MdHXK1 is involved in glucose-mediated improvement of tolerance to high salinity by interacting with and phosphorylating the tonoplast Na+/H+ exchanger MdNHX1.
Abstract
Glc regulates many vital processes, including plant growth, development, metabolism, and responses to biotic and abiotic stress. However, the molecular mechanism by which Glc acts as a signal to regulate salinity tolerance remains unclear. In this study, we found that the apple (Malus domestica Borkh.) Glc sensor hexokinase1 (MdHXK1) contributes to Glc-mediated salinity tolerance. A combination of split ubiquitin system, pull-down, co-immunoprecipitation, and bimolecular fluorescence complementation assays demonstrated that MdHXK1 interacts with and phosphorylates the Na+/H+ exchanger MdNHX1 at its Ser-275 residue. Phosphorylation improved the stability of MdNHX1 and enhanced its Na+/H+ transport activity in MdNHX1 overexpression transgenic apple and yeast complementation cells. Furthermore, Ser-275 of MdNHX1 was found to be crucial for MdHXK1-mediated phosphorylation. Finally, a series of transgenic analyses demonstrated that salt tolerance mediated by MdHXK1 partially depended on MdNHX1. Overall, our findings provide insights into how sugar recruits and regulates MdNHX1 in response to high salinity in plants.
Sugars are a primary energy source in plants and influence almost all stages of the plant lifecycle, including seeding germination, photosynthesis, flower growth and pigmentation, fruit development, and senescence (Ohto et al., 2001). Sugar and energy status mediate metabolic, environmental, and developmental cues pertaining to physiological and developmental plasticity. In response to external abiotic stresses, sugars regulate osmotic potential by providing osmotic driving force for cell expansion and act as signaling molecules to regulate gene expression (Moore et al., 2003; Rolland et al., 2006; Rosa et al., 2009).
Abiotic stresses such as drought, salinity, low temperature, and flooding usually lead to sugar accumulation (Krasensky and Jonak, 2012). It has been reported that the accumulation of Glc, Suc, and Fru under high salinity plays a very important role in carbon storage, osmotic regulation, and homeostasis, as well as scavenging of free radicals (Singh et al., 2015). Exogenous Glc application delimits chlorophyll destruction, increases dry weight, maintains ionic homeostasis, enhances Pro accumulation, prevents water loss, inhibits lipid peroxidation, and activates antioxidant enzyme activity (Hu et al., 2012). In rice (Oryza sativa), Glc and Fru are involved in maintaining osmotic potential and scavenging free radicals (Pattanagul and Thitisaksakul, 2008). Furthermore, soluble sugars are also involved in ROS anabolism and catabolism, such as the oxidative pentose P pathway associated with ROS scavenging (Couée et al., 2006). In wheat (Triticum) seedlings, exogenous application of Glc delimits Na+ accumulation but stimulates K+ uptake under salt stress, thus maintaining ionic homeostasis under salinity stress (Nemati et al., 2011).
Soluble sugars can be sensed to transduce specific signaling pathways and to elicit sugar responses in plant growth and metabolism (Rolland et al., 2006). Suc and hexoses (mainly Glc and Fru) are recognized as primary messengers that control sugar signaling by regulating the expression of different genes and enzyme activities (Rosa et al., 2009). Unfavorable environmental factors sponsor metabolic changes, which lead to an expressional change of several carbohydrate metabolism genes, including ADP-Glc pyrophosphorylase in starch biosynthesis, Suc synthase, Suc P synthase, and invertase in Suc metabolism (Mita et al., 1995; Ohto et al., 1995; Toroser and Huber, 1997). In addition, expression of the Arabidopsis (Arabidopsis thaliana) bZIP transcription factor gene ATB2 is suppressed by Suc-specific signaling pathways (Rook et al., 1998). When Glc levels exceed their demand, α-amylase gene expression is down-regulated by a process that involves sugar sensing (Yu et al., 1996).
Furthermore, the sugar signaling pathway often communicates between stresses and plant hormones (Gazzarrini and McCourt, 2001). For example, ABA inhibitory effects on seed germination can be relieved by exogenous application of a low concentration of Glc (Finkelstein and Lynch, 2000). However, a high concentration of Glc increases ABA accumulation, thereby resulting in seed germination delay (Arenas-Huertero et al., 2000; Gill et al., 2003). Water stress also causes a noticeable accumulation of hexoses, which inhibits the activity of cell wall invertase in maize (Zea mays) ovaries but increases the activity of vacuolar invertase in mature leaves (Zinselmeier et al., 1995). Biotic and abiotic stresses such as pathogen infection and salt stress always result in an up-regulation of extracellular invertase (Roitsch et al., 2003). Higher accumulation of soluble sugars in roots makes mycorrhizal plants more resistant to salt-induced osmotic stress (Feng et al., 2002). Elevating trehalose accumulation by either internal overproduction or acquisition from an exogenous trehalose supply is sufficient to improve the plants tolerance to drought and salt stress (Redillas et al., 2012). In summary, sugar signaling is involved in the regulation of plant growth and development, as well as biotic and abiotic stress responses. However, the exact molecular mechanism by which sugar signal influences these processes is unclear.
Among the various sugars, Glc is one of the main products of photosynthesis and is widely recognized as a signal molecule that regulates plant growth and development (Gibson, 2005; Lastdrager et al., 2014). In many organisms, hexokinases (HXKs) are considered as the most ancient, conserved sugar sensor (Claeyssen and Rivoal, 2007). Currently, the Arabidopsis AtHXK1 is the best characterized Glc sensor in plants (Rolland and Sheen, 2005). Its sensor function was identified by loss-of-function mutant Glc insensitive2 (gin2) in Arabidopsis (Moore et al., 2003). In addition, HXK1 is also reported to be involved in different abiotic and biotic stress tolerances. For example, virus-induced gene silencing of HXK1 results in apoptotic cell death in leaves, indicating that HXK1 depletion activates programmed cell death (Kim et al., 2006). Overexpression of Arabidopsis HXK1 or HXK2 enhances tolerance to MV-induced oxidative stress and pathogen infection (Sarowar et al., 2008). The HXK1 deletion strain ∆hxk in Fusarium verticillioides impairs utilization of hexose sugars and reduced osmotic stress tolerance (Kim et al., 2011).
In Arabidopsis, hexokinase1 (HXK1) is a dual-function enzyme localized to both the cytosol and nucleus. It carries out catalytic activity in the glycolytic pathway and also functions in the sugar sensing and signaling pathways (Cho et al., 2006; Cheng et al., 2011). HXK1 mutations (AtHXK1S177A and AtHXK1G104D) with little or no catalytic activity have been shown to mediate diverse Glc responses (Harrington and Bush, 2003). Furthermore, the HXKs localize to the nucleus and can modulate specific target gene transcription independent of Glc metabolism (Cho et al., 2006). Most recently, we report that apple MdHXK1 interacts with and phosphorylates MdbHLH3 to promote anthocyanin accumulation (Hu et al., 2016). However, it remains unclear if and how HXK-mediated Glc signaling is involved during plant salt stress responses.
In this study, we report that exogenous Glc application improved salt tolerance, and this process depended on Glc sensor MdHXK1. Furthermore, it was found that a tonoplast-localized Na+/H+ exchanger MdNHX1 interacted with MdHXK1 and acted as its phosphorylation target. Subsequently, the function of MdHXK1 and MdNHX1 in response to salt stress was characterized. Finally, the relationship between sugar signaling and abiotic stress was also discussed.
RESULTS
Glc Improves Salt Tolerance in an HXK-Dependent Manner
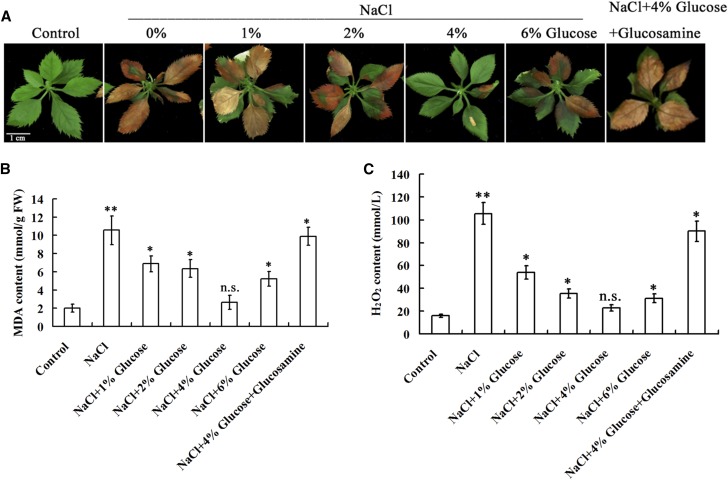
To detect if exogenous application of Glc influences salt tolerance, apple ‘Gala’ plantlets were allowed to grow on normal medium for 3 weeks and then transferred to medium containing 200 mM NaCl alone or 200 mM NaCl plus different concentrations of Glc (1%, 2%, 4%, 6%, w/v) for 14 d. The result demonstrated that Glc application noticeably enhanced the tolerance of plantlets to salt stress. Plantlets exhibited the greatest salt tolerance when 4% Glc was used (Fig. 1A), and produced the lowest malondialdehyde (MDA) and H2O2 contents under salt stress (Fig. 1, B and C). Meanwhile, 4%(w/v) mannitol was used in a separate treatment as a control for the possible osmotic effect caused by 4% Glc (Supplemental Fig. S1).
