The Arabidopsis receptor-like cytoplasmic kinase BSK1 phosphorylates MAPKKK5 to regulate plant defense responses.
Abstract
Arabidopsis (Arabidopsis thaliana) immune receptor FLAGELLIN SENSING2 (FLS2) rapidly forms a complex to activate pathogen-associated molecular pattern-triggered immunity (PTI) upon perception of the bacterial protein flagellin. The receptor-like cytoplasmic kinase BRASSINOSTEROID-SIGNALINGKINASE1 (BSK1) interacts with FLS2 and is critical for the activation of PTI. However, it is unknown how BSK1 transduces signals to activate downstream immune responses. We identified MEK Kinase5 (MAPKKK5) as a potential substrate of BSK1 by whole-genome phosphorylation analysis. In addition, we demonstrated that BSK1 interacts with and phosphorylates MAPKKK5. In the bsk1-1 mutant, the Ser-289 residue of MAPKKK5 was not phosphorylated as it was in the wild type. Similar to the bsk1 mutant, the mapkkk5 mutant displayed enhanced susceptibility to virulent and avirulent strains of the bacterial pathogen Pseudomonas syringae pv tomato DC3000, and to the fungal powdery mildew pathogen Golovinomyces cichoracearum. Phosphorylation of the Ser-289 residue is not involved in MAPKKK5-triggered cell death but is critical for MAPKKK5-mediated resistance to both bacterial and fungal pathogens. Furthermore, MAPKKK5 interacts with multiple MAPK kinases, including MKK1, MKK2, MKK4, MKK5, and MKK6. Overall, these results indicate that BSK1 regulates plant immunity by phosphorylating MAPKKK5 and suggest a direct regulatory mode of signaling from the immune complex to the MAPK cascade.
Plants use complex immune response to defend against pathogen infection (Jones and Dangl, 2006), including the immunity mediated by the pattern recognition receptors on plant surfaces that recognize conserved components of pathogenic microbes (i.e. pathogen-associated molecular patterns, PAMPs), such as flagellin and elongation factor Tu from bacteria, and chitin from fungi (Gómez-Gómez and Boller, 2000; Gómez-Gómez et al., 2001; Kunze et al., 2004; Chinchilla et al., 2006; Zipfel et al., 2006; Miya et al., 2007). This perception induces PAMP-triggered immunity (PTI), including callose deposition, a burst of reactive oxygen species (ROS), induced transcription of defense-related genes, and activation of mitogen-activated protein kinases (MAPKs; Couto and Zipfel, 2016; Tang et al., 2017).
A conserved 22-amino acid epitope of the N-terminal region of bacterial flagellin (flg22) is recognized by FLAGELLIN SENSING2 (FLS2), a well-studied receptor kinase in plants (Gómez-Gómez and Boller, 2000). The activation and transduction of FLS2 is tightly regulated, as activation of PTI leads to defects in growth and development (Couto and Zipfel, 2016; Shen et al., 2017; Stegmann et al., 2017; Tang et al., 2017). Once bound to the flg22 region, FLS2 rapidly recruits BRI1-ASSOCIATED RECEPTOR KINASE (BAK1) and phosphorylates downstream receptor-like cytoplasmic kinases (RLCKs) to induce PTI responses (Chinchilla et al., 2007; Heese et al., 2007; Schulze et al., 2010). RLCKs are the key components of receptor kinase complexes in plants due to their important role in regulating phospho-relay immune responses by detecting extracellular ligands (Liu et al., 2011; Lin et al., 2013a, 2013b; Yamaguchi et al., 2013). For instance, BOTRYTIS-INDUCED KINASE1 (BIK1), a member of the Arabidopsis (Arabidopsis thaliana) RLCK subfamily VII, interacts with FLS2 and BAK1 in the absence of elicitation by flg22. Upon perception of flg22, FLS2 associates with BAK1, and then BIK1 is phosphorylated and disassociates from the immune complex to transduce the signal downstream (Lu et al., 2010; Zhang et al., 2010). BIK1 directly phosphorylates the NADPH oxidase RbohD, thereby regulating the burst of ROS triggered by perception of flg22 (Kadota et al., 2014; Li et al., 2014).
BRASSINOSTEROID-SIGNALINGKINASE1 (BSK1), a member of the RLCK XII subfamily, plays an important role in the brassinosteroid (BR) signaling pathway (Tang et al., 2008) and is a key component of PTI. BSK1 physically associates with FLS2 and mediates a series of responses upon exposure to flg22 (Shi et al., 2013). The bsk1-1 mutant is more susceptible than wild type to the fungal powdery mildew pathogen Golovinomyces cichoracearum, and to virulent and avirulent strains of the bacterium Pseudomonas syringae pv tomato (Pto) DC3000. Powdery mildew resistance in enhanced disease resistance1 (edr1) and edr2 is dependent on BSK1 (Shi et al., 2013). Similar to BIK1, BSK1 associates with FLS2 in the absence of flg22, but disassociates from FLS2 and transduces the signal upon perception of PAMPs (Shi et al., 2013). Nevertheless, it is unknown how BSK1 transduces the signals from the FLS2 complex to induce downstream responses.
MAPK cascades are highly conserved signaling modules that transduce various extracellular stimuli to downstream targets during plant growth, development, abiotic stress, and defense responses (Kiegerl et al., 2000; Mockaitis and Howell, 2000; Nishihama et al., 2001; Asai et al., 2002; Tena et al., 2011; Meng and Zhang, 2013). The activation of MAPK cascades is one of the first events in plant immunity (Meng and Zhang, 2013). For instance, elicitation by flg22 leads to the activation of MKK4/MKK5 and MPK3/MPK6, and subsequently induces the expression of downstream defense-related genes (Asai et al., 2002). However, it is not well understood how the immune signal is transduced from the PAMP reception sites at the cell surface to the downstream MAPK pathway. Arabidopsis RLCK PBL27 positively regulates MAPKKK5 and MKK4/5 to specifically mediate chitin-triggered MPK activation (Yamada et al., 2016). The pbl27 mutant displayed weaker MPK3/6 activation in response to chitin, but stronger MPK3/6 activation in response to flg22 compared to the wild type (Shinya et al., 2014). In rice (Oryza sativa), OsRLCK185 (a rice ortholog of PBL27) transmits chitin signaling from OsCERK1 to an MAPK signaling cascade by interacting with several MAPKKKs, including OsMAPKKKε (Wang et al., 2017). Overexpression of OsMAPKKKε leads to enhanced chitin-induced MPK3/6 activation, whereas knockdown of OsMAPKKKε reduces chitin-induced MPK3/6 activation and enhances susceptibility to the rice blast fungus. OsRLCK185 physically interacts with and phosphorylates OsMAPKKKε, suggesting that OsCERK1, OsRLCK185, and a MAPK cascade mediate chitin signaling transduction in rice (Wang et al., 2017). By contrast, Yamada et al. (2017) demonstrated that OsMAPKKK18, a homolog of Arabidopsis MAPKKK5, functions in the rice chitin signaling pathway. Whether OsMAPKKK18 and OsMAPKKKε play redundant roles in chitin signaling pathway remains to be determined. The MAPKKKs are also reported to play a negative role in plant immunity. For example, EDR1, a Raf-like MAPKKK, physically associates with MKK4/MKK5 and negatively regulates the MAPK cascade to fine-tune plant defense responses and cell death (Zhao et al., 2014). Plus, MAPKKK7 was shown to interact with FLS2 to negatively regulate flg22-triggered MAPK activation by an unknown mechanism (Mithoe et al., 2016).
BSK1 is a key component in the FLS2 immune complex that plays a crucial role in plant immunity; however, the substrate of BSK1 is not known. To understand how BSK1 transduces the signals from the FLS2 complex to downstream components, we conducted a genomewide phosphorylation analysis using mass spectrometry with Arabidopsis wild-type and bsk1-1 mutant plants. In this assay, we found that the Ser-289 residue of MAPKKK5 was not phosphorylated in the bsk1-1mutant. Furthermore, we discovered that BSK1 directly interacts with MAPKKK5. Overexpression of MAPKKK5 in Nicotiana benthamiana induced cell death in the injected leaves. The mapkkk5 mutant showed enhanced susceptibility to Pto DC3000 and G. cichoracearum. The phosphorylation ofSer-289 on MAPKKK5 by BSK1 is critical for MAPKKK5 immune function. Our results indicate that MAPKKK5 is the phosphorylation substrate of BSK1, and BSK1 regulates plant immunity by modulating MAPKKK5.
