The Arabidopsis VOZ proteins interact physically with CONSTANS to modulate its function and promote photoperiod-dependent flowering.
Abstract
In plants, endogenous and environmental signals such as light control the timing of the transition to flowering. Two phytochrome B-interacting transcription factors, VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2, redundantly promote flowering in Arabidopsis (Arabidopsis thaliana). In the voz1 voz2 mutant, the expression of FLOWERING LOCUS C (FLC) was up-regulated and that of FLOWERING LOCUS T (FT) was down-regulated, which was proposed to be the cause of late flowering in voz1 voz2. However, the detailed mechanism by which the VOZ genes promote flowering is not well understood. Here, we show that neither the reduced FT expression nor the late-flowering phenotype of voz1 voz2 is suppressed in the voz1 voz2 flc triple mutant. Genetic interaction experiments between voz1 voz2 and constans-2 (co-2) mutants reveal that the VOZs and CO work in the same genetic pathway. Using in vitro pull-down, electrophoretic mobility shift, and bimolecular fluorescence complementation assays, we show that VOZ1 and VOZ2 interact with CO. The voz1 voz2 35S::CO:YFP plants show suppression of the early-flowering phenotype induced by CO overexpression, suggesting that CO requires VOZ for the induction of flowering. Determination of the VOZ consensus-binding site followed by genome-wide sequence analysis failed to identify any VOZ-binding sites near known flowering time genes. Together, these results indicate that the VOZ genes regulate flowering primarily through the photoperiod pathway, independent of FLC, and suggest that VOZs modulate CO function to promote flowering.
In plants, the onset of flowering is an important developmental decision that governs reproductive success. Therefore, plants have evolved several strategies to regulate the timing of this transition so that flowering occurs at the most optimal time of the year. Molecular and genetic analyses of flowering time mutants in Arabidopsis (Arabidopsis thaliana) and other model plants have identified distinct genetic pathways that integrate cues from various endogenous and environmental factors to regulate the onset of flowering (Koornneef et al., 1991; Simpson, 2004; Srikanth and Schmid, 2011; Song et al., 2013a, 2015).
Light (especially daylength and spectral light quality) is an important environmental factor that affects flowering time. Plants can sense and adapt to seasonal fluctuations in daylength (photoperiod), and based on their photoperiod requirement, plants can be classified as long day (LD), short day (SD), or day neutral. Daylength is perceived in the leaves, resulting in the production of a mobile flowering signal historically called florigen, which is then transported to the shoot apical meristem to initiate flowering (Zeevaart, 1976; Corbesier et al., 1996). Arabidopsis is a facultative LD plant that flowers earlier in inductive LDs compared with noninductive SDs.
Central to this daylength discrimination is the regulation of CONSTANS (CO), a B-box-type zinc finger transcription factor that promotes flowering in response to increasing daylengths (Koornneef et al., 1991; Putterill et al., 1995). Under the control of the circadian clock, the CO mRNA shows a biphasic diurnal oscillation pattern with one peak of expression toward the end of the photoperiod and a second in the night under LDs, whereas under SDs, the CO mRNA peaks only in the night (Suárez-López et al., 2001; Yanovsky and Kay, 2002). Two circadian clock-controlled proteins, GIGANTEA and FLAVIN-BINDING, KELCH REPEAT, F-BOX1, work as a heterodimeric complex in a blue light-dependent manner to degrade CYCLING DOF FACTOR1, a repressor of CO expression, late in the afternoon of LDs, resulting in a daytime peak in CO expression (Imaizumi et al., 2003, 2005; Sawa et al., 2007). Furthermore, regulation of the CO protein plays a key role in the LD-induced flowering transition. Light-mediated regulation of CO through photoreceptors results in stabilization of the CO protein at the end of LDs (Valverde et al., 2004). In the dark, the CO protein is degraded rapidly by the E3 ligase COP1 (Jang et al., 2008). As a consequence, the CO protein accumulates only under LDs and, in turn, activates the expression of FLOWERING LOCUS T (FT) in leaf phloem cells (Kardailsky et al., 1999; Kobayashi et al., 1999; Suárez-López et al., 2001; Yanovsky and Kay, 2002; An et al., 2004). Research in the last decade has established the FT protein as the main component of florigen, which moves from the leaf to the shoot apical meristem (Corbesier et al., 2007; Tamaki et al., 2007). At the shoot apex, FT forms a complex with FD, a bZIP transcription factor, and the FT-FD complex activates floral meristem identity genes such as APETALA1 to induce flowering (Abe et al., 2005; Wigge et al., 2005).
FT is not regulated exclusively by the photoperiod pathway. The expression of FT and SUPPRESSOR OF OVEREXPRESSOR OF CONSTANS1 (SOC1), collectively known as the floral integrators, is suppressed directly by the MADS domain-containing transcription factor FLOWERING LOCUS C (FLC), the major floral repressor in Arabidopsis (Michaels and Amasino, 1999; Helliwell et al., 2006; Searle et al., 2006). In the winter-annual accessions of Arabidopsis, FLC expression is induced to very high levels by FRIGIDA (FRI), thereby causing extremely late flowering in the absence of vernalization (Sanda et al., 1997; Levy and Dean, 1998; Michaels and Amasino, 1999; Le Corre, 2005). In response to vernalization, the FLC locus is epigenetically silenced by the well-orchestrated action of noncoding RNAs and protein factors, including VERNALIZATION INSENSITIVE3, VERNALIZATION1 (VRN1), and VRN2, rendering the plants competent to flower (Gendall et al., 2001; Levy et al., 2002; Sung and Amasino, 2004; Swiezewski et al., 2009; Heo and Sung, 2011; Song et al., 2013a). In contrast, the summer-annual varieties do not have fully functional FRI or FLC alleles and, therefore, flower early even without vernalization, in contrast to their FRI/FLC-positive counterparts (Johanson et al., 2000; Gazzani et al., 2003; Michaels et al., 2003; Shindo et al., 2005). Besides FRI and vernalization, FLC also is regulated by the autonomous pathway, which acts to suppress FLC as a function of plant age to control flowering time (Koornneef et al., 1991; Simpson, 2004). Consequently, mutations in genes in the autonomous pathway cause elevated FLC levels, conferring a late-flowering phenotype in both LDs and SDs.
Research by several groups in diverse model plant systems has led to the identification of a plethora of genes that affect flowering time and has helped to build a molecular and genetic framework of pathways that regulate flowering (Koornneef et al., 1991; Simpson, 2004; Srikanth and Schmid, 2011; Song et al., 2013a, 2013b, 2015). Despite the vast knowledge, our understanding of flowering time control is still incomplete, and more genes involved in these pathways are being discovered. Two such genes, VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2, were found to promote flowering in Arabidopsis (Yasui et al., 2012; Celesnik et al., 2013). VOZ1 and VOZ2 encode zinc finger transcription factors and have been shown to interact with and function downstream of phyB to regulate FT expression and, thereby, flowering transition (Mitsuda et al., 2004; Yasui et al., 2012). It was also demonstrated that the late-flowering phenotype of the voz1 voz2 double mutant plants was specific to LDs and that VOZ2 protein abundance in the nucleus was regulated by light quality in a phytochrome-dependent manner, suggesting a role for the VOZ genes in the light-dependent regulation of flowering (Yasui et al., 2012; Celesnik et al., 2013). Contrary to this, it was also reported that FLC expression was up-regulated in the voz1 voz2 mutant and that the late-flowering phenotype of the voz1 voz2 mutant was partially rescued in the flc-3 mutant background (Yasui et al., 2012; Celesnik et al., 2013; Yasui and Kohchi, 2014). Moreover, both the late-flowering phenotype and the elevated FLC expression of the voz1 voz2 mutant could be rescued by vernalization treatment (Celesnik et al., 2013). Thus, it is unclear whether the control of flowering by VOZ genes is light mediated or through the FLC-mediated pathway. Also, it remains unanswered what genes act downstream of VOZ1 and VOZ2 to control FT expression and, therefore, flowering.