Figure 1.
Glc improves salt tolerance of ‘Gala’ apple plantlets. A, Salt tolerance of apple plantlets in the presence of 0mM NaCl, 200 mm NaCl, or 200 mm NaCl plus Glc at different concentrations of 200 mm NaCl plus 4% (w/v) and 100 mm glucosamine. B and C, MDA (B) and H2O2(C) contents of apple plantlets treated as in (A). Each value represents the mean ± sd from three separate experiments. Statistical significance was determined and compared to the control value using Student’s t test. n.s., P > 0.01; *, P < 0.01; **, P < 0.001.
To determine if a Glc sensor and the key metabolic enzyme HXK mediates this process, HXK inhibitor glucosamine was added to the medium with 4% Glc and 200 mM NaCl (Zheng et al., 2009). The result indicated that glucosamine treatment reduced salt tolerance, indicated by enhanced plantlet damage and MDA and H2O2 content increases compared with plantlets exposed to 4% Glc and 200 mm NaCl (Fig. 1). These results demonstrated that exogenous Glc improved salt tolerance at least partially in an HXK-dependent manner.
The apple MdHXK1 gene encodes a hexokinase (Hu et al., 2016). Its expression was positively induced by Glc treatment (Supplemental Fig. S2A) and MdHXK1 is required for Glc-mediated enhancement of salt tolerance (Supplemental Fig. S3). Moreover, MdHXK1 improved salt tolerance depending on both catalytic activity and signaling function under low-Glc conditions, but mainly in the signaling pathway under high-Glc conditions (Supplemental Fig. S4).
MdHXK1 interacts with MdNHX1
To reveal the regulatory mechanism by which MdHXK1 is involved in salt tolerance, possible MdHXK1-interacting proteins were screened through by liquid chromatography/mass spectrometry (LC/MS; Hu et al., 2016). The 35S::MdHXK1-Myc vector was constructed and used for genetic transformation of apple plants. Total proteins were extracted from MdHXK1 transgenic apple plants, then used for coimmunoprecipitation (Co-IP) against the monoclonal anti-Myc antibody. LC/MS analysis showed that a salt tolerance-associated Na+/H+ exchanger was a candidate. Phylogenetic tree analysis showed that the putative Na+/H+ exchanger closely clustered with AtNHX1, thereby being named as “MdNHX1” hereafter (Supplemental Fig. S5). Expression analysis demonstrated that MdNHX1 transcripts increased with 4% Glc treatment (Supplemental Fig. S2B).
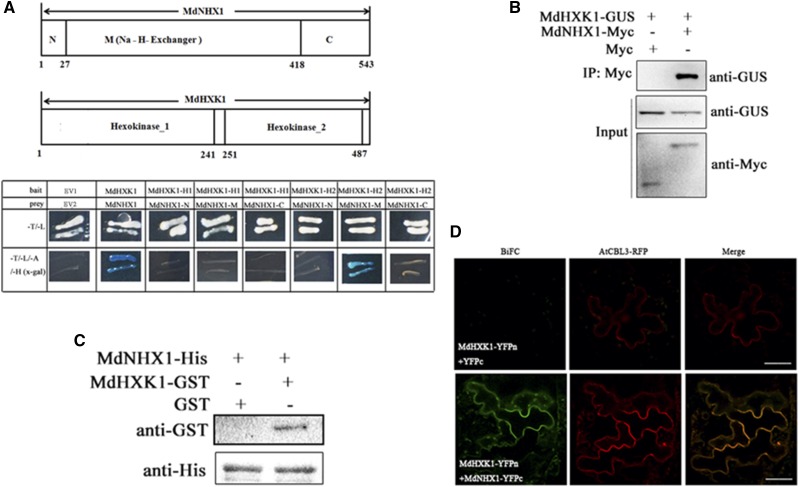
The split-ubiquitin membrane-based yeast two-hybrid system was used to confirm the interaction between MdHXK1 and membrane protein MdNHX1. The result showed that MdHXK1 interacted with MdNHX1 (Fig. 2A). MdHXK1 contains two hexokinase domains Hexokinase_1 and Hexokinase_2, whereas MdNHX1 contains a short N terminus, a Na_H_Exchanger domain, and a long C terminus. It was found that the Na_H_Exchanger domain of MdNHX1 and the Hexokinase_2 domain of MdHXK1 are crucial for their interaction (Fig. 2A).
Figure 2.
MdHXK1 interacts with MdNHX1 in vivo and in vitro. A, Split-ubiquitin membrane-based yeast two-hybrid assays for interaction between MdHXK1 and MdNHX1. The hexokinase_2 domain of MdHXK1 and Na_H_Exchanger domain of MdNHX1 are crucial for their interaction. B, Interaction between MdHXK1 and MdNHX1 proteins in vivo. Total protein extracted from 35S::MdHXK1-GUS + 35S::MdNHX1-Myc or 35S::MdHXK1-GUS + 35S::Myc double transgenic apple calli were immunoprecipitated with anti-Myc. The proteins from crude lysates (input) were detected with anti-GUS antibody and anti-Myc antibody. The immunoprecipitated proteins (output) were detected with anti-GUS antibody. C, Interaction between MdHXK1 and MdNHX1 proteins in vitro. MdNHX1-His proteins were incubated with immobilized MdHXK1-GST or GST, and the proteins immunoprecipitated with His-beads were detected using anti-GST antibody. D, BiFC analysis of the interaction between MdHXK1 and MdNHX1 in N. benthamiana. Arabidopsis CBL3 was fused to RFP and used as a vacuole-located marker. Scale bars represent 50 μm.
co-immunoprecipitation(Co-IP) assay was performed to verify if MdHXK1 interacts with MdNHX1 in vivo. First, a 35S::MdHXK1-GUS vector was constructed and used for genetic transformation into apple calli. Then, 35S::MdHXK1-GUS transgenic apple calli was transformed with 35S::Myc or 35S::MdNHX1-Myc to generate 35S::MdHXK1-GUS/35S::Myc or 35S::MdHXK1-GUS/35S::MdNHX1-Myc coexpressed apple calli. Two double transgenic calli were used for Co-IP assays. The result showed that the MdNHX1-Myc fusion protein but not Myc coimmunoprecipitated by anti-GUS antibody (Fig. 2B). To confirm the in vitro interaction between MdHXK1 and MdNHX1, an affinity pull-down assay was carried out using His-labeled MdNHX1. It was incubated with GST-labeled MdHXK1 protein or GST alone. GST-labeled MdHXK1 protein but not GST alone was pulled down by an anti-His antibody, demonstrating that MdHXK1 physically interacted with MdNHX1 in vitro (Fig. 2C). Furthermore, a bimolecular fluorescence complementation (BiFC) assay demonstrated that MdHXK1-MdNHX1 interaction occurred on the tonoplast of tobacco epidermal cells (Fig. 2D). These results collectively indicated that MdHXK1 interacted with MdNHX1 in vitro and in vivo.
MdHXK1 Phosphorylates MdNHX1 Protein at Ser-275
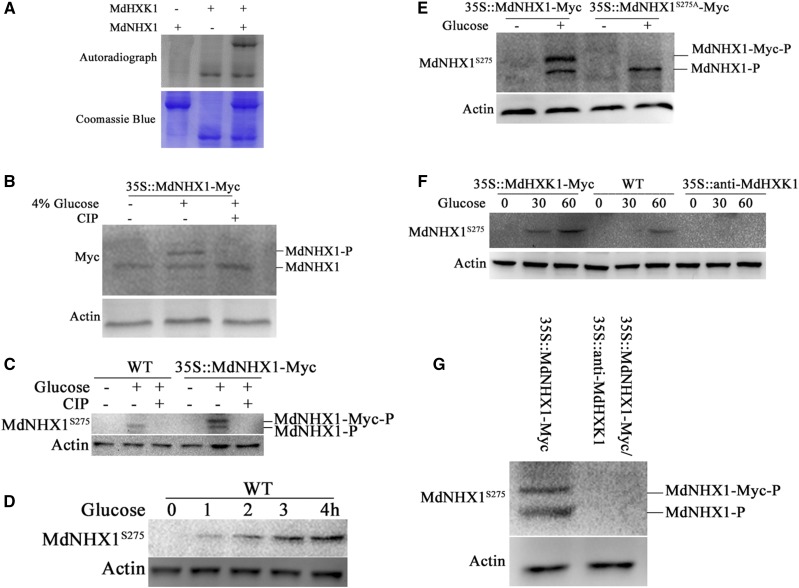
Because MdHXK1 interacted with MdNHX1, it is reasonable to hypothesize that MdHXK1 may act as a protein kinase to phosphorylate MdNHX1. To verify this hypothesis, in vitro phosphorylation assays were performed using recombinant proteins MdNHX1-GST and MdHXK1-His to examine if MdNHX1 is a direct substrate of MdHXK1. The result showed that the MdNHX1-GST fusion was phosphorylated by MdHXK1-His (Fig. 3A), indicating that MdNHX1 protein is a direct substrate of MdHXK1 protein kinase.
