Abstract
The lncRNA HOTAIR has been implicated in several human cancers. Here, we evaluated the molecular alterations and upstream regulatory mechanisms of HOTAIR in glioma, the most common primary brain tumors, and its clinical relevance. HOTAIR gene expression, methylation, copy-number and prognostic value were investigated in human gliomas integrating data from online datasets and our cohorts. High levels of HOTAIR were associated with higher grades of glioma, particularly IDH wild-type cases. Mechanistically, HOTAIR was overexpressed in a gene dosage-independent manner, while DNA methylation levels of particular CpGs in HOTAIR locus were associated with HOTAIR expression levels in GBM clinical specimens and cell lines. Concordantly, the demethylating agent 5-Aza-2′-deoxycytidine affected HOTAIR transcriptional levels in a cell line-dependent manner. Importantly, HOTAIR was frequently co-expressed with HOXA9 in high-grade gliomas from TCGA, Oncomine, and our Portuguese and French datasets. Integrated in silico analyses, chromatin immunoprecipitation, and qPCR data showed that HOXA9 binds directly to the promoter of HOTAIR. Clinically, GBM patients with high HOTAIR expression had a significantly reduced overall survival, independently of other prognostic variables. In summary, this work reveals HOXA9 as a novel direct regulator of HOTAIR, and establishes HOTAIR as an independent prognostic marker, providing new therapeutic opportunities to treat this highly aggressive cancer.
Keywords: HOTAIR, glioblastoma, glioma, prognosis, HOXA9
INTRODUCTION
Gliomas encompass the majority of primary central nervous system tumors, and have been historically classified based on their histopathological features, according to four grades of malignancy (World Health Organization – WHO – grade I-IV). Glioblastoma (GBM; WHO grade IV) represents the most common and malignant primary adult brain tumor [1], with a median patient overall survival (OS) of approximately 15 months [2]. The remarkable tumor heterogeneity, invasive phenotype, and resistance to radio- and chemotherapy are main determinants of this poor prognosis [3–5]. Additionally, several molecular alterations (epigenetic, genetic, transcriptional and proteomic) contribute to GBM tumorigenesis and may represent putative biomarkers of GBM prognosis and aggressiveness [6].
Recent studies have explored the complexity of the transcriptome and reported the existence of a large number of transcripts that do not code for a protein, which nonetheless have the critical ability to regulate gene expression [7]. Among others, these transcripts include long non-coding RNAs (lncRNA) [8, 9] that are defined as spliced, polyadenylated and >200 nucleotides long RNAs that modify the genome in a highly tissue-specific manner [9, 10]. LncRNA have also emerged as new players in cancer development and progression, with recent studies highlighting the importance of both tumor suppressor and oncogenic lncRNAs in the deregulation of cellular pathways and gene expression programs [11]. The HOX Transcript Antisense Intergenic RNA (HOTAIR) is a trans-acting lncRNA, involved in the genome-wide reprogramming of the chromatin [10]. HOTAIR is transcribed at the HOXC locus in chromosome 12 (between HOXC11 and HOXC12), and its 5′-domain interacts with the Polycomb Repressor Complex 2 (PRC2), and the 3′-domain interacts with the lysine specific demethylase 1/REST corepressor 1/RE1-silencing transcription factor (LSD1/CoREST/REST) complex. Thus, HOTAIR can act as modular scaffold, with different binding sites for harboring demethylase (LSD1, that mediates the enzymatic demethylation of histone H3 dimethyl lysine 4, H3K4me2) and methylase (PRC2 and the histone H3 lysine 27 – H3K27 – methylase EZH2) complexes, resulting in the transcriptional inhibition of the HOXD locus on chromosome 2 by epigenetic control of the chromatin structure [10, 12].
HOTAIR has also been shown to have functional roles in a variety of human cancers (reviewed in [13–15]), being associated with patients’ decreased OS, increased metastatic potential, tumor recurrence and chemotherapy resistance. In glioma, HOTAIR was reported to be involved in proliferation, invasion, cell cycle and colony formation ability, in vivo tumor growth, and GBM patients’ OS [16–20]. However, the molecular mechanisms underlying its aberrant activation in glioma remain elusive. In this context, we aimed to investigate the integrated molecular status (gene copy number, expression, and DNA methylation levels) of HOTAIR in glioma samples, unravel the underlying mechanisms regulating HOTAIR expression in these tumors, and assess its clinical significance in independent large cohorts of GBM.
RESULTS
HOTAIR expression is associated with high-grade gliomas
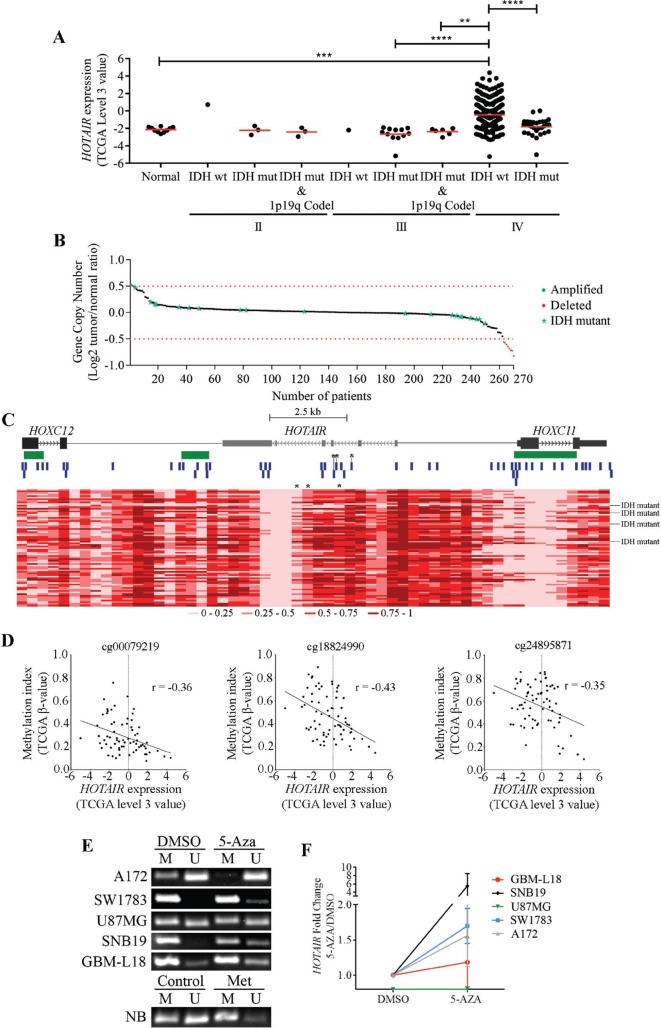
In a first approach, we analyzed the expression levels of HOTAIR in grades II and III gliomas, and in grade IV GBM patients using gene expression microarray data and RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA). Globally, HOTAIR was highly expressed in GBM, particularly in IDH-wt GBM, as compared to lower-grade gliomas and normal brain (Figure 1A and Supplementary Figure 1A). Of note, in the RNA-seq data, IDH-wt grade III gliomas presented higher HOTAIR expression than the remaining grade II and III gliomas entities (Supplementary Figure 1A). Concordantly, four additional datasets from the Oncomine database (Murat [21], Freije [22], Sun [23], and Phillips [24]) and two independent Portuguese and French glioma series, confirmed that HOTAIR expression is far more frequent in grades III and IV than in grade II gliomas and normal brain (Supplementary Table 1 and Supplementary Figure 1B). In all tested glioma datasets, HOTAIR expression was not significantly associated with any other clinicopathological feature (patient age, sex, Karnofsky performance status [KPS], and exposure to radiotherapy or chemotherapy, except for associations with patient age in Phillips [24], Repository of Molecular Brain Neoplasia Data (REMBRANDT) and the French datasets, and associations with IDH mutation status in the French datatset; data not shown). Moreover, HOTAIR expression was not associated with any particular GBM molecular subtype as defined by Wang et al. [25] (Supplementary Figure 1C and 1D; p > 0.9999 for microarray and RNA-seq data). Together, these data suggest that HOTAIR is highly expressed in high-grade gliomas, being particularly frequent in IDH-wt cases.
Figure 1. Molecular characterization of HOTAIR in gliomas.