In this study, we describe the roles of VOZ1 and VOZ2 in the photoperiodic regulation of flowering in Arabidopsis. As reported previously, we found that the voz1 voz2 double mutant exhibits LD-specific late flowering, elevated FLC expression, and down-regulated FT expression. Here, we demonstrate genetically that neither reduced FT expression nor the late-flowering phenotype of voz1 voz2 is suppressed by the flc mutation. We further show that, although VOZ and CO do not affect each other’s expression, they act in the same genetic pathway and interact physically with each other in vitro and in vivo. Although the determination of VOZ consensus DNA-binding sequences combined with microarray analysis failed to identify any known flowering time genes as direct transcriptional targets of VOZ, a genetic interaction between 35S::CO:YFP and voz1 voz2 showed that VOZs are required to manifest the early-flowering phenotype in CO-overexpressing plants. Based on these results, we propose a model where modulation of the function of the CO protein by VOZs contributes to promoting FT expression and photoperiodic flowering.
RESULTS
Establishment of voz1 and voz2 Mutants
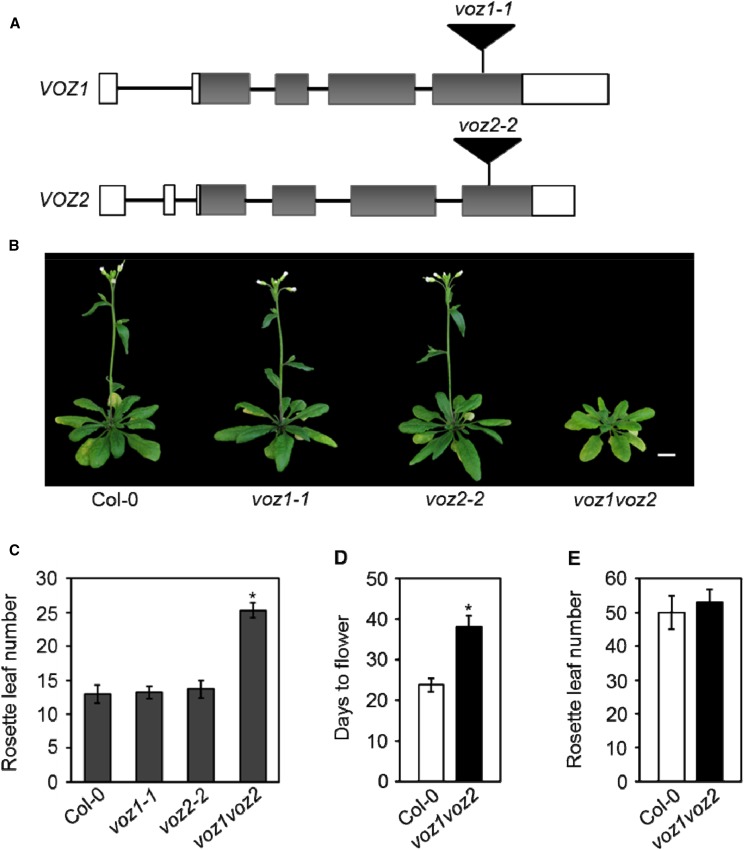
The VOZ1 and VOZ2 genes of Arabidopsis have been implicated in promoting the floral transition. We had previously established two homozygous T-DNA insertion lines, voz1-1 and voz2-2, to study the function of the VOZ genes (Fig. 1A). The precise positions of the insertions within the genes were determined by DNA sequencing, which showed that the VOZ1 and VOZ2 loci are disrupted by T-DNA at the fourth exon in the voz1-1 and voz2-2 alleles, respectively (Fig. 1A; Supplemental Fig. S1A). Since these two genes share a high level of sequence identity (Mitsuda et al., 2004), we also crossed voz1-1 with voz2-2 and generated the voz1-1 voz2-2 double mutant (referred to as voz1 voz2 hereafter) and tested the functional redundancy of the VOZ genes. To determine whether the transcripts of these genes are altered in the mutant lines, reverse transcription (RT)-PCR analysis was performed on the RNA samples isolated from Columbia-0 (Col-0), voz1-1, voz2-2, and voz1 voz2 plants and compared. While a full-length VOZ1 transcript was detected in Col-0 and voz2-2, no VOZ1 transcript could be detected either in voz1-1 or voz1 voz2 (Supplemental Fig. S1B). Similarly, the full-length VOZ2 transcript was absent from voz2-2 and voz1 voz2 mutants (Supplemental Fig. S1B). These results suggest that voz1-1 and voz2-2 are null alleles. Our results are in accordance with previously published results in the voz mutants (Yasui et al., 2012).
Figure 1.
VOZ1 and VOZ2 redundantly promote flowering only under LDs. A, Schematic showing the T-DNA (black triangles) insertions in the VOZ1 and VOZ2 loci. Allele names are shown above the respective insertions. White rectangles represent 5′ and 3′ untranslated regions (UTRs), gray rectangles represent exons, and black lines represent introns. B, Five-week-old plants of the indicated genotypes grown under LDs. Bar = 1 cm. C, Number of rosette leaves at bolting of the indicated genotypes grown under LDs. Data are shown as means ± sd (n ≥ 16). D, Days from germination to flowering of Col-0 and voz1 voz2 plants grown under LDs. Data are shown as means ± sd (n = 28). E, Number of rosette leaves at bolting of Col-0 and voz1 voz2 plants grown under SDs. Data are shown as means ± sd (n = 21). Asterisks in C and D indicate significant differences from Col-0 (P < 0.001, unpaired Student’s t test).
VOZ1 and VOZ2 Redundantly Promote Flowering in the Photoperiod Pathway
Under normal growth conditions (LDs), none of the mutant lines showed any striking morphological differences. However, the voz1 voz2 double mutant showed a considerable delay in flowering compared with Col-0 (Fig. 1B). A detailed examination of flowering time revealed that, while the voz1-1 and voz2-2 single mutants flowered at a similar time to Col-0, the voz1 voz2 double mutant showed a significant delay in flowering under LDs, when measured as either the number of rosette leaves at bolting or as days to flower (Fig. 1, C and D). To examine whether the voz1 voz2 late-flowering phenotype is photoperiod dependent, we studied flowering time under the SD condition and observed that the voz1 voz2 double mutant did not exhibit any significant difference in flowering time compared with the wild-type plants (Fig. 1E). These data suggest that VOZ1 and VOZ2 act redundantly to promote flowering specifically in the photoperiod pathway. These findings are consistent with other recently published studies on VOZ1 and VOZ2 (Yasui et al., 2012; Celesnik et al., 2013).
VOZ1 and VOZ2 Act Independently of FLC to Regulate the Floral Transition
In Arabidopsis, the photoperiodic induction of flowering is mediated by the activation of FT expression in leaves under LDs (Kardailsky et al., 1999; Corbesier et al., 2007; Tamaki et al., 2007). Therefore, to assess the role of VOZs in the photoperiodic regulation of flowering, we analyzed the expression of FT in the voz1 voz2 double mutant. As reported previously (Yasui et al., 2012), we found that FT expression was reduced significantly in voz1 voz2 compared with the wild type under LDs (Fig. 2A), suggesting that VOZ1 and VOZ2 (VOZs) activate FT expression to promote flowering.
Figure 2.
