SUMMARY
Brown adipose tissue (BAT) adaptively transfers energy from glucose and fat into heat by inducing a gene network that uncouples mitochondrial electron transport. However, the innate transcription factors that enable the rapid adaptive response of BAT are unclear. Here, we identify estrogen-related receptor gamma (ERRγ) as a critical factor for maintaining BAT identity. ERRγ is selectively expressed in BAT versus WAT, in which, in the absence of PGC1α, it drives a signature transcriptional network of thermogenic and oxidative genes in the basal (i.e., thermoneutral) state. Mice lacking ERRγ in adipose tissue (ERRγKO mice) display marked downregulation of BAT-selective genes that leads to a pronounced whitening of BAT. Consistent with the transcriptional changes, the thermogenic capacity of ERRγKO mice is severely blunted, such that they fail to survive an acute cold challenge. These findings reveal a role for ERRγ as a critical thermoneutral maintenance factor required to prime BAT for thermogenesis.
In Brief
Ahmadian et al. find that estrogen-related receptor gamma (ERRγ) is a critical factor for maintaining brown fat identity. They find that ERRγ controls a transcriptional network of thermogenic genes in the basal state. Mice lacking ERRγ exhibit a whitening of brown fat and are unable to survive an acute cold challenge.
INTRODUCTION
Brown adipose tissue (BAT) and white adipose tissue (WAT) differ in their gene expression signatures, morphologies, and physiological functions (Rosen and Spiegelman, 2014). In contrast to WAT, BAT expresses high levels of genes involved in fatty acid oxidation and thermogenesis, is rich in mitochondria, and has numerous small lipid droplets compared with the unilocular droplets of WAT (Cannon and Nedergaard, 2004; Cinti, 2012). Because the major function of WAT is to store and release lipid, it is well equipped to adapt to fluctuations in nutrient availability. BAT, on the other hand, is specialized to burn lipids and glucose in response to the need for extra heat, such as reduced ambient temperature or arousal from hibernation. Although foundational links among WAT, peroxisome proliferator-activated receptor gamma (PPARγ), nutrient storage, and adiposity have been extensively studied, much less is known about BAT. Indeed, with so much focus on WAT and the obesity epidemic, it has only recently been appreciated that adult humans can display cold-activated browning and that relative BAT mass inversely correlates with obesity (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009).
Two distinguishable thermogenic adipocytes have been described in rodents with distinct developmental and anatomical features: classic brown adipocytes located in dedicated BAT depots and beige adipocytes, which reside mainly in subcutaneous WAT. Adult human BAT has been shown to have characteristics of both rodent classic brown adipocytes and beige adipocytes, so understanding both of these cell types is critical (Jespersen et al., 2013; Lidell et al., 2013; Cypess et al., 2013). In contrast to beige adipocytes that arise postnatally in response to external cues such as chronic cold exposure, brown adipocytes express relatively high levels of thermogenic genes prior to an adaptive challenge and thus remain “primed” for thermogenesis, even in the basal (i.e., non-stimulated) state (Kajimura et al., 2015). Although transcription factors and co-regulators, such as PPARγ coactivator 1-alpha (PGC1α), have been shown to mediate adaptive changes in BAT in response to cold, how thermogenic capacity is established and maintained prior to challenge and prior to PGC1 induction is less understood (Villena et al., 2007; Dempersmier et al., 2015).
The estrogen-related receptor gamma (ERRγ, encoded by Esrrg) is an orphan nuclear receptor (NR) and a member of the subfamily of estrogen-related receptors that also includes ERRα and ERRβ (Giguère et al., 1988; Deblois and Giguère, 2011; Eichner and Giguère, 2011). In muscle, ERR isoforms variously contribute to aspects of mitochondrial biogenesis, mitochondrial maintenance, vascularity, energy expenditure, and mitochondrial uncoupling. Although the activity of ERRα is dependent on PGC1α/β induction, ERRγ is sufficient to orchestrate oxidative functions in highly energetic tissues such as type 1 muscle fibers and pancreatic beta cells (Pei et al., 2015; Narkar et al., 2011; Dufour et al., 2007; Kida et al., 2015; Wang et al., 2015; Alaynick et al., 2007, 2010, Yoshihara et al., 2016). In muscle, ERRγ functions to maintain a highly oxidative basal state even in the absence of exercise, and transgenic expression is sufficient to promote type 1 fiber-type switching (Narkar et al., 2011). In the pancreas, ERRγ is required for beta cell maturation and glucose-stimulated insulin secretion (GSIS), and its loss, by targeted knockout, leads to glucose intolerance (Yoshihara et al., 2016). Although ERRγ is also highly expressed in BAT, its contribution to BAT identity or function is not known.
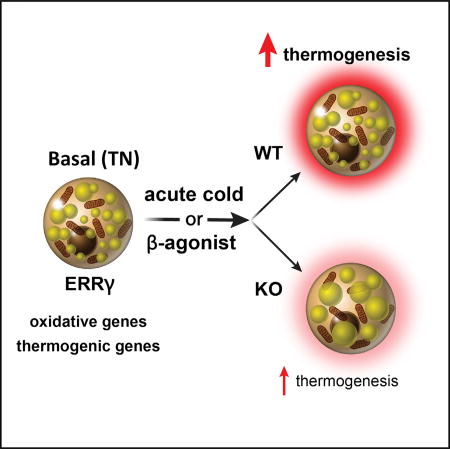
Here, using a combination of biochemical, genetic, and genomic approaches, we uncover an innate or pre-demand function for BAT ERRγ referred to as “priming.” Essentially, transcriptional priming by ERRγ allows the establishment of the appropriate chromatin template needed to facilitate the rapid response to cold exposure. Thus, loss of ERRγ in BAT results in decreased expression of BAT signature genes under thermoneutral (TN) conditions, an unusual “whitening” in the appearance of the BAT organ, and an inability of knockout (KO) mice to survive acute cold exposure.