(A) Expression levels of HOTAIR in 424 gliomas, stratified according to WHO grade, IDH and 1p/19q codeletion statuses (1 IDH-wt, 3 IDH-mut, and 3 IDH-mut and 1p/19q codeleted grade II gliomas; 1 IDH-wt, 12 IDH-mut, and 6 IDH-mut and 1p/19q codeleted grade III gliomas; 368 IDH-wt and 30 IDH-mut glioblastomas (GBM); and 10 unmatched normal brains from the TCGA microarray data). HOTAIR is highly expressed (TCGA data “level 3” values ≥ 0) in 34.2% (n = 126) of IDH-wt GBM samples and in 1 IDH-mut GBM (3%) and 1 IDH-wt grade II glioma (100%). (B) HOTAIR gene copy number status in 270 GBMs (250 IDH-wt and 20 IDH-mut) from TCGA. HOTAIR is amplified (Log2 Copy Number Tumor/Normal ≥ 0.5) in 0.8% (n = 2; green dots), and deleted (Log2 Copy Number Tumor/Normal ≤ –0.5) in 3.2% (n = 8; red dots) of IDH-wt GBM samples. Red dashed lines represent the normal copy number interval. (C) Heatmap representations of DNA methylation levels (TCGA β-values) of the chromosomal region encompassing HOTAIR and the 2 closest genes (HOXC12 and HOXC11) in 74 GBMs (70 IDH-wt and 4 IDH-mut) from TCGA. A total of 56 methylation probes (vertical blue bars) were assessed. CpG islands > 300bp are represented in green. *indicate probes whose methylation indexes are significantly inversely correlated with HOTAIR expression levels (probes cg00079219, cg18824990 and cg24895871). The color code (grades of red color corresponding to different methylation indexes) is shown below the heatmap. Each column corresponds to a probe and each row to a patient. (D) Correlation graphs between HOTAIR expression levels (TCGA “level 3” value) and DNA methylation indexes (TCGA β-values) in 70 GBM samples. Only probes whose methylation values are significantly inversely correlated with HOTAIR expression are shown (cg00079219, cg18824990 and cg24895871; marked with *in C). (E–F) Glioma cell lines were treated with 5 µM 5-Aza for 72 hours, upon which promoter methylation status (E) and HOTAIR expression levels (F) were evaluated. 5-Aza treatment promoted HOTAIR promoter demethylation (E) that associated with its increased expression in a cell line-dependent manner (F). qPCR levels were normalized to the expression of HPRT1 and are presented as fold-changes; methylation-specific PCR was controlled by blood DNA (NB) untreated (Control) or in vitro methylated (Met). No detectable HOTAIR expression was found for U87 (untreated or 5-Aza-treated). The results are representative of at least 2 replicates (mean ± SD). *p < 0.05; NB - DNA from normal blood.
DNA methylation regulates HOTAIR levels in glioma
In order to understand the molecular mechanisms underlying HOTAIR increased expression in GBMs, we analyzed gene copy number aberrations (Figure 1B) and DNA methylation levels in the HOTAIR locus (Figure 1C). In 250 IDH-wt GBMs analyzed for gene copy number, HOTAIR amplification and deletion (either focal or chromosomal) were found in 0.8% (2/250) and in 3.2% of the samples (8/250; Figure 1B), respectively, while no amplifications or deletions were found in 20 IDH-mut GBMs (Figure 1B). While HOTAIR levels were high in 3 of the 5 HOTAIR-amplified tumors, it was also present in 1 tumor with gene deletion and in 116 without copy number aberrations (Figure 1B). These data show HOTAIR copy number aberrations are rare in GBM, and unlikely associated with its increased expression levels (p = 0.182). Analysis of 56 methylation probes spanning from HOTAIR’s most proximal genes (HOXC11 and (HOXC12) revealed a wide spectrum of methylation levels across different GBM samples (Figure 1C). Of note, some probes consistently presented similar methylation levels across all samples (e.g., several probes within the HOTAIR intragenic region consistently showed low methylation levels), suggesting those regions are not prone to de novo methylation in GBM. In a subset of 70 GBMs (66 IDH-wt and 4 IDH-mut) for which both gene expression and methylation data were available, we found the methylation levels of 3 probes located in the intragenic region of HOTAIR (cg00079219, cg18824990 and cg24895871) to be significantly inversely correlated with HOTAIR expression levels (r = –0.36, p = 0.002; r = –0.43, p = 0.0002; and r = –0.35, p = 0.003, respectively; Figure 1D). Of note, performing the same analysis exclusively for the IDH-wt GBMs showed that the methylation levels of the probes cg00079219 and cg18824990 were inversely correlated with HOTAIR expression (r = –0.39, p = 0.0002; r = –0.39, p = 0.0014), while methylation levels at the probe cg24895871 lost their association with expression (r = –0.26, p = 0.167). We further corroborated the relevance of these associations in IDH-wt GBM patients in the French dataset as the 3 intragenic methylation probes were also found to inversely correlate with HOTAIR expression (cg00079219 r = –0.53, p = 0.016; cg18824990 r = –0.523, p = 0.017; cg24895871 r = –0.521, p = 0.018), and in the combination of IDH-wt glioma grade III and IDH-wt GBM patients (cg00079219 r = -0.43, p = 0.004; cg18824990 r = –0.453, p = 0.002; cg24895871 r = −0.431, p = 0.004).
To further explore the relevance of DNA methylation in HOTAIR regulation, we treated glioma cell lines with the global DNA demethylating agent 5-Aza-2′-deoxycytidine (5-Aza) followed by methylation-specific PCR (MSP) and quantitative PCR (qPCR) analyses (Figure 1E and 1F). Interestingly, HOTAIR intragenic CpGs were found to be mostly methylated in untreated glioma cells. In addition, 5-Aza treatment resulted in DNA demethylation (Figure 1E) and associated increase in HOTAIR transcriptional levels in most but not all of the cell lines (Figure 1F), suggesting a cell line-dependent regulation. Together, our data suggest an association between methylation and HOTAIR’s expression in GBM patients and glioma cell lines, but other mechanisms may be crucial in reactivating HOTAIR in these tumors.
HOXA9 directly binds the HOTAIR promoter in GBM cells
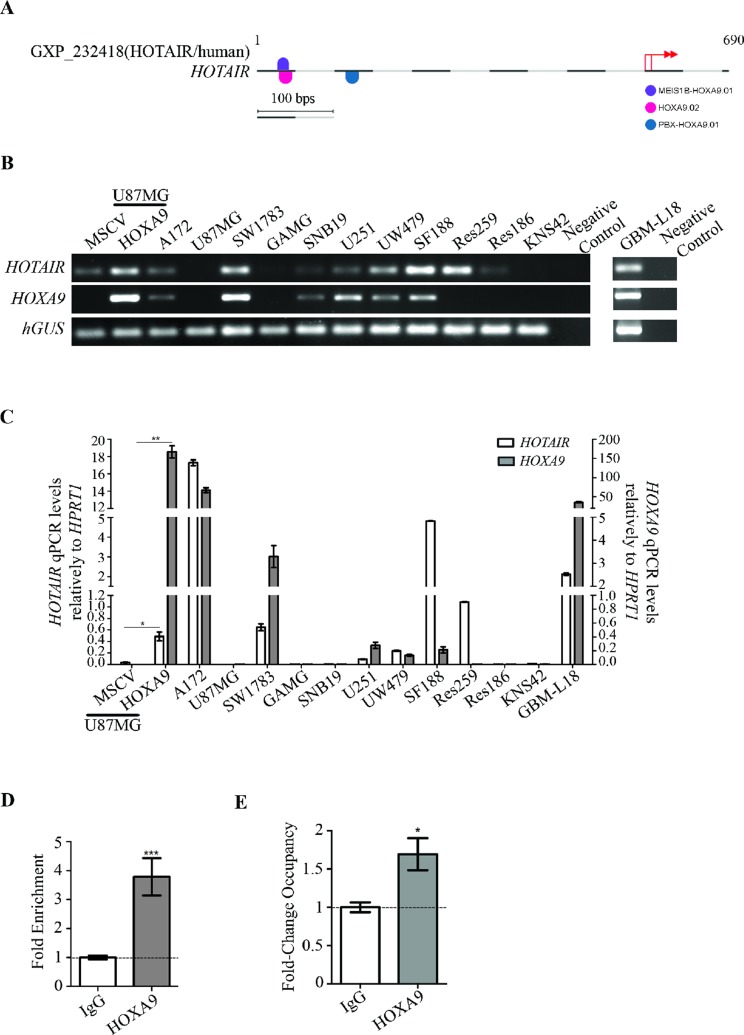
Previously published transcriptomic data from our group (GEO accession number GSE56517) [26, 27] suggested a possible regulation of HOTAIR by the homeoprotein HOXA9, an important protein in the aggressiveness, chemotherapy resistance, and prognosis of GBM [21, 26, 28, 29]. Specifically, GBM cell lines with overexpression or silencing of HOXA9 presented increased or reduced expression of HOTAIR, respectively; raising the hypothesis that HOXA9 may directly regulate HOTAIR expression. Indeed, an in silico analysis using MatInspector (Genomatix) revealed 3 putative binding sites for HOXA9 with high matrix similarities to HOXA9.02 (0.899), PBX-HOXA9.01 (0.849), and MEIS1B-HOXA9.01 (0.837) matrixes (Figure 2A), suggesting that HOXA9 may bind to the promoter region of HOTAIR and regulate its transcription. To test this, we studied the expression pattern of HOTAIR and HOXA9 in adult and pediatric glioma-derived cell lines by semi-quantitative reverse transcription-PCR (RT-PCR) and qPCR, and found these genes to be frequently co-expressed in glioma (Figure 2B and 2C). Moreover, U87MG-HOXA9 cells (with exogenous overexpression of HOXA9) presented significantly increased expression of HOTAIR as compared to their HOXA9-negative counterpart (U87MG-MSCV; Figure 2B and 2C). We then performed anti-HOXA9 chromatin-immunoprecipitation (ChIP) on U251 GBM cell line (endogenously expressing HOTAIR and HOXA9), U87MG-HOXA9 (HOXA9 overexpression models) and their HOXA9-negative counterparts. In both U251 and U87MG-HOXA9, ChIP-qPCR revealed significantly increased HOXA9 occupancy of the promoter region of HOTAIR in comparison to their respective controls (Figure 2D and 2E; p < 0.0002 for U251; p = 0.0148 for U87MG-HOXA9), confirming that HOXA9 directly interacts with the promoter region of HOTAIR. Together, these results indicate that HOXA9 is a direct activator of HOTAIR expression in GBM cells, thus supporting the strong co-expression of these genes observed in glioma cell lines.