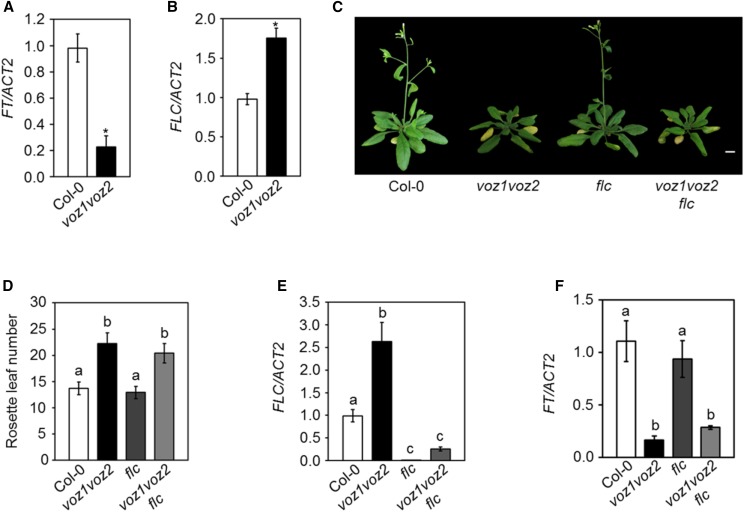
VOZ1 and VOZ2 regulate flowering independently of FLC. A and B, Relative transcript levels of FT (A) and FLC (B) analyzed by RT-quantitative PCR (qPCR). Asterisks indicate significant differences from Col-0 (P < 0.001, unpaired Student’s t test). C, Five-week-old plants of the indicated genotypes under LDs. Bar = 1 cm. D, Number of rosette leaves at bolting of the indicated genotypes under LDs. Data are shown as means ± sd (n = 15). Letters shared between the genotypes indicate no significant difference (P < 0.001, one-way ANOVA, Tukey’s multiple comparison test). E and F, Relative transcript levels of FLC (E) and FT (F) analyzed by RT-qPCR. Letters shared between the genotypes indicate no significant difference (P < 0.05, one-way ANOVA, Tukey’s multiple comparison test). For RT-qPCR, total RNA was isolated at zeitgeber time (ZT)-16 (A and F) and ZT-8 (B and E) from 14-d-old seedlings grown under LDs. RNA extraction was performed three times independently. The transcript levels were normalized to ACTIN2 (ACT2). Data are shown as means ± sd (n = 3).
Previously, it was reported that the late-flowering phenotype of the voz1 voz2 mutant was the result of increased FLC expression (Yasui et al., 2012). Since, in this study, we used a different combination of voz1 and voz2 single mutants to generate the double mutant, we reanalyzed the expression of FLC in the voz1 voz2 line. Consistent with the previous reports (Yasui et al., 2012; Celesnik et al., 2013), we found that FLC expression was up-regulated in the voz1 voz2 mutant as compared with the wild type (Fig. 2B), implying that VOZs repress FLC and, thereby, activate FT to promote flowering. On the contrary, the voz1 voz2 mutant exhibits late flowering specifically under LDs, and given the fact that FLC is a target of the autonomous pathway, it is expected that an increased FLC expression would cause a delay in flowering in both LDs and SDs. Therefore, to determine if the delay in voz1 voz2 flowering is indeed due to increased FLC expression, we used a genetic approach where voz1 voz2 was introduced into an flc mutant background that harbors a T-DNA insertion in the first intron. Interestingly, the voz1 voz2 flc triple mutant flowered at the same time as the voz1 voz2 double mutant under LDs (Fig. 2, C and D) under our growth conditions. The expression of FLC in voz1 voz2 flc was comparable to that of the flc single mutant (Fig. 2E). The low FT transcript level observed in the voz1 voz2 double mutant also was maintained in the voz1 voz2 flc triple mutant (Fig. 2F). Taken together, our results strongly suggest that the VOZ genes regulate FT expression and flowering independently of FLC.
VOZ1 and VOZ2 Act Together with CO in the Photoperiod Pathway
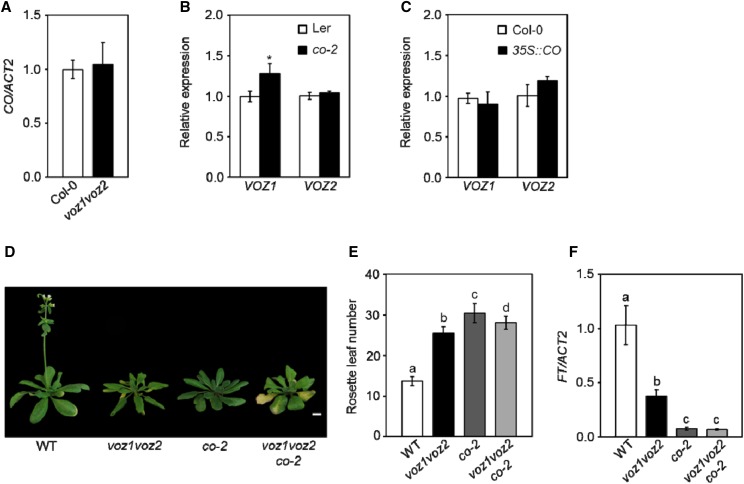
The observation that the voz1 voz2 mutant shows late flowering only in LDs suggests that VOZ1 and VOZ2 function in the photoperiod pathway. CO is central to the photoperiodic control of flowering and directly activates FT expression in the phloem companion cells specifically at the end of LDs (Yanovsky and Kay, 2002; An et al., 2004; Tiwari et al., 2010). Both CO and VOZs are expressed in the leaf vasculature (An et al., 2004; Mitsuda et al., 2004) and code for transcription factors that positively regulate FT expression under LDs (Putterill et al., 1995; Samach et al., 2000; Mitsuda et al., 2004; Yasui et al., 2012). Therefore, to understand the relation between VOZs and CO, we measured their transcript levels in co-2 and voz1 voz2 mutant lines, respectively. We found no significant difference in CO expression in the voz1 voz2 double mutant compared with the wild type (Fig. 3A), which is consistent with earlier reports (Yasui et al., 2012). Also, while FT was down-regulated significantly in the co-2 mutant (Supplemental Fig. S2A), the expression of VOZ1 and VOZ2 was largely unchanged in co-2 when compared with the wild type (Fig. 3B). To further strengthen this finding, we examined VOZ1 and VOZ2 expression in a CO-overexpressing line (35S::CO) that shows highly up-regulated CO and FT transcripts (Supplemental Fig. S2B) and, consequently, flowers early (Supplemental Fig. S2C). We found no significant difference in VOZ expression in 35S::CO compared with the wild type (Fig. 3C). Altogether, these results indicate that VOZs do not regulate CO transcription and vice versa.
Figure 3.
VOZs and CO function together in the photoperiod pathway to promote flowering. A, Relative CO transcript levels as analyzed by RT-qPCR. B, Relative VOZ1 and VOZ2 transcript levels as analyzed by RT-qPCR. The asterisk indicates a significant difference from Ler (P < 0.05, unpaired Student’s t test). C, Relative VOZ1 and VOZ2 transcript levels as analyzed by RT-qPCR. D, Five-week-old plants of the indicated genotypes grown under LDs. Bar = 1 cm. E, Number of rosette leaves at bolting of the indicated genotypes under LDs. Data are shown as means ± sd (n ≥ 15; P < 0.01, one-way ANOVA, Tukey’s multiple comparison test). F, Relative transcript levels of FT as analyzed by RT-qPCR. Letters shared between the genotypes indicate no significant difference (P < 0.05, one-way ANOVA, Tukey’s multiple comparison test). For RT-qPCR, total RNA was isolated at zeitgeber time (ZT)-12 (A), ZT-8 (B and C), and ZT-16 (F) from 14-d-old seedlings (A–C) and 11-d-old seedlings (F) grown under LDs. RNA extraction was performed three times independently. The transcript levels were normalized to ACT2. Data are shown as means ± sd (n = 3). WT, Wild type.
To test the interaction between VOZs and CO at the genetic level, we introduced the co-2 mutation into the voz1 voz2 double mutant background by genetic crossing and examined the flowering time of the voz1 voz2 co-2 triple mutant. Since co-2 is in Landsberg erecta (Ler) and voz1 voz2 is in the Col-0 background, we established wild-type, co-2, and voz1 voz2 homozygous plants from the co-2 × voz1 voz2 F2 individuals to serve as more appropriate controls, all of them being in a mixed Col-0/Ler background, for flowering time measurement. Flowering time analysis revealed that the co-2 mutant showed a strong late-flowering phenotype and flowered later than the voz1 voz2 mutant (Fig. 3, D and E). The voz1 voz2 co-2 triple mutant also flowered at a similar time to the parental genotypes (Fig. 3, D and E). Consistent with the above finding, the reduction in FT expression also was similar in co-2 and voz1 voz2 co-2 plants (Fig. 3F). The fact that the voz1 voz2 co-2 triple mutant did not display an additive late-flowering phenotype compared with the parents strongly implies that VOZ1 and VOZ2 act together with CO in the photoperiodic control of flowering.