RESULTS
ERRγ Is Constitutively Expressed in Mature Brown Adipocytes
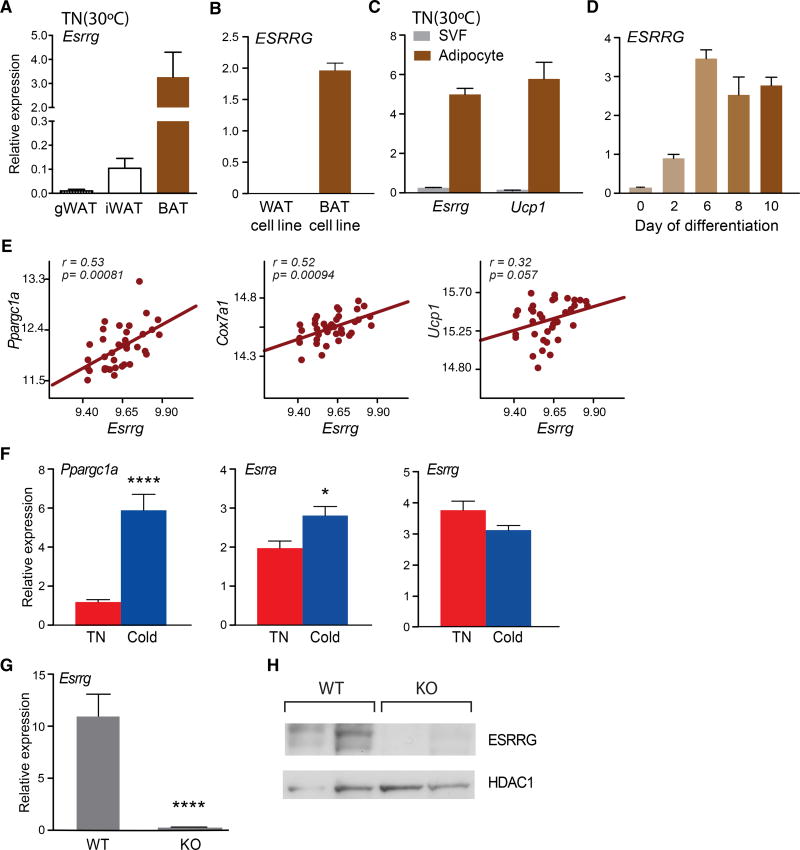
Errγ is highly expressed in BAT versus WAT under basal, TN conditions, similar to the profile observed in human adipocyte cell lines (Figures 1A and 1B) (Kazantzis et al., 2012). Notably, at TN, Errγ is found in the mature adipocyte fraction but not in the stromal vascular fraction (SVF), suggestive of a role in brown adipocyte function (Figure 1C). Consistent with this notion, we find ERRγ to be induced late during differentiation of the PAZ-6 human brown adipose cell line (Kazantzis et al., 2012) (Figure 1D). Furthermore, Errγ expression in BAT positively correlates with important BAT genes, including Ppargc1a, Cox7a1, and Ucp1 in 37 diverse strains of mice from the BXD genetic reference population (Andreux et al., 2012; Wu et al., 2014) (Figure 1E). However, in contrast to the marked induction upon chronic cold exposure of Ppargc1a and Esrra (encoding ERRα), Errγ expression was unaffected, consistent with a role in BAT priming (Figure 1F) (Villena et al., 2007). In order to investigate the role of ERRγ in BAT function, we generated ERRγ adipose conditional KO (ERRγKO) mice and confirmed its selective deletion in BAT (Figures 1G and S1A). Consistent with this, ERRγ protein was not detectable in BAT from ERRγKO mice (Figure 1H). ERRγKO mice were born at a normal Mendelian ratio and exhibited no obvious phenotypic abnormalities under standard animal housing conditions (Figures S1B–S1F).
Figure 1. ERRγ Is Constitutively Expressed in Mature Brown Adipocytes but Is Not Induced by Chronic Cold.
(A) Relative Esrrg mRNA levels in gonadal WAT (gWAT), inguinal WAT (iWAT), and BAT of mice housed at thermoneutrality (TN) (30°C) (n = 10).
(B) Relative ESRRG mRNA expression in differentiated white adipose-derived stem cells (ADSCs, WAT cell line) and PAZ-6 human brown adipocytes (BAT cell line, n = 6).
(C) Relative Esrrg and Ucp1 mRNA levels in the stromal vascular fraction (SVF) and mature adipocyte fraction of BAT from mice housed at TN (n = 3).
(D) Relative Essrg mRNA levels during the differentiation of PAZ-6 human brown adipocytes (n = 4–6).
(E) Pearson correlation of the expression of Ppargc1a, Cox7a1, and Ucp1 with Essrg in BAT from 37 strains of BXD genetic reference population mice.
(F) Relative mRNA levels of Ppargc1a, Esrra, and Esrrg in BAT at TN or chronic cold conditions (1 week 18°C, followed by 1 week 4°C) (n = 10).
(G) Relative Esrrg mRNA levels from BAT of control flox/flox (WT) and ERRγKO (KO) mice (n = 4–7).
(H) Western blot for ESRRG and HDAC1 from nuclear extracts of WT and KO BAT.
Data represent mean ± SEM. *p < 0.05 and ****p < 0.0001, Student’s unpaired t test. See also Figure S1.
ERRγ Drives Thermogenic Gene Expression under Basal Conditions
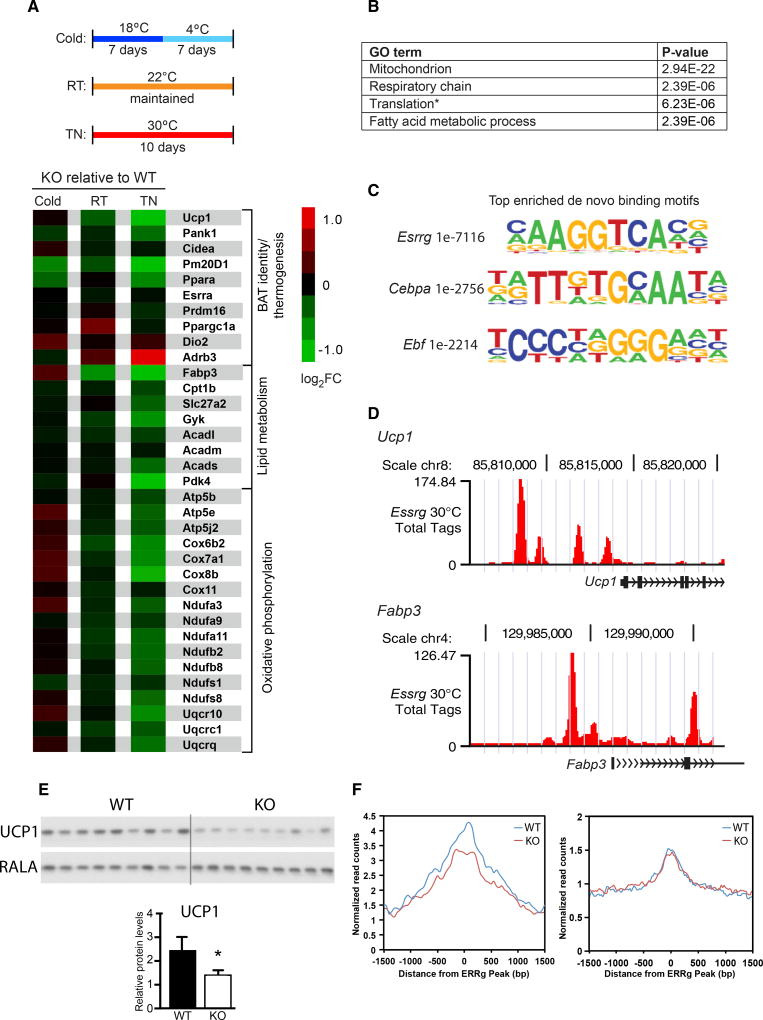
In order to investigate the function of ERRγ in BAT metabolism, ERRγKO mice and control flox/flox littermates were housed under chronic cold acclimated conditions (4°C; see Figure 2A, top), mild cold stress (22°C, room temperature [RT]), or at thermoneutrality (30°C, TN), and their BAT gene signatures were compared using RNA sequencing (RNA-seq). Two hundred ninety-one genes were dysregulated in ERRγKO mice under chronic cold conditions (149 up- and 142 downregulated) and 71 genes at RT (34 up- and 37 downregulated) (Figure S2A). Strikingly, 380 expression changes were observed between mice housed at TN, at which 156 and 224 genes were up- and downregulated, respectively (Figure S2A). Notably, there was minimal overlap in dysregulated genes among the three temperature conditions (Figure S2A). We next examined the expression of genes important for BAT identity and thermogenesis, lipid metabolism, and oxidative phosphorylation in BAT from ERRγKO mice relative to control flox/flox littermates. Indeed, we found that the majority of these key BAT genes were dysregulated at TN, while there were minimal changes under cold or RT conditions (Figure 2A, bottom). In addition, Gene Ontology (GO) analysis of all significantly downregulated genes in ERRγKO BAT under TN conditions identified mitochondrion, respiratory chain, translation (the majority of genes were involved in mitochondrial translation), and fatty acid metabolic process as the predominant dysregulated pathways and processes (Figure 2B).
