SUMMARY
The synthesis of active Klebsiella aerogenes urease via an 18-subunit enzyme apoprotein-accessory protein pre-activation complex has been well-studied biochemically, but thus far this complex has remained refractory to direct structural characterization. Using ion mobility-mass spectrometry, we characterized several protein complexes between the core urease apoprotein and its accessory proteins, including the 610 kDa (UreABC)3(UreDFG)3 complex. Using our recently-developed computational modeling workflow, we generated ensembles of putative (UreABC)3(UreDFG)3 species consistent with experimental restraints and characterized the structural ambiguity present in these models. By integrating structural information from previous studies, we increased the resolution of the ion mobility-mass spectrometry derived models substantially, and we observe a discrete population of structures consistent with all of the available data for this complex.
eTOC Blurb
Eschweiler et. al. demonstrate the use of ion mobility-mass spectrometry data for molecular modeling of the 18-subunit urease pre-activation assembly. By incorporating collision cross sections and other experimental datasets, the authors define a discrete population of putative structures consistent with all available experimental data.
INTRODUCTION
Protein-protein interactions are critical to nearly all complex cellular processes, thus making structural characterization of such interactions imperative to our understanding of biology (Robinson et al., 2007, Marsh and Teichmann, 2015). These interactions range from discrete protein dimers and other small oligomers (Venkatakrishnan et al., 2010) to large, labile interaction networks comprised of dozens of protein chains (Perkins et al., 2010). One such multi-protein system, the urease activation complex from Klebsiella aerogenes, features a wide range of protein subunit sizes, interaction strengths, and stable subcomplexes within a putative 18-subunit network that has been the focus of diverse structural biology efforts (Carter et al., 2009, Farrugia et al., 2013b, Chang et al., 2004, Quiroz-Valenzuela et al., 2008). Despite the presence of numerous structural datasets for this system, few direct measurements of its higher-order complexes have been made, and thus relatively little is known about the structure of the urease activation complex or its mode of action in vivo.
Ureases are an important class of enzymes responsible for the hydrolysis of urea to ammonia and carbamate in some bacteria, fungi, algae, and plants (Mobley et al., 1995, Mazzei et al., 2017). Significant attention has been paid to ureases due to their impacts on human health (Collins and Dorazio, 1993) and agriculture (Bremner, 1995). K. aerogenes urease, the subject of this study, is composed of three protein chains, UreA, UreB, and UreC, which form a trimer of trimers (UreABC)3 with a molecular weight of around 250 kDa (Farrugia et al., 2013b, Carter et al., 2011). X-ray crystallography has elucidated the details of the quaternary and tertiary structure for this enzyme, including details about its dinuclear Ni2+ active site featuring a carbamylated lysine metal ligand (Pearson et al., 1997). In contrast, less is known about the GTP and CO2-dependent assembly of the urease active site by the K. aerogenes urease accessory proteins UreD, UreE, UreF, and UreG (Farrugia et al., 2013b). Biochemical studies of these accessory proteins have provided insight into their specific roles in urease activation. Briefly, UreD is a relatively insoluble protein that binds directly to urease (Park et al., 1994), but is not competent for urease activation without the other accessory proteins (Carter and Hausinger, 2010). Recent studies have provided experimental evidence for a Ni2+ transport channel through UreD, highlighting its unique role in supplying Ni2+ to urease apoprotein (Farrugia et al., 2015). UreF, a similarly insoluble protein, binds to the UreD:urease apoprotein complex and acts as a GTPase modulator to the GTPase UreG (Boer and Hausinger, 2012). The latter protein is soluble, its GTPase activity is essential for urease activation, and it binds Ni2+ (Boer et al., 2010, Moncrief and Hausinger, 1997). Finally, UreE is a nickel-binding metallochaperone protein that delivers the metal to UreG (Boer et al., 2010).
Early hypotheses for urease activation proposed the sequential binding of UreD, UreF, and UreG to the urease apoprotein at each of its three nascent active sites to form an octadecameric pre-activation complex, (UreABC)3(UreDFG)3, that could accept Ni2+ ions from UreE before performing a GTP-dependent Ni2+ insertion event (Carter et al., 2009, Farrugia et al., 2013b). More recently, we characterized a soluble version of a stable complex of (UreDFG)2 that binds to the urease apoprotein and directly forms the (UreABC)3(UreDFG)3 complex (Farrugia et al., 2013a). Helicobacter pylori forms a similar complex, named (UreHFG)2, that was structurally characterized (Fong et al., 2013). UreE is likely to deliver Ni2+ to UreG prior to its incorporation into the (UreABC)3(UreDFG)3 complex (Yang et al., 2015, Hausinger, 2017).
Despite its importance, the (UreABC)3(UreDFG)3 pre-activation complex has eluded detailed structural characterization. One factor precluding such analysis is the lability of this complex, which results in multiple coexisting subcomplexes that make interpretation of any dataset extremely difficult without a high-resolution separation step. Importantly, several subcomplexes have been identified by native mass spectrometry (MS) (Farrugia et al., 2013a) and chemical crosslinking (Farrugia et al., 2013a, Chang et al., 2004), including (UreABC)3(UreDFG), (UreABC)3(UreDFG)2, (UreABC)3(UreD)3, and (UreABC)3(UreDFF). Detailed analysis of crosslinked peptides by tandem MS revealed putative interaction sites of urease with UreD (Chang et al., 2004), which have also been supported by small-angle X-ray scattering (SAXS) datasets for samples containing (UreABC)3(UreD)3 (Quiroz-Valenzuela et al., 2008). These datasets were integrated with molecular docking to provide the most comprehensive picture of the (UreABC)3(UreDFG)3 structure to date (Ligabue-Braun et al., 2013); however, without direct observation of the complex it is difficult to assign a confidence level to the model produced.