Figure 3.
MdHXK1 mediates phosphorylation of MdNHX1 in response to Glc. A, In vitro phosphorylation assay of MdNHX1 by protein kinase MdHXK1. SDS-PAGE gel with Coomassie Blue-stained MdHXK1-His, MdNHX1-GST proteins (bottom panel); autoradiograph showing MdNHX1 phosphorylation by MdHXK1 (top panel, top band) and MdHXK1 autophosphorylation (top panel, bottom bands). B, Gel-shift assay using 35S::MdNHX1-Myc transgenic apple calli. Two-week-old 35S::MdNHX1-Myc apple calli were treated with 4%(w/v) Glc or together with CIP for 1 h. MdNHX1-Myc protein was extracted and then subjected to immunoblot analysis using a Myc antibody. C, Western blot detected phosphorylation of MdNHX1 protein in the wild-type and 35S::MdNHX1-Myc transgenic calli using a specific anti-MdNHX1S275 antibody against MdNHX1 phosphorylation site at residue 275. D, Glc-induced phosphorylation of MdNHX1 depends on Glc treatment time. The wild-type apple calli were treated with 4% Glc for different times (0 min, 20 min, 40 min, or 60 min). E, The Glc-induced phosphorylation of MdNHX1 disappeared in MdNHX1S275A-Myc transgenic apple calli using anti-MdNHX1S275 antibody. F and G, MdHXK1 was required for Glc-induced phosphorylation of MdNHX1. Western blot detected the phosphorylation of MdNHX1 in the wild-type, 35S::MdHXK1-Myc, and 35S::anti-MdHXK1 transgenic calli using anti-MdNHX1S275 antibody (F). Western blot detected the phosphorylation of MdNHX1 in 35S::MdNHX1-Myc and 35S::MdNHX1-Myc+35S::anti-MdHXK1 transgenic calli using anti-MdNHX1S275 antibody (G). For western-blot assays, actin was used as a loading control to ensure identity in the amounts of protein.
Furthermore, 35S::MdNHX1-Myc transgenic apple calli were treated with or without 4% Glc and used for western blotting with a Myc antibody. The result showed that a large band appeared in 35S::MdNHX1-Myc transgenic apple calli upon 4% Glc treatment, indicating that Glc induced posttranslational modification for MdNHX1 (Fig. 3B). After treatment with calf intestine alkaline phosphatase (CIP), however, the large band disappeared, indicating that MdNHX1 undergoes posttranslational modification by phosphorylation (Fig. 3B).
To identify the phosphorylation sites of MdNHX1, total proteins were extracted from 35S::MdNHX1-Myc transgenic calli treated with 4% Glc. The liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis showed residue Ser at 275 site (Ser-275) was identified as a putative phosphorylation site (Supplemental Fig. S6). A monoclonal antibody against MdNHX1 phosphorylation site at this Ser-275 residue was made and named as the anti-MdNHX1S275 antibody. It successfully detected phosphorylated MdNHX1 in the wild-type calli and both phosphorylated MdNHX1 and MdNHX1-Myc in MdNHX1-Myc transgenic calli (Fig. 3C). Furthermore, CIP treatment abolished phosphorylation modification of MdNHX1 and MdNHX1-Myc proteins (Fig. 3C). In addition, it was found that the phosphorylation level of MdNHX1 protein was enhanced with Glc treatment duration (Fig. 3D).
To determine whether Ser-275 is essential for Glc-induced phosphorylation of MdNHX1, it was artificially mutated to Ala. Expression vector 35S::MdNHX1S275A-Myc was constructed and transformed into apple calli. The resultant transgenic calli 35S::MdNHX1S275A-Myc, together with 35S::MdNHX1-Myc, were then used to detect phosphorylation modification for MdNHX1 protein using an anti-MdNHX1S275-specific antibody. The result showed that the Ser-275-to-Ala-275 mutation completely abolished modification of MdNHX1S275A-Myc protein, but did not influence MdNHX1 phosphorylation (Fig. 3E), indicating Ser-275 is a crucial target for Glc-induced phosphorylation of MdNHX1.
Subsequently, western blotting with a phospho-specific anti-MdNHX1S275 antibody was performed using the wild-type control, 35S::MdHXK1, and 35S::antiMdHXK1 transgenic apple calli treated with Glc (Supplemental Fig. S7). The result showed that phosphorylation levels of MdNHX1 were higher in 35S::MdHXK1-overexpressing calli, and lower in 35S::antiMdHXK1 calli, compared to the wild-type control (Fig. 3F). Meanwhile, calli coexpressing 35S::MdNHX1-Myc + 35S::antiMdHXK1 were obtained and used to detect the phosphorylation level of MdNHX1. The result showed that the phosphorylation modification disappeared when MdHXK1 expression was suppressed (Fig. 3G).
MdHXK1 Enhances Stability and Transport Activity of MdNHX1 Protein
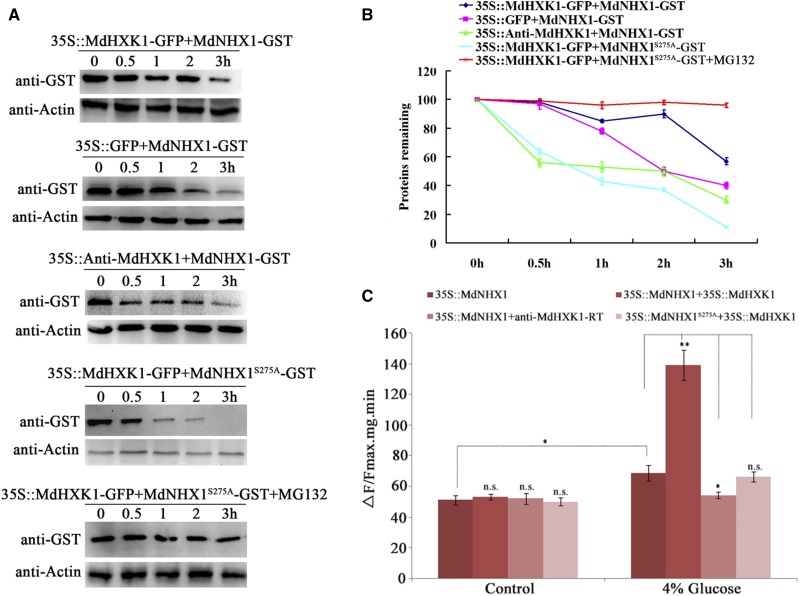
To examine if MdHXK1 affects the stability of MdNHX1 protein, cell-free degradation assays were performed by incubating recombinant proteins MdNHX1-GST and MdNHX1S275A-GST with total proteins extracted from MdHXK1 overexpression (35S::MdHXK1-GFP) and suppression (35S::anti-MdHXK1) transgenic calli, respectively. MdNHX1-GST degraded much more slowly in protein extracts of 35::MdHXK1-GFP calli but faster in those of 35S::anti-MdHXK1 apple calli than in the control protein samples extracted from 35S::GFP (Fig. 4, A and B), indicating that MdHXK1 promoted protein stability for MdNHX1. In contrast, MdNHX1S275A-GST protein degraded much faster than MdNHX1-GST in protein samples extracted from 35::MdHXK1-GFP calli (Fig. 4, A and B). These data indicated that MdHXK1 promoted protein stability for MdNHX1 and that Ser-275 played an important role in this process. In addition, MG132 treatment noticeably inhibited degradation of MdNHX1 (Fig. 4, A and B).
Figure 4.
MdHXK1 improves stability and Na+/H+ exchanger activity of MdNHX1. A, Cell-free degradation assay of recombinant protein MdNHX1-GST or MdNHX1S275A-GST in the protein extract of transgenic apple calli as labeled. Protein samples of each transgenic apple calli were extracted in degradation buffer. The levels of MdNHX1-GST or MdNHX1S275A-GST were visualized by immunoblotting using an anti-GST antibody. B, Quantification of MdNHX1-GST protein or MdNHX1S275A-GST levels as in (A). C, Na+/H+ exchanger activity of vacuolar membrane proteins in 35S::MdNHX1 and 35S::MdNHX1S275A transgenic calli after transformation with 35S::MdHXK1 or anti-MdHXK1-RT vector. Each value represents the mean ± sd from three separate experiments. n.s., P > 0.01; *, P < 0.01; **, P < 0.001.
Subsequently, a protoplast transient expression assay was performed to examine if MdHXK1 influences the transport activity of MdNHX1. First, two kinds of MdNHX1 transgenic apple calli, i.e. 35S::MdNHX1 and 35S::MdNHX1S275A, were obtained. Then, the sense overexpression vector (35S::MdHXK1) and the antisense suppression vector (35S::anti-MdHXK1) were transformed into protoplasts of 35S::MdNHX1 and 35S::MdNHX1S275A transgenic calli, respectively. Vacuolar membrane proteins were isolated to measure Na+/H+ exchanger activity. The result demonstrated that MdHXK1 overexpression increased Na+/H+ exchanger activity of 35S::MdNHX1 transgenic protoplasts upon 4% Glc treatment. When MdHXK1 was suppressed in 35S::MdNHX1 transgenic protoplast, however, Na+/H+ exchanger activity was not enhanced with Glc treatment. Additionally, when the phosphorylation site of MdNHX1 was mutated, expression change of MdHXK1 gene failed to influence Na+/H+ exchanger activity of 35S::MdNHX1S275A transgenic protoplasts (Fig. 4C), indicating that the phosphorylation site Ser-275 was important for this process. These results suggested that MdHXK1 improved Na+/H+ exchanger activity of MdNHX1 by phosphorylating it at Ser-275.
