This study describes a class of stress-related diterpenoids, termed dolabralexins, in maize and identifies diterpene synthases and a cytochrome P450 involved in their biosynthesis.
Abstract
Terpenoids are a major component of maize (Zea mays) chemical defenses that mediate responses to herbivores, pathogens, and other environmental challenges. Here, we describe the biosynthesis and elicited production of a class of maize diterpenoids, named dolabralexins. Dolabralexin biosynthesis involves the sequential activity of two diterpene synthases, ENT-COPALYL DIPHOSPHATE SYNTHASE (ZmAN2) and KAURENE SYNTHASE-LIKE4 (ZmKSL4). Together, ZmAN2 and ZmKSL4 form the diterpene hydrocarbon dolabradiene. In addition, we biochemically characterized a cytochrome P450 monooxygenase, ZmCYP71Z16, which catalyzes the oxygenation of dolabradiene to yield the epoxides 15,16-epoxydolabrene (epoxydolabrene) and 3β-hydroxy-15,16-epoxydolabrene (epoxydolabranol). The absence of dolabradiene and epoxydolabranol in Zman2 mutants under elicited conditions confirmed the in vivo biosynthetic requirement of ZmAN2. Combined mass spectrometry and NMR experiments demonstrated that much of the epoxydolabranol is further converted into 3β,15,16-trihydroxydolabrene (trihydroxydolabrene). Metabolite profiling of field-grown maize root tissues indicated that dolabralexin biosynthesis is widespread across common maize cultivars, with trihydroxydolabrene as the predominant diterpenoid. Oxidative stress induced dolabralexin accumulation and transcript expression of ZmAN2 and ZmKSL4 in root tissues, and metabolite and transcript accumulation were up-regulated in response to elicitation with the fungal pathogens Fusarium verticillioides and Fusarium graminearum. Consistently, epoxydolabranol significantly inhibited the growth of both pathogens in vitro at 10 µg mL−1, while trihydroxydolabrene-mediated inhibition was specific to F. verticillioides. These findings suggest that dolabralexins have defense-related roles in maize stress interactions and expand the known chemical space of diterpenoid defenses as genetic targets for understanding and ultimately improving maize resilience.
Plants produce diverse arrays of specialized metabolites that govern interactions with, and adaptation to, their biotic and abiotic environments (Pichersky and Gershenzon, 2002; Hartmann, 2007; Pichersky and Lewinsohn, 2011; Tholl, 2015). Among these specialized metabolites, diterpenoids are a large class of more than 10,000 known compounds with diverse physiological and biological functions. Plants typically harbor both a widely conserved subset of diterpenoids such as the GAs, a class of phytohormones mediating developmental processes, and myriad specialized compounds with diverse roles in chemical ecology and plant adaptation (Keeling and Bohlmann, 2006; Gershenzon and Dudareva, 2007; Chaturvedi et al., 2012; Schmelz et al., 2014; Hedden and Sponsel, 2015). Because of their agronomic importance as two of the world’s major food crops, maize (Zea mays) and rice (Oryza sativa) have been studied extensively for their terpenoid-based defense systems (Degenhardt, 2009; Schmelz et al., 2014). In rice, a suite of momilactone, oryzalexin, and phytocassane diterpenoids confer defense against several major pathogens, such as rice blast (Magnaporthe oryzae; Peters, 2006; Kato-Noguchi and Kobayashi, 2009; Toyomasu et al., 2014). As components of rice root exudates, momilactones also can exhibit allelopathic activity, suppressing the development of neighboring plants of other species (Kato-Noguchi and Peters, 2013). These findings exemplify the diverse, protective biological roles of diterpenoids in Poaceae crops. However, the potential applications of these chemical defenses in agricultural improvement have been limited, in part, by knowledge gaps in understanding identities, biosynthesis, regulation, and ecological roles in different crop species.
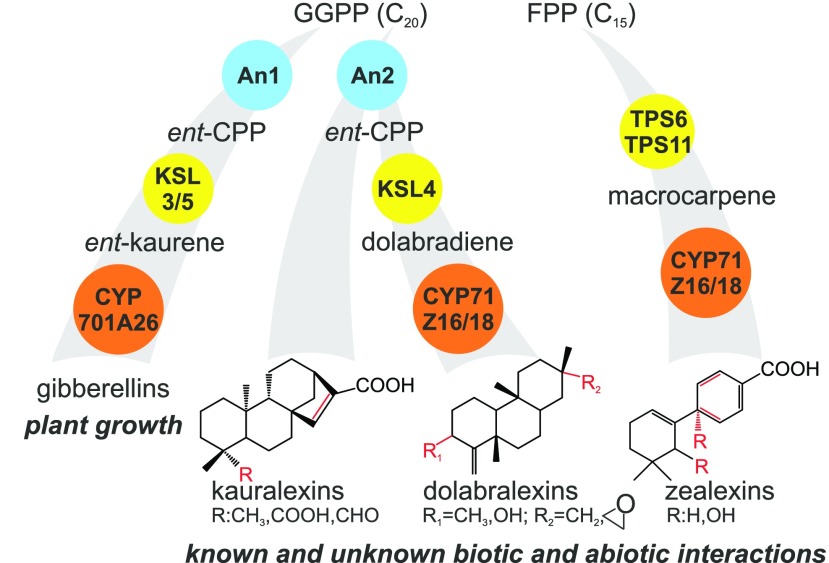
In maize, two groups of acidic terpenoids, the kauralexin class of diterpenoids and the zealexin class of sesquiterpenoids (Fig. 1), are part of a complex repertoire of defense-related specialized metabolites, such as phenylpropanoids, oxylipins, benzoxazinoids, and volatile terpenoids (Schnee et al., 2006; Köllner et al., 2008a; Santiago and Malvar, 2010; Ahmad et al., 2011; Huffaker et al., 2011; Schmelz et al., 2011; Christensen et al., 2015; Richter et al., 2016; Wouters et al., 2016). Inducible zealexin and kauralexin production contributes to defense, both aboveground and belowground, against pathogenic fungi, including Fusarium, Aspergillus, and Colletotrichum spp. (Huffaker et al., 2011; Schmelz et al., 2011; Vaughan et al., 2015). Kauralexins also function as insect feeding deterrents (Schmelz et al., 2011) and accumulate in response to salt and drought stress (Vaughan et al., 2015). These properties are consistent with diverse roles in stress resilience that extend well beyond antimicrobial activity.
Figure 1.
Modular systems of TPS and P450 enzymes in the biosynthesis of maize GAs, kauralexins, dolabralexins, and zealexins. Maize deploys functionally diverse class II diTPSs (blue), class I diTPSs (yellow), and P450 enzymes (orange) to form an array of biologically active terpenoid metabolites that function as GA phytohormones and in the response to both biotic and abiotic stress. The zealexin sesquiterpenoids and kauralexin diterpenoids were identified previously as components of the maize defense system (Huffaker et al., 2011; Schmelz et al., 2011). The stress-inducible dolabralexins represent an additional family of defense-related maize diterpenoids. Structural variations of the individual terpenoid groups are depicted as R. FPP, Farnesyl diphosphate; GGPP, geranylgeranyl diphosphate.
Much of the diterpenoid chemical diversity of plants is produced through the sequential activity of enzymes encoded by two functionally diverse gene families, the diterpene synthases (diTPSs) and the cytochrome P450 monooxygenases (P450s). Plants employ variable combinations of different diTPSs and P450s to transform a few ubiquitous diterpene precursors into thousands of different diterpenoid metabolites, many of which are of limited taxonomic distribution (Peters, 2010; Zerbe and Bohlmann, 2015). Modular enzyme networks largely determine the variations of diterpene profiles in closely and distantly related plant species (Peters, 2010; Hamberger and Bak, 2013; Zerbe and Bohlmann, 2015). Naturally occurring modularity in diterpenoid biosynthesis further enables the engineering of combinatorial diTPS and P450 expression systems in microbial and plant host systems to accelerate the discovery of terpenoid pathways (Brückner and Tissier, 2013; Lange and Ahkami, 2013; Kitaoka et al., 2015; Zerbe and Bohlmann, 2015).
In monocots, diterpenoid pathway networks comprise sets of diTPSs with conserved and species-specific functions in wheat (Triticum aestivum) and rice (Prisic et al., 2004; Xu et al., 2004, 2007a; Morrone et al., 2006; Wu et al., 2012; Zhou et al., 2012; Fu et al., 2016). The ensuing diTPS products often undergo further structural modifications through the activity of P450 enzymes, as exemplified in rice by the P450-catalyzed formation of several bioactive diterpenoid metabolites (Swaminathan et al., 2009; Wang et al., 2011, 2012a, 2012b). Comparatively, less was known about the complexity of maize diterpenoid biosynthesis. Characterized maize diterpenoid enzymes include the ENT-COPALYL DIPHOSPHATE (CPP) SYNTHASE ZmAN1 (Bensen et al., 1995), the ENT-KAURENE SYNTHASES ZmTPS1, ZmKLS3, and ZmKSL5 (Fu et al., 2016), and the ENT-KAURENE OXIDASE CYP701A26 (Mao et al., 2017; Fig. 1). All of these enzymes have either validated or predicted roles in GA metabolism. However, the frequently observed substrate promiscuity of characterized enzymes (Peters, 2010; Morrone et al., 2011; Zerbe and Bohlmann, 2015) also may enable roles in defense-related specialized metabolism. In the absence of mutant analyses, the maize kauralexin and zealexin metabolic pathways have remained largely unproven, with the exception of the ent-CPP synthase ZmAN2, which has been empirically demonstrated to function in kauralexin formation (Harris et al., 2005; Vaughan et al., 2015). The β-macrocarpene synthases ZmTPS6, ZmTPS11, and ZmCYP71Z18 have been assigned to the zealexin pathway based on predictions from in vivo biochemical assays (Köllner et al., 2008b; Mao et al., 2016).
In this study, we functionally characterized ZmAN2, ZmKSL4, and ZmCYP71Z16, which together form a biosynthetic branch of diterpenoids, termed dolabralexins. Patterns of inducible dolabralexin accumulation and gene expression in maize roots exposed to different stresses, combined with the antifungal bioactivity of 3β-hydroxy-15,16-epoxydolabrene (epoxydolabranol) and 3β,15,16-trihydroxydolabrene (trihydroxydolabrene), support diverse roles for dolabralexins in the maize defensive arsenal.
