Abstract
Venom peptides are known to have strong antimicrobial activity and anticancer properties. King cobra cathelicidin or OH-CATH (KF-34), banded krait cathelicidin (BF-30), wolf spider lycotoxin I (IL-25), and wolf spider lycotoxin II (KE-27) venom peptides were found to strongly inhibit E. coli membrane bound F1Fo ATP synthase. The potent inhibition of wild-type E. coli in comparison to the partial inhibition of null E. coli by KF-34, BF-30, Il-25, or KE-27 clearly links the bactericidal properties of these venom peptides to the binding and inhibition of ATP synthase along with the possibility of other inhibitory targets. The four venom peptides KF-34, BF-30, IL-25, and KE-27, caused ≥85% inhibition of wild-type membrane bound E.coli ATP synthase. Venom peptide induced inhibition of ATP synthase and the strong abrogation of wild-type E. coli cell growth in the presence of venom peptides demonstrates that ATP synthase is a potent membrane bound molecular target for venom peptides. Furthermore, the process of inhibition was found to be fully reversible.
Keywords: E. coli F1Fo ATP synthase, F1-ATPase, Venom peptides, OH-CATH, cathelicidin, lycotoxin I, lycotoxin II
Introduction
The ubiquitous enzyme ATP synthase or complex V of the respiratory chain is a molecular machine comprised of an ion pump and a catalytic nanomotor [1]. The ion pump (Fo-sector) uses a proton gradient to rotate and generate conformational changes in the catalytic nanomotor (F1-sector). Thus allowing the binding of ADP and Pi to produce ATP [2]. This terminal enzyme of oxidative phosphorylation is also the smallest known biological nanomotor [3, 4]. The universally accepted function of ATP synthase is the generation and hydrolysis of ATP via the proton-motive force. The well-known primary locations of ATP synthase are the inner membranes of mitochondria, plasma membrane of bacteria, and membranes of chloroplast thylakoid. Multiple studies have confirmed the presence of ATP synthase on the plasma membrane of several eukaryotic cell types, including endothelial cells [5, 6], keratinocytes [7], adipocytes [8], hepatocytes [9], and cancer cells [10, 11].
In view of the increasing microbial resistance against many known antibiotics, it is of paramount importance to find alternative approaches to combat microbial resistance. ATP is indispensable for the proper growth of cells. About 95% ATP, the required energy of cells, ATP, is generated by the enzyme ATP synthase. Thus, inhibition of ATP synthase is expected to deprive cells of ATP, resulting in cell death. ATP synthase is a valued molecular drug target in many disease conditions such as cancer, tuberculosis, obesity, and microbial infections [12–14]. A wide variety of natural and synthetic compounds, including peptides, are known to potently and selectively inhibit the ATP synthase [12, 15–19].
Venom from animals such as snakes, spiders, wasps, bees, scorpions, and toads is a mixture of biologically active molecules along with peptides. Throughout the animal kingdom, venom peptides have evolved to interact with the specific molecular targets on the intended prey. The combination of high potency, efficiency, and target selectivity makes venom peptides effective drugs against multiple disease conditions including microbial infections and cancer. The successful survival story of reptiles in diverse microbe-filled environmental conditions is owed to the presence of antimicrobial peptides (AMPs). AMPs are an integral part of their innate immune system [20]. Venom peptide cathelicidins are a major class of antimicrobial peptides in higher eukaryotes. Cathelicidin peptides from the snake family are well characterized and are a diverse group of peptides. [20–22]. Cathelicidins are cationic host-defense peptides and contain three discrete motifs, (i) the N-terminal signal peptide motif, (ii) the conserved cathelin motif, and (iii) the C-terminal antimicrobial motif [23].
The King Cobra (Ophiophagus hannah) venom peptide OH-CATH (KF-34) is an excellent antimicrobial agent and shows broad-spectrum activity against multi-drug resistant bacterial strains. KF-34 is an α-helical 34 residue long amphipathic cationic peptide (Table 1 and Fig. 1). It potently inhibits a wide variety of human pathogenic bacteria and causes little or no hemolytic activity against erythrocytes up to [200] µg/mL, suggesting low cytotoxicity of the peptide to eukaryotic cells [24]. The OH-CATH derivative OH-CATH30 also possesses broad-spectrum, salt-independent antimicrobial activity against antibiotic resistant bacteria without hemolytic activity [23, 25].
Table 1.
Amino acid sequence, length, and source of venom peptides
| Peptide | Sequence | Length | Species |
|---|---|---|---|
| OH-CATH (KF-34) | KRFKKFFKKLKNSVKKRAKKFFKKPRVIGVSIPF | 34 | Ophiophagus hannah |
| Cathelicidin (BF-30) | KFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | 30 | Bungarus fasciatus |
| Lycotoxin I (IL-25) | IWLTALKFLGKHAAKHLAKQQLSKL | 25 | Lycosa carolinensis |
| Lycotoxin II (KE-27) | KIKWFKTMKSIAKFIAKEQMKKHLGGE | 27 | Lycosa carolinensis |
Fig. 1.
