Despite its critical roles in plant heat stress responses, plastid translation factor EF-Tu rapidly becomes insoluble at high temperatures, which leads to its inactivation.
Abstract
Translation elongation factor Tu (EF-Tu) is a conserved GTP-binding protein essential for protein translation in prokaryotes and in eukaryotic mitochondria and plastids. EF-Tu also has a GTP/GDP-independent chaperone activity that may function in acclimation to heat. Here, we report that the Arabidopsis (Arabidopsis thaliana) plastid EF-Tu, Rabe1b, rapidly becomes insoluble at temperatures as low as 35°C in vitro and 41°C in vivo, with more than 90% aggregation after 9 h at 45°C in vivo. Based on its established function in protein translation, heat-induced aggregation likely inactivates Rabe1b. To determine the effect of heat-induced aggregation, we isolated an Arabidopsis rabe1b knockdown mutant and discovered it to be highly compromised in heat tolerance. Overexpression of constitutive GTP- or GDP-bound mutant forms of Rabe1b in Arabidopsis and virus-induced silencing of Rabe1b in tomato (Solanum lycopersicum) also reduced heat tolerance. Compromised heat tolerance in the Arabidopsis rabe1b mutant and in the lines overexpressing constitutive GTP- or GDP-bound mutant Rabe1b proteins was associated with reduced plastid translation under heat stress. The Arabidopsis rabe1b mutant also showed compromised heat-induced expression of HsfA2 and its target genes. Constitutive overexpression of HsfA2 activated its target genes but only partially restored the heat tolerance of the rabe1b mutant. These results strongly suggest that the plastid protein EF-Tu is heat sensitive and acts as a critical limiting factor in plant heat stress responses, primarily functioning in plastid protein translation but also in protein folding and retrograde signaling of nuclear heat-responsive gene expression.
In plants, moderate heat disturbs cellular homeostasis, causing inhibition of growth and development; moreover, severe heat can cause irreversible damage and death. Chloroplasts are highly sensitive to heat, which affects chlorophyll biosynthesis, photochemical reactions, electron transport, thylakoid membrane fluidity, photophosphorylation, and CO2 assimilation (Wahid et al., 2007). Heat impairment of chloroplast functions often is the result of the inactivation of heat-sensitive proteins (e.g. oxygen-evolving complex of PSII and Rubisco activase) and the down-regulation of important chloroplast components (e.g. enzymes involved in tetrapyrrole metabolism), which can lead to decreased photosynthesis, redox imbalance, other associated damage, or even cell death (Eamus et al., 1995; Klimov et al., 1997; Salvucci and Crafts-Brandner, 2004a, 2004b; Allakhverdiev et al., 2008).
Because heat is such a fundamental threat to all life, heat stress responses characterized by rapid expression of heat shock proteins (HSPs) are found universally (Craig et al., 1993; Georgopoulos and Welch, 1993; Jakob et al., 1993; Arrigo, 2005; Latijnhouwers et al., 2010; Xu et al., 2012; Reddy et al., 2014). In plants, heat stress rapidly induces the production of well-characterized HSPs, including Hsp101, Hsp70, and small HSPs, which act as molecular chaperones in protein quality control by promoting the folding and refolding of nonnative proteins (Baniwal et al., 2007; Kotak et al., 2007; von Koskull-Döring et al., 2007; Schramm et al., 2008). Protein chaperones also are involved in monitoring misfolded/damaged proteins and targeting their degradation by the ubiquitin proteasome system, autophagy, and other pathways (Arias and Cuervo, 2011; Amm et al., 2014). A great deal has been learned about the signaling network of heat shock-responsive gene expression and protein quality control in important cellular compartments such as the cytosol and endoplasmic reticulum (Brodsky and McCracken, 1999; Schäfer and Wolf, 2005; Kruse et al., 2006; Radke et al., 2008; Liu and Howell, 2010; Okiyoneda et al., 2011; Needham and Brodsky, 2013).
Heat shock transcription factors (HSFs) are the terminal regulators of heat stress signal transduction, mediating the expression of HSPs and other heat-induced genes (Kotak et al., 2007; von Koskull-Döring et al., 2007). In plants, HSFs are encoded by a multigene family with more than 20 members in Arabidopsis (Arabidopsis thaliana) and can be grouped into three evolutionary classes: A, B, and C (Nover et al., 2001). Studies in plants including Arabidopsis and tomato (Solanum lycopersicum) have shown that several HSFs, including HsfA1a, HsfA2, and HsfB1, play critical roles in the transcriptional regulatory network responsible for the expression of nuclear heat-responsive genes (Kotak et al., 2007). HsfA1a is constitutively expressed and regulates the heat-induced expression of HsfA2 and HsfB1 (Mishra et al., 2002). HsfA2, which is highly responsive to heat stress, is a major HSF in the heat-induced expression of nuclear heat-responsive genes (Kotak et al., 2007). HsfA1a can function as a coactivator of HsfA2 by forming an HsfA1a-HsfA2 protein complex (Kotak et al., 2007). Disruption of Arabidopsis HsfA2 decreased plant thermotolerance, whereas its overexpression increased thermotolerance in transgenic plants (Li et al., 2005; Nishizawa et al., 2006; Charng et al., 2007; Ogawa et al., 2007).
It has long been recognized that heat stress responses are associated with altered protein biosynthesis, a fundamental biological process that provides the driving force for the growth and development of all living organisms (Matts and Hurst, 1992; Matts et al., 1992; Shalgi et al., 2013). Protein translation is accomplished by the combined actions of the ribosomes and ancillary proteins such as initiation, elongation, and termination factors. A central step in protein biosynthesis is the elongation cycle. In this process, the elongation factors EF-Tu (in bacteria) and eEFIA (in eukarya) play pivotal roles by carrying the aminoacyl-tRNAs (aa-tRNAs) to the ribosomes (Brisio et al., 2010). EF-Tu and EFIA belong to the large family of GTP-binding proteins, or G-proteins, that catalyze the binding of an aa-tRNA to the ribosome (Brisio et al., 2010). The ribosome has three sites for tRNA binding: the aminoacyl/acceptor or A site, the peptidyl or P site, and the exit or E site. In prokaryotes, GTP-bound EF-Tu forms complexes with aa-tRNA and transports the correct aa-tRNA to the A site of the ribosome, where the anticodon of the tRNA binds to the codon of the mRNA (Krab and Parmeggiani, 1998, 2002). Following the correct anticodon/codon binding, the ribosome changes configuration to activate the GTPase of EF-Tu, resulting in the hydrolysis of bound GTP to GDP. GTP hydrolysis results in a drastic change in the conformation of EF-Tu, causing its dissociation from the aa-tRNA and ribosome complex (Krab and Parmeggiani, 2002). This allows the aa-tRNA to fully enter the A site, where its amino acid is brought near the P site’s polypeptide so that the ribosome can catalyze the covalent transfer of the polypeptide onto the amino acid (Krab and Parmeggiani, 2002). Upon leaving the ribosome in the cytoplasm, the deactivated GDP-bound EF-Tu is activated by the elongation factor Ts (EF-Ts), which belongs to the family of guanine nucleotide exchange factors, resulting in the restoration of its GTP-bound form, which can then associate with another aa-tRNA to start the next cycle (Krab and Parmeggiani, 2002).
Apart from the pivotal role in transporting aa-tRNA to the ribosome in protein biosynthesis, EF-Tu contributes to translational accuracy through several mechanisms (Krab and Parmeggiani, 2002). In addition, EF-Tu can act as a protein chaperone that promotes the refolding of a number of denatured proteins in vitro. Two earlier studies suggested that the chaperone activity of a bacterial EF-Tu was enhanced by GTP (Kudlicki et al., 1997; Caldas et al., 2000). However, those studies were not reproducible, and subsequent studies with bacterial, mitochondrial, and chloroplast EF-Tu proteins have established that the chaperone activity of EF-Tu is independent of GTP or GDP (Malki et al., 2002; Rao et al., 2004; Ristic et al., 2007; Suzuki et al., 2007). In plants, genes encoding EF-Tu are induced by abiotic stresses (Bhadula et al., 2001; Momcilovic and Ristic, 2007; Ristic et al., 2008), and the levels of plant plastid EF-Tu in plants have been correlated with the levels of heat tolerance (Ristic et al., 2004; Fu et al., 2008; Fu and Ristic, 2010). It is unclear whether the role of the plant EF-Tu protein in heat tolerance is mediated by its role as a protein translation factor in protein biosynthesis, as a protein chaperone in protein refolding, or unknown activities.
We have studied the protein degradation pathways, including the 26S proteasome, autophagy, and multivesicular bodies, in the degradation of nonnative proteins under biotic and abiotic stresses, including heat stress (Zhou et al., 2013, 2014b; Wang et al., 2014, 2015). We became interested in plastid EF-Tu from our research on the mechanisms by which autophagy targets the degradation of ubiquitinated protein aggregates derived from heat-denatured proteins (Zhou et al., 2013, 2014b). Through proteomic profiling of heat-induced protein aggregates accumulated under heat stress, we discovered that the Arabidopsis plastid EF-Tu, Rabe1b, was one of the most abundant aggregates under heat stress (Zhou et al., 2014b). Here, we report that Arabidopsis Rabe1b proteins rapidly became insoluble both in vitro and in vivo during heat stress. The heat-induced aggregation of Rabe1b would inactivate the elongation factor based on its established mode during the elongation cycle of protein translation. To determine the impact of reduced levels of soluble, active plastid EF-Tu, we isolated an Arabidopsis rabe1b knockdown mutant and discovered that it was highly compromised in heat tolerance. Overexpression of constitutive GTP- and GDP-bound mutant forms of Rabe1b in Arabidopsis and virus-induced silencing of Rabe1b in tomato also led to reduced heat tolerance. We also found that the compromised heat tolerance of the Arabidopsis rabe1b mutant and overexpression lines for the constitutive GTP- and GDP-bound mutant Rabe1b was associated with reduced plastid translation under heat stress, and the Arabidopsis rabe1b knockdown mutant also was compromised in heat-induced expression of HsfA2 and its target genes. Overexpression of HsfA2 could partially restore the heat tolerance of the rabe1b mutant, with activated expression of HsfA2 target genes. Based on these results, we propose that heat-sensitive plastid EF-Tu is a critical limiting factor of plant heat stress responses primarily as a plastid translation factor but also through positive roles in protein folding and retrograde signaling of nuclear heat-responsive gene expression.
RESULTS
Heat-Induced Rapid Aggregation of AtRabe1b Both in Vitro and in Vivo
In our previously reported proteomic profiling of insoluble protein aggregates in heat-treated plants, we discovered that the Arabidopsis plastid EF-Tu, Rabe1b, is one of the most abundant aggregates under heat stress (Zhou et al., 2014b). Other abundant protein aggregates include Rubisco activase and catalases (Zhou et al., 2014b), which are known to be heat sensitive and aggregate prone (Feinstein et al., 1967; Chen et al., 1993a, 1993b; Salvucci et al., 2001). The heat hypersensitivity of Rubisco activase and catalases contributes to reduced photosynthetic efficiency and increased reactive oxygen species (ROS) accumulation under heat stress, respectively (Rizhsky et al., 2002; Kurek et al., 2007). The aggregate proneness of plastid EF-Tu raises important questions about its structural and functional properties under heat stress.
To confirm and characterize the possible aggregate-prone nature of the Arabidopsis plastid EF-Tu, we generated a myc-tagged AtRabe1b gene construct under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed it into Arabidopsis Columbia-0 (Col-0) wild-type plants. As Rabe1b is a G protein, we were also interested in determining whether its GTP and GDP binding and exchange affect temperature-induced aggregation. Therefore, we also made myc-tagged gene constructs for two mutant Rabe1b proteins: AtRabe1bH152L and AtRabe1bN203I. AtRabe1bH152L is a constitutive GTP-bound form of AtRabe1b (AtRabe1b-GTP) due to its GTPase deficiency as a result of the substitution of the catalytic residue His-152, which corresponds to His-84 in Escherichia coli EF-Tu (Fig. 1; Daviter et al., 2003). Indeed, recombinant AtRabe1bN203I produced in E. coli showed deficiency in its GTPase activity (Supplemental Fig. S1A). AtRabe1bN203I is a constitutive GDP-bound form of AtRabe1b (AtRabe1b-GDP) due to the substitution at Asn-203, which corresponds to the Asn-135 in E. coli EF-Tu (Fig. 1) that is required for binding to the guanine nucleotide exchange factor EF-Ts (Hwang et al., 1992). We confirmed using yeast two-hybrid assays that both wild-type AtRabe1b and the GTP-bound AtRabe1bH152L mutant protein, but not the GDP-bound AtRabe1bN203I mutant protein, interacted with the plastid EF-Ts guanine nucleotide exchange factor (Supplemental Fig. S1B). The AtRabe1b-GTP-myc and AtRabe1b-GDP-myc constructs under the control of the CaMV 35S promoter also were transformed into Arabidopsis. For each construct, we generated more than 30 independent transformants. Surprisingly, more than 70% of T1 transformants for each construct developed bleached leaves (Supplemental Fig. S2, A and B). The bleached areas often expanded to stem, inflorescence, and other tissues to various degrees (Supplemental Fig. S2A). Both reverse transcription quantitative PCR (RT-qPCR) and immunoblotting analysis (Supplemental Fig. S2C) indicated that the bleaching phenotype of the transformants was strictly correlated with silencing of the Rabe1b gene. From the remaining transgenic plants with no detectable bleaching phenotype, we identified two lines for each transgene with similar levels of AtRabe1b transgene transcripts based on RT-qPCR (Supplemental Fig. S3) and AtRabe1b-myc proteins based on immunoblotting using an anti-myc antibody. These lines were used to investigate the temperature-induced aggregation of the plastid translation elongation factor.
Figure 1.
Sequence comparison between Arabidopsis chloroplast EF-Tu and E. coli E-Tu proteins. Identical amino acid residues are in red. Previously established GTPase-deficient, constitutive GTP-bound E. coli EF-TuH84L and constitutive GDP-bound EF-TuN135I mutant proteins are indicated. The corresponding amino acid residues His-84 and Asn-135 of E. coli EF-Tu are His-152 and Asn-135, respectively, in Arabidopsis plastid EF-Tu, and their constitutive GTP- and GDP-bound mutant forms also are indicated.
