Abstract
There are extensive long-distance chromosomal interactions in eukaryotic genomes, but to what extent these interactions affect gene expression is not clear. Recent works have identified several cases where clustering of co-regulated genes leads to enhanced gene expression in budding yeast. Similar phenomenon was also observed in mammalian cells. These results challenge widely held views of gene regulation in yeast and further our understanding of how the 3D organization of the genome contribute to gene regulation in eukaryotes.
Keywords: Long-distance gene regulation, Chromosome configuration capture, Long-distance chromosomal interaction, 3D clustering
Transcription regulation in eukaryotes is a complex process that involves coordination at multiple levels. Besides cis-elements (such as transcription factor binding site and TATA box) and nucleosome structure, 3D organization of chromosome also plays an important role in transcription regulation (Babu et al. 2008; Lanctot et al. 2007; Rowley and Corces 2016). The recent advances in chromosome conformation capture techniques (3C, 4C, Hi-C, etc.) allow researchers to study the spatial organization of the genome with unprecedented resolution and output (Dekker et al. 2002; Sandhu et al. 2012; van Berkum et al. 2010; Zhao et al. 2006). Application of these techniques in yeast, flies, and mammalian cells have revealed numerous intra- and inter-chromosomal interactions (Dixon et al. 2012; Duan et al. 2010; Ghavi-Helm et al. 2014; Jin et al. 2013). However, it is important to point out that these methods detect interactions based on their physical proximity, but not on their functional consequences. As a result, among the millions of interactions found in these experiments, it is hard to tell which ones carry active regulatory functions, and which ones are passive consequences of chromosome folding. In fact, computational models of yeast chromosome as polymers with structural constraints can reproduce the DNA contact frequency measured by the Hi-C experiment to a large degree (Tjong et al. 2012; Wong et al. 2012). For mammalian cells, different cell types often yield very similar interaction maps in spite of their significantly different transcription programs (Dixon et al. 2012; Mifsud et al. 2015; Won et al. 2016). These results suggest that a large fraction of the Hi-C signals may have simple physical bases rather than regulatory roles. Therefore, identifying long-distance chromosomal interactions that regulate gene expression and understanding the underlying mechanism will be one of the main focuses of this field in the coming years.
The relation between long-distance chromosomal interaction and gene regulation is particularly intriguing in budding yeast because it is traditionally thought as a species that lacks gene regulation over long distance. In the yeast genome, regulatory regions and their targeted genes tend to be located closely within a few hundred base pairs (Erb and van Nimwegen 2011; Yan et al. 2015). In addition, by artificially placing the upstream activating sequences (UASs) further and further away from a core promoter, it was shown that UASs quickly lose their ability to activate transcription from that promoter (Dobi and Winston 2007). The idea here is that the constraint of long-distance activation in yeast is essential to ensure UAS-core promoter specificity in its compact genome (Dobi and Winston 2007). Nevertheless, Hi-C experiment in yeast revealed extensive long-distance interactions between sites >20 kb away from each other (Duan et al. 2010). The domain-like configuration of these interactions (Eser et al. 2017), as well as the interaction density (number of interactions per DNA fragment), are similar to those found in mammalian cells. More importantly, statistical analysis of the Hi-C data showed that co-regulated yeast loci tend to cluster, and physically proximal genes tend to co-express (Ben-Elazar et al. 2013; Capurso et al. 2016; Homouz and Kudlicki 2013). These observations strongly raise the possibility that some longdistance interactions play a role in gene regulation in yeast. In particular, a “gene proximity model” has been proposed that the aggregation of specific transcription factors within the nucleus space might function as a recruiter to draw their target genes close in space and probably to nearby transcription factories for coordinated expression (Li and Heermann 2013). Indeed, experimental evidence from a few recent studies support this idea.
