Abstract
The circadian clock is an endogenous time keeping system shared by most organisms. In mammals, a master pacemaker in the hypothalamus orchestrates temporal alignment of behavior and physiology by transmitting daily signals to multiple clocks in peripheral tissues. Disruption of this communication has a profound affect on human health and has been linked to diverse pathogenic conditions, including cancer. At the center of the molecular circadian machinery is a set of clock genes, generating rhythmic oscillations on a cellular level. In the past several years, research from different fields has revealed the complexity and ubiquitous nature of circadian regulation, uncovering intriguing associations between clock components and cellular pathways implicated in tumorigenesis. In this review, we discuss the emerging role of circadian genes in hematological and hormone-related malignances. These new insights suggest that manipulating circadian biology as a way to fight cancer, as well as, other life threatening diseases is within the realm of possibility.
Keywords: circadian, hematopoietic system, breast cancer, prostate cancer, tumorigenesis
Introduction
For most organisms, the ability to adapt to their environment enhances overall well-being and provides a selective advantage. Indeed, organisms ranging from cyanobacteria (among the earliest cellular life forms) to mammals developed an internal time keeping system—termed circadian—allowing them to anticipate one of the most profound environmental signals, the daily cycle of light/dark (translated from Latin, circa diem: about a day). For ancient life forms, restricting the UV-sensitive S-phase of the cell cycle to nighttime, thereby avoiding strong irradiation-induced DNA damage, may have had a selective value, what is known as the “escape from UV” hypothesis.1 From this humble beginning, circadian influence in modern organisms has found its way to an unparalleled array of biological activities synchronizing cells, organs and organisms with earth’s daily rotation around its axis. In fact, the ability to keep time is a basic characteristic of life on plant earth.
The first report of endogenous rhythms is attributed to the French astronomer de Mairan who, in 1729 demonstrated that daily leaf movements of a plant persists in constant darkness, and remarkably went on, to propose that this phenomenon is related to sleep patterns of bedridden patients.2 Major advances in elucidating the molecular intricacies underlying circadian rhythms began in the early 70s with the discovery of clock genes in Drosophila. However, it was the decade of the 90s that saw an explosion of data in the field, leading to the current model of the clock mechanisms.3 Research over the last ten years added to these discovers and revealed the complexity and extensiveness of circadian regulation not only within the clock machinery but also as it relates to other molecular networks. The challenge in the coming decade is to convert these molecular insights to benefit life in the real world.
Although the clock components are not conserved between kingdoms, the molecular mechanisms underlying circadian rhythms are universal in all model systems. In mammals this system consists of a central pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus, input pathways transmitting temporal information (primarily light) to the pacemaker, and output pathways conveying information from the central pacemaker to organs throughout the body.4-8 At the core of the molecular circadian machinery are interlocked transcriptional/translational feedback loops generated by a set of clock genes.9 The heterodimeric transcription factor complex, Clock/ Bmal1 binds to E-boxes in the promoter of Period (Per) and Cryptochrome (Cry) genes. Per and Cry proteins heterodimers and upon reaching a critical concentration, attenuate Clock/ Bmal1 activity, thereby forming the major negative feedback loop. A secondary feedback loop is composed of two subfamilies of nuclear hormone receptors, Rev-erb and Ror, which directly regulate the core feedback loop, increasing its robustness. Further levels of regulation are achieved by posttranslational modification and chromatin remolding that are crucial for maintaining circadian rhythmicity.10-12
The same clock genes which make up the SCN oscillator are expressed, and, moreover, exhibit temporal oscillations in most peripheral tissues.4,5 Circadian oscillations can be detected even in established cell lines.13,14 The peripheral clocks are synchronized by the central clock and also have inherent circadian properties. Remarkably, the clock transcriptional machinery is not restricted to generate temporal expression of clock genes, but extends to regulate hundreds of non-clock genes in a tissue specific manner. Expression profiling studies demonstrated that in any given tissue, up to 15% of transcripts are clock controlled genes.15,16 The Clock/Bmal1 complex directly controls the expression of some of these genes, whereas others are indirectly regulated through circadian expression of relevant transcription factors. Subsequently, these rhythmically expressed genes integrate circadian information into multiple organismal and cellular processes such as hormone secretion, aging, metabolic pathways, DNA damage response and cell cycle progression. The importance of this multifaceted system to human physiology implies that disruption of circadian function will inevitably lead to disease.8,17 Indeed, circadian deregulation has been linked to an impressive array of pathologic conditions including diabetes, cardiovascular disease, depression and cancer. Herein, we review some of the recent findings linking the circadian system to cancer and focus primarily on hematopoietic and hormone-related malignances.