VOZ1 and VOZ2 Interact Physically with CO
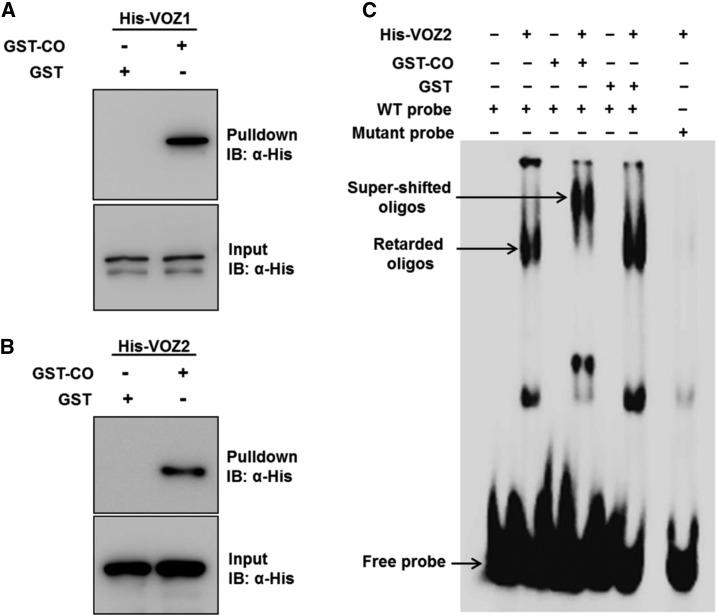
The above results show that VOZs and CO work in association in the photoperiod pathway to promote FT expression but do not affect each other’s expression, suggesting a physical interaction between these two proteins. This is supported by the spatial and temporal overlap in their expression patterns as demonstrated by reporter analysis (An et al., 2004; Mitsuda et al., 2004; Yasui et al., 2012). Furthermore, our in silico analysis using the Diurnal tool (Mockler et al., 2007) showed a correlated temporal expression of the VOZ1, VOZ2, and CO transcripts under LDs (Supplemental Fig. S3, A and B) and SDs (Supplemental Fig. S3, C and D). We tested the VOZ-CO interaction using an in vitro pull-down assay wherein recombinant His-VOZ1, His-VOZ2, GST-CO, and GST proteins were expressed in Escherichia coli. GST-CO or GST lysates were mixed with equal amounts of either His-VOZ1 or His-VOZ2 and incubated with glutathione-Sepharose beads in separate columns. The bead-bound proteins were eluted, separated by SDS-PAGE, and subjected to immunoblot analysis with an anti-His antibody (Fig. 4, A and B). Immunoblotting detected distinct bands corresponding to His-VOZ1 (Fig. 4A) and His-VOZ2 (Fig. 4B) in the proteins eluted from the GST-CO columns but not from the GST columns (Fig. 4, A and B), showing that both VOZ1 and VOZ2 interact with CO in vitro.
Figure 4.
VOZ1 and VOZ2 interact with CO in vitro. A and B, GST pull-down assay showing the interaction of VOZ1 and VOZ2 with CO. His-VOZ1 (A) or His-VOZ2 (B) proteins were incubated with GST or GST-CO together with glutathione-Sepharose beads in a column. The bead-bound proteins were subjected to 10% (w/v) SDS-PAGE and detected by immunoblot (IB) analysis using anti-His antibody. For the input blots, 0.5% (v/v) input extracts were loaded to detect His-VOZ1 or His-VOZ2. C, Electrophoretic mobility shift assay (EMSA) showing the interaction of VOZ1 and VOZ2 with CO. [γ-32P]ATP-labeled wild-type (WT) probe containing the VOZ2-binding site (GCGTGTGATACACGT) was incubated with His-VOZ2 alone or in combination with GST-CO or GST. As controls, GST-CO and GST alone were incubated separately with the wild-type probe. Additionally, His-VOZ2 was incubated with a probe containing a mutant VOZ2-binding site (tCGTGTGATACACGT).
To examine the VOZ-CO interaction further, we exploited the DNA-binding property of VOZ proteins. Previously, VOZ2 was shown to be capable of binding to the GCGT(N)7ACGT sequence (Mitsuda et al., 2004). A [γ-32P]ATP-labeled oligonucleotide containing the VOZ2-binding site (GCGTGTGATACACGT) was incubated with His-VOZ2 alone or in combination with GST-CO or GST (Fig. 4C). While His-VOZ2 was able to retard the wild-type probe, neither GST-CO nor GST alone bound to the wild-type probe (Fig. 4C). His-VOZ2, however, did not bind to the probe containing a mutant VOZ2-binding site (tCGTGTGATACACGT) used as a control (Fig. 4C), demonstrating that the interaction between VOZ2 and the wild-type probe is specific. His-VOZ2 in combination with GST-CO further retarded the wild-type probe (seen as a supershift) but not when used in combination with GST (Fig. 4C). The supershift observed in the mobility of the His-VOZ2-wild-type probe complex caused by GST-CO (Fig. 4C) further confirms the interaction of VOZ2 and CO in vitro.
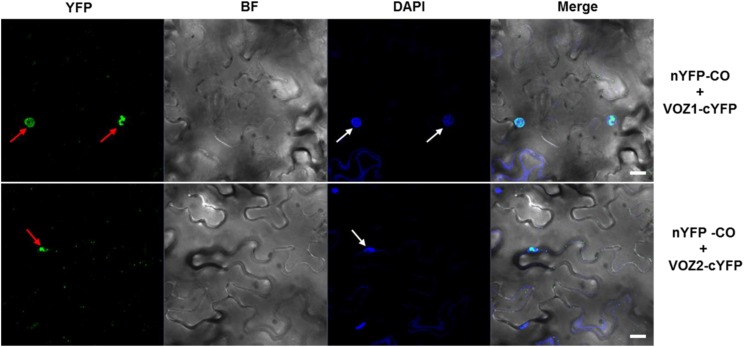
To test whether VOZs and CO interact in planta, we performed a bimolecular fluorescence complementation (BiFC) assay. For this, the N-terminal half of the yellow fluorescent protein (YFP)-coding gene was fused to CO and the C-terminal half to VOZ1 or VOZ2, and the resulting nYFP-CO and VOZ1-cYFP or VOZ2-cYFP constructs, respectively, were coexpressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens-mediated infiltration. Confocal microscopy revealed a distinct, reconstituted YFP signal in the nucleus of epidermal cells in leaves that coexpressed nYFP-CO/VOZ1-cYFP or nYFP-CO/VOZ2-cYFP (Fig. 5). However, no YFP fluorescence was detected in the nuclei that coexpressed nYFP-CO/cYFP, nYFP/VOZ1-cYFP, or nYFP/VOZ2-cYFP, which were used as negative controls (Supplemental Fig. S4). Taken together, these results show that both VOZ1 and VOZ2 interact physically with CO in vitro as well as in vivo.
Figure 5.
VOZ1 and VOZ2 interact with CO in vivo. BiFC analysis shows the interaction of VOZ1 (top) and VOZ2 (bottom) proteins with CO in N. benthamiana epidermal cells. Red arrows indicate YFP signal, and white arrows indicate the nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI). BF, Bright field; Merge, merge of YFP, BF, and DAPI. Bars = 20 µm.
Transcripts of Known Flowering Time Genes Remain Unaltered in the voz1 voz2 Mutant
Although the VOZ transcription factors are known to regulate the flowering transition as well as biotic and abiotic stress responses in Arabidopsis (Nakai et al., 2013a, 2013b), their direct transcriptional targets have not been reported so far. In an attempt to identify the VOZ targets that may mediate the control of flowering time, global transcriptomic analysis of the voz1 voz2 double mutant was performed using a DNA microarray. Comparative transcriptomic analysis revealed that 699 genes were up-regulated and 1,278 genes were down-regulated in the voz1 voz2 double mutant compared with Col-0 (Supplemental Data Set S1). Although genes involved in several important biological processes, such as stress regulation, energy metabolism, development, and signaling (Supplemental Fig. S5A), were deregulated, we did not detect any known flowering time genes that can be implicated in the late-flowering phenotype of voz1 voz2.