The first line of evidence comes from the interaction and regulatory effect between homologous alleles in somatic diploid yeast cells (Fig. 1a). Pairing between homologous chromosomes has been observed in diploid budding yeast for over a decade (Burgess et al. 1999), but its functional significance in gene expression has begun to unravel in recent years only. The studies so far all use the classic activation system, GAL1 promoter, as the model. It was first found that β-estradiol-induced GAL1 promoters at allelic locations could form significantly stronger transinteractions under the activating condition (Mirkin et al. 2013), suggesting that this interaction is intricately related to gene expression. Extending from this observation, Zhang et al. showed that two allelic reporters, one driven by wild-type GAL1 promoter and the other by a mutated GAL1 promoter with delayed response to galactose induction, physically associate upon induction. The wild-type GAL1pr triggers synchronized firing of the defective promoter and accelerates its activation without affecting its steady-state expression level (Zhang and Bai 2016). Importantly, the same reporters located at non-allelic locations do not show such interaction and trans-regulatory effect. Brickner et al. also reported that the wild-type GAL1-10 alleles in diploid yeast cells cluster upon induction, and a cis-element in the GAL1-10 promoter, GRS4, is critical for promoting the interaction. Again, this clustering contributes to stronger expression of GAL1 and GAL10 by increasing the fraction of cells that respond to the inducer (Brickner et al. 2016).
Fig. 1.
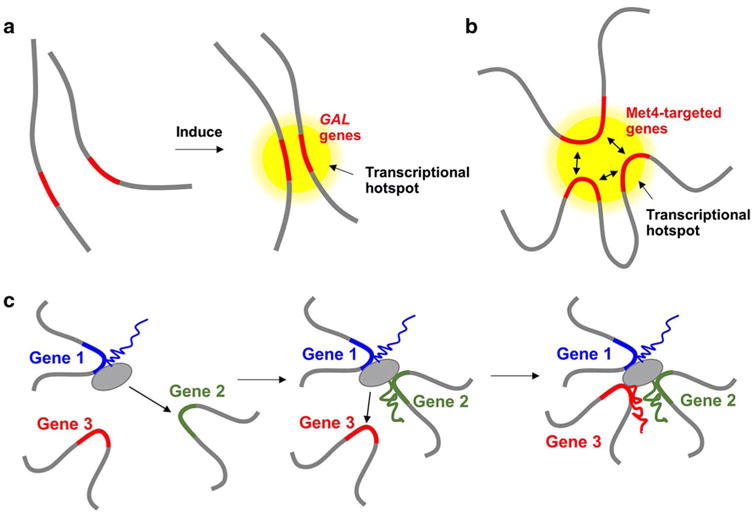
3D clustering of co-regulated genes. a Two copies of GAL1 genes (red) at allelic locations in diploid yeast cluster together in a transcriptional hotspot (yellow circle) upon galactose induction. b A fraction of Met4-targeted genes (red) form 3D cluster in a transcriptional hotspot. Double arrows indicate the physical interactions between different genes in the cluster. c Three NFκB-regulated genes (Gene 1, 2, and 3 shown in blue, green, and red, respectively) form a multigene complex with hierarchy: transcription of Gene 1 is required for the recruitment and expression of Gene 2, and is then followed by the recruitment and expression of Gene 3. Gray circles represent the transcription machineries
The second line of evidence comes from a recent study of the interaction between non-homologous loci and their function in gene regulation. Du et al. developed a medium-throughput assay to screen for functional long-distance interactions that affect the expression of a reporter gene in the budding yeast genome (Du et al. 2017). An insulated MET3 promoter flanked by ∼1 kb invariable sequences was integrated into thousands of genomic loci, allowing it to make contacts with different parts of the genome. The idea is that, if the MET3 promoter activity changes, it has to be caused by mechanisms that initiate more than 1 kb away. Their data suggest that a subset of MET3 co-regulated genes on different chromosomes can physically associate and form 3D clusters, and the activity of the MET3 promoter increases when inserted near these genes (Fig. 1b). The same phenomenon was also observed for MET13, an endogenous gene in the cluster. When translocated to a different genomic locus, MET13 loses the interactions with other genes in the cluster and shows lower expression, indicating that the endogenous genes also benefit from the cluster for higher activity.