Hematopoietic System
Circadian rhythmicity in hematopoietic cell numbers was reported as early as the 50s. Later studies revealed expression of clock genes in multiple hematopoietic cell types.18-23 Furthermore, circadian genes appear to be temporally expressed and developmentally regulated in various murine and human hematopoietic lineages including stem/progenitor cells. Additionally, circulating levels of hematopoietic growth factors such as granulocyte-colony stimulating factor (G-CSF), granulocyte-monocyte colony stimulating factor (GM-CSF), tumor necrosis factor (TNF), and interleukins-2, -6 and -10 also display daily oscillations. At the cellular level, and similar to other tissues, the circadian apparatus and clock specific proteins regulate cell cycle and apoptotic pathways in hematopoietic cells.24 Hematopoiesis also does not escape circadian regulation at the systematic level (as discussed in more details below).25
Circadian regulation is critical to normal hematopoiesis; pari passu, disruption of circadian function has been implicated in hematopoietic neoplasms. The significance of proper circadian regulation to cell proliferation and neoplastic transformation was illustrated by studies in mice containing core clock gene mutations (Table 1). When kept in constant darkness Per2 mutant mice become arrhythmic; remarkably, Per2 deficient mice are also prone to develop cancer.26 In response to radiation these mice have a 10-fold increased incidence of lymphomas relative to wild-type controls. Temporal expression of genes involved in cell cycle regulation and tumor suppression, such as Cyclin D1 and A, c-Myc, Mdm2 and Gadd45α, is deregulated in Per2 mutant mice. In particular, c-Myc is under direct circadian control through a clock-responsive E-box in its promoter. In additional studies, after partial hepatectomy, livers from Cry deficient mice were found to regenerate slower than those from normal mice. In this model, the circadian clock controls the G2/M transition by directly regulating Wee1 kinase, an inhibitor of the cdc2/cyclin B1 complex, the major cyclin complex governing the G2/M transition.27
Table 1.
Disruption of circadian clock genes associated with dysregulated cell proliferation and cancer in murine models
| Mouse genotype | Mouse phenotype | Reference |
|---|---|---|
| Clock-deficient | High apoptotic and low proliferation rate in lymphocytes and thymocytes; low proliferation rate in embryo-derived fibroblasts; accelerated aging in response to radiation; reduced hematopoietic repopulation after chemotherapy | 38, 51, 82 |
| Bmal1 KO | Premature aging associated with increased levels of reactive oxygen | 83 |
| Per2-deficient | Increase in tumor development and reduced apoptosis in thymocytes in response to radiation; develop intestinal polyps | 26, 84 |
| Apc-mutant/Per2-deficient | Increase in intestinal polyps compared with ApcMin/+ mice | 84 |
| Cry1,2 KO | Delayed liver regeneration by controlling G2/M transition | 27 |
| p53 KO/Cry1/2 KO | Reduced cancer risk compared with p53 KO mice | 85 |
Abbreviations: KO, knockout.
Analyzing primary patient samples shows that Per gene expression is downregulated in various types of leukemias and lymphomas (Table 2). For example, Per gene expression is reduced in peripheral blood of chronic myeloid leukemia (CML) patients compared with healthy individuals.28 Furthermore, CpG sites of Per2 and Per3 promoters are often methylated in CML patients, with a significant increase frequency in Per3 methylation in patients during blast crisis compared with those in the chronic phase of the disease. We analyzed Per2 expression in a large number of leukemia29 and lymphoma (Thoennissen and Koeffler HP, unpublished) patient samples, as well as control normal mononuclear bone marrow and tonsils. Per2 levels were markedly down-regulated in acute myeloid leukemia (AML) and diffuse large B cell lymphoma (DLBCL). Genetic variants of the clock genes are associated with a risk for non-Hodgkin’s lymphoma (NHL),30,31 and epidemiological studies noted that men who work at night have an increased risk for NHL.32 In vitro cell culture experiments showed that forced expression of Per2 in human and murine AML and pro-B lymphoid cell lines led to growth inhibition, cell cycle arrest, apoptosis and loss of clonogenic ability.29
Table 2.