Figure 2. HOXA9 transcriptionally activates HOTAIR via direct interaction with its promoter region.
(A) MatInspector representation of specific binding sites for HOXA9 matrix in HOTAIR promoter region. Each matrix match is represented by a half round symbol and each color symbolizes a matrix family. Pink, blue and violet colors represent the matrixes HOXA9.02 (matrix sim = 0.899, sequence: ctgtgacaTAAAattgg and family ABDB), PBX-HOXA9.01 (matrix sim = 0.849, sequence: gaggTGGTttatgagct and family HOXC) and MEIS1B-HOXA9.01 (matrix sim = 0.837, sequence: TGCCaattttatgtc and family HOXH), respectively. Basepairs in italic appear in a position with a high conservation profile in the matrix (ci-value > 60). Basepairs in capital letters represent the core sequence used by the program. Matches represented on the top of the sequence line were found on the positive strand, while below the sequence line reside matches found on the negative strand. The red arrows represent HOTAIR putative transcription start sites (TSS). (B–C) HOTAIR and HOXA9 expression were evaluated by reverse-transcriptase PCR (B) and quantitative PCR (C) in a panel of adult and pediatric glioma-derived cell lines, and in the GBM-L18 primary GBM-derived cell culture. GBM cell line U87MG-MSCV does not present detectable levels of endogenous HOTAIR expression, which were significantly increased upon retrovirally-mediated HOXA9 overexpression (U87MG-HOXA9). White and grey bars represent HOTAIR and HOXA9 expressions, respectively. qPCR levels were normalized to the expression of HPRT1. The results are representative of triplicates (mean ± SD). *p = 0.029; **p = 0.0088. (D–E) The putative binding of HOXA9 protein to the promoter region of HOTAIR was assessed by chromatin immunoprecipitation (ChIP) analysis followed by quantitative PCR in U251 cells (D), and U87MG-HOXA9 and their HOXA9-negative counterparts (E). IgG was used as negative control for the ChIP. Chromatin immunoprecipitated with an anti-HOXA9 antibody shows direct binding of HOXA9 to the HOTAIR promoter. Relative enrichment is normalized to input DNA (not subjected to immunoprecipitation) and to the IgG background signal (D), whereas fold change occupancy is normalized to input, IgG and HOXA9-negative cells (E), from three independent experiments (mean ± SD). (D) ***p = 0.0002; (E) *p = 0.0148.
Co-expression of HOTAIR and HOXA9 is a frequent event in clinical specimens of glioma
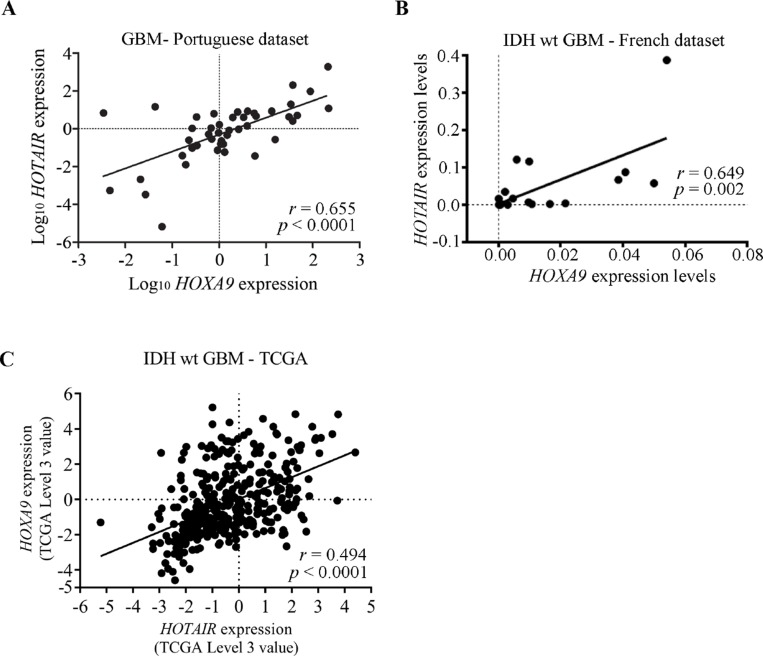
To verify if the frequent co-expression of HOTAIR and HOXA9 in glioma cell lines is present in glioma clinical specimens, we evaluated their expression levels and potential correlations in the Portuguese, French, and TCGA datasets. We found that HOTAIR and HOXA9 frequently have a coherent transcriptional activation status in grade IV glioma (Portuguese dataset Pearson’s r = 0.642, p < 0.0001; IDH-wt GBM in French dataset Pearson’s r = 0.649, p = 0.002; IDH-wt GBM in TCGA microarray dataset Pearson’s r = 0.494, p < 0.0001; IDH-wt GBM in TCGA RNA-seq dataset Pearson’s r = 0.525, p < 0.0001; Figure 3A–3C, Supplementary Figure 2H and Table 1). Similarly, significant correlations between HOTAIR and HOXA9 expression were also observed in specific combinations of glioma grades (Portuguese dataset WHO grades II+III, III, III+IV and II+III+IV; French dataset IDH-wt WHO grade III and grade IV; Table 1 and Supplementary Figure 2A–2G). While HOTAIR and HOXA9 co-expression did not reach statistical significance for grade II gliomas (n = 6; Supplementary Figure 2A), this is likely due to the small sample size, and the same trend for co-expression was observed; indeed, all of these tumors were either double-positive or double-negative (Table 1).
Figure 3. Expression levels of HOTAIR and HOXA9 are significantly correlated in GBM clinical specimens.
(A–C) Correlation graphs of HOTAIR and HOXA9 expression levels in GBMs from the Portuguese dataset (A), and IDH-wt GBMs from the French dataset (B), and IDH-wt GBMs the TCGA dataset (microarray data) (C), showing statistically significant correlations between their expressions in all datasets. (A) Pearson’s r = 0.655, p < 0.0001; (B) Pearson’s r = 0.649; p = 0.002; (C) Pearson’s r = 0.494; p < 0.0001.