In this study, we use ion mobility-mass spectrometry (IM-MS) to characterize samples relating to a derivative of the (UreDFG)2 complex as well as complexes formed between this version of UreDFG and the urease apoprotein. IM-MS is a tandem methodology that separates protein and protein complex ions produced by using nano-electrospray ionization (nESI) under native conditions, first by size using IM and then by m/z using MS. IM-based size separations can be calibrated to produce orientationally-averaged collision cross section (CCS) values that can be used, along with connectivity information recovered from native MS, to restrain modeling efforts (Eschweiler et al., 2017). We utilize a previously-reported maltose binding protein-UreD (MBP-UreD) fusion protein to increase the solubility of the system, while still allowing for formation of key protein complexes that are competent activators of urease (Carter and Hausinger, 2010). Furthermore, we report direct mass spectrometric observation of the hypothesized (UreABC)3(MBP-UreDFG)3 complex and, using IM-derived CCS information (Ruotolo et al., 2008), we develop a method for coarse-grained modeling (Politis et al., 2010, Hall et al., 2012) of (UreABC)3(MBP-UreDFG)3 and its subcomplexes that allows us to characterize the conformational space of (UreABC)3(UreDFG)3 in a manner that is consistent with all available structural data.(Chang et al., 2004, Quiroz-Valenzuela et al., 2008, Farrugia et al., 2013a)
RESULTS
M-MS of (MBP-UreDFG)2 and (UreABC)3(MBP-UreDFG)3 Containing Samples
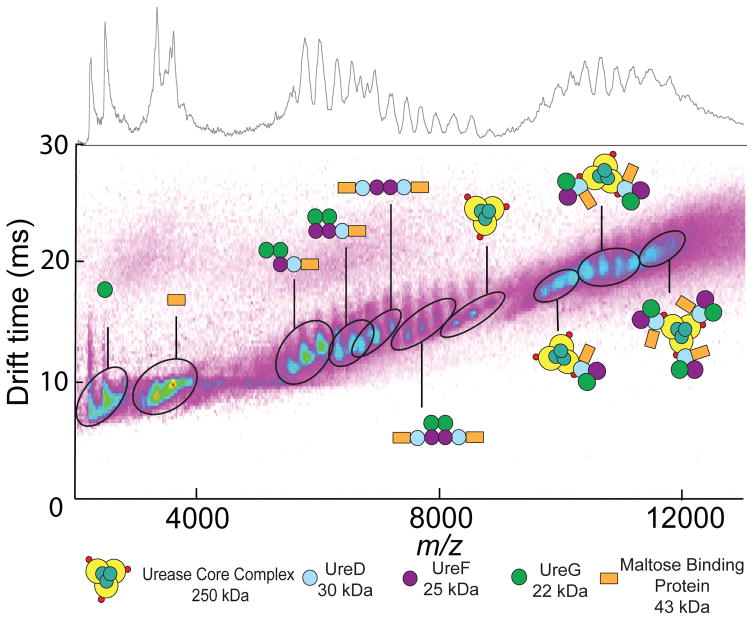
IM-MS analysis of samples containing the fusion protein MBP-UreD, UreF, and UreG revealed the predicted (MBP-UreDFG)2 complex plus a number of subcomplexes consistent with our previous study (Farrugia et al., 2013a). Analysis of the CCS of each subcomplex revealed values consistent with those predicted from the homologous H. pylori urease accessory protein complex, (UreHFG)2, for which a crystal structure exists (Fong et al., 2013), indicating a high degree of topological agreement between these two complexes. After evaluation of the complexes derived from (MBP-UreDFG)2, we incubated this complex with the (UreABC)3 apoprotein before subsequent IM-MS analysis. The resulting dataset (Figure 1) featured many of the subcomplexes found in the (MBP-UreDFG)2 sample, plus a host of complexes derived from interactions between MBP-UreDFG and (UreABC)3. Notably, in this dataset we also observe a new complex, the (MBP-UreDF)2 assembly, which was not observed in the absence of urease apoprotein; however, the corresponding (UreHF)2 analog was observed in the H. pylori urease activation pathway (Fong et al., 2013). In contrast to our previous study, these new datasets also contained convoluted signals at high molecular weight, presumed to be related to complexes of the urease core with accessory proteins. After developing a model of the data based on expected masses of predicted urease complexes, we deconvoluted the data to reveal direct evidence of the fully assembled (UreABC)3(MBP-UreDFG)3 complex, as well as the subcomplexes (UreABC)3(MBP-UreDFG)2 and (UreABC)3(MBP-UreDFG) (Figure S2) (McKay et al., 2006). These results provide further evidence for the modular addition of MBP-UreDFG to urease to form the pre-activation complex, and also provide structural information for the (UreABC)3(MBP-UreDFG)3 complex..
Figure 1. IM-MS analysis of (UreABC)3(MBP-UreDFG)3 and its subcomplexes.