RESULTS
MAPKKK5 Displays Differential Phosphorylation in the Wild Type and bsk1-1 Mutant
BSK1 encodes a RLCK, which functions in both BR signaling and plant immunity by associating with the BR receptor BRI1 and the PAMP receptor FLS2 (Tang et al., 2008; Shi et al., 2013). To understand the molecular mechanism of BSK1-mediated resistance, we conducted a genomewide phosphorylation analysis. This analysis identified a number of peptides that were differentially phosphorylated between the wild-type and bsk1-1mutant plants, which carry a point mutation (R443Q) in the conserved tetratricopeptide repeat domain in BSK1 (Shi et al., 2013; Supplemental Table S1). BSK1 is a functional protein kinase (Shi et al., 2013); therefore, these differentially phosphorylated proteins might include the substrate of BSK1. Phosphorylation at the Ser-230 residue of BSK1, which is the major phosphorylation site for the BR receptor BRI1 (Tang et al., 2008), was found in the wild type but not in bsk1-1, indicating that the whole-genome phosphorylation analysis was an effective approach. We also found that phosphorylation was absent at the Ser-289 residue in MAPKKK5 in the bsk1-1 mutant. MAPKKK5 is a member of the MEKK subfamily (Ichimura et al., 2002), which is phosphorylated by PBL27 and plays a role in chitin-triggered MAPK activation (Yamada et al., 2016). Because MAPK pathways are important signaling modules that play pivotal roles in PTI (Meng and Zhang, 2013), we focused on MAPKKK5 in this study.
MAPKKK5 Interacts with BSK1
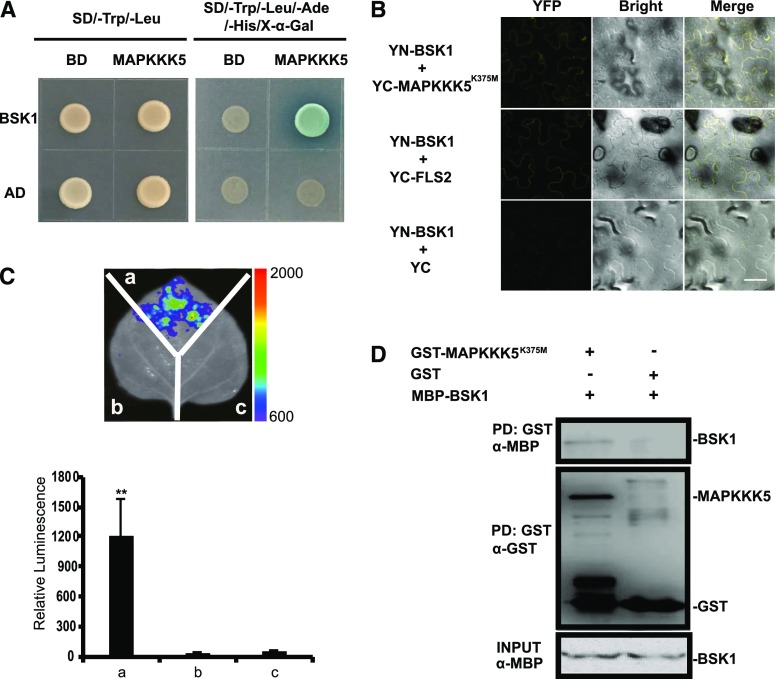
Because phosphorylation was lacking at the Ser-289 residue of MAPKKK5 in the bsk1-1 mutant, we hypothesized that MAPKKK5 may be a substrate of BSK1. To test this hypothesis, we first investigated, by several independent experiments, whether these two proteins physically interact. In a yeast two-hybrid assay, we cotransformed pGADT7-BSK1 and pGBKT7-MAPKKK5, with the corresponding empty vectors as the negative controls, into yeast strain AH109. As shown in Figure 1A, BSK1 interacted with MAPKKK5 in yeast.
Figure 1.
MAPKKK5 interacts with BSK1. A, MAPKKK5 interacted with BSK1 in the yeast two-hybrid assay. Yeast cells containing the indicated plasmids were spotted onto the SD/-Trp/-Leu or SD/-Trp/-Leu/-Ade/-His/X-α-Gal medium, as indicated. Photos were taken after 3 d of incubation. B, The interaction between BSK1 and MAPKKK5 was examined with the BiFC assay in N. benthamiana. BSK1 and MAPKKK5K375M were fused to the N terminus or C terminus of the YFP fragment, respectively. YFP fluorescence was detected by confocal microscopy. Bar = 50 μm. The combination of YN-BSK1 and YC-FLS2 was used as the positive control. C, The interaction between BSK1 and MAPKKK5 was examined with the LUC assay. Agrobacterium tumefaciens strain GV3101 containing the indicated construct pairs was coinfiltrated into N. benthamiana leaves. The infiltrated leaves were removed and sprayed with 1 mm luciferin and the fluorescence signal was captured by a CCD camera at 2 d after injection. Left panel, a, Cluc-BSK1 + MAPKKK5K375M-Nluc; b, empty vector (Cluc) + MAPKKK5K375M-Nluc; c, Cluc-BSK1 + empty vector (Nluc). The color bar at the right side of the left panel displays the relative luminescence units. Right panel, the relative luminescence unit in the histogram was measured 3 d after injection. Data represent mean ± sd. Two asterisks indicate statistically significant differences [n ≥ 8; P < 0.01; one-way ANOVA (ANOVA)]. D, The interaction between BSK1 and MAPKKK5 was examined with the in vitro pull-down assay. MBP-tag fused BSK1 protein was incubated with GST-bound beads and GST or GST-fused MAPKKK5K375M. Protein was detected by an immunoblot assay with α-GST or α-His antibodies, respectively. AD, pGADT7; BD, pGBKT7.
To confirm the interaction between MAPKKK5 and BSK1 in planta, we performed bimolecular fluorescence complementation (BiFC) assays. The kinase-inactive version of MAPKKK5 (MAPKKK5K375M) has a disrupted ATP binding site and was used in this experiment because the expression of native MAPKKK5 in N. benthamiana leaves caused extensive cell death (Yamada et al., 2016). We fused MAPKKK5K375M with the C terminus of YFP (YC), fused BSK1 with the N terminus of YFP (YN), and then transiently expressed the fused proteins in N. benthamiana leaves by Agrobacterium injection. We used the YN-BSK1 and YC-FLS2 pair as a positive control, and YN-BSK1 with YC as a negative control. We observed the YFP signal in leaves that expressed YN-BSK1 and YC-MAPKKK5K375M as well as the positive control leaves, but the signal was absent in the negative control leaves (Fig. 1B). This indicated that BSK1 interacted with MAPKKK5 in N. benthamiana.
Additionally, we performed a firefly split-luciferase complementation assay in N. benthamiana leaves to investigate the interaction. In this assay, the N terminus (Nluc) of firefly luciferase was fused to MAPKKK5K375M, whereas the C terminus (Cluc) was fused to BSK1. We transiently expressed MAPKKK5K375M-Nluc and BSK1-Cluc, as well as MAPKKK5K375M-Nluc and Cluc, plus BSK1-Cluc and Nluc as negative controls. A strong fluorescence signal was detected in leaves that expressed both MAPKKK5K375M-Nluc and BSK1-Cluc, but not in the negative controls (Fig. 1C), indicating that MAPKKK5K375M associated with BSK1 in N. benthamiana.
We then performed a pull-down assay to confirm the interaction between MAPKKK5 and BSK1. Because it was not possible to express and purify wild-type MAPKKK5 protein in Escherichia coli, we instead expressed and purified the MAPKKK5K375M and GST fusion protein. This was followed by an in vitro GST pull-down assay by incubating GST-MAPKKK5K375M or GST with MBP-BSK1. As shown in Figure 1D, MBP-BSK1 was detected when incubated with GST-MAPKKK5K375M but not in the negative control, indicating that BSK1 interacted with MAPKKK5K375M in vitro. Taken together, these observations indicate that BSK1 and MAPKKK5 physically associate.