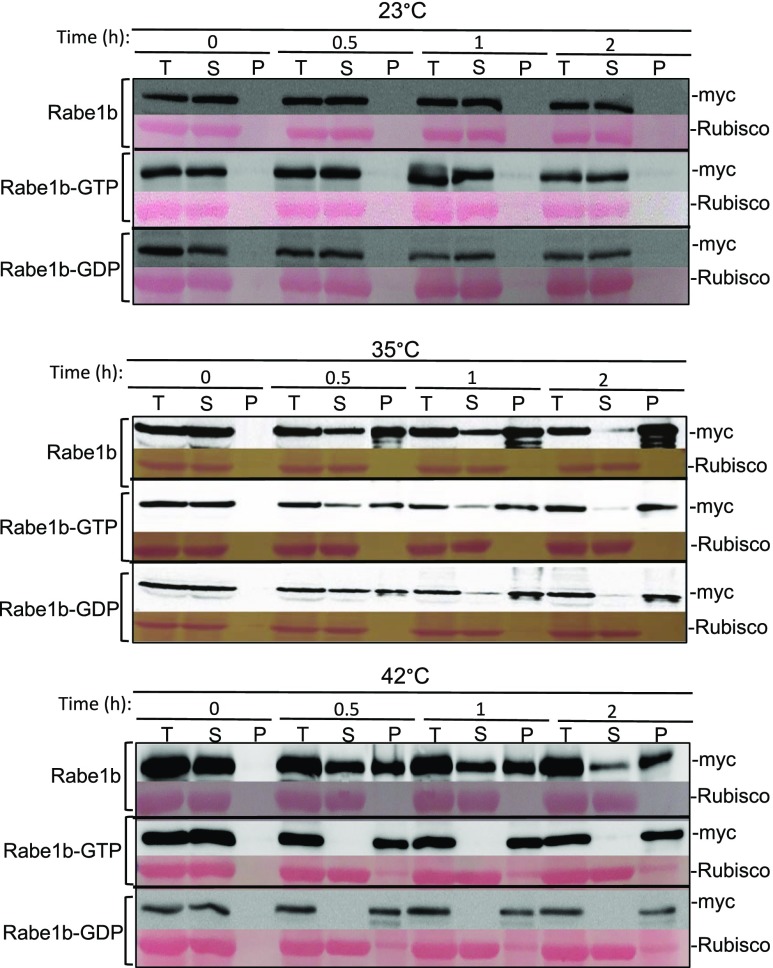
We first examined the temperature-induced aggregation of AtRabe1b in vitro. Total soluble proteins were extracted from transgenic Arabidopsis plants overexpressing myc-tagged AtRabe1b (AtRabe1b-OE), AtRabe1b-GTP (AtRabe1b-GTP-OE), and AtRabe1b-GDP (AtRabe1b-GDP-OE). The extracted proteins were incubated at different temperatures for 0.5, 1, and 2 h. Soluble and insoluble proteins in the protein extracts were then separated by low-speed centrifugation, and the relative amounts of AtRabe1b-myc in the soluble and insoluble fractions were analyzed by immunoblotting. When incubated at 23°C, AtRab1b-myc and its constitutive GTP- and GDP-bound mutant forms remained soluble, with little aggregation detected after the 2-h incubation (Fig. 2). At 35°C, on the other hand, incubation for as short as 0.5 h caused aggregation of more than 50% of the myc-tagged AtRabe1b and its GTP- and GDP-binding mutant forms (Fig. 2). Extended incubation at 35°C led to a further increase in the aggregated form of the proteins (Fig. 2). After a 2-h incubation at 35°C, more than 80% of myc-tagged AtRabe1b proteins became insoluble (Fig. 2). Interestingly, when incubated at 42°C, the rates of insolubilization of wild-type Rabe1b proteins were not substantially different from those at 35°C. However, insolubilization of the GTP- and GDP-bound forms of the plastid EF-Tu were enhanced substantially when the temperature increased from 35°C to 42°C (Fig. 2). Almost all the GTP- and GDP-bound forms of Rabe1b became aggregated after incubation for as short as 0.5 h at 42°C (Fig. 2). Inclusion of 20 μm GTP in the protein extracts had no detectable effect on the heat-induced aggregation of either the wild-type or mutant form of Rabe1b at 42°C (Supplemental Fig. S4). By contrast, less than 5% of the total proteins in the extract became aggregated in the insoluble fraction after incubation at 42°C for 2 h. For comparison, Rubisco large subunit proteins, which were used as the loading control in immunoblotting, remained mostly soluble after the 2-h incubation at both 35°C and 42°C (Fig. 2). These results indicated that AtRabe1b proteins were highly prone to aggregation at elevated temperatures in vitro. In fact, a substantial percentage of Rabe1b proteins became aggregated when incubated for 2 h at a temperature as low as 25°C in vitro (Supplemental Fig. S5).
Figure 2.
Heat-induced rapid aggregation of Arabidopsis plastid EF-Tu in vitro. Total proteins (T) were extracted from transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE Arabidopsis plants and were incubated at the indicated temperatures for the indicated hours. Soluble proteins in the supernatants (S) and insoluble proteins in the pellets (P) were separated by low-speed centrifugation, and the relative amounts of myc-tagged AtRabe1b, AtRabe1b-GTP, and AtRabe1b-GDP proteins in the protein fractions were analyzed by immunoblotting using an anti-myc monoclonal antibody. Rubisco large subunit proteins stained with Ponceau S are shown as the loading control.
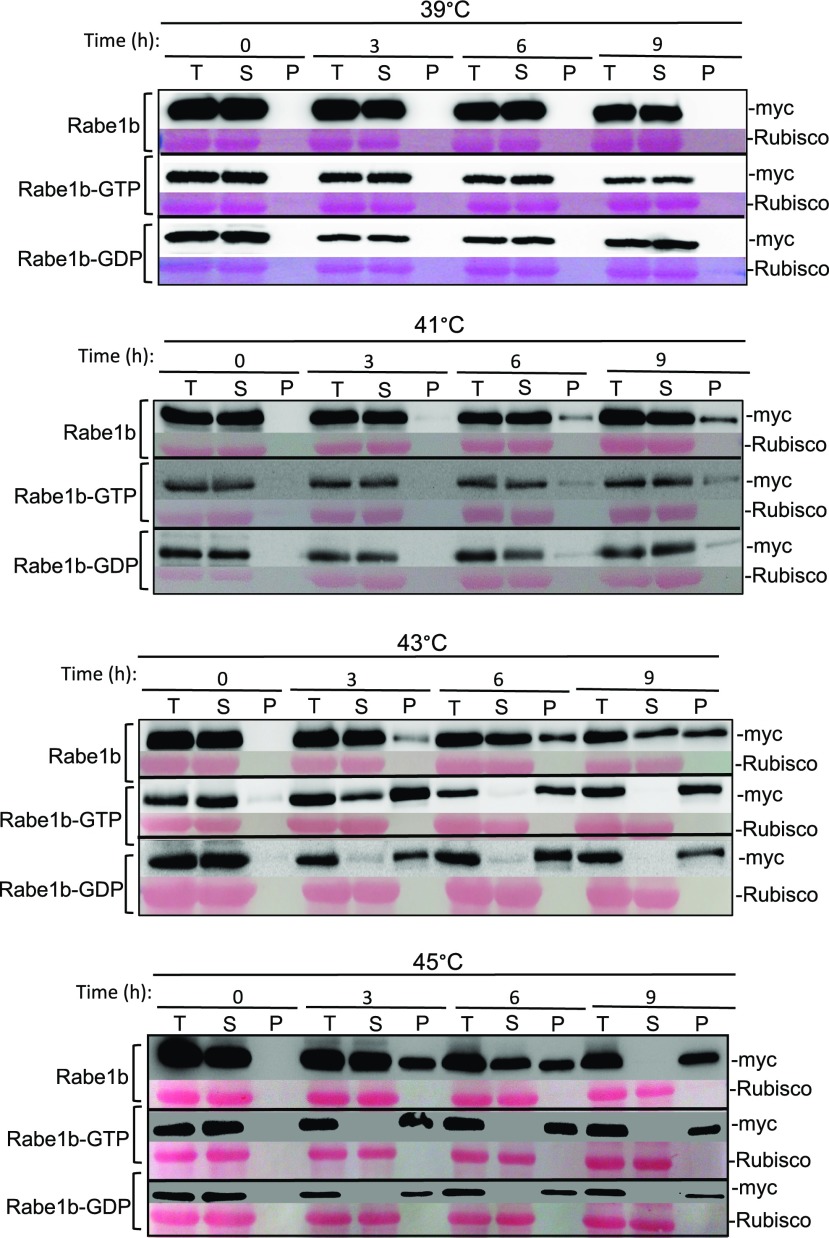
We also examined the temperature-induced aggregation of AtRabe1b in vivo. Stable transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE plants were treated at different temperatures for 3, 6, and 9 h, and total proteins were extracted and separated into soluble and insoluble fractions by low-speed centrifugation. As shown in Figure 3, treatment of these transgenic plants at a temperature up to 39°C for up to 9 h caused little accumulation of Rabe1b aggregates. When incubated at 41°C, no detectable levels of aggregates of Rabe1b and its GTP- and GDP-binding forms were observed during the first 3 h (Fig. 3). Extension of the incubation time at 41°C to 6 and 9 h, however, resulted in detectable accumulation of Rabe1b protein aggregates (Fig. 3). Increasing the temperature to 43°C and 45°C further increased the aggregation of the Rabe1b proteins (Fig. 3). Again, the constitutive GTP- and GDP-bound mutant forms of Rabe1b were substantially more prone to aggregation than the wild-type Rabe1b proteins (Fig. 3). For example, at 43°C, almost all constitutive GTP- and GDP-bound mutant forms of Rabe1b became aggregated after a 6-h incubation but less than 50% of the wild-type Rabe1b proteins were in the insoluble fraction (Fig. 3). At 45°C, incubation of only 3 h caused complete aggregation of the GTP- and GDP-bound forms of Rabe1b (Fig. 3). On the other hand, complete aggregation of wild-type Rabe1b proteins occurred after 9 h at 45°C (Fig. 3). It should be noted again that, even after a 9-h incubation at 45°C, when almost all Rabe1b proteins became insoluble, only a small percentage of the total proteins (less than 5%) became aggregated in the insoluble fraction. Also, little aggregation of Rubisco large subunit proteins, which were used as the loading control in immunoblotting, was detected after a 9-h incubation of the transgenic plants at 45°C (Fig. 3). Thus, even though Rabe1b proteins were more stable in vivo than in vitro, they were still exceptionally heat sensitive and prone to aggregation in vivo at high temperatures.
Figure 3.
Heat-induced rapid aggregation of Arabidopsis plastid EF-Tu in vivo. Transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE Arabidopsis plants were treated at the indicated temperatures for the indicated hours. Total proteins (T) were extracted from treated transgenic plants and separated by low-speed centrifugation into soluble proteins in the supernatants (S) and insoluble proteins in the pellets (P). Myc-tagged AtRabe1b, AtRabe1b-GTP, and AtRabe1b-GDP proteins in the total, soluble, and insoluble fractions were analyzed by immunoblotting. Rubisco large subunit proteins stained with Ponceau S are shown as the loading control.
Heat-Induced Aggregation of Rabe1b Was Irreversible in Vitro and Likely in Vivo as Well
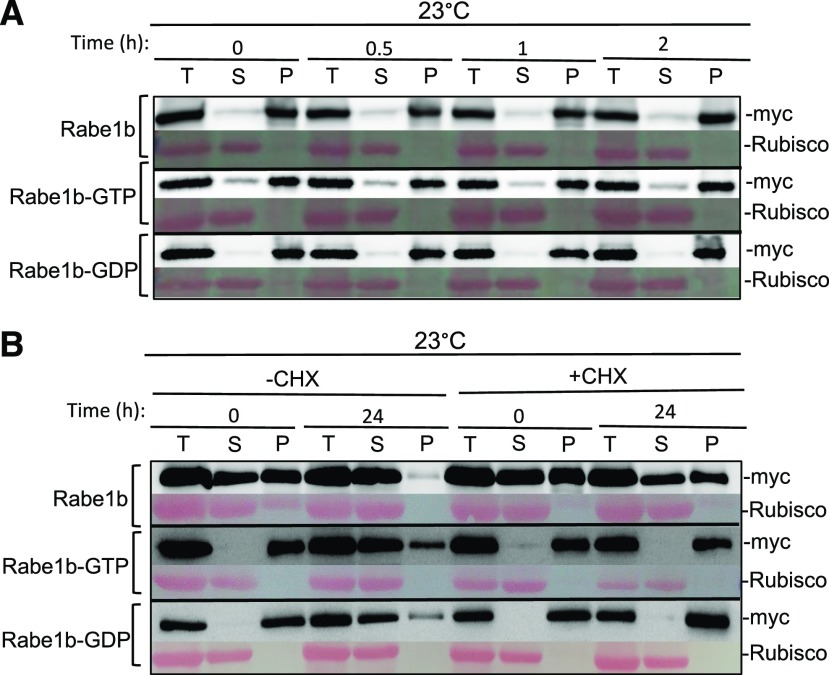
To determine whether the heat-induced aggregation of Rabe1b proteins is reversible in vitro, we first extracted total proteins from transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE plants and incubated the proteins at 42°C for 2 h to induce Rabe1b aggregation. The heat-treated protein extracts were then incubated at a low temperature (23°C) for various amounts of time and analyzed for possible solubilization of Rabe1b aggregates. As shown in Figure 4A, incubation at 23°C for 2 h did not lead to a reduction in Rabe1b aggregates or an increase in the soluble forms. Further increase in the incubation time at the low temperature for up to 24 h did not increase the amount of soluble Rabe1 proteins either, even though protein degradation became apparent after the extended incubation in vitro (data not shown). Thus, heat-induced aggregation of Rabe1b proteins was irreversible in vitro.
Figure 4.
Analysis of heat-induced Rabe1b aggregates during recovery. A, Heat-induced Rabe1b aggregates during recovery in vitro. Total proteins were extracted from transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE Arabidopsis plants incubated at 42°C for 2 h to induce Rabe1b aggregation and then placed at 23°C for the indicated hours. Total proteins (T), soluble proteins in the supernatants (S), and insoluble proteins in the pellets (P) were separated, and the amount of myc-tagged AtRabe1b proteins in the fractions was analyzed by immunoblotting. Rubisco proteins stained with Ponceau S are shown as the loading control. B, Effects of a protein synthesis inhibitor on heat-induced Rabe1b aggregates during recovery in vivo. Transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE Arabidopsis plants were treated with the protein synthesis inhibitor cycloheximide (CHX; 100 μm) and then incubated at 45°C for 6 h. The plants were then transferred to 23°C for recovery. Total proteins were extracted from treated plants at 0 and 24 h of recovery. Soluble proteins in the supernatants and insoluble proteins in the pellets were separated, and myc-tagged AtRabe1b proteins were analyzed by immunoblotting. Rubisco proteins stained with Ponceau S are shown as the loading control.
We also analyzed the fate of heat-induced Rabe1b protein aggregates in vivo after recovery at a low room temperature. Transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE plants were first treated for 6 h at 45°C to induce Rabe1b aggregation and then were transferred to 23°C. After a 24-h recovery at 23°C, total proteins were extracted and the relative amounts of soluble and insoluble Rabe1b proteins were analyzed by low-speed centrifugation and immunoblotting. As shown in Figure 4B, recovery of heat-treated transgenic plants at 23°C for 24 h led to an increase in soluble Rabe1b and a corresponding reduction in insoluble Rabe1b proteins. However, it was unclear whether the increased accumulation of soluble Rabe1b during the recovery at the low temperature after heat treatment was due to the solubilization of heat-induced Rabe1b aggregates or to new Rabe1b synthesis. Likewise, reduction of heat-induced Rabe1b aggregates during the recovery at 23°C could result from their solubilization or from their degradation. To examine these possibilities, we treated the transgenic plants first with the protein synthesis inhibitor CHX before heat treatment at 45°C. After the 6-h heat treatment, the plants were again transferred to 23°C and Rabe1b proteins were analyzed after the 24-h recovery at low temperature. Interestingly, we observed that CHX treatment prevented not only an increase in soluble Rab1b proteins but also a reduction of heat-induced Rabe1b aggregates during the 24-h recovery at 23°C. The blocking of the reduction of heat-induced Rabe1b aggregates during the recovery by a protein synthesis inhibitor would argue against their ability to spontaneously refold into soluble forms at room temperature. The observed reduction of heat-induced Rabe1b aggregates during the recovery in the absence of CHX likely could result from their degradation by autophagy or other protein degradation pathways whose induction or activation may be dependent on protein synthesis. In addition, some heat-induced Rabe1b aggregates may undergo facilitated, not spontaneous, refolding into soluble forms with the aid of other proteins, such as protein chaperones, whose synthesis is sensitive to CHX.