IM-MS analysis of (UreABC)3(MBP-UreDFG)3 is presented as a plot of drift time vs m/z, with the standard m/z dimension projected on top of the figure. These data reveal the masses and collision cross sections of many subunits, and subcomplexes that comprise the 610 kDa (UreABC)3(MBP-UreDFG)3 octadecamer (Figure S2). IM-MS datasets for other urease-related subcomplexes can be found in Figure S3–S4
IM-MS-Based Modeling of (UreABC)3(MBP-UreDFG)3
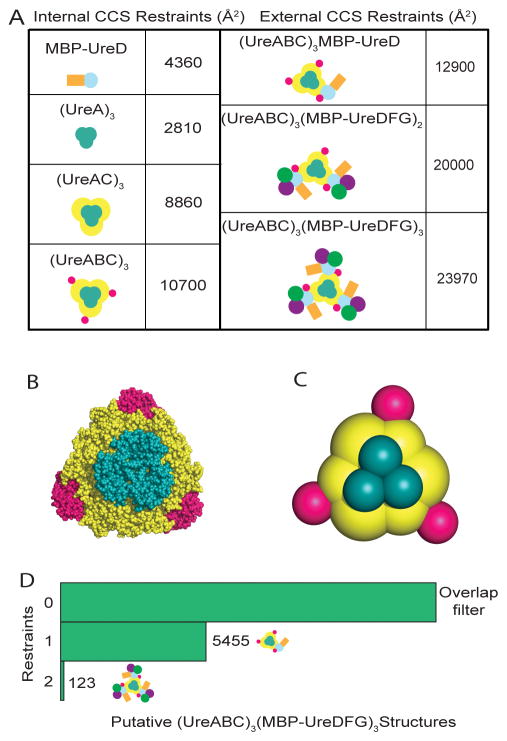
After deconvolution of mass spectral data, we calculated CCS values for (UreABC)3(MBP-UreDFG)3 and its subcomplexes based only on charge states where minimal or no interference was observed from neighboring signals in the mass spectrum (Figure S2). In addition, we extracted CCS data for the urease core, specifically (UreA)3, (UreAC)3, and (UreABC)3, and for (UreABC)3-(MBP-UreD) from other datasets. (Figure 2A and supplementary Figure S3–S4). We hypothesized that these data would be sufficient to restrain a coarse-grained model of the (UreABC)3(MBP-UreDFG)3 complex and began our modeling by representing each protein within the complex as a sphere with radius corresponding to its measured or calculated CCS. Specifically, we derived CCS values for UreA, UreC, UreF, and UreD from the trajectory method approximation in IMPACT, while deriving values for UreB, UreG, and MBP from calibrated experimental drift times. In terms of the urease apoprotein core complex (UreABC)3, our experimental CCS values were in close agreement with trajectory method approximations from IMPACT, with an error of −1.4%. We leveraged these data, as well as experimental data for (UreA)3 and (UreAC)3 to build a coarse-grained model of the urease core that matched our experimental dataset with errors <1% (Figure 2B and 2C). Importantly, we found it was necessary to model UreC as two spheres, representing each domain within the protein chain, as it was otherwise impossible to accurately recapitulate the shape and CCS of the complex.
Figure 2. IM-MS restraints for building molecular models of (UreABC)3(MBP-UreDFG)3.
(A) Left panel: CCS restraints used to build the urease core scaffold (UreABC)3 and to restrain the interaction between maltose binding protein (MBP) and UreD. Right panel: CCS restraints for target complexes used to filter large ensembles of structures generated using a Monte Carlo search in IM-MS_modeler. (B) The X-ray structure of (UreABC)3 compared to (C) a coarse-grained model generated with IM-MS data that was used as a scaffold for modeling of (UreABC)3(MBP-UreDFG)3. (D) Results of filtering an ensemble of 54,000 putative structures of (UreABC)3(MBP-UreDFG)3 based on biophysical and experimental data. “0” restraint corresponds to filtering only by spherical overlap parameters related to the interaction geometries of proteins. “1” and “2” restraints incorporate filters for the experimental CCS values +/− 3% for (UreABC)3(MBP-UreD) and (UreABC)3(MBP-UreDFG)3, respectively.
Once an accurate model of the urease core was developed using gas-phase restraints, we developed a restraint-based scoring function for a Monte-Carlo search for an ensemble of representative structures of the complete pre-activation complex that agreed with our experimental IM-MS data. The restraints in our initial scoring function included a rigidly restrained model of MBP-UreD, however all other restraints were defined simply by connectivity, which allows subunits to adopt a range of distances and orientations as long as they remain in contact. Some of these connectivity restraints were derived from previous studies which demonstrated connectivity between UreC:UreD, UreB:UreD, UreD:UreF, and UreF:UreG (Chang et al., 2004, Boer and Hausinger, 2012). Although detailed structural information is available for the UreH(~UreD):UreF and UreF:UreG interactions found in homologous complexes (Fong et al., 2013), we chose to use this data to assign only protein-protein connectivity, rather than rigid distance restraints to avoid biasing the model toward interactions that may not be present in the (UreABC)3(MBP-UreDFG)3 structure. In general, our Monte Carlo search optimized the following scoring function from a random set of coordinates while considering only C3 symmetric structures around the rigidly-restrained UreABC core:
Where H is a harmonic function on the distance between the first two arguments and centered around the third argument; and SQ is a square function defining the center-to-center distance between the two arguments must be between 15% and 45% of the sum of their radii (Hall et al., 2012). In this scoring function, all of the square functions must have scores of 0 for the model to be accepted, while simple, linear harmonic functions are minimized to near-zero scores by an arbitrary force constant. We intend the simplicity of this restraint function to limit biases during Monte Carlo search steps. Overall, We generated 54,000 possible conformations for the complex of MBP, UreD, UreF, and UreG with respect to the urease core structure, with the same C3 symmetry enforced as found in the core (Pearson et al., 1997). Since the scoring function did not contain penalties for unphysical levels of overlap between components without explicitly-defined pairwise restraints, a final filtering step was implemented to ensure that no components overlapped with each other beyond 45% of their summed radii, resulting in a much smaller ensemble of 14,073 putative structures, which we subjected to the next round of filtering based on agreement with experimental CCS data.