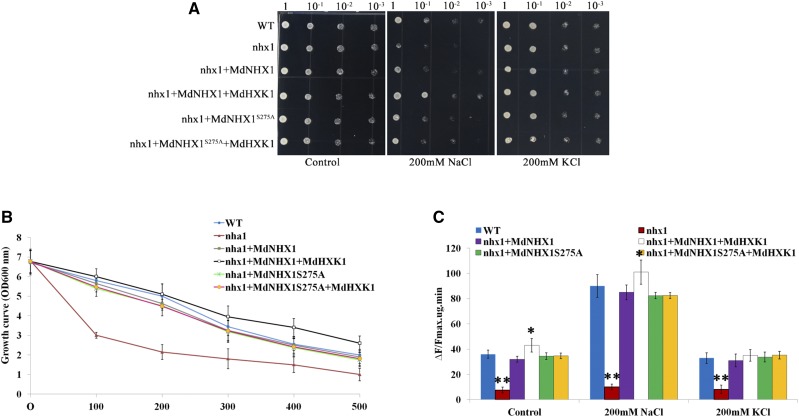
To further confirm that MdHXK1-mediated phosphorylation of MdNHX1 contributes to improved salt tolerance, the yeast mutant strain YDR456W bearing a disrupted endogenous sodium transporter NHA1 (nhx1) was used. MdNHX1, MdNHX1S275A, MdHXK1, and their combinations were genetically transformed into strain YDR456W. Under control conditions without salt stress, all yeast cells tested normally grew on selective Hartwell’s complete (SCU) solid medium with Glc (Fig. 5A). When shifted to NaCl-containing medium, however, YDR456W cells transformed with empty vector pYES2 were sensitive to 200 mM NaCl, whereas YDR456W cells transformed with MdNHX1-pYES2 exhibited recovery growth (Fig. 5A). In addition, MdHXK1-pYES2 was genetically transformed into MdNHX1 complementary transformants. The resultant double transgenic yeast cells exhibited the best growth among all kinds of cells tested (Fig. 5B). Furthermore, MdHXK1- and MdNHX1-coexpressed yeast cells exhibited higher Na+/H+ transport activity than the wild-type and MdNHX1 complementary cells growing in NaCl-containing liquid medium for 48 h (Fig. 5C). In addition, it was found that the salt sensitivity of YDR456W yeast cells was also rescued by a transformation with MdNHX1S275A-pYES2, but transformation of MdHXK1 into a MdNHX1S275A complementary strain failed to further improve salt tolerance. As controls, all yeast cells tested showed similar growth under conditions with 200 mM KCl treatment.
Figure 5.
MdHXK1 improves salt tolerance in yeast. A, MdHXK1 enhances salt tolerance of transgenic yeast cells expressing MdNHX1, but not MdNHX1S275A. Yeast cells of wild-type, mutant (nhx1), and recombinant strains were grown under the same concentration (OD value = 1) in SCU medium containing 4% (w/v) Glc. Two microliters of serial decimal dilutions were spotted onto plates of SCU medium supplemented with 0 mm NaCl (CK), 200 mm NaCl, and 200 mm KCl as indicated. Plates were incubated at 28°C for 4 d. B, The growth curve of each of the yeast cells in (A) under 0 mM to 500 mm NaCl treatment. C, The Na+/H+ transport activity of each of the transgenic yeast cells. The results were analyzed statistically using the two-tailed Student’s t test. Values are the mean of three replicates. Statistical significance was determined and compared to the control value using Student’s t test. n.s., P > 0.01; *, P < 0.01; **, P < 0.001. nhx1, mutant strain transformed with plasmid pYES2 empty vector; WT, wild-type strain transformed with plasmid pYES2 empty vector; nha1+ MdNHX1, mutant strain transformed with plasmid pYES2-MdNHX1; nha1 + MdNHX1+ MdHXK1, mutant strain transformed with pYES2-MdNHX1 and pYES2-MdHXK1.
MdHXK1 Improves Salt Tolerance At Least Partially in an MdNHX1-Dependent Manner
To verify the function of MdHXK1 in salt tolerance, 35S::MdHXK1 and 35S::anti-MdHXK1 apple calli were treated with 100 mm NaCl. The result showed that 35S::MdHXK1 transgenic calli exhibited higher salt tolerance, whereas 35S::anti-MdHXK1 showed lower salt tolerance compared to the wild-type control. To investigate whether MdHXK1-mediated salt tolerance depends on MdNHX1, a suppression vector 35S::anti-MdNHX1 was genetically transformed into 35S::MdHXK1 transgenic calli. The resultant double transgenic calli 35S::MdHXK1/35S::anti-MdNHX1 were treated with high salinity. The result showed that 35S::MdHXK1/35S::anti-MdNHX1 double transgenic calli were more sensitive to salt stress than 35S::MdHXK1 calli (Supplemental Fig. S8).
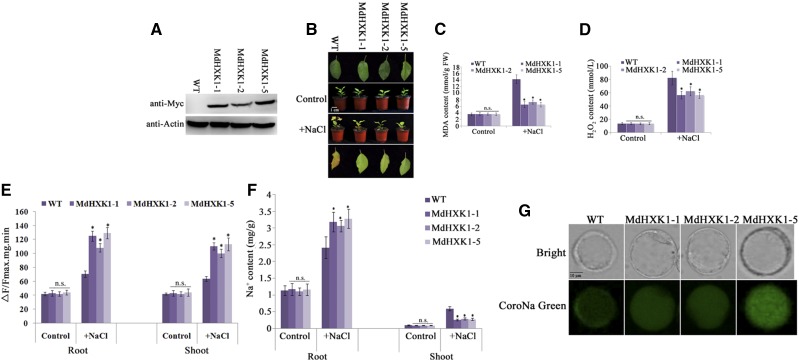
To further characterize its function in salt tolerance in planta, MdHXK1 was inserted into the pCXMyc-P plant expression vector downstream of CaMV 35S promoter to be fused with a Myc tag. The resultant expression vector was then genetically transformed into apple culitvar ‘Gala’. Five independent transgenic lines were obtained. Quantitative real-time PCR (qRT-PCR) analysis showed that transcript levels of three transgenic lines (MdHXK1-1/2/5) produced many more MdHXK1 transcripts than the ‘Gala’ control and were chosen for further investigation (Supplemental Fig. S9). Western blotting using a Myc antibody indicated that MdHXK1-Myc was detected in the transgenic plants (Fig. 6A). Three transgenic lines showed higher salt tolerance and accumulated less MDA and H2O2 under NaCl treatment than the wild-type control (Fig. 6, B to D). Subsequently, vacuolar membrane proteins were isolated from leaf and root, respectively, and used to detect Na+/H+ exchanger activity. The result showed that three transgenic lines showed much higher Na+/H+ exchanger activity than the wild-type control both in root and shoot (Fig. 6E). Meanwhile, it was found that root Na+ content was higher, whereas shoot Na+ content lower, in three transgenic lines compared to the wild-type control (Fig. 6F). Furthermore, root protoplasts were used for CoroNa Green fluorescent dye (Thermo Fisher Scientific) to detect Na+ accumulation in root cells. The result showed that strong fluorescence was observed in the cytosol of wild-type protoplasts, whereas transgenic protoplasts showed strong fluorescence in the vacuole of (Fig. 6G), indicating that MdHXK1 overexpression promoted Na+ accumulation in the vacuole.
Figure 6.
MdHXK1 overexpression improves salt tolerance in apple plants. A, Western-blotting assay of MdHXK1 protein abundance in ‘Gala’ and 35S:: MdHXK1-Myc transgenic plants using anti-Myc antibody. B, Salt tolerance of three transgenic lines i.e. MdHXK1-1, MdHXK1-2, and MdHXK1-5, compared to ‘Gala’. Plants were grown in soil for 2 months under normal condition and used for NaCl treatment. C and D, MDA content (C), H2O2 (D) of apple plants as treated as in (B). E, Na+/H+ exchange activity in the root and shoot of wild type and three transgenic lines. F, Na+ content in the root and shoot of wild type and three transgenic lines. G, CoroNa Green fluorescent dye showed Na+ accumulation in the protoplasts isolated from roots of wild-type and three MdHXK1 transgenic plants. Each value represents the mean ± sd from three separate experiments. Statistical significance was determined and compared to the control value using Student’s t test. n.s., P > 0.01; *, P < 0.01; **, P < 0.001.