Overexpression of Constitutive GTP- and GDP-Bound Mutant Rabe1b Proteins Compromised Heat Tolerance
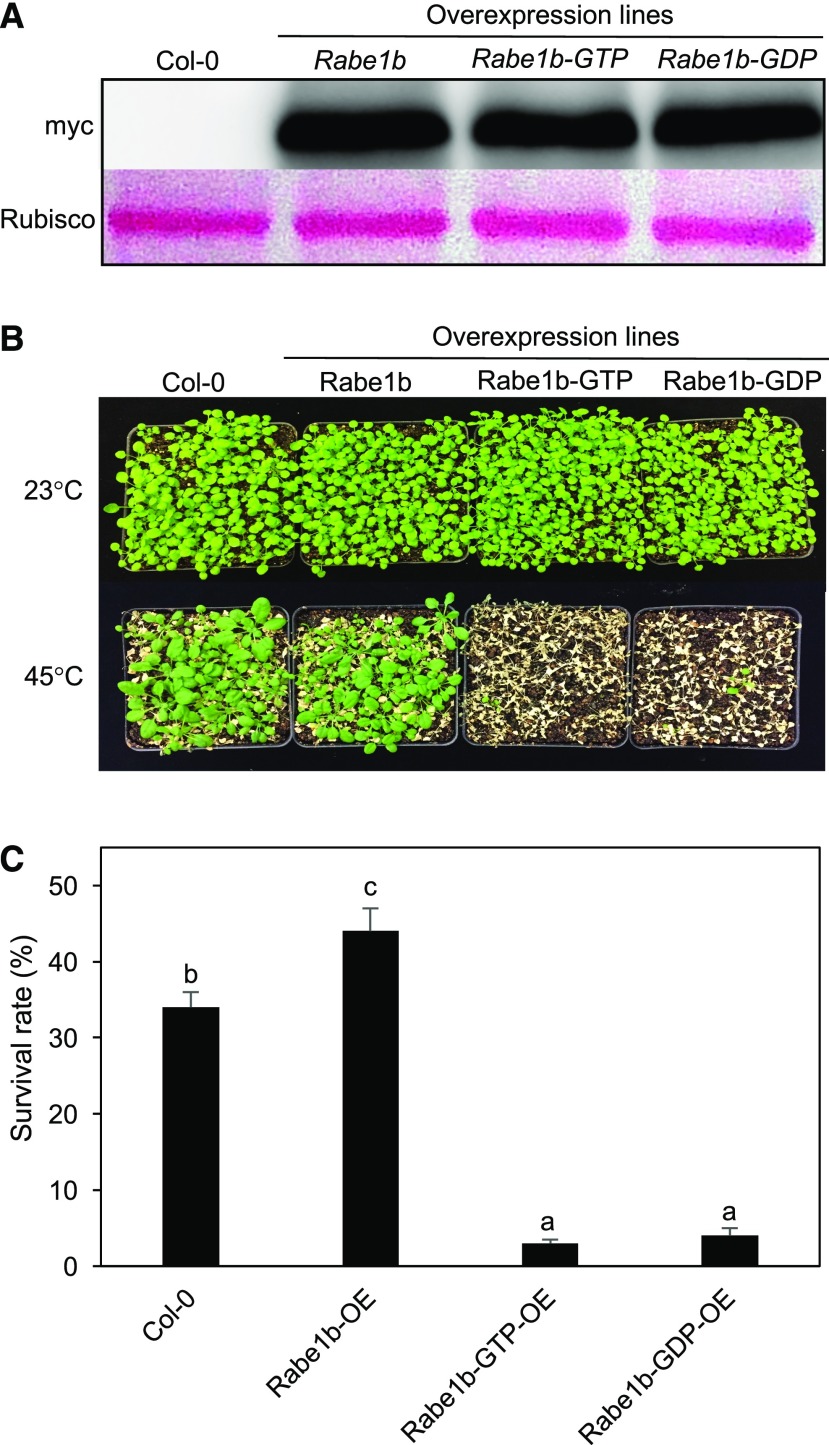
As a translation factor, Rabe1b plays an essential role in plastid protein synthesis, and its rapid aggregation under heat stress could have important effects on the growth and survival of plants under heat stress. To determine this possibility, we analyzed the effects of the overexpression of wild-type Rabe1b and its constitutive GTP- and GDP-bound mutant forms on plant heat tolerance. During the elongation phase of protein translation, EF-Tu alternates between GTP- and GDP-bound forms, which are required for its dynamic interactions with other factors, including aa-tRNAs, ribosomes, and other proteins (Krab and Parmeggiani, 2002). Unlike wild-type EF-Tu, constitutive GTP- and GDP-bound forms of EF-Tu would interact with only certain cellular factors but not with other cellular factors required for protein elongation in a dynamic manner and, therefore, could act as dominant negative proteins that interfere with plastid translation. To determine heat tolerance, we used the transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE lines with similar levels of the expressed AtRabe1b proteins (Fig. 5A), placed 3-week-old seedlings in a 45°C growth chamber for 9 h, and scored for survival rates after recovery for 5 d at room temperature. As shown in Figure 5, B and C, more than 30% of wild-type Col-0 seedlings survived after the heat stress. Overexpression of wild-type Rabe1b significantly increased heat tolerance, as their survival rates were about 10% higher than those of wild-type plants (Fig. 5, B and C). By contrast, the survival rates of transgenic AtRabe1b-GTP-OE and AtRabe1b-GDP-OE lines were less than 5% after heat stress (Fig. 5, B and C).
Figure 5.
Effects of overexpression of Rabe1b, Rabe1b-GTP, and Rabe1b-GDP on the heat tolerance of Arabidopsis seedlings. A, Immunoblotting of myc-tagged AtRabe1b, AtRabe1b-GTP, and AtRabe1b-GDP proteins in transgenic plants. Total proteins were extracted from transgenic AtRabe1b-OE, AtRabe1b-GTP-OE, and AtRabe1b-GDP-OE Arabidopsis plants with similar levels of the transgene transcripts and analyzed by immunoblotting using an anti-myc antibody. Rubisco large subunit proteins stained with Ponceau S are shown as the loading control. B, Assays of young seedling heat tolerance. Two-week-old seedlings were placed in a 45°C growth chamber for 9 h. The heat-treated plants were then moved to a 23°C growth chamber for recovery. The photograph was taken 5 d after the heat treatment. C, Survival rates of heat-stressed young seedlings determined after treatment at 45°C for 9 h followed by 23°C for 5 d. Means and se were calculated from survival rates determined from three experiments with approximately 100 seedlings per experiment for each genotype. According to Duncan’s multiple range test (P < 0.01), mean survival rates do not differ significantly if they are indicated with the same letter.
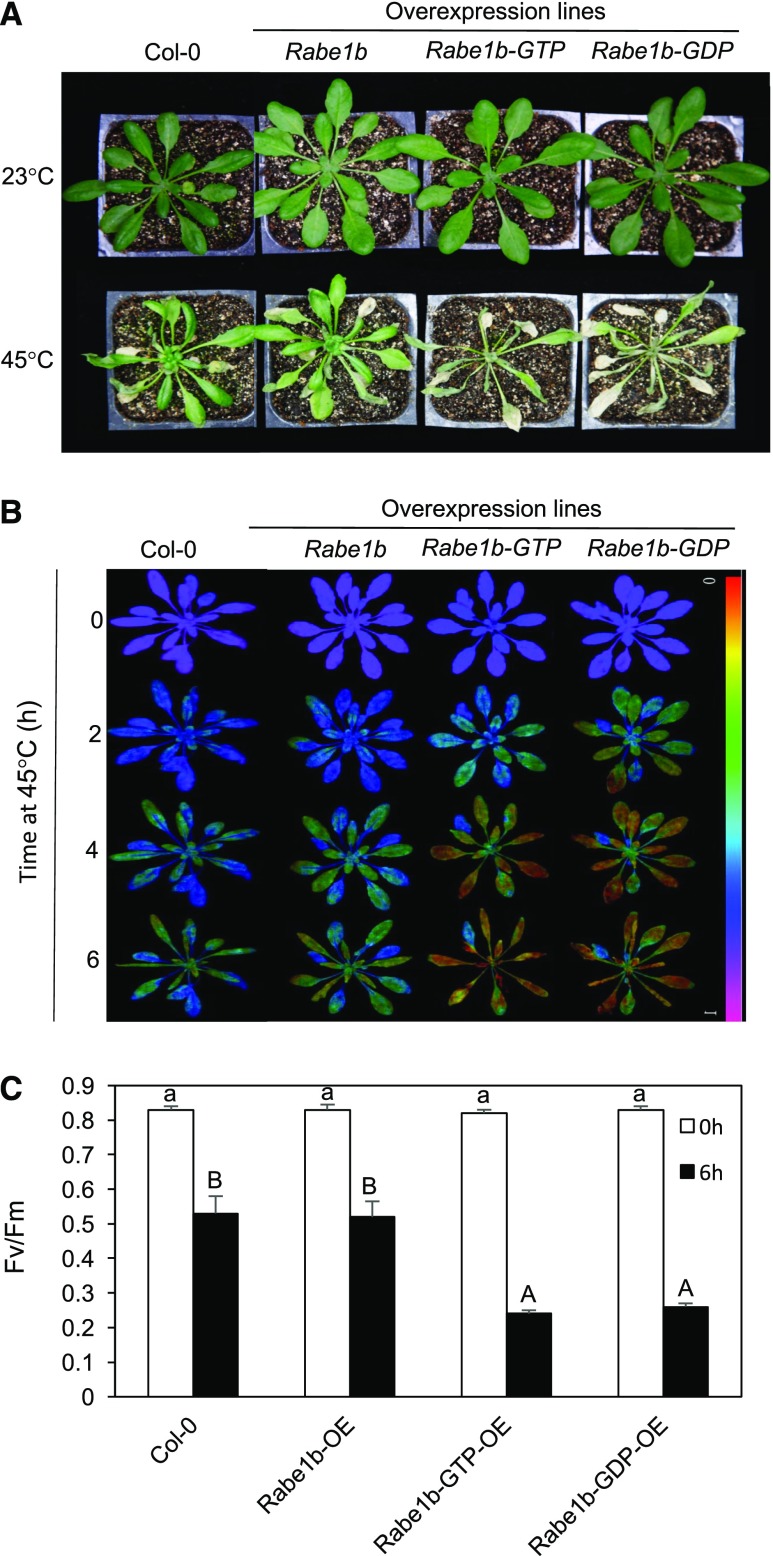
We also compared mature plants of the wild type and overexpression lines for heat tolerance. Five-week-old mature plants were treated at 45°C for 10 h and observed for growth and survival after 5 d of recovery at room temperature. As shown in Figure 6A, while Col-0 wild-type and transgenic AtRabe1b-OE plants survived after heat stress, most AtRabe1b-GTP-OE and AtRab1lb-GDP-OE plants displayed wilting symptoms during heat stress and died during the recovery at room temperature. At the biochemical level, heat stress causes damage to the light-harvesting complexes of PSII and inhibition of PSII (Allakhverdiev et al., 1996, 2008). Therefore, we compared the ratio of variable chlorophyll fluorescence to maximum fluorescence (Fv/Fm), indicative of the maximum quantum yield of PSII, in fully expanded leaves of the overexpression lines with those of wild-type plants immediately before and after 2, 4, and 6 h of heat treatment at 45°C. As shown in Figure 6, B and C, no significant difference in Fv/Fm was observed among the wild type and the Rabe1b-overexpressing lines prior to the heat treatment. After the 6-h heat treatment, both wild-type and AtRabe1b-OE plants displayed a reduction of about 35% in Fv/Fm when compared with those before heat treatment (Fig. 6, B and C). On the other hand, AtRabe1b-GTP-OE and AtRabe1b-GDP-OE plants showed a greater than 70% reduction in Fv/Fm from heat treatment (Fig. 6, B and C). Thus, overexpression of constitutive GTP- and GDP-bound mutant Rabe1b compromised heat tolerance in transgenic Arabidopsis plants.
Figure 6.
Effects of overexpression of Rabe1b, Rabe1b-GTP, and Rabe1b-GDP on the heat tolerance of Arabidopsis mature plants. A, Six-week-old mature plants were placed in a 45°C growth chamber for 10 h. The heat-treated plants were then moved to a 23°C growth chamber for recovery. The photograph was taken 5 d after the heat treatment. B, Fv/Fm images of fully expanded leaves determined immediately after the indicated hours (h) at 45°C heat treatment. The color code in the images ranged from 0 (black) to 1 (purple). C, Fv/Fm values of fully expanded leaves determined immediately after the indicated hours of heat treatment at 45°C. Means and se were calculated from average Fv/Fm values determined from three experiments with 10 leaves per experiment for each genotype. According to Duncan’s multiple range test (P < 0.01), means of Fv/Fm at 0 h do not differ significantly if they are indicated with the same lowercase letter; means of Fv/Fm at 6 h do not differ significantly if they are indicated with the same uppercase letter.
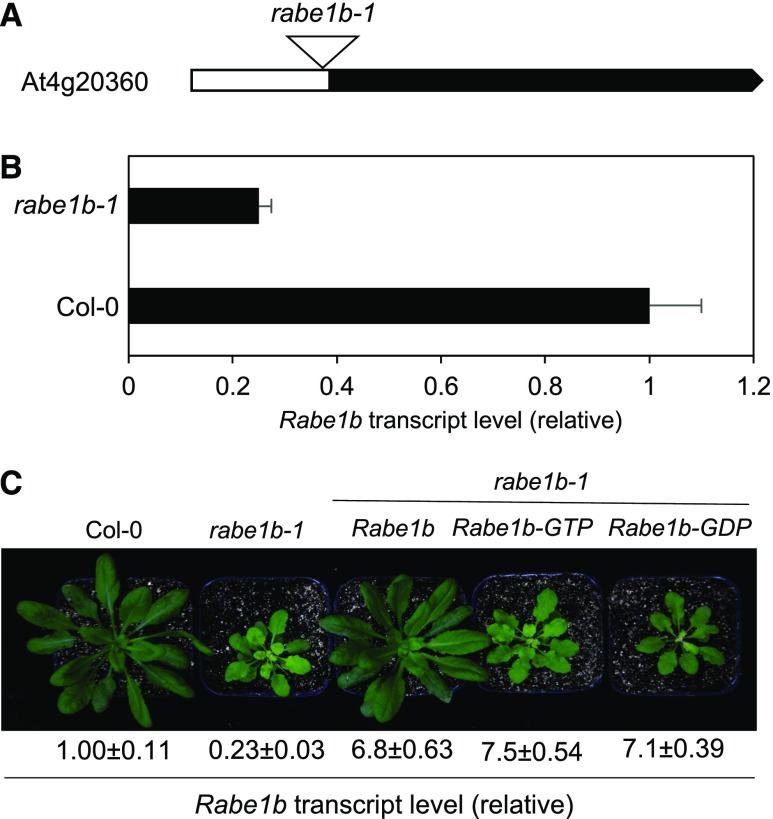
Knockdown of Rabe1b Increased Heat Sensitivity
To further analyze the role of Rabe1b in plant heat tolerance, we isolated T-DNA insertion mutants for the gene. Two T-DNA insertion lines, SAIL_415_E05 and SAIL_659_G09, were screened using PCR with primers flanking the insertion sites. SAIL_415_E05 contains a T-DNA insertion in the middle of the single-exon gene, but a PCR screen failed to identify homozygous plants, most likely due to their lethal phenotype. SAIL_659_G09 contains a T-DNA insertion in the 5′ region of the gene, approximately 120 nucleotides upstream of its translation start codon (Fig. 7A). RT-qPCR showed that homozygous SAIL_659_G09 contains about 25% of the wild-type levels of the Rabe1b transcripts (Fig. 7B). Therefore, SAIL_659_G09 is a knockdown mutant and was named rabe1b-1 (Fig. 7A). The rabe1b-1 mutant plants were reduced in size and were slightly pale green when grown at normal conditions (Fig. 7C). To determine that the growth phenotype of the rabe1b-1 mutant was due to the reduced expression of Rabe1b, we transformed myc-tagged AtRabe1b, AtRabe1b-GTP, and AtRabe1b-GDP genes into the mutant. As observed in the wild type, a substantial percentage of transformants displayed bleached phenotypes due to cosuppression. Among those lines that expressed the transgenes based on both RT-qPCR and immunoblotting, we observed that myc-tagged AtRabe1b, but not its constitutive GTP- or GDP-bound mutant form, could complement the mutant (Fig. 7C). This result indicates that the constitutive GTP- and GDP-bound mutant forms of Rabe1b are inactive as translation factors.