RESULTS
The Maize Diterpene Synthase Family
The maize genome sequence (B73 RefGen_v3) encodes a group of four class II diTPSs (ZmAN1, ZmAN2, ZmCPS3, and ZmCPS4) and six class I diTPSs (ZmTPS1 and ZmKSL1–ZmKSL5). Of the identified class I diTPSs, three (ZmTPS1, ZmKSL3, and ZmKSL5) have been characterized as ent-kaurene synthases (Fu et al., 2016) and three members (ZmKSL1, ZmKLS2, and ZmKSL4) are of unknown function. Phylogenetic analysis placed ZmKSL1, ZmKLS2, and ZmKSL4 adjacent to rice diTPSs of specialized metabolism (Fig. 2). Most members of the maize diTPS gene family are dispersed across the genome (Supplemental Fig. S1), which differs from the clustering of genes of specialized diterpenoid pathways in rice (Schmelz et al., 2014). However, the maize ent-CPP synthases, ZmAN1, which functions in GA biosynthesis (Bensen et al., 1995), and ZmAN2, which has functions in specialized metabolism (Harris et al., 2005), colocalize with ZmKSL4 on chromosome 1, with distances of ∼4 and ∼159 Mb, respectively.
Figure 2.
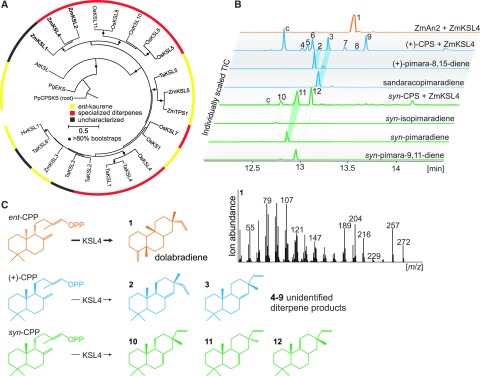
Functional characterization of the maize diterpene synthase ZmKSL4. A, Maximum likelihood phylogenetic tree of select monocot class I diTPSs (Supplemental Table S2). The tree is rooted with the ancestral Physcomitrella patens ent-kaurene synthase. Circles indicate bootstrap support of greater than 80% (500 repetitions), and the functional association with the biosynthesis of ent-kaurene (yellow) or specialized diterpenoids (red) is highlighted. B, Total ion chromatograms (TIC) of reaction products resulting from E. coli coexpression assays of ZmKSL4 with ZmAN2 CPP synthase (Harris et al., 2005), rice CPS4 syn-CPP synthase (Xu et al., 2004), and the grand fir abietadiene synthase variant D621A producing (+)-CPP (Peters and Croteau, 2002) as compared with available authentic standards for product identification. Compound c represents a contamination product from the engineered E. coli system. C, Major products of ZmKSL4 from prenyl diphosphate intermediates of ent-, normal (+)-, and syn-stereochemistry. The mass spectrum of dolabradiene 1 resulting from the coupled activity of ZmAN2 and ZmKSL4 is shown, and the structure of dolabradiene is depicted as verified by NMR analysis.
ZmKSL4 Produces an Unusual Diterpene Scaffold
To test for the activity of ZmKSL4 in specialized diterpenoid metabolism, we utilized the combinatorial functionality of class II and class I diTPSs of different plant species, which can be exploited to probe diTPS functions (Brückner and Tissier 2013; Zerbe et al., 2013; Kitaoka et al., 2015). An established Escherichia coli coexpression system (Morrone et al., 2010) was used to analyze the coupled activities of ZmKSL4 with (1) the ent-CPP synthase ZmAN2, (2) the rice syn-CPP synthase (OsCPS4; Xu et al., 2004), and (3) the grand fir (Abies grandis) abietadiene synthase variant D621A that produces CPP of normal [i.e. (+)] stereochemistry (Morrone et al., 2010). These three different combinations were selected, since ent-, syn-, and (+)-CPP represent the known stereochemical variations of class II diTPS products occurring naturally in many monocots (Peters, 2010; Schmelz et al., 2014). When coexpressed with ZmAN2, ZmKSL4 converted ent-CPP into a single product with an unusual fragmentation pattern showing major mass ions of mass-to-charge ratio (m/z) 204, 189, and 216, in addition to the mass ions m/z 257 and 272 that are characteristic of labdane diterpenoid structures (Fig. 2, compound 1). Preparative enzymatic synthesis and purification of this product by silica chromatography and semipreparative HPLC enabled 1D and 2D NMR analyses and identified the ZmAN2/ZmKSL4 product as dolabradiene (Fig. 2; Supplemental Fig. S2). In addition, ZmKSL4 was active in combination with (+)-CPP to yield low amounts of several other diterpenoids (compounds 2–9), of which (+)-pimara-8,15-diene 2 and (+)-sandaracopimaradiene 3 could be identified by comparison with authentic standards (Fig. 2; Supplemental Fig. S3). Similarly, ZmKLS4 showed activity with syn-CPP, resulting in the formation of syn-isopimara-7,15-diene 10, syn-pimara-7,15-diene 11, and syn-pimara-9(11),15-diene 12.
ZmAN2, ZmKSL4, and ZmCYP71Z16 Function Together to Form 15,16-Epoxydolabrene
To gain insight into the structural elaboration of the predominant product of ZmAN2/ZmKSL4, namely dolabradiene, we tested its further modification by maize P450s. To this end, we performed a BLAST search of the B73 RefGen_v3 genome against monocot P450s of the CYP71, CYP76, and CYP99 families known to function in terpenoid metabolism. This search identified a member of the CYP71 family, ZmCYP71Z16, which showed high protein sequence similarity (89%) to the recently reported maize ZmCYP71Z18 involved in zealexin biosynthesis (Mao et al., 2016). Phylogenetic analysis placed ZmCYP71Z16 adjacent to ZmCYP71Z18 within a clade that also contained the rice P450s CYP71Z1, CYP71Z6, and CYP71Z7, of which CYP71Z6 and CYP71Z7 catalyze reactions in oryzalide and phytocassane biosynthesis (Wu et al., 2011; Fig. 3).
Figure 3.
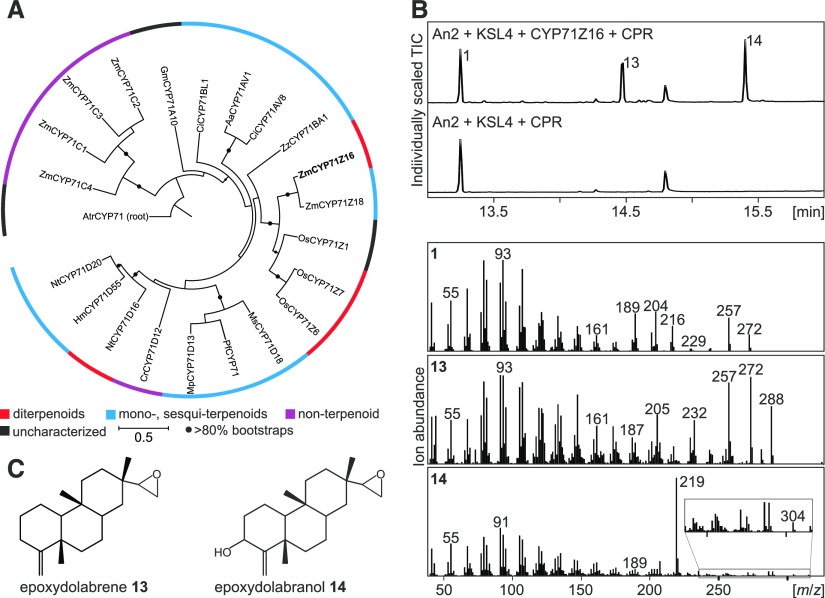
Functional characterization of ZmCYP71Z16. A, Maximum likelihood phylogenetic tree of select P450 proteins of the CYP71 family (Supplemental Table S2). The tree is rooted with an uncharacterized CYP71 of Amborella trichopoda. Circles indicate bootstrap support of greater than 80% (500 repetitions). Demonstrated activities in diterpenoid (red), monoterpenoid/sesquiterpenoid (blue), and other specialized metabolic pathways (purple) are highlighted. B, Total ion chromatograms (TIC) and mass spectra of products 1, 13, and 14 derived from microbial coexpression of ZmAN2, ZmKSL4, ZmCYP71Z16, and ZmCPR2. C, Structures of epoxydolabrene 13 and epoxydolabranol 14 as verified by NMR analysis.
To biochemically characterize ZmCYP71Z16, a codon-optimized and N-terminally modified coding sequence was synthesized and coexpressed in E. coli with ZmAN2 and ZmKSL4 as well as the maize cytochrome P450 reductase ZmCPR2. Coexpression of the two diTPSs and ZmCPR2 without ZmCYP71Z16 served as a negative control. When all four enzymes were coexpressed, the presence of ZmCYP71Z16 resulted in the conversion of dolabradiene into two new products with fragmentation patterns featuring m/z 288 (compound 13) and m/z 304 (compound 14) mass ions that are characteristic of diterpenoid scaffolds containing one and two oxygen atoms, respectively (Fig. 3). High-resolution liquid chromatography-mass spectrometry (LC-MS) analysis identified the exact mass of compound 14 as m/z 304.2478, consistent with the molecular formula C20H32O2. Subsequent HPLC purification of compounds 13 and 14 followed by NMR analysis verified the structures as 15,16-epoxydolabrene (epoxydolabrene) and epoxydolabranol, respectively (Fig. 3; Supplemental Fig. S4). The biochemical characterization of ZmCYP71Z16 adds a previously unknown function to the diverse activities of members of the plant CYP71 family, facilitating the regiospecific oxygenation of dolabradiene at C-3 and C-16, presumably in a sequential reaction process. The formation of the monoepoxide 13, in the absence of detectable formation of a monohydroxyl intermediate, is consistent with a P450 reaction sequence of epoxidation at C-16 prior to hydroxylation at C-3. To further investigate the catalytic activity of ZmCYP71Z16, we performed E. coli coexpression assays with ZmCPR2 and fed 10 µm purified epoxydolabrene and epoxydolabranol to the culture 5 h post induction. Expectedly, no new products were identified when feeding epoxydolabranol to the culture. In contrast, epoxydolabrene was converted to the hydroxylated derivative (Supplemental Fig. S5), supporting our proposed reaction sequence from epoxydolabrene to epoxydolabranol.