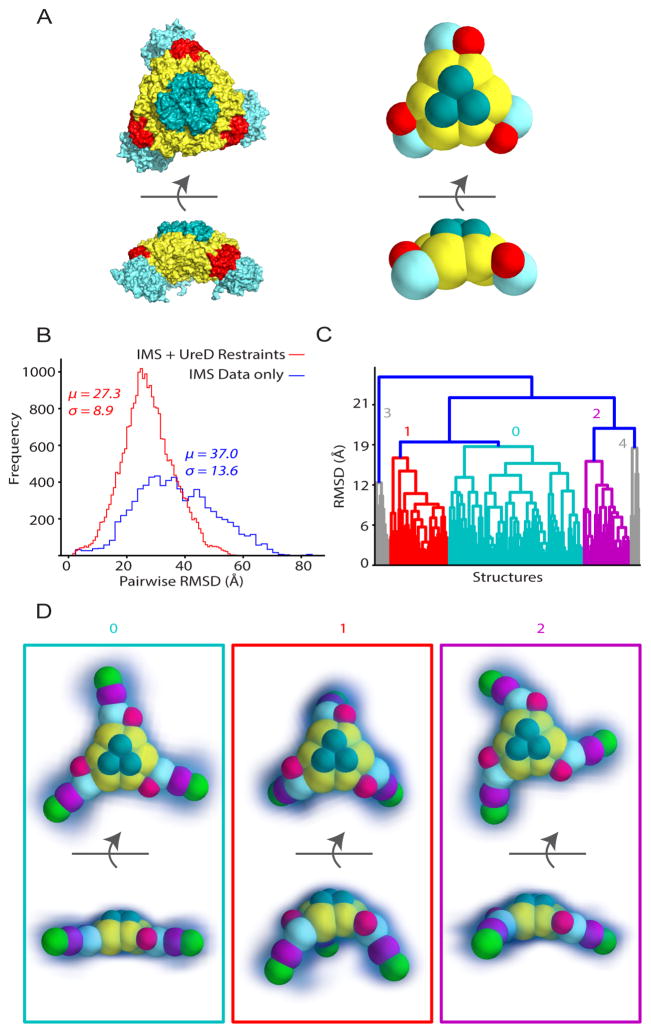
In this step, we used IMPACT to calculate CCS values for each model, as well as subcomplexes within a given model. Models were passed into the filtered ensemble if they agreed with the experimental CCS values for (UreABC)3(MBP-UreDFG)3, (UreABC)3(MBP-UreDFG)2, and (UreABC)3(MBP-UreD) within +/− 3%. We chose this value based on literature precedence as well as our own observations of dozens of urease-related CCS measurements that showed standard deviations no greater than 2% (Figure S1) (Bush et al., 2010). It is important to point out that while the above error description accurately reflects the uncertainty associated with experimental CCS values, it is not indicative of the error in our modeling protocol, which produces coarse-grained structures of high precision (<1%) owing to the direct relationship between CCS values and the spherical representations of subunits used in our models. In Figure 2D, filtering the ensemble based on CCS restraints extracted from (UreABC)3(MBP-UreD) and (UreABC)3(MBP-UreDFG)3 each decreased the size of the ensemble, indicating that each restraint added unique information about the structure of the pre-activation complex. We subjected the structures in the resulting ensemble, matching all biophysical and experimental CCS restraints, to structural analysis by hierarchical clustering. The resulting dendrogram (Figure 3A) reveals the presence of several structural families within the ensemble. Previous work has indicated that the relative abundance of a structural family within an ensemble is a good indicator of significance, and that extremely minor (<5% relative abundance) families are in many cases spurious and unlikely to represent important structural classes. (Hall et al. 2012, Eschweiler et al. 2017) Therefore, in this study we did not consider in detail those clusters with less than 5% abundance, and moved forward with a thorough structural analysis of only the major structural families in the ensemble, denoted as cluster 0, cluster 1, and cluster 2. (See Figure S5A for structural data pertaining to minor clusters and Table S1 and S2 for CCS values).
Figure 3. Hierarchical clustering reveals ambiguity in under-restrained models.
(A) An ensemble of 123 structures that agree with all experimental restraints was subjected to hierarchical clustering analysis to identify structural families within the group. In this example, the ensemble clustered into 3 well-defined structural families, shown by clusters 0, 1, and 2. Clusters 3, 4, and 5 are not considered because of <5% abundance and, thus, grayed out. (B) Each structural family is represented by the median structure and the kernel density function (blue densities) estimated from the structural ensemble. CCS values of the median structures shown ranged from 23706 Å2 to 24095 Å2. (See Figure S5A for structures of clusters 3, 4, and 5 and Tables S1 and S2 for CCS data)
In Figure 3B, each cluster is visualized by plotting the median structure of each cluster plus the kernel density function that represents the probability density of structures around the median. Visual analysis of these clusters reveals key ambiguities that are present in our data. First, the position of the MBP (brown spheres) cannot be resolved, which is expected because it can adopt many orientations around UreD (light blue spheres) within the model. Next, the position of UreD itself is changed substantially between clusters, anchoring the UreDFG assembly to opposite sides of UreB (red spheres) in cluster 1 and cluster 2, and in front of UreB in cluster 0. Our models suggest that this positioning plays a role in the possible configurations for UreF (purple spheres) and UreG (green spheres) within the ensemble, where the clusters with UreD in plane with UreB and UreC (clusters 1 and 2) adopt similar configurations for UreF and UreG, and are in contrast to the conformations found in cluster 0.
Although structural ambiguity was present in these models, we were encouraged by the clustering of the ensemble into distinct groups having structural differences that were easily assessed qualitatively. Since the major sources of ambiguity for non-MBP urease accessory proteins hinged on the positioning of UreD relative to UreB and UreC, we sought to incorporate data from other sources into our model to improve the confidence in our structure assignment for the pre-activation urease complex.