Table 1. Analysis of HOTAIR and HOXA9 expression correlations in human glioma samples from the Portuguese dataset.
| Grade II | Grade III | Grade IV | ||||||
|---|---|---|---|---|---|---|---|---|
| HOTAIR | HOTAIR | HOTAIR | ||||||
| + | − | + | − | + | − | |||
| HOXA9 | + | 1 | 0 | 4 | 4 | 8 | 6 | |
| − | 0 | 5 | 1 | 5 | 5 | 34 | ||
|
r = 0.774 p = 0.07 |
r = 0.546 p = 0.030 |
r = 0.655 p < 0.0001 |
||||||
|
r = 0.619 p = 0.005 |
||||||||
|
r = 0.640 p < 0.0001 |
||||||||
|
r = 0.642 p < 0.0001 |
||||||||
*Pearson’s correlation.
Further analysis in 4 independent datasets showed a statistically significant correlation between the expression of HOTAIR and HOXA9 in all glioma sets (Table 2). Furthermore, in IDH-wt (n = 55) and IDH-mut (n = 137) grades II and III glioma with available RNA-seq data from TCGA, a statistically significant correlation between the expression of HOTAIR and HOXA9 was also found (IDH-wt Spearman’s r = 0.65, p < 0.0001, and IDH-mut r = 0.21, p = 0.01; data not shown), while in 1p19q codeleted and IDH-mut grades II and III glioma no correlation was found (n = 85, r < 0.01, p = 0.999; data not shown). Interestingly, no significant correlations between HOTAIR and HOXA9 were found for other cancer types, including lung, leukemia, colorectal, and breast cancers (Table 2). Taken together, our results strongly suggest that HOTAIR and HOXA9 are concomitantly co-expressed in human gliomas, but not in other cancer types.
Table 2. Analysis of HOTAIR co-expression with HOXA9 in glioma datasets and in other cancer types (lung, leukemia, colorectal, and breast cancer datasets from TCGA) available at Oncomine(1).
| Datasets | Correlation HOTAIR/HOXA9 | p-value |
|---|---|---|
| Glioma, NOS | ||
| Portuguese dataset | 0.642 | <0.0001 |
| French dataset | 0.516 | <0.001 |
| TCGA, microarray data | 0.545 | <0.0001 |
| TCGA, RNA-seq data | 0.675 | <0.0001 |
| Oncomine: | ||
| Freije et al. (Freije, et al. 2004) | 0.372 | <0.001 |
| Murat (Murat, et al. 2008) | 0.293 | 0.008 |
| Sun (Sun, et al. 2006) | 0.487 | <0.0001 |
| Phillips (Phillips, et al. 2006) | 0.309 | 0.001 |
| Other cancers from Oncomine: | ||
| TCGA Lung | 0.046 | 0.555 |
| TCGA Leukemia | –0.019 | 0.791 |
| TCGA Colorectal | 0.012 | 0.854 |
| TCGA Breast | –0.051 | 0.215 |
(1)Microarray data from Oncomine dataset, microarray or RNA-seq data from TCGA dataset, quantitative PCR data from the Portuguese dataset (relative gene expression normalized to hGUS or TBP levels), and microfluidic-based qPCR analysis (Fluidigm) data from the French dataset were used to evaluate the Pearson’s correlation.
NOS – not otherwise specified.
High levels of HOTAIR expression associate with shorter survival in patients with malignant glioma
To validate the prognostic value of HOTAIR expression reported in GBM patients from the Chinese population [16], we firstly analyzed two independent datasets of GBM patients from TCGA and REMBRANDT. In 554 GBM patients from TCGA with available survival data, a statistically significant decrease in OS was observed in patients with high HOTAIR expression (n = 177, median OS = 383 days) as compared to patients whose GBMs presented low HOTAIR levels (n = 377, median OS = 447 days; p = 0.026 Log-rank test; Figure 4A). Similarly, associations were found in the subset of 387 IDH-wt GBMs (HOTAIR-high n = 126, median OS = 370 days and HOTAIR-low n = 241, median OS = 447 days; p = 0.032 Log-rank test; Figure 4B and Supplementary Table 2). As expected, classic prognostic factors including patient age at diagnosis, KPS, and use of radio-chemotherapy, were significantly associated with OS (p < 0.05, Log-rank test, Supplementary Table 2). Importantly, HOTAIR expression was significantly associated with shorter OS of IDH-wt GBM patients in a multivariable Cox model (p = 0.036), independently of other prognostic factors (age, gender, KPS, use of chemoradiotherapy, and treatment with additional chemoradiotherapy; Supplementary Table 2). Consistently, in an independent dataset of 67 GBM patients from the REMBRANDT dataset, high HOTAIR expression was present in 72% of patients (n = 48) who had a significantly shorter OS (median OS = 15.8 months) than those with low expression (median OS = 37.4 months), both in univariable and multivariable analyses (p = 0.005 Log-rank test; p = 0.034 Cox regression, Figure 4C and Supplementary Table 3). Since HOXA9 expression is a known prognostic factor in GBM patients [26, 28, 29], we investigated in the large TCGA dataset whether HOTAIR was still prognostically valuable in the subset of HOXA9-negative IDH-wt GBM patients. Indeed, high expression of HOTAIR was able to identify a subgroup of HOXA9-negative IDH-wt GBM patients that presented a significantly shorter OS (median OS = 375 days) than those with low HOTAIR levels (median OS = 463 days; Supplementary Figure 3; Log-rank p = 0.037).
Figure 4. High levels of HOTAIR expression are significantly associated with shorter survival in GBM patients.
Kaplan–Meier overall survival curves of HOTAIR prognostic value in (A) 554 GBM patients from TCGA, (B) 367 IDH-wt GBM patients from TCGA and (C) 67 GBM patients from REMBRANDT dataset, showing patients whose tumors present high HOTAIR expression have a statistically significant shorter overall survival compared to those with HOTAIR-low tumors, in both independent datasets (A, p = 0.026; B, p = 0.032; C, p = 0.005; Log-rank tests).
Additionally, we tested the prognostic value of HOTAIR expression among 28 glioma grade III patients from the French dataset with available survival data. Similarly to GBM, we found for the first time an association between high HOTAIR expression levels and shorter OS in grade III glioma patients with high HOTAIR expression (p = 0.002, Log-rank test; Supplementary Table 4). Multivariable Cox analysis further showed that high HOTAIR expression is a poor prognostic factor (p = 0.022) independently of other known prognostic factors such as age at diagnosis, surgery (partial or complete resection), KPS, treatment, and IDH and 1p19q status. The median survival is 1.4 years [0.7–2.5] in the group of 28 grades III. Survival medians are respectively of 2 years in the group of patients with HOTAIR-lower expression (n = 16, median HOTAIR expression = 0.0002 [2.96e-6 – 0.0053]) and of 0.7 years in the group of patients with HOTAIR-higher expression (n = 12, median HOTAIR expression = 0.0387 [6.4e-6 – 0.0332]).
Finally, we found that high HOTAIR expression is also associated with shorter overall survival in grade II (n = 226) and grade III (n = 240) glioma patients in the RNA-seq data from the larger TCGA dataset (Supplementary Tables 5 and 6; p = 0.020 and p < 0.0001, respectively; Log-rank test), and independently of patient age, gender, and molecular subtype (IDH and 1p/19q status) in the case of grade II gliomas (p = 0.032; Cox regression model), but not for grade III gliomas (p = 0.395; Cox regression model), in which high HOTAIR levels were highly correlated with wild-type IDH.
Taken together, our findings establish HOTAIR as a clinically-relevant biomarker of prognosis in malignant glioma patients.
DISCUSSION
An increasing focus has been given to lncRNAs as key regulators of a range of biological functions and as crucial players in the etiology of disease, especially in cancer [8, 9]. The relevance of HOTAIR in cancer was first reported by an association between HOTAIR expression and increased metastatic potential and diminished survival of breast cancer patients [30]. Later studies evaluated the roles of HOTAIR in several tumor types (reviewed in [13–15]), globally highlighting oncogenic roles of HOTAIR. Likewise, HOTAIR expression was associated with more aggressive disease in glioma patients [16–19]. While some oncogenic functions of HOTAIR have been reported in glioma [17, 31, 32], the underlying mechanisms responsible for HOTAIR activation in such a devastating disease have remained unknown thus far. Here, we explore the multi-layered complexity of HOTAIR activation by integrating molecular (genetic and epigenetic) analyses, in silico approaches, and data from glioma patients and cell lines. Additionally, we further investigated the relevance of HOTAIR as a prognostic biomarker in GBM, and in malignant WHO grade II and III glioma.