Circadian clock genes abnormalities in cancer
| Abnormality | Cancer type | Reference |
|---|---|---|
| Per gene mutation | breast, colorectal | 62 |
| Deregulation of Per gene expression | breast, colorectal, endometrial, lung, CLL, DLBL, AML, CML | 28, 29, 59-61, 86-89 |
| Single nucleotide polymorphisms in circadian genes associated with cancer risk and patient survival | breast, prostate, NHL | 30, 31, 65-67 |
| Per1 translocation | CML | 90 |
Abbreviations: CLL, Chronic lymphocytic leukemia; DLBCL, Diffuse large B-cell lymphoma; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; NHL, non-Hodgkin’s lymphoma.
Tissue specific transcription factors acting as oncogenes or tumor suppressors may also interfere with proper clock function. CCAAT/enhancer binding proteins (C/EBPs) are a family of transcription factors critical for proliferation and differentiation in multiple cell types including hematopoietic cells. Deregulation of C/EBPs plays a role in a number of hematologic malignancies, with C/EBPα being a bona fide tumor suppressor in leukemia.33 Using gene expression profiles, we found that the core clock genes Per2 and Rev-Erbα, as well as Dbp (albumin site d-binding protein, a circadian output gene), are C/EBP targets.29 In vitro studies in human leukemic cell lines suggest that induction of Per2 by C/EBPα contributes to, at least some of the tumor suppressive properties C/EBPα. Interestingly, C/EBPs themselves are rhythmically expressed in various tissues and have been proposed to act as circadian transcriptional regulators,34 that may form an additional circadian feedback loop (Fig. 1). C/EBP family members are also key regulators of G-CSF, the most important cytokine regulating neutrophil production. A recent study showed that C/ EBPs regulate G-CSF-induced myeloid differentiation via a feedback loop involving the NAD+-Sirt1 metabolic pathway,35 which is under direct circadian control36,37 (as discussed below). These new discoveries together with bioinformatics and genome-wide screening approaches,15,16,38-44 reveal an ever expanding network connecting the circadian clock genes to a multitude of cellular pathways.
Figure 1.
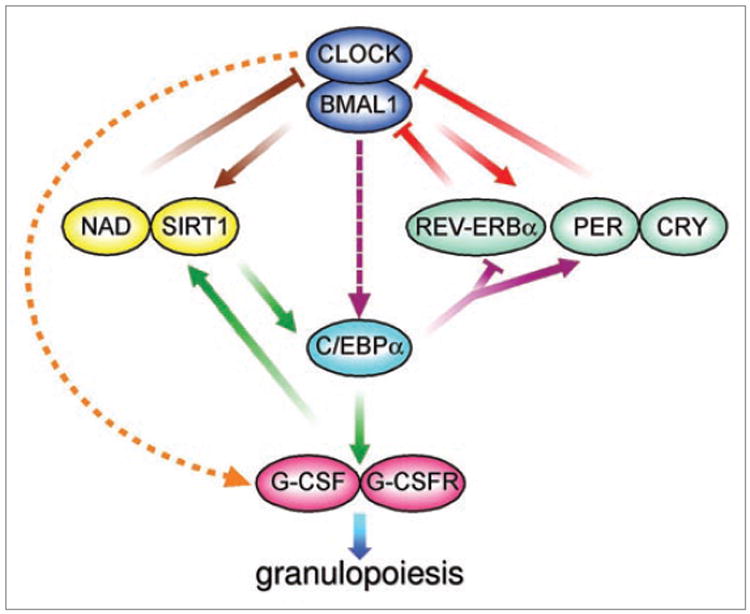
Connections between circadian cycles, C/EBPs and the NAD+-Sirt1 pathway may affect G-CSF induced granulopoiesis. The circadian oscillator is composed of transcriptional feedback loops (Red). Clock and Bmal1 drive expression of core clock genes Per, Cry and Rev-Erbα which in turn, inhibit Clock/Bmal1 activity. Throughout the body, these clock cycles are embedded into tissue-specific pathways to sustain local rhythmic physiologies. The Clock/NAD+-Sirt1 feedback loop (brown) couples circadian oscillations to metabolism. Diurnal cytokine patterns link circadian regulation to hematopoietic processes (orange). C/EBP transcription factors may form a feedback loop with the circadian clock (purple) as they regulate core clock genes and are themselves rhythmically expressed in some tissues. In myeloid cells, C/EBPs are part of a feedback loop (green) that regulates granulocyte colony stimulating factor (G-CSF) and G-CSF receptor (G-CSFR) in a NAD+-Sirt1 dependent manner, leading to induction of granulopoiesis.