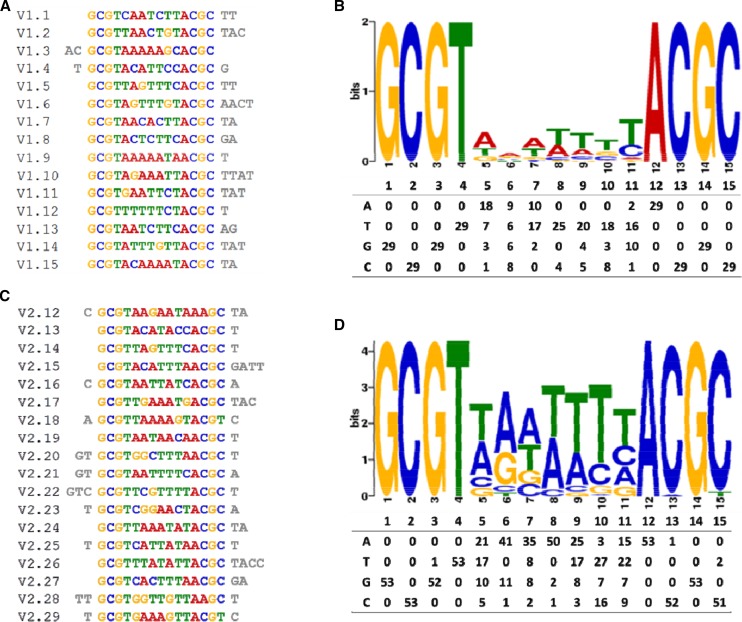
Therefore, we performed a systematic evolution of ligands by exponential enrichment (SELEX) experiment to determine the consensus DNA-binding elements of VOZ1 and VOZ2 (Pollock and Treisman, 1990). Analysis of the sequences obtained for VOZ1 and VOZ2 by SELEX (Fig. 6; Supplemental Tables S1 and S2) yielded a total of five possible consensus VOZ-binding sequences (Supplemental Fig. S5B). In a previous study (Mitsuda et al., 2004), VOZ1 and VOZ2 were shown to bind strongly to GCGT(N)7ACGC, while VOZ2 also bound to GCGT(N)7ACGT, albeit weakly, supporting the results from our SELEX analysis. To identify the genes that contain the VOZ consensus motifs in their regulatory regions and, therefore, are potentially the targets of the VOZ transcriptional activity, we scanned the Arabidopsis genome for the presence of all five VOZ consensus-binding sequences (Supplemental Fig. S5B) using the PatMatch service (https://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). The analysis revealed that a total of 774 genes contain at least one VOZ consensus motif within 1 kb of the upstream regulatory region (Supplemental Data Set S2) and 76 genes contain the motif within the 5′ UTR (Supplemental Data Set S3). At least one of these putative cis-elements (in the AT1G32640 locus) containing the GCGT(N)7ACGT motif bound to the VOZ2 protein efficiently and specifically, as determined by EMSA (Fig. 4C; Supplemental Fig. S5C). However, out of the 845 genes that contained VOZ consensus sites, we did not find any gene that can explain the LD-specific late-flowering phenotype of the voz1 voz2 mutant (see “Discussion”). The flowering integrator FT, whose transcript level was reduced in the voz1 voz2 double mutant (Fig. 2A), also did not have any VOZ-binding sites in its upstream regulatory region, suggesting an indirect control of FT by the VOZ proteins.
Figure 6.
A, 15 representative unique sequences obtained for VOZ1 SELEX. B, 18 representative unique sequences obtained for VOZ2 SELEX. C, Consensus logo for VOZ1 (top) obtained from the software tool MEME and the count matrix (bottom) showing the occurrence of each base. D, Consensus logo for VOZ2 (top) obtained from the software tool MEME and the count matrix (bottom) showing the occurrence of each base.
Therefore, we hypothesized that the regulation of flowering by VOZs is mediated by protein-protein interactions with CO. To test this, we crossed the early-flowering 35S::CO:YFP plants with the late-flowering voz1 voz2 double mutant and analyzed the flowering transition of the voz1 voz2 35S::CO:YFP plants. While 35S::CO-YFP plants flowered much earlier than the wild type and made only approximately eight leaves as compared with ∼16 leaves at bolting by Col-0 under LDs, the voz1 voz2 35S::CO:YFP plants flowered significantly later (Fig. 7). To test whether the elevated CO expression is maintained in the voz1 voz2 35S::CO:YFP plants, we determined the CO transcript abundance in these seedlings by RT-qPCR analysis. Even though the CO level was reduced compared with that in 35S::CO:YFP plants, the voz1 voz2 35S::CO:YFP seedlings showed a greater than 4-fold increase in CO transcript compared with Col-0 (Supplemental Fig. S6A). It has been demonstrated that a moderate 2- to 3-fold increase in CO expression is sufficient to cause a corresponding increase in FT expression and early flowering (Fornara et al., 2009). However, the 4-fold higher CO transcript level failed to promote FT expression in the voz1 voz2 35S::CO:YFP plants compared with Col-0 (Supplemental Fig. S6A). Taken together, this suggests that CO requires VOZ proteins for FT activation and, thus, its flowering-promoting effect.
Figure 7.
Genetic interaction between 35S::CO:YFP and voz1 voz2. A, Five-week-old plants of the indicated genotypes under LDs. B, Number of rosette leaves at bolting of the indicated genotypes under LDs. Data are shown as means ± sd (n = 15). Letters shared between the genotypes indicate no significant difference (P < 0.001, one-way ANOVA, Tukey’s multiple comparison test).
DISCUSSION
The role of VOZ genes in regulating flowering time has been reported previously (Yasui et al., 2012; Celesnik et al., 2013). However, the mechanism of this regulation and the components acting downstream to VOZs are not clearly known. In this study, we confirm that these genes promote the floral transition specifically through the photoperiod pathway. Using genetics, we show that VOZs act independently of FLC but in association with CO in the control of the floral transition. In vitro GST pull-down, EMSA, and BiFC analyses demonstrate that VOZ1 and VOZ2 interact physically with CO in vitro as well as in planta.
VOZs Regulate Flowering Independently of FLC
Results from previous studies (Yasui et al., 2012; Celesnik et al., 2013; Yasui and Kohchi, 2014) and from our analysis here show that the voz1 voz2 mutant has elevated FLC expression (Fig. 2C). Since FLC suppresses flowering by directly repressing FT, it could be presumed that the reduced FT expression and the late-flowering phenotype of the voz1 voz2 plants are due to higher FLC levels. Considering this possibility, the voz1 voz2 double mutant was expected to exhibit a late-flowering phenotype under both LDs and SDs like the autonomous pathway mutants, which show a photoperiod-independent late flowering due to increased FLC expression (Koornneef et al., 1991; Simpson, 2004). Contrary to this, voz1 voz2 plants show an LD-specific delay in floral transition (Fig. 1E), suggesting that the voz1 voz2 double mutant is defective in photoperiod sensing and that VOZs function in the photoperiod pathway. In fact, our results show that neither the down-regulation in FT expression nor the voz1 voz2 late flowering could be rescued in the voz1 voz2 flc triple mutant (Fig. 2, D and F). In contrast to our findings, recently it was shown that the voz1 voz2 late-flowering phenotype could be partially rescued in the flc-3 mutant background (Yasui and Kohchi, 2014). We speculate that the difference in the results could be due to the different flc allele used in our study. Recent studies also reported that the late flowering of the voz1 voz2 plants could be rescued by a vernalization treatment (Celesnik et al., 2013; Yasui and Kohchi, 2014). These contradictions can be explained by the fact that vernalization can promote flowering not only by repressing FLC but also via FLC-independent mechanisms (Michaels and Amasino, 2001). Thus, the up-regulation of FLC in the voz1 voz2 background seems to have only a minor role, if any, in the late flowering of the voz1 voz2 mutant and could not explain the LD-specific delay in the floral transition. Therefore, we conclude that VOZs promote flowering specifically in the photoperiod pathway largely independently of FLC.