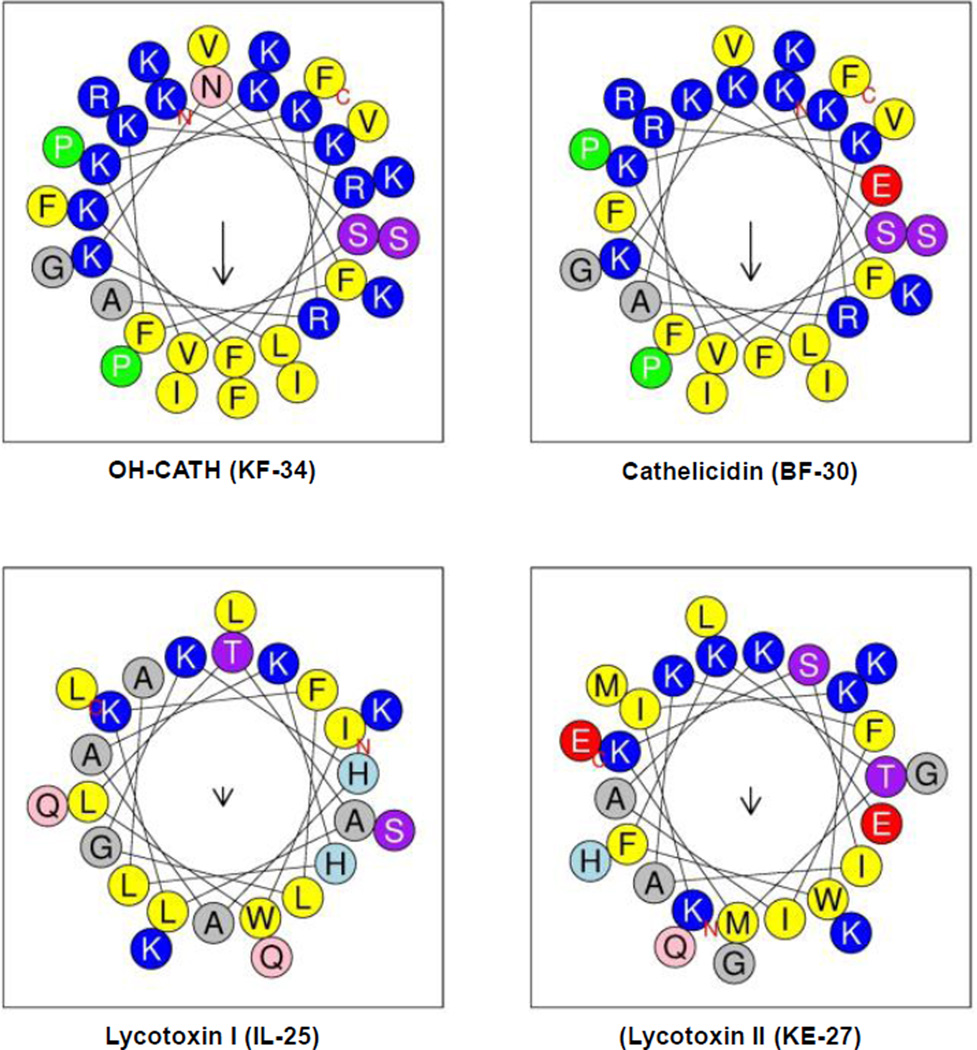
Helix-wheel plots of venom peptides OH-CATH, (KF-34), cathelicidin (BF-30), lycotoxin I (IL-25), or lycotoxin II (KE-27). The size of the downward arrow ↓ indicates the level of hydrophobicity. Hydrophobic and hydrophilic faces of KF-34, BF-30, IL-25, and KE-27 in the helix wheel plots are shown in yellow and blue colors respectively.
The Banded krait (Bungarus fasciatus) venom peptide cathelicidin (BF-30) is a 30 residue long peptide (Table 1). It is an amphipathic α-helical peptide with broad-spectrum antimicrobial activity (Fig. 1). BF-30 was found to be effective against drug-resistant Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus. In comparison to gentamicin, ampicillin, or bacitracin, BF-30 showed stronger antimicrobial activity against a broad spectrum of microbes. BF-30 was shown to kill P. aeruginosa and S. aureus rapidly in less than 120 minutes. BF-30 was also found to be effective in burns and acute infections. In rat models, BF-30 significantly reduced the P. aeruginosa colony formation and prevented subsequent infection and inflammation. Moreover, this observed wide-spectrum antimicrobial activity against drug resistant microbes was with no obvious hemolytic or cytotoxic activity [26, 27]. Fragmented derivatives of BF-30 also possess significant anti-microbial activity both in vitro and in vivo [20, 28].
Throughout the world there has been a steady growth in cancer related deaths. The ability of carcinogenic cells to evade the host immune systems allows them to replicate aggressively and metastasize violently. Chemo, radiation, or surgical procedures used for killing or removing the tumor cells result in severe side effects and toxicities [29]. Therefore, it is essential to find and develop therapies which specifically target tumor cells with the least amount of toxicity and side effects. Targeting cell surface proteins in cancer cells by peptides could help in eliminating cancer cells with the least amount of side effects [30]. Selective molecular targeting of ATP synthase by venom peptides provides great opportunity to kill cancer cells. Cathelicidin BF-30 possesses outstanding anti-tumor activity. It was shown to selectively inhibit melanoma cell proliferation suggesting BF-30 as a potential candidate against malignant melanoma [31].