The Kinase Domain Is Essential for the Interaction Between MAPKKK5 and BSK1
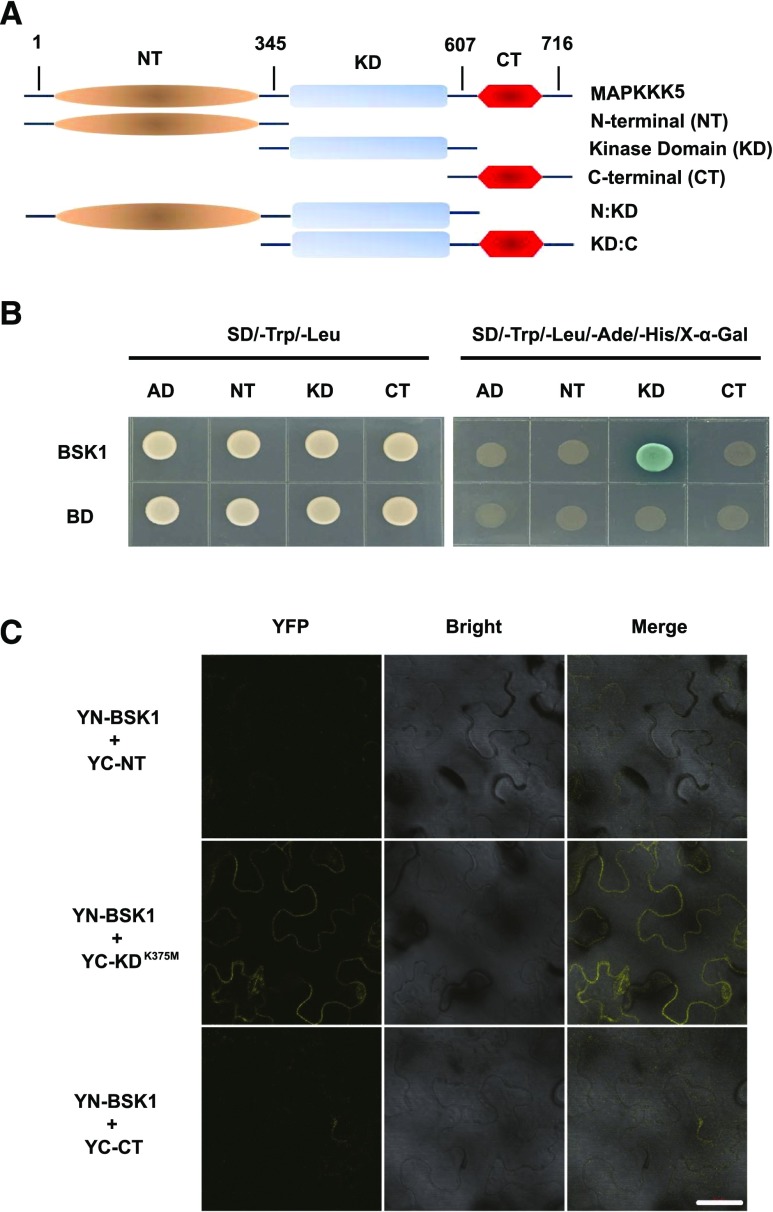
MAPKKK5 contains unknown N-terminal and C-terminal structural domains, and a kinase domain in the middle (Fig. 2A). To investigate which domain of MAPKKK5 is responsible for the interaction with BSK1, we generated several truncated forms of MAPKKK5 including the N terminus (NT), the kinase domain (KD), and the C terminus (CT) (Fig. 2A) and performed yeast two-hybrid and BiFC assays. Interestingly, although the Ser-289 residue is in the NT domain, the potential phosphorylation site in MAPKKK5 for BSK1, only yeast cotransformed with BD-BSK1 and AD-MAPKKK5KD could grow on the synthetic dropout (SD) selective medium (Fig. 2B). This suggested that the KD of MAPKKK5, but not the NT or CT domains, could interact with BSK1. Similarly, in the BiFC assay, the YFP signal was detected only in N. benthamiana leaves expressing YN-BSK1 and YC-MAPKKK-KDK375M, but not in other combinations (Fig. 2C). Therefore, these experiments indicated that the KD of MAPKKK5 is necessary and sufficient for its physical interaction with BSK1.
Figure 2.
The kinase domain of MAPKKK5 is essential for the interaction with BSK1. A, The different domains ofMAPKKK5 are shown. MAPKKK5 contains a kinase domain in the middle, in addition to N-terminal and C-terminal domains. The numbers indicate the positions of amino acid sequences. B, The kinase domain of MAPKKK5 interacted with BSK1 in the yeast two-hybrid assay. Corresponding constructs were cotransformed into yeast strain AH109. A 10 μL suspension of each pair was dropped on the indicated SD medium. C, The kinase domain of MAPKKK5 interacted with BSK1 in the BiFC assay. Leaves of N. benthamiana were infiltrated with A. tumefaciens strain GV3101 containing the indicated construct pairs. BSK1 and truncated MAPKKK5 proteins were fused to the YN or YC. YFP signal was detected by confocal microscopy. Bar = 50 μm. AD, pGADT7; BD, pGBKT7; CT, C-terminal domain; KD, kinase domain; KD:C, MAPKKK5 lacking the N terminus of 345 amino acids; N:KD, MAPKKK5 lacking the C terminus of 109 amino acids; NT, N-terminal domain.
BSK1 Directly Phosphorylates MAPKKK5 In Vitro
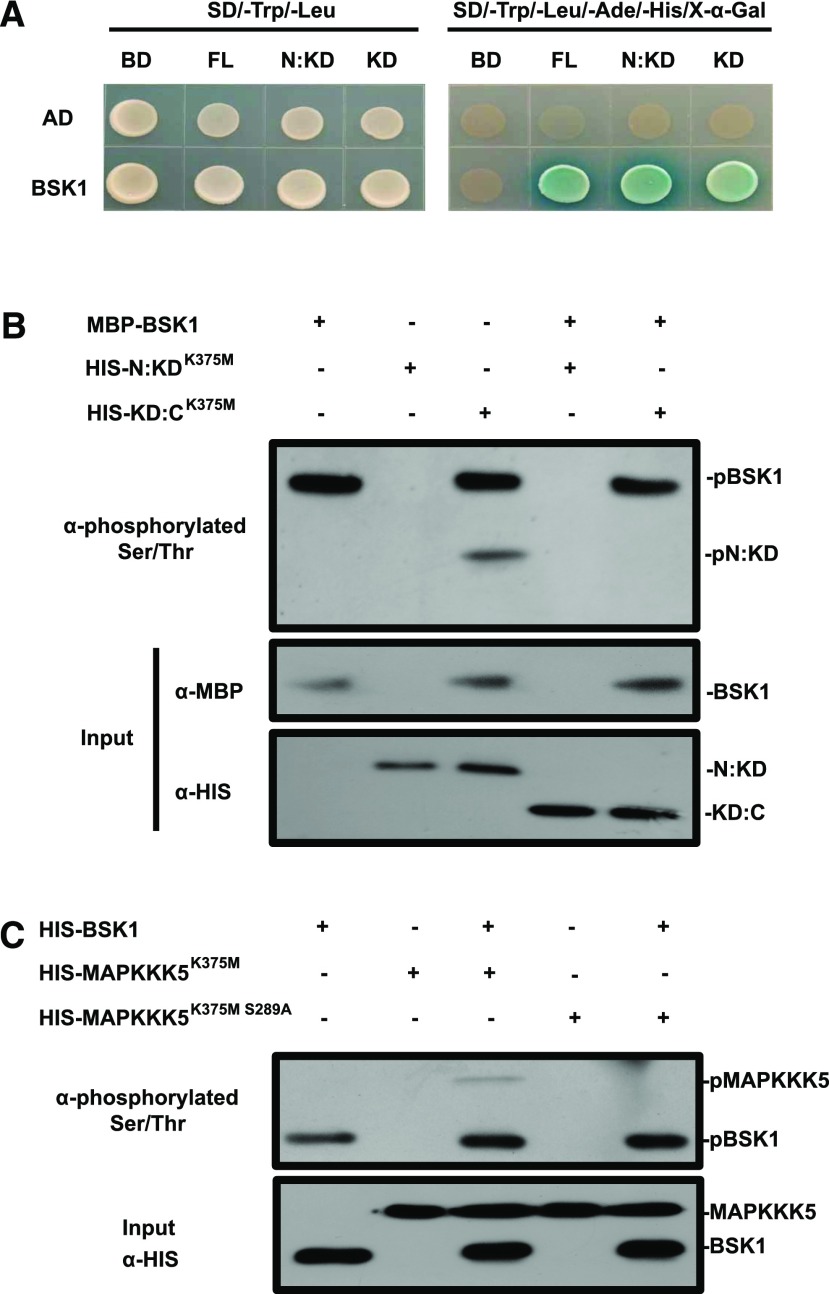
In the yeast two-hybrid assay, BSK1 interacted with the full-length and the truncated forms of MAPKKK5 that contained the kinase domain (Fig. 3A). Because MAPKKK5 in the bsk1-1 mutant lacked phosphorylation at the Ser-289 residue, and MAPKKK5 physically associated with BSK1, we hypothesized that BSK1 directly phosphorylates MAPKKK5. To test this hypothesis, we performed an in vitro kinase assay with purified recombinant MBP-BSK1 and His-N:KDK375M or His-KD:CK375M of MAPKKK5, respectively. As demonstrated in Figure 3B, autophosphorylated MBP-BSK1 (100 kD) and phosphorylated N:KD of MAPKKK5 (70 kD) were detected by an antibody that could recognize the phosphorylated Ser/Thr. However, no phosphorylation was detected with the His-KD:CK375M of MAPKKK5 (50 kD), indicating that BSK1 directly phosphorylates the N terminus of MAPKKK5 in vitro. In addition, no obvious phosphorylation of MAPKKK5 by BSK1 was detected when the 289 Ser of MAPKKK5 was changed to Ala (Fig. 3C), suggesting that BSK1 mainly phosphorylates Ser-289 of MAPKKK5.