Integration of Additional Structural Data for Improved Structural Resolution
To resolve the structural ambiguity within our IM-MS-derived models, we looked to previously reported structural data to incorporate into our model. Ligabue-Braun and colleagues [21] have previously reported a model for the (UreABC)3(UreDFG)3 complex based on molecular docking that broadly agrees with existing crosslinking (Chang et al., 2004) and SAXS (Quiroz-Valenzuela et al., 2008) datasets relating to the (UreABC)3(UreD)3 complex. Specifically, the positioning of UreD in the Ligabue-Braun (UreABC)3(UreD)3 model is broadly consistent with chemical crosslinks between UreC K401 and the UreD N-terminus, UreB K76 and the UreD N-terminus, and with deactivation of UreC K515 crosslinking upon binding of UreD. The simulated SAXS profile for the (UreABC)3(UreD)3 conformation is also in agreement with experimental data. Since our IM-MS derived model is of too low resolution to incorporate atomic-level distance restraints derived from chemical crosslinks, we used the (UreABC)3(UreD)3 structure put forth by Ligabue-Braun to restrain the position of UreD in our coarse-grained model. Although Ligabue-Braun and colleagues provided a model for the fully assembled (UreABC)3(UreDFG)3 complex, we chose not include any restraints related to higher order complexes into our model, as we could find little experimental support for the positioning of UreF and UreG within the (UreABC)3(UreDFG)3 complex. When we implemented these new restraints into the (UreABC)3(MBP-UreD)3 subcomplex, the resulting structures were largely in agreement with the UreB:UreC:UreD configuration in cluster 2 from our initial modeling effort (Figure 3B). MBP-UreD is found proximal to the nascent active site, on the side of UreB, and oriented toward the back of the urease complex (Figure 3 A–B).
Using this new model as a scaffold (Figure 4A), we repeated our Monte-Carlo search to generate 52,000 possible structures that represent the conformational space available to UreF and UreG within this restraint space.. Filtering this ensemble by the previously discussed biophysical restraints resulted in 17,069 candidate structures, which we filtered by agreement with experimental CCS to yield a population of 226 structures. In this ensemble, we identify three major clusters that appear to define a discrete range of conformations for the complex (Figure 4C; note that spheres corresponding to MBP have been removed for clarity). Indeed, the RMSD distribution between models within this new ensemble is centered around 27.3 Å compared to 37 Å in the unrestrained ensemble, and the distribution also has a lower standard deviation, indicating that restraining UreD resulted in a model with significantly less ambiguity. (Figure 4B). Visualization of these structural families revealed that the experimental data define a discrete structural space that lies between a largely planar and extended structure and a slightly more compact structure featuring UreDFG modules directed toward the back of (UreABC)3 (Figure 4D) (See Figure S5B for structural data pertaining to minor clusters and Table S3 and S4 for CCS values).
Figure 4. Resolving ambiguity by integration of new data.
(A) A previously published all-atom model of (UreABC)3(UreD)3 was used to restrain the position of UreD in our coarse-grained scaffold structure. (B) Resolution of the model is significantly increased as seen by the decrease in the mean (μ) and standard deviation (σ) of the pairwise RMSD distribution between models generated with (red) and without (blue) the additional restraint. (C) Hierarchal clustering of the new ensemble reveals three distinct structural families, shown by clusters 0, 1, and 2. The grey clusters, 3 and 4, are not considered because of low abundance.. (D) Median structures and kernel density estimates (blue densities) for the two structural families identified. The CCS values for median structures shown ranged from 20949Å2 to 21158 Å2 (See Figure S5B for structures of clusters 3 and 4 and Tables S3 and S4 for CCS data).
Comparison with Previous Models
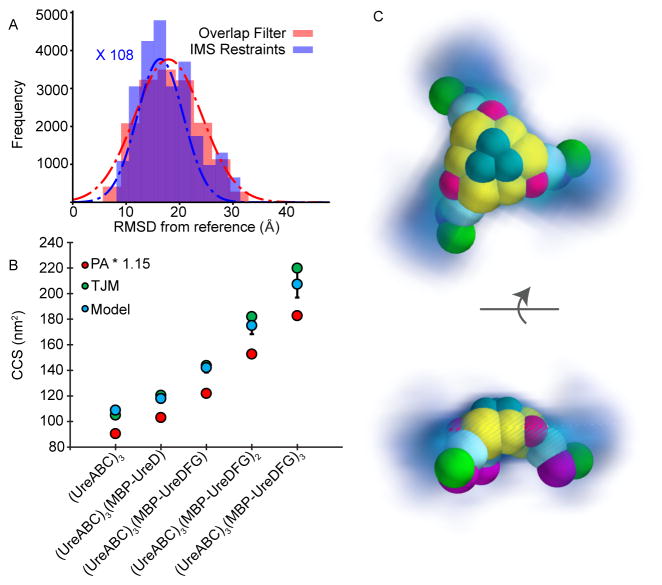
Although our coarse-grained model incorporated some elements of the model put forward by Ligabue-Braun [21], a detailed analysis of our IM-MS derived structure ensemble reveals more potential conformations for the urease (UreABC)3(MBP-UreDFG)3 complex considering all available data. Figure 5A shows the RMSD distributions for two structural ensembles derived from the coarse-grained version of the Ligabue-Braun model. The first ensemble, represented by a red histogram with a red Gaussian fit, incorporates no experimental CCS data, and represents the entirety of conformational space that can be adopted by UreF and UreG on a scaffold of (UreABC)3(MBP-UreD)3. The same ensemble after filtering by experimental CCS restraints is represented in blue with a blue Gaussian fit. (Frequency axis is scaled to allow for unbiased comparison). This result reveals that an ensemble filtered by our experimental CCS data is only minimally enriched for models akin to the Ligabue-Braun model, indicating only limited alignment between our experimental data and the model. Since a direct comparison of experimental CCS values with those calculated from the Ligabue-Braun model is difficult due to the presence of the MBP tag in our experimental data, we also compared the CCS values for our entire ensemble of experimentally-restrained models (with MBP removed) with CCS values calculated from the Ligabue-Braun model. In Figure 5B, we compare CCS values for our experimentally restrained models with theoretical CCS values computed from the all-atom structures proposed by Ligabue-Braun. CCS values for our experimentally restrained models are shown as blue dots, with error bars representing 2 standard deviations within the ensemble. CCS