The phylogenetic relatedness of ZmCYP71Z16 and the previously reported zealexin-forming ZmCYP71Z18 (Mao et al., 2016) indicated that ZmCYP71Z16 may be active on other substrates beyond a role in epoxydolabranol biosynthesis. To test this hypothesis, we conducted E. coli coexpression assays of ZmCYP71Z16 with ZmCPR2 and ZmTPS11, which form the zealexin precursor β-macrocarpene (Köllner et al., 2008b; Huffaker et al., 2011). This enzyme combination resulted in the formation of zealexin A1 (Supplemental Fig. S6). Reciprocal E. coli coexpression assays of ZmCYP71Z18 with ZmAN2 and ZmKSL4 did not result in the detectable formation of epoxydolabrene, epoxydolabranol, or other diterpenoid products. Therefore, we tested the activity of ZmCYP71Z18 using yeast in vivo feeding assays in the strain BY4741 expressing ZmCPR2 and ZmCYP71Z18. After feeding 25 µm purified dolabradiene to the culture, partial conversion into epoxydolabranol was observed (Supplemental Fig. S6).
ZmAN2 Is Required for Dolabralexin Biosynthesis in Vivo
The above in vitro coexpression assays demonstrated that ZmAN2 and ZmKSL4 can function together as a duo of class II and class I diTPSs to afford the dolabradiene scaffold and its downstream derivatives (Fig. 2). To validate if ZmAN2 provides the ent-CPP intermediate converted by ZmKSL4 into dolabradiene in vivo, we measured the abundance of dolabradiene and epoxydolabranol in the Zman2 mutant, which is deficient in kauralexins (Vaughan et al., 2015), and compared the metabolite levels with those in wild-type plants of the near-isogenic W22 inbred line. Consistent with a function of ZmAN2 in the formation of dolabralexins, no epoxydolabranol or dolabradiene was detectable in the Zman2 mutant line, while dolabralexins were abundant in root tissue of wild-type plants (Supplemental Fig. S7). Thus, both kauralexins and dolabralexins require ZmAN2 activity in vivo.
Dolabralexins Are Abundant in Maize Root Tissue
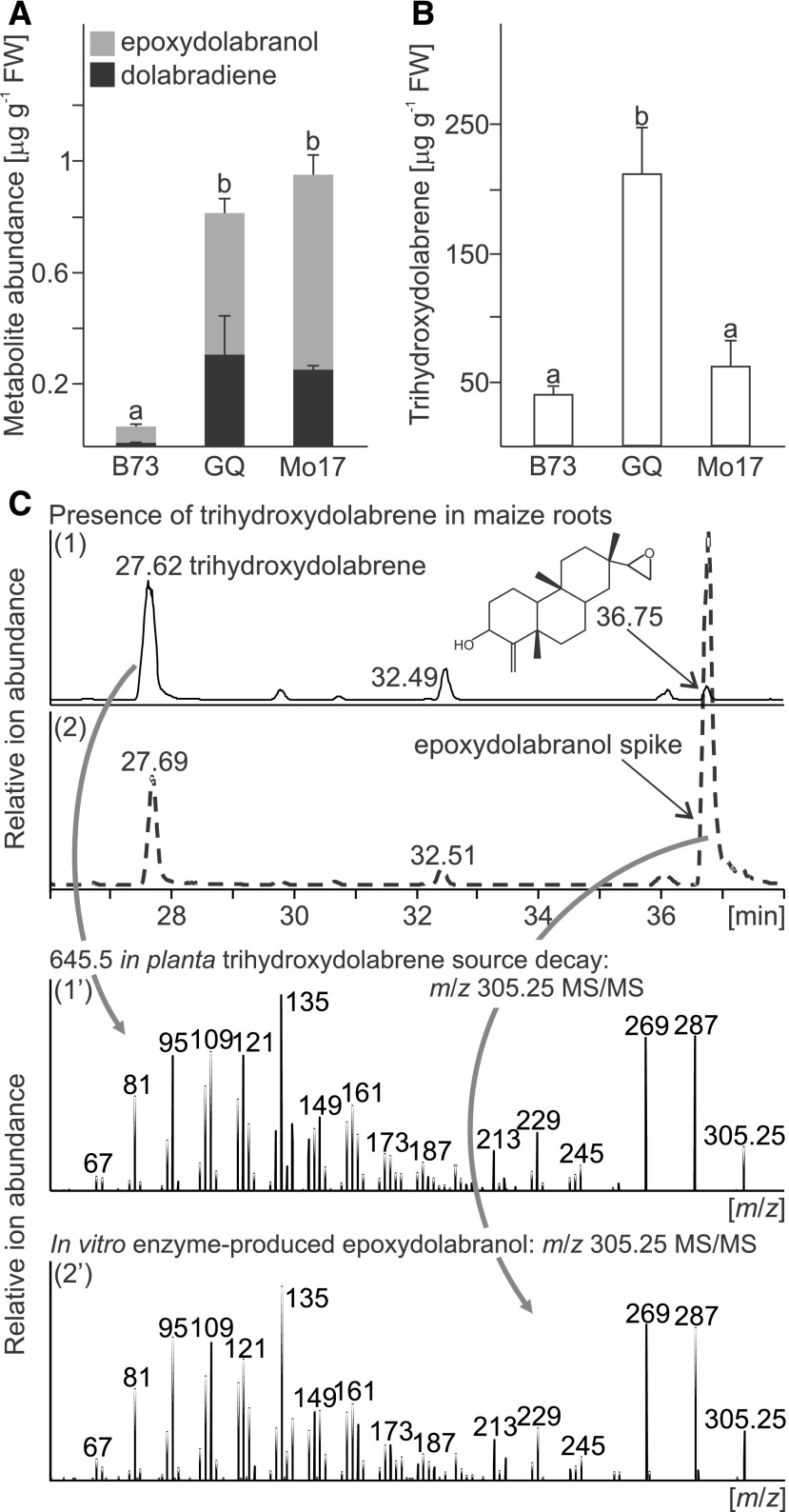
To investigate if dolabralexins, derived from the coupled activity of ZmAN2, ZmKSL4, and ZmCYP71Z16, occur as abundant metabolites in planta, terpenoid metabolite profiling was performed on different maize varieties, including the inbred lines B73 and Mo17 and a commercial hybrid sweet corn (Golden Queen). LC-MS analysis of field-grown 70-d-old plants showed that both dolabradiene and epoxydolabranol were abundant in the roots of all tested genotypes (Fig. 4). Concentrations of both compounds differed between genotypes; however, epoxydolabranol was consistently more abundant than dolabradiene and displayed greater abundance in Mo17 (700 ± 68 ng g−1 fresh weight) and Golden Queen (507 ± 52 ng g−1 fresh weight) compared with B73 (62 ± 7 ng g−1 fresh weight).
Figure 4.
Occurrence of dolabralexins in planta. A and B, Average quantities of dolabradiene and epoxydolabranol (A) and trihydroxydolabrene (B) in field-grown roots of B73, Mo17, and hybrid sweet corn (Golden Queen [GQ]). Error bars represent propagated se values across four biological replicates. Letters represent significant differences at P < 0.05 as measured using ANOVA and Tukey’s tests to correct for multiple comparisons between control and treatments. FW, Fresh weight. C, Analysis of trihydroxydolabrene extracted from roots using LC-MS/MS analysis, with traces representing combined extracted ion chromatograms (m/z 305–305.5 + 645.25–645.75). In-source decay of the [2M+H]+ m/z 645.5 trihydroxydolabrene parent ion yields [M-H2O+H]+ m/z 305.25, occurring at a retention time of 27.6 min. MS/MS analyses of the trihydroxydolabrene-derived m/z 305.25 results in highly similar fragmentation patterns as compared with the enzyme-produced epoxydolabranol analytical standard (retention time = 36.7 min).
Alongside epoxydolabranol, we observed an additional earlier eluting and predictably more polar metabolite at high abundance in maize roots, with concentrations of 60 ± 18.5 μg g−1 fresh weight in Mo17, 212 ± 34 μg g−1 fresh weight in Golden Queen, and 41 ± 4 μg g−1 fresh weight in B73 (Fig. 4; Supplemental Figs. S8 and S9). High-resolution LC-tandem mass spectrometry (MS/MS) analysis of this metabolite defined an accurate parent mass [2M+H]+ of m/z 645.5. In addition, LC-MS and LC-MS/MS analyses revealed two notable features. First, the presence of substantial in-source decay led to a charged molecule of [M-H2O+H]+ m/z 305.25 and [M-2H2O+H]+ m/z 287.09 with an MS/MS fragmentation pattern similar to epoxydolabranol (Fig. 4). Second, MS/MS of the [2M+H]+ m/z 645.5 parent ion yielded closely related spectra m/z 305.25 and 287.09, as seen for epoxydolabranol (Supplemental Fig. S8). Under markedly different LC-MS conditions, an alternative predominant candidate parent ion [M+Na+H]+ m/z 345 was obtained (Supplemental Fig. S9). To elucidate the precise identity of the dolabradiene-related metabolite, a large-scale methanol (MeOH) extraction of mature field-grown maize roots was subjected sequentially to C18 flash chromatography and semipreparative HPLC with LC-MS-based monitoring of factions using the [M+Na+H]+ m/z 345 candidate parent ion (Supplemental Fig. S9). A purified 300-µg sample dissolved in deuterated dimethyl sulfoxide (d6-DMSO) was used for 1H-NMR, and carbon chemical shift assignments were based on heteronuclear single quantum correlation (HSQC) and heteronuclear multiple bond correlation (HMBC) analyses (Supplemental Fig. S10). Collectively, all data are consistent with trihydroxydolabrene (C20H34O3; exact mass, m/z 322.25) representing a hydrolysis of the epoxy group of epoxydolabranol, which accumulates as the dominant dolabralexin pathway metabolite. Predictably, trihydroxydolabrene was present at only trace levels in Zman2 mutant plants as compared with control plants (Supplemental Fig. S7).