Lycotoxin I (IL-25) and Lycotoxin II (KE-27), spider wolf (Lycosa carolinensis) venom peptides, are amphipathic α-helical cationic peptides containing 25 and 27 amino acids respectively (Table 1 and Fig. 1). They occur at a very high concentration, in the range of 1–5 mM, in the venom and possess potent anti-microbial activity against Escherichia coli and yeast Candida glabrata at micromolar concentrations [32]. The lycotoxins were shown to both disable the prey and protect the spider from possible infections from the ingestion of prey. Lycotoxins were also suggested to have the potential use as bio-insecticidal agents [33]. Thus, spider venom represents a potential new source of novel antimicrobial agents with important medical significances.
Anti-microbial peptides, including venom peptides, are a highly abundant and diverse group of molecules found in wide variety animal species [34]. One of the proposed mechanisms of action of AMPs for bactericidal activity is the increased permeability and loss of the membrane barrier as a result of damage to the cytoplasmic membrane [35]. The second is bactericidal effect through the inhibition of ATP synthase. It was found that amphibian peptides and melittin, honey bee venom peptide, potently inhibit E. coli ATP synthase leading to the abrogation of E. coli growth [18, 19]. In this study, we examined the inhibitory effects of venom peptides KF-34, BF-30, IL-25, and KE-27 on membrane bound F1Fo ATP synthase and on the growth of E. coli cells. Our results show that all four venom peptides strongly inhibit ATPase activity and bacterial growth, suggesting that the beneficial antimicrobial activities of venom peptides are at least in part linked to the inhibition of ATP synthase.
Materials and Methods
Venom peptides and other chemicals
King cobra OH-CATH (KF-34), banded krait cathelicidin (BF-30), wolf spider lycotoxin I (IL-25), and wolf spider lycotoxin II (KE-27) venom peptides were purchased from Biomatik (http://www.biomatik.com). Amino acid sequences are presented in Table 1. All peptides were greater than 95% pure determined by HPLC. Lyophilized powder was stored at −20 °C upon receipt and resuspended in distilled deionized water for use as needed. All other chemicals used in this study were ultra-pure analytical grade purchased either from Sigma–Aldrich Chemical Company or Fisher Scientific Company.
Wild type and null E. coli strains; growth in limiting glucose; preparation of membrane bound E. coli F1Fo ATP synthase; measurement of ATPase activity of membranes
Wild type membrane bound F1Fo ATP synthase (membrane) was isolated from the pBWU13.4/DK8 E. coli strain [36]. Our null strain is pUC118/DK8 from where ATPase gene has been deleted. Null strain usually grows 40–50% with respect to wild-type due to glycolytic pathway to generate ATP. Wild-type can use both glycolysis and oxidative phosphorylation. Oxidative and substrate level phosphorylation in limiting glucose (3 mM) was measured as in [37]. Membrane bound F1Fo ATP synthase were prepared as in [38] using three washes of the initial membrane pellets. The first wash by buffer containing 50 mM TES pH 7.0, 15% glycerol, 40 mM 6-aminohexanoic acid, 5 mM p-aminobenzamidine is followed by two additional washes by buffer containing 5 mM TES pH 7.0, 15% glycerol, 40 mM 6-aminohexanoic acid, 5 mM p-aminobenzamidine, 0.5 mM DTT, 0.5 mM EDTA. Membrane bound F1Fo were washed twice more by resuspension and ultracentrifugation in 50 mM TrisSO4 pH 8.0, 2.5 mM MgSO4 before the experiments.
ATPase activity was measured in 1 ml ATP cocktail (ATP assay buffer) containing 10 mM NaATP, 4 mM MgCl2, 50 mM TrisSO4, pH 8.5 at 37 °C. Reactions were initiated by the addition of 1 ml ATP cocktail to membranes and stopped by the addition of 1 ml SDS to a final concentration of 3.3%. Pi (inorganic phosphate) release was measured by the Taussky and Shorr method [39]. 20µg wild-type membrane was used in the ATPase assay with a reaction time of 5 min. Reactions were found to be linear with time and protein concentration. SDS-gel electrophoresis (10% acrylamide) and immunoblotting with rabbit polyclonal anti-F1-α and anti-F1-β antibodies was used to check the integrity and purity of protein [40].
Inhibition of membrane bound F1Fo ATPase activity by OH-CATH (KF-34), cathelicidin (BF-30), lycotoxin I (IL-25), or lycotoxin II (KE-27) venom peptides
Membrane bound wild-type F1Fo membranes were incubated with different concentrations of venom peptides for 60 minutes at room temperature (RT) in 50 mM TrisSO4 at pH 8.0. ATP cocktail was used to measure the enzyme activity. The reaction was stopped by the addition of SDS. Addition of Taussky and Shorr reagent generated a blue color which was measured at OD700. Venom peptide induced inhibition profiles were generated with Sigma plot 10.0. Relative ATPase activity was calculated from the absolute values of ATPase activity in the absence of venom peptides taken as 100%.