Figure 3.
BSK1 phosphorylates the N terminus of MAPKKK5 in vitro. A, The interaction of the truncated versions of MAPKKK5 with BSK1 was examined in the yeast two-hybrid assay. Corresponding constructs were cotransformed into yeast strain AH109. A 10 μL suspension of each pair was dropped on the indicated SD medium. Pictures were taken 3 d after incubation. B, BSK1 phosphorylates the N terminus of MAPKKK5 in vitro. The in vitro phosphorylation was revealed by an antibody that could specifically detect phosphorylated Ser and Thr. The phosphorylated proteins were detected by an immunoblot assay with the indicated antibodies. The phosphorylated N:KD fragment of MAPKKK5 and BSK1 was labeled. C, BSK1 phosphorylates the Ser-289 of MAPKKK5 in vitro. The phosphorylated proteins were detected by an immunoblot assay with the antibodies that could specifically detect phosphorylated Ser and Thr. The phosphorylated MAPKKK5 and BSK1 were labeled. AD, pGADT7; BD, pGBKT7; KD, kinase domain; KD:C, MAPKKK5 lacking the N terminus of 345 amino acids; N:KD, MAPKKK5 lacking the C terminus of 109 amino acids; NT, N-terminal domain.
The mapkkk5 Mutant Displays Enhanced Susceptibility to Pto DC3000 and G. cichoracearum
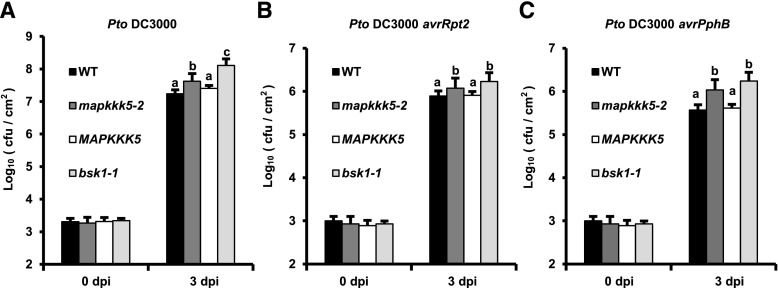
To gain insight into the role of MAPKKK5 in plant immunity, we examined the phenotypes of the loss-of-function mapkkk5-2 mutant in response to different pathogens. First, the mapkkk5-2 mutants were infected with virulent and avirulent strains of Pto DC3000 that carried different effector genes. The mapkkk5-2 mutant was very similar to the bsk1-1 mutant in that it showed enhanced susceptibility to both strains and supported significantly more growth of the bacteria than the wild type at 3 d post inoculation (dpi) (Fig. 4). Molecular complementation of the mapkkk5-2 mutant with the wild type MAPKKK5 rescued the enhanced susceptibility to Pto DC3000, Pto DC3000 (avrRpt2), and Pto DC3000 (avrPphB), indicating that MAPKKK5 plays an important role in resistance to both virulent and avirulent strains of Pto DC3000 (Fig. 4).
Figure 4.
The mapkkk5 mutant displays enhanced susceptibility to P. syringae. Four-week-old plants were inoculated with P. syringae Pto DC3000 (A), Pto DC3000 avrRpt2 (B), and Pto DC3000 avrPphB (C). Plants were inoculated with bacterial suspensions at OD600 = 5 × 10−4. Bacterial growth was monitored at 0 and 3 dpi. Data represent mean ± sd. Different lower-case letters indicate statistically significant differences (n = 9; P < 0.05; one-way ANOVA). cfu, colony-forming units; WT, wild type.
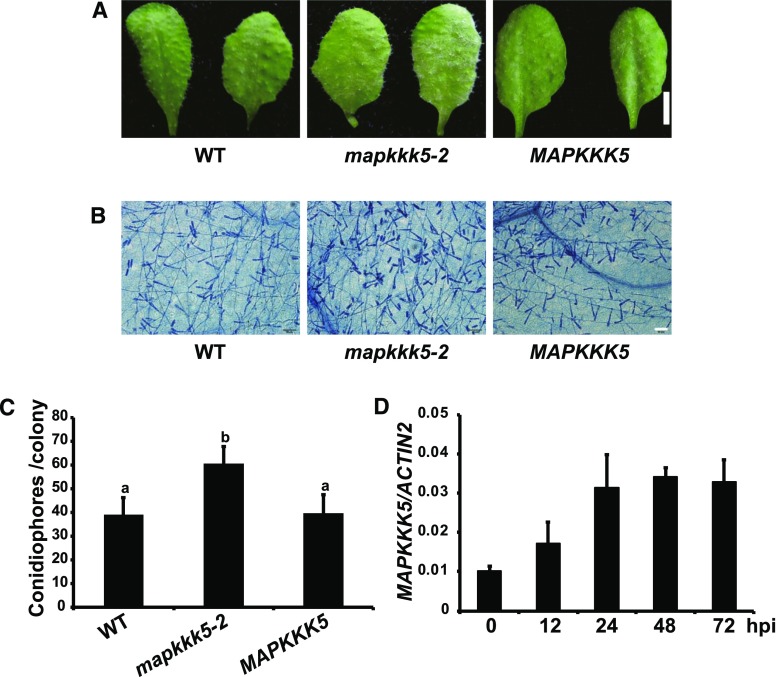
The bsk1-1 mutant displayed enhanced susceptibility to the powdery mildew fungus G. cichoracearum compared with wild type (Shi et al., 2013). To investigate if MAPKKK5 plays a role in resistance to powdery mildew, we also infected wild-type and mapkkk5 mutant plants with G. cichoracearum. After infection, the mapkkk5-2 mutant accumulated lower levels of defense-related gene transcripts, including PATHOGENESIS-RELATED GENE1 (PR1), PR2, FLAVIN-DEPENDENT MONOOXYGENASE1, and SALICYLIC ACID INDUCTION DEFICIENT2, compared to wild type (Supplemental Fig. S1). The mapkkk5-2 mutant also showed enhanced susceptibility and supported higher levels of fungal growth, which was rescued by complementation with the wild type MAPKKK5 (Fig. 5, A to C). The transgenic line that accumulated significantly higher levels of MAPKKK5 transcripts (Supplemental Fig. S2) showed enhanced resistance to powdery mildew (Supplemental Fig. S3), further confirming the role of MAPKKK5 in promoting immunity. Consistent with its role in powdery mildew resistance, the expression of MAPKKK5 was upregulated in the wild type after infection with G. cichoracearum (Fig. 5D).
Figure 5.
The mapkkk5 mutant shows increased susceptibility to G. cichoracearum. A, Four-week-old plants were infected with G. cichoracearum and the leaves were photographed at 8 dpi. The mapkkk5-2 mutants displayed enhanced susceptibility compared to the wild type. The transgenic complementation plants expressing the genomic MAPKKK5 gene in mapkkk5-2 showed wild-type-like phenotypes. Bar = 0.5 cm. B, The infected leaves were stained with Trypan Blue to observe fungal structures and dead plant cells at 8 dpi. More spores were produced in the mapkkk5-2 mutant than in the wild type. Bar = 50 µm. C, Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. The mapkkk5-2 mutants supported significantly more fungal growth than the wild type. Data represent mean ± sd. Different letters indicate statistically significant differences (n = 25; P < 0.05; one-way ANOVA). The experiment was repeated three times with similar results. D, MAPKKK5 expression was up-regulated in the wild-type plants after inoculation with G. cichoracearum. The transcript accumulation was examined by RT-qPCR at the indicated time points. ACTIN2 was used as an internal control. Data represent mean ± sd (n = 3). WT, wild type.