Figure 2. ERRγ Maintains Thermogenic Gene Expression at Thermoneutrality.
(A) Top: schematic of acclimation protocol for flox/flox (WT) and ERRγKO (KO) mice housed under thermoneutral (TN; 30°C), room temperature (RT; 22°C), and cold (18°C, 4°C) conditions. Bottom: heatmap of selected BAT signature genes in BAT from KO relative to WT mice.
(B) Gene Ontology (GO) of enriched pathways or processes in significantly downregulated genes in KO BAT at TN.
(C) Top enriched de novo binding motifs from ERRγ ChIP-seq in BAT of WT mice acclimated to TN.
(D) ERRγ occupancy at the promoters of Ucp1 and Fabp3 in BAT from WT mice acclimated to TN.
(E) Western blot (top) and quantitation (bottom) for UCP1 from WT and KO BAT of mice housed at TN. RALA was used as a loading control.
(F) Average ATAC-seq signals at ERRγ binding sites within ±5 kb of promoters of ERRγ-regulated (left) and non-regulated genes (right) in BAT isolated from WT and ERRγKO mice.
Data represent mean ± SEM. *p < 0.05, Student’s unpaired t test. See also Figure S2.
Interestingly, loss of ERRγ led to a reduction in BAT Pparα expression at TN (Figure 2A, bottom). As PPARα controls genes involved in fatty acid use and thermogenesis in BAT (Ahmadian et al., 2011), we explored whether the downregulation of BAT signature genes in ERRγKO mice was secondary to decreased PPARα expression. Comparative gene expression analysis of BAT from Pparα-null mice under TN conditions revealed that PPARα and ERRγ control largely unique gene sets, with minimal overlap in downregulated genes (Figures S2B and S2C). These findings indicate that the reduction in Pparα expression is not causative in the altered ERRγKO gene signature.
To explore the mechanisms through which ERRγ controls the expression of metabolic genes in the basal state, we determined its genome-wide binding sites in BAT of control mice acclimated to TN. ERRγ-bound regions identified by ChIP-seq were significantly enriched in ERRγ motifs, consistent with direct DNA binding (Figure 2C). In addition, bound regions were enriched in CEBPA and EBF motifs, suggesting co-localization of ERRγ with other important regulators of thermogenesis (Figure 2C) (Inagaki et al., 2016). Although the majority of binding sites were in intergenic and intronic regions, as reported for many transcription factors (Figure S2D), we found ERRγ directly bound to promoter regions of key BAT genes downregulated in ERRγKO mice, including Ucp1 and Fabp3 (Figure 2D). Accordingly, we found a ∼50% reduction in UCP1 protein in ERRγKO BAT (Figure 2E) supporting a central role for ERRγ in BAT function. GO analysis of the subset of genes bound by ERRγ (±1 kbof transcription start site [TSS]) and downregulated in ERRγKO BAT at TN revealed the same pathways and processes as those identified when all downregulated genes in ERRγKO BAT were analyzed, supporting a direct role for ERRγ in regulating the expression of these key genes involved in mitochondrial function at TN (Figures S2E and 2B).
A complementary story was recently published identifying a role for HDAC3 in establishing a transcriptional landscape in BAT required for the rapid response to acute cold challenge (Emmett et al., 2017). Dating mining of RNA-seq from HDAC3 KO BAT (Emmett et al., 2017) and comparison with RNA-seq from ERRγKO BAT (both performed at TN) revealed commonly downregulated genes in HDAC3KO and ERRγKO BAT, including the key genes UCP1 and Cox7a1, raising the logical possibility of cooperation between HDAC3 and ERRγ in BAT (Figure S2F). However, the majority of the downregulated genes were unique to the specific model, consistent with predominantly parallel and possibility complementary regulatory pathways.
We next investigated if ERRγ binding affects the accessibility of chromatin by performing assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in BAT from ERRγKO and flox/flox control mice that had been acclimated to TN. Notably, we observed higher chromatin accessibility at promoter proximal regions (±5 kb of TSS) of ERRγ-regulated genes compared to non-ERRγ-regulated genes (Figure 2F). Additionally, the accessibility was reduced upon ERRγ knockout, suggesting that ERRγ further induces the open chromatin environment (Figure 2F, left). Although we observed less ATAC-seq signal around non-ERRγ-regulated genes, no changes were observed upon loss of ERRγ (Figure 2F, right). Taken together, these findings indicate that ERRγ acts to prime TN BAT for thermogenesis by promoting DNA accessibility around key genes needed for the rapid response to cold exposure.