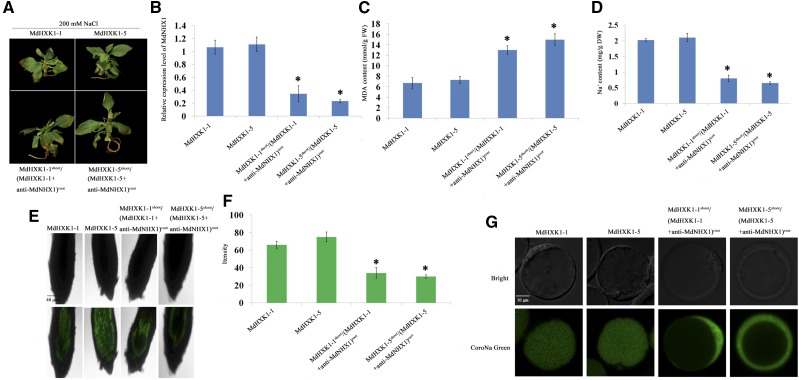
To examine if MdNHX1 is required for MdHXK1 function in salt tolerance, Agrobacterium rhizogenes MSU440 containing a specific antisense MdNHX1 fragment was used to quickly induce regeneration of MdNXK1-silenced hairy roots in two MdHXK1 transgenic lines (MdHXK1-1/5).The successfully transformed roots were distinguished by their exogenously expressed red fluorescence protein (RFP). Strong RFP signal was observed in transformant roots, demonstrating that the suppression vector with the MdNHX1 gene was successfully transformed and functional in hairy roots (Supplemental Fig. S10). As a result, chimeric transgenic plants MdHXK1-3shoot/(MdHXK1-3+anti-MdNHX1)root and MdHXK1-5shoot/(MdHXK1-5+anti-MdNHX1)root were obtained. They exhibited MdHXK1 overexpression in shoot and MdHXK1 overexpression plus MdNHX1 suppression in root. Subsequently, the wild-type, two MdHXK1 transgenic lines (MdHXK1-1/5), and two chimeric transgenic plants were treated with 200 mm NaCl for 2 weeks. MdNHX1 suppression in the root reduced salt tolerance of MdHXK1 transgenic lines (indicated by wrinkled leaves), increased MDA and H2O2, and decreased Na+ content (Fig. 7, A to D). Confocal imaging using CoroNa Green fluorescent dye demonstrated that two chimeric transgenic plants accumulated less Na+ than MdHXK1-1 and MdHXK1-5 transgenic plants in their roots (Fig. 7, E and F). Furthermore, the protoplasts were isolated for CoroNa Green fluorescent dye analysis. The result showed that a strong fluorescence signal was observed in the vacuole of MdHXK1 transgenic root, indicating intracellular Na+ was mainly transported and accumulated in the vacuole. However, when MdNHX1 expression was suppressed, fluorescence signal was observed mainly in the cytoplasm (Fig. 7G).
Figure 7.
MdHXK1 enhances salt tolerance via MdNHX1. A, Salt tolerance of two MdHXK1 transgenic lines (MdHXK1-1/5) and two chimeric transgenic plants MdHXK1-3shoot/(MdHXK1-3+anti-MdNHX1)root and MdHXK1-5shoot/(MdHXK1-5+anti-MdNHX1)root. These plants were treated with 200 mM NaCl for 1 week. Scale bar represents 1 cm. B, Relative expression levels of MdNHX1 in two MdHXK1 transgenic lines and two chimeric transgenic plants. For qRT-PCR analysis, samples were assayed in triplicate and normalized to 18S RNA. C and D, MDA content (C) and Na+ content (D) of apple plants treated as in (A). E, Bright-field images and CoroNa Green fluorescent dye images in roots of two MdHXK1 transgenic lines and two chimeric transgenic plants after treatment with 100 mm NaCl for 3 d. All images were taken using the same settings and exposure times to enable direct comparisons. Scale bar represents 40 μm. F, Mean CoroNa Green fluorescence intensity. G, CoroNa Green fluorescent dye of protoplasts isolated from the root of two MdHXK1 transgenic lines and two chimeric transgenic plants. Scale bar represents 10 μm. Values shown are means ± se (n = 10). Statistical significance was determined and compared to the control value using Student’s t test. n.s., P > 0.01; *, P < 0.01; **, P < 0.001.
A transient expression method using leaf showed the same result (Supplemental Fig. S11). These findings indicate that MdHXK1 promotes Na+ accumulation in the vacuole and enhances salt tolerance at least partially in a MdNHX1-dependent manner.
DISCUSSION
High salinity in soils severely affects crop quality and production. Here, it was found that Glc improved salt tolerance, which is mediated by a regulatory module composed of a Glc sensor MdHXK1 and a vacuolar Na+/H+ transporter MdNHX1. Our findings shed light on how plant cells enhance salt tolerance by improving their ability to sequestrate more Na+ into the vacuole in response to a Glc signal.
In addition to working as an energy material, osmotic protectant, and synthetic substrate for various compounds, Glc also acts as a direct and central signaling molecule in plants. It has emerged as a key regulator of many vital processes by Glc sensing and signaling mechanisms. The specific function of hexokinase1 (HXK1) in the plant Glc-signaling network is defined by loss-of-function mutant gin2 in Arabidopsis (Rolland et al., 2006). Arabidopsis HXK1 is found to mediate multiple functions in Glc repression and Glc promotion of transcription and growth. It has been documented that sugar signals including Glc appear to positively modulate salt and osmotic-stress responsive regulation of ABA biosynthesis and signaling (Gazzarrini and McCourt, 2001; León and Sheen, 2003). In this study, it was found that 4% exogenous Glc improved high salinity tolerance in apple (Fig. 1). HXK inhibitor glucosamine and MdHXK1-TRV infection demonstrated that Glc-mediated improvement of salt tolerance is dependent at least partially, if not completely, on the activity of MdHXK1 (Fig. 1; Supplemental Fig. S3).
HXK is a bifunctional protein possessing metabolic and signaling functions (Harrington and Bush, 2003). As is well known, amino acid residues G104 and S177 are crucial for its metabolic function. Both in Arabidopsis and apple, mutations to two key residues, i.e. G104D and S177A, were obtained. Two mutants successfully abolish the Glc phosphorylation activity but retain signaling functions. They exhibit a similar Glc signaling function as the wild-type HXK1 in coordinating nutrient, light, and hormonal signaling (Gazzarrini and McCourt, 2001; Rolland et al., 2006), demonstrating that Glc signaling is uncoupled from Glc metabolism. HXK catalyzes the first step of hexose metabolism (Claeyssen and Rivoal, 2007). Under unfavorable growth conditions, plants must change their metabolic processes to produce more sugars or sugar-deprived compounds to maintain proper energy and osmotic levels for countering stress (Hasegawa et al., 2000). Under low-Glc (i.e. 1% in this study) conditions, plants must produce more Glc to meet the preliminary requirement for survival under normal and stress conditions. Therefore, S177A and G104D mutations decreased salt tolerance due to their reduced metabolic activity (Supplemental Fig. S4). In contrast, they failed to influence salt tolerance under high-Glc (4%) conditions. In our previous report, it is also found that Glc-induced anthocyanin accumulation is mediated by both Glc signaling and metabolic functions of MdHXK1 (Hu et al., 2016). Taken together, our findings indicate that HXK1 mainly functions as an enzyme hexokinase to provide biosynthetic substrates and energy for plants via metabolic pathways under Glc-limited conditions, while also functioning as a Glc sensor in various processes such as anthocyanin biosynthesis and salinity response via a signaling pathway under Glc sufficient conditions.
In plants, HXK1 is the most ancient and evolutionarily conserved sugar sensor. Generally, upon sensing changing Glc levels, HXK1 transmits a signal to the nucleus and controls the expression of Glc-responsive genes (Rolland et al., 2001; Rolland and Sheen, 2005). In Arabidopsis, AtHXK1 forms a core complex with two HXK1 unconventional partners, i.e. the plant vacuolar H+-ATPase (VHA-B1/HUP1) and the 19S regulatory particle (RPT5B/HUP2) of the proteasome complexes in nucleus. This complex specifically binds to cis-regulatory elements upstream of CAB2 (Chl a/b binding protein 2) and CAB3 coding regions (Cho et al., 2006). Besides transcriptional regulation, it was also found through maize and the Arabidopsis mesophyll protoplasts system that Glc induces proteasome-dependent degradation of ETHYLENE INSENSITIVE3 (EIN3) proteins at the posttranslational level in the nucleus in a HXK1-dependent manner (Yanagisawa et al., 2003). In addition, MdbHLH3 is a key transcription factor involved in regulating anthocyanin biosynthesis (Xie et al., 2012). MdHXK1 interacts with and phosphorylates MdbHLH3 to modulate anthocyanin accumulation (Hu et al., 2016). Similarly, it was found in this study that MdHXK1 interacted with and phosphorylated a Na+/H+ exchanger MdNHX1 by posttranslationally modulating its protein abundance and transport activity (Figs. 3 and 4), thereby helping to maintain ion homeostasis in cells under salt stress. This work provides solid evidence for HXK1 acting as a Glc sensor to exert its signaling function by transcriptionally and posttranslationally regulating its targets, as well as for why sugars are conducive to salt tolerance.