The mapkkk5-2 and bsk1-1 mutants displayed enhanced susceptibility to powdery mildew, but the bsk1-1 mapkkk5-2 double mutant was very similar to the bsk1-1 single mutant in its response to infection with G. cichoracearum (Supplemental Fig. S4). Furthermore, the expression of defense-related genes in the bsk1-1 mapkk5-2 double mutant was very similar to that of the bsk1-1 mutant (Supplemental Fig. S5), suggesting that MAPKKK5 and BSK1 likely act in the same genetic pathway.
The Ser-289 Residue of MAPKKK5 Is Critical for its Function in Plant Immunity
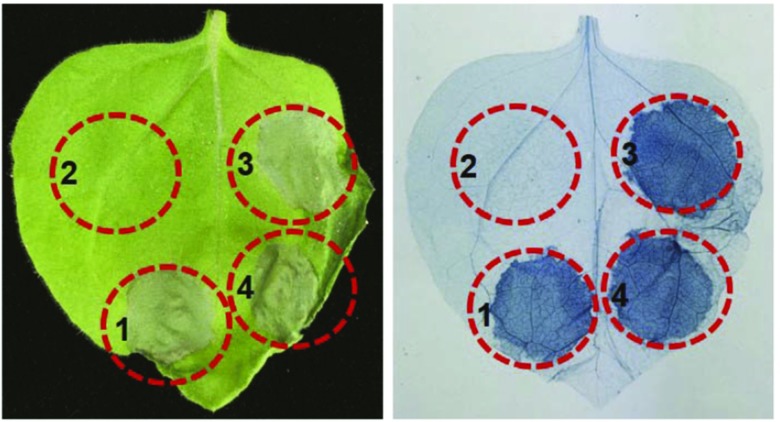
MAPKKK5 induces hypersensitive response (HR)-like cell death in N. benthamiana (Yamada et al., 2016). To investigate if the phosphorylation site Ser-289 is critical for MAPKKK5-induced cell death, we transiently overexpressed two MAPKKK5 variants in N. benthamiana; S289A causes loss of phosphorylation and S289D mimics constitutive phosphorylation. Both MAPKKK5S289A and MAPKKK5S289D induced HR-like cell death, and no obvious difference was observed between MAPKKK5 and the two variants (Fig. 6). This suggests that phosphorylation at Ser-289 might not be important for MAPKKK5-induced cell death.
Figure 6.
MAPKKK5-induced cell death is independent of Ser-289 phosphorylation. Different MAPKKK5 variants were transiently expressed in N. benthamiana. The infiltrated leaves were photographed (left) and then stained with Trypan Blue (right) for observation of dead cells at 3 dpi. Similar cell death was induced in the leaves expressing MAPKKK5-YFP-HA, MAPKKK5S289A-YFP-HA, or MAPKKK5S289D-YFP-HA, but not in the leaves expressingMAPKKK5K375M-YFP-HA. 1, MAPKKK5-YFP-HA; 2, MAPKKK5K375M-YFP-HA; 3, MAPKKK5S289A-YFP-HA; 4, MAPKKK5S289D-YFP-HA.
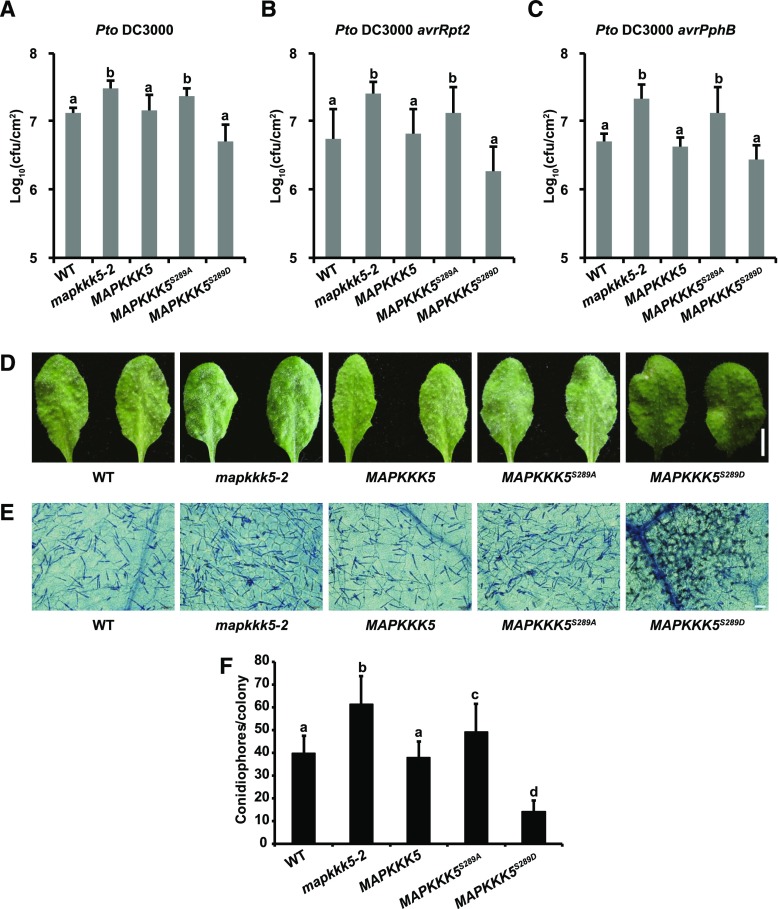
To further investigate the role of Ser-289 phosphorylation in plant immunity, we introduced the MAPKKK5S289A or MAPKKK5S289D variants into the mapkkk5-2 mutant, and then assessed resistance to the different strains of Pto DC3000 and to G. cichoracearum. As presented in Figure 7, A to C, MAPKKK5S289A could not complement the enhanced susceptible phenotype of the mapkkk5-2 mutant to the different strains of Pto DC3000. By contrast, MAPKKK5S289D was able to complement the mapkkk5-2 mutant phenotype. Similarly, MAPKKK5S289A could not fully complement the enhanced susceptibility phenotype of the mapkkk5-2 mutant to powdery mildew, whereas MAPKKK5S289D increased resistance to powdery mildew compared to wild type (Fig. 7, D to F). The expression of MAPKKK5, MAPKK5S289A, and MAPKKK5S289D was detected at similar levels in the different transgenic lines (Supplemental Fig. S6). Together, these observations indicated that the Ser-289 residue of MAPKKK5 is critical for its function in resistance to both bacterial and fungal pathogens.
Figure 7.
The phosphorylation of the Ser-289 residue contributes to MAPKKK5-mediated resistance to Pto DC3000 and G. cichoracearum. Four-week-old plants were inoculated with indicated pathogens. Wild-type, mapkkk5-2, and the mapkkk5-2 transgenic plants carrying wild-type MAPKKK5, MAPKKK5S289A, or MAPKKK5S289D, respectively, were used. Pto DC3000 (A), Pto DC3000 avrRpt2 (B), Pto DC3000 avrPphB (C), or G. cichoracearum (D). A to C, Plants were inoculated with bacterial suspensions at OD600 = 5 × 10−4. Bacterial growth was monitored at 0 dpi and 3 dpi. Data represent mean ± sd. Different lower-case letters indicate statistically significant differences (n = 9; P < 0.05; one-way ANOVA). D, Plants were inoculated with G. cichoracearum and leaves were removed and photographed at 8 dpi. Bar = 0.5 cm. E, The infected leaves were stained with Trypan Blue for observation of fungal structures and dead plant cells at 8 dpi. Bar = 50 µm. F, Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Data represent mean ± sd. Different lower-case letters indicate statistically significant differences (n = 25; P < 0.05; one-way ANOVA). The experiments were performed three times with similar results. WT, wild type.