Although the studies above focus on different types of interactions, three common themes emerged. First, co-regulated genes can interact with each other, but only when they are located in certain genomic loci. For example, among the insertion sites tested, GAL1 reporters only interact when they are at allelic locations, or on non-homologous chromosomes but have equal distance to centromere (Mirkin et al. 2013; Zhang and Bai 2016). Similarly, only a small fraction of Met4-targeted genes seems to cluster, and the MET3 reporter makes contacts with the cluster only when inserted into certain loci (Du et al. 2017). These results indicate that the search for interaction partners is constrained to a nuclear sub-volume imposed by the chromosome context (Noordermeer et al. 2011). Second, the intensity of the interaction changes with transcriptional status. For both GAL1 and MET3 reporters, the interaction becomes stronger under the activating condition, suggesting that the interactions may be mediated by transcription-related proteins or RNA transcripts. In fission yeast, it was proposed that condensin is used to connect actively transcribed genes (Iwasaki and Noma 2016; Robellet et al. 2016). Third, genes at the cluster show higher expression. The detailed mechanism is not clear, but a simple model is that the clustering may generate a “transcriptional hotspot” with high local concentration of related factors, allowing co-regulated neighboring genes to fire with more strength. Also, factors can quickly bind and rebind among these spatially co-localized genes, making the transcription process more efficient.
Clustering of co-regulated genes has also been observed in the mammalian system, and sometimes referred to as “multigene complexes.” Genomic loci from multigene complexes were shown to associate with Pol II foci or “specialized transcription factories,” suggesting that it may provide a structural framework for co-transcription (Li et al. 2012; Papantonis et al. 2012; Schoenfelder et al. 2010). Consistent with this idea, a few studies have demonstrated that the formation of multigene complexes coincides with alterations in gene expression (Apostolou and Thanos 2008; Fullwood et al. 2009; Sandhu et al. 2012; Spilianakis et al. 2005). For example, during the differentiation of naïve T cells to effector T-helper cells, there is a drastic change of chromosomal interactions made by two loci, Ifng and TH2 LCR, and this switch is thought to be critical for establishing the T-helper cell identity (Spilianakis et al. 2005). Most of the studies showed correlations between chromosomal interaction and gene expression, but some works also investigated the causal relationship between them. In Fanucchi et al., for instance, they perturbed the site of contact in the NFκB-regulated multigene complex, and showed that it reduces the transcription of other interacting genes (Fanucchi et al. 2013). Interestingly, some genes in the complex play more “dominant” roles, and their transcriptions are required for other members to interact and express (Fig. 1c). Collectively, these results show that transcriptional co-association is a wide-spread phenomenon that occurs in many transcriptional programs and has regulatory functions.
Despite recent progress, many important questions about the 3D clustering and transcription regulation require further elucidation. Do all regulons of different transcription factors experience 3D clustering? Among one regulon, how many genes come together? What is special about these genes? What factors are mediating the clustering? At the cluster, how are different genes arranged and orientated? Is there significant cell-to-cell variability of these clusters? Is there a causal relationship between clustering and enhanced gene activity? What is the molecular mechanism underlying the enhanced gene activity? To address these questions, we need more efficient methods to map the 3D localization of all the regulons in the genome, preferably in a more targeted fashion. We also need new methods to selectively perturb the chromosome configuration and examine the biological consequences.