Figure 7.
Identification and growth of Arabidopsis rabe1b-1 mutants. A, Diagram of the Rabe1b gene and the T-DNA insertion site. B, Transcript levels of Rabe1b in the Col-0 wild type and the rabe1b-1 mutant as determined using RT-qPCR. Means and se were calculated from three replicates. C, Growth of Col-0, the rabe1b-1 mutant, and the rabe1b-1 mutant complemented with the wild-type Rabe1b or its constitutive GTP- and GDP-bound mutant forms. The photograph was taken 4 weeks after germination. Means and se of the relative Rabe1b transcript levels in the plants determined by RT-qPCR from three replicates also are shown at the bottom.
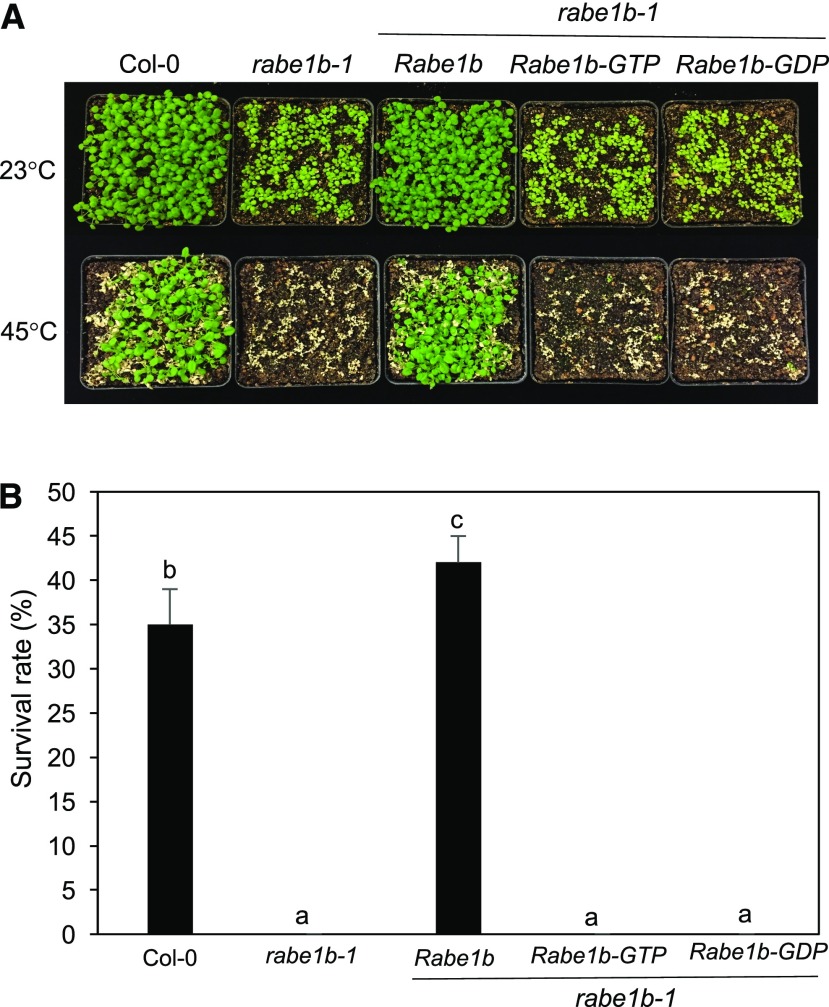
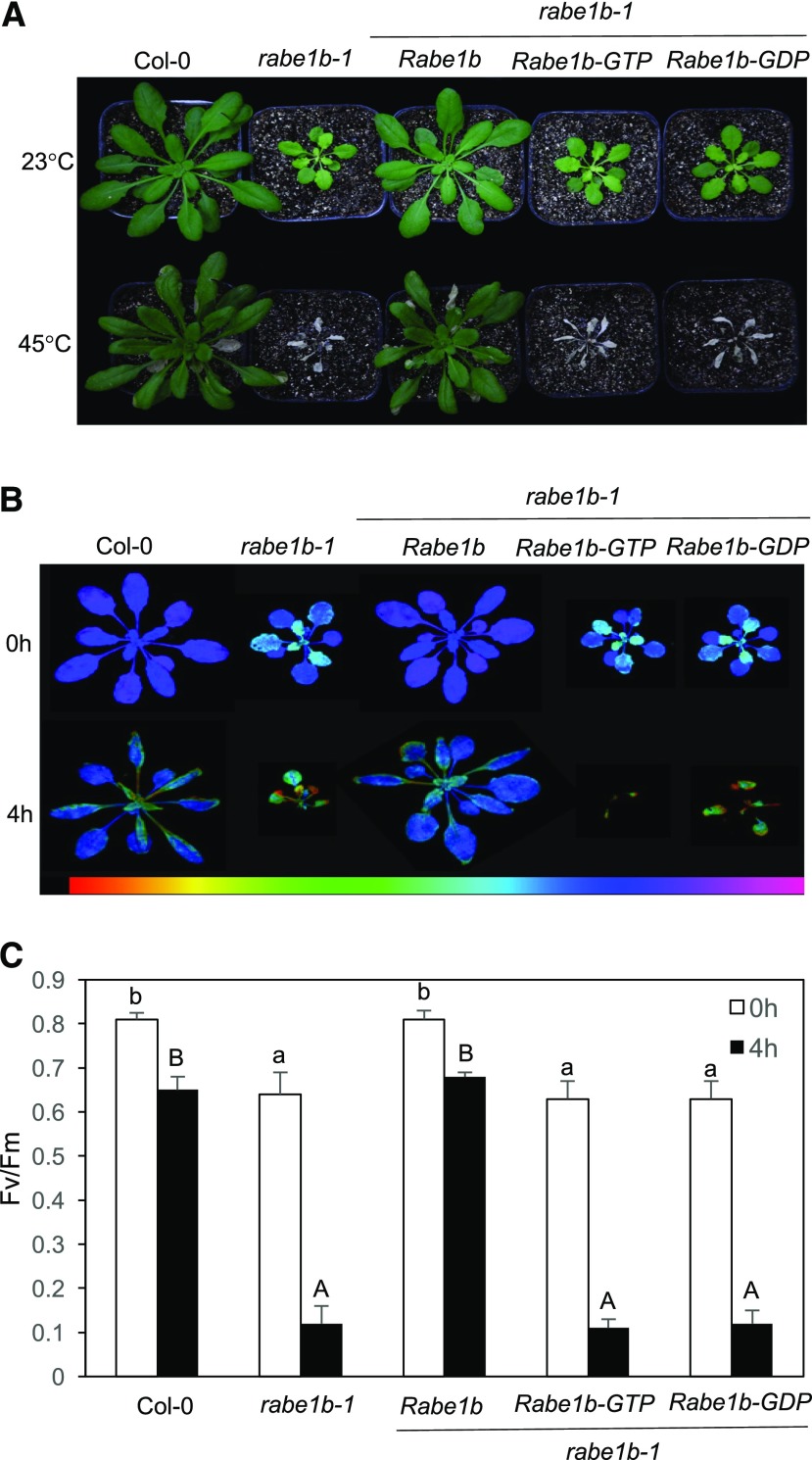
To compare the wild type and the rabe1b mutant for heat tolerance, we again tested both seedlings and mature plants for survival rates after heat treatment. As shown in Figure 8, about 35% of wild-type seedlings survived after heat treatment at 45°C for 9 h. By contrast, none of the rabe1b-1 mutant seedlings survived the same heat treatment (Fig. 8). Introduction of the myc-tagged AtRabe1b, but not the AtRabe1b-GTP or AtRabe1b-GDP transgene, could completely restore the heat tolerance of the rabe1b mutant seedlings (Fig. 8). Similar results also were obtained with mature plants based on an analysis of the survival and the Fv/Fm ratios after heat treatment (Fig. 9). Thus, the rabe1b knockdown mutant, despite the substantial residual levels of the Rabe1b transcripts, is highly compromised in heat tolerance.
Figure 8.
Functional analysis of Arabidopsis Rabe1b in the heat tolerance of Arabidopsis seedlings. A, Assays of the heat tolerance of young seedlings. Approximately 50 2-week-old seedlings of wild-type Col-0, the rabe1b-1 mutant, and the rabe1b mutant complemented with the Rabe1b, Rabe1b-GTP, or Rabe1b-GDP transgene were placed in a 45°C growth chamber for 9 h. The heat-treated plants were then moved to a 23°C growth chamber for recovery. The photograph was taken 5 d after the heat treatment. B, Survival rates of heat-stressed young seedlings determined after treatment at 45°C for 9 h followed by 23°C for 5 d for recovery. Means and se were calculated from survival rates determined from three experiments with approximately 100 seedlings per experiment for each genotype. According to Duncan’s multiple range test (P < 0.01), means of survival rates do not differ significantly if they are indicated with the same letter.
Figure 9.
Functional analysis of Arabidopsis Rabe1b in the heat tolerance of mature plants. A, Six-week-old mature plants of wild-type Col-0, the rabe1b-1 mutant, and the rabe1b mutant complemented with the Rabe1b, Rabe1b-GTP, or Rabe1b-GDP transgene were placed in a 45°C growth chamber for 10 h. The heat-treated plants were then moved to a 23°C growth chamber for recovery. The photograph was taken 5 d after the heat treatment. B, Fv/Fm images of fully expanded leaves determined immediately after 0 and 4 h at 45°C. The color code in the images ranged from 0 (black) to 1 (purple). C, Fv/Fm values of fully expanded leaves determined immediately after the indicated hours of heat treatment at 45°C. Means and se were calculated from five replicates. According to Duncan’s multiple range test (P < 0.01), means of Fv/Fm at 0 h do not differ significantly if they are indicated with the same lowercase letter; means of Fv/Fm at 4 h do not differ significantly if they are indicated with the same uppercase letter.
We also transformed the rabe1b mutant with the wild-type Rabe1b gene under the control of its native promoter. Again, we observed a substantial percentage of transformants displaying bleached phenotypes due to cosuppression. The remaining lines that expressed the transgene based on both RT-qPCR and immunoblotting were indistinguishable from the wild type (Supplemental Fig. S6). Furthermore, both the heat tolerance (Supplemental Fig. S6) and heat induction of HsfA2 and its target genes (data not shown) of the rabe1b mutant were fully restored to that of wild-type plants. Thus, the rabe1b mutant could be fully complemented by the wild-type Rabe1b gene under the control of its native promoter.
Silencing of Rabe1b Compromised Heat Tolerance in Tomato
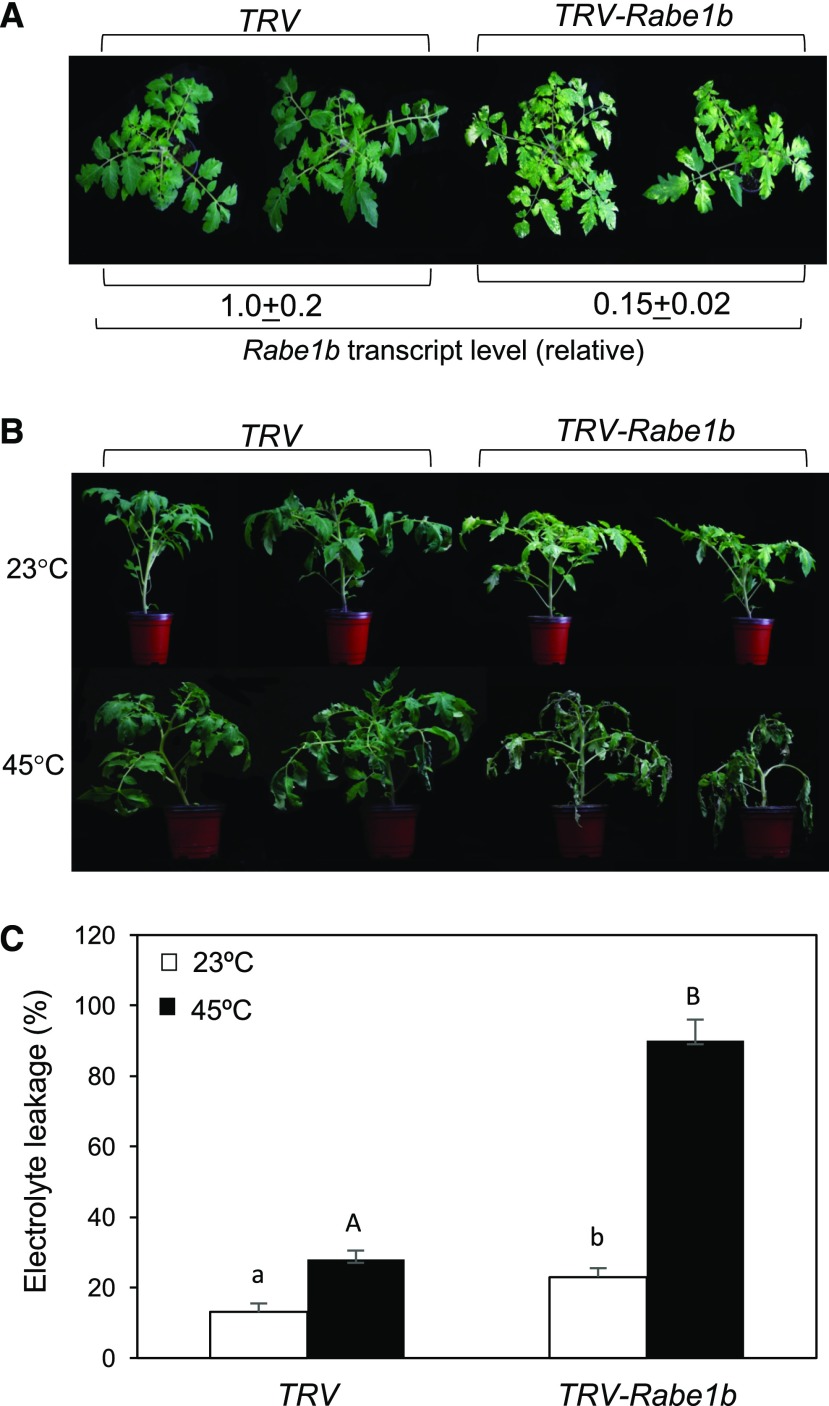
To determine whether the critical role of plastid EF-Tu in plant heat tolerance is evolutionarily conserved, we extended our functional analysis to tomato using virus-induced gene silencing (VIGS). From the sequenced tomato genome, we identified two tomato Rabe1b genes, SlRabe1b-1 (Solyc03g112150) and SlRabe1b-2 (Solyc06g071790). Tomato Rabe1b-1 and Rabe1b-2 genes encode two proteins of 477 and 478 amino acid residues, respectively, that share approximately 88% sequence identify with each other and about 85% with Arabidopsis Rabe1b, with most of the variable amino acid residues in the N-terminal plastid-targeting signal peptides (Supplemental Fig. S7). For VIGS, we cloned a DNA fragment specific to the tomato plastid Rabe1b genes into the pTRV vector and infiltrated Agrobacterium tumefaciens cells harboring the VIGS vector into tomato cotyledons. Approximately 3 weeks after the infiltration, tomato plants infiltrated with the pTRV-Rabe1b vector started to develop pale green leaves with bleached patches (Fig. 10A). No such phenotypes were observed in the tomato plants infiltrated with the pTRV empty vector (Fig. 10A). RT-qPCR analysis showed that transcript levels of the tomato Rabe1b genes in the plants infiltrated with pTRV-Rabe1b were reduced by about 85% when compared with those in the control plants infiltrated with the pTRV empty vector (Fig. 10). To assess the effect of silencing of the Rabe1b genes on heat tolerance, we placed the tomato plants in a 45°C growth chamber for 8 h and then moved them to room temperature for a 3-d recovery. For heat-treated pTRV control plants, we observed few symptoms after recovery (Fig. 10B). On the other hand, almost all expanded leaves of pTRV-Rabe1b plants displayed wilting after recovery (Fig. 10B). The severe wilting symptoms in Rabe1b-silenced tomato plants after heat stress were consistent with the drastic increase in electrolyte leakage in the silenced plants relative to that in the pTRV control plants (Fig. 10C). These results indicated that the plastid Rabe1b proteins also play a critical role in tomato heat tolerance.