The ubiquitous nature of cellular and systemic circadian regulation suggests that understanding circadian intricacies may offer new therapeutic avenues. Recently, chronotherapy, i.e., administration of treatment in coordination with circadian rhythms, has been attracting attention, particularly in the field of oncology.45,46 Conventional cancer therapeutics usually target proliferating cells and these drugs are often limited by their toxicity to normal tissues. Cancer chronotherapy is based on asynchronies in cell proliferation and drug metabolic rhythms between normal and tumor cells, with the aim of minimizing the damage to normal tissues and maximizing drug efficacy in malignant cells. This approach has been used successfully in several animal models, as well as in patients with advanced stage cancers.
Under physiological conditions, hematopoietic stem cells (HSC) continuously move between the bone marrow and the bloodstream. Artificially induced mobilization of HSC (by cytotoxic drugs or cytokines) from the bone marrow into the peripheral blood along with engraftment of intravenously injected HSC into the bone marrow, are the basis for HSC transplantations. Murine models show that unforced, physiological release of HSC is influenced by circadian fluctuations in expression levels of CXCL12, a chemokine essential for HSC mobilization and its receptor, CXCR4.25 Neural signals transmitted from the SCN to stromal cells in the bone marrow regulate circadian CXCL12 expression, while rhythmic expression of CXCR4 on HSC is controlled by core clock genes. Furthermore, by controlling the expression of the CXCL12/CXCR4 in the bone marrow niche, circadian rhythms continue to affect HSC mobilization even in the presence of pharmacological agents like the CXCR4 antagonist, AMD3100 or G-CSF, the most common mobilizer used in the clinic.47 Additionally, circadian variations of bone marrow engraftability were described in a congenic murine transplant model.48 These findings suggest that simple adjustments in timing of stem cell harvest and infusion may result in higher yields and greater engraftment, respectively. The bone marrow microenvironment is required for growth and survival of not only normal HSC but also that of leukemic stem cells (LSC). Furthermore, the hematopoietic niche may offer protection to LSC during chemotherapy and by doing so contribute to chemoresistance and disease relapse. Disrupting CXCL12/CXCR4 signaling has been shown to overcome resistance to chemotherapy and to kinase inhibitors in AML.49,50 In future studies, it will be interesting to determine if chronotherapy can be applied to maximize efficacy of drugs disrupting LSC/niche interactions, with minimum affect on HSC.
Local circadian control in hematopoietic cells may also modulate circadian susceptibility to chemotherapy. A recent study found that mice with different core clock gene mutations show variations in sensitivity to the chemotherapeutic agent cyclophosphamide.51 Interestingly, the differences were not attributed to changes in drug metabolic pathways but to changes in hematopoietic cells recovery rate, which was dependent on the functional status of Clock/Bmal1. Although neutrophil counts varied only slightly, the survival/recovery of B cells in response to cyclophosphamide showed significant differences between wild-type and core clock gene mutant mice. These findings demonstrate a mechanistic link between core clock factors expressed by immune cells and the sensitivity to conventional cytotoxic treatments.
Hormone-Related Breast and Prostate Cancers
The secretion of many endocrine and metabolic hormones is controlled by the day/night cycle, and reciprocally circulating levels of hormones influence circadian rhythms. While this bidirectional flow of information is critical for normal physiology, a growing number of studies suggest that circadian variation is particularly relevant to endocrine malignances. A remarkable example is the finding that overnight shift workers have an increase in hormone-related breast and prostate cancers, as well as other types of malignancies (Table 3).52-58
Table 3.
Abnormal circadian rhythms associated with cancer in animal models and humans
| Abnormality | Outcome | Cancer type | References | |
|---|---|---|---|---|
| Mice | Disruption of circadian rhythm either by SCN ablation or chronic jet lag | accelerated malignant growth and shortened survival | transplantable osteosarcoma and pancreatic adenocarcinoma | 91, 92 |
| Human | Disruption of circadian rhythms in night-shift workers | independent risk factor for cancer | NHL, breast, colorectal, endometrial, and prostate cancers | 32, 52-58 |
| Flattened or abnormal diurnal cortisol rhythms | predictor of survival | metastatic breast cancer | 93 | |
| Poor 24-h rest/activity rhythms | independent predictor for overall survival | metastatic colorectal cancer patients receiving chemotherapy | 94 |
Abbreviations: NHL, non-Hodgkin’s lymphoma.