Whitening of BAT in ERRγKO Mice
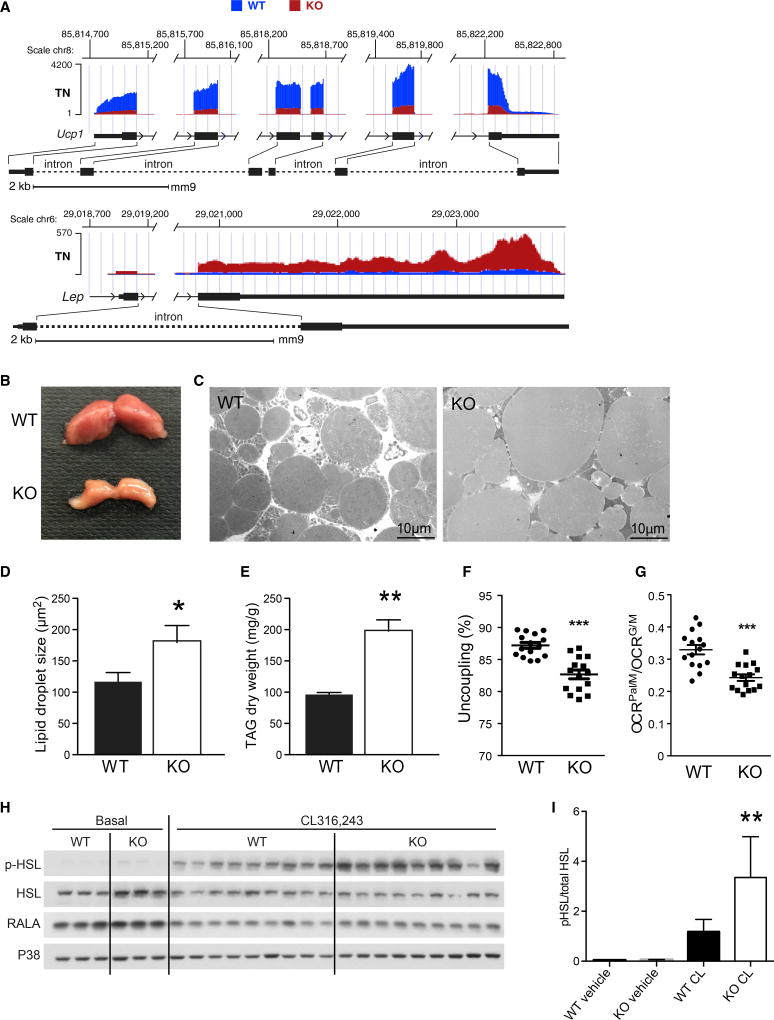
The marked decrease in Ucp1 and unexpected increase in Lep expression in ERRγKO mice suggested that ERRγ is required to maintain BAT functionality under TN conditions (Cinti et al., 1997) (Figure 3A). We attribute these changes to the regulation of BAT-selective genes, as ERRγ deficiency did not affect the expression of and was not found on the promoters of WAT-selective genes at TN (Figure S3A; data not shown). These findings indicate that ERRγ is required for maintaining the robustness of the brown adipose phenotype by promoting the expression of key BAT-selective genes. In addition to these tran-scriptional changes, ERRγKO BAT was visibly smaller and paler than in control flox/flox mice (Figures 3B and S3B), and increases in lipid droplet size and triacylglycerol (TAG) levels were observed (Figure 3C–3E). Furthermore, although mitochondria number and size were unchanged (Figures S3C–S3D), the percentage of uncoupled respiration was reduced in mitochondria isolated from ERRγKO BAT (Figure 3F), consistent with the lower levels of UCP1 (Figure 2E). Moreover, fatty acid metabolism was compromised in ERRγKO BAT mitochondria, as evidenced by reduced palmitate-fueled relative to glutamate-fueled oxygen consumption rate (OCR) (Figure 3G), in agreement with the observed changes in gene expression (Figures 2A and 2B).
Figure 3. ERRγKO Mice Exhibit a “Whitening” of BAT.
(A) Representative RNA-seq browser tracks for Ucp1 and Lep from BAT from flox/flox (WT) and ERRγKO (KO) mice housed at thermoneutral (TN) conditions.
(B) Representative photograph of BAT from 10-week-old WT and KO mice housed at TN since weaning.
(C) Transmission electron microscopy of WT and KO BAT, focusing on the lipid droplets.
(D) Quantitation of lipid droplet size in BAT.
(E) Quantitation of TAG content in BAT (n = 3).
(F) Percentage uncoupling from isolated mitochondria of WT and KO BAT of mice housed at TN.
(G) Oxygen consumption rate (OCR) of palmitate/malate relative to glutamate/malate in isolated mitochondria of WT and KO BAT of mice housed at TN.
(H) Western blots of hormone-sensitive lipase (HSL), phosphorylated HSL (pHSL), RALA, and P38 in BAT lysates 5 min after injection of the β3 agonist CL316,243 or PBS (basal) into WT and KO mice housed at TN.
(I) Quantitation of pHSL from western blot in (H).
Data represent mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, Student’s unpaired t test. See also Figure S3.
Interestingly, although genes important for BAT function were downregulated, expression of the β3 adrenoreceptor Adrb3 was significantly upregulated in ERRγKO BAT (Figure 2A). To explore whether this change was functionally relevant, catecholamine signaling was compared in wild-type (WT) and ERRγKO mice. Although cAMP levels were similar in BAT lysates from the two genotypes after treatment with the β3 agonist CL316,243, increased levels of hormone-sensitive lipase (HSL) phosphorylation (p-HSL) were found in ERRγKO BAT, indicating enhanced catecholamine sensitivity (Figures 3H, 3I, and S3E). Taken together, these findings suggest that ERRγKO mice may upregulate catecholamine signaling as a compensatory mechanism for their defective basal thermogenic program.
ERRγ Is Required to Survive Acute Cold Exposure
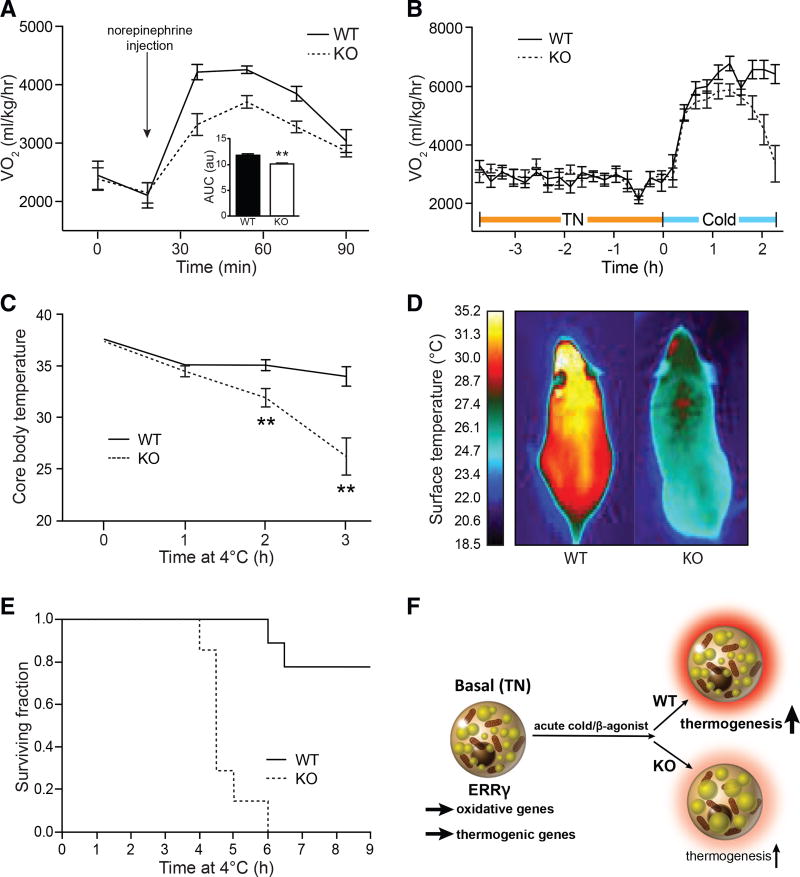
To determine the physiological consequence of the loss of ERRγ in BAT at TN, mice were maintained on standard chow or challenged with a high-fat diet (HFD). Over a period of 14 weeks, the weight gain of ERRγKO mice was similar to control mice on both diets (Figure S4A), and no differences in body composition, serum parameters, or insulin and glucose tolerance were seen (Figures S4B–S4F). Furthermore, with the exception of BAT, the gross appearance and weights of the major metabolic organs in ERRγKO mice were indistinguishable from control mice (data not shown). Notably, these findings indicate that loss of BAT identity due to lack of ERRγ does not predispose mice to diet-induced obesity or insulin resistance when housed at TN. Additionally, there were no changes in browning of WAT (Figures S3F and S3G), significant differences in RER, VO2, physical activity, or food intake between WT and ERRγKO mice upon gradual acclimation to cold (Figures S3H–S3K).