Dolabralexins Are Induced in Biotic and Abiotic Stress Responses
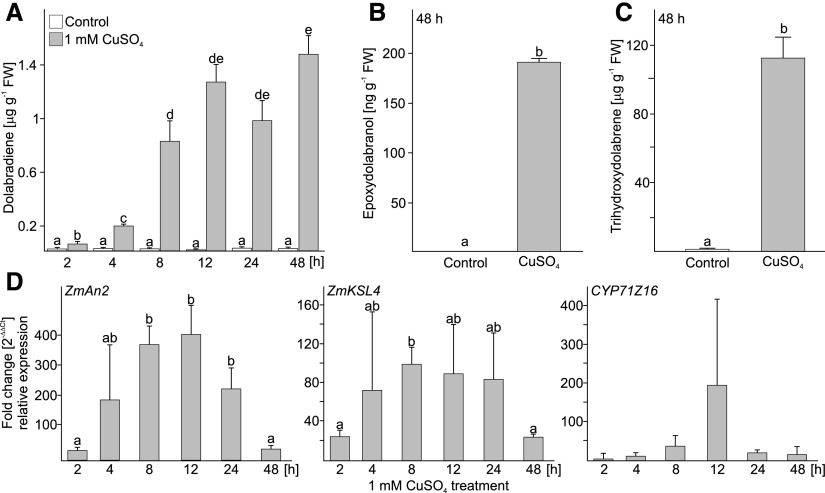
Following the detection of dolabradiene, epoxydolabranol, and trihydroxydolabrene in planta, we quantified the accumulation of all three compounds in roots of 16-d-old Golden Queen seedlings under abiotic stress conditions. Root treatments with 1 mm CuSO4 were used as a proxy for oxidative stress compared with controls treated with water only. The accumulation of dolabradiene and epoxydolabranol was detected between 4 and 48 h post CuSO4 treatment and increased to a concentration of ∼160 ng g−1 fresh weight epoxydolabranol at 48 h (Fig. 5). Notably, trihydroxydolabrene was detected as the dominant metabolite in CuSO4-challenged roots, reaching concentrations greater than 100 µg g−1 fresh weight. Consistent with inducible dolabralexin accumulation, gene expression analysis via quantitative real-time PCR of ZmAN2, ZmKSL4, and ZmCYP71Z16 revealed increased transcript levels of all three genes from 2 to 12 h, with ZmAN2 showing the most significant up-regulation of ∼400-fold (Fig. 5).
Figure 5.
Accumulation of dolabralexins in response to abiotic stress. A to C, Average quantities of dolabradiene (A), epoxydolabranol (B), and trihydroxydolabrene (C) in maize (Golden Queen) roots treated with either water (Control) or 1 mm CuSO4 over a period of 48 h. FW, Fresh weight. D, Fold change (2−ΔΔCt) of the transcript abundance of ZmAN2, ZmKSL4, and ZmCYP71Z16 in the same root tissues used for diterpenoid analyses. Gene expression was measured by quantitative real-time PCR and normalized to the internal reference EF1-α. Primer efficiency was verified by dissociation curves and sequence verification of representative products. Letters (a–e) represent significant differences at P < 0.05 as measured using ANOVA and Tukey’s tests to correct for multiple comparisons between control and treatments. Error bars represent propagated se values (n = 4).
We next investigated the accumulation of dolabralexins in the response of maize to elicitation with Fusarium verticillioides and Fusarium graminearum, which are causal agents of seedling blights, stalk rots, ear rots, and mycotoxin contamination (Munkvold, 1997; Goswami and Kistler, 2004; Baldwin et al., 2014). For this purpose, roots of 53-d-old Mo17 plants were inoculated with live F. verticillioides or F. graminearum spores and harvested for the analysis of both metabolites and transcripts 7 d later. Controls were performed with roots wounded and treated with water alone. Upon inoculation with F. verticillioides or F. graminearum spores, dolabradiene accumulation was induced 6- or 45-fold, respectively, compared with wounded plants (Fig. 6). Similarly, epoxydolabranol accumulation was increased 55- and 79-fold, respectively, in response to the two Fusarium species. Moreover, in response to F. verticillioides and F. graminearum, trihydroxydolabrene accumulated 14- and 32-fold greater compared with control plants treated by wounding only. Trihydroxydolabrene reached concentrations of up to 225 µg g−1 fresh weight in elicited root tissue. By comparison, kauralexin A- and B-series metabolites accumulated to approximately 9 µg g−1 fresh weight in response to pathogen infection in the same mature root tissues. Consistent with the pathogen-inducible accumulation of dolabralexins, the transcript abundance of ZmAN2 and ZmKSL4 increased upon fungal elicitation while ZmCYP71Z16 showed no up-regulated gene expression at this 7-d time point (Fig. 6).
Figure 6.
Accumulation of dolabralexins in response to biotic stress. A, Average (n = 4) quantities of dolabradiene, epoxydolabranol, and trihydroxydolabrene in Mo17 roots damaged (Dam) and treated with water or inoculated separately with F. verticillioides (F.v.) or F. graminearum (F.g.). Treatments occurred in 53-d-old plants with tissue harvests 7 d later. B, Quantification of A-series (KA) and B-series (KB) kauralexin metabolites in the same tissue samples. Error bars represent propagated se values. Letters represent significant differences at P < 0.05 as measured using ANOVA and Tukey’s tests to correct for multiple comparisons between control and treatments. FW, Fresh weight. C, Fold change (2−ΔΔCt) of the transcript abundance of ZmAN2, ZmKSL4, and ZmCYP71Z16 in the corresponding Mo17 roots normalized to the internal reference gene EF1-α. Error bars represent propagated se values of the biological replicates. Asterisks indicate significant changes compared with wounded tissue at P < 0.0005 (***), P < 0.01 (**), and P < 0.1(*).
Dolabralexins Are Active against Fungal Pathogens
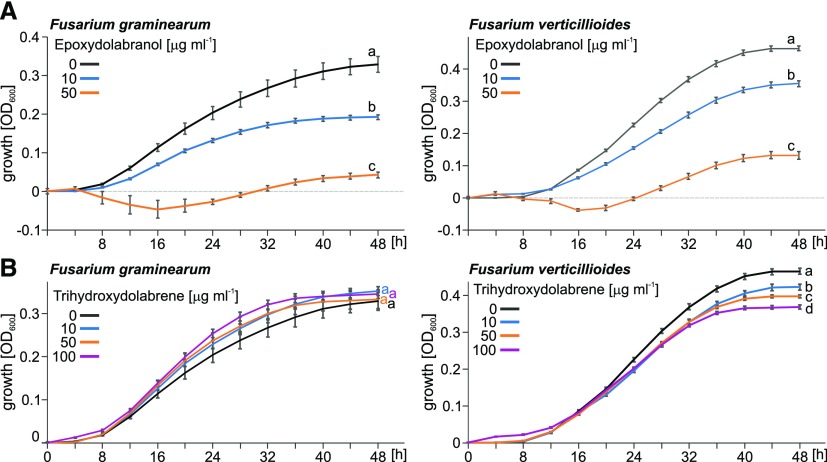
To further assess the predicted defense-related roles for dolabralexins, in vitro antimicrobial assays were performed with F. verticillioides and F. graminearum. The growth of fungal hyphae in the presence and absence of purified epoxydolabranol and trihydroxydolabrene was measured in defined liquid medium using an established 96-well microtiter assay (Schmelz et al., 2011). Based on previous maize diterpenoid bioassays and the quantified in vivo abundance of epoxydolabranol and trihydroxydolabrene (Figs. 4–6), we utilized 10 and 50 μg mL−1 as relevant local tissue concentrations to test for antimicrobial activity. After 48 h, epoxydolabranol at 10 and 50 μg mL−1 substantially reduced the growth of F. graminearum by 41% and 87%, respectively, and likewise inhibited the growth of F. verticillioides by 24% and 71%, respectively (Fig. 7). We further assessed the antimicrobial efficacy of trihydroxydolabrene at concentrations of 10 to 100 μg mL−1, well below those detectable in planta. In contrast to epoxydolabranol, trihydroxydolabrene had no inhibitory activity on the growth of F. graminearum but resulted in a dose-dependent growth reduction of F. verticillioides at concentrations of 10 μg mL−1 (9%), 50 μg mL−1 (15%), and 100 μg mL−1 (21%).
Figure 7.
Antifungal activity of dolabralexins. Average growth (OD600) of F. graminearum and F. verticillioides is shown in the absence and presence of purified epoxydolabranol (A) and trihydroxydolabrene (B) measured over a 48-h time course in defined minimal broth medium using a microtiter plate assay. Error bars represent propagated se values (n = 6), and letters (a–d) represent significant differences at P < 0.05 as measured using ANOVA and Tukey’s tests to correct for multiple comparisons between control and treatments.
DISCUSSION
The combination of extreme and shifting biotic and abiotic stresses can overwhelm the natural defense systems of plants and result in substantial yield losses in staple crops (Chakraborty and Newton, 2011; de Sassi and Tylianakis, 2012). An improved understanding of the inherent strengths and weaknesses that underlie mechanisms of crop resilience can mitigate yield loss. For example, detailed knowledge of terpenoid metabolism, the underlying genes, and their contribution to maize stress resilience could provide useful resources for improving crop traits (Degenhardt et al., 2009). This study highlights the utility of integrating genomics and metabolomics with both in vivo and in vitro biochemical approaches to elucidate defense pathways in well-studied crop plants. We identified functions for the combined pathway of the maize diTPSs ZmAN2 and ZmKSL4 and the cytochrome P450 ZmCYP71Z16 that fuel diterpenoid metabolism and the production of dolabralexins. Dolabralexins and corresponding pathway genes are strongly stress inducible, consistent with roles in protecting and buffering against the negative impacts of biotic and abiotic stresses.