Among the cancers known to be impacted by the circadian clock, breast cancer has received the most attention. In addition to epidemiological studies showing a clear correlation between disruption of circadian rhythms and breast cancer risk, a number of studies reported that Per1 and Per2 expression in both sporadic and familial primary breast tumors is significantly decreased compared to normal breast tissues.59-61 Expression of Per genes also correlates with expression of other genes implicated in breast cancers including Her2, BRACA1 and estrogen receptor. The deregulation of Per is most frequently associated with methylation of regions of the Per gene promoters. However, a large screen of human breast cancer genomes identified mutations in both Per1 and Per2 genes,62 supporting their role as tumor suppressors.
Compared with the relative large number of studies addressing circadian function in breast cells, fewer studies have explored the existence of circadian oscillations in the prostate, and molecular understanding of a possible interplay between circadian regulation and prostate cancer is largely unknown. We and others demonstrated circadian expression of the core clock genes in prostate and other male reproductive tissues in mice entrained to 12 hours light/dark cycles.63,64 On the other hand, rhythmic expression of core clock genes is severely impaired in serum-synchronized human prostate cancer cell lines. Additionally, meta-in-silico analysis showed that Per1 and Per2 expression is significantly lower in prostate carcinoma patients compared with normal controls, and that Per1/2 levels decreased with progression to metastatic disease. Population-based studies found associations between genetic variants in clock genes and prostate/breast cancer risk and survival.65-67
Forced overexpression of either Per1 or Per2 in breast and prostate cancer cells inhibits their proliferation in culture, while silencing of Per1/2 expression by siRNA accelerates their growth.68,69 Although the molecular mechanisms linking Per genes to breast and prostate cancers are still elusive and likely involve multiple cellular pathways, we recently showed that Per genes link the circadian system to estrogen and androgen signaling, providing at least one possible explanation for the strong association between hormonal related cancers and the circadian clock. The gonadal steroid hormones estrogen and androgen are essential to normal physiology and are important factors in breast and prostate tumorigenesis, respectively. These hormones mediate their affects mainly through the estrogen and androgen receptors (ER and AR) which are members of the nuclear receptor (NR) superfamily. Recent observations demonstrated intimate relationships between NR and central as well as peripheral circadian clocks; this is most notable in metabolic physiology.70,71 While several steroid receptors and their hormones have been linked to both circadian and reproductive rhythms, the significance of these connections to cancer is largely unknown. We found that Per2 interacts with ERα, and suppresses estrogen-mediated transcription of ER target genes in breast cells.69 In turn, Per2 itself is an estrogen-inducible gene, suggesting a feedback mechanism that couples the circadian clock to the estrogen pathway. Likewise, we showed that Per1 physically binds to AR.64 Furthermore, forced expression of Per1 in prostate cancer cells inhibits AR transcriptional activity, including DHT stimulation of PSA. Conversely, silencing of Per1 expression using siRNA is associated with increased transcriptional activity of AR. In turn, activated AR increases the transcription of Per1 associated with the recruitment of AR to an androgen response element in the Per1 promoter. Our results suggest that Per1 acts as an AR co-repressor coupling circadian rhythms to AR signaling in prostate cells (Fig. 2). The finding that Per1/2 are estrogen-and androgen-responsive genes provides a direct link between core circadian components and the ER/AR pathways. Temporal expression of all 49 murine NR was surveyed in murine liver, skeletal muscle and adipose tissues.70 Although AR was expressed in these metabolic tissues, its level did not show circadian expression. Because circadian regulation of gene expression is tissue-specific, we examined whether AR is rhythmically expressed in the prostate. Remarkably, AR mRNA levels oscillate in prostate tissue of mice entrained to 12-hours light/dark cycles, suggesting novel roles of AR in coupling the peripheral clocks to hormonal regulation.64 More generally, these studies point to an important role for the circadian regulation in proper prostate function, both locally and in the hypothalamic-pituitary-prostate axis.
Figure 2.