It is well known that disruption of ion homeostasis is one of the consequences of high salt stress. Various ion transporters are key determinants of ionic homeostasis (Park et al., 2016). Among them, Salt Overly Sensitive1 (SOS1) is a plasma membrane-localized Na+/H+ antiporter. It mediates Na+ efflux from roots and loads Na+ ions into the xylem (Shi et al., 2000). In Arabidopsis, the SOS signaling pathway has been well characterized and is known to mediate ionic homeostasis and Na+ tolerance. Salt stress-generated Ca2+ signals activate the SOS3-SOS2 complex, which phosphorylates and activates the transport activity of SOS1 to mediate Na+ efflux from the cytoplasm (Ji et al., 2013). In addition to SOS1, the Na+ transporter high-affinity K+ transporter1 (HKT1) is another salt tolerance determinant that coordinates with SOS1 to regulate salt stress response and tolerance (Rus et al., 2001, 2004). AtHKT1 is predominantly localized in the plasma membrane of phloem cells and is involved in recirculating Na+ from the shoot to the root by unloading Na+ from the xylem and decreasing Na+ movement from the xylem into leaf cells (Rus et al., 2004).
Sequestering excess Na+ into the vacuole has long been proposed as an important mechanism for establishing ionic balance. Na+/H+ antiporters appear to form a multigene family that might show different temporal or spatial expression of various isoforms (Yokoi et al., 2002). The importance of vacuolar transporters for plant salt tolerance is underscored by the finding that overexpression of AtNHX1 appears to substantially improve plant salt tolerance (Bassil et al., 2012). In Arabidopsis, AtNHX1 comprises nine transmembrane domains, which is distinct from the human Na+/H+ antiporter NHE1 or any known Na+/H+ antiporter. Its N terminus faces the cytosol and a hydrophilic C-terminal hydrophilic region resides close to the vacuolar lumen (Yamaguchi et al., 2003). In Avicennia marina, NO enhances Na+ sequestration by stimulating protein expression of the vacuolar H+-ATPase and NHX1 (Chen et al., 2010). Apple tonoplast-located Na+/H+ exchanger MdNHX1 is a homolog of Arabidopsis AtNHX1 and positively regulates salt tolerance (Li et al., 2010; Sun et al., 2017).
The important role of vacuolar Na+/H+ antiporters in plant salt tolerance has been well highlighted by gene overexpression in a serious of plant species (Bassil and Blumwald, 2014). However, little is known about the regulatory pathway from various signals to Na+/H+ antiporters. It has already been reported that AtNHX1 activity is regulated by a calmodulin-like protein AtCaM15, which binds to its C terminus in response to stress-induced rises in intracellular Ca2+. This binding modifies the Na+-K+ selectivity of antiporter and decreases the Na+-K+ transport ratio (Yamaguchi et al., 2005). Besides the second messenger Ca2+, sugar also influences salt tolerance (Fig. 1; Dubey and Singh, 1999; Feng et al., 2002). Here, it is found that MdNHX1 is a target of Glc sensor kinase MdHXK1 for posttranslational phosphorylation modification (Fig. 3). It seems that it is a common regulatory module composed of protein kinases (such as SOS3-SOS2 complex and HXK1) and ion antiporters/exchangers (such as SOS1 and NHX1) that controls ion homeostasis in response to salt stress. In addition, it was found that Ser-275 is an important phosphorylation site for MdNHX1. As such, a Ser-275-to-Ala-275 (S275A) mutation abolished the MdHXK1-mediated MdNHX1 phosphorylation (Fig. 3E). In yeast cells, ectopic expression of MdNHX1S275A successfully recovered growth of the nhx1 mutant, indicating that MdNHX1S275A still has Na+/H+ exchange activity and functions as a Na+/H+ exchanger. However, MdHXK1 failed to further improve Na+/H+ exchange activity of MdNHX1S275A and enhance salt tolerance of yeast cells (Fig. 5), suggesting that MdNHX1 phosphorylation at site Ser-275 is crucial for HXK1-mediated salt tolerance in response to a sugar signal.
Based on a series of transgenic analyses, we found that MdHXK1 regulated salt tolerance at least partially in an MdNHX1-dependent manner. However, we cannot rule out the possibility that other genes may also participate in this salinity response. For example, MdHXK1 positively regulates the biosynthesis of anthocyanins that are active ROS scavengers (Hu et al., 2016). HXK1-promoted anthocyanin accumulation is conducive to a plant defense against ROS stress sponsored by high salinity. Taken together, our findings yield insight into the molecular mechanism by which the HXK1-NHX1 regulatory module maintains ion homeostasis toward Glc-induced salt tolerance.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Salt Treatments
Plantlets of Malus × domestic cv ‘Royal Gala’ apple were used. ‘Gala’ and transgenic apple shoot cultures were grown on Murashige & Skoog (MS) medium containing 0.6 mg l-1 6-BA, 0.2 mg l-1 IAA, and 0.1 mg l-1 GA and rooted in MS medium containing 0.15 mg l-1 NAA at 25°C under long-day conditions (16 h light/8 h dark). For normal soil growth, apple plantlets were transferred to pots containing a mixture of soil/perlite (1:1) and grown in the greenhouse under a 16-h light/8-h dark and 25°C/20°C d/n cycle.
‘Orin’ apple (M. domestica Borkh.) calli were used for genetic transformation. The calli were grown on MS medium supplemented with 0.5 mg l-1 IAA, 1.5 mg l-1 6-BA, and 3% (w/v) Suc at 25°C in the dark. The apple calli were subcultured three times at 15-d intervals before being used for genetic transformation and in other assays.
For phenotypic analyses of salt tolerance, apple ‘Gala’ plantlets were grown MS medium supplemented with 200 mM NaCl and different concentrations of Glc (1%, 2%, 4%, 6%, w/v) or 4% (w/v) mannitol. For salt treatment of apple plantlets grown in soil, plants were flooded every 2 d with NaCl solution, incrementally increasing with each successive watering from 100, 150, 200, and 250 to a final concentration of 300 mm NaCl. The apple calli was grown on solid medium containing 100 mM NaCl for 3 weeks.
Plasmid Construction and Genetic Transformation
Sense full-length sequences of MdHXK1 and MdNHX1 were amplified to construct sense overexpression vectors. Antisense partial 5′ UTR sequences of MdHXK1 and MdNHX1 were amplified to construct antisense suppression vectors. The resulting PCR products were inserted into the pCXSN vector under the control of the 35S promoter. These vectors were transformed into the ‘Gala’ cultures or ‘Orin’ calli using Agrobacterium (GV3101)-mediated transformation as described in Hu et al. (2016). All of the primers used in this study are listed in Supplemental Table S1, unless otherwise indicated.
RNA Extraction and qRT-PCR Assays
Total RNA was extracted with the Trizol Reagent (Invitrogen Life Technologies). A quantity of 2 μg of total RNA was used to synthesize first-strand cDNA with a PrimeScript First Strand cDNA Synthesis Kit (TaKaRa).
For qRT-PCR analysis, the reactions were performed with iQ SYBR Green Supermix in an iCycler iQ5 system (Bio-Rad) following the instructions of the manufacturer. The relative quantification of specific mRNA levels was performed using the cycle threshold (Ct) 2-DDCt method. For all analyses, the signal obtained for a gene of interest was normalized against the signal obtained for the 18S gene. All of the samples were tested in at least three biological replicates.
Protein Extraction and Western Blotting
A quantity of 2 g of apple calli or apple plants for each sample was ground in buffer containing: 20 mm Tris (pH 7.4), 100 mm NaCl, 0.5% Nonidet P-40 (v/v), 0.5 mm EDTA, 0.5 mm PMSF, and 0.5% (v/v) Protease Inhibitor Cocktail (Sigma-Aldrich). Protein extracts were separated on a 12% SDS-PAGE gel and transferred to PVDF membranes (Roche) using an electro-transfer apparatus (Bio-Rad). The membranes were incubated with MYC, GFP, GUS, or MdHXK1 (Sigma-Aldrich) primary antibodies and then peroxidase-conjugated secondary antibodies (Abcam) before visualization of immunoreactive proteins using an ECL Detection Kit (Millipore). Actin served as a protein loading control.
The Split-Ubiquitin Membrane-Based Yeast Two-Hybrid System
The DUAL membrane system is based on the split-ubiquitin mechanism to detect the interaction between an integral membrane protein and its interaction partners (Thaminy et al., 2003). MdNHX1 was fused to the C terminus of ubiquitin with an artificial transcription factor LexA-VP16 as bait, whereas MdHXK1 was fused with the N-terminal part of ubiquitin as prey. Interaction was determined by growth of yeast colonies on medium lacking -T/-l/-H/-A and by X-Gal assay.
In Vitro Pull-down Assay
The MdNHX1-coding sequence was cut with BamHI and SalI double digestion and cloned into pET32a with a His tag, and full-length cDNAs of MdHXK1 were cut by EcoRI and SalI double digestion and cloned into pGEX with a GST tag. Plasmids pGEX-MdHXK1 and pET32a-MdNHX1 were transformed into Escherichia coli BL21 (DE3; Transgene), respectively. For pull-down analysis, MdHXK1-GST proteins were first eluted from glutathione-agarose beads before being incubated with MdNHX1-His, which remained attached to the tetradentated-chelated nickel resin. In general, proteins were incubated at least 4 h at 4°C under shaking conditions before being centrifuged. Precipitates were washed no fewer than three times to remove unspecific bindings and boiled (10 min, 100°C). Then, the precipitates were further analyzed with SDS-PAGE and protein gel blotting.