We show that HOTAIR is overexpressed in a subset of high-grade gliomas, particularly in GBMs. Of note, our study includes data from independent patient datasets, and obtained with different complementary techniques, including qPCR, expression microarrays, microfluidic-based qPCR analysis (Fluidigm) and RNA-Seq data. To understand the molecular mechanisms regulating the expression of HOTAIR, we evaluated copy number aberrations and DNA methylation levels, and searched for putative transcription factors with binding sites in HOTAIR’s promoter region. Copy number alterations in HOTAIR locus were rare and did not associate with its expression levels, suggesting this is not a prominent underlying mechanism in GBM. Contrarily, modulation of DNA methylation levels in GBM cell lines by the demethylating agent 5-Aza indicated that intragenic CpGs in HOTAIR locus affected its transcriptional levels in a cell line-dependent manner (globally, most cell lines increased HOTAIR levels upon demethylation, while U87MG did not, possibly attributable to different kinetics of 5-Aza response and/or lack of expression of all necessary HOTAIR transcriptional regulators). Moreover, methylation probes covering CpG islands in HOTAIR locus showed significant inverse correlations between HOTAIR’s intragenic DNA methylation and expression levels in GBM specimens. In fact, it was previously shown that intragenic DNA methylation is associated with decreased gene expression by altering the chromatin structure and reducing the efficiency of RNA Pol II elongation [33, 34]. Although these results indicate that DNA methylation may regulate HOTAIR expression in GBM, this association was not universal, so other transcriptional regulation mechanisms are involved in glioma. Previous observations from our group suggested HOXA9 as a possible regulator of HOTAIR in glioma [26]. HOXA9 is an oncogenic transcription factor whose expression was described in a subset of highly aggressive GBMs, and associated with patients’ decreased OS [26, 28]. Interestingly, we found HOTAIR and HOXA9 are frequently co-expressed in gliomas, but this was not observed in patients with lung, leukemia, colorectal, and breast cancers, suggesting that this co-expression may be an exclusive event of gliomas. Indeed, our ChIP data indicates a direct regulation of HOTAIR by HOXA9, which binds to HOTAIR promoter region to activate its transcription. While HOXA9 and other HOX genes have been shown to be transcriptionally regulated by HOTAIR [10, 35] in fibroblasts and breast cancer cell lines, it is currently unknown whether a similar regulation of HOXA9 may also occur in glioma. Indeed, the expression of HOXA9 in GBM is known to be epigenetically regulated by the PI3K pathway via inhibition of EZH2-mediated H3K27 trimethylation at the HOXA9 locus [36], and HOTAIR is known to interact with PRC2 complex (containing EZH2 and other subunits) to guide it to target loci [10, 13, 37]. Therefore, it is conceptually feasible that HOTAIR might regulate HOXA9 expression, which together with other transcription factors can regulate the expression of HOTAIR, in a putative loop mechanism. Of note, the roles of HOXA9 in glioma and in breast cancer are very distinct: while in the first it displays a variety of oncogenic functions, associated with tumor aggressiveness, higher grades of malignancy, tumor implantation in in vivo models, therapy resistance and shorter survival of patients [26, 38–40], in breast cancer HOXA9 is a tumor suppressor gene whose expression is frequently lost, leading to cell growth, survival, invasiveness and changes in morphogenesis [41]. These studies highlight the importance of cell context for the biological effects mediated by HOXA9, which do not thus preclude the possibility that the transcriptional regulatory mechanisms between HOTAIR and HOXA9 may also be distinct in glioma and breast cancer. Although conceptually possible, this hypothesis has not been tested in GBM, and warrants further elucidation in new studies. We report here for the first time the ability of HOXA9, a homeoprotein, to directly regulate the expression of HOTAIR. While we provide strong indications that HOTAIR-positive GBMs may result from altered intragenic DNA methylation levels and/or HOXA9 binding, other mechanisms cannot be excluded. For example, the CT variant of the functional single nucleotide polymorphism rs12826786 in HOTAIR locus was associated with its higher intratumoral expression in glioma [42], and would require further addressing. Thus, our work provides novel insights on HOTAIR’s transcriptional regulators, widening our understanding of the HOTAIR-associated mechanisms of aggressiveness and malignancy in gliomas. Interpreting our findings at the light of other studies further supports the classic tissue-specific roles and regulatory mechanisms of lncRNAs [43].
The prognostic value of HOTAIR that we observed in 3 independent datasets further supports the reported clinical value of HOTAIR in glioma [16]. Critically, this is the first study establishing a prognostic value for HOTAIR in lower grade glioma (grades II and III). Importantly, we also found that HOTAIR expression was able to identify a subset of GBM patients with poor prognosis even in the universe of HOXA9-negative GBM patients, indicating that the combinatorial evaluation of HOTAIR and HOXA9 expression levels may prove advantageous towards a molecularly-based stratification of GBM patients. Testing how the concomitant inhibition of HOTAIR and HOXA9 may affect the behavior of GBM in vitro and in vivo will be interesting and warrants further investigation in future studies. Furthermore, the clinical importance of HOTAIR suggests it as a putative therapeutic target. While drugs directly interfering with HOTAIR function are currently being studied [44–47] but still not clinically used in glioma, it may be interesting to test pharmacological approaches that act upstream of HOTAIR activation, namely PI3K inhibitors (e.g., PI-103 [48] and LY294002 [49]) that have been shown to silence HOXA9 transcription, and might thus result in HOTAIR downregulation. In addition, part of HOTAIR effects in glioma may also be limited by using drugs directed to PRC2 components, as the EZH2 inhibitor DZNep [50], or drugs that specifically target LSD1 activity and have been tested in cancer [51, 52].
In summary, our study shows that (i) HOTAIR is highly expressed in a subset of malignant gliomas, independently of gene copy number alterations; (ii) epigenetic marks at the level of DNA methylation in particular CpG sites associate with HOTAIR levels in GBM; (iii) co-expression of HOTAIR and HOXA9 occurs frequently in high-grade glioma, but not in other cancer types; (iv) HOTAIR is prognostically valuable in malignant glioma patients; and (v) HOTAIR and HOXA9 may be useful biomarkers to integrate a molecularly-based stratification of GBM patients. We anticipate that HOTAIR may mediate some of the effector mechanisms by which HOXA9 creates a more aggressive and therapy-resistant form of GBM. Further studies are warranted to better identify HOTAIR downstream target genes at the genome-wide level in GBM, in an attempt to better understand the mechanisms by which HOTAIR affects survival of patients, and ultimately investigate new therapeutic opportunities.
MATERIALS AND METHODS
Glioma primary samples and cell lines
Glioma tumor specimens were obtained from “Hospital Braga” (n = 34) and “Hospitais da Universidade de Coimbra” (n = 39), Portugal, and from the Clermont-Ferrand University Hospital Center (n = 51), France (Supplementary Table 1), upon approval by each Hospital’s ethical committee and patients signed a written individual informed consent according to institutional guidelines. Tumor tissues were snap-frozen in dry ice or isopentane cooled in liquid nitrogen directly from the operating room, and stored at –80° C. In the French dataset, 57% of the samples were IDH1 genotyped by EpigenDx (Worcester, MA) using pyrosequencing (assays ADS1703 and ADS1704), while the other 43% by analyzing the most frequent IDH1 mutations in exon 4 (codon 132) using RFLP [53]. Regarding 1p19q samples characterization, the analysis was performed using the Vysis 1p36/1q25 and 19q13/19p13 FISH probe kit (4N6020, Abbott), according to manufacturer’s instructions. For some cases lacking 1p19q characterization, internexin A (INA) immunohistochemical analyses were carried out on paraffin sections using antibodies directed against internexin neuronal intermediate filament protein alpha (INA). Since INA expression was reported to be a surrogate marker of 1p19q codeletion [54, 55]), its expression was assessed to indirectly inquire about the 1p19q deletion status. Pediatric glioma cell lines KNS42, Res259, Res186, SF188, and UW479 were cultured in DMEM/F12 (Gibco, Grand Island, NY, USA) supplemented with 10% Fetal Bovine Serum (FBS; Biochrom, Berlin, DE), and 1% Penicillin-Streptomycin (Pen-Strep; Invitrogen, Grand Island, NY, USA) as previously described [56]. Commercially-available adult glioma cell lines A172, GAMG, SNB19, SW1783, U251, and U87MG were originally purchased from ATCC (Rockville, MD, USA), and the primary GBM-derived culture GBM-L18 (established in our lab) were cultured in DMEM (Gibco, Grand Island, NY, USA) supplemented with 10% FBS, and 1% Pen-Strep. U87MG (which do not express HOXA9 endogenously) were previously [28] genetically-engineered with murine stem cell virus (MSCV) retroviral vectors containing the HOXA9 coding region to obtain HOXA9 overexpressing cells (U87MG-HOXA9) or with control empty vector (U87MG-MSCV). Selection of retrovirally-infected cells was maintained with 500 μg/mL G418 (Sigma-Aldrich, St. Louis, MO, USA). Incubations were performed at 37° C, in a humidified atmosphere containing 5% CO2.