These results led us to consider the possibility that ERRγ might be required to direct the many changes needed for TN mice to activate BAT and respond to an acute cold challenge. To initially explore this notion, the thermogenic response of mice to a norepinephrine (NE) injection was monitored by adaptive changes in Vo2. Although control mice increased their metabolic rate upon NE injection by 2,400 ml/kg/hr, ERRγKO mice managed only a 1,200 ml/kg/hr increase, indicating impaired thermogenic capacity at TN (Figure 4A). A similar blunted VO2 response was observed in response to the β3 agonist CL316,243 without any change in RER (Figures S4G and S4H). As catecholamine signaling is not compromised in ERRγKO mice (Figures 3H and 3I), we attribute their impaired thermogenic capacity to reduced basal expression of oxidative and thermogenic genes.
Figure 4. BAT ERRγ Is Required for Survival during Acute Cold Exposure.
(A) Oxygen consumption (Vo2) in flox/flox (WT) and ERRγKO (KO) mice after intraperitoneal injection of NE bitartrate (1 mg/kg). Mice were housed at thermoneutrality (TN) (n = 4–7).
(B) Adaptive changes in Vo2 of WT and KO mice housed at TN when transferred to cold conditions (4°C) (n = 6).
(C) Body temperature of WT and KO mice exposed to cold (n = 8 or 9).
(D) Representative infrared image of WT and KO mice after exposure to cold.
(E) Kaplan-Meier survival curve of WT and KO mice during cold exposure (n = 7–9).
(F) Model depicting the role of ERRγ in maintaining basal BAT thermogenic capacity.
Data represent mean ± SEM. **p < 0.01, Student’s unpaired t test. See also Figure S4.
We next challenged ERRγKO mice with acute cold exposure. Although the initial response to cold exposure was similar between genotypes, the high metabolic rate needed to fend off cold dropped precipitously in ERRγKO mice (Figure 4B). Accordingly, although control mice maintained their body temperature upon exposure to acute cold, ERRγKO mice reached life-threatening hypothermia after only 3 hr exposure (Figure 4C). Furthermore, thermographic imaging revealed that BAT surface temperature was rapidly decreased in ERRγKO mice upon exposure to cold (Figures 4D and S4I). Importantly, the inability of ERRγKO mice to maintain their body temperature severely compromised their survival. Although ∼80% of control mice withstood ≥6 hr of acute cold exposure, all of the ERRγKO mice died, indicating the key survival role of ERRγ in this type of acute cold challenge (Figure 4E).
DISCUSSION
This study uncovers a mechanism for maintaining BAT structure and functional readiness in the TN state. Surprisingly, in TN mice with ERRγ deficiency, the BAT depot whitens, shrivels, and functionally disintegrates, resulting in a physically lethal hypothermia if exposed to sudden cold. The intrinsic transcriptional activity of ERRγ allows it to prime TN BAT in the absence of inducible drivers such as PGC1α. By maintaining thermogenic genes in an accessible state, BAT remains robust and ready for thermogenesis, in turn allowing a TN host to respond quickly to acute cold exposure.
BAT is inherently different than WAT, even under basal conditions. Our studies here identify ERRγ as an important factor in maintaining its gene expression signature, morphology, and physiological function. Understanding how these fundamental differences between BAT and WAT are maintained is important to understand BAT biology. Indeed, we show that ERRγ drives a transcriptional program that includes key genes necessary for proper mitochondrial function in BAT, including genes involved in FA use, uncoupling, mitochondrial translation, and oxidative phosphorylation. Mice lacking ERRγ in BAT have dysfunctional BAT mitochondria and are unable to appropriately respond to an acute cold challenge (Figure 4F). We found that ERRγ is required to maintain DNA accessibility around key BAT genes at TN. ERRγ likely allows the establishment of the appropriate chromatin template needed to facilitate the rapid response to cold exposure, effectively priming BAT for thermogenesis.
A logical extension of this work is the suggestion that thermogenesis may not be a single or monolithic state but may occur in diverse ways to anticipate different challenges. For example, the adaptive induction of PGC1α allows this cofactor to become gradually induced under progressive cold challenge (such as the cool state of RT or a gradual reduction from RT to life-threatening cold (e.g., 4°C). In this context, ERRα dormant isoform exploits PGC1α induction at RT and below to enhance mitochondrial biogenesis and thermogenic capacity in an adaptive fashion. In contrast, sudden cold exposure from the TN state apparently depends on a baseline thermogenic capacity, which is highly compromised in ERRγKO mice. This supports the argument that there are mechanistically two distinct types of thermogenic challenge and that the differing dependences on ERRα, ERRβ, and ERRγ and their need for PGC1α exploits this potential. This duality holds some analogy to the roles of ERRs and PGCs in skeletal muscle in which ERRγ is intrinsically high in the mitochondria-rich and highly vascularized type I muscle fiber, while ERRα dominates in the PGC1α-dependent type II fiber.
With this proposed “two-state” paradigm for BAT in mind, we note that ERRγ has been shown to work independently of PGC1α not only in type I muscle but also in brown adipocytes in vitro to induce Ucp1 and fatty acid oxidation (Dixen et al., 2013). Interestingly, Gadd45γ has been identified as a cold-inducible activator of ERRγ in BAT (Gantner et al., 2014), suggesting the model that baseline expression of ERRγ promotes depot resilience in the TN state effectively priming BAT for thermogenesis with Gadd45γ amplifying the response upon acute cold exposure. These results, combined with the finding that ERRγ maintains a highly oxidative state in muscle in the absence of exercise (Narkar et al., 2011), implicate ERRγ in controlling metabolism under innate rather than adaptive conditions. A complementary story was recently published identifying a role for HDAC3 in establishing a transcriptional landscape in BAT required for the rapid response to acute cold challenge (Emmett et al., 2017). Future studies elucidating the unique and overlapping functions of ERRγ and HDAC3 in BAT will an important avenue of future research.