MAPKKK5 Interacts with Multiple MAPKKs
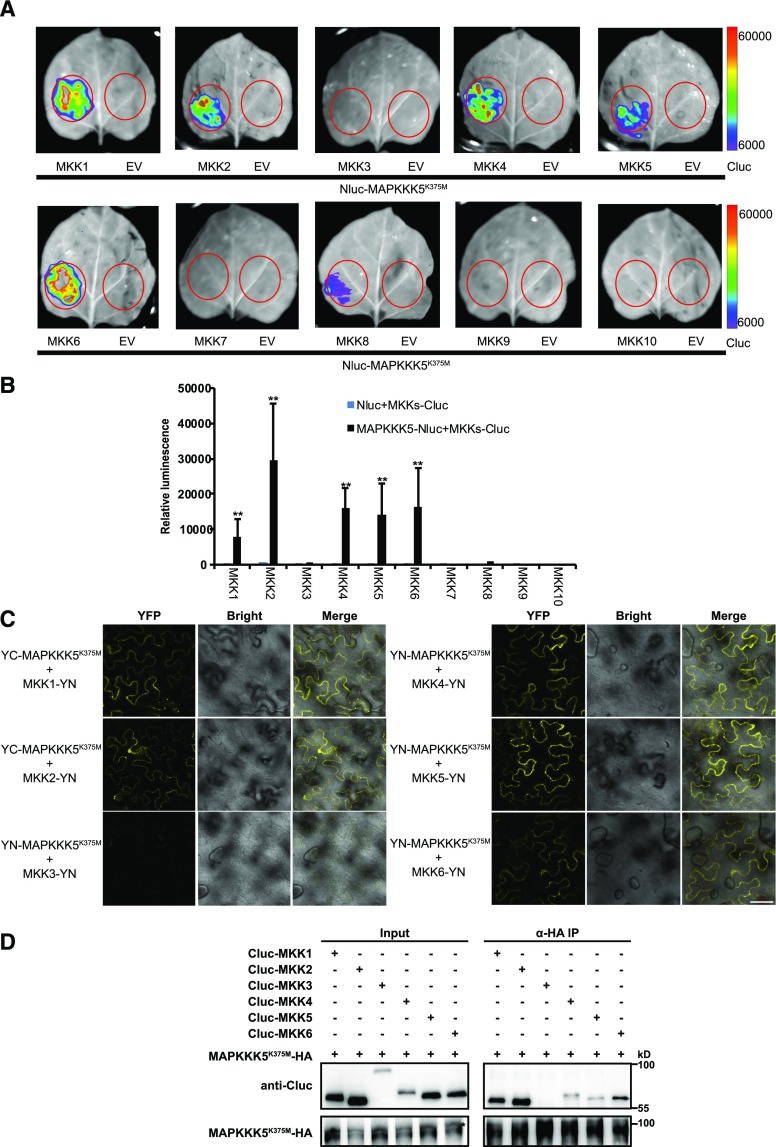
The Arabidopsis Col-0 genome encodes 10 MAPKKs (Ichimura et al., 2002). MAPKKK5 interacts with MKK4/5 and modulates PTI triggered by chitin (Yamada et al., 2016). To examine whether MAPKKK5 associates with other MAPKKs, we examined the interactions between MAPKKK5 and all 10 annotated MAPKKs in the Arabidopsis Col-0 genome. Using a split-luciferase complementation assay in N. benthamiana leaves, we transiently expressed MAPKKK5K375M-Nluc and Cluc-MKKs, or MAPKKK5K375M-Nluc and Cluc. Strong signals were observed in the coexpression of MAPKKK5 with MKK1, 2, 4, 5, and 6, indicating that MAPKKK5 interacts with each of these MKKs (Fig. 8, A and B). The interactions of MAPKKK5 with MKK1, MKK2, MKK4, MKK5, and MKK6 were also validated by the BiFC assay (Fig. 8C). To further confirm the interaction of MAPKKK5 and MKKs, we performed a Co-IP assay by coexpressing MAPKKK5K375M-HA and Cluc-MKK1/2/3/4/5/6 in Arabidopsis protoplasts. Total protein was extracted from the protoplasts and then MAPKKK5 was immunoprecipitated with HA antibody and examined to determine if Cluc-MKKs coimmunoprecipitated with MAPKKKK5. MKK1/2/4/5/6 but not MKK3 were detected with anti-Cluc antibody from the protoplasts that coexpressed both MAPKKK5K375M-HA and Cluc-MKK1/2/4/5/6 (Fig. 8D), indicating that MAPKKK5 and MKK1/2/4/5/6 form a protein complex in Arabidopsis.
Figure 8.
MAPKKs interact with MAPKKK5. A, The interaction between 10 MKKs and MAPKKK5 was examined by the luciferase complementation imaging assay. MAPKKK5K375M and MKKs were fused to the N-terminal or C-terminal fragment of firefly luciferase. Bars on the right display the relative luminescence units. B, Quantitative analysis of the interaction of MAPKKK5 and MKKs in (A). Constructs were transiently expressed in N. benthamiana leaves and the relative luminescence unit was measured 3 d after injection. Data represent mean ± sd. An empty vector was used as a control. Two asterisks indicate statistically significant differences (n ≥ 8; P < 0.01; one-way ANOVA). C, The interaction between MAPKKK5 and MKKs was examined with the BiFC assay in N. benthamiana. MAPKKK5K375M and MKKs were fused to the YN and YC, respectively. YFP signal was detected by confocal microscopy. Bar = 50 μm. D, MAPKKK5 interacted with MKK1/2/4/5/6 in Arabidopsis protoplasts. Co-IP of MKKs and MAPKKK5K375M from Arabidopsis protoplasts transiently expressing Cluc-MKK1/2/3/4/5/6 and MAPKKK5K375M-HA. The MAPKKK5K375M protein was immunoprecipitated by HA antibody, followed by immunoblot analysis with anti-Cluc antibody. Asterisk indicates MAPKKK5K375M-HA. EV, empty vector.
DISCUSSION
To understand how BSK1 regulates plant immunity and to identify potential substrates of BSK1, we conducted a genomewide phosphorylation study using mass spectrometry with Arabidopsis wild-type and bsk1-1 mutant plants. In this analysis, we found that the Ser-289 residue of MAPKKK5 was not phosphorylated in the bsk1-1 mutant. We showed that BSK1 directly interacts with and phosphorylates MAPKKK5, and the phosphorylation of Ser-289 on MAPKKK5 contributes to plant immunity. In addition, MAPKKK5 associates with multiple MAPKKs, suggesting that MAPKKK5 plays an important role in the MAPK cascade. Taken together, our results indicate that BSK1 may transduce the immune signal from the cell’s surface immune complexes to MAPKKK5.
The Arabidopsis Col-0 genome contains 62 MAPKKKs. MAPKKK5 belongs to the MAPKKK A2 subfamily, which includes two close homologs of MAPKKK5, MAPKKKα (MAPKKK3), and YODA (MAPKKK4; Ichimura et al., 2002; Meng and Zhang, 2013). The mapkkk5 mutant is more susceptible to Pto DC3000 and powdery mildew compared to the wild type, indicating that MAPKKK5 plays critical roles in plant immunity. Consequently, overexpression of MAPKKK5 leads to increased resistance to powdery mildew.
PBL27 interacts with and phosphorylates the C terminus of MAPKKK5. This phosphorylation is reported to be important for chitin-induced MAPK activation (Yamada et al., 2016). Although both PBL27 and BSK1 phosphorylate MAPKKK5, the phosphorylation sites are different: PBL27 phosphorylates the CT of MAPKKK5, whereas BSK1 phosphorylates the NT of MAPKKK5 at Ser-289. The phosphorylation of Ser-289 appears to be critical for conferring immunity, as the MAPKKK5S289A variant was unable to complement the susceptible phenotype of the mapkkk5-2 mutant to Pto DC3000 and powdery mildew, whereas the MAPKKK5S289D variant conferred enhanced resistance to powdery mildew compared to the wild type. It is possible that the phosphorylation of different sites of MAPKKK5 by different RLCKs helps plants to respond to different stresses. It would be interesting to examine whether the site of phosphorylation by PBL27 is important in a plant’s response to Pto DC3000 and G. cichoracearum, and how plants differentially phosphorylate distinct MAPKKK5 sites to respond to different stimuli.
