CNSs) in higher plants. Curr Opin Plant Biol 12: 126–132 [DOI] [PubMed] [Google Scholar]
- Friedli M, Trono D (2015) The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 31: 429–451 [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30: 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. ; modENCODE Consortium (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD (2007) FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K (2009) Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217-218: 109–119 [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CD, Springer NM (2017) Transposable element influences on gene expression in plants. Biochim Biophys Acta 1860: 157–165 [DOI] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Jacques PE, Jeyakani J, Bourque G (2013) The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9: e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. (2015) The ‘dark matter’ in the plant genomes: non-coding and unannotated DNA sequences associated with open chromatin. Curr Opin Plant Biol 24: 17–23 [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P (1996) CENSOR--a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20: 119–121 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002) Selection in the evolution of gene duplications. Genome Biol 3: research0008.0001 [DOI] [PMC free article] [PubMed]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, Xiao H, Song R (2015) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell 27: 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Eichten SR, Hermanson PJ, Zaunbrecher VM, Song J, Wendt J, Rosenbaum H, Madzima TF, Sloan AE, Huang J, et al. (2014) Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. (2013) How important are transposons for plant evolution? Nat Rev Genet 14: 49–61 [DOI] [PubMed] [Google Scholar]
- Lockton S, Gaut BS (2005) Plant conserved non-coding sequences and paralogue evolution. Trends Genet 21: 60–65 [DOI] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D (2007) Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci USA 104: 8005–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M (2008) The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop Plant Biol 1: 181–190 [Google Scholar]
- Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11: e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Zhang T, Zhu B, Jiang J (2017) Towards genome-wide prediction and characterization of enhancers in plants. Biochim Biophys Acta 1860: 131–139 [DOI] [PubMed] [Google Scholar]
- Mariño-Ramírez L, Lewis KC, Landsman D, Jordan IK (2005) Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet Genome Res 110: 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. ; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team (2007) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177 [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, Falcone Ferreyra L, Mejía-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, et al. (2012) A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nishihara H, Smit AFA, Okada N (2006) Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res 16: 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Spinger-Verlag, New York [Google Scholar]
- Ohno S. (1972) So much “junk” DNA in our genome. Brookhaven Symp Biol 23: 366–370 [PubMed] [Google Scholar]
- Orgel LE, Crick FHC (1980) Selfish DNA: the ultimate parasite. Nature 284: 604–607 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. (2014) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler M, Eveland AL, LaRue T, Yang F, Weeks R, Lunde C, Je BI, Meeley R, Komatsu M, Vollbrecht E, Sakai H, Jackson D (2015) FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27: 104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Li R, Zhang S, Wang K, Xu M, Li J, Du Y, Yu H, Cui X (2016) Identification of regulatory DNA elements using genome-wide mapping of DNase I hypersensitive sites during tomato fruit development. Mol Plant 9: 1168–1182 [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL (2012) Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42 [DOI] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Vera DL, Bass HW, Buckler ES (2016) Open chromatin reveals the functional maize genome. Proc Natl Acad Sci USA 113: E3177–E3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel PJ, Vitte C (2009) The LTR-retrotransposons of maize. In: Bennetzen JL, Hake S, eds Handbook of Maize: Genetics and Genomics. Springer, New York, pp 307–328 [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM (2013) Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17: 505–512 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, et al. (2009) Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE (2010) DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2010: t5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E, Wachsman G, Benfey PN (2013) Spatiotemporal signalling in plant development. Nat Rev Genet 14: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. ; Mouse ENCODE Consortium (2012) An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E (2013) Determinants of nucleosome positioning. Nat Struct Mol Biol 20: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030 [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T (2014) Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14(10A): 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_ENCODE_Project_Consortium (2004) The ENCODE (ENCylopedia Of DNA Elements) project. Science 306: 636–640 [DOI] [PubMed]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompana M, Buck MJ (2014) Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco G, Schnable JC, Pedersen B, Freeling M (2013) Automated conserved non-coding sequence (CNS) discovery reveals differences in gene content and promoter evolution among grasses. Front Plant Sci 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D (2007) Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA 104: 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2006) Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation Machinery. Plant Physiol 141: 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang W, Jiang J (2015) Genome-wide mapping of DNase I hypersensitive sites in plants. Methods Mol Biol 1284: 71–89 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wu Y, Schnable JC, Zeng Z, Freeling M, Crawford GE, Jiang J (2012a) High-resolution mapping of open chromatin in the rice genome. Genome Res 22: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang T, Wu Y, Jiang J (2012b) Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HN, Zhu XB, Wang K, Gent JI, Zhang WL, Dawe RK, Jiang JM (2016) Gene expression and chromatin modifications associated with maize centromeres. G3 (Bethesda) 6: 183–192 [DOI] [PMC free article] [PubMed]
- Zhu B, Zhang W, Zhang T, Liu B, Jiang J (2015) Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27: 2415–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]