Interestingly, loss of ERRγ in BAT does not predispose mice to systemic metabolic dysregulation on a HFD. This finding is reminiscent of mice lacking PRDM16 in BAT (Myf5-Cre PRDM16f/f mice). PRDM16 has been shown to be a critical component of the transcriptional network that drives and maintains BAT identity (Harms and Seale, 2013; Harms et al., 2014). Similar to ERRγKO mice, Myf5-Cre PRDM16f/f mice do not gain more weight than WT mice despite a severely blunted thermogenic capacity. Interestingly, brown adipocytes derived from PRDM16-knockout BAT have decreased expression of Errγ as well as BAT-selective ERRγ target genes, including Ucp1, Coxa1, and Pparα- It is possible that some PRDM16 effects are mediated through ERRγ and also that these factors may synergize to maintain BAT identity. Further studies, including comparison of binding sites between ERRγ and PRDM16, as well as other BAT transcription factors at TN, will be important to unravel the mechanistic hierarchy of factors that cooperate to drive transcription in brown adipocytes.
With the realization that adult humans have BAT and that its quantity is inversely correlated with obesity and increasing age, BAT has emerged as a potential therapeutic target. Because humans are generally at TN, studying the TN BAT state in mice may have increased relevance to BAT in humans (Harms and Seale, 2013). In rodents, age-associated decline in thermogenesis has been associated with BAT atrophy and loss of UCP1 activity (Saely et al., 2012). On the other hand, activation of BAT and browning of WAT have been shown to be key events in the development of cancer-associated cachexia, a lethal syndrome characterized by systemic inflammation, atrophy of fat, and muscle wasting (Saely et al., 2012). Therefore, modulation of ERRγ activity may represent an important therapeutic strategy for treating these physiologic and pathologic conditions.
EXPERIMENTAL PROCEDURES
Animal Studies
ERRγ flox/flox mice were crossed with adiponectin-Cre mice (Jackson Laboratory) to generate ERRγKO mice. ERRγKO mice and flox/flox control littermates received a standard chow diet (MI laboratory rodent diet 5001; Harlan Teklad) or high-fat (60%) diet (F3282; Bio-Serv) and water ad libitum. All mice used for studies were male and housed at thermoneutrality (30°C), unless otherwise indicated. All procedures involving animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) and Animal Resources Department of the Salk Institute for Biological Studies.
RNA Extraction and Gene Expression Analysis
Total RNA was isolated from mouse tissue and cells using TRIzol reagent (Invitrogen). cDNA was synthesized from 1 µg of DNase-treated total RNA using SuperScript II reverse transcriptase (Invitrogen). mRNA levels were quantified by qPCR with SYBR Green (Invitrogen). Samples were run in technical triplicates, and relative mRNA levels were calculated by using the standard curve methodology and normalized against cyclophilin (mouse) or 36B4 (human) mRNA levels in the same samples.
RNA-Seq and Analysis
Ten-week-old male chow-fed WT and ERRγKO mice were housed at 30°C for 10 days or at 18°C for 1 week, followed by 4°C for an additional week, for TN and cold acclimation (Lim et al., 2012). Mice were sacrificed at 2 p.m. in the ad libitum state. RNA was isolated from mouse tissues, with biological triplicates for all conditions. Total RNA was extracted using TRIzol (Invitrogen) and the RNeasy Mini Kit (QIAGEN). RNA purity and integrity were confirmed using an Agilent Bioanalyzer. Libraries were prepared from 100 ng total RNA (TrueSeq version 2; Illumina), and single-ended sequencing was performed on the Illumina HiSeq 2500, using bar-coded multiplexing and a 100 bp read length, yielding a median of 34.1 million reads per sample. Read alignment and junction finding were accomplished using STAR (Dobin et al., 2013) and differential gene expression with Cuffdiff 2 (Trapnell et al., 2013), using University of California, Santa Cruz (UCSC), mm9 as the reference sequence.
ChIP-Seq and Analysis
Pooled BAT depots from 20 mice pre-conditioned at 30°C were minced into ∼1 × 1 mm fragments in PBS prior to fixation with disuccinimidyl glutarate (DSG; 2 mM final concentration for 30 min at RT). Tissues were subsequently cross-linked with paraformaldehyde (1% final concentration for an additional 10 min) and then quenched with glycine (1/20 volume of 2.5 M glycine). Minced tissue was then centrifuged (2,500 rpm for 5 min at RT) and the upper adipose tissue layer collected, washed twice with 10 mL ice-cold PBS, then centrifuged at 2,500 rpm at 4°C for 5 min. Three milliliters of pre-chilled nuclei isolation buffer (20 mM Tris [pH 8], 85 mM KCl, 0.5% NP-40, and protease inhibitor cocktail) was added, and the tissue was disrupted with a Teflon dounce at 4°C. Cell homogenates were allowed to swell in nuclei isolation buffer for a further 10 min on ice, then filtered using a 100 mm cell strainer to remove large debris. Adipose nuclei were pelleted by centrifugation (2,500 rpm for 5 min at 4°C), washed twice with nuclei isolation buffer, then sonicated with a Biorupter in sonication buffer (1% SDS, 50 mM Tris [pH 7.5], 10 mM EDTA, and protease inhibitor cocktail). Sonicated BAT chromatin was diluted 10× in dilution buffer (16.7 mM Tris [pH 8.0], 0.01%SDS, 1.1 % Triton X-100,1.2 mM EDTA, 167 mM NaCl, and protease inhibitor) prior to incubation with ERRγ antibody (10 mg overnight at 4°C). Chromatin was immunoprecipitated with magnetic protein G beads (50 mL for 2 hr at 4°C), and collected beads were washed once by low-salt wash buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 20 mM Tris [pH 8.0], and 0.1% Na-deoxycholate), twice by high-salt wash buffer (1% Triton X-100, 0.1 % SDS, 500 mM NaCl, 1 mM EDTA, 20 mM Tris [pH 8.0], and 0.1% Na-deoxycholate), once by LiCl wash buffer (250 mM LiCl, 0.5% NP-40, 1 mM EDTA, 20 mM Tris [pH 8.0], and 0.5% Na-deoxycholate), and twice by TE wash buffer (10 mM Tris [pH 8.0] and 1 mM EDTA). Chromatin immunocomplexes were eluted (2× 200 mL elution buffer [0.1 M NaHCO3 and 1% SDS] at RT with rotation for 15 min) and associated protein digested (0.2 mg/ mL Proteinase K overnight at 55°C). DNA was subsequently extracted with phenol/chloroform and quantified using a Qubit fluorometer. ChIP-seq libraries were prepared using the NEXTflex Illumina ChIP-Seq Library Prep Kit (Bioo Scientific). High-throughput sequencing was performed on the Illumina HiSeq 2500 System with a 50 bp read length. Sequencing reads were aligned against mouse reference genome mm9 by Bowtie2 (Langmead and Salzberg, 2012) with default parameters. The aligned reads were converted to UCSC track files, and ChIP-seq peaks were called and annotated using the Homer (Heinz et al., 2010) program with default parameters for transcription factor. Each ERRγ binding peak was assigned to the nearest gene.