Reversal of venom peptide induced inhibition of ATPase activity
Reversibility was measured by dilution of the membranes. Membranes were reacted with maximal inhibitory concentrations of venom peptides for 60 min at room temperature. These concentrations were based on the maximal inhibition of the membrane bound F1Fo ATP synthase (Fig. 2). Then, 50 mM TrisSO4 pH 8.0 buffer was added to reduce the venom peptide concentration to near non-inhibitory levels and incubation continued for an additional 60 min at room temperature before the ATPase assay. Control samples without venom peptides were incubated for the same time periods as the samples with venom peptides.
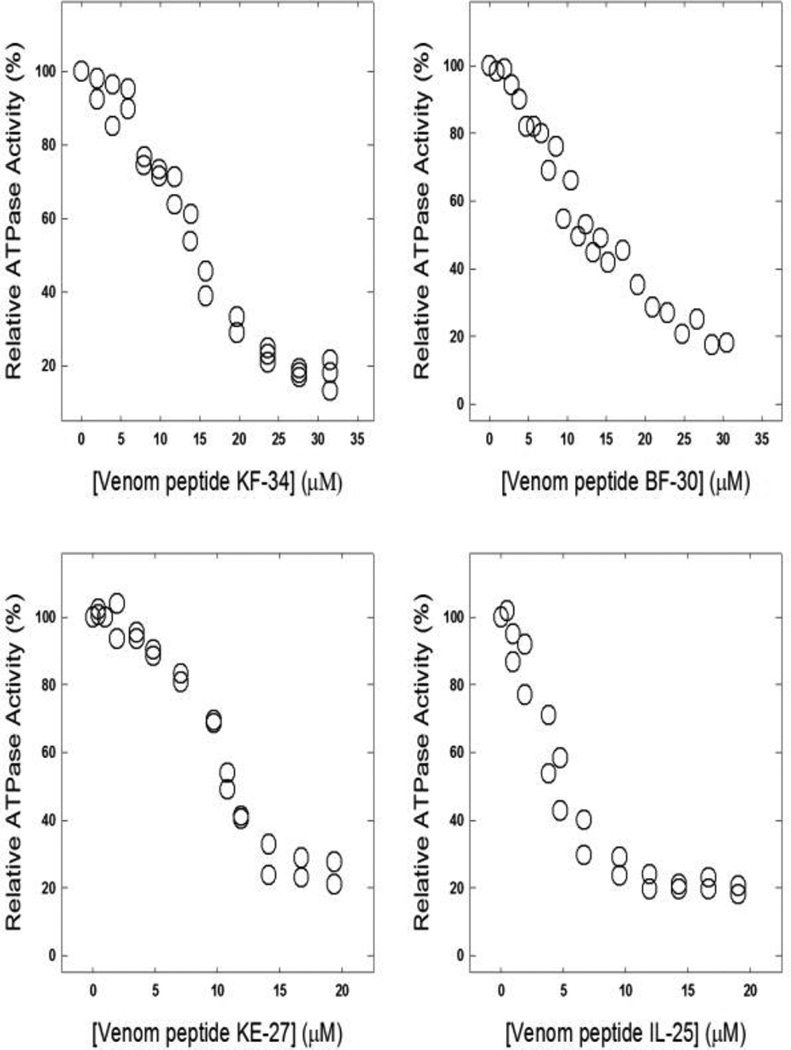
Fig. 2. Inhibition of membrane bound wild-type F1Fo ATP synthase by venom peptides.
Membrane bound F1Fo ATP synthase was preincubated for 60 min at room temperature with varied concentrations of venom peptides, OH-CATH, (KF-34), cathelicidin (BF-30), lycotoxin I (IL-25), or lycotoxin II (KE-27). The Materials and Methods section contains the detailed procedure. Data points are the average of three to four experiments, using 2–3 independent membrane preparations. Results agreed within ± 5%.
Generation of helix-wheel plots
Expert Protein Analysis System (ExPASy) software package (http://heliquest.ipmc.cnrs.fr/) was used to generate the helix wheel plots in Figure 1 [41].
Results
Structural characteristics of KF-34, BF-30, IL-25, and KE-27 venom peptides
Amino acid sequences of KF-34, BF-30, IL-25, and KE-27 venom peptides are shown in Table 1. Figure 1 shows the helix-wheel plots of these cationic amphipathic peptides. The α-helical structures can be seen segregating into hydrophilic charged residues on one side and hydrophobic residues on the other side.
Venom peptide KF-34, BF-30, IL-25, and KE-27 induced inhibition of wild-type membrane bound E. coli F1Fo ATP synthase
Previously, several peptides were shown to bind and inhibit E. coli ATP synthase [18, 19, 42, 43]. The lack of desired success in the clinical development of combinatorial chemistry and small molecule based therapies has brought the increased interest in AMPs including venom peptides [44]. Venom peptides evidently offer important therapeutic viable alternatives to conventional antibiotics [25, 45]. For this reason we embarked on to study the venom peptide induced inhibition of ATP synthase. As shown in Fig. 2, KF-34, BF-30, IL-25, and KE-27 caused strong inhibition of ATP synthase. The maximal inhibition induced by KF-34 was ~90%, by BF-30 was ~85%, by IL-25 was ~85%, and by KE-27 was ~88%.