Figure 10.
Functional analysis of tomato Rabe1b in tomato heat tolerance using TRV-mediated gene silencing. A, Tomato plants infiltrated with A. tumefaciens cells harboring the empty pTRV vector or the silencing pTRV-Rabe1b vector were placed in a 23°C growth room. Photographs of the representative plants were taken 4 weeks after A. tumefaciens infiltration. Tomato Rabe1b transcript levels in tomato plants infiltrated with A. tumefaciens cells harboring the empty pTRV vector or the pTRV-Rabe1b silencing vector were determined by RT-qPCR analysis using total RNA isolated from the terminal leaflets of the fifth leaves of A. tumefaciens-infiltrated tomato plants and are listed below the images. Means and se were calculated from approximately 10 leaflets from five plants. B, A. tumefaciens-infiltrated tomato plants were placed in a 23°C or 45°C growth chamber for 8 h. Photographs of the whole plants were taken after the 5-d recovery. C, Electrolyte leakage (EL) of the terminal leaflets of the fifth leaves was determined immediately after 8 h at 23°C or 45°C heat treatment. Means and se were calculated from average EL values determined from three experiments with10 leaves per experiment for each genotype. According to Duncan’s multiple range test (P < 0.01), means of EL at 23°C do not differ significantly if they are indicated with the same lowercase letter; means of EL at 45°C do not differ significantly if they are indicated with the same uppercase letter.
Knockdown of Rabe1b Inhibits Nuclear Heat-Responsive Gene Expression
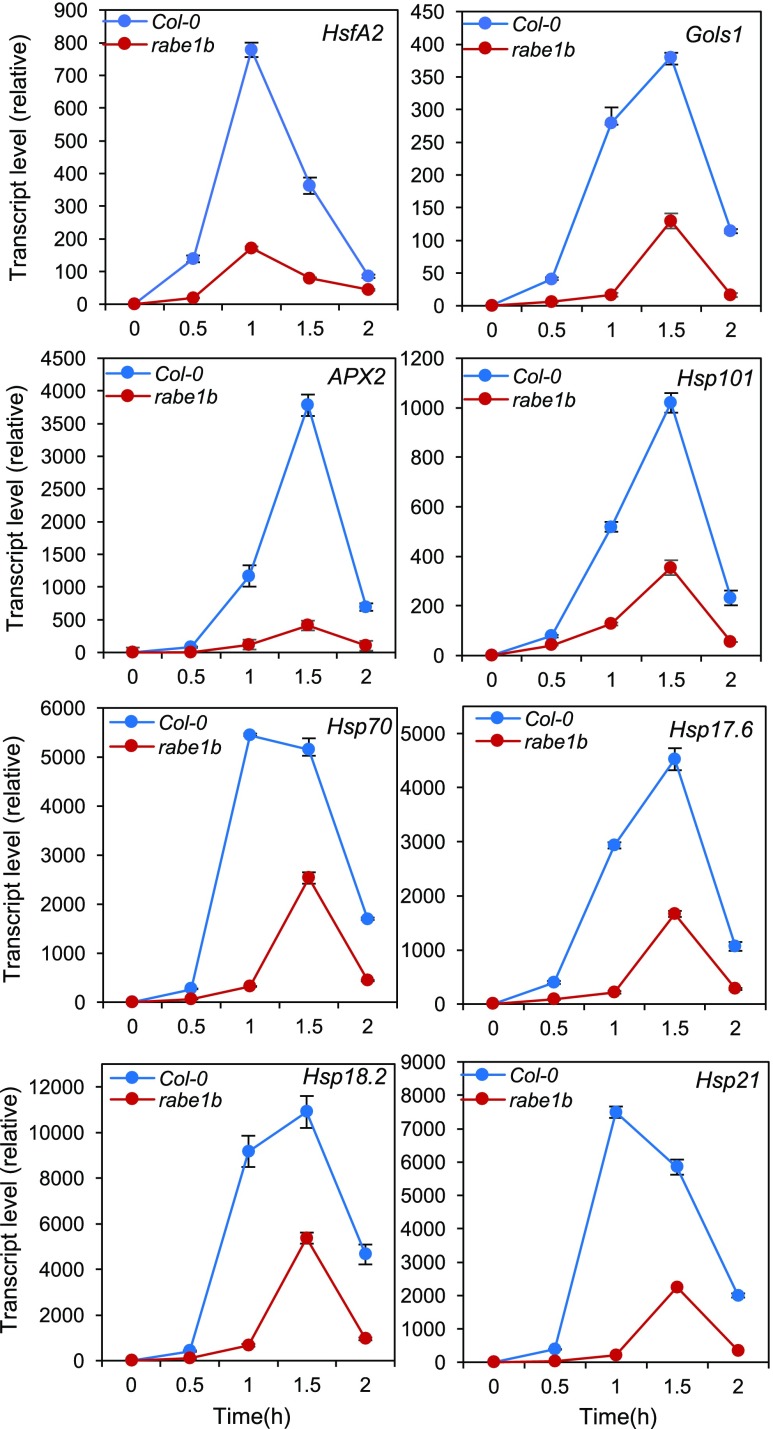
In plants, heat stress rapidly induces the production of well-characterized HSPs, including Hsp101, Hsp70, and small HSPs, which act as molecular chaperones in protein quality control by promoting the folding and refolding of nonnative proteins (Whitley et al., 1999). The regulation of HSP gene expression is mediated by the conserved HSFs. In Arabidopsis, HsfA2 plays a key role in the triggering and amplification of plant heat stress responses (Kotak et al., 2007). Interestingly, the basal transcript levels of Arabidopsis HsfA2 in the Arabidopsis rabe1b-1 mutant were only about 30% of those in the wild type in the absence of heat stress. At 45°C, the transcript levels of HsfA2 in the wild type increased rapidly, with an approximately 800-fold induction at the 1-h peak point under heat stress (Fig. 11). In the rabe1b mutant, the induction of HsfA2 gene expression was inhibited, with less than a 200-fold induction at the 1-h peak point during heat treatment (Fig. 11). We also analyzed a subset of HsfA2 target genes for their expression before and after heat treatment. These target genes encode APX2, GolS1, and several HSPs (Hsp17.6, Hsp18.2, Hsp21, Hsp70, and Hsp101). We again observed that the basal levels of these HsfA2 target genes in the absence of heat stress were all reduced by 60% to 80% when compared with those in the wild type. All these HsfA2 target genes were strongly induced in the wild type, where the transcript levels peaked 1 to 2 h after heat stress (Fig. 11). As with HsfA2, the heat induction of these HsfA2 target genes was all reduced substantially in the rabel1-1 knockdown mutant relative to that in the wild type (Fig. 11). Thus, the reduced expression of Rabe1b in the rabe1b mutant caused substantial inhibition of both basal and induced expression of HsfA2 and its target genes.
Figure 11.
Induction of Arabidopsis HsfA2 and its target genes by heat stress. Five-week-old Arabidopsis Col-0 wild-type and rabe1b-1 mutant plants were placed in a 45°C growth chamber, and total RNA was isolated from leaf samples collected at the indicated times. Transcript levels were determined using RT-qPCR. Means and se were calculated from three replicates.
Constitutive Expression of HsfA2 Partially Restored the Heat Tolerance of rabe1b-1
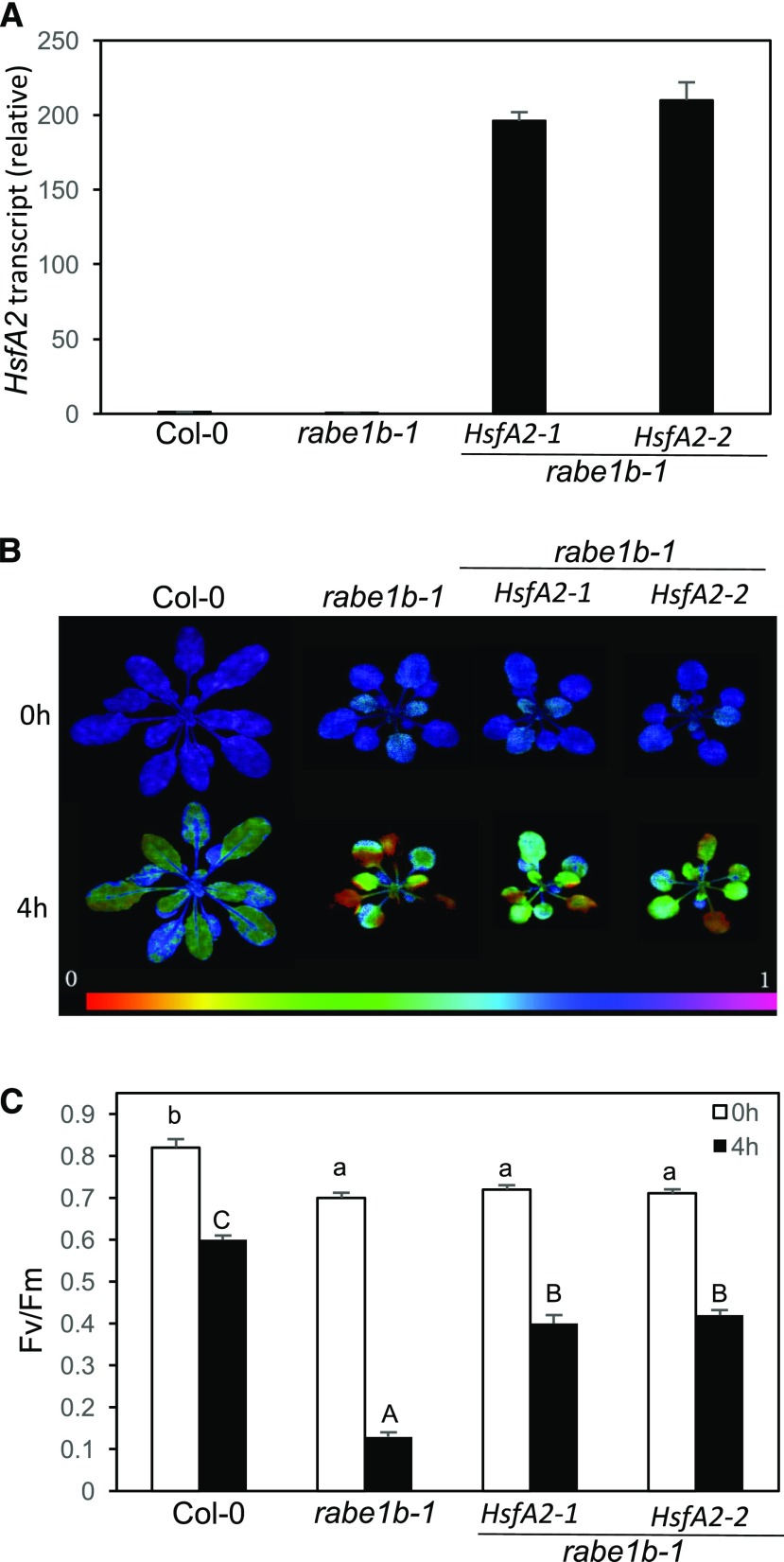
HsfA2 regulates the expression of a variety of genes involved in stress responses with protective roles in scavenging ROS, production of osmolytes, and protein quality control (Shigeoka et al., 2002; Taji et al., 2002; Nishizawa et al., 2006). Reduced basal and induced expression of HsfA2 and its target genes in rabe1b-1 could lead to reduced capacities of these protective mechanisms and, consequently, growth defects and compromised heat tolerance in the mutant. To determine this possibility, we generated transgenic rabe1b mutant plants harboring an HsfA2 transgene under the control of the constitutive CaMV 35S promoter. Transgenic lines constitutively expressing the HsfA2 gene were obtained based on RT-qPCR (Fig. 12A). Notably, constitutive expression of HsfA2 in the rabe1b-1 mutant did not rescue the phenotypes of reduced size and pale green leaves of the mutant (Fig. 12B).
Figure 12.
Effects of the constitutive expression of HsfA2 on the growth and heat tolerance of rabe1b-1. A, Constitutive expression of the HsfA2 transgene in transgenic rabe1b plants. Total RNA was isolated from Arabidopsis Col-0 wild type, the rabe1b mutant, and two independent lines of transgenic rabe1b mutant plants harboring the HsfA2 transgene under the control of the CaMV 35S promoter. HsfA2 transcript levels were determined using RT-qPCR. Means and se were calculated from three replicates. B, Fv/Fm images of fully expanded leaves determined immediately after 0 and 4 h at 45°C. The color code in the images ranged from 0 (black) to 1 (purple). C, Fv/Fm values of fully expanded leaves determined immediately after the indicated hours of heat treatment at 45°C. According to Duncan’s multiple range test (P < 0.01), means of Fv/Fm at 0 h do not differ significantly if they are indicated with the same lowercase letter; means of Fv/Fm at 4 h do not differ significantly if they are indicated with the same uppercase letter.