TCGA data meta-analysis in glioma patients
HOTAIR gene expression, copy number, DNA methylation and clinical data were downloaded from TCGA; available from: http://cancergenome.nih.gov/, accessed 2017) [57].
TCGA Agilent’s G4502A 244K gene expression profiles from 572 GBMs, 27 grades II and III gliomas (Supplementary Table 1), and 10 unmatched normal samples were analyzed, and “level 3” values of HOTAIR (probe A_32_P168442) and HOXA9 (probe A_23_P500998) were used. HOTAIR-high was considered when “level 3” value ≥0, and HOXA9-high when ≥2. RNA-seq data from additional 161 GBMs, 511 grades II and III glioma patients and 5 unmatched normal samples were also collected (Supplementary Table 1). These data were evaluated using Illumina HiSeq 2000 sequencing system. Genes with a “level 3” value = 0 were considered to have undetectable levels of mRNA expression. Cases were classified based on WHO malignancy grade and considering genetic information according to latest WHO recommendations [1] to: IDH-wt; IDH-mut (without 1p/19q codeletion), and IDH-mut and 1p/19q codeleted.
Gene copy number data were assessed by Affymetrix Genome-Wide Human SNP Array 6.0 in 372 GBM samples. Gene amplifications or deletions were considered for Log2 copy number tumor/normal ≥0.5 (gene copy number ≥3) or ≤–0.5 (gene copy number ≤1), respectively.
Tumor DNA methylation profiles were detected by Illumina Infinium Human DNA Methylation 450 array and include the methylation status of 74 GBM samples. We evaluated 56 probes that span from the HOXC12 (upstream of HOTAIR) to HOXC11 (downstream of HOTAIR). High methylation was considered for β-values > 0.5.
Analysis of HOTAIR expression in Oncomine and REMBRANDT
The human cancer microarray database Oncomine (www.oncomine.com; Oncomine™, Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization [58] of HOTAIR expression (probe 239153_at) in 45 WHO grade II, 98 WHO grade III, and 296 primary WHO grade IV gliomas (Supplementary Table 1). Categorization of HOTAIR-positive and HOTAIR-negative glioma patients was based on the Log2 median-centered intensity values of HOTAIR probe. Log2 median-centered intensity values >0 correspond to high HOTAIR expression, and Log2 values ≤0 correspond to low/negative HOTAIR expression. The Oncomine database was also used to identify genes frequently co-expressed with HOTAIR in gliomas and other cancer datasets [lung (n = 167), breast (n = 593), leukemia (n = 197), and colorectal cancer (n = 237)]. Details on the normalization techniques and statistical calculations can be found on the website (www.oncomine.com) [58].
The REMBRANDT platform (National Cancer Institute REMBRANDT, accessed 2013 June; available now at https://gdoc.georgetown.edu/gdoc/) was queried to evaluate 67 GBM patients (Supplementary Table 1); the cut-off for HOTAIR high expression was established as ≥4-fold than HOTAIR expression in non-tumor samples.
RNA extraction, cDNA synthesis, and gene expression analyses in primary glioma samples and cell lines
Total RNA was extracted from glioma patient samples and cell lines using the TRIzol method (Invitrogen, Carlsbad, CA, USA) for the Portuguese dataset or the RNeasy Mini Kit (Qiagen, Hamburg, GmbH) for the French dataset, and cDNA was synthesized using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol.
For the Portuguese dataset, gene expression levels were assessed by RT-PCR (AmpliTaq Gold 360, Applied Biosystems, Foster City, CA, USA) and quantitative reverse transcriptase real-time PCR (qPCR; KAPA Biosystems, Foster City, CA, USA) according to manufacturer’s instructions (primer sequences in Supplementary Table 7). Housekeeping genes used to normalize gene expression levels were hGUS, TBP, HPRT1, or ACTIN, as stated on each analysis. For all tested genes, the standard PCR parameters were as follows: 4 minutes at 94° C, 35 cycles of denaturation for 30 seconds for the RT-PCR or 3 seconds for the qPCR at 94° C, annealing for 30 seconds (Supplementary Table 7), extension at 72° C for 30 seconds, and final extension at 72° C for 8 minutes for conventional PCR, or increments of 1° C each 5 seconds from 65 to 95° C for the qPCR. The cut-off for high expression of both HOTAIR and HOXA9 was established as >5% of relative expression (upon normalization to housekeeping gene).
In the French dataset, microfluidic-based quantitative analysis (Fuidigm) was used to assess HOTAIR and HOXA9 genes expression levels. cDNA was preamplified for 14 cycles with the pool of primers available in Supplementary Table 7 by using the TaqMan PreAmplification Master Mix (Applied Biosystems). qRT-PCRs were next conducted and validated on Fluidigm 96.96 Dynamic Arrays using the Biomark HD system (Fluidigm Corp.) according to the manufacturer’s instructions. The relative quantification in gene expression was determined after normalization to a geometrical mean of housekeeping genes expression, namely PPIA, HPRT1 and TBP. For each condition, data presented in this study are obtained from two independent experiments, each analyzed in duplicate.
5-Aza-2′-deoxycytidine treatment, DNA isolation, bisulfite conversion and HOTAIR methylation-specific PCR
Glioma cell lines were treated with 5 µM 5-Aza (Sigma-Aldrich®, St. Louis, MO, USA) or dimethyl sulfoxide (DMSO; Sigma-Aldrich®, St. Louis, MO, USA) for 72 hours with daily renewal. Cells were collected by trypsinization, and DNA and RNA were extracted by the TRIzol method. RNA was used to assess HOTAIR expression levels as previously described, and DNA was subjected to sodium bisulfite conversion using the EZ DNA Methylation-Gold™ Kit (Zymo Research, Foster City, CA, USA), according to the manufacturer’s protocol. Touchdown MSPs for HOTAIR promoter were performed (AmpliTaq Gold 360; 3 cycles at 62° C with decrement of 1° C; 6 cycles at 59.5° C with decrement of 0.5° C; and 29 cycles at 57° C; primers in Supplementary Table 7). Blood DNA of a control subject (NB569) was bisulfite treated and used as an unmethylated control for MSP reactions. The same DNA was in vitro methylated (CpG Methyltransferase M.SssI; New England Biolabs Inc.) according to manufacturer’s protocol, followed by sodium bisulfite treatment, and used as a methylated control.
In the French dataset, HOTAIR DNA methylation profile was obtained by Illumina Infinium Human DNA Methylation 450 array performed on the 43 glioma samples. We evaluated 26 probes that cover the HOTAIR locus between HOXC11 and HOXC12 (chr12:54,355,686–54,369,516). High methylation was considered for β-values > 0.5.
In silico analysis of transcription factor binding sites in HOTAIR promoter by Genomatix
MatInspector from Genomatix software [59] (www.genomatix.de) was used to investigate putative transcription factors binding sites in HOTAIR gene. This in silico tool identifies transcription factor binding sites in nucleotide sequences based in a large library of weight matrices [59, 60]. A perfect match gets a matrix similarity of 1 when the tested sequence corresponds to the most conserved nucleotide at each position of the matrix. A good match to the matrix was considered when matrix similarity >0.80.