values for the Ligabue-Braun model were calculated using both a linearly scaled projection approximation method (PA*1.15) as well as the trajectory method estimation method (TJM) within IMPACT in red and green circles respectively. Interestingly, although the TJM values agree very well with our experimental measurements overall, we note increasing deviation between these values as additional subunits are added. In contrast, the scaled projection approximation values for the high-resolution models put forth by Ligabue-Braun are consistently 11% to 17% lower than the CCS values associated with models generated by IM-MS. To understand these deviations from model CCS values, we built a coarse-grained model based on Ligabue-Braun’s structure and superimposed it into the density cloud calculated from our IM-MS restrained ensemble (Figure 5C). This representation reveals that the ensemble restrained by our experimental CCS generally adopts a more extended conformation than the previously-reported model. Although a portion of the experimentally-derived models do agree well with the Ligabue-Braun model, specifically those in cluster 1 shown within Figure 4, it is clear that our IM-MS experiments restrain a conformational space containing alternative conformations.
Figure 5. Comparing IM-MS-derived models with structures from molecular docking.
(A) RMSD distributions of IM-MS derived models from a reference model generated using molecular docking and integrative modeling. The red distribution (red Gaussian fit) represents an ensemble of structures generated from the (UreABC)3(UreD)3 scaffold with no filtering by experimental CCS restraints. The blue ensemble (blue Gaussian fit) represents the same ensemble filtered by experimental CCS (frequency axis multiplied by 108 to have similar amplitude as red Gaussian fit). (B) Experimental and predicted CCS values for several urease complexes. Blue dots indicate IM-MS-derived models where the error bars represent two standard deviations, green and red dots represent calculated CCS values for the reference structure by the trajectory method approximation and scaled projection approximation, respectively. (C) Qualitative comparison of the kernel density function of an IM-MS-derived ensemble with a coarse-grained representation of the reference structure.
DISCUSSION
In this study, we characterized the 610 kDa, 18-subunit urease apoprotein pre-activation complex using IM-MS. To our knowledge, this complex, among the largest heterocomplexes to be characterized by IM-MS, and has not been directly observed by any other method. We used CCS values derived from ion mobility drift times of the fully assembled complex as well as several subcomplexes to build coarse-grained models revealing possible gas-phase structures of the complex. Our IM-MS data alone were not sufficient for unambiguous structural assignment, but when combined with data from chemical crosslinking, SAXS, and molecular modeling, we were able to define a narrow population of possible structures falling within our experimental restraints. Our model shares major structural features with a model proposed by computational docking, however it differs in the angle of UreDFG modules relative to the urease core structure. By estimating kernel density functions for ensembles of experimentally-restrained structures, we visualized the discrepancies between our experimental data and the previously reported model. These discrepancies may be due to gas-phase rearrangements of the (UreABC)3(MBP-UreDFG)3 complex, they may be representative of how the complex can alter its structure under different experimental conditions, or they may relate to the scarcity of experimental data incorporated into previous models. We note that due to the size and putative structures of the proteins involved, a scenario that rationalized the above-described differences based solely on a gas-phase rearrangement is unlikely, and that the relative flexibility of the urease pre-activation complex has been discussed in detail previously (Quiroz-Valenzuela et al., 2008, Ligabue-Braun et al., 2013). It is also worth noting that the flatter arrangement of accessory proteins on the urease core depicted in Figure 4D cluster 0 are comparable with analogous interactions presumed to occur on the spherical urease cores of ((UreAB)3)4 H. pylori protein (PDBID 1E9Z) (Ha et al., 2001) or the back-to-back arrangement of the jack bean enzyme (PDBID 3LA4) (Balasubramanian and Ponnuraj, 2010).
In summary, the model of the urease pre-activation complex presented in this report represents the most restrained structure of the assembly to date, representing a consensus of datasets acquired through IM-MS, chemical cross-linking, and SAXS experiments reported from multiple laboratories. Our modeling approach relies upon experimental and rigorously-defined theoretical CCS values mapped directly into our models as spheres. Based on the simplicity of the model components and its scoring function, we assume the errors associated with inefficient or biased sampling and the incorporation of inaccurate CCS values are small compared to the structural variability defined by the data and the associated experimental errors. Clearly, urease activation includes additional steps and protein binding events in order to load the enzyme with its required dinuclear Ni2+ metallocenter, but given the information content presented in this report we expect that our model will drive new discussions surrounding the role of this activation complex in the context of current urease activation mechanisms. Furthermore, the lability and size of this complex represents a frontier for the IM-MS technique in terms of its capabilities to build structural models of such large protein hetero-oligomers, and points to a bright future for the tool in similar structural biology efforts.