ATAC-Seq
ATAC-seq was performed as described previously (Buenrostro et al., 2013) with a minor modification. Pooled BAT depots from four mice pre-conditioned at 30°C were minced into ∼1 × 1 mm fragments in PBS. Minced tissue was then centrifuged (2,500 rpm for 5 min at RT) and the upper adipose tissue layer collected, washed twice with 10 mL ice-cold PBS, then centrifuged at 2,500 rpm at 4°C for 5 min. Three milliliters of pre-chilled nuclei isolation buffer (20 mM Tris [pH 8], 85 mM KCl, 0.5% NP-40, and protease inhibitor cocktail) was added, and the tissue was disrupted with a Teflon dounce at 4°C. Cell homogenates were allowed to swell in nuclei isolation buffer for a further 10 min on ice, then filtered using a 100 mm cell strainer to remove large debris. Adipose cells were pelleted by centrifugation (2,500 rpm for 5 min at 4°C). About 50,000 cells were washed with PBS twice and permeabilized with lysis buffer (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.05% NP-40) for 10 min. Roughly isolated nuclei were pelleted, incubated with Tn5 Transposase (Nextera kit; Illumina) for 30 min at 37°C, and purified using PCR purification kit (QIAGEN). ATAC-seq libraries were PCR amplified with custom indexed primers as described previously (Buenrostro et al., 2013) and sequenced as paired-end reads using Illumina NextSeq 500 System. Sequencing reads were mapped to mouse reference genome (mm9) using bowtie2 (Langmead and Salzberg, 2012). ATAC-seq peaks were called using Homer software (Heinz et al., 2010). A subset of peaks located within 5 kb from TSS was used to determine the changes in accessibility sites.
Western Analyses
Tissues were homogenized in lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 250 mM sucrose, 1% NP-40, 2 mM DTT, 1 mM sodium vanadate, 100 mM NaF, 10 mM Na4P2O7, and freshly added protease inhibitor tablet), then incubated for 1 hr at 4°C. Crude lysates were then centrifuged at 14,000 × g for 15 min, and the protein concentration was normalized using Bio-Rad Protein Assay Reagent; this step was repeated to improve precision (Bio-Rad, Hercules, CA). Samples were diluted in SDS sample buffer and boiled for 5 min at 95°C. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). Individual proteins were detected with the specific antibodies and visualized on film using horseradish peroxidase-conjugated secondary antibodies (Bio-Rad, Hercules, CA) and Western Lightning Enhanced Chemiluminescence (PerkinElmer Life Sciences, Waltham, MA). Nuclear lysates (15 mg) were subjected to 10% SDS-PAGE, transferred to nitrocellulose membranes, and probed with rabbit anti-(Santa Cruz), anti-UCP-1 (Sigma-Aldrich), ERRγ, and HDAC1 primary antibodies followed by horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Blots were visualized using enhanced chemiluminescence substrate (PerkinElmer), and images were captured.
Correlation and Network Analysis
Transcriptomics data from BAT of BXD genetics reference population (GRP) was used for network and correlation analysis as described earlier (Williams et al., 2016; Wu et al., 2014). In brief, 37 BXD stains on chow diet underwent extensive phenotyping tests as described previously (Williams et al., 2016). Mice were sacrificed at 29 weeks of age, and various tissues including BAT were used for transcriptomics analysis. Correlation and network analysis was done in R. For network analysis, we used a custom package, imsbInfer, currently on GitHub (https://github.com/wolski/imsbInfer) but in the process of being added to Bioconductor.
Cold Exposure
Mice were acclimated to thermoneutrality (30°C) for 10 days and then transferred to cold (4°C). Food was removed at the beginning of acute cold exposure.
Cell Culture
PAZ-6 human brown adipocytes were differentiated as previously described (Kazantzis et al., 2012).
Lipid Droplet Size
Lipid droplet size was calculated from electron microscopy images of BAT using ImageJ software.
TAG Dry Weight
BAT was homogenized in lysis buffer (50 mM Tris [pH 8], 100 mM NaCl, and 0.1% NP-40), and lysate was dried in SpeedVac and then extracted with chloroform/methanol (2:1 v/v). Dried lipids were resuspended in chloroform and quantitated with TAG assay reagent (Infinity).
Electron Microscopy
Freshly dissected brown fat was immersion fixed in ice-cold fixative consisting of 2.5% paraformaldehyde, 3% glutaraldehyde, and 0.02% picric acid in 0.1 M cacodylate buffer for 1 week at 4°C. The tissues were then buffer washed, post-fixed in buffered 2% osmium tetroxide, and subsequently dehydrated in graded ethanol series, transitioned in propylene oxide and embedded in Spurr resin (Electron Microscopy Sciences, Hatfield PA). Thick sections (1 mm) were cut, mounted on glass slides, and stained in toluidine blue for general assessment in the light microscope. Subsequently, 70 nm thin sections were cut with a diamond knife (Diatome, Hatfield PA), mounted on copper slot grids coated with parlodion, and stained with uranyl acetate and lead citrate for examination on a Philips CM100 electron microscope (FEI, Hillsbrough OR). Images were documented using a Megaview III charge-coupled device (CCD) camera (Olympus Soft Imaging Solutions, Münster, Germany).
Gradual Cold Acclimation and Metabolic Measurements
Mice
VO2 was measured using the Comprehensive Laboratory Animal Monitoring System (CLAMS; Columbus, OH). Data were normalized to body weight. Body temperatures were assessed using a RET-3 rectal probe for mice (Physitemp). CL316,243 (Sigma-Aldrich) and NE (Sigma-Aldrich) were intraperitoneally injected into mice at 1 mg/kg body weight.
Mitochondrial Isolation and Seahorse Bioanalyzer Analysis
Mitochondria were isolated from freshly dissected interscapular BAT using a protocol adapted from a previous study (Fan et al., 2017). The entire isolation procedure was performed at 4°C. Briefly, BAT from each mouse were homogenized in 12 mL cold homogenization buffer (H-buffer; 210 mM mannitol, 70 mM sucrose, 1 mM EGTA, 1% BSA [fatty acid free], and 5 mM HEPES [pH 7.2]) in a motor-driven 15 mL polytetrafluoroethylene (PTFE) tissue grinder. Homogenate was next centrifuged at 1,000 × g for 20 min. The supernatant was collected and centrifuged at 10,000 × g for 15 min. The resulting mitochondrial pellet was resuspended in 500 µL H-buffer, of which 20 µL is used to determine protein concentration. Mitochondrial respiration analysis was performed with the Seahorse XF96 bioanalyzer (Agilent) following the manufacturer’s protocol. Briefly, 5 µg of freshly isolated mitochondria in 75 µL respiration buffer (R-buffer; 225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris-HCl, 5 mM KH2PO4 [pH 7.2]) was loaded into each well of a XF96 cell culture plate pre-treated with Cell-Tak adhesive (Corning). The plate was then centrifuged at 3,000 × g for 1 hr at 4°C. Malate and glutamate (10 mM each) or malate and palmitoyl-L-carnitine (2 mM and 40 µM, respectively) were added as mitochondrial substrates. Oligomycin (Olig; 5 µM) and rotenone and antilycin A (R&A; 1 µM each) are injected during the experiment. Uncoupling rate was calculated using malate and glutamate as substrates using the following formula: (OCROlig - OCRR&A)/(OCRbasal – OCRR&A).