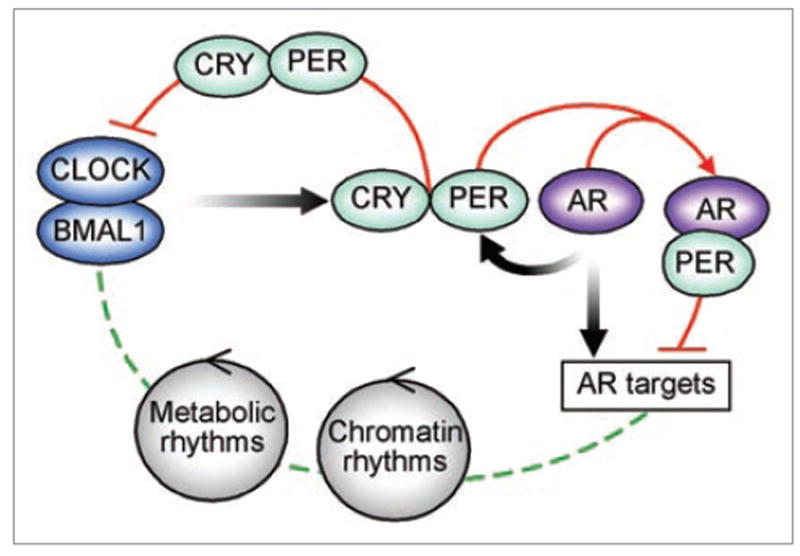
Per couples the circadian cycle to the androgen response. Clock and Bmal1 activate the transcription of core clock genes (e.g., Per and Cry) as well as tissue specific rhythmic genes (e.g., AR). Per and Cry heterodimerize and inhibit the Clock/Bmal1 complex. Tissue specific transcription factors like AR, additionally regulate the expression of core clock genes. In turn, Per binds to AR and inhibits AR transcriptional activity. Metabolic and chromatin remodeling cycles further integrate into the circadian control of normal cellular function.
Recently, acetylation and other chromatin modifications were discovered to be an intrinsic part of circadian function. The Clock protein acts as a histone acetyltransferase (HAT) that acetylates histones, as well as nonhistone proteins; and Sirt1, best known for its potential anti-aging properties, is a histone deacetylase (HDAC) that counteracts Clock.72,73 Additionally, activation of Hdac3 by the NR co-repressor, NcoR1, is a key step coupling epigenetic regulation to circadian and metabolic physiology.74 AR transcriptional activity is regulated by posttranslational modifications including acetylation and both Sirt1 and Ncor1 function as AR co-repressors.75-77 Sirt1 also regulates prostate cancer cell growth and chemoresistance in androgen-refractory cells.78 A potentially fruitful area of future studies is to examine whether circadian mediated modifications of acetylation plays a role in AR-dependent and -independent prostate cancer cell growth and drug sensitivity.
Any discussion of circadian hormone-related cancer interplay cannot be completed without the mention of melatonin, the main hormone relaying the circadian rhythm to the peripheral organs. In addition to its well-known role in circadian physiology, recent studies revealed that melatonin also has anti-tumor activities in various human cancers, in particular breast, colon, lung, melanoma and leukemia.79 A variety of mechanisms have been suggested to explain how reduced melatonin production (occurring artificially upon exposure to light at night, and physiologically with aging) may exaggerate cancer risk. That notwithstanding, melatonin inhibition of breast and prostate cancer cell growth has been attributed at least in part, to its interaction with estrogen and androgen signaling pathways.80,81 For example, in human breast cancer cells melatonin reduces the expression and activity of aromatase, a key enzyme in estrogen biosynthesis. In human prostate cancer cell lines melatonin induces nuclear exclusion of AR. Small-scale studies showed that combination of melatonin and tamoxifen (used to treat estrogen-positive breast cancer) can have modest synergistic benefits, and that melatonin may enhance the effects of some chemotherapeutic drugs used to treat breast and prostate cancer patients. ER and AR are already targets of widely used drugs; understanding their circadian regulation may therefore have tangible clinical implications.
In summary, over the last decade a wealth of data from interdisciplinary fields revealed that circadian regulation is hardwired into almost every aspect of human physiology. Though much remains unanswered, the notion that pharmacological manipulation of circadian genes can one day be applied to help prevent and treat complex multistep diseases like cancer, may be more than just a dream away.