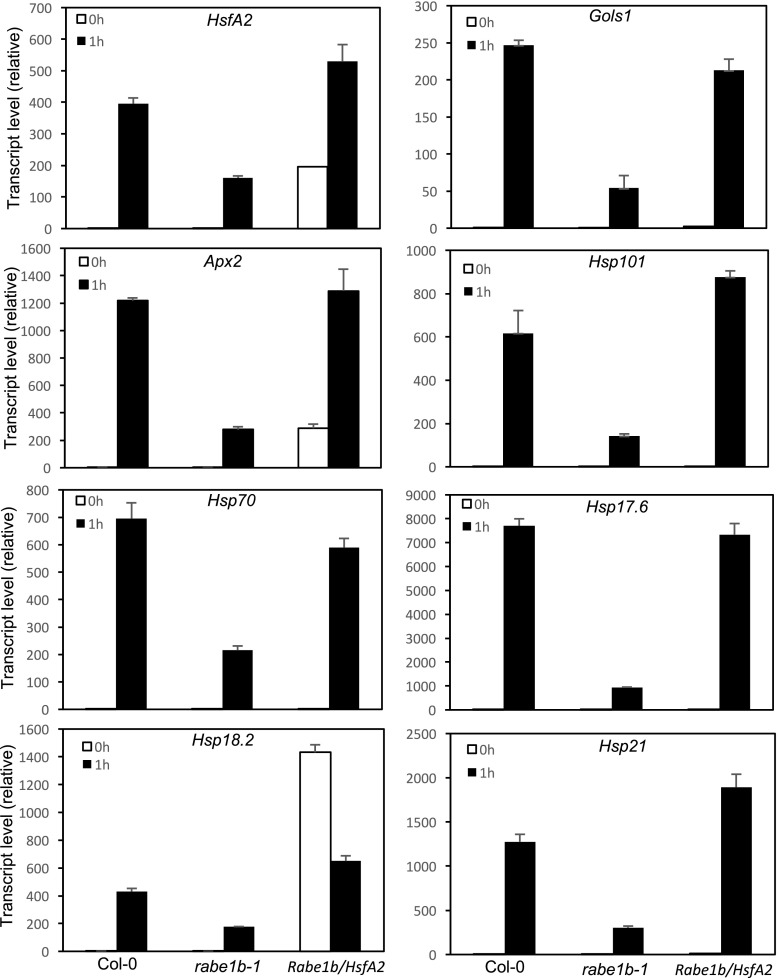
To determine the impact of the constitutive expression of HsfA2 on the heat tolerance of the rabe1b-1 mutant, we compared Fv/Fm values before and after heat treatment at 45°C for 10 h among wild-type and rabe1b mutant plants with or without the HstA2-myc transgene. The Fv/Fm value for the rabe1b mutant was more than 10% lower than that for the wild type even when grown at normal conditions (Fig. 12, B and C). Constitutive expression of HsfA2 in the rabe1b mutant did not significantly affect the Fv/Fm value of the mutant prior to heating. After the 4-h heat treatment at 45°C, the Fv/Fm value of the rabe1b mutant was reduced by more than 85%, compared with less than a 30% reduction in wild-type plants (Fig. 12, B and C). In the rabe1b plants constitutively expressing HsfA2, the Fv/Fm value was reduced by approximately 50% after 4 h at 45°C. RT-qPCR analysis, on the other hand, showed that constitutive expression of HsfA2 in the rabe1b mutant increased both the basal and heat-induced expression of HsfA2 and HsfA2 target genes to levels close to or even higher than those in wild-type plants (Fig. 13). Thus, constitutive expression of HsfA2 had no significant effect on the growth of the rabe1b mutant and only partially restored the heat tolerance of the mutant despite fully restored induction of HsfA2 and HsfA2 target genes.
Figure 13.
Effects of the constitutive expression of HsfA2 on HsfA2 and HsfA2 target genes. Five-week-old Arabidopsis wild-type Col-0, the rabe1b-1 mutant, and the rabe1b mutant constitutively expressing HsfA2 were treated by heat stress at 45°C for 0 and 1 h. Total RNA was isolated from leaf samples, and transcript levels of the indicated genes were determined using RT-qPCR. Means and se were calculated from three replicates.
Activity of Rabe1b Proteins in Plastid Protein Translation and Protein Folding
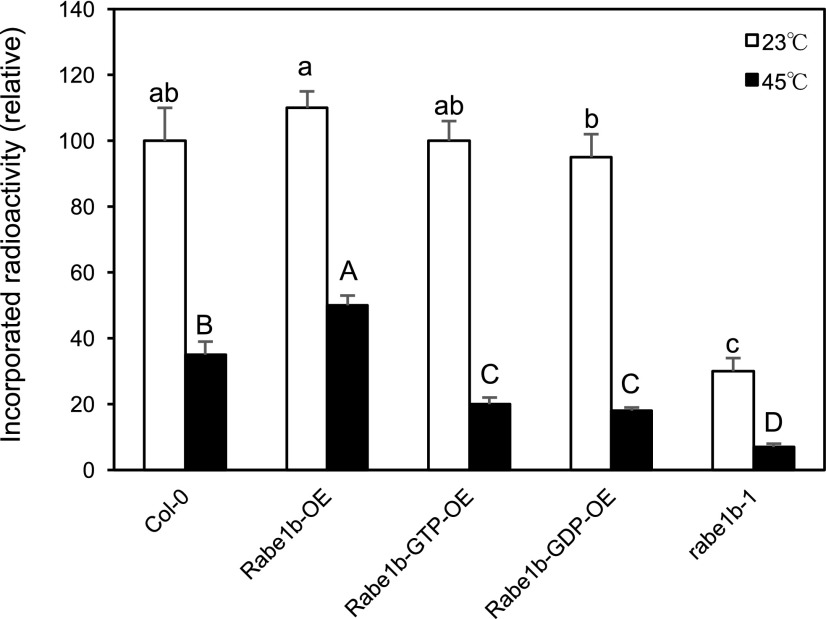
To further study how Arabidopsis Rabe1b plays such a critical role in plant heat stress responses, we examined the effects of the rabe1b-1 mutation and the overexpression of wild-type Rabe1b and its constitutive GTP- and GDP-bound mutant forms on plastid protein translation at normal conditions and during heat stress. Plastids were isolated from the Arabidopsis wild type, transgenic Rabe1b-OE, Rabe1b-GTP-OE, and Rabe1b-GDP-OE plants, and the rabe1b-1 mutant, and plastid protein translation was assayed in isolated plastids using [35S]Met. As shown in Figure 14, at normal conditions, overexpression of wild-type Rabe1b increased plastid protein synthesis by about 10%, whereas overexpression of the constitutive GTP- and GDP-bound mutant forms has no significant effect on plastid protein synthesis. After 6 h at 45°C, protein synthesis in plastids from wild-type plants was reduced by about 65% when compared with that at normal conditions (Fig. 14). In transgenic plants overexpressing wild-type Rabe1b, the heat treatment also reduced plastid translation by approximately 55% and, as a result, plastid protein synthesis in the transgenic plants was about 40% higher than that in wild-type plants after heat treatment (Fig. 14). In transgenic plants overexpressing the constitutive GTP- and GDP-bound mutant Rabe1b forms, heat stress reduced plastid protein translation by about 80% and, as a result, plastid protein synthesis in these transgenic plants was about 40% lower than that in wild-type plants after heat treatment (Fig. 14). In the rabe1b-1 mutant, plastid protein synthesis was reduced to about 30% of the wild-type level at normal conditions (Fig. 14). After heat treatment at 45°C for 6 h, plastid protein synthesis in the rabe1b-1 mutant was reduced further to about 20% of the wild-type level (Fig. 14). Thus, the levels of plastid protein synthesis under heat stress were highly associated with the levels of heat tolerance among the wild type, the rabe1b mutant, and the three types of transgenic overexpression lines (Figs. 5, 6, 8, and 9).
Figure 14.
Effects of Rabe1b proteins and heat stress on plastid protein translation. Plastids were isolated from leaves harvested from plants grown under normal conditions (23°C) or after 6 h of heat treatment (45°C). Protein synthesis was assayed in isolated plastids with [35S]Met. The relative levels of protein synthesis were represented by the relative amount of incorporated [35S]Met radioactivity on an equal number of isolated plastids. Means and se were calculated from three replicates. According to Duncan’s multiple range test (P < 0.05), means of the relative amount of incorporated radioactivity in plastid protein synthesis from plants grown at normal conditions (23°C) do not differ significantly if they are indicated with the same lowercase letter; means of incorporated radioactivity in plastids isolated from heat-treated plants (45°C) do not differ significantly if they are indicated with the same uppercase letter.
To examine how wild-type Rabe1b and the constitutive GTP- and GDP-bound mutant forms differed in their abilities to promote heat tolerance, we were interested in determining their effects on plastid protein folding and stability, which, however, is currently still not feasible in vivo. Therefore, we compared the chaperone activity of the three proteins in vitro. We generated Rabe1b proteins in E. coli and assayed their activity in protecting the highly heat-sensitive malate dehydrogenase (MDH) at 43°C, which has been widely used to determine protein chaperone activity. MDH was incubated with Rabe1b or bovine serum albumin (BSA) at 43°C for 30 min and then analyzed immediately for residual MDH activity. As shown in Supplemental Figure S8, wild-type Rabe1 and its constitutive GTP- and GDP-bound mutant forms had similar levels of activity, protecting heat-sensitive MDH at the high temperature when compared with the BSA control. This result is consistent with the previous reports that EF-Tu proteins from bacteria, mitochondria, and plastids act as protein chaperones in a GTP/GDP-independent manner (Malki et al., 2002; Rao et al., 2004; Ristic et al., 2007; Suzuki et al., 2007). Thus, despite their differential ability in promoting plant heat tolerance, wild-type Rabe1b and its constitutive GTP- and GDP-bound mutant forms appeared to have similar levels of protein chaperone activity.
DISCUSSION
As a translation factor, EF-Tu plays an essential role in protein biosynthesis in prokaryotes and in eukaryotic organelles of mitochondria and plastids. EF-Tu also can act as a protein chaperone, and chloroplast EF-Tu from plants plays a role in plant heat tolerance based on the expression of their corresponding genes and changed heat tolerance from altered levels of the translation elongation factor (Bhadula et al., 2001; Rao et al., 2004; Ristic et al., 2004, 2007, 2008; Momcilovic and Ristic, 2007; Fu et al., 2008; Brisio et al., 2010; Fu and Ristic, 2010). Here, we extended these studies by providing important information about the dynamic states of chloroplast EF-Tu under heat stress and its important role and possible mode of action in plant responses to heat stress.
Heat-Induced Aggregation of Chloroplast EF-Tu
We reported previously from proteomic profiling that the Arabidopsis plastid EF-Tu, Rabe1b, accumulates as aggregates in heat-treated plants (Zhou et al., 2014b). Through comprehensive analysis of its total, soluble, and insoluble forms using immunoblotting, we confirmed that aggregation of Arabidopsis Rabe1b proteins occurred in vitro at a temperature as low as 25°C (Supplemental Fig. S5) and that more than 50% of the proteins became insoluble in vitro after 30 min at 35°C (Fig. 2). In vivo, the plastid EF-Tu also aggregated when the plants were treated with a temperature above 41°C (Fig. 3). Thus, plastid Rabe1b proteins were highly prone to aggregate at elevated temperatures. In vitro and in vivo protein aggregation is a topic of high importance to the understanding of protein structures and functions, production of recombinant proteins in biotechnology, and human health (Philo and Arakawa, 2009). Among the possible mechanisms of protein aggregation, the predominant one involves conformational changes that expose hydrophobic portions of the proteins, which may interact with the exposed hydrophobic patches of other proteins (Philo and Arakawa, 2009). The EF-Tu from prokaryotes, mitochondria, and plastids is a triangular three-domain protein that undergoes substantial conformational changes during the elongation cycle of protein translation (Brisio et al., 2010). The GTP-bound EF-Tu forms a ternary complex with aa-tRNA, and upon codon-anticodon recognition and GTP hydrolysis to GDP, EF-Tu can undergo a conformational change necessary for the release of the aa-tRNA and dissociation from the ribosome in the EF-Tu-GDP form (Brisio et al., 2010). Biochemical and structural analyses of EF-Tu from mesophilic bacteria like E. coli have revealed that the translation factor, particularly its N-terminal GTP/GDP-binding domain, is prone to secondary structure shift (Brisio et al., 2010). On the other hand, EF-Tu proteins from hyperthermophilic bacteria are highly structured and more resistant to high temperatures (Katava et al., 2016).
During the translation elongation, EF-Tu undergoes dynamic association with aa-tRNAs, ribosomes, and other proteins, including guanidine exchange factor EF-Ts (Brisio et al., 2010). Therefore, aggregation of EF-Tu at high temperatures would inactivate its activity as a translation elongation factor. The high propensity of plastid EF-Tu to aggregate in vitro even under slightly elevated temperatures above 25°C would be highly detrimental if it was also the case in vivo, given its essential role in plastid protein synthesis. Apparently, there are mechanisms that promote stability and reduce the aggregation of plastid Rabe1b proteins in plant cells, as evident from the substantially higher temperatures required to induce Rabe1b protein aggregation in vivo than in vitro (Figs. 2 and 3). One possible mechanism could be the active engagement of soluble Rabe1b proteins in protein translation with dynamic interactions with GTP, GDP, aa-tRNA, ribosomes, and other proteins, which would thermodynamically increase the resistance to forming insoluble aggregates by reducing their interactions with unfolded or partially unfolded proteins under heat stress. This possible mechanism is consistent with the observation that constitutive GTP- and GDP-bound forms of Rabe1b proteins were more prone to aggregate, possibly due to their lack of dynamic interactions with different guanine nucleotides, aa-RNA, ribosomal, and other proteins. In addition, the stronger in vivo stability of plastid Rabe1b proteins could be aided by the protective chaperone proteins in chloroplasts. We have observed previously that the Rabe1b protein aggregates accumulate to higher levels in the mutants for the CARBOXY TERMINUS OF HSC70 INTERACTING PROTEIN (CHIP) ubiquitin E3 ligase and the autophagy-selective receptor NEIGHBOR OF BREAST CANCER1 (NBR1; Zhou et al., 2014b), suggesting that Rabe1b protein aggregates are targeted for degradation by the CHIP-mediated 26S proteasome system and NBR1-mediated selective autophagy. Likewise, aggregates of heat-sensitive Rubisco activase and catalase proteins also accumulated at higher levels in the chip nbr1 mutant than in wild-type plants (Zhou et al., 2014b). Interestingly, CHIP and NBR1 deficiency in the chip nbr1 mutant also increased total catalase and Rubisco protein levels but reduced the levels of their soluble forms (Zhou et al., 2014b). Thus, CHIP- and NBR-mediated degradation pathways not only remove insoluble protein aggregates but also protect soluble native proteins. By promoting the removal of aggregated forms, these pathways could possibly reduce the interactions of protein aggregates with soluble proteins, thereby decreasing the inactivation and aggregation of soluble proteins. CHIP- and NBR1-mediated pathways could play a similar role in protecting plastid Rabe1b proteins from aggregation by removing highly interactive plastid protein aggregates.
The Role of Plastid EF-Tu in Plant Growth, Photoprotection, and Heat Tolerance
In this study, we have comprehensively analyzed the biological functions of plastid EF-Tu in plant growth and heat tolerance using gene silencing in both Arabidopsis and tomato, overexpression of constitutive GTP- or GDP-bound mutant Rabe1b genes, and a T-DNA insertion mutant in Arabidopsis. As the only gene encoding an important chloroplast translation factor in Arabidopsis, Rabe1b is likely to be essential for plant growth based on the failure to obtain a homozygous T-DNA knockout mutant. Cosuppression and VIGS of Rabe1b in Arabidopsis and tomato, respectively, led to the photobleaching of green tissues, strikingly similar to the phenotype caused by the silencing of plant PHYTOENE DESATURASE (PDS). Plant PDS encodes an enzyme in the biosynthesis of carotenoids, which act as antioxidants in photooxidation by quenching single-oxygen and triple sensitizers generated during photosynthesis (Wang et al., 2005; Romero et al., 2011). The bleaching of green tissues associated with the silencing of chloroplast EF-Tu indicates that a high capacity of chloroplast protein biosynthesis is required for photoprotection during photosynthesis. Upon triggering of silencing of the Rabe1b gene and inhibition of chloroplast protein translation, photooxidation could be induced due to increased production of ROS as a result of the imbalance between light harvesting and subsequent photochemical reactions caused by the reduced synthesis of specific components in chloroplasts. In addition, increased photooxidation in Rabe1b-silenced green tissues could be caused by reduced levels of antioxidants and, consequently, reduced capacity of photoprotection in chloroplasts.