Statistical Analyses
Data are expressed as mean ± SEM. Statistically significant differences between two groups were assessed using Student’s t test. Survival curves were plotted using the Kaplan-Meier estimate and analyzed for significance between groups with the Log-Rank test.
Supplementary Material
Highlights.
ERRγ is a critical factor for maintaining BAT identity
ERRγ drives a transcriptional network of thermogenic genes in the basal state
Adipose ERRγKO mice exhibit decreased thermogenic gene expression at thermoneutrality
Adipose ERRγKO mice fail to survive an acute cold challenge
Acknowledgments
We thank J. Alvarez, S. Kaufman, B. Collins, and H. Juguilon for technical assistance; L. Ong and C. Brondos for administrative assistance; Muriel Strosberg for the PAZ-6 human brown adipocytes; Melissa Kazantzis for assistance culturing PAZ-6 human brown adipocytes; and Jamie Simon for graphical assistance. M.A. is supported by NIH grant 1K01DK113065. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology and is supported by NIH grants (DK057978, HL088093, and HL105278), the Glenn Foundation for Medical Research, the Leona M. and Harry B. Helmsley Charitable Trust (2017-PG-MED001), Ipsen/Biomeasure, the California Institute for Regenerative Medicine, The Ellison Medical Foundation, and a gift from Steven and Lisa Altman. C.L. and M.D. are funded by grants from the National Health and Medical Research Council of Australia (project grants 512354, 632886, and 1043199). A.R.S. is supported by the NIH (grants RO1DK060591 and RO1DK100319) and the DRC (grant P30DK063491). S.M.R. is supported by the NIH (grant K01DK105075) and the DRC (grant P30DK063491). This work was supported by the NGS Core Facility of the Salk Institute with funding from an NIH National Cancer Institute (NCI) Cancer Center Support Grant (CCSG) (P30 014195), the Chapman Foundation and the ´ Leona M. and Harry B. Helmsley Charitable Trust. J.A. is supported by École Polytechnique Fe´ de´ rale de Lausanne (EPFL), the Velux Stiftung (1019), and the Swiss National Science Foundation (31003A-140780). Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the NIH under award number P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is SRA: SRP063705.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.061.
AUTHOR CONTRIBUTIONS
M.A., S.L., S.M.R., N.H., P.J., W.F., R.T.Y., A.R.A., J.A., A.R.S., M.D., and R.M.E. designed the studies, supervised the research, and wrote the manuscript. M.A., S.L., S.M.R., N.H., P.J., C.D.D.M.F, W.F., E.Y., S.J., A.V.G., and Y.D. performed the experiments. M.A., S.L., S.M.R., N.H., E.Y., P.J., W.F., C.D.D.M.F., R.T.Y., and C.L. analyzed the data.
DECLARATION OF INTERESTS
R.M.E., M.D., A.R.A., R.T.Y., and M.A. are co-inventors of methods of using ERRγ.
References
- Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13:739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguère V, Evans RM. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, Yu R, Jonker JW, Holt JA, Rajpal DK, et al. ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 2010;24:299–309. doi: 10.1210/me.2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cinti S. The adipose organ at a glance. Dis. Model. Mech. 2012;5:588–594. doi: 10.1242/dmm.009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S, Frederich RC, Zingaretti MC, De Matteis R, Flier JS, Lowell BB. Immunohistochemical localization of leptin and uncoupling protein in white and brown adipose tissue. Endocrinology. 1997;138:797–804. doi: 10.1210/endo.138.2.4908. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G, Giguère V. Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim. Biophys. Acta. 2011;1812:1032–1040. doi: 10.1016/j.bbadis.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, Kwan HY, Kang C, Wong RH, Sul HS. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell. 2015;57:235–246. doi: 10.1016/j.molcel.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixen K, Basse AL, Murholm M, Isidor MS, Hansen LH, Petersen MC, Madsen L, Petrovic N, Nedergaard J, Quistorff B, Hansen JB. ERRγ enhances UCP1 expression and fatty acid oxidation in brown adipocytes. Obesity (Silver Spring) 2013;21:516–524. doi: 10.1002/oby.20067. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR:ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguère V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Giguère V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- Emmett MJ, Lim HW, Jager J, Richter HJ, Adlanmerini M, Peed LC, Briggs ER, Steger DJ, Ma T, Sims CA, et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546:544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Waizenegger W, Lin CS, Sorrentino V, He MX, Wall CE, Li H, Liddle C, Yu RT, Atkins AR, et al. PPARδ promotes running endurance by preserving glucose. Cell Metab. 2017;25:1186–1193. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner ML, Hazen BC, Conkright J, Kralli A. GADD45γ regulates the thermogenic capacity of brown adipose tissue. Proc. Natl. Acad. Sci. U S A. 2014;111:11870–11875. doi: 10.1073/pnas.1406638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, Kurokawa M, Won KJ, Seale P. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supra-clavicular region of adult humans. Cell Metab. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Spiegelman BM, Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantzis M, Takahashi V, Hinkle J, Kota S, Zilberfarb V, Issad T, Abdelkarim M, Chouchane L, Strosberg AD. PAZ6 cells constitute a representative model for human brown pre-adipocytes. Front. Endocrinol. (Lausanne) 2012;3:13. doi: 10.3389/fendo.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida YS, Kawamura T, Wei Z, Sogo T, Jacinto S, Shigeno A, Kushige H, Yoshihara E, Liddle C, Ecker JR, et al. ERRs mediate a metabolic switch required for somatic cell reprogramming to pluripotency. Cell Stem Cell. 2015;16:547–555. doi: 10.1016/j.stem.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Lim S, Honek J, Xue Y, Seki T, Cao Z, Andersson P, Yang X, Hosaka K, Cao Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat. Protoc. 2012;7:606–615. doi: 10.1038/nprot.2012.013. [DOI] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. Exercise and PGC-1a–independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Mu Y, Leblanc M, Alaynick W, Barish GD, Pankratz M, Tseng TW, Kaufman S, Liddle C, Yu RT, et al. Dependence of hippocampal function on ERRγ-regulated mitochondrial metabolism. Cell Metab. 2015;21:628–636. doi: 10.1016/j.cmet.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saely CH, Geiger K, Drexel H. Brown versus white adipose tissue: a mini-review. Gerontology. 2012;58:15–23. doi: 10.1159/000321319. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Villena JA, Hock MB, Chang WY, Barcas JE, Giguère V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. U S A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerbäck S, Nuutila P. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Wang T, McDonald C, Petrenko NB, Leblanc M, Wang T, Giguere V, Evans RM, Patel VV, Pei L. Estrogen-related receptor a (ERRα) and ERRγ are essential coordinators of cardiac metabolism and function. Mol. Cell. Biol. 2015;35:1281–1298. doi: 10.1128/MCB.01156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara E, Wei Z, Lin CS, Fang S, Ahmadian M, Kida Y, Tseng T, Dai Y, Yu RT, Liddle C, et al. ERRγ is required for the metabolic maturation of therapeutically functional glucose-responsive b cells. Cell Metab. 2016;23:622–634. doi: 10.1016/j.cmet.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.