Upon the perception of the bacterial protein flagellin, FLS2-mediated PTI is activated, which includes rapid activation of the MAPK cascade comprising the MKK4/MKK5-MPK3/MPK6 module. However, it is unknown how plants transduce the signal from the FLS2 immune complex to the MAPK cascade. MAPKKK5 was recently reported to be upstream of this module in chitin-triggered immunity (Yamada et al., 2016). Upon chitin treatment, MPK3/6 activation was strongly reduced in the mapkkk5 mutant. By contrast, the mapkkk5 mutation was reported to significantly enhance flg22-induced MPK3/6 activation. However, we observed a very weak reduction of MPK3/6 activation upon treatment with flg22 in the mapkkk5-2 mutant (Supplemental Fig. S7). The overexpression of MAPKKK5 could moderately enhance the flg22-induced MPK3/6 activation (Supplemental Fig. S8). In addition, we found there was no obvious difference in activation of MPK3/6 between mapkkk5-2 and the wild type when treated with chitin (Supplemental Fig. S9). The discrepancy between these two studies is unresolved but may have been caused by different growth conditions and/or different types of chitin. It is unknown why the mapkkk5 mutant did not display a strong reduction of MPK3/6 activation when treated with flg22. One possibility is redundancy with close homologs of MAPKKK5, such as MAPKKKα and YODA. Arabidopsis MAPKKKα is an ortholog of NbMAPKKKα, which plays an important role in regulating the cell death associated with plant immunity (Pozo et al., 2004; Oh et al., 2010). Hence, it is possible that MAPKKKα may have functional redundancy with MAPKKK5 in immunity and downstream activation of MAPKs. YODA plays a key role in the early development of Arabidopsis embryos, and acts as an indispensable component of the stomatal development regulatory pathway. YODA was shown to be upstream of the MKK4/MKK5-MAP3/6 module to regulate stomatal development and patterning; however, the yoda mutant did not show defects in PAMP-triggered MAP3/6 activation (Lukowitz et al., 2004; Wang et al., 2007). The wild-type MAPKKK5 interacted with MKK4 and MKK5, but the mapkkk5-2 mutant showed a relatively weak phenotype in MPK3/6 activation upon treatment with flg22. It would be interesting to investigate the double and triple mutants of mapkkk5, mapkkk3, and yoda. The double or triple mutants of those genes may display more severe phenotypes in MPK3/6 activation. Consistent with this, rice OsRLCK185 interacts with several MAPKKKs, suggesting a redundant role of these MAPKKKs in chitin signaling (Wang et al., 2017).
BSK1 associates with and phosphorylates MAPKKK5 at the Ser-289 residue. Therefore, BSK1 may transduce the immune signal to the MAPK cascade by phosphorylating MAPKKK5. However, in the bsk1 mutant, the defects of MPK3 and MPK6 activation were not observed, suggesting that the disease resistance phenotype mediated by BSK1 is independent of MPK3/6 activation. As BSK1 belongs to a small RLCK family, we cannot exclude the possibility of redundancy between BSK1 and other members of this RLCK family. Multiple RLCKs may work together to regulate MAPK activation. In this scenario, loss of BSK1 alone would not lead to defects in MAPK activation. Notably, BSK1 was previously shown to be required for edr1-mediated powdery mildew resistance. EDR1 is a Raf-like MAPKKK that associates with MKK4/5 and negatively regulates plant immunity (Zhao et al., 2014). As the bsk1 mutant did not show defects in MPK3/6 activation, it remains to be determined why edr1-mediated powdery mildew resistance requires BSK1.
In this study, we demonstrated that BSK1 regulates plant immunity by phosphorylating MAPKKK5. It is worth mentioning that we only conducted whole-genome phosphorylation analysis with untreated wild-type and bsk1-1 mutant plants. As the immune response is usually induced by pathogen infection, we expect that phosphorylation of many proteins may not have occurred or occurred at very low levels in the absence of pathogens or elicitors.
In conclusion, RLCK BSK1 regulates plant immunity by phosphorylating MAPKKK5 at the Ser-289 residue. Because BSK1 is a key component in the cell surface immune complex, this work may indicate a direct link of signaling from the immune complex to the MAPK cascade through a RLCK.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study included the wild-type Col-0 ecotype, the bsk1-1 mutant (Shi et al., 2013), the fls2 mutant (Xiang et al., 2008), and the T-DNA insertion mutant mapkkk5-2 (Salk_122847) (Yamada et al., 2016), obtained from the Arabidopsis Biological Resource Center (www.Arabidopsis.org). The plants were grown in a growth room at 20°C to 22°C under a 9-h-light/15-h-dark cycle for phenotyping or a 16-h-light/8-h-dark cycle for seed set, as described in Wu et al. (2015).
Phosphorylated-Mass Spectrometric Analysis
To identify the different phosphorylated proteins between bsk1-1 and the wild type in Arabidopsis, leaves of 4-week-old plants including bsk1-1 and the wild type were picked off. Then the leaves were ground to powder (about 4 g to 5 g) in liquid nitrogen. The leaf powder was treated with 3 volumes of protein extraction buffer containing 10% (v/v) of TCA and 0.07% β-mercaptoethanol in acetone. The samples were left overnight at −20°C,then centrifuged at 12,000 rpm at 4°C for 30 min, and the supernatant discarded. The same volume of ice-cold acetone containing 0.07% (v/v) β-mercaptoethanol was added, and the pellet was resuspended with gentle shaking, followed by centrifugation at 12,000 rpm at 4°C for 30 min. This step was repeated twice, and then the precipitate was freeze-dried with a SpeedVac concentrator (Thermo Fisher Scientific) at 10,000 rpm at 4°C for 10 min. The proteins were resuspended in 8 m urea buffer (50 mm NH4HCO3, 8 m urea, 1 mm sodium orthovanadate, 1 mm sodium fluoride, 2.5 mm sodium pyrophosphate, 1 mm B-glycerophosphate, 1× protease inhibitor cocktail; Sigma-Aldrich), reduced with 10 mm DTT at 37°C for 1 h and alkylated with 25 mm iodoacetamide at room temperature for 1 h in the dark. The sample was then diluted with 50 mm NH4HCO3 to lower than 2 m urea, and subsequently digested with trypsin (Sigma-Aldrich) at 37°C for 18 h using a trypsin/substrate ratio of 1:50. The resulting peptides were desalted and dried by vacuum centrifugation. Phosphopeptides were further enriched by IMAC beads (Phos-Select Iron Affinity Gel; Sigma-Aldrich). Briefly, each sample was resuspended in 500 μL wash buffer [250 mm acetic acid, 30% acetonitrile (ACN)] and incubated with the IMAC beads for 60 min. The bound phosphopeptides were then eluted from the beads using a buffer containing 150 mm ammonium hydroxide and 25% acetonitrile, and subsequently dried by vacuum centrifugation.
For MS analyses, peptides were resuspended in 0.1% formic acid (FA) and analyzed with a LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) coupled online to an Easy-nLC 1000 (Thermo Fisher Scientific) in the data-dependent mode. The peptides were separated by reverse phase LC with a 75 μm (ID) × 150 mm (length) analytical column packed with C18 particles of 5 μm in diameter. The mobile phases for the LC contained buffer A (2% ACN, 0.1% FA) and buffer B (98% ACN, 0.1% FA), and a linear gradient of buffer B from 3% to 30% for 90 min was used for the separation. The mass spectrometer was operated to acquire CID MS/MS scans after each MS1 scan on the 25 most abundant ions with MSA central losses of m/z 98, 49, and 32.6.
The data were analyzed using a prerelease version of the software Proteome Discoverer version 1.3 (Thermo Fisher Scientific). The proteome sequences for Arabidopsis from UNIPROT were used for the database searching and the mass tolerance was set to 0.05 D. The maximum number of missed cleavages was set at two, the minimum peptide length was set at six amino acids, and the maximum peptide length was set at 144 amino acids. The false discovery rate was set at 0.01 for peptide and protein identifications. Ser, Thr, and Tyr phosphorylation, and Met oxidation were included in the search as the variable modifications. Cys carbamidomethylation was set as stable modification.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed as described in Zhao et al. (2015). pGADT7-BSK1 was cotransformed into Saccharomyces cerevisiae strain AH109 with MAPKKK5 or N:KD (1 amino acids to 607 amino acids) of MAPKKK5 fused with pGBKT7. The truncated versions of MAPKKK5, including NT (1 amino acids to 345 amino acids), KD (346 amino acids to 607 amino acids), and CT (608 amino acids to 716 amino acids) were fused with pGADT7 and cotransformed into AH109 with pGBKT7-BSK1. A single colony for each transformant grown on SD/-Leu/-Trp plates was incubated in liquid media, and a 10 μL drop of dilution was plated on SD/-Ade/-His/-Leu/-Trp plates to assess the interaction at 3 d after plating.