Star Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brandon Ruotolo (bruotolo@umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Escherichia coli BL21-Gold(DE3) cells co-expressing pEC005 (with ureFG cloned into pACT3) and pEC002 (with ureD cloned into pASKIBA3plus) generated the heterotrimeric (MBP-UreDFG)2 species containing a maltose binding protein (MBP) fusion of UreD (MBP-UreD) along with UreF and UreG (Farrugia et al., 2013b) The cells were grown in lysogeny broth (LB) supplemented with 300 μg/mL ampicillin and 50 μg/mL chloramphenicol with shaking at 37 °C until the OD600 reached ~0.4. Protein production was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG); the cultures were incubated at 28 °C with shaking for 16 h, and cells were harvested by centrifugation. E. coli BL21-Gold(DE3) harboring pUreAC were used to produce (UreAC)2 (Carter et al., 2011) The cells were grown in LB supplemented with 300 μg/mL ampicillin with shaking at 37 °C until they re ached an OD600 of ~0.5. Protein production was induced by the addition of 1 mM IPTG; the cultures were incubated under the same conditions for 3 h, and cells were harvested by centrifugation. E. coli BL21-Gold(DE3) cells transformed with pKK17 were used for isolation of urease apoprotein (Carter et al., 2011) The cells were grown with shaking in LB supplemented with 300 μg/ml ampicillin at 37°C until the cells reached an OD600 of ~0.5, at which point the culture was induced with 0.1 mM IPTG and allowed to incubate overnight until the cells were harvested.
METHOD DETAILS
Sample Preparation
The (MBP-UreDFG)2, (UreAC)3, and (UreABC)3 samples were purified as reported previously (Farrugia et al., 2013, Carter and Hausinger, 2010, Carter et al., 2011). In all cases, cell pellets containing the desired protein were resuspended in PEB buffer (20 mM Na-phosphate buffer, pH 7.4, 1 mM EDTA, 1 mM 2-mercaptoethanol) and disrupted by sonication. Soluble cell-free extracts were obtained by centrifugation at 100,000 g for 1 h at 4°C. The (MBP-UreDFG) 2 protein was isolated by sequential chromatography on amylose, DEAE-Sepharose, and Superdex 200 columns. The (UreAC)3 and (UreABC)3 proteins were isolated by applying the appropriate cell-free extracts to DEAE-Sepharose columns preequilibrated with PEB buffer and running a linear gradient from 0 M to 1 M KCl in PEB buffer. The fractions containing the appropriate protein were pooled, adjusted to 1.5 M KCl, and applied to phenyl-Sepharose CL-4B (GE Healthcare) columns preequilibrated with PEB buffer containing 1.5 M KCl. The columns were washed with equilibration buffer, and bound proteins were step-eluted with PEB buffer containing no KCl. Fractions containing pure proteins were pooled and dialyzed against TEB buffer containing 25 mM NaCl. (UreABC)3(MBP-UreDFG)x samples were prepared by incubating (UreABC)3 with a 3-fold excess of (MBP-UreDFG)2 for 30 min before flash freezing and storage at −80 °C.
IM-MS
Samples were buffer exchanged into 200 mM ammonium acetate using Micro Bio-Spin P-30 columns (Bio-Rad, Hercules, CA) at an initial concentration of ~1 μM. The final concentrations of the samples ranged from 100 to 900 nM, based on expected losses during buffer exchange. We performed IM-MS experiments on a Synapt G2 IM-MS platform (Waters Corp., Milford, MA) equipped with a nESI source. Briefly, the capillary voltage was set to 1.5 kV, with sampling and extraction cone voltages set to 0 V to preserve noncovalent interactions. The trap and transfer collision energies were both set to 4 V. Optimal mobility parameters were as previously published (Zhong et al., 2011), with IM gas pressure set at approximately ~4 mBar with the wave height and wave velocity set to 15 V and 150 m/s, respectively. Data were processed using Masslynx and Driftscope (Waters Corp., Milford, MA). Mass assignments were calculated using the maximum entropy method as implemented in ESIprot (Winkler, 2010).
Analysis of Urease complexes mass spectrum
In order to assign the charge states and Urease complex species to the mass spectrum in Figure 1, we followed a previously reported procedure to analyze heterogeneous complexes’ mass spectrum (McKay et al., 2006). Briefly, Gaussian curves were fitted to determine the centroid m/z values of the peaks in 9500 to 12000 m/z region. 21 different charge states were assigned to the base peak to determine the mean molecular mass and standard deviation. Base peak of +41 charge state yielded the lowest standard deviation. However, the measured mass did not correspond to any Urease complexes’ anticipated mass. This, along with broad and unresolved peaks, indicated that multiple species are present but not resolved within the spectrum.
In order to deconvolute the data, we modeled the mass spectrum as a combination arising from (UreABC)3(MBP-UreDFG)1, (UreABC)3(MBP-UreDFG)2, and (UreABC)3(MBP-UreDFG)3. All possible charge states spanning over the m/z space for all species were screened. The individual spectra were summed, and the intensities and peak widths were adjusted to minimize the sum of residual squares. Only charge states with significant intensity were include for each species. With the select charge state distributions for each species, the intensities and peak width were adjusted iteratively until convergence providing the summed spectrum that agrees very well with the experimental spectrum (Figure S2). The charge state assignments shown in Figure S2 were used for CCS calculations.
CCS Calibration
CCS values were determined as previously described (Ruotolo et al., 2008, Bush et al., 2010). cytochrome c, avidin, alcohol dehydrogenase, and glutamate dehydrogenase were used as calibrants for the wide range of CCS values observed. Typical calibration curves produced correlation coefficients of greater than 0.99.