The dolabralexin precursor, dolabradiene, was reported previously in only a few coniferous trees of the Araucariaceae and Cupressaceae families (Brophy et al., 2000; Takahashi et al., 2001), and its biological role in these species is unknown. Thus, based on current knowledge and the absence of dolabralexin in other major Poaceae crops, such as rice and wheat, the inferred emergence of the dolabralexin biosynthetic pathway occurred after the evolutionary separation of maize from wheat and rice approximately 50 million years ago (Wolfe et al., 1989; Grass Phylogeny Working Group II, 2012).
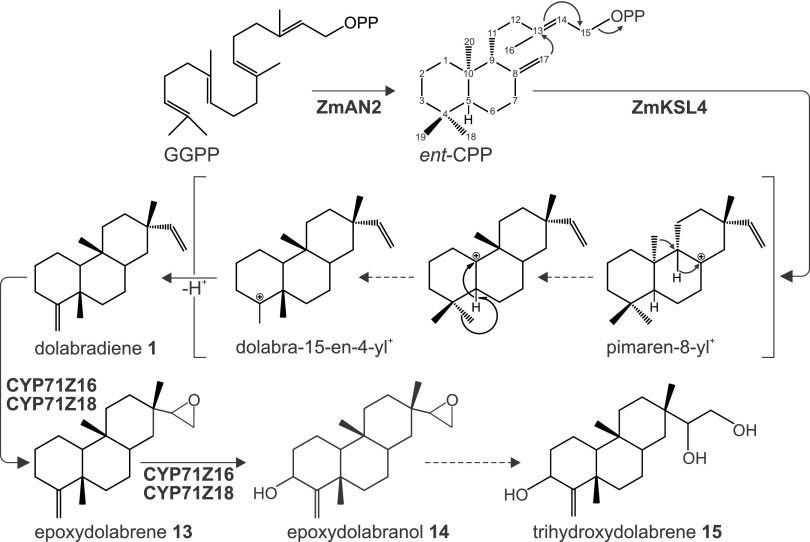
Mechanistically, we propose a pathway for the biosynthesis of epoxydolabranol that proceeds through ZmKSL4-catalyzed ionization-dependent cyclization of ent-CPP via an ent-dolabra-15-en-4-yl+ carbocation to afford dolabradiene and subsequent P450-catalyzed functional modification of the latter (Fig. 8). Dolabradiene production via microbial coexpression of ZmAN2 and ZmKSL4 combined with the absence of dolabralexins in the established Zman2 mutant substantiates our hypothesis that ZmAN2 provides the ent-CPP substrate for dolabradiene formation. This also is consistent with previous work demonstrating a role of ZmAN2 in specialized metabolism (Harris et al., 2005). From ent-CPP, the reaction will inevitably proceed via the common pimaren-8-yl+ carbocation, as shown for other labdane-related diterpene scaffolds (Peters, 2010). Dolabradiene formation then can be achieved through sequential C-9,8-hydride shift, methyl migration from C-10 to C-9, C-5,10-hydride shift, and another methyl transfer from C-18 to C-5 to yield the ent-dolabra-15-en-4-yl+ intermediate (Peters, 2010), deprotonation of which gives rise to dolabradiene (Fig. 8). By contrast, the formation of ent-kaurene and ent-iso-kaurene, as precursors of GAs and kauralexins, respectively, would proceed through secondary cyclization of the pimaren-8-yl+ carbocation followed by ring rearrangement to afford the tetracyclic kauranyl scaffold (Xu et al., 2007b). Subsequent to ent-dolabra-15-en-4-yl+ formation, ZmCYP71Z16-enabled sequential epoxidation at C-16 and hydroxylation at C-3 yields epoxydolabranol. The detection of epoxydolabrene and epoxydolabranol, but not a 3β-hydroxydolabrene intermediate, combined with the ZmCYP71Z16-catalyzed conversion of epoxydolabrene to epoxydolabranol in in vitro feeding assays, supports a reaction sequence where the epoxide group is formed prior to hydroxylation. Epoxydolabranol is further converted into trihydroxydolabrene via either enzymatic or spontaneous hydrolysis. The presence of trihydroxydolabrene at high concentrations suggests that the characterized products of ZmAN2/ZmKSL4/ZmCYP71Z16 do not represent dominant pathway end products but, instead, may serve as intermediates to trihydroxydolabrene and possibly more complex and increasingly polar maize diterpenoids.
Figure 8.
Biosynthesis of maize dolabralexins. Shown is a proposed biosynthetic pathway of dolabralexins through the sequential activity of ZmAN2, ZmKSL4, and ZmCYP71Z16.
Previous studies demonstrated that Zman2 lacks pathogen-elicited diterpenoids of the kauralexin class (Vaughan et al., 2015). Our results demonstrate that this mutant also is deficient in dolabralexin biosynthesis (Supplemental Fig. S7) and suggest a biosynthetic interconnectivity and potential biosynthetic competition between the kauralexin and dolabralexin pathways. ZmAN2 was first described in a differential display analysis of highly elicited transcripts following F. graminearum challenge of maize silks (Harris et al., 2005). Following the first description of ZmAN2, pathogen-elicited transcripts have been observed and reported in a large number of maize studies with diverse microbes (Huffaker et al., 2011; Schmelz et al., 2011, 2014; van der Linde et al., 2011; Vaughan et al., 2014; Christie et al., 2017). The nearly ubiquitous presence of ZmAN2 as a pathogen defense transcript marker is consistent with the essential biosynthetic role in the modular formation of multiple specialized diterpenoid metabolites, including not only A- and B-series kauralexins (Schmelz et al., 2011) but also dolabralexins. Notably, ZmAN2 does not cluster with any class I diTPSs in the maize genome, illustrating a different genomic organization of diterpenoid metabolism in maize as compared with rice and tomato (Solanum lycopersicum), where several specialized diterpenoid pathways form functional biosynthetic clusters (Matsuba et al., 2013; Nützmann et al., 2016). However, the presence of the uncharacterized class II diTPSs ZmCPS3 and ZmCPS4 suggests that specialized diterpenoid metabolism in maize likely follows the common modular blueprint of combining different diTPS and P450 enzymes, as shown for rice, wheat, and various other species across the plant kingdom (Xu et al., 2007a; Zhou et al., 2012; Hall et al., 2013; Cui et al., 2015; Zerbe and Bohlmann, 2015). This hypothesis is substantiated by (1) the in vitro substrate promiscuity of ZmKSL4 with CPP of ent-, syn-, and (+)-stereochemistry and (2) the capacity of ZmCYP71Z16 and ZmCYP71Z18 (previously reported to function in zealexin biosynthesis; Mao et al., 2016) to convert both sesquiterpenoid and diterpenoid intermediates (Supplemental Fig. S6). Consistent with their largely redundant substrate promiscuity, the ZmCYP71Z16 and ZmCYP71Z18 genes are located directly adjacent to each other on chromosome 5, suggesting that these P450s emerged from a more recent gene duplication event observed frequently in specialized diterpenoid metabolism (Zi et al., 2014) and may have overlapping or complementary functions in vivo.
The occurrence of dolabralexins in field-grown maize roots and their accumulation in response to abiotic and biotic stresses encourage a more extensive exploration of biological roles in planta. In the context of biotic stress, epoxydolabranol (albeit at moderately higher concentrations than observed in planta) inhibits the growth of two major maize Fusarium spp. pathogens with a comparable potency, as demonstrated earlier for the kauralexins (Schmelz et al., 2011). These findings are supported by a recent study demonstrating increased disease susceptibility to F. verticillioides in Zman2 mutants. However, due to the critical role of ZmAN2 in kauralexin and dolabralexin biosynthesis, this result does not address the specific roles of kauralexin and dolabralexin pathway branches (Christensen et al., 2018). The apparent yet lower antimicrobial potency of trihydroxydolabrene at concentrations below those observed in elicited plant tissues, and the specificity of trihydroxydolabrene activity to F. verticillioides, highlight the importance of the C-15,16 epoxy group for bioactivity. These results suggest distinct protective functions and biological roles for individual dolabralexins. The inducible accumulation of dolabralexins in response to oxidative and pathogen stress is consistent with dual functionality in abiotic and biotic stress responses, similar to the kauralexin and zealexin pathways (Huffaker et al., 2011; Schmelz et al., 2011; Vaughan et al., 2015). Recent findings enabled by mutant analyses highlight an important yet thus far rarely proven role of diterpenoids in protecting roots against environmental perturbations. For example, Zman2 mutant plants were more susceptible to drought stress than the corresponding wild-type plants (Vaughan et al., 2015). While this impact was attributed to kauralexin deficiency, the discovery of the dolabralexins now forces the consideration of multiple ZmAN2-dependent pathway branches to understand responses to drought and belowground stresses. This hypothesis is supported further by the quantification of predominant dolabralexins accumulating in challenged roots. For example, in pathogen-challenged roots, trihydroxydolabrene is present at 20-fold greater levels than established kauralexins. In contrast, the induced accumulation of kauralexins was shown previously in stems and scutella tissues to exceed 100 µg g−1 fresh weight as compared with the 10 µg g−1 fresh weight currently detected in mature roots (Schmelz et al., 2011). Notably, ZmAN2 is located proximal to a quantitative trait locus mapped to bin 1.03 that is associated with root growth (Rahman et al., 2011), and ZmKSL4 colocates in bin 1.08 with quantitative trait loci associated with both drought tolerance (Tuberosa et al., 2002) and abscisic acid biosynthesis, which can further mediate drought-induced phytoalexin biosynthesis in maize roots (Vaughan et al., 2015). Our findings here contribute to a growing body of knowledge demonstrating roles for root diterpenoids, including momilactone phytoalexins in rice (Toyomasu et al., 2008), the antiherbivory activity of rhizathalene in Arabidopsis (Arabidopsis thaliana; Vaughan et al., 2013), and drought tolerance mediated by isorosmanol in rosemary (Rosmarinus officinalis; Munné-Bosch and Alegre, 2000).
The existence of an abundant novel maize root defense, as well as significant and distinct antifungal activities for epoxydolabranol and trihydroxydolabrene in vitro, merit a closer examination of the precise ecological functions of dolabralexins. Such studies will ultimately require multiple genetic mutants of ZmKSL4 and ZmCYP71Z16/18 in future work. While historically recalcitrant to discovery, the complex modular networks of diterpenoid defense pathways in maize, rice, and possibly other cereal crops are import to understanding ecological interactions and the genetic basis of crop stress resilience (Schmelz et al., 2014).