The comprehensive functional analysis through molecular genetic approaches supported the critical role of plastid EF-Tu in plant heat tolerance. The Arabidopsis rabe1b-1 knockdown mutant was highly compromised in heat tolerance (Figs. 8 and 9). Likewise, tomato plants with silenced Rabe1b genes also became highly sensitive to high temperature (Fig. 10). The reduced expression of Rabe1b in the rabe1b-1 knockdown mutant and in Rabe1b-silenced tomato plants was associated with other phenotypes, including pale greening, bleaching, and reduced growth (Figs. 7 and 10; Supplemental Fig. S2). Therefore, it could be argued that the role of Rabe1b in plant heat tolerance might be through its general effects on photosynthesis, chloroplast functions, and, ultimately, plant growth. The evidence against such an argument comes from the transgenic Arabidopsis plants overexpressing the constitutive GTP- or GDP-bound mutant Rabe1b form. Overexpressed GTP- and GDP-bound mutant Rabe1b forms can potentially interfere with the endogenous Rabe1b, thereby inhibiting plastid protein translation. Surprisingly, these transgenic plants were indistinguishable from wild-type plants in morphology and growth (Fig. 6A). They also were normal in photosynthetic efficiency of PSII when grown at normal temperatures (Fig. 6, B and C). The small effect of overexpressed GTP- and GDP-bound mutant Rabe1b forms on plant growth under normal conditions could be due to two reasons. First, plastid EF-Tu is a relatively abundant protein, based on our comparison of transgenic plants expressing myc-tagged AtRabe1b under the control of the strong CaMV 35S promoter versus its native promoter. Second, the dominant negative activity of the GTP- and GDP-bound mutant forms of Rabe1b may be relatively weak. Thus, the relative abundance of native Rabe1b proteins in these transgenic plants under normal conditions would make Rabe1b very unlikely to be a rate-limiting factor, with a relatively weak dominant effect from the overexpressed GTP- and GDP-bound mutant forms of Rabe1b. This interpretation is consistent with the finding that overexpression of GTP- and GDP-bound forms does not significantly affect plastid protein translation in transgenic plants under normal conditions (Fig. 14). However, transgenic plants overexpressing the GTP- and GDP-bound mutant forms displayed reduced heat tolerance at both their seedling and mature stages (Figs. 5 and 6). This effect of the GTP- and GDP-bound mutant forms on plant heat tolerance also could result from two possible reasons. First, due to heat-induced aggregation at high temperatures, the soluble, active form of the endogenous Rabe1b is reduced substantially, making it more likely to be a rate-limiting factor in plastid translation. Second, at high temperatures, the genes encoding the GTP- and GDP-bound mutant forms are under the control of a strong constitutive promoter; therefore, they are continuously synthesized, become misfolded, aggregate, and eventually degrade. Misfolded proteins are known to interact nonspecifically with other proteins, promote their misfolding, or interfere with their functions. Therefore, GTP- and GDP-bound Rabel1b mutant proteins could still interfere with the activity of endogenous Rabe1b or promote their aggregation through nonspecific interactions prior to or during their rapid aggregation. This interpretation is consistent with the finding that overexpression of the GTP- and GDP-bound mutant forms substantially reduces plastid protein translation at 45°C but not at room temperature (Fig. 14). Therefore, high levels of active chloroplast Rabe1b proteins appear to be particularly important for plant heat tolerance.
Both the Arabidopsis rabe1b-1 knockdown mutant and Rabe1b-silenced tomato plants were severely compromised in heat tolerance, despite their residual levels of Rabe1b transcripts, which were approximately 15% to 25% of wild-type levels (Figs. 6 and 10). In vivo, a significant percentage of plastid Rabe1b proteins became insoluble at 41°C, and most aggregated within several hours at 43°C (Fig. 3). Thus, at a temperature above 43°C, the rapid aggregation of plastid Rabe1b proteins would lead to reduction of the soluble active form of the translation factor to a level that severely compromises plant heat tolerance. In other words, the high heat sensitivity of plastid EF-Tu likely makes it a critical limiting factor of plant heat stress responses. As a result, EF-Tu would likely be a target for evolutionary selection and genetic engineering for increased heat tolerance. In prokaryotes, the EF-Tu proteins from thermophiles have evolved to be more heat stable than their counterparts in mesophiles (Katava et al., 2016). In plants, genes encoding plastid EF-Tu are induced by heat and other stresses for the elevation of protein levels, and manipulation of the Rabe1b protein levels through molecular and genetic means could lead to altered heat tolerance in plants (Bhadula et al., 2001; Ristic et al., 2004, 2007, 2008; Momcilovic and Ristic, 2007; Fu et al., 2008). In light of the heat-sensitive nature of the plastid translation factor, a more effective way to improve plant heat tolerance would be to enhance not only the protein levels but also the heat stability of plastid EF-Tu.
Possible Mechanisms by Which Chloroplast EF-Tu Promotes Plant Heat Tolerance
Prokaryotic, mitochondrial, and plastid EF-Tu proteins not only function as essential translation factors in protein biosynthesis but also act as protein chaperones that promote the renaturation of denatured proteins and prevent their aggregation. Studies with EF-Tu proteins from prokaryotes, mammalian mitochondria, and plant plastids have concluded that EF-Tu proteins can promote the refolding of denatured proteins. The chaperone activities of EF-Tu proteins are widely believed to be primarily responsible for their positive role in plant heat tolerance (Fu et al., 2012). Unlike their activity as a protein translation elongation factor, which is dependent on dynamic GTP and GDP binding, the chaperone activity of EF-TU is not dependent on GTP or GDP (Malki et al., 2002; Rao et al., 2004; Ristic et al., 2007; Suzuki et al., 2007). In this study, we generated transgenic Arabidopsis plants that overexpressed plastid EF-Tu and its constitutive GTP- and GDP-bound mutant forms, which had similar levels of chaperone activity (Supplemental Fig. S8). If the chaperone activity of EF-Tu is primarily responsible for its positive role in plant heat tolerance, we expected that overexpression of Rabe1b and its GTP- and GDP-bound mutant forms would have similar positive effects on the heat tolerance of the transgenic plants. Importantly, we observed significantly increased heat tolerance only in transgenic plants overexpressing the wild-type Rabe1b proteins but not the constitutive GTP- or GDP-bound mutant forms (Figs. 5 and 6). In fact, the constitutive GTP- or GDP-bound forms of Rabe1b substantially reduced plant heat tolerance when overexpressed (Figs. 5 and 6), most likely by acting as dominant negative proteins that interfere with wild-type Rabe1b in protein translation. Therefore, while the chaperone activities of plastid EF-Tu could play a role in heat tolerance, the high activity of EF-Tu as a protein translation factor is crucial for the growth and survival of plants under heat stress. It is noteworthy that the transgenic plants overexpressing the constitutive GTP- and GDP-bound mutant forms of plastid EF-Tu were normal in growth and photosynthesis under normal growth conditions but were highly compromised in plant heat tolerance (Figs. 5 and 6). Furthermore, the levels of plastid protein synthesis were strongly associated with the levels of heat tolerance among the wild type, the rabe1b mutant, and transgenic plants overexpressing wild-type Rabe1b or its constitutive GTP- and GDP-bound mutant forms (Fig. 14). These findings again indicated that sufficiently high levels of plastid EF-Tu activity as a protein translation factor are particularly important for plants under heat stress. This could arise from the need for increased synthesis of heat-induced plastid proteins with important roles in heat stress responses. Increased synthesis also would be necessary for heat-labile proteins important for chloroplast and other cellular functions under heat stress.
The compromised heat tolerance of the rabe1b knockdown mutant was associated with inhibited induction of HsfA2 and its target genes under heat stress (Fig. 11). Thus, plastid EF-Tu also acts as a positive regulator of chloroplast retrograde signaling regulating the heat-induced expression of nuclear heat-responsive genes. It has been reported previously that an Arabidopsis knockdown mutant for CHLOROPLAST RIBOSOMAL PROTEIN S1 (RPS1) also exhibited reduced growth, pale green leaves, and compromised heat tolerance associated with reduced induction of HsfA2 and its target genes under heat stress (Yu et al., 2012). Importantly, constitutive overexpression of HsfA2 in the rps1 knockdown mutant could restore not only the induction of the HsfA2 target genes but also the growth and heat tolerance of the mutant (Yu et al., 2012). The authors proposed that the reduced growth, destabilized thylakoid membranes, and compromised heat tolerance are caused by reduced production of ROS-scavenging enzymes and other protective mechanisms as a result of inhibited HsfA2 expression in the rps1 mutant (Yu et al., 2012). Intriguingly, constitutive overexpression of HsfA2 in the rabe1b knockdown mutant led to elevated expression of some but not all of its target genes at normal conditions, as has been found previously in the rps1 mutant (Yu et al., 2012). This could be due to the possibility that HsfA2 is inactive or not fully active in the absence of heat stress or other cofactors that are not overexpressed in the transgenic plants or activated at normal conditions. Constitutive overexpression of HsfA2 activated the expression of HsfA2 target genes at high temperature but only partially restored the heat tolerance of the mutant (Figs. 12 and 13). More importantly, the reduced growth and pale green leaves of the rabe1b mutant were not rescued by the constitutive overexpression of HsfA2 (Fig. 12). Likewise, the reduced size and pale green leaf phenotype of the rabe1b-1 mutant were not rescued by the GTP- or GDP-bound mutant of Rabe1b (Fig. 7). Thus, inhibited HsfA2 only contributed in part to the compromised heat tolerance of the rabe1b mutant but was not responsible for the growth phenotype of the mutant. More likely, the reduced growth phenotype of the rabe1b mutant is due to the reduced protein level of the plastid translation factor and, consequently, the reduced plastid protein translation. These findings further highlight the importance of the levels of active Rabe1b proteins as a core plastid translation factor in determining the extent of plant growth, photoprotection, and heat tolerance.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) materials were in the Col-0 background. The homozygous Arabidopsis rabe1b-1 (SAIL_659_G09) mutant was identified by PCR using primers flanking the insertion site (5′-GATTTCCTCCACGGCATTAG-3′/5′-CGGGACAGAGAGAAGGAGAA-3′). Arabidopsis plants were grown in growth chambers at 23°C on a photoperiod of 12 h of light and 12 h of dark. Tomato (Solanum lycopersicum ‘Alisa Craig’) seeds were germinated in a growth medium filled with a mixture of peat and vermiculite (7:3, v/v) in trays. When the first true leaf was fully expanded, seedlings were transplanted into plastic pots containing the same medium.
Generation of Rabe1b and HsfA2 Constructs
To generate the myc-tagged wild-type Arabidopsis Rabe1b gene for overexpression in Arabidopsis, we amplified its coding sequence using a pair of AtRabe1b-specific primers (5′-AGCCTCGAGATGGCGATTTCGGCTCCA-3′ and 5′-AGCTTAATTAATTCGAGGATCGTCCCAATAA-3′). Coding sequences for the constitutive GTP- and GDP-bound forms of AtRabe1b were generated by overlapping PCR using substitution-specific primers (5′-CCTGGTCTCGCTGATTACGTTAA-3′/5′-TTAACGTAATCAGCGAGACCAGG-3′ for Rabe1bH152L and 5′-GGTTGTGTTTCTTATCAAAGAGGATCAAGT-3′/5′-ACTTGATCCTCTTTGATAAGAAACACAACC-3′ for Rabe1bN203I). The amplified AtRabe1b fragments were fused with a 4×myc tag sequence in a binary vector. The coding sequence of HsfA2 was PCR amplified using a pair of gene-specific primers (5′-AGCCTCGAGATGGAAGAACTGAAAGTGGAAATGG-3′ and 5′-AGCTTAATTAAAGGTTCCGAACCAAGAAAACCCAT-3′) and cloned into a binary vector behind the CaMV 35S promoter. The AtRabe1b gene also was cloned behind its native promoter of ∼2 kb, which was PCR amplified from Arabidopsis genomic DNA using the following primers: 5′-AGCGGATCCGGGAGAAACCTGGTTTTTAT-3′ and 5′-AGCGTCGACGGGAAGATGGAATTGGAGAG-3′. The resulting constructs were introduced into Agrobacterium tumefaciens and transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transformants were identified by glufosinate resistance, and expression of the transgenes was analyzed by RT-qPCR or immunoblotting using an anti-myc monoclonal antibody (Sigma-Aldrich).
Yeast Two-Hybrid Assays
For assays of AtRabe1b protein interaction with plastid EF-Ts (AT4G29060) in yeast, the plastid EF-Ts coding sequence was PCR amplified using gene-specific primers (5′-AGCGGATCCGCTGAAAGCATCAAAGGCA-3′/5′-AGCGTCGACTCAGTTATCTTCTCCAAGAGTAAACTTC-3′) and cloned into a pBD-Gal4 vector. Full-length coding sequences of AtRabe1b and its GTP- and GDP-bound mutant genes were cloned into the pAD-Gal4 vector. Various combinations of bait and prey constructs were cotransformed into yeast cells, and interactions were analyzed by plating onto selection medium lacking Trp, Leu, and His.
RT-qPCR
Total plant RNA isolation and reverse transcription were performed as described previously (Zhou et al., 2013, 2014a). RT-qPCR was performed using the iCycler iQ real-time PCR detection system (Bio-Rad), and relative gene expression was calculated as described previously (Livak and Schmittgen, 2001). Actin genes from Arabidopsis and tomato were used as internal controls. Gene-specific primers for RT-qPCR are listed in Supplemental Table S1.
Separation of Soluble and Insoluble Proteins and Western Blotting
Separation and analysis of soluble and insoluble proteins were performed as described previously (Zhou et al., 2013, 2014b). Briefly, Arabidopsis leaves were collected before and after heat treatment, ground in liquid nitrogen, and homogenized in a detergent containing extraction buffer (100 mm Tris-HCl, pH 8, 10 mm NaCl, 1 mm EDTA, 1% Triton X-100 (v/v), and 0.2% β-mercaptoethanol (v/v)). The homogenates were filtered through a 300- and a 100-μm nylon mesh to obtain the total proteins. The total proteins were clarified by centrifugation at 2,200g for 5 min, and the supernatants were kept as soluble proteins. The pellets were resuspended in the same buffer and clarified by the same low-speed centrifugation. The process was repeated twice, and the final pellets were resuspended in the extraction buffer as insoluble proteins. Protein fractionation by SDS-PAGE and immunoblotting for the detection of myc-tagged Rabe1b proteins were performed using an anti-myc antibody. The antigen-antibody complexes were detected by enhanced chemiluminescence using luminal as a substrate.
Tomato VIGS
The TRV VIGS constructs for the silencing of tomato Rabe1b genes were generated by PCR amplification using gene-specific primers (5′-AGCGAATTCCGCCCCAGAAGAGCGTGCTC-3′/5′-AGCCTCGAGGAATATCATCACCAGGGAAC-3′), digested with EcoRI and XhoI restriction enzymes, and ligated into the same sites of pTRV2 (Liu et al., 2002). The resulting plasmids were transformed into A. tumefaciens strain GV3101. A. tumefaciens-mediated virus infection was performed as described previously (Ekengren et al., 2003). Plants were then kept at 23°C for 4 weeks before they were used for the experiments (Kandoth et al., 2007). Terminal leaflets of the fifth fully expanded leaves, which showed 15% to 20% transcript levels of the control plants, were used.