Reversal of membrane bound F1Fo ATPase activity from venom peptide KF-34, BF-30, IL-25, and KE-27 inhibition
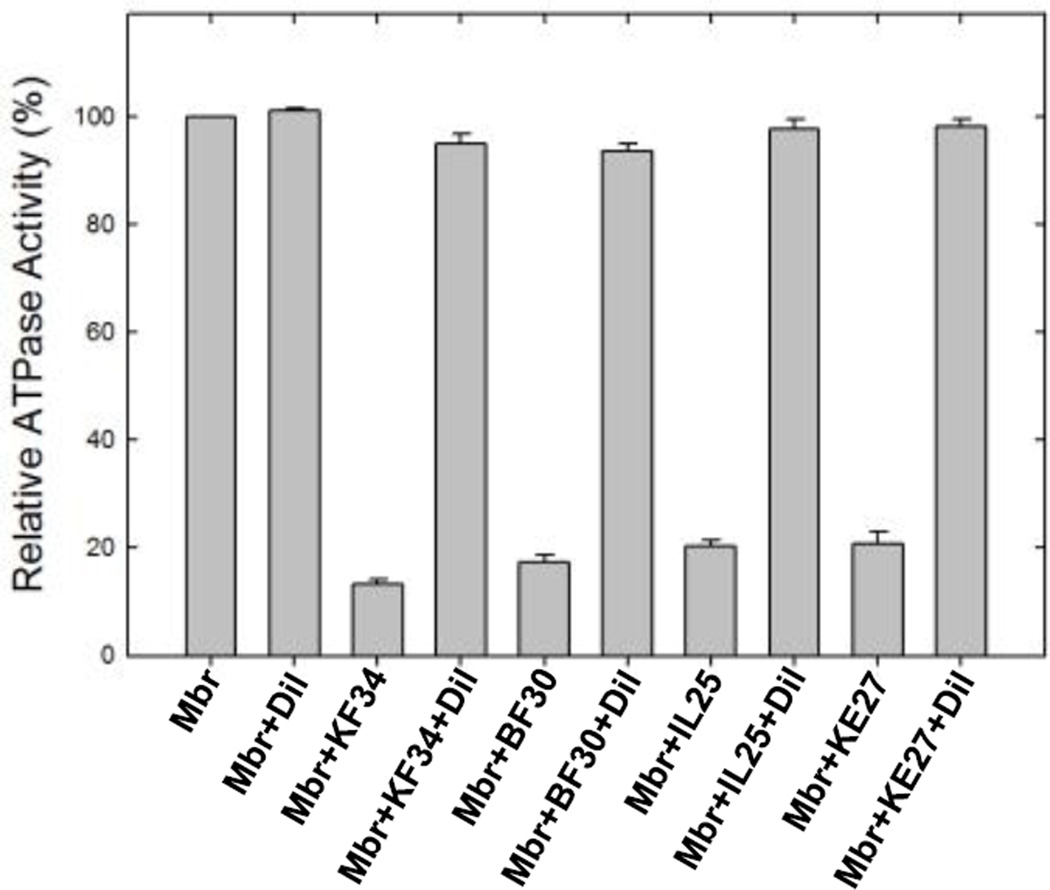
Venom peptide KF-34, BF-30, IL-25, and KE-27 induced inhibition of ATP synthase was found to be fully reversible (Fig. 3) Membrane bound F1Fo ATP synthase was inhibited with the maximum inhibitory concentrations of venom peptides for one hour at RT as in Fig. 2. Samples were then diluted to a non-inhibitory concentration by adding TrisSO4 pH 8.0 buffer before the measurement of ATPase activity. Reversibility data indicates that the observed inhibition is not the result of protein denaturation and that the ATP synthase reactivates after dilution of the venom peptides, suggesting non-covalent interaction between venom peptides and ATP synthase.
Fig. 3. Reversal of ATPase activity from venom peptide induced inhibition.
Membrane bound F1Fo ATP synthase (Mbr) was inhibited with inhibitory concentrations of venom peptides as shown in the figure for 60 min under conditions as described in Fig 2 and in the Materials and Methods section. TrisSO4 pH 8.0 buffer was added to bring back the venom peptide concentrations to a lower level and activity was measured. Nomenclature used are: Mbr, membranes; Dil, dilution; KF34, OH-CATH; BF30, cathelicidin; IL25, lycotoxin I; IL27, lycotoxin II.
Inhibition of E. coli cell growth in the presence of venom peptide KF-34, BF-30, IL-25, or KE-27
Both wild-type and mutant E. coli strains were grown on LB, succinate, and limiting glucose media in the presence or absence of venom peptides KF-34, BF-30, IL-25, or KE-2. (Table 2). While the venom peptides potently inhibited the growth of wild-type E. coli strain (pBWU13.4/DK8), the null strain (pUC118) was only partially inhibited.
Table 2.