References
- 1.Rosato E, Kyriacou CP. Origins of circadian rhythmicity. J Biol Rhythms. 2002;17:506–11. doi: 10.1177/0748730402238232. [DOI] [PubMed] [Google Scholar]
- 2.Roenneberg T, Merrow M. Circadian clocks—the fall and rise of physiology. Nat Rev Mol Cell Biol. 2005;6:965–71. doi: 10.1038/nrm1766. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Okamura H. Clock genes in cell clocks: roles, actions and mysteries. J Biol Rhythms. 2004;19:388–99. doi: 10.1177/0748730404269169. [DOI] [PubMed] [Google Scholar]
- 7.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–56. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–9. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 10.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–90. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belden WJ, Loros JJ, Dunlap JC. CLOCK leaves its mark on histones. Trends Biochem Sci. 2006;31:610–3. doi: 10.1016/j.tibs.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2009;41:81–6. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 13.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–81. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 14.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 16.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 17.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 18.Chen YG, Mantalaris A, Bourne P, Keng P, Wu JH. Expression of mPer1 and mPer2, two mammalian clock genes, in murine bone marrow. Biochem Biophys Res Commun. 2000;276:724–8. doi: 10.1006/bbrc.2000.3536. [DOI] [PubMed] [Google Scholar]
- 19.Smaaland R, Sothern RB, Laerum OD, Abrahamsen JF. Rhythms in human bone marrow and blood cells. Chronobiol Int. 2002;19:101–27. doi: 10.1081/cbi-120002594. [DOI] [PubMed] [Google Scholar]
- 20.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–5. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 21.Tsinkalovsky O, Smaaland R, Rosenlund B, Sothern RB, Hirt A, Steine S, et al. Circadian variations in clock gene expression of human bone marrow CD34+ cells. J Biol Rhythms. 2007;22:140–50. doi: 10.1177/0748730406299078. [DOI] [PubMed] [Google Scholar]
- 22.Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354:924–8. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 23.Gimble JM, Floyd ZE, Bunnell BA. The 4th dimension and adult stem cells: Can timing be everything? J Cell Biochem. 2009;10:569–78. doi: 10.1002/jcb.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granda TG, Liu XH, Smaaland R, Cermakian N, Filipski E, Sassone-Corsi P, et al. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19:304–6. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- 25.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–7. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 26.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 28.Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler H. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–36. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman AE, Zheng T, Stevens RG, Ba Y, Zhang Y, Leaderer D, et al. Clock-cancer connection in non-Hodgkin’s lymphoma: a genetic association study and pathway analysis of the circadian gene cryptochrome 2. Cancer Res. 2009;69:3605–13. doi: 10.1158/0008-5472.CAN-08-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, Boyle P, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–5. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahti TA, Partonen T, Kyyrönen P, Kauppinen T, Pukkala E. Night-time work predisposes to non-Hodg-kin lymphoma. Int J Cancer. 2008;123:2148–51. doi: 10.1002/ijc.23566. [DOI] [PubMed] [Google Scholar]
- 33.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–28. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozek K, Relógio A, Kielbasa SM, Heine M, Dame C, Kramer A, et al. Regulation of clock-controlled genes in mammals. PLoS One. 2009;4:4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skokowa J, Lan D, Thakur BK, Wang F, Gupta K, Cario G, et al. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat Med. 2009;15:151–8. doi: 10.1038/nm.1913. [DOI] [PubMed] [Google Scholar]
- 36.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 38.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104:3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4:4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–18. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–63. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sathyanarayanan S, Zheng X, Kumar S, Chen CH, Chen D, Hay B, et al. Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 2008;22:1522–33. doi: 10.1101/gad.1652308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;10:7–52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, et al. A Genome-wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 46.Lévi F. Chronotherapeutics: the relevance of timing in cancer therapy. Cancer Causes Control. 2006;17:611–21. doi: 10.1007/s10552-005-9004-7. [DOI] [PubMed] [Google Scholar]
- 47.Lucas D, Battista M, Shi PA, Isola L, Frenette PS. Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell. 2008;3:364–6. doi: 10.1016/j.stem.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Hondt L, McAuliffe C, Damon J, Reilly J, Carlson J, Dooner M, et al. Circadian variations of bone marrow engraftability. J Cell Physiol. 2004;200:63–70. doi: 10.1002/jcp.20032. [DOI] [PubMed] [Google Scholar]