References
- Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Babu MM, Janga SC, de Santiago I, Pombo A. Eukaryotic gene regulation in three dimensions and its impact on genome evolution. Curr Opin Genet Dev. 2008;18:571–582. doi: 10.1016/j.gde.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Ben-Elazar S, Yakhini Z, Yanai I. Spatial localization of co-regulated genes exceeds genomic gene clustering in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2013;41:2191–2201. doi: 10.1093/nar/gks1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell. 2016;27:2980–2993. doi: 10.1091/mbc.E16-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurso D, Bengtsson H, Segal MR. Discovering hotspots in functional genomic data superposed on 3D chromatin configuration reconstructions. Nucleic Acids Res. 2016;44:2028–2035. doi: 10.1093/nar/gkw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Zhang Q, Bai L. Three distinct mechanisms of longdistance modulation of gene expression in yeast. PLoS Genet. 2017;13:e1006736. doi: 10.1371/journal.pgen.1006736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb I, van Nimwegen E. Transcription factor binding site positioning in yeast: proximal promoter motifs characterize TATA-less promoters. PLoS One. 2011;6:e24279. doi: 10.1371/journal.pone.0024279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U, Chandler-Brown D, Ay F, Straight AF, Duan Z, Noble WS, Skotheim JM. Form and function of topologically associating genomic domains in budding yeast. Proc Natl Acad Sci USA. 2017;114:E3061–E3070. doi: 10.1073/pnas.1612256114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155:606–620. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- Homouz D, Kudlicki AS. The 3D organization of the yeast genome correlates with co-expression and reflects functional relations between genes. PLoS One. 2013;8:e54699. doi: 10.1371/jour-nal.pone.0054699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Noma KI. Condensin-mediated chromosome organization in fission yeast. Curr Genet. 2016;62:739–743. doi: 10.1007/s00294-016-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Li S, Heermann DW. Transcriptional regulatory network shapes the genome structure of Saccharomyces cerevisiae. Nucleus. 2013;4:216–228. doi: 10.4161/nucl.24875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GL, Ruan XA, Auerbach RK, Sandhu KS, Zheng MZ, Wang P, Poh HM, Goh Y, Lim J, Zhang JY, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong SZ, Zhang ZZ, Landt S, Raha D, Euskirchen G, Wei CL, Ge WH, Wang HE, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan YJ. extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, Herman B, Happe S, Higgs A, LeProust E, Follows GA, Fraser P, Luscombe NM, Osborne CS. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Chang FS, Kleckner N. Dynamic trans interactions in yeast chromosomes. PLoS One. 2013;8:e75895. doi: 10.1371/journal.pone.0075895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–U161. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Kohro T, Baboo S, Larkin JD, Deng BW, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, Li GL, Poh HM, Ruan XA, Aburatani H, Ruan YJ, Kodama T, Wada Y, Cook PR. TNF alpha signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J. 2012;31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X, Vanoosthuyse V, Bernard P. The loading of condensin in the context of chromatin. Curr Genet. 2016 doi: 10.1007/s00294-016-0669-0. [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. The three-dimensional genome: principles and roles of long-distance interactions. Curr Opin Cell Biol. 2016;40:8–14. doi: 10.1016/j.ceb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Li G, Poh HM, Quek YL, Sia YY, Peh SQ, Mulawadi FH, Lim J, Sikic M, Menghi F, Thalamuthu A, Sung WK, Ruan X, Fullwood MJ, Liu E, Csermely P, Ruan Y. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep. 2012;2:1207–1219. doi: 10.1016/j.celrep.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp. 2010 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C, Eskin E, Voineagu I, Ernst J, Geschwind DH. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Marie-Nelly H, Herbert S, Carrivain P, Blanc H, Koszul R, Fabre E, Zimmer C. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr Biol. 2012;22:1881–1890. doi: 10.1016/j.cub.2012.07.069. [DOI] [PubMed] [Google Scholar]
- Yan C, Zhang DY, Garay JAR, Mwangi MM, Bai L. Decoupling of divergent gene regulation by sequence-specific DNA binding factors. Nucleic Acids Res. 2015;43:7292–7305. doi: 10.1093/nar/gkv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Bai L. Interallelic interaction and gene regulation in budding yeast. Proc Natl Acad Sci USA. 2016;113:4428–4433. doi: 10.1073/pnas.1601003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]