QUANTIFICATION AND STATISTICAL ANALYSIS
IM-MS Data Processing
Data were processed using Masslynx and Driftscope (Waters Corp., Milford, MA). Mass assignments were calculated using the maximum entropy method as implemented in ESIprot (Winkler, 2010), or using a more advanced model as discussed above. Measured drift time values were converted to collision cross sections based on the calibration curve mentioned above, where all calibrant ion drift times are related to absolute collision cross section measurements made on drift tube instruments.(Ruotolo et al., 2008)
Coarse-Grained Modeling
Our general method for coarse-grained modeling is similar to previously described protocols (Politis et al., 2010) and adds modules associated with assessing the ambiguity of the models produced (Eschweiler et al. JASMS 2017). Using the Integrative Modeling Platform library in Python (Russel et al., 2012), we represented individual protein subunits as spheres with radii corresponding to their experimental or calculated CCS. We restrained higher-order complexes by specific geometric constraints or more ambiguous connectivity restraints as defined by the experimental data acquired. We generated an ensemble of putative models by repeated optimization of a scoring function built from the above restraints. Briefly the Monte Carlo parameters used in the study are:
(initial_temperature = 1000, final_temperature = 100, Temp_gradient = 0.999, mc_cool_cycles = 500, mc_cool_steps = 5000, mc_cycles = 1000, mc_steps = 1000, optimization_cycles = 10)
We note that our Monte Carlo algorithm is split into two stages, a cooling step and a constant temperature step, as noted in these parameters. Typical ensembles ranged from 50,000 to 100,000 structures, but this number was drastically reduced in size by filtering these model pools using experimental and biophysical restraints not included in the original scoring function, like CCS and spherical overlap (Politis et al., 2010). CCSs for each model were determined using the projection approximation function in IMPACT (Marklund et al., 2015), and CCS values with an uncertainty of +/− 3% were used as an experimental filter for the ensemble. After CCS filtering, models that agree with biophysical and experimental data were analyzed using hierarchical clustering to determine the predominant structural families present in the ensemble. Ensembles or subsets thereof were represented by kernel-density functions, with the median structure shown for reference. These same groups were also characterized by the average RMSD from the mean, which provided an estimate of the relative resolutions between ensembles.
Data and software availability
The software and datasets used and generated in this work for modeling and analysis is freely available at https://sites.lsa.umich.edu/ruotolo/software/im-ms-modeler/
Supplementary Material
HIGHLIGHTS.
Direct observation of the 18-subunit urease pre-activation assembly by IM-MS
Collision cross sections define possible architectures for the assembly
Integrative modeling defines ambiguity in multiple structural datasets
Acknowledgments
Protein topology modeling efforts in the Ruotolo lab are supported through the National Institute of General Medical Sciences, National Institutes of Health (R01 GM095832), and work in the Hausinger lab was partly supported by the National Institutes of Health (DK04586). We also acknowledge Prof. Aaron Frank (University of Michigan) for his continued support of our computational modeling efforts.
Footnotes
Author Contributions
B.T.R. and R.P.H designed the project. M.A.F. expressed and purified the protein complexes. J.D.E. collected and analyzed IM-MS data, performed coarse-grained modeling, and wrote the manuscript. S.M.D. modeled mass spectra and analyzed data.
Declaration of Interests
The authors declare no competing interests.
Supplemental data can be found in the supporting document (insert supporting info document link here)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balasubramanian A, Ponnuraj K. Crystal Structure of the First Plant Urease from Jack Bean: 83 Years of Journey from Its First Crystal to Molecular Structure. Journal of Molecular Biology. 2010;400:274–283. doi: 10.1016/j.jmb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Boer JL, Hausinger RP. Klebsiella aerogenes UreF: Identification of the UreG Binding Site and Role in Enhancing the Fidelity of Urease Activation. Biochemistry. 2012;51:2298–2308. doi: 10.1021/bi3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer JL, Quiroz-Valenzuela S, Anderson KL, Hausinger RP. Mutagenesis of Klebsiella aerogenes UreG To Probe Nickel Binding and Interactions with Other Urease-Related Proteins. Biochemistry. 2010;49:5859–5869. doi: 10.1021/bi1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JM. Recent research on problems in the use of urea as a nitrogen fertilizer. Fertilizer research. 1995;42:321–329. [Google Scholar]
- Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82:9557–65. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- Carter EL, Boer JL, Farrugia MA, Flugga N, Towns CL, Hausinger RP. Function of UreB in Klebsiella aerogenes Urease. Biochemistry. 2011;50:9296–9308. doi: 10.1021/bi2011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, Hausinger RP. Characterization of the Klebsiella aerogenes Urease Accessory Protein UreD in Fusion with the Maltose Binding Protein. Journal of Bacteriology. 2010;192:2294–2304. doi: 10.1128/JB.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Kuchar J, Hausinger RP. Chemical Cross-linking and Mass Spectrometric Identification of Sites of Interaction for UreD, UreF, and Urease. Journal of Biological Chemistry. 2004;279:15305–15313. doi: 10.1074/jbc.M312979200. [DOI] [PubMed] [Google Scholar]
- Collins CM, Dorazio SEF. Bacterial Ureases - Structure, Regulation Of Expression And Role In Pathogenesis. Molecular Microbiology. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- Eschweiler JD, Frank AT, Ruotolo BT. Coming to Grips with Ambiguity: Ion Mobility-Mass Spectrometry for Protein Quaternary Structure Assignment. J Am Soc Mass Spectrom. 2017;28:1991–2000. doi: 10.1007/s13361-017-1757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia MA, Han L, Zhong Y, Boer JL, Ruotolo BT, Hausinger RP. Analysis of a Soluble (UreD:UreF:UreG)2 Accessory Protein Complex and its Interactions with Klebsiella aerogenes Urease by Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2013a;24:1328–1337. doi: 10.1007/s13361-013-0677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the Urease Metallocenter. Journal of Biological Chemistry. 2013