MATERIALS AND METHODS
Plant and Fungal Materials
Seeds of hybrid maize (Zea mays variety Golden Queen; Southern States Cooperative), landrace inbreds (B73, Mo17, and W22; National Genetic Resources Program, Germplasm Resources Information Network), and a Ds insertion mutant, Zman2 (Vaughan et al., 2015), were germinated in MetroMix 200 (Sun Gro Horticulture) supplemented with 14-14-14 Osmocote (Scotts Miracle-Gro) and grown as described previously (Schmelz et al., 2009). Field-challenged roots from B73, Mo17, Ky21, and hybrid sweet corn grown at the Biology Field Station at the University of California, San Diego, during the summer of 2016 were recovered 75 d after planting, washed, frozen in liquid N2, ground to a fine powder, and ultimately used for metabolite analysis. Fungal stock cultures of Fusarium verticillioides (Northern Regional Research Laboratory; NRRL stock no. 7415) and Fusarium graminearum (NRRL stock no. 31084) were grown on V8 agar for 2 to 3 weeks before the quantification and use of spores (Huffaker et al., 2011).
Isolation and Cloning of cDNAs
Total RNA was extracted from ground tissue of young maize (B73) seedlings as described previously (Chourey et al., 2010). For cloning of ZmKSL4, ZmCYP71Z18, and ZmTPS11, 5 μg of total RNA was reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences) followed by PCR amplification of the target genes with gene-specific oligonucleotides (Supplemental Table S1). The amplified full-length genes of ZmTPS11 and ZmCYP71Z18 and an N-terminally truncated form of ZmKSL4 (ZmKSL4Δ106, lacking the predicted plastidial transit peptide) were ligated with the zero Blunt II-TOPO vector (Invitrogen) and transformed into TOP 10 chemically competent cells for plasmid isolation and sequence verification. ZmKSL4 was subcloned further into the expression vectors pET28b(+) and pCOLA-DUET (Novagen/EMD) for coexpression in Escherichia coli. ZmTPS11 was inserted into the vector pCOLA-DUET for expression in E. coli. ZmCYP71Z18 was subcloned into pET28b(+) and pCOLA-DUET, as well as pESC-Leu-2D (Stratagene), for expression in E. coli and Saccharomyces cerevisiae, respectively. In addition, codon-optimized genes for the full-length sequences of maize FARNESYL DIPHOSPHATE SYNTHASE3 (ZmFPPS3) and ZmCPR2, as well as a modified version of ZmCYP71Z16 (32 N-terminal residues replaced with the leader sequence MAKKTSSKGK), were obtained from the Department of Energy Joint Genome Institute through Community Science Program grant WIP2568 (Supplemental Table S1). ZmCYP71Z16 and ZmCPR2 were inserted into the multiple cloning sites of the pET-DUET1 vector (Novagen/EMD) to generate the plasmid pET-DUET1:ZmCPR2/ZmCYP71Z16 for expression in E. coli. ZmFPPS3 was subcloned into the vector pCOLA-DUET together with ZmTPS11 for expression in E. coli.
Quantitative Real-Time PCR
Gene expression analyses of ZmAN2, ZmKSL4, and ZmCYP71Z16 were performed using the same CuSO4-treated or Fusarium spp.-inoculated roots as described for terpenoid profiling. Total RNA was isolated as described previously (Kolosova et al., 2004). First-strand cDNA was synthesized using the SuperScript III First-Strand Synthesis Kit (Invitrogen) and oligo(dT) primers according to the manufacturer’s instructions. Transcript levels were quantified with a Bio-Rad C1000 Touch Thermo Cycler interfaced with a CFX96 Real-Time System and iTaq Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s protocols with 5 ng μL−1 cDNA and 300 nm oligonucleotides. Mean cycle threshold values of two technical and three biological replicates were normalized using elongation factor EF1-α as a reference gene. Fold change values were calculated using the 2−ΔΔCt method. Gene-specific oligonucleotides are listed in Supplemental Table S1.
Combinatorial Expression in E. coli
The functional coexpression of enzymes was carried out using a previously described E. coli system engineered for enhanced diterpenoid production (Morrone et al., 2010; Kitaoka et al., 2015). For biochemical characterization of ZmKSL4, the N-terminally truncated gene in the expression vector pET28b(+) or pCOLA-DUET was cotransformed in E. coli BL21DE3-C41 cells (Lucigen) with a plasmid carrying a geranylgeranyl diphosphate synthase and constructs of class II diTPSs with different products: ZmAN2 forming ent-CPP (pGGeC), rice (Oryza sativa) CPS4 producing syn-CPP (pGGsC), or a variant of grand fir (Abies grandis) abietadiene synthase forming (+)-CPP (pGGnC; Morrone et al., 2010). For additional coexpression of ZmCYP71Z16, the codon-optimized construct pET-DUET1:ZmCPR2/ZmCYP71Z16 was coexpressed with pGGeC and pET28b:ZmKSL4Δ106. For analysis of zealexin formation, the construct pET-DUET1:ZmCPR2/ZmCYP71Z16 was coexpressed with ZmFPPS3 and the β-macrocarpene synthase TPS6 (Köllner et al., 2008b; Richter et al., 2015) as well as the enhancer plasmid pIRS (Morrone et al., 2010). All coexpression assays were performed in E. coli BL21DE3-C41 and grown in 45 mL of Terrific Broth medium to an OD600 of ∼0.6 at 37°C. Cultures were cooled to 16°C before induction with 1 mm isopropyl-thiogalactoside and incubation for 72 h with supplements of 1 mm MgCl2 and 25 mm sodium pyruvate. For P450 coexpression, cultures were supplemented further with 4 mg L−1 riboflavin and 75 mg L−1 δ-aminolevulinic acid. Enzyme products were extracted with 50 mL of 100% hexane or 1:3 ethyl acetate:hexane, concentrated under an N2 stream, and resuspended in 1 mL of hexane for gas chromatography (GC)-MS analysis.
Yeast Whole-Cell Activity Assays
To analyze the catalytic specificity of ZmCYP71Z18 (Mao et al., 2016), the full-length construct was coexpressed with ZmCPR2 in S. cerevisiae strain BY4741 (Jensen et al., 2011; Mao et al., 2016). Whole-cell assays were conducted as reported earlier (Pompon et al., 1996). In brief, cells were grown in 50 mL of selective dropout medium (−Leu, with 2% [w/v] dextrose) at 30°C to an OD600 of ∼0.6, then transferred to selective dropout medium with 2% (w/v) Gal with further incubation for 5 h. Subsequently, cultures were supplemented with 25 µm dolabradiene or macrocarpene 1:1 MeOH:DMSO and incubated for 24 h prior to cell harvest and product extraction with diethyl ether (cell pellet) or ethyl acetate (supernatant). Extracts were dried, resuspended in 200 μL of MeOH, and derivatized with 10 μL of tetramethylsilane-diazomethane (Sigma-Aldrich) for 2 h. After drying under an N2 stream and redissolving in hexane, samples were analyzed via GC-MS as described below.
GC-MS Analysis
GC-MS analysis of enzyme products was performed on an Agilent 7890B gas chromatograph with a 5977 Extractor XL MS Detector at 70 eV and 1.2 mL min−1 He flow, using an HP5-MS column (30 m, 250 µm i.d., 0.25 µm film) with a sample volume of 1 µL and the following GC parameters: pulsed splitless injection at 250°C and 50°C oven temperature; held at 50°C for 3 min, 20°C min−1 to 300°C, and held for 3 min. MS data from 90 to 600 m/z were collected after a 10-min solvent delay. For the analysis of dolabradiene present in plant samples, vapor phase extraction was used (Schmelz et al., 2004) for sample enrichment, and GC-MS analyses were conducted on an Agilent 6890 Series gas chromatograph coupled to an Agilent 5973 MS Detector. Separation was achieved on an Agilent DB-35MS column (30 m × 0.25 mm × 0.25 µm). Samples were introduced in splitless injection mode with an initial oven temperature of 45°C. The temperature was held for 2.25 min, then increased to 300°C with a gradient of 20°C min−1, and held at 300°C for 5 min (interface temperature, 250°C; mass temperature, 150°C; source temperature, 230°C; electron energy, 70 eV). GC-(electron impact)-MS-based quantification of plant endogenous dolabradiene was based upon the slope of an external standard curve constructed from enzyme-produced and chemically purified dolabradiene using the diagnostic m/z 216 fragment ion. The identification of dolabradiene included the comparison of retention time (13.88 min) with the standard and the comparison of mass spectra with the Wiley, National Institute of Standards and Technology, and Adams libraries.
LC-MS Analysis of Dolabralexin Pathway Metabolites
Plant samples were ground to a fine powder with liquid N2 and weighed out in 50-mg aliquots. LC-MS analyses of diterpenoids were performed as described elsewhere (Ding et al., 2017). In brief, tissue samples were sequentially and additively bead homogenized in (1) 100 μL of 1-propanol:acetonitrile (ACN):formic acid (1:1:0.01), (2) 250 μL of ACN:ethyl acetate (1:1), and (3) 100 μL of water. Aliquots were analyzed via LC-MS using an Agilent 1260 Infinity Series HiP Degasser (G4225A) with a 1260 binary pump (G1312B) and a 1260 autosampler (G1329B) with a binary gradient mobile phase consisting of 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in MeOH on an Agilent Zorbax Eclipse Plus C18 Rapid Resolution HD column (1.8 μm, 2.1 × 50 mm; 0.35 mL min−1 flow rate). Eluted analytes underwent electrospray ionization via an Agilent Jet Stream Source with thermal gradient focusing using the following parameters: nozzle voltage (500 V), N2 nebulizing gas (flow, 12 L min−1; 55 p.s.i., 225°C), and sheath gas (350°C, 12 L min−1). The transfer inlet capillary was 3,500 V, and both MS1 and MS2 heaters were at 100°C. Positive ionization [M+H]+ mode scans (0.1-amu steps, 2.25 cycles s−1) from m/z 100 to 1,000 were acquired. While the positive ion [M+H]+ parent mass of epoxydolabranol was detectable with m/z 305, the predominant signal was consistent with a loss of water [M-H2O+H]+, namely m/z 287, with a stable retention time of 16.15 min. Absolute concentrations of compounds were calculated using external calibration curves of purified epoxydolabranol at concentrations ranging from 0.123 to 10 ng µL−1. To estimate the quantities of trihydroxydolabrene, the predominant sodium adduct parent ion [M+Na+H]+ m/z 345 was utilized, which additionally contained in-source decay fragment ions [M-H2O+H]+, [M-2H2O+H]+, and [M-3H2O+H]+ of m/z 305, 287, and 269, respectively. The complete ion series was functionally absent in all elicited Zman2 samples, supporting parent-fragment interrelationships. Using the above instrument conditions, m/z 345 was used for quantification purposes in concert with an external standard curve of HPLC-purified trihydroxydolabrene.