Co-IP Assay
For IP assays, 1 mg of freshly extracted protein was precleaned with 30 mL of Protein A agarose beads (4 h, 4°C). The beads were centrifuged and the supernatant was transferred into a fresh tube and incubated with Myc/GUS antibody (overnight, 4°C). After brief centrifugation, four washing steps followed, after which loading buffer was added to the precipitates and boiled as described above. The precipitates were further analyzed by SDS-PAGE and western blotting using standard procedures.
BiFC Assay
The coding regions of MdHXK1 and MdNHX1 genes were cloned into pSPYNE-35S and pSPYCE-35S, which encode the N- or C-terminal regions of YFP (YFPN or YFPC), respectively, to generate plasmids MdHXK1-YFPN and MdbNHX1-YFPC as described by Xie et al. (2012). Arabidopsis CBL3 was fused to RFP and used as a vacuole-located marker. These constructs were transformed into the Agrobacterium tumefaciens GV3101 strain through electroporation.
A. tumefaciens strains carrying the fluorescent protein-fused constructs were infiltrated into Nicotiana benthamiana leaves and incubated at 24°C for 48 h before detection of YFP fluorescence. Imaging was performed using a laser scanning confocal microscope (LSM 510 Meta; Carl Zeiss) with excitation at 488 nm and the emission at 510 nm to 520 nm.
Identification of Phosphorylation Sites Using LC-MS/MS
MdNHX1 proteins were immunoprecipitated with Myc antibody-conjugated agarose beads and then separated with SDS-PAGE and stained with Coomassie Brilliant Blue. The band containing MdNHX1 protein was cut from the stained SDS-PAGE gel. Protein digestion, phosphopeptide enrichment, mass spectrometry data acquisition, data analysis, and label-free quantitation were carried out as described in Wang et al., (2013).
Detection of Phosphoproteins
A quantity of 1 g of wild-type or transgenic apple calli was preincubated in 10 mL MS liquid medium plus Glc (4%, w/v) for 1 h. Subsequently, the treated apple calli samples were ground for protein extraction. Additionally, a portion of extracted protein samples was treated with 5 U of CIP. Then, these protein samples were used for western blotting with a Myc antibody. Actin was served as a protein-loading control.
In Vitro Phosphorylation Assay
A total of 0.2 μg of recombinant protein kinase MdHXK1-His and 1 μg of MdNHX1-GST fusion proteins was incubated in 25 μL of reaction buffer [20 mm Tris (pH 7.5), 5 mm MgCl2, 10 mm NaCl and 2 mm DTT] with 100 μm ATP and [λ-32P] ATP (0.2 mCi per reaction) at room temperature for 30 min. The phosphorylated proteins were visualized using autoradiography after separation on a 12% SDS-PAGE gel.
Cell-free Degradation
Cells (E. coli, BL21) were induced by 0.1 mm IPTG and allowed to grow for 12 h at 16°C. MdNHX1-GST or MdNHX1S275A-GST were eluted from glutathione-agarose beads. The total proteins of the transgenic apple calli were subsequently extracted in degradation buffer containing 25 mm Tris-HCl (pH 7.5), 10 mm NaCl, 10 mm MgCl2, 4 mm PMSF, 5 mm DTT, and 10 mm ATP. The supernatant was collected and the protein concentration was determined by the Bradford assay reagent (Bio-Rad). Each reaction mix contained 100 ng of MdNHX1-GST protein or MdNHX1S275A-GST and 500 µg of total protein from transgenic apple calli. The reaction mixes were incubated at 22°C, and were stopped by the addition of SDS-PAGE sample buffer and boiled for 10 min. The results were quantified using Quantity One 1-D Analysis Software (Bio-Rad).
Construction of Viral Vectors and Transient Expression in Apple Leaves
To construct antisense expression viral vectors, a specific sequence in the 5′ UTR of MdHXK1 and MdNHX1 was cloned into the tobacco rattle virus (TRV) vector in the antisense orientation under the control of the dual 35S promoter. The vectors were named “TRV-MdHXK1” and “TRV-MdNHX1”. The antisense expression viral vectors were transformed into A. tumefaciens strain GV3101 for inoculation. The injected apple leaves were cultured on the medium without or with NaCl treatment in the dark at room temperature for 2 days. Then infected apple leaves were used for gene expression analysis and MDA, H2O2, and Na+ content determination.
Transformation System with A. rhizogenes MSU440
A specific cDNA fragment of MdNHX1 was amplified and cloned into the pK7GWIWG2 vector. The pK7GWIWG2 vector contained a ubiquitin promoter-driven red fluorescent protein gene and a spectinomycin resistance gene. The resulting vector was transformed into A. rhizogenes MSU440 and used for root infection. The resulting MSU440 can constitutively express red fluorescent protein, which can be used as a marker for successful transformation. The shoot cultures of apple ‘Gala’ cultivar with the same growth state were used for infection. Then, the infected shoot cultures were grown in MS + IAA (0.2 mg L−1) medium to induce roots.
CoroNa Green Dye Na+ Analysis
The Na+ content in apple roots was quantified using the green fluorescent Na+ dye CoroNa Green acetoxymethyl ester (Molecular Probes) as described in Wu et al. (2015). The CoroNa Green indicator stock was added to 10 mL of measuring buffer (10 mm KCl and 5 mm Ca2+-MES, pH 6.1) and diluted to a final concentration of 15 mM. The apple root tip was incubated in a solution containing 20 mM CoroNa Green for 2 h in the dark. Protoplasts isolated from roots of wild-type and transgenic plants were incubated in a solution containing 20 mM CoroNa Green for 1 h in the dark. After incubation, the samples were rinsed in a buffered MES solution and examined using confocal microscopy. Confocal imaging was performed using a laser scanning confocal microscope (LSM 510 Meta; Carl Zeiss). The excitation wavelength was set at 488 nm and the emission was detected at 510 nm to 520 nm.
Determination of Malondialdehyde, Na+, and H2O2 Contents
The determinations of MDA and H2O2 were performed as described in Ma et al. (2017). For Na+ content measurement, a SP-3500AA Series Atomic Absorption Spectrophotometer (Shanghai Spectrum Instrument) was used. Na+ contents were performed as in Dong et al. (2011).
Functional Identify of MdNHX1 and MdHXK1 in Yeast
The following Saccharomyces cerevisiae strain was used in this study: nhx1 mutant yeast, YDR456w (MAT a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0); and wild-type yeast, BY4741 (MAT a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0). The yeast cells were grown at 30°C in YPD medium adding 4% (w/v) Glc. Transformation of yeast cells was performed using the lithium acetate method. The transferred strains were grown on SCU medium.
Full length MdHXK1, MdNHX1, and MdNHX1S275A was inserted into the yeast expression vector pYES2. All plasmids were transformed into the yeast strain YDR456w for functional complementation. For the stress tolerance assays, yeast cells were normalized upon the absorbance of 0.12 at a wavelength of 600 nm. Four-mL aliquots of each 10-fold serial dilution were spotted onto SCU plates supplemented with KCl, or NaCl as indicated, and incubated at 30°C for 3 d. The empty vector pYES2 was transformed into the same yeast strains as a control.
Isolation of Tonoplast Vesicles and Na+/H+ Exchange Assays
After 4% (w/v) Glc treatment for 1 h, 2 g of apple calli were collected from the solid medium and washed twice by 1×PBS. As a control, apple calli were treated with 4% (w/v) mannitol to eliminate the effects of osmotic pressure. A quantity of 2 g of yeast cells was centrifuged at 5000g for 5 mins and washed twice by 1×PBS. Tonoplast vesicles were isolated from each sample according to previous methods (Blumwald and Poole, 1985). After purification, the Na+/H+ exchange activities were measured by monitoring the fluorescence recovery rates of acridine orange using a spectrofluorometer. A quantity of 50 μg of tonoplast vesicles was added to 1 mL reaction buffer including: 10 μM acridine orange, 300 mm Suc, 200 mm KCl, 2 mm NaN3. The reaction was started by adding 5 mm ATP-Tris and stopped by adding 250 nm bafilomycin. Then, different concentrations of NaCl were added. The Na+/H+ exchange activities were calculated as F (the fluorescence recovery) / (Fmax−Fmin) × μg protein/min.
Statistical Analysis
Samples were analyzed in triplicate, and the data are expressed as the mean ± sd unless noted otherwise. Statistical significance was determined using Student’s t test. A difference at P < 0.01 was considered significant, and P < 0.001 was considered extremely significant.
Accession Numbers
AtNHX1 (AT5G27150), AtNHX2 (AT3G05030), AtNHX3 (AT5G55470), AtNHX4 (AT3G06370), AtNHX5 (AT1G54370), AtNHX6 (AT1G79610), AtNHX7 (AT2G01980), AtNHX8 (AT1G14660), MdNHX1 (MDP0000041327), MdHXK1(MDP0000309677).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Salt tolerance of apple plantlets in medium plus 0 mM NaCl, 200 mM NaCl, and 200 mM NaCl plus 4% (w/v) mannitol, respectively.