Effect of venom peptides on the growth of Escherichia coli cells
| Presence/absence of venom peptides |
aGrowth on limiting glucose (%) |
bGrowth on succinate (%) |
|---|---|---|
| WT (pBWU13.4) | 100 | 100 |
| Null (pUC118) | 46±4 | 3±2 |
| WT+KF-34 | 0±3 | 1±3 |
| Null + KF-34 | 24±4 | 2±2 |
| WT + BF-30 | 2±4 | 2±3 |
| Null + BF-30 | 23±3 | 3±3 |
| WT + IL-25 | 1±4 | 1±2 |
| Null + IL-25 | 24±4 | 3±3 |
| WT + IL-27 | 0±3 | 1±3 |
| Null + IL-27 | 22±4 | 3±2 |
Growth yield on limiting glucose was measured as OD595 after ~20 hours growth at 37 °C.
Growth on succinate medium after 72 hours was determined by OD595
All experiments were done at least three times at 37 °C. Individual experimental points are average of duplicate assays.
Discussion
Venom and its components have a long history of use in traditional medicines. Venom from reptiles has been used for the treatment of arthritis, gastrointestinal ailments, asthma, cancer, polio, multiple sclerosis, rheumatism and pain [46, 47]. The success of the venom peptide based anti-hypertensive drug captopril has brought the venom peptide to the forefront of research and drug discovery [48]. Moreover, the high rate of microbial resistance against common and widely used antibiotics necessitates the development of alternative approaches to combat microbial infections. Higher chemical and thermal stability of venom peptides along with selectivity and potency makes them valuable lead molecules for the development of novel therapeutics.
Interest in the use of peptides as antimicrobial and antitumor agents is steadily growing. Several peptides including venom peptides are being used or are under clinical trials as potential anti-tumor or antimicrobial agents [23, 26–29, 31, 32, 49]. The goal of this study was to determine if the antimicrobial properties of venom peptides are linked to the inhibition of ATP synthase. Therefore, we examined the inhibitory effects of KF-34, BF-30, IL-25, and KE-27 on ATPase activity and on growth of E. coli strains.
Previously, a number of α-helical amphipathic cationic peptides were shown to bind and inhibit ATPase activity to variable degrees [13, 18, 43, 50, 51]. It was also documented that peptides bind at the highly conserved βDELSEED-motif of ATP synthase [19]. All four venom peptides used in this study potently inhibited membrane bound F1Fo ATP synthase (between 85–90%, Fig. 2). It is interesting to note that in previous studies, many natural and synthetic molecules including peptides caused nil, partial, or total inhibition of ATP synthase [18, 52, 53]. To make sure that the maximal observed inhibition is the true inhibition in the presence of venom peptides, the maximally inhibited samples were re-inhibited. Additional inhibition of already inhibited membrane bound F1Fo by KF-34 (30µM), BF-30 (30µM), IL-25 (20µM), or KE-27 (20 µM) did not change the extent of inhibition to a meaningful level. This suggests that the left over 10–15% residual activity was not the result of uninhibited enzyme or due to the degradation of the venom peptides during the process of inhibition.
KF-34, BF-30, IL-25, and KE-27 induced inhibition of membrane bound F1Fo ATP synthase was found to be fully reversible. After having been inhibited with venom peptides at higher concentration, membrane bound enzyme regained activity once returned to lower concentration of venom peptide by dilution with buffer. This process of reversibility shows that the venom peptides bind non-covalently to the peptide binding site.
Venom peptides have been shown to potently disrupt the growth of many drug-resistant pathogenic bacterial species, such as Salmonella typhi, Bacillus subtilis, Enterobacter cloacae Escherichia coli, Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus, [25, 54] and yeast (Candida albicans, Candida glabrata) [54, 55]. Damage to cytoplasmic membranes [25], blocking of membrane bound sodium, calcium, potassium, and chloride channels, nicotinic acetylcholine receptors, or noradrenaline transporters [49], and inhibition of membrane bound F1Fo ATP synthase [18] were proposed and shown to be the cause for the bactericidal activity. The cationic amphipathic α-helical structure of venom peptides, with all charged residues on one side and hydrophobic residues on the other side, is highly suitable to interact simultaneously with the βDELSEED-motif from one side and to the membrane from other side (Fig 1).
Our growth inhibition assay of wild-type (pBWU13.4/DK8) and null (pUC118/DK8) E. coli strains (Table 2) provides a strong and interesting argument regarding possibility of more than one target and the synergetic mode of action for the bactericidal effect of venom peptides. It can be seen in Table 2 that while wild-type E. coli is potently inhibited by venom peptides, the null E. coli strain still grows ~23±1% in limiting glucose in presence of venom peptides which is ~50% of the total growth of null strain in limiting glucose. In limiting glucose, null strain normally grows in between 40–50% in comparison to the wild-type. This is due to the use of glycolytic pathway to generate ATP. As can be seen in Table 2, the null strain shows no significant growth in the nonfermentable carbon source succinate due to the absence of ATP synthase. Wild-type on the other hand, uses glycolysis, TCA, and oxidative phosphorylation, and therefore grows well in both the fermentable carbon source glucose and non-fermentable carbon source succinate.