For MS/MS analyses of trihydroxydolabrene, an Agilent 1100 HPLC device and locally made analytical column (using a custom pressure cell and 2.5-µm BEH C18 particle packed into fused silica capillary tubing with 200 µm i.d., 360 µm o.d., and 20 cm length) were integrated with a custom nanospray tip with an i.d. of less than 1 µm. Separation was achieved using a 60-min reverse-phase gradient: 0 to 1 min at 100% A (water and 0.1% formic acid), 1 to 26 min at 95% A and 5% B (ACN and 0.1% formic acid), 26 to 30 min at 5% A and 95% B(all w/v), and 31 to 60 min at 100% A. A splitter was used to split the flow rate from 0.2 to 0.6 mL min−1. Spectra were acquired on a Q Exactive HF Orbitrap mass spectrometer (Thermo Electron), which was operated in positive ion mode with a spray voltage of 300 V, a source temperature of 275°C, and an S-lens radio frequency (RF) level set to 50%. A combined Top-N data-dependent scan and data-independent scan method was used to acquire high-resolution MS data. For data-dependent scans, each MS scan was followed by 10 MS/MS scans of the most intense ions from the parent MS scan. Mass resolutions of 60,000 and 15,000 were used for MS and MS/MS modes, respectively. An isolation window of 1 D and a normalized collision energy (NCE) of 30 were used for both data-dependent and data-independent scans.
NMR Analyses
For structural verification via NMR analysis, enzyme products were prepared as described above but using 500-mL cultures. Extracts were dried by rotary evaporation and resuspended in suitable volumes of hexane prior to chromatographic purification by HPLC using an Agilent 1100 Series instrument with a diode array UV detector and an Agilent ZORBAX Eclipse Plus-C8 column (4.6 × 150 mm, 5 μm) at a 0.5 mL min−1 flow rate and a water/ACN gradient as mobile phase. Purified products were then dissolved in 0.5 mL of deuterated chloroform (Sigma-Aldrich), and NMR spectra were acquired at 25°C on a Bruker Avance III 800 spectrometer equipped with a 5-mm triple resonance cryo probe (CPTCI). Chemical shifts were calculated by reference to known deuterated chloroform (13C 77.23 ppm, 1H 7.24 ppm) signals offset from tetramethylsilane. All spectra were acquired using standard experiments on Bruker TopSpin 3.2 software, including 1D 1H, 2D HSQC, correlation spectroscopy (COSY), and HMBC (600 MHz), and 1D 13C spectra (201 MHz). For the purification of trihydroxydolabrene, 420 g of 75-d-old field-grown Ky21 root tissue was ground to a powder in liquid N2, extracted with 500 mL of MeOH, filtered, and dried using a rotary evaporator. The resulting oily residue was then separated by preparative flash chromatography (CombiFlashRf; Teledyne ISCO) on a 15.5-g C18 (RediSepRf High Performance GOLD) column. The mobile phase consisted of solvent A (100% MilliQ water) and solvent B (100% ACN) with a continuous gradient of 0% to 100% B from 1 to 50 min using a flow rate of 19 mL min−1. One-minute fractions were collected and analyzed by LC-MS. At 22 min (48% [v/v] ACN), a fraction highly enriched in trihydroxydolabrene was obtained. This fraction was purified further by HPLC using repeated 1-mg injections on a Zorbax RX-C18 (250 × 4.6 mm, 5 μm; Agilent) column and a mobile phase consisting of solvent A (ACN:water, 1:4) and solvent B (100% ACN) with a continuous gradient of A to B from 0 to 27 min using a flow rate of 1 mL min−1. The recollected fractions spanning 15 to 16 min contained trihydroxydolabrene at approximately 85% purity and were used to generate samples for NMR, external standard curves for quantification, and antifungal bioassays. Purified trihydroxydolabrene was dissolved in d6-DMSO (Cambridge Isotope Laboratories), and NMR spectra were acquired on a Bruker 600-MHz spectrometer equipped with a 1.7-mm CPTCI cryoprobe. Chemical shifts were calculated by reference to known d6-DMSO (13C 39.52 ppm, 1H 2.50 ppm) signals. All spectra were acquired using standard experiments on a Bruker Avance III console and TopSpin 2.1.6 software, including 1D 1H and 2D HSQC, COSY, and HMBC (600 MHz).
Abiotic Elicitation of Maize Roots with CuSO4
Seeds of maize (Golden Queen) were germinated on wetted paper for 4 d at 23°C in the dark. Seedlings were then transferred to a hydroponic medium (Schmelz et al., 2001) and grown for 12 d under 16/8 h of light/darkness (28°C), light intensity of 180 µmol photons m−2 s−1, and ∼60% relative humidity. For the controlled exposure to oxidative stress, 1 mm CuSO4 was added to the hydroponic medium. Root samples were collected at the indicated time points after treatment and frozen in liquid N2 for metabolite and gene expression analyses.
Fungal Elicitation of Mature Maize Roots
For the root elicitation assays with live fungal pathogens (Fusarium spp.), large nodal roots (2 mm or greater diameter) of 53-d-old greenhouse plants, grown in separate 10-L pots and supplemented with 14-14-14 Osmocote (Scotts Miracle-Gro) fertilizer, were punctured with a blunt-ended circular steel pin (0.6 mm diameter) at 1-cm intervals and inoculated with 10 µL of either water or 1 × 107 conidia mL−1 F. verticillioides or F. graminearum at each wound site. In order to avoid mechanical damage to intentionally untreated tissues, treatments were limited to exposed roots growing along the outer edge of the soil in close contact with the vertical wall of the plastic pot. Seven days after inoculation, root samples were collected and frozen in liquid N2 for metabolite and gene expression analysis.
In Vitro Antifungal Assays with Epoxydolabranol and Trihydroxydolabrene
Maize antifungal assays using epoxydolabranol and trihydroxydolabrene were performed using the Clinical and Laboratory Standards Institute M38-A2 guidelines as described previously (Schmelz et al., 2011). Briefly, fungal growth at 30°C in broth medium was monitored using a Synergy4 (BioTech Instruments) reader with a 96-well microtiter plate-based method through periodic measurements of changes in OD600. Each well contained 200 µL of initial fungal inoculum (2.5 × 104 conidia mL−1) with 0.5 µL of either pure DMSO or DMSO containing dilutions of HPLC-purified epoxydolabranol from in vitro assays and root-derived trihydroxydolabrene.
Phylogenetic Analysis
Protein sequence alignments (Supplemental Table S2) were performed using the CLCBio software package and curated with G-blocks (Talavera and Castresana, 2007). Maximum-likelihood phylogenetic trees were generated using PhyML version 3.0 (Guindon et al., 2010) with four rate substitution categories, LG substitution model, BIONJ starting tree, and 500 bootstrap repetitions.
Accession Numbers
Nucleotide sequences of characterized enzymes are available at the GenBank/EBI Data Bank with accession numbers DAA49845 (ZmKSL4), AFW68701 (ZmCYP71Z16), and AFW59698 (ZmCPR2) or the MaizeGDB server with accession numbers GRMZM2G061922 (ZmKSL4), GRMZM2G067591 (ZmCYP71Z16), and GRMZM2G104294 (ZmCPR2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chromosomal locations of maize diTPS and P450 genes.
Supplemental Figure S2. NMR analysis of dolabradiene.
Supplemental Figure S3. Mass spectra of ZmKSL4 reaction products.
Supplemental Figure S4. NMR analysis of the ZmCYP71Z16 reaction products.
Supplemental Figure S5. Feeding assays of ZmCYP71Z16.
Supplemental Figure S6. Substrate specificity of ZmCYP71Z16 and ZmCYP71Z18.
Supplemental Figure S7. Absence of dolabralexins in the maize Zman2 mutant.
Supplemental Figure S8. LC-MS/MS spectra of trihydroxydolabrene.
Supplemental Figure S9. Routine analysis of trihydroxydolabrene.
Supplemental Figure S10. NMR elucidation of trihydroxydolabrene.
Supplemental Table S1. Oligonucleotides and plasmid constructs used in this study.
Supplemental Table S2. Protein sequences used in phylogenetic analyses.
Acknowledgments
We thank Dr. David Nelson (University of Tennessee) for assistance with the annotation of ZmCYP71Z16 and Dr. Reuben Peters (Iowa State University) for providing the pIRS, pGGxC, and ZmAN2 constructs and select authentic standards.
Footnotes
Articles can be viewed without a subscription.