VOZs Function with CO in the Photoperiod Pathway
Daylength is perceived in the leaves by CO, which activates FT expression in the phloem companion cells in response to long photoperiods (Suárez-López et al., 2001; Yanovsky and Kay, 2002; An et al., 2004). Similar to CO and FT, VOZ1 and VOZ2 are expressed strongly in the leaf vasculature (Mitsuda et al., 2004; Yasui et al., 2012), suggesting that the VOZ genes are involved in the photoperiodic control of flowering. In fact, similar to loss-of-function mutants of CO, the voz1 voz2 double mutant exhibits an LD-specific late-flowering phenotype, a characteristic feature of photoperiod pathway mutants (Koornneef et al., 1991). Our results demonstrate that, although FT expression is reduced in the voz1 voz2 background, CO does not affect VOZ expression and vice versa (Fig. 3, A–C). However, the genetic interaction between voz1 voz2 and co-2 mutants reveals that VOZs work together with CO to control FT expression in the photoperiod pathway (Fig. 3, D–F).
VOZ1 and VOZ2 Interact Physically with CO
The posttranscriptional regulation of CO by light is central to photoperiod-dependent flowering control (Valverde et al., 2004). A plethora of factors are known to interact with the CO protein to modulate its stability such that CO only accumulates under LDs toward the end of the day, which, in turn, activates FT to induce flowering (Song et al., 2015). CO and VOZs do not regulate each other transcriptionally and yet work in the same genetic pathway (Fig. 3), suggesting the possibility that VOZ proteins interact with CO. Our pull-down assays using recombinant VOZ1, VOZ2, and CO proteins clearly show that both VOZ1 and VOZ2 interact physically with CO in vitro (Fig. 4, A and B). The VOZ2-CO interaction was further demonstrated by EMSA (Fig. 4C). Furthermore, BiFC analyses demonstrate that VOZ1 and VOZ2 interact with CO in planta and that this interaction is observed specifically in the nucleus (Fig. 5).
VOZs Likely Promote Flowering Independently of Transcriptional Activity
The VOZ proteins are DNA-binding transcription factors capable of activating the transcription of target genes that contain the VOZ-binding sites (Mitsuda et al., 2004), yet the known flowering time genes are likely not the direct targets of their transcriptional activity (Supplemental Fig. S5; Supplemental Data Sets S1–S3). Of the 845 genes that contain VOZ consensus sites, four genes, namely EARLY FLOWERING4 (ELF4), DAY NEUTRAL FLOWERING (DNF), SUPPRESSOR OF AUXIN RESISTANCE3 (SAR3), and PHYTOCHROME AND FLOWERING TIME1 (PFT1), are known regulators of flowering time (Supplemental Table S3). Since ELF4 and DNF suppress the flowering transition by repressing CO transcription (Morris et al., 2010; Kim et al., 2013) and because CO transcript levels remain unaffected in the voz1 voz2 mutant (Fig. 3A), we inferred that VOZs regulate the flowering transition independently of ELF4 and DNF. Since SAR3 mutants flower early (Zhang and Li, 2005; Parry et al., 2006) and affect flowering strongly in the FRI-containing background (Jacob et al., 2007), the LD-specific late flowering of the voz1 voz2 mutant cannot be explained by SAR3 activity. Since PFT1 acts downstream of phyB and promotes flowering via CO-dependent and CO-independent pathways (Iñigo et al., 2012), it can potentially mediate VOZ-dependent flowering time control. To test this, we analyzed PFT1 expression in the voz1 voz2 mutant. However, no difference was observed in the PFT1 transcript level in the voz1 voz2 mutant compared with the wild type (Supplemental Fig. S6B).
The flowering time integrator FT is activated directly by CO and also is down-regulated in the voz1 voz2 double mutants. Even though overexpression of CO caused early flowering in the 35S::CO:YFP plants, it failed to exert its effect to the same extent in the absence of functional VOZ genes (Fig. 7). Thus, promotion of flowering by CO requires functional VOZ proteins. Since VOZ and CO form a stable complex (Figs. 4 and 5), it is possible that the CO protein is either stabilized or made functional by interacting with VOZ. It has been reported previously that overexpression of VOZ2 has no effect on FT or FLC expression beyond their respective levels in the wild type (Yasui et al., 2012). It is possible that VOZs exert their effect on FT expression, and thereby flower promotion, by associating with CO, the level of which acts as a limiting factor when VOZ is overexpressed.
Therefore, we speculate that the VOZ-CO interaction stabilizes CO and, therefore, the loss-of-function voz1 voz2 plants would have a reduced abundance of CO protein and, hence, reduced FT transcript, resulting in a delay in flowering. It is known that phyB destabilizes CO and that this effect is counteracted by PHYTOCHROME DEPENDENT LATE FLOWERING (PHL), which interacts with both phyB and CO (Valverde et al., 2004; Yasui et al., 2012; Endo et al., 2013). We show that VOZ proteins interact with CO (Figs. 4 and 5), and it was shown recently that VOZs also interact with phyB (Yasui et al., 2012). Therefore, it is possible that, similar to PHL, VOZs also inhibit phyB-mediated CO destabilization. Nevertheless, we do not rule out the possibility that VOZs may mediate CO stabilization independently of phyB. Additionally, while our SELEX and microarray experiments failed to identify direct transcriptional targets of VOZ1/VOZ2 that could be implicated in the control of flowering, it is still possible that VOZs regulate flowering also via yet unidentified target proteins.
CONCLUSION
In summary, our study provides new insights into the role of VOZ genes in the photoperiodic regulation of flowering. Our results reveal that VOZs work together with CO in the photoperiod pathway to promote flowering. Moreover, we demonstrate that VOZ1 and VOZ2 interact physically with CO in vitro as well as in planta. Finally, we show that VOZs are required by CO to promote flowering. Therefore, we propose that VOZs modulate CO function to regulate flowering by promoting FT induction. However, further research is required to determine the exact mechanism by which the VOZ-CO interaction affects CO activity.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) accessions Col-0 and Ler were used as wild-type controls. The voz1-1 (GABI_418B02), voz2-2 (SALK_115813), flc (SALK_003346), and co-2 (GABI_363408) mutants were obtained from the Arabidopsis Biological Resource Center (http://arabidopsis.org/). The voz1-1, voz2-2, and flc mutants are in the Col-0 background, whereas co-2 is in the Ler background. The voz1 voz2 (voz1-1 voz2-2), voz1 voz2 flc, and voz1 voz2 co-2 mutants were generated by crossing. The seeds of 35S::CO and 35S::CO:YFP lines were a kind gift from George Coupland. Plants were grown on Murashige and Skoog agar medium (Himedia), stratified for 3 d in dark at 4°C, and then transferred to LDs (16 h of light/8 h of dark) or SDs (8 h of light/16 h of dark) at 22°C. Light intensity was ∼130 μmol m−2 s−1.
Flowering Time Measurement
Flowering time was measured by recording the number of days from the day of sowing to the day when the flowering shoot grew ∼1 cm long and also by counting the number of rosette leaves at bolting. The number of cauline leaves also was counted separately in some cases.
Genotyping of Mutant Lines
Specific combinations of gene-specific and insertion-specific primers were used to genotype the voz1-1, vlb2-2, and flc mutants. To genotype the co-2 double mutant, a combination of gene-specific forward and gene-specific reverse primers were used to PCR amplify a product specific to the mutant co-2 allele. The product was digested with the PscI restriction enzyme (Thermo Scientific) and analyzed on an agarose gel for the presence of a co-2-specific banding pattern (324-, 292-, and 114-bp fragments). The wild-type CO allele produced a different banding pattern (616- and 114-bp fragments). A list of the primers used is presented in Supplemental Table S1.