Supplemental Figure S2. Relative expression of MdHXK1 and MdNHX1 in response to 4% (w/v) Glc treatment.
Supplemental Figure S3. MdHXK1 is required for the Glc-induced salt tolerance in apple.
Supplemental Figure S4. MdHXK1 is involved in Glc-mediated salt tolerance through the Glc metabolic and signaling pathways.
Supplemental Figure S5. Phylogenetic tree analysis of MdNHX1.
Supplemental Figure S6. Collision-induced dissociation mass spectrum showed the phosphorylation site was Ser (Thr) at residue 275 (Thr-275) of MdNHX1.
Supplemental Figure S7. Transcript level of MdHXK1 in 35S::Anti-MdHXK1 transgenic apple calli and the WT control.
Supplemental Figure S8. Salt tolerance of 35S::MdHXK1, Anti-MdHXK1-RT, and 35S::MdHXK1+Anti-MdNHX1.
Supplemental Figure S9. Transcription level of MdHXK1 in the overexpression lines compared to ‘Gala’ determined by qPCR.
Supplemental Figure S10. Root induction in MdHXK1 overexpression lines (MdHXK1-1/5) by MSU440 transformed with the antisense suppression vector of MdNHX1.
Supplemental Figure S11. Reliance on MdHXK1 on MdNHX1 for salt resistance in apple plant leaves.
Footnotes
This work was supported by grants from the Natural Science Foundation of China (31430074, 31601728), the Ministry of Education of China (IRT15R42), Shandong Province Government (SDAIT-06-03), Ministry of Agriculture of China (CARS-28), and Young Scientists Funds of Shandong Agricultural University (564024).
Articles can be viewed without a subscription.
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Blumwald E (2014) The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr Opin Plant Biol 22: 1–6 [DOI] [PubMed] [Google Scholar]
- Bassil E, Coku A, Blumwald E (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot 63: 5727–5740 [DOI] [PubMed] [Google Scholar]
- Blumwald E, Poole RJ (1985) Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol 78: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xiao Q, Wu F, Dong X, He J, Pei Z, Zheng H (2010) Nitric oxide enhances salt secretion and Na+ sequestration in a mangrove plant, Avicennia marina, through increasing the expression of H+-ATPase and Na+/H+ antiporter under high salinity. Tree Physiol 30: 1570–1585 [DOI] [PubMed] [Google Scholar]
- Cheng W, Zhang H, Zhou X, Liu H, Liu Y, Li J, Wang Y (2011) Subcellular localization of rice hexokinase (OsHXK) family members in the mesophyll protoplasts of tobacco. Biol Plant 55: 173–177 [Google Scholar]
- Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Claeyssen E, Rivoal J (2007) Isozymes of plant hexokinase: occurrence, properties and functions. Phytochemistry 68: 709–731 [DOI] [PubMed] [Google Scholar]
- Couée I, Sulmon C, Gouesbet G, El Amrani A (2006) Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot 57: 449–459 [DOI] [PubMed] [Google Scholar]
- Dong QL, Liu DD, An XH, Hu DG, Yao YX, Hao YJ (2011) MdVHP1 encodes an apple vacuolar H(+)-PPase and enhances stress tolerance in transgenic apple callus and tomato. J Plant Physiol 168: 2124–2133 [DOI] [PubMed] [Google Scholar]
- Dubey RS, Singh AK (1999) Salinity induces accumulation of soluble sugars and alters the activity of sugar metabolising enzymes in rice plants. Biol Plant 42: 233–239 [Google Scholar]
- Feng G, Zhang FS, Li XL, Tian CY, Tang C, Rengel Z (2002) Improved tolerance of maize plants to salt stress by Arbuscular mycorrhiza is related to higher accumulation of soluble sugars in roots. Mycorrhiza 12: 185–190 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol 122: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Gibson SI. (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gill PK, Sharma AD, Singh P, Bhullar SS (2003) Changes in germination, growth and soluble sugar contents of Sorghum bicolor (L.) Moench seeds under various abiotic stresses. Plant Growth Regul 40: 157–162 [Google Scholar]
- Harrington GN, Bush DR (2003) The bifunctional role of hexokinase in metabolism and glucose signaling. Plant Cell 15: 2493–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hu DG, Sun CH, Zhang QY, An JP, You CX, Hao YJ (2016) Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet 12: e1006273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Shi Z, Zhang Z, Zhang Y, Li H (2012) Effects of exogenous glucose on seed germination and antioxidant capacity in wheat seedlings under salt stress. Plant Growth Regul 68: 177–188 [Google Scholar]
- Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant 6: 275–286 [DOI] [PubMed] [Google Scholar]
- Kim H, Smith JE, Ridenour JB, Woloshuk CP, Bluhm BH (2011) HXK1 regulates carbon catabolism, sporulation, fumonisin B1 production and pathogenesis in Fusarium verticillioides. Microbiology 157: 2658–2669 [DOI] [PubMed] [Google Scholar]
- Kim M, Lim JH, Ahn CS, Park K, Kim GT, Kim WT, Pai HS (2006) Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63: 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S (2014) Sugar signals and the control of plant growth and development. J Exp Bot 65: 799–807 [DOI] [PubMed] [Google Scholar]
- León P, Sheen J (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Feng F, Liang D, Cheng L, Ma F, Shi S (2010) Overexpression of a Malus vacuolar Na+/H+ antiporter gene (MdNHX1) in apple rootstock M.26 and its influence on salt tolerance. Plant Cell Tiss Org 102: 337–345 [Google Scholar]
- Ma QJ, Sun MH, Lu J, Liu YJ, You CX, Hao YJ (2017) An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell Environ 40: 2207–2219 [DOI] [PubMed] [Google Scholar]
- Mita S, Suzuki-Fujii K, Nakamura K (1995) Sugar-inducible expression of a gene for beta-amylase in Arabidopsis thaliana. Plant Physiol 107: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Nemati I, Moradi F, Gholizadeh S, Esmaeili MA, Bihamta MR (2011) The effect of salinity stress on ions and soluble sugars distribution in leaves, leaf sheaths and roots of rice (Oryza sativa L.) seedlings. Plant Soil Environ 57: 26–33 [Google Scholar]
- Ohto MA, Hayashi K, Isobe M, Nakamura K (1995) Involvement of Ca2+ signalling in the sugar‐inducible expression of genes coding for sporamin and β‐amylase of sweet potato. Plant J 7: 297–307 [Google Scholar]
- Ohto MA, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim WY, Yun DJ (2016) A new insight of salt stress signaling in plant. Mol Cells 39: 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanagul W, Thitisaksakul M (2008) Effect of salinity stress on growth and carbohydrate metabolism in three rice (Oryza sativa L.) cultivars differing in salinity tolerance. Indian J Exp Biol 46: 736–742 [PubMed] [Google Scholar]
- Redillas MC, Park SH, Lee JW, Kim YS, Jeong JS, Jung H, Kim JK (2012) Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant Biotechnol Rep 6: 89–96 [Google Scholar]
- Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinha AK (2003) Extracellular invertase: key metabolic enzyme and PR protein. J Exp Bot 54: 513–524 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Sheen J (2005) Sugar sensing and signalling networks in plants. Biochem Soc Trans 33: 269–271 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal M, Prado FE (2009) Soluble sugars metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal Behav 4: 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Lee BH, Muñoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136: 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarowar S, Lee JY, Ahn ER, Pai HS (2008) A role of hexokinases in plant resistance to oxidative stress and pathogen infection. J Plant Biol 51: 341–346 [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Kumar J, Singh S, Singh VP, Prasad SM (2015) Roles of osmoprotectants in improving salinity and drought tolerance in plants: a review. Rev Environ Sci Bio 14: 407–426 [Google Scholar]
- Sun MH, Ma QJ, Liu X, Zhu XP, Hu DG, Hao YJ (2017) Molecular cloning and functional characterization of MdNHX1 reveals its involvement in salt tolerance in apple calli and Arabidopsis. Sci Hortic (Amsterdam) 215: 126–133 [Google Scholar]
- Thaminy S, Auerbach D, Arnoldo A, Stagljar I (2003) Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res 13: 1744–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser D, Huber SC (1997) Protein phosphorylation as a mechanism for osmotic-stress activation of sucrose-phosphate synthase in spinach leaves. Plant Physiol 114: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shabala L, Liu X, Azzarello E, Zhou M, Pandolfi C, Chen ZH, Bose J, Mancuso S, Shabala S (2015) Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front Plant Sci 6: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit coloration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E (2005) Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci USA 102: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100: 12510–12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30: 529–539 [DOI] [PubMed] [Google Scholar]
- Yu SM, Lee YC, Fang SC, Chan MT, Hwa SF, Liu LF (1996) Sugars act as signal molecules and osmotica to regulate the expression of α-amylase genes and metabolic activities in germinating cereal grains. Plant Mol Biol 30: 1277–1289 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Tian L, Liu H, Pan Q, Zhan J, Huang W (2009) Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul 58: 251–260 [Google Scholar]
- Zinselmeier C, Westgate ME, Schussler JR, Jones RJ (1995) Low water potential disrupts carbohydrate metabolism in maize (Zea mays L.) ovaries. Plant Physiol 107: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]