References
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. (2011) Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol 157: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin TT, Zitomer NC, Mitchell TR, Zimeri AM, Bacon CW, Riley RT, Glenn AE (2014) Maize seedling blight induced by Fusarium verticillioides: accumulation of fumonisin B1 in leaves without colonization of the leaves. J Agric Food Chem 62: 2118–2125 [DOI] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy JJ, Goldsack RJ, Wu MZ, Fookes CJ, Forster PI (2000) The steam volatile oil of Wollemia nobilis and its comparison with other members of the Araucariaceae (Agathis and Araucaria). Biochem Syst Ecol 28: 563–578 [DOI] [PubMed] [Google Scholar]
- Brückner K, Tissier A (2013) High-level diterpene production by transient expression in Nicotiana benthamiana. Plant Methods 9: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Newton AC (2011) Climate change, plant diseases and food security: an overview. Plant Pathol 60: 2–14 [Google Scholar]
- Chaturvedi R, Venables B, Petros RA, Nalam V, Li M, Wang X, Takemoto LJ, Shah J (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J 71: 161–172 [DOI] [PubMed] [Google Scholar]
- Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, Brodie EL, Hettich RL (2010) Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res 9: 6615–6622 [DOI] [PubMed] [Google Scholar]
- Christensen SA, Huffaker A, Kaplan F, Sims J, Ziemann S, Doehlemann G, Ji L, Schmitz RJ, Kolomiets MV, Alborn HT, et al. (2015) Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc Natl Acad Sci USA 112: 11407–11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SA, Sims J, Vaughan M, Hunter C, Block A, Willett D, Alborn HT, Huffaker A, Schmelz EA (2018) Commercial hybrids and mutant genotypes reveal complex protective roles for inducible terpenoid defenses. J Exp Bot doi.org/10.1093/jxb/erx495 [DOI] [PubMed] [Google Scholar]
- Christie N, Myburg AA, Joubert F, Murray SL, Carstens M, Lin YC, Meyer J, Crampton BG, Christensen SA, Ntuli JF, et al. (2017) Systems genetics reveals a transcriptional network associated with susceptibility in the maize-grey leaf spot pathosystem. Plant J 89: 746–763 [DOI] [PubMed] [Google Scholar]
- Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, Snyder JH, Song J, Chen S, Huang L, et al. (2015) Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol 169: 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J. (2009) Indirect defense responses to herbivory in grasses. Plant Physiol 149: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Hiltpold I, Köllner TG, Frey M, Gierl A, Gershenzon J, Hibbard BE, Ellersieck MR, Turlings TCJ (2009) Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc Natl Acad Sci USA 106: 13213–13218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sassi C, Tylianakis JM (2012) Climate change disproportionately increases herbivore over plant or parasitoid biomass. PLoS ONE 7: e40557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Huffaker A, Köllner TG, Weckwerth P, Robert CAM, Spencer JL, Lipka AE, Schmelz EA (2017) Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol 175: 1455–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Ren F, Lu X, Mao H, Xu M, Degenhardt J, Peters RJ, Wang Q (2016) A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol 170: 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Goswami RS, Kistler HC (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5: 515–525 [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II (2012) New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol 193: 304–312 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hall DE, Zerbe P, Jancsik S, Quesada AL, Dullat H, Madilao LL, Yuen M, Bohlmann J (2013) Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol 161: 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59: 881–894 [DOI] [PubMed] [Google Scholar]
- Hartmann T. (2007) From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry 68: 2831–2846 [DOI] [PubMed] [Google Scholar]
- Hedden P, Sponsel V (2015) A century of gibberellin research. J Plant Growth Regul 34: 740–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Kaplan F, Vaughan MM, Dafoe NJ, Ni X, Rocca JR, Alborn HT, Teal PE, Schmelz EA (2011) Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol 156: 2082–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen NB, Zagrobelny M, Hjernø K, Olsen CE, Houghton-Larsen J, Borch J, Møller BL, Bak S (2011) Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat Commun 2: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H, Kobayashi K (2009) Jasmonic acid, protein phosphatase inhibitor, metals and UV-irradiation increased momilactone A and B concentrations in the moss Hypnum plumaeforme. J Plant Physiol 166: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Peters RJ (2013) The role of momilactones in rice allelopathy. J Chem Ecol 39: 175–185 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J (2006) Diterpene resin acids in conifers. Phytochemistry 67: 2415–2423 [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Lu X, Yang B, Peters RJ (2015) The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol Plant 8: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Held M, Lenk C, Hiltpold I, Turlings TC, Gershenzon J, Degenhardt J (2008a) A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Li S, Svatos A, Schneider B, Gershenzon J, Degenhardt J (2008b) Protonation of a neutral (S)-beta-bisabolene intermediate is involved in (S)-beta-macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J Biol Chem 283: 20779–20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Miller B, Ralph S, Ellis BE, Douglas C, Ritland K, Bohlmann J (2004) Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques 36: 821–824 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ahkami A (2013) Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes: current status and future opportunities. Plant Biotechnol J 11: 169–196 [DOI] [PubMed] [Google Scholar]
- Mao H, Liu J, Ren F, Peters RJ, Wang Q (2016) Characterization of CYP71Z18 indicates a role in maize zealexin biosynthesis. Phytochemistry 121: 4–10 [DOI] [PubMed] [Google Scholar]
- Mao H, Shen Q, Wang Q (2017) CYP701A26 is characterized as an ent-kaurene oxidase with putative involvement in maize gibberellin biosynthesis. Biotechnol Lett 39: 1709–1716 [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schäfer P, Kudrna D, Wing RA, Bolger AM, et al. (2013) Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25: 2022–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Hillwig ML, Mead ME, Lowry L, Fulton DB, Peters RJ (2011) Evident and latent plasticity across the rice diterpene synthase family with potential implications for the evolution of diterpenoid metabolism in the cereals. Biochem J 435: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Jin Y, Xu M, Choi SY, Coates RM, Peters RJ (2006) An unexpected diterpene cyclase from rice: functional identification of a stemodene synthase. Arch Biochem Biophys 448: 133–140 [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol 85: 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold GA. (1997) Fumonisins in maize: can we reduce their occurrence? Plant Dis 81: 556–565 [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S, Alegre L (2000) Changes in carotenoids, tocopherols and diterpenes during drought and recovery, and the biological significance of chlorophyll loss in Rosmarinus officinalis plants. Planta 210: 925–931 [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Huang A, Osbourn A (2016) Plant metabolic clusters: from genetics to genomics. New Phytol 211: 771–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ. (2006) Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317 [DOI] [PubMed] [Google Scholar]
- Peters RJ. (2010) Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 27: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ, Croteau RB (2002) Abietadiene synthase catalysis: mutational analysis of a prenyl diphosphate ionization-initiated cyclization and rearrangement. Proc Natl Acad Sci USA 99: 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H, Pekic S, Lazic-Jancic V, Quarrie SA, Shah SM, Pervez A, Shah MM (2011) Molecular mapping of quantitative trait loci for drought tolerance in maize plants. Genet Mol Res 10: 889–901 [DOI] [PubMed] [Google Scholar]
- Richter A, Schaff C, Zhang Z, Lipka AE, Tian F, Köllner TG, Schnee C, Preiß S, Irmisch S, Jander G, et al. (2016) Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 28: 2651–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Seidl-Adams I, Köllner TG, Schaff C, Tumlinson JH, Degenhardt J (2015) A small, differentially regulated family of farnesyl diphosphate synthases in maize (Zea mays) provides farnesyl diphosphate for the biosynthesis of herbivore-induced sesquiterpenes. Planta 241: 1351–1361 [DOI] [PubMed] [Google Scholar]
- Santiago R, Malvar RA (2010) Role of dehydrodiferulates in maize resistance to pests and diseases. Int J Mol Sci 11: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Alborn HT, Tumlinson JH (2001) The influence of intact-plant and excised-leaf bioassay designs on volicitin- and jasmonic acid-induced sesquiterpene volatile release in Zea mays. Planta 214: 171–179 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH III, Teal PE (2009) Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA 106: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79: 659–678 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Kaplan F, Huffaker A, Dafoe NJ, Vaughan MM, Ni X, Rocca JR, Alborn HT, Teal PE (2011) Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci USA 108: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Held M, Turlings TC, Gershenzon J, Degenhardt J (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA 103: 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nagahama S, Nakashima T, Suenaga H (2001) Chemotaxonomy on the leaf constituents of Thujopsis dolabrata Sieb. et Zucc.: analysis of neutral extracts (diterpene hydrocarbon). Biochem Syst Ecol 29: 839–848 [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577 [DOI] [PubMed] [Google Scholar]
- Tholl D. (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148: 63–106 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H (2008) Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci Biotechnol Biochem 72: 562–567 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, Otomo K, Hirose Y, Miyao A, Hirochika H, Okada K, Shimizu T, Koga J, et al. (2014) Reverse-genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol Plant 150: 55–62 [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S, Sanguineti MC, Landi P, Maccaferri M, Conti S (2002) Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann Bot 89: 941–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K, Kastner C, Kumlehn J, Kahmann R, Doehlemann G (2011) Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol 189: 471–483 [DOI] [PubMed] [Google Scholar]
- Vaughan MM, Christensen S, Schmelz EA, Huffaker A, McAuslane HJ, Alborn HT, Romero M, Allen LH, Teal PE (2015) Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ 38: 2195–2207 [DOI] [PubMed] [Google Scholar]
- Vaughan MM, Huffaker A, Schmelz EA, Dafoe NJ, Christensen S, Sims J, Martins VF, Swerbilow J, Romero M, Alborn HT, et al. (2014) Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides. Plant Cell Environ 37: 2691–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O’Donnell C, et al. (2013) Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ (2012a) Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem 287: 6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Peters RJ (2011) CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J 65: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Wu Y, Peters RJ (2012b) CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol 158: 1418–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86: 6201–62052762323 [Google Scholar]
- Wouters FC, Blanchette B, Gershenzon J, Vassão DG (2016) Plant defense and herbivore counter-defense: benzoxazinoids and insect herbivores. Phytochem Rev 15: 1127–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hillwig ML, Wang Q, Peters RJ (2011) Parsing a multifunctional biosynthetic gene cluster from rice: biochemical characterization of CYP71Z6 & 7. FEBS Lett 585: 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou K, Toyomasu T, Sugawara C, Oku M, Abe S, Usui M, Mitsuhashi W, Chono M, Chandler PM, et al. (2012) Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39: 309–318 [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ (2007a) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68: 312–326 [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Peters RJ (2007b) Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc Natl Acad Sci USA 104: 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe P, Bohlmann J (2015) Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol 33: 419–428 [DOI] [PubMed] [Google Scholar]
- Zerbe P, Hamberger B, Yuen MM, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Bach SS, Bohlmann J (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol 162: 1073–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Xu M, Tiernan M, Xie Q, Toyomasu T, Sugawara C, Oku M, Usui M, Mitsuhashi W, Chono M, et al. (2012) Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol 65: 259–286 [DOI] [PMC free article] [PubMed] [Google Scholar]