RNA Isolation and Expression Analysis
Plants were grown at 22°C under LDs with fluorescent white light, and then whole seedlings were harvested at the indicated times (Figs. 2 and 3; Supplemental Figs. S2 and S6) for RNA isolation. Total RNA was extracted using the Tri Reagent (Sigma). After treatment with RNase-free DNase (Thermo Scientific), first-strand cDNA was synthesized from 2 µg of total RNA with oligo(dT18) primers using the SuperScript III reverse transcriptase (Invitrogen). For RT-qPCR, the cDNA samples were diluted with distilled water (1:10). Gene expression was monitored using 1 μL of the diluted cDNA in the SYBR-qPCR master mix (KAPA Biosystems), according to the manufacturer’s instructions. The following thermal profile was used for all RT-qPCRs: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. To determine the specificity of the reaction, a melt-curve analysis of the product was performed immediately after the final PCR cycle by gradually increasing the temperature from 60°C to 95°C at 0.05°C s−1. RT-qPCR was performed with the 7900HT real-time PCR detection system (ABI systems). Data analysis was performed using ABI Prism 7900HT SDS software (Applied Biosystems), and the relative expression was calculated using the equation 2−ΔΔCt, where ∆∆Ct = [(Ct gene of interest − Ct internal control) sample A − (Ct gene of interest − Ct internal control) sample B] (Schmittgen and Livak, 2008). Quantification of three independent biological replicates was performed for each sample. Expression of ACT2 or TUBULIN2 (TUB2) was used for normalization. RT-PCR was performed using Taq DNA polymerase (Thermo Scientific). A 20-µL PCR amplification was performed with respective primers, and after 30 cycles, the PCR products were visualized on an ethidium bromide-stained 1% (w/v) agarose gel. UBIQUITIN10 (UBQ10) was used as an internal control. Each reaction was repeated at least three times, and one representative result is shown. A list of the primers used is presented in Supplemental Table S1.
Plasmid Construction
For the GST pull-down assay and EMSA, the complete coding sequences of CO, VOZ1, and VOZ2 were PCR amplified using specific primers and cloned into the pGEMT-Easy vector (Promega) as described previously (Protocols and Applications Guide, online edition, 2005, Promega). The CO-pGEMT-Easy clone was restriction digested with BamHI and SalI, and the resulting fragment was ligated in frame into the pGEX-4T-1 vector to generate the pGEX-4T-1-CO fusion construct. The VOZ1-pGEMT-Easy and VOZ2-pGEMT-Easy clones were restriction digested with EcoRI, and the fragments were ligated in frame into pRSETB and pRSETC to generate pRSETB-VOZ1 and pRSETC-VOZ2 fusion constructs. For BiFC, the complete coding sequences of CO, VOZ1, and VOZ2 were first PCR amplified using specific primer combinations and then cloned into the pGEMT-Easy vector (Promega). The CO-pGEMT-Easy clone was restriction digested with BamHI and KpnI, and the fragment was ligated in frame into pSPYNE(R)173 vector (Waadt et al., 2008) to generate the nYFP-CO fusion construct. VOZ1-pGEMT-Easy and VOZ2-pGEMT-Easy clones were restriction digested with SpeI/KpnI and SalI/SmaI, respectively, and the fragments were ligated in frame into the pSPYCE(M) vector (Waadt et al., 2008) to generate VOZ1-cYFP and VOZ2-cYFP fusion constructs. For the SELEX experiment, the complete coding sequences of VOZ1 and VOZ2 were PCR amplified using specific primers and cloned in frame with the MBP coding sequence between the EcoRI and SalI sites of the pMALc2x vector to generate the pMALc2x-VOZ1 and pMALc2x-VOZ2 constructs. The primers used for the plasmid construction are listed in Supplemental Table S4.
Protein Purification
Competent Escherichia coli BL21 (DE3, pLysS) cells were transformed with the pGEX-4T-1 (GST), pGEX-4T-1-CO (GST-CO), pRSETB-VOZ1 (His-VOZ1), and pRSETC-VOZ2 (His-VOZ2) constructs. Primary cultures were prepared by inoculating 5 mL of Luria-Bertani broth, containing 100 μg mL−1 ampicillin and 25 μg mL−1 chloramphenicol, with a single colony of transformed bacteria using a sterile tip and incubated overnight in a shaking incubator (180 rpm) at 37°C. A total of 100 mL of prewarmed Luria-Bertani broth with appropriate antibiotics was inoculated with 1% (v/v) inoculum from the primary cultures. Protein expression was induced at 0.4 to 0.6 OD600 with 0.6 mm isopropylthio-β-galactoside at 18°C for 12 h, and the cells were harvested by centrifugation at 6,000 rpm for 10 min at 4°C. Cell pellets were resuspended in 5 mL of lysis buffer (50 mm Tris-Cl [pH 7.4], 150 mm NaCl, 1 mm DTT, 0.1% [v/v] Tergitol-type NP-40 [NP-40], and 0.2 mm phenylmethylsulfonyl fluoride [PMSF]). Cells were disrupted by sonication on ice, and the lysate was cleared by centrifugation at 13,000 rpm for 30 min at 4°C. The GST and GST-CO proteins were purified from the supernatant using GST-bind resin in a column (as described by the manufacturer [Novagen, Merck Millipore]). GST and GST-CO proteins were eluted with 10 mm reduced glutathione in 0.5-mL fractions, and His-VOZ1 and His-VOZ2 proteins were purified from the supernatant using Ni-NTA His-bind resin in a column (as described by the manufacturer [Novagen, Merck Millipore]). His-VOZ1 and His-VOZ2 proteins were eluted with 500 mm imidazole in 0.5-mL fractions. The purified GST, GST-CO, His-VOZ1, and His-VOZ2 proteins were dialyzed in 1,000 mL of dialysis buffer (50 mm Tris-Cl [pH 7.4], 150 mm NaCl, 1 mm DTT, 0.1% [v/v] NP-40, 0.2 mm PMSF, and 10% [v/v] glycerol) for 6 h. Protein concentrations were determined by a Bradford assay (Bradford, 1976), and the purities were analyzed by running the samples on 10% (w/v) SDS-PAGE.
In Vitro Pull-Down Assay
For in vitro pull-down assays, a combination of 20 µg each of either His-VOZ1 or His-VOZ2 and CO-GST proteins was incubated with 50 µL of glutathione-Sepharose beads (Novagen, Merck Millipore) in lysis buffer (50 mm Tris-Cl [pH 7.4], 150 mm NaCl, 1 mm DTT, 0.1% [v/v] NP-40, and 0.2 mm PMSF) for 2 h at 4°C. As a control, a combination of 20 µg each of either His-VOZ1 or His-VOZ2 and GST proteins was incubated with 50 µL of glutathione-Sepharose in lysis buffer for 2 h at 4°C. The samples were then resolved by 10% (w/v) SDS-PAGE and analyzed by immunoblotting using an anti-His antibody (1:5,000; Sigma) followed by an anti-mouse IgG secondary antibody (1:10,000; Sigma). For the input blots, 0.5% (v/v) input extracts were loaded to detect His-VOZ1 or His-VOZ2.
EMSA
Oligonucleotides were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Thermo Scientific). Binding reactions were performed in a 25-µL reaction volume containing oligonucleotide probe (5 × 104 cpm), binding buffer (10 mm HEPES [pH 7.8], 50 mm KCl, 5 mm MgCl2, 0.1% [w/v] BSA, 10 ng of herring sperm DNA, 10% [v/v] glycerol, and 5 mm DTT), and 0.5 µg of purified recombinant proteins. Binding reaction mixtures were incubated for 30 min at 4°C and resolved on a 6% (w/v) native polyacrylamide gel in 0.5× TBE buffer (45 mm Tris-borate and 1 mm EDTA). For the competition experiment, multifold excesses of either wild-type or mutant oligonucleotides were used along with the labeled wild-type probe. Electrophoresis was conducted at 4 V cm−1 for 1 h at 4°C. The gel images were analyzed with a BAS-2000 system (Fuji Film). The oligonucleotides used for this experiment are listed in Supplemental Table S4.