Analysis of Fv/Fm and Electrolyte Leakage
Chlorophyll fluorescence was measured using an Imaging-PAM Chlorophyll Fluorometer equipped with a computer-operated PAM-control unit (IMAG-MAXI; Heinz Walz). The plants were maintained in the dark for more than 30 min before the measurements were performed. The intensities of the actinic light and saturating light were 280 and 2,500 μmol m−2 s−1 photosynthetic photon flux density, respectively. Fv/Fm was measured and calculated as described previously (Zhou et al., 2013). Three replicates for each treatment were used with 12 plants for each replicate. For the determination of electrolyte leakage caused by high temperature, the terminal leaflets of the fifth fully expanded leaves were measured after heat stress as described previously (Huang et al., 2010).
Plastid Protein Translation
Six-week-old Arabidopsis plants were treated with heat stress at 45°C for 0 and 6 h. Approximately 20 g of leaves was used for plastid isolation by Percoll gradient centrifugation as described previously (Klein and Mullet, 1986). The number of plastids was determined using a hemocytometer with a photomicroscope. Assays of protein synthesis in isolated plastids were performed largely as described previously (Klein and Mullet, 1986). Briefly, the protein synthesis mixture (50 mL) contained 0.33 m sorbitol, 50 mm HEPES-KOH (pH 8), 40 μm of each amino acid (minus Met), 10 mm DTT, 25 μCi of [35S]Met (Perkin Elmer), 5 mm ATP, and 5 mm MgC12. Isolated plastids were added to a final concentration of 5 × 106 per assay. The mixture was incubated at 23°C for 20 min before adding unlabeled Met to a final concentration of 8.5 mm. Sorbitol-HEPES was then added to terminate the reaction. Extraction and TCA precipitation of plastid proteins were performed, and TCA-insoluble radioactivity was determined as described previously (Klein and Mullet, 1986).
Assays of GTPase and Chaperone Activities of Recombinant Arabidopsis Rabe1b Proteins
The full-length coding sequences for Arabidopsis wild-type Rabe1b and the constitutive GTP- and GDP-bound mutant proteins were cloned into the expression vector pET-32a (Novagen) and transformed into Escherichia coli strain BL21(DE3). Induced expression and purification of recombinant proteins were performed according to the manual provided by Novagen. GTPase activity of the Rabel1b proteins was determined with a GTPase assay kit (BioLegend) following the manufacturer’s instructions. Assays of the chaperone activity of the recombinant proteins were performed as described previously (Rao et al., 2004). Briefly, the assays (1 mL) containing 0.5 μm MDH (Sigma-Aldrich), 2 μm recombinant Rabe1b proteins, or BSA and other appropriate components were incubated at 43°C for 30 min. The MDH activities before and after incubation were measured as described (Rao et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data library under the following accession numbers: AT4G20360 (AtRabe1b), AT2G26150 (AtHsfA2), AT1G74310 (AtHsp101), AT1G16030 (AtHsp70b), AT4G27670 (AtHsp21), AT5G59720 (AtHsp18.2), AT5G12030 (AtHsp17.6), AT2G47180 (AtGolS1), AT3G09640 (AtAPX2), Solyc03g112150 and Solyc06g071790 (SlRabe1b), and AT4G29060 (AtEF-Ts).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Assays of AtRabe1b GTPase activity and interaction with plastid EF-Ts.
Supplemental Figure S2. Cosuppression of Arabidopsis Rabe1b in transgenic plants.
Supplemental Figure S3. RT-qPCR analysis of Rabe1b transgene transcripts in transgenic Rabe1b, Rabe1b-GTP, and Rabe1b-GDP Arabidopsis plants.
Supplemental Figure S4. Effect of GTP on heat-induced aggregation of Arabidopsis plastid EF-Tu in vitro.
Supplemental Figure S5. Heat-induced rapid aggregation of Arabidopsis plastid EF-Tu in vitro.
Supplemental Figure S6. Restoration of growth and heat tolerance of the rabe1b-1 mutant by the Rabe1b gene under the control of its native promoter.
Supplemental Figure S7. Sequence comparison between Arabidopsis and tomato mature chloroplast Rabe1b proteins.
Supplemental Figure S8. Effect of recombinant Arabidopsis Rabe1b proteins on the activity of MDH before and after incubation at 43°C.
Supplemental Table S1. Primers for RT-qPCR.
Acknowledgments
We thank the Arabidopsis Resource Center at Ohio State University for the Arabidopsis mutants.
Footnotes
This work is supported by the U.S. National Science Foundation (grant nos. IOS-0958066 and IOS1456300), the Natural Science Foundation of China (grant nos. 31771698, 31401923, and 31470368), and the Department of Science and Technology of Zhejiang Province of China (2013TD05).
Articles can be viewed without a subscription.
References
- Allakhverdiev SI, Feyziev YM, Ahmed A, Hayashi H, Aliev JA, Klimov VV, Murata N, Carpentier R (1996) Stabilization of oxygen evolution and primary electron transport reactions in photosystem II against heat stress with glycinebetaine and sucrose. J Photochem Photobiol B 34: 149–157 [DOI] [PubMed] [Google Scholar]
- Allakhverdiev SI, Kreslavski VD, Klimov VV, Los DA, Carpentier R, Mohanty P (2008) Heat stress: an overview of molecular responses in photosynthesis. Photosynth Res 98: 541–550 [DOI] [PubMed] [Google Scholar]
- Amm I, Sommer T, Wolf DH (2014) Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta 1843: 182–196 [DOI] [PubMed] [Google Scholar]
- Arias E, Cuervo AM (2011) Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 23: 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo AP. (2005) [Heat shock proteins as molecular chaperones]. Med Sci (Paris) 21: 619–625 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Chan KY, Scharf KD, Nover L (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282: 3605–3613 [DOI] [PubMed] [Google Scholar]
- Bhadula SK, Elthon TE, Habben JE, Helentjaris TG, Jiao S, Ristic Z (2001) Heat-stress induced synthesis of chloroplast protein synthesis elongation factor (EF-Tu) in a heat-tolerant maize line. Planta 212: 359–366 [DOI] [PubMed] [Google Scholar]
- Brisio R, Ruggiero A, Vitagliano L (2010) Elongation factors EF1A and EF-Tu: their role in translation and beyond. Isr J Chem 50: 71–79 [Google Scholar]
- Brodsky JL, McCracken AA (1999) ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol 10: 507–513 [DOI] [PubMed] [Google Scholar]
- Caldas T, Laalami S, Richarme G (2000) Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J Biol Chem 275: 855–860 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ricigliano JW, Klessig DF (1993a) Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc Natl Acad Sci USA 90: 9533–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig DF (1993b) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262: 1883–1886 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Craig EA, Gambill BD, Nelson RJ (1993) Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev 57: 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV (2003) Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332: 689–699 [DOI] [PubMed] [Google Scholar]
- Eamus D, Duff GA, Berryman CA (1995) Photosynthetic responses to temperature, light flux-density, CO2 concentration and vapour pressure deficit in Eucalyptus tetrodonta grown under CO2 enrichment. Environ Pollut 90: 41–49 [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917 [DOI] [PubMed] [Google Scholar]
- Feinstein RN, Sacher GA, Howard JB, Braun JT (1967) Comparative heat stability of blood catalase. Arch Biochem Biophys 122: 338–343 [DOI] [PubMed] [Google Scholar]
- Fu J, Momcilović I, Clemente TE, Nersesian N, Trick HN, Ristic Z (2008) Heterologous expression of a plastid EF-Tu reduces protein thermal aggregation and enhances CO2 fixation in wheat (Triticum aestivum) following heat stress. Plant Mol Biol 68: 277–288 [DOI] [PubMed] [Google Scholar]
- Fu J, Momcilovic I, Prasad PVV (2012) Roles of protein synthesis elongation factor EF-Tu in heat tolerance in plants. J Bot 2012: 1–8 [Google Scholar]
- Fu J, Ristic Z (2010) Analysis of transgenic wheat (Triticum aestivum L.) harboring a maize (Zea mays L.) gene for plastid EF-Tu: segregation pattern, expression and effects of the transgene. Plant Mol Biol 73: 339–347 [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Welch WJ (1993) Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol 9: 601–634 [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YW, Carter M, Miller DL (1992) The identification of a domain in Escherichia coli elongation factor Tu that interacts with elongation factor Ts. J Biol Chem 267: 22198–22205 [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268: 1517–1520 [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katava M, Kalimeri M, Stirnemann G, Sterpone F (2016) Stability and function at high temperature: what makes a thermophilic GTPase different from its mesophilic homologue. J Phys Chem B 120: 2721–2730 [DOI] [PubMed] [Google Scholar]
- Klein RR, Mullet JE (1986) Regulation of chloroplast-encoded chlorophyll-binding protein translation during higher plant chloroplast biogenesis. J Biol Chem 261: 11138–11145 [PubMed] [Google Scholar]
- Klimov VV, Baranov SV, Allakhverdiev SI (1997) Bicarbonate protects the donor side of photosystem II against photoinhibition and thermoinactivation. FEBS Lett 418: 243–246 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Krab IM, Parmeggiani A (1998) EF-Tu, a GTPase odyssey. Biochim Biophys Acta 1443: 1–22 [DOI] [PubMed] [Google Scholar]
- Krab IM, Parmeggiani A (2002) Mechanisms of EF-Tu, a pioneer GTPase. Prog Nucleic Acid Res Mol Biol 71: 513–551 [DOI] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA (2006) Autophagy: an ER protein quality control process. Autophagy 2: 135–137 [DOI] [PubMed] [Google Scholar]
- Kudlicki W, Coffman A, Kramer G, Hardesty B (1997) Renaturation of rhodanese by translational elongation factor (EF) Tu: protein refolding by EF-Tu flexing. J Biol Chem 272: 32206–32210 [DOI] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19: 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, Xu XM, Møller SG (2010) Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 232: 567–578 [DOI] [PubMed] [Google Scholar]
- Li C, Chen Q, Gao X, Qi B, Chen N, Xu S, Chen J, Wang X (2005) AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China C Life Sci 48: 540–550 [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malki A, Caldas T, Parmeggiani A, Kohiyama M, Richarme G (2002) Specificity of elongation factor EF-TU for hydrophobic peptides. Biochem Biophys Res Commun 296: 749–754 [DOI] [PubMed] [Google Scholar]
- Matts RL, Hurst R (1992) The relationship between protein synthesis and heat shock proteins levels in rabbit reticulocyte lysates. J Biol Chem 267: 18168–18174 [PubMed] [Google Scholar]
- Matts RL, Xu Z, Pal JK, Chen JJ (1992) Interactions of the heme-regulated eIF-2 alpha kinase with heat shock proteins in rabbit reticulocyte lysates. J Biol Chem 267: 18160–18167 [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic I, Ristic Z (2007) Expression of chloroplast protein synthesis elongation factor, EF-Tu, in two lines of maize with contrasting tolerance to heat stress during early stages of plant development. J Plant Physiol 164: 90–99 [DOI] [PubMed] [Google Scholar]
- Needham PG, Brodsky JL (2013) How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim Biophys Acta 1833: 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Okiyoneda T, Apaja PM, Lukacs GL (2011) Protein quality control at the plasma membrane. Curr Opin Cell Biol 23: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo JS, Arakawa T (2009) Mechanisms of protein aggregation. Curr Pharm Biotechnol 10: 348–351 [DOI] [PubMed] [Google Scholar]
- Radke S, Chander H, Schäfer P, Meiss G, Krüger R, Schulz JB, Germain D (2008) Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J Biol Chem 283: 12681–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D, Momcilovic I, Kobayashi S, Callegari E, Ristic Z (2004) Chaperone activity of recombinant maize chloroplast protein synthesis elongation factor, EF-Tu. Eur J Biochem 271: 3684–3692 [DOI] [PubMed] [Google Scholar]
- Reddy PS, Kavi Kishor PB, Seiler C, Kuhlmann M, Eschen-Lippold L, Lee J, Reddy MK, Sreenivasulu N (2014) Unraveling regulation of the small heat shock proteins by the heat shock factor HvHsfB2c in barley: its implications in drought stress response and seed development. PLoS ONE 9: e89125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z, Bukovnik U, Momcilović I, Fu J, Vara Prasad PV (2008) Heat-induced accumulation of chloroplast protein synthesis elongation factor, EF-Tu, in winter wheat. J Plant Physiol 165: 192–202 [DOI] [PubMed] [Google Scholar]
- Ristic Z, Momcilović I, Fu J, Callegari E, DeRidder BP (2007) Chloroplast protein synthesis elongation factor, EF-Tu, reduces thermal aggregation of Rubisco activase. J Plant Physiol 164: 1564–1571 [DOI] [PubMed] [Google Scholar]
- Ristic Z, Wilson K, Nelsen C, Momcilovic I, Kobayashi S, Meeley R, Muszynski M, Habben J (2004) A maize mutant with decreased capacity to accumulate chloroplast protein synthesis elongation factor (EF-Tu) displays reduced tolerance to heat stress. Plant Sci 167: 1367–1374 [Google Scholar]
- Rizhsky L, Hallak-Herr E, Van Breusegem F, Rachmilevitch S, Barr JE, Rodermel S, Inzé D, Mittler R (2002) Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J 32: 329–342 [DOI] [PubMed] [Google Scholar]
- Romero I, Tikunov Y, Bovy A (2011) Virus-induced gene silencing in detached tomatoes and biochemical effects of phytoene desaturase gene silencing. J Plant Physiol 168: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004a) Inhibition of photosynthesis by heat stress: the activation state of Rubisco as a limiting factor in photosynthesis. Physiol Plant 120: 179–186 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004b) Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiol 134: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Osteryoung KW, Crafts-Brandner SJ, Vierling E (2001) Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiol 127: 1053–1064 [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Wolf DH (2005) Endoplasmic reticulum-associated protein quality control and degradation: genome-wide screen for ERAD components. Methods Mol Biol 301: 289–292 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Doring P (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol Cell 49: 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- Suzuki H, Ueda T, Taguchi H, Takeuchi N (2007) Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J Biol Chem 282: 4076–4084 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61: 199–223 [Google Scholar]
- Wang F, Shang Y, Fan B, Chen Z (2014) Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog 10: e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang Y, Wang Z, Zhou J, Fan B, Chen Z (2015) A critical role of Lyst-Interacting Protein5, a positive regulator of multivesicular body biogenesis, in plant responses to heat and salt stresses. Plant Physiol 169: 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Iyer LM, Pancholy R, Shi X, Hall TC (2005) Assessment of penetrance and expressivity of RNAi-mediated silencing of the Arabidopsis phytoene desaturase gene. New Phytol 167: 751–760 [DOI] [PubMed] [Google Scholar]
- Whitley D, Goldberg SP, Jordan WD (1999) Heat shock proteins: a review of the molecular chaperones. J Vasc Surg 29: 748–751 [DOI] [PubMed] [Google Scholar]
- Xu Q, Metzler B, Jahangiri M, Mandal K (2012) Molecular chaperones and heat shock proteins in atherosclerosis. Am J Physiol Heart Circ Physiol 302: H506–H514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HD, Yang XF, Chen ST, Wang YT, Li JK, Shen Q, Liu XL, Guo FQ (2012) Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genet 8: e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xia XJ, Zhou YH, Shi K, Chen Z, Yu JQ (2014a) RBOH1-dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J Exp Bot 65: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang Y, Qi J, Chi Y, Fan B, Yu JQ, Chen Z (2014b) E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet 10: e1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]