Chromatin-immunoprecipitation analysis
ChIP experiments were done as previously described [61]. In brief, U251, U87MG-MSCV and U87MG-HOXA9 cells were cross-linked with 1.42% formaldehyde for 15 minutes, followed by quenching with 125 mM glycine for 5 minutes. Cells were then lysed with immunoprecipitation buffer (150 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA, 0.5% NP-40, 1% Triton X-100) and chromatin was sheared by sonication (Sonics Vibra Cell VC70T, 21 cycles for 15 seconds) to obtain DNA fragments of approximately 0.5–1kb. The volume of sheared chromatin equivalent to 2 million cells was incubated with the required antibody in an ultrasonic bath for 15 minutes, followed by incubation with protein A-sepharose beads (Amersham, Uppsala, SE) and Chelex 100 (Bio-Rad, Hercules, CA, USA). The following antibodies were used per immunoprecipitation: 4 μg anti-HOXA9 (Santa Cruz Biotechnologies, Santa Cruz, CA), and 3 μg anti-Immunoglobulin G (IgG; Sigma-Aldrich, St. Louis, MO, USA) as ChIP negative control. The input represents a control for the amount of DNA used in precipitations. DNA amplification was done by qPCR (KAPA SYBR FAST, KAPA Biosystems) according to manufacturer’s instructions with primers designed to amplify a portion of the HOTAIR promoter region that spans from bp -991 to -826 from the HOTAIR transcriptional start site (Supplementary Table 7). The qPCR parameters were as follows: 4 minutes at 94° C, 35 cycles of denaturation for 3 seconds, annealing for 30 seconds at 60° C, extension at 72° C for 30 seconds, and final extension consisting in increments of 1° C each 5 seconds from 65 to 95° C. The U251 fold enrichment of HOTAIR promoter over IgG, and the fold change occupancy of HOTAIR promoter in HOXA9-overexpressing over their negative counterparts for U87MG, were calculated for each experiment using the ΔΔCt method as described previously [62]. Three biological replicates were tested, and each qPCR experiment was performed in triplicates.
Statistical analyses
Statistical analyses were performed using the SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA) for the TCGA and Portuguese datasets, and the Stata software, version 13 (StataCorp, College Station, TX, US) for the French dataset. Tests were two-sided, with a type I error set at α = 0.05. Due to more limited sample size in the French dataset, HOTAIR threshold has been determined according to ROC curve analysis, and estimation of several indexes recommended in literature [63–65] and biological relevance. Univariable survival analyses to assess the prognostic value of HOTAIR and of other clinicopathological features (patient age, gender, KPS, use of chemoradiation therapy, and institution where the patients were treated) were performed by the Log-rank test whenever these data were available. Additionally, the independent prognostic value of HOTAIR was further analyzed by a multivariable Cox proportional hazard model adjusted for those potential confounding variables. The Chi-square test was used to assess differences between the distributions of tumors with high and low HOTAIR expression, stratified for methylation levels (β-value, TCGA). Correlation between the expression levels of HOTAIR and HOXA9 were calculated by Pearson’s or Spearman’s correlation as indicated throughout the text, and according to data normality. HOTAIR expression level differences between histological groups was assessed by one-way ANOVA (Kruskal-Wallis test) for the TCGA dataset. The qPCR differences in ChIP experiments and between histological groups in the French dataset, and genes expression were calculated by Mann-Whitney test and unpaired Student’s t-test, respectively, using Prism GraphPad software (version 6.0a, San Diego, CA, USA). In Oncomine [58], each gene was evaluated for differential expression using Student’s t-test in the case of two-class analyses (e.g. tumor tissue versus respective normal tissue); for multiclass analyses (e.g. grade II, III, and IV gliomas) Pearson’s correlation was used. The association between methylation indexes of HOTAIR and its expression levels in the TCGA dataset was measured by the Pearson’s correlation (r) using SPSS 22.0 software.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
The authors would like to extend their appreciation to Sandro Queirós for helpful assistance regarding data processing from the TCGA.
Abbreviations
- 5-Aza
5-Aza-2′-deoxycytidine
- ChIP
Chromatin-immunoprecipitation
- FBS
fetal bovine Serum
- GBM
glioblastoma
- HR
Hazard Ratio
- H3K27
histone H3 lysine 27
- H3K4me2
histone H3 dimethyl lysine 4
- HOTAIR
HOX transcript antisense intergenic RNA
- IDH
isocitrate dehydrogenase
- IgG
Immunoglobulin G
- lncRNA
long non-coding RNAs
- KPS
Karnofsky Performance Score
- LSD1/CoREST/REST
lysine specific demethylase 1/REST corepressor 1/RE1-silencing transcription factor
- MSCV
murine stem cell virus
- MSP
methylation-specific PCR
- mut
mutant
- OS
overall survival
- Pen-Strep
penicillin-streptomycin
- PRC2
polycomb repressor complex 2
- qPCR
quantitative PCR
- REMBRANDT
Repository of Molecular Brain Neoplasia Data
- RT-PCR
reverse transcription-PCR
- TCGA
The Cancer Genome Atlas
- WHO
World Health Organization
- wt
wildtype
Author contributions
A.X-M. and C.S.G. designed, conducted and analyzed experiments, and wrote the manuscript. A.F. and B.P. conducted experiments and analyzed data from the French dataset. A.F., T.L. and P.A. assisted in manuscript writing. T.L. and M.P. assisted in obtaining the glioma specimens and generation of DNA and RNA, and provided intellectual input. M.C.L., I.C., O.R. and H.T. provided the samples from the Hospital Coimbra dataset. J.L., R.M. and A.A.P. provided the samples from the Hospital Braga dataset. M.R., C.J., R.M.R, J.F.C., P.A., and N.S. assisted during data analysis and provided intellectual input and valuable discussion. B.M.C. designed and supervised experiments, analyzed data, and wrote the manuscript.
CONFLICTS OF INTEREST
The authors disclose no potential conflicts of interest.
FUNDING
Fundação Para A Ciência e Tecnologia (PTDC/SAU-GMG/113795/2009; SFRH/BPD/33612/2009 and IF/00601/2012 to B.M.C.; SFRH/BD/88220/2012 to A.X.M.; SFRH/BD/92786/2013 to C.S.G; SFRH/BD/81042/2011 to M.P.; and SFRH/BD/51996/2012 to T.L.), Project co-financed by Programa Operacional Regional do Norte (ON.2 – O Novo Norte), Quadro de Referência Estratégico Nacional (QREN), Fundo Europeu de Desenvolvimento Regional (FEDER); Fundação Calouste Gulbenkian (B.M.C.); and Liga Portuguesa Contra o Cancro, Portugal (B.M.C.). This article has been developed under the scope of the projects NORTE-01-0246-FEDER-000012, NORTE-01-0145-FEDER-000023 and NORTE-01-0145-FEDER-000013, supported by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER). This work has been funded by FEDER funds, through the Competitiveness Factors Operational Programme (COMPETE), and by National funds, through the Foundation for Science and Technology (FCT), under the scope of the project POCI-01-0145-FEDER-007038. C.J. acknowledges NHS funding to the Biomedical Research Centre. P.A. acknowledges the Plan Cancer-INSERM (CS14085CS‘Gliobiv’, PA), the Cancéropole CLARA (Oncostarter «Gliohoxas»; PA), Fonds de dotation Patrick Brou de Lauriére (PA).