Venom peptides reduced wild-type growth from ~100% to nearly 0% in both limiting glucose and succinate media, but the null strain growth is reduced from 46% to only ~23%. Potent inhibition of growth in the wild-type can be attributed to the loss of oxidative phosphorylation through inhibition of ATP synthesis and damage to the membrane by venom peptides. In the absence of ATP synthase venom peptides cause only limited damage to cell membrane. Thus, partial growth retention in the null E. coli strain clearly suggests that ATP synthase is a molecular target for the venom peptides. Thus, the complete bactericidal effect requires inhibition of ATP synthase. These results establish that the venom peptide induced inhibition of bacteria is at least in part through their binding and inhibition of ATP synthase.
Peptides are an essential part of natural defense mechanisms for many animals and microorganisms. Cationic amphipathic 10 to 50 amino acid residues long AMPs have been observed to possess broad spectrum antimicrobial activity [56]. It was also demonstrated that the peptides bind at the βDELSEED-motif of ATP synthase [18, 19]. Based on the KF-34, BF-30, IL-25, and KE-27 induced inhibition of membrane bound F1Fo ATP synthase and potent inhibition of bacterial growth we conclude that the bactericidal properties of venom peptides are linked to their inhibitory effects on ATP synthase. Moreover, targeting ATP synthase by venom peptides seems to be an effective alternative approach to combat drug resistant bacterial strains.
Acknowledgments
This work was supported by the National Institutes of Health Grant GM085771 to ZA. ZA is grateful to Dr. Margaret Wilson, dean of KCOM, ATSU for the funding to purchase French Press.
Abbreviations used
- KF34
OH-CATH
- BF30
cathelicidin
- IL25
lycotoxin I
- KE27
lycotoxin II
- Membrane or Mbr
membrane bound F1Fo ATP synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Senior AE. Cell. 2007;130:220–221. doi: 10.1016/j.cell.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Senior AE. J Biol Chem. 2012;287:30049–30062. doi: 10.1074/jbc.X112.402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber J, Senior AE. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad Z, Cox JL. The Scientific World Journal. 2014;2014:10. [Google Scholar]
- 5.Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV. Proc Natl Acad Sci U S A. 2001;98:6656–6661. doi: 10.1073/pnas.131067798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moser TL, Stack MS, Asplin I, Enghild JJ, Hojrup P, Everitt L, Hubchak S, Schnaper HW, Pizzo SV. Proc Natl Acad Sci U S A. 1999;96:2811–2816. doi: 10.1073/pnas.96.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrell HE, Wlodarski B, Foster BJ, Buckley KA, Sharpe GR, Quayle JM, Simpson AW, Gallagher JA. J Biol Chem. 2005;280:29667–29676. doi: 10.1074/jbc.M505381200. [DOI] [PubMed] [Google Scholar]
- 8.Kim BW, Choo HJ, Lee JW, Kim JH, Ko YG. Exp Mol Med. 2004;36:476–485. doi: 10.1038/emm.2004.60. [DOI] [PubMed] [Google Scholar]
- 9.Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezon E, Champagne E, Pineau T, Georgeaud V, Walker JE, Terce F, Collet X, Perret B, Barbaras R. Nature. 2003;421:75–79. doi: 10.1038/nature01250. [DOI] [PubMed] [Google Scholar]
- 10.Huang TC, Chang HY, Hsu CH, Kuo WH, Chang KJ, Juan HF. J Proteome Res. 2008;7:1433–1444. doi: 10.1021/pr700742h. [DOI] [PubMed] [Google Scholar]
- 11.Chang HY, Huang HC, Huang TC, Yang PC, Wang YC, Juan HF. Cancer Res. 2012;72:4696–4706. doi: 10.1158/0008-5472.CAN-12-0567. [DOI] [PubMed] [Google Scholar]
- 12.Hong S, Pedersen PL. Microbiol Mol Biol Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad Z, Laughlin TF. Curr Med Chem. 2010;17:2822–2836. doi: 10.2174/092986710791859270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SP, Alonso S, Rand L, Dick T, Pethe K. Proc Natl Acad Sci U S A. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Ramirez VD. Br J Pharmacol. 2000;130:1115–1123. doi: 10.1038/sj.bjp.0703397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen PL. J Bioenerg Biomembr. 2007;39:1–12. doi: 10.1007/s10863-007-9070-5. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Vik SB, Tu Y. J Nutr Biochem. 2012;23:953–960. doi: 10.1016/j.jnutbio.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laughlin TF, Ahmad Z. Int J Biol Macromol. 2010;46:367–374. doi: 10.1016/j.ijbiomac.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad Z, Tayou J, Laughlin TF. Int J Biol Macromol. 2015;75:37–43. doi: 10.1016/j.ijbiomac.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoek ML. Pharmaceuticals (Basel) 2014;7:723–753. doi: 10.3390/ph7060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Li X, Wang Z. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen OE, Borregaard N. Comb Chem High Throughput Screen. 2005;8:273–280. doi: 10.2174/1386207053764602. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhao H, Yu GY, Liu XD, Shen JH, Lee WH. Peptides. 2010;31:1488–1493. doi: 10.1016/j.peptides.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Gan TX, Liu XD, Jin Y, Lee WH, Shen JH, Zhang Y. Peptides. 2008;29:1685–1691. doi: 10.1016/j.peptides.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Li SA, Lee WH, Zhang Y. Antimicrob Agents Chemother. 2012;56:3309–3317. doi: 10.1128/AAC.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Hong J, Liu X, Yang H, Liu R, Wu J, Wang A, Lin D, Lai R. PLoS One. 2008;3:e3217. doi: 10.1371/journal.pone.0003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Dou J, Wang J, Chen L, Wang H, Zhou W, Li Y, Zhou C. Peptides. 2011;32:1131–1138. doi: 10.1016/j.peptides.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Li B, Li Y, Dou J, Hao Q, Tian Y, Wang H, Zhou C. Arch Pharm Res. 2014;37:927–936. doi: 10.1007/s12272-013-0248-6. [DOI] [PubMed] [Google Scholar]
- 29.Wu D, Gao Y, Qi Y, Chen L, Ma Y, Li Y. Cancer Lett. 2014;351:13–22. doi: 10.1016/j.canlet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Shadidi M, Sioud M. Drug Resist Updat. 2003;6:363–371. doi: 10.1016/j.drup.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Ke M, Tian Y, Wang J, Li B, Wang Y, Dou J, Zhou C. Eur J Pharmacol. 2013;707:1–10. doi: 10.1016/j.ejphar.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Yan L, Adams ME. J Biol Chem. 1998;273:2059–2066. doi: 10.1074/jbc.273.4.2059. [DOI] [PubMed] [Google Scholar]
- 33.Hughes SR, Dowd PF, Johnson ET. Pharmaceuticals (Basel) 2012;5:1054–1063. doi: 10.3390/ph5101054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brogden KA. Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 35.Vaara M. Curr Opin Pharmacol. 2009;9:571–576. doi: 10.1016/j.coph.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Ketchum CJ, Al-Shawi MK, Nakamoto RK. Biochem. J. 1998;330:707–712. doi: 10.1042/bj3300707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senior AE, Latchney LR, Ferguson AM, Wise JG. Arch Biochem Biophys. 1984;228:49–53. doi: 10.1016/0003-9861(84)90045-6. [DOI] [PubMed] [Google Scholar]
- 38.Senior AE, Langman L, Cox GB, Gibson F. Biochem. J. 1983;210:395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taussky HH, Shorr E. J Biol Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- 40.Ahmad Z, Laughlin TF, Kady IO. PLoS One. 2015;10:e0127802. doi: 10.1371/journal.pone.0127802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier R, Douguet D, Antonny B, Drin G. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 42.Ashby M, Petkova A, Hilpert K. Curr Opin Infect Dis. 2014;27:258–267. doi: 10.1097/QCO.0000000000000057. [DOI] [PubMed] [Google Scholar]
- 43.Lu J, Chen ZW, Wu Y, Zhang M, Ding JP, Cederlund E, Jornvall H, Bergman T. Biochem Biophys Res Commun. 2014;446:519–522. doi: 10.1016/j.bbrc.2014.02.138. [DOI] [PubMed] [Google Scholar]
- 44.Lewis RJ, Dutertre S, Vetter I, Christie MJ. Pharmacol Rev. 2012;64:259–298. doi: 10.1124/pr.111.005322. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Parente J, Harris SM, Woods DE, Hancock RE, Falla TJ. Antimicrob Agents Chemother. 2005;49:2921–2927. doi: 10.1128/AAC.49.7.2921-2927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King GF. Expert Opin Biol Ther. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- 47.Reid PF. Crit Rev Immunol. 2007;27:291–302. doi: 10.1615/critrevimmunol.v27.i4.10. [DOI] [PubMed] [Google Scholar]
- 48.Cushman DW, Ondetti MA. Hypertension. 1991;17:589–592. doi: 10.1161/01.hyp.17.4.589. [DOI] [PubMed] [Google Scholar]
- 49.Lewis RJ, Garcia ML. Nat Rev Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 50.Ahmad Z, Okafor F, Azim S, Laughlin TF. Curr Med Chem. 2013;20:1956–1973. doi: 10.2174/0929867311320150003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bullough DA, Ceccarelli EA, Roise D, Allison WS. Biochim Biophys Acta. 1989;975:377–383. doi: 10.1016/s0005-2728(89)80346-9. [DOI] [PubMed] [Google Scholar]
- 52.Ahmad Z, Ahmad M, Okafor F, Jones J, Abunameh A, Cheniya RP, Kady IO. Int J Biol Macromol. 2012;50:476–486. doi: 10.1016/j.ijbiomac.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Int J Biol Macromol. 2010;46:478–486. doi: 10.1016/j.ijbiomac.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen W, Yang B, Zhou H, Sun L, Dou J, Qian H, Huang W, Mei Y, Han J. Peptides. 2011;32:2497–2503. doi: 10.1016/j.peptides.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Adao R, Seixas R, Gomes P, Pessoa JC, Bastos M. J Pept Sci. 2008;14:528–534. doi: 10.1002/psc.993. [DOI] [PubMed] [Google Scholar]
- 56.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]