BiFC
The BiFC assay was performed as described in Wu et al. (2015). Briefly, BSK1 and truncated MAPKKK5 versions were cloned into two intermediate vectors, pSY736 and pSY735, in-frame with the N terminus and C terminus of the yellow fluorescent protein (YFP). Then, the fusion sequences were amplified and cloned into the binary vector pMDC32 for expressing the fusion protein in planta. Agrobacterium strain GV3101 containing the respective plasmids was injected into the leaves of 4-week-old Nicotiana benthamiana plants. YFP signal was detected by confocal microscopy (LSM 710 NLO microscope, with ZEN 2009 software; Carl Zeiss) at 3 d after injection. The excitation and emission wavelengths for YFP were 514 nm and bright field was used to observe the cell contour of epidermis of N. benthamiana. Images of every single filter and merged images (YFP and bright field) were displayed.
Split-Luciferase Complementation Assay
The assay was performed as described in Chen et al. (2008). A low-light cooled CCD imaging apparatus (NightOWL II LB983 with Indigo software; Berthold Technologies) was used to capture luciferase (LUC) images. The leaves were smeared evenly with 1 mM luciferin, and then placed in darkness for 5 min before detection. Relative LUC activity per cm2 of infiltrated leaf area was calculated. Each data point contained at least eight replicates that used at least eight different leaves from different plants grown in different pots at the same time.
In Vitro Kinase Assay and GST Pull-Down Assay
MBP-fused and GST-fused proteins were expressed by transforming the pMAL-c2G and pGEX-4T-1 expression vectors into Escherichia coli strain BL21, respectively. Expression was induced by 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 18°C for 12 h. Recombinant proteins were then purified using amylose resin (New England Biolabs) or GST binding resin (Novagen). The in vitro kinase assay and GST pull-down assay were performed as described in Li et al. (2014). We used an antibody that could detect phosphorylated Ser and Thr to examine the phosphorylation.
Hypersensitive Response Assay
Agrobacterium strain GV3101 was injected into 4-week-old N. benthamiana leaves. The injected leaves were removed at 3 dpi and photographed, and then stained using Trypan Blue to examine cell death and photographed.
Pathogen Infection
The powdery mildew fungus Golovinomyces cichoracearum strain UCSC1 was used to infect plants as described in Liu et al. (2017). Fungal structures and dead plant cells were stained using Trypan Blue (Frye and Innes, 1998). Infection with Pto DC3000 was performed as described in Tang et al. (2005). Briefly, 4-week-old plants were inoculated with Pto DC3000, Pto DC3000 avrRpt2, or Pto DC3000 avrPphB by infiltration with suspensions at OD600 = 5 × 10−4. Bacterial growth was monitored at 3 dpi.
Reverse Transcription Quantitative PCR
Reverse transcription quantitative PCR (RT-qPCR) was performed as described in Nie et al. (2012). Briefly, total RNA was isolated and then first-strand cDNA was synthesized by M-MLV reverse transcriptase (Promega). Quantitative PCR was performed using SYBR Green Premix Extaq (Takara).
MAPK Activation
Eight-d-old seedlings grown on solid 1/2 Murashige & Skoog (MS) medium were gently moved to liquid 1/2 MS medium for 3 h. Then, seedlings were treated with 1/2 MS liquid medium supplemented with 100 nM flg22 for 5 min and 10 min. Seedlings were ground to a fine powder in liquid nitrogen and solubilized in MAPK buffer (Schwessinger et al., 2011). The phosphorylated MPK3 and MPK6 were detected by immunoblot analysis with α-p42/44 antibodies (1:2000; Cell Signaling Technology) as described in Zhao et al. (2014).
Coimmunoprecipitation Assay
A total of 1 mL of protoplasts was transfected with the indicated plasmids and incubated for 16 h at 23°C. Total protein was extracted with 800 μL extraction buffer (50 mm Tris-HCl at pH 7.5, 150 mm KCl, 1 mm EDTA, 1 mm DTT, 0.3% Trition-X 100, 1× proteinase inhibitor cocktail; Sigma-Aldrich). For anti-HA IP, total protein was incubated with 8 μL anti-HA antibody (Abmart) for 4 h at 4°C with gentle rotation. Then 50 μL of a 50% (v/v) protein G agarose bead (Millipore) slurry was added and incubated for another 3 h. The beads were washed three times with PBS containing 0.08% (v/v) IGEPAL CA-630 (Sigma-Aldrich) and once with PBS. After washing, the protein was separated by SDS-PAGE and detected by anti-HA and corresponding antibody immunoblot.
Accession Numbers
Sequence data can be found in The Arabidopsis Information Resource under the following numbers: At4g35230 (BSK1), At5g66850 (MAPKKK5), At4g26070 (MKK1), At4g29810 (MKK2), At5g40440 (MKK3), At1g51660 (MKK4), At3g21220 (MKK5), AT5G56580 (MKK6), At1g18350 (MKK7), At3g06230 (MKK8), At1g73500 (MKK9), AT1G32320 (MKK10), AT3g45640 (MPK3), and AT2g43790 (MPK6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The expression of defense-related genes is reduced in the mapkkk5-2 mutant when infected with G. cichoracearum.
Supplemental Figure S2. The expression of MAPKKK5.
Supplemental Figure S3. Overexpression of MAPKKK5 significantly enhances resistance to G. cichoracearum.
Supplemental Figure S4. The bsk1-1 mapkkk5-2 double mutant displays similar susceptibility to G. cichoracearum as the bsk1-1 mutant.
Supplemental Figure S5. The expression of defense-related genes in the bsk1-1 mapkkk5-2 double mutants is similar to the bsk1-1 mutant upon G. cichoracearum infection.
Supplemental Figure S6. The expression of MAPKKK5 in the wild type, the mapkkk5-2 mutant, and the transgenic plants.
Supplemental Figure S7. The mapkkk5-2 mutant shows a weak reduction in MAP kinase activation and marker gene expression in response to treatment with flg22.
Supplemental Figure S8. Overexpression of MAPKKK5 moderately enhances the flg22-induced MAP kinase activation.
Supplemental Figure S9. The mapkkk5-2 mutant shows wild-type-like MAP kinase activation in response to treatment with chitin.
Supplemental Table S1. Identified phosphorylated proteins.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank the ABRC stock center for the mapkkk5-2 mutant seeds.
Footnotes
T.D., Y.H., and Z.Y. designed the experiments; Y.H. and Z.Y. performed most of the experiments; S.H. and L.J. assisted in materials preparation and data analysis; W.Y. analyzed the data from the MS assay; T.D., Y.H., and Z.Y. wrote the article.
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020100), the Ministry of Science and Technology of China (2015CB910202 and 2014DFA31540), and the National Science Fund for Distinguished Young Scholars of China (31525019) to D.T.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gómez-Gómez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, Heberle-Bors E, Ellis BE, et al. ; MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, Zipfel C (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklöf S, Till S, Bögre L, Hirt H, Meskiene I (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, Chen S, Zhou JM (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L (2013a) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA 110: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ma X, Shan L, He P (2013b) Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol 55: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hake K, Wang W, Zhao T, Romeis T, Tang D (2017) CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1-mediated immunity. Plant Cell 29: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Mithoe SC, Ludwig C, Pel MJ, Cucinotta M, Casartelli A, Mbengue M, Sklenar J, Derbyshire P, Robatzek S, Pieterse CM, Aebersold R, Menke FL (2016) Attenuation of pattern recognition receptor signaling is mediated by a MAP kinase. EMBO Rep 17: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Howell SH (2000) Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J 24: 785–796 [DOI] [PubMed] [Google Scholar]
- Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158: 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y (2001) The NPK1 mitogen-activated protein kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev 15: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo dO, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Bourdais G, Pan H, Robatzek S, Tang D (2017) Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc Natl Acad Sci USA 114: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, Maeda K, Suzuki M, Tanimoto T, Takeda J, Nakashima M, Funama R, et al. (2014) Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J 79: 56–66 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW (2005) Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J 44: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang G, Zhang C, Zhu P, Dai H, Yu N, He Z, Xu L, Wang E (2017) OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol Plant 10: 619–633 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Liu S, Zhao Y, Wang W, Kong Z, Tang D (2015) ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, Kobayashi Y, Matsui H, et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35: 2468–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Yoshimura S, Terauchi A, Kawasaki T (2017) Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol 58: 993–1002 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Kawasaki T (2013) Receptor-like cytoplasmic kinases are pivotal components in pattern recognition receptor-mediated signaling in plant immunity. Plant Signal Behav 8: 4161, 25662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, Mengiste T, Zhang Y, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhao C, Nie H, Shen Q, Zhang S, Lukowitz W, Tang D (2014) EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet 10: e1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, Tang D (2015) A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet 11: e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]