- 49.Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–24. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–14. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/ BMAL1 transactivation complex. Proc Natl Acad Sci USA. 2005;102:3407–12. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–7. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 53.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 54.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–62. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 55.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Night-shift work and risk of colorectal cancer in the nurses’ health study. Natl Cancer Inst. 2003;95:825–8. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 56.Megdal SP, Kroenke CH, Laden F, Pukkala E, Schernhammer ES. Night work and breast cancer risk: a systematic review and meta-analysis. Eur J Cancer. 2005;41:2023–32. doi: 10.1016/j.ejca.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Kubo T, Ozasa K, Mikami K, Wakai K, Fujino Y, Watanabe Y, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549–55. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 58.Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007;67:10618–22. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 59.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 60.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuo SJ, Chen ST, Yeh KT, Hou MF, Chang YS, Hsu NC, et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454:467–74. doi: 10.1007/s00428-009-0761-7. [DOI] [PubMed] [Google Scholar]
- 62.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 63.Bebas P, Goodall CP, Majewska M, Neumann A, Giebultowicz JM, Chappell PE. Circadian clock and output genes are rhythmically expressed in extratesticular ducts and accessory organs of mice. FASEB J. 2009;23:523–33. doi: 10.1096/fj.08-113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, Koeffler HP. A role for the clock gene, Per1 in prostate cancer. Cancer Res. 2009;69:7619–25. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu LW, Zhu Y, Yu K, Zheng T, Yu H, Zhang Y, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–8. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi C, Mu L, de la Longrais IA, Sochirca O, Arisio R, Yu H, et al. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–20. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Downregulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423–31. doi: 10.1007/s10549-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 70.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 71.Teboul M, Gréchez-Cassiau A, Guillaumond F, Delaunay F. How nuclear receptors tell time. J Appl Physiol. 2009;107:1965–71. doi: 10.1152/japplphysiol.00515.2009. [DOI] [PubMed] [Google Scholar]
- 72.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 73.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–4. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–35. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–21. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol Endocrinol. 2002;16:1492–501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- 78.Kojima K, Ohhashi R, Fujita Y, Hamada N, Akao Y, Nozawa Y, et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373:423–8. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 79.Ravindra T, Lakshmi NK, Ahuja YR. Melatonin in pathogenesis and therapy of cancer. Indian J Med Sci. 2006;60:523–35. [PubMed] [Google Scholar]
- 80.Cos S, González A, Martínez-Campa C, Mediavilla MD, Alonso-González C, Sánchez-Barceló EJ. Estrogen-signaling pathway: a link between breast cancer and melatonin oncostatic actions. Cancer Detect Prev. 2006;30:118–28. doi: 10.1016/j.cdp.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Srinivasan V, Spence DW, Pandi-Perumal SR, Trakht I, Cardinali DP. Therapeutic actions of melatonin in cancer: possible mechanisms. Integr Cancer Ther. 2008;7:189–203. doi: 10.1177/1534735408322846. [DOI] [PubMed] [Google Scholar]
- 82.Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE, et al. Period 2 mutation accelerates ApcMin/+ tumorigen-esis. Mol Cancer Res. 2008;6:1786–93. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106:2841–6. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog. 2006;45:732–40. doi: 10.1002/mc.20198. [DOI] [PubMed] [Google Scholar]
- 87.Krugluger W, Brandstaetter A, Kállay E, Schueller J, Krexner E, Kriwanek S, et al. Regulation of genes of the circadian clock in human colon cancer: reduced period-1 and dihydropyrimidine dehydrogenase transcription correlates in high-grade tumors. Cancer Res. 2007;67:7917–22. doi: 10.1158/0008-5472.CAN-07-0133. [DOI] [PubMed] [Google Scholar]
- 88.Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 89.Eisele L, Prinz R, Klein-Hitpass L, Nückel H, Lowinski K, Thomale J, et al. Combined PER2 and CRY1 expression predicts outcome in chronic lymphocytic leukemia. Eur J Haematol. 2009 doi: 10.1111/j.1600-0609.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 90.Penas EM, Cools J, Algenstaedt P, Hinz K, Seeger D, Schafhausen P, et al. A novel cryptic translocation t(12;17)(p13;p12-p13) in a secondary acute myeloid leukemia results in a fusion of the ETV6 gene and the antisense strand of the PER1 gene. Genes Chromosomes Cancer. 2003;37:79–83. doi: 10.1002/gcc.10175. [DOI] [PubMed] [Google Scholar]
- 91.Filipski E, Delaunay F, King VM, Wu MW, Claustrat B, Gréchez-Cassiau A, et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64:7879–85. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 92.Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–7. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 93.Sephton SE, Sapolsky RM, Kraemer HC, Spiegel D. Diurnal cortisol rhythm as a predictor of breast cancer survival. J Natl Cancer Inst. 2000;92:994–1000. doi: 10.1093/jnci/92.12.994. [DOI] [PubMed] [Google Scholar]
- 94.Innominato PF, Focan C, Gorlia T, Moreau T, Garufi C, Waterhouse J, et al. Chronotherapy Group of the European Organization for Research and Treament of Cancer. Circadian rhythm in rest and activity: a biological correlate of quality of life and a predictor of survival in patients with metastatic colorectal cancer. Cancer Res. 2009;69:4700. doi: 10.1158/0008-5472.CAN-08-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]