BiFC Assay
Agrobacterium tumefaciens (strain EHA-105) cultures harboring the BiFC constructs were grown overnight at 28°C in 10 mL of Luria-Bertani broth in the presence of appropriate antibiotics. The cells were harvested, resuspended in infiltration medium (10 mm MgCl2, 150 mm acetosyringone, and 10 mm MES, pH 5.6) to an OD600 of 1, incubated for 2 to 3 h at room temperature, and then infiltrated into 3- to 4-week-old Nicotiana benthamiana leaves grown under LDs. At 30 h post A. tumefaciens infiltration, the N. benthamiana leaves were infiltrated with 0.1% (v/v) DAPI solution for 5 min to visualize the nuclei. Leaf discs were cut and imaged for YFP fluorescence (excitation, 488 nm; emission, 520–545 nm) and for DAPI (excitation, 405 nm; emission, 410–474 nm) using a confocal laser scanning microscope (Zeiss; LSM 880).
Microarray Experiment
Total RNA was isolated from 14-d-old LD-grown Col-0 and voz1 voz2 seedlings using the Spectrum Plant Total RNA Isolation Kit (Sigma) according to the manufacturer’s instructions. Two biological replicates each were performed for Col-0 and voz1 voz2. The quality of the RNA samples was assessed with an Agilent Bioanalyzer (Agilent Technologies). RNA samples were labeled with a single-color dye (Cy3) using Agilent’s Quick-Amp labeling kit and hybridized onto an 8x60 K Arabidopsis Agilent microarray chip (Agilent Technologies). Data analysis and normalization were done using GeneSpring GX 12.0 Software. Genes whose expression changed by 2-fold or greater and had P ≤ 0.05 were considered to be up-/down-regulated.
SELEX Assay
The pMALc2x-VOZ1 and pMALc2x-VOZ2 constructs were transformed into BL21 (DE3, pLysS) E. coli cells. Five-milliliter cultures from the transformed cells were grown to 0.4 OD600 in the appropriate antibiotics (100 μg mL−1 ampicillin and 25 μg mL−1 chloramphenicol) and induced with 1 mm isopropylthio-β-galactoside. The cells were harvested and lysed by sonication in column buffer (20 mm Tris, pH 8, and 200 mm NaCl). The lysate was centrifuged, and the supernatant was used for SELEX.
The SELEX method was modified from Schommer et al. (2008). Briefly, MBP-VOZ1 and MBP-VOZ2 protein lysates were first bound to amylose beads on ice for 30 min in column buffer. After washing away the unbound protein, the beads were equilibrated with binding buffer: 20 mm Tris-Cl, pH 8, 50 mm NaCl, 5% (v/v) glycerol, 5 mm MgCl2, 1 mm DTT, and fresh herring sperm DNA (200 μg mL−1). A total of 200 ng of random double-stranded oligonucleotides (R704), consisting of the sequence 5′-GGAAACAGCTATGACCATG[N]18GTAAAACGACGGCCAGT-3′, was incubated with the protein-bead mix on ice for 30 min. After washing away the unbound oligonucleotides, the protein-DNA complex was eluted in 100 μL of water by boiling for 10 min. Selected oligonucleotides were amplified with primers complementary to the 19-bp flanking sequences of R704 (forward, 5′-GGAAACAGCTATGACCATG-3′; reverse, 5′-GTAAAACGACGGCCAGT-3′). The obtained PCR product was purified, and 10 μL was subjected to another round of selection with R704. With each round of selection, the number of PCR cycles was reduced as the fraction of high-affinity binding sites increased. Purified products after multiple rounds of selection were cloned into the pGEMT-Easy vector and sequenced.
Bioinformatic Analysis of the VLB-Binding Site
The PatMatch service of the TAIR Web site (http://www.arabidopsis.org) was used to identify VLB-binding sites in the upstream region of all Arabidopsis genes. The annotation of each gene was cited from the TAIR Web site.
Accession Numbers
Sequence data for the genes mentioned in this article can be found in the Arabidopsis Genome Initiative with the following accession numbers: At1g28520 (VOZ1), At2g42400 (VOZ2), At3g18780 (ACT2), At1g65480 (FT), At5g15840 (CO), At5g10140 (FLC), At1g32640 (MYC2), At5g62690 (TUB2), At1g25540 (PFT1), At4g05320 (UBQ10), At2g40080 (ELF4), At3g19140 (DNF), and At1g80680 (SAR3). Microarray data (accession no. GSE111342) have been deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Establishment of voz mutants.
Supplemental Figure S2. Relative expression of FT and CO.
Supplemental Figure S3. In silico analysis output of the diurnal expression of VOZ1, VOZ2, and CO.
Supplemental Figure S4. VOZ1 and VOZ2 interact with CO in vivo.
Supplemental Figure S5. Analysis of microarray results and VOZ-binding sequences.
Supplemental Figure S6. Relative expression of CO, FT, and PFT1.
Supplemental Table S1. Unique DNA-binding sequences obtained for VOZ1 by SELEX.
Supplemental Table S2. Unique DNA-binding sequences obtained for VOZ2 by SELEX.
Supplemental Table S3. List of flowering time genes containing VOZ consensus sequences in their upstream regulatory regions.
Supplemental Table S4. List of primers used in this study.
Supplemental Data Set S1. List of genes differentially expressed in the voz1 voz2 double mutant.
Supplemental Data Set S2. List of genes that contain a VOZ-binding site in the 1-kb upstream region.
Supplemental Data Set S3. List of genes that contain a VOZ-binding site in the 5′-UTR.
Acknowledgments
We thank George Coupland (Max Planck Institute for Plant Breeding Research) for 35S::CO and the 35S::CO:YFP lines, Ashis Nandi (Jawaharlal Nehru University) for the flc line, and the Arabidopsis Biological Resource Center for T-DNA insertion lines.
Footnotes
S.K. was supported by a fellowship from the Council of Scientific and Industrial Research, Government of India; P.C. and M.G. were supported by fellowships from MHRD, Government of India; the authors thank DST-FIST, UGC Centre for Advanced Study, and DBT-IISc Partnership Program for funding and infrastructure support.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Celesnik H, Ali GS, Robison FM, Reddy AS (2013) Arabidopsis thaliana VOZ (Vascular plant One-Zinc finger) transcription factors are required for proper regulation of flowering time. Biol Open 2: 424–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Gadisseur I, Silvestre G, Jacqmard A, Bernier G (1996) Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J 9: 947–952 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A (2013) PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci USA 110: 18017–18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, Peacock WJ, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD (2012) PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD (2007) The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol 144: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim Y, Lim J, Yeom M, Kim H, Kim J, Wang L, Kim WY, Somers DE, Nam HG (2013) ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep 3: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Le Corre V. (2005) Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol Ecol 14: 4181–4192 [DOI] [PubMed] [Google Scholar]
- Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10: 1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297: 243–246 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hisabori T, Takeyasu K, Sato MH (2004) VOZ: isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Morris K, Thornber S, Codrai L, Richardson C, Craig A, Sadanandom A, Thomas B, Jackson S (2010) DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 22: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Fujiwara S, Kubo Y, Sato MH (2013a) Overexpression of VOZ2 confers biotic stress tolerance but decreases abiotic stress resistance in Arabidopsis. Plant Signal Behav 8: e23358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakahira Y, Sumida H, Takebayashi K, Nagasawa Y, Yamasaki K, Akiyama M, Ohme-Takagi M, Fujiwara S, Shiina T, et al. (2013b) Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J 73: 761–775 [DOI] [PubMed] [Google Scholar]
- Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M (2006) The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Treisman R (1990) A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res 18: 6197–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sanda S, John M, Amasino R (1997) Analysis of flowering time in ecotypes of Arabidopsis thaliana. J Hered 88: 69–72 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. (2004) The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C (2013a) Remembering the prolonged cold of winter. Curr Biol 23: R807–R811 [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013b) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yasui Y, Kohchi T (2014) VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 repress the FLOWERING LOCUS C clade members to control flowering time in Arabidopsis. Biosci Biotechnol Biochem 78: 1850–1855 [DOI] [PubMed] [Google Scholar]
- Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T (2012) The phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24: 3248–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. (1976) Physiology of flower formation. Annu Rev Plant Physiol Plant Mol Biol 27: 321–348 [Google Scholar]
- Zhang Y, Li X (2005) A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]