REFERENCES
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 2.Preusser M, de Ribaupierre S, Wohrer A, Erridge SC, Hegi M, Weller M, Stupp R. Current concepts and management of glioblastoma. Ann Neurol. 2011;70:9–21. doi: 10.1002/ana.22425. https://doi.org/10.1002/ana.22425. [DOI] [PubMed] [Google Scholar]
- 3.Bai RY, Staedtke V, Riggins GJ. Molecular targeting of glioblastoma: Drug discovery and therapies. Trends Mol Med. 2011;17:301–12. doi: 10.1016/j.molmed.2011.01.011. https://doi.org/S1471-4914(11)00012-810.1016/j.molmed.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–80. doi: 10.1002/glia.21136. https://doi.org/10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 5.Hadziahmetovic M, Shirai K, Chakravarti A. Recent advancements in multimodality treatment of gliomas. Future Oncol. 2011;7:1169–83. doi: 10.2217/fon.11.102. https://doi.org/10.2217/fon.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27. doi: 10.1007/s11060-011-0793-0. https://doi.org/10.1007/s11060-011-0793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. https://doi.org/10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. https://doi.org/10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. https://doi.org/10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. https://doi.org/S0092-8674(07)00659-910.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–61. doi: 10.1093/hmg/ddq353. https://doi.org/ddq35310.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. https://doi.org/science.119200210.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–64. doi: 10.1016/j.bbcan.2015.07.001. https://doi.org/10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. https://doi.org/10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol Med Rep. 2015;12:5611–8. doi: 10.3892/mmr.2015.4161. https://doi.org/10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 16.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, Jiang T, Kang CS, Chinese Glioma Cooperative Group HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15:1595–603. doi: 10.1093/neuonc/not131. https://doi.org/10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, Xue YX. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–49. doi: 10.18632/oncotarget.4290. https://doi.org/10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Li Z, Tian N, Han L, Fu Y, Guo Z, Tian Y. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol Lett. 2016;12:879–86. doi: 10.3892/ol.2016.4743. https://doi.org/10.3892/ol.2016.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, Wei J, Ren Y, Yang J, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6:537–46. doi: 10.18632/oncotarget.2681. https://doi.org/10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang K, Sun J, Yang C, Wang Y, Zhou B, Kang C, Han L, Wang Q. HOTAIR upregulates an 18-gene cell cycle-related mRNA network in glioma. Int J Oncol. 2017 Mar 7 doi: 10.3892/ijo.2017.3901. https://doi.org/10.3892/ijo.2017.3901. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. Journal of clinical oncology. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. https://doi.org/10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 22.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Gene expression profiling of gliomas strongly predicts survival. Cancer research. 2004;64:6503–10. doi: 10.1158/0008-5472.CAN-04-0452. https://doi.org/10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. https://doi.org/10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. https://doi.org/S1535-6108(06)00056-010.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, Barthel F, Cho HJ, Lin YH, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. https://doi.org/10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pojo M, Goncalves CS, Xavier-Magalhaes A, Oliveira AI, Goncalves T, Correia S, Rodrigues AJ, Costa S, Pinto L, Pinto AA, Lopes JM, Reis RM, Rocha M, et al. A transcriptomic signature mediated by HOXA9 promotes human glioblastoma initiation, aggressiveness and resistance to temozolomide. Oncotarget. 2015;6:7657–74. doi: 10.18632/oncotarget.3150. https://doi.org/10.18632/oncotarget.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goncalves CS, Xavier-Magalhaes A, Pojo M, Oliveira AI, Correia S, Reis RM, Sousa N, Rocha M, Costa BM. Transcriptional profiling of HOXA9-regulated genes in human glioblastoma cell models. Genom Data. 2015;5:54–8. doi: 10.1016/j.gdata.2015.05.010. https://doi.org/10.1016/j.gdata.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, Zardo G, Nigro J, James CD, Fridlyand J, Reis RM, Costello JF. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer research. 2010;70:453–62. doi: 10.1158/0008-5472.CAN-09-2189. https://doi.org/10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson AD, Reis RM, Hargrave D, Workman P, Jones C. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer research. 2010;70:9243–52. doi: 10.1158/0008-5472.CAN-10-1250. https://doi.org/10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. https://doi.org/nature0897510.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L, Kong L, Wei J, Chen L, Yang J, Wang Q, Zhang J, Yang Y, et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget. 2015;6:8353–65. doi: 10.18632/oncotarget.3229. https://doi.org/10.18632/oncotarget.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian EB, Ma CC, He XJ, Wang C, Zong G, Wang HL, Zhao B. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget. 2016;7:30610–25. doi: 10.18632/oncotarget.8895. https://doi.org/10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. https://doi.org/nature0916510.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature structural & molecular biology. 2004;11:1068–75. doi: 10.1038/nsmb840. https://doi.org/10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC, Sarver A, Koller A, Zhi J, Ma Y, Yu J, Chen J, Huang H. BRCA1 is a negative modulator of the PRC2 complex. The EMBO journal. 2013;32:1584–97. doi: 10.1038/emboj.2013.95. https://doi.org/10.1038/emboj.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. https://doi.org/10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. https://doi.org/10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, Zardo G, Nigro J, James CD, Fridlyand J, Reis RM, Costello JF. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–62. doi: 10.1158/0008-5472.CAN-09-2189. https://doi.org/0008-5472.CAN-09-218910.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. https://doi.org/26/18/301510.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 40.Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson AD, Reis RM, Hargrave D, Workman P, Jones C. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–52. doi: 10.1158/0008-5472.CAN-10-1250. https://doi.org/0008-5472.CAN-10-125010.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, Benezra M, Licht JD, Boudreau NJ, Tsai KK, Welm AL, Feldman MD, Weber BL, et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535–50. doi: 10.1172/JCI39534. https://doi.org/10.1172/JCI39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xavier-Magalhaes A, Oliveira AI, de Castro JV, Pojo M, Goncalves CS, Lourenco T, Viana-Pereira M, Costa S, Linhares P, Vaz R, Nabico R, Amorim J, Pinto AA, et al. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J Neurooncol. 2017;132:27–34. doi: 10.1007/s11060-016-2345-0. https://doi.org/10.1007/s11060-016-2345-0. [DOI] [PubMed] [Google Scholar]
- 43.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–21. doi: 10.1073/pnas.0706729105. https://doi.org/10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad NG, Wahlestedt C. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112:8326–31. doi: 10.1073/pnas.1424220112. https://doi.org/10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Zhang Q, Bao J, Du C, Wang J, Tong Q, Liu C. Schisandrin B inhibits the proliferation and invasion of glioma cells by regulating the HOTAIR-micoRNA-125a-mTOR pathway. Neuroreport. 2017;28:93–100. doi: 10.1097/WNR.0000000000000717. https://doi.org/10.1097/WNR.0000000000000717. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Lin C, Yong W, Ye Y, Huang Z. Calycosin and genistein induce apoptosis by inactivation of HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells. Cell Physiol Biochem. 2015;35:722–8. doi: 10.1159/000369732. https://doi.org/10.1159/000369732. [DOI] [PubMed] [Google Scholar]
- 47.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, Saini S, Chang I, Tanaka Y, Enokida H. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PloS One. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Yamano M, Okada M, Ohta M, Tsukamoto S, Raynaud FI, Workman P, Waterfield MD, Parker P. Synthesis and biological evaluation of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorganic & medicinal chemistry letters. 2007;17:2438–42. doi: 10.1016/j.bmcl.2007.02.032. https://doi.org/10.1016/j.bmcl.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 49.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) The Journal of biological chemistry. 1994;269:5241–8. [PubMed] [Google Scholar]
- 50.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–63. doi: 10.1101/gad.1524107. https://doi.org/10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio. 2013;4:e00558–12. doi: 10.1128/mBio.00558-12. https://doi.org/10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Jr, Marton L, Woster P, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nature medicine. 2012;18:605–11. doi: 10.1038/nm.2661. https://doi.org/10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elsayed GM, Nassar HR, Zaher A, Elnoshokaty EH, Moneer MM. Prognostic value of IDH1 mutations identified with PCR-RFLP assay in acute myeloid leukemia patients. J Egypt Natl Canc Inst. 2014;26:43–9. doi: 10.1016/j.jnci.2013.11.001. https://doi.org/10.1016/j.jnci.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Ducray F, Criniere E, Idbaih A, Mokhtari K, Marie Y, Paris S, Navarro S, Laigle-Donadey F, Dehais C, Thillet J, Hoang-Xuan K, Delattre JY, Sanson M. alpha-Internexin expression identifies 1p19q codeleted gliomas. Neurology. 2009;72:156–61. doi: 10.1212/01.wnl.0000339055.64476.cb. https://doi.org/10.1212/01.wnl.0000339055.64476.cb. [DOI] [PubMed] [Google Scholar]
- 55.Ducray F, Mokhtari K, Criniere E, Idbaih A, Marie Y, Dehais C, Paris S, Carpentier C, Dieme MJ, Adam C, Hoang-Xuan K, Duyckaerts C, Delattre JY, et al. Diagnostic and prognostic value of alpha internexin expression in a series of 409 gliomas. Eur J Cancer. 2011;47:802–8. doi: 10.1016/j.ejca.2010.11.031. https://doi.org/10.1016/j.ejca.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 56.Bax DA, Little SE, Gaspar N, Perryman L, Marshall L, Viana-Pereira M, Jones TA, Williams RD, Grigoriadis A, Vassal G, Workman P, Sheer D, Reis RM, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PloS One. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. https://doi.org/10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. https://doi.org/10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. https://doi.org/10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 60.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–84. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–85. doi: 10.1038/nprot.2006.27. https://doi.org/10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. https://doi.org/10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Liu X. Classification accuracy and cut point selection. Stat Med. 2012;31:2676–86. doi: 10.1002/sim.4509. https://doi.org/10.1002/sim.4509. [DOI] [PubMed] [Google Scholar]
- 64.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. https://doi.org/10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2–3. [DOI] [PubMed] [Google Scholar]
- 65.Hajian-Tilaki K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J Intern Med. 2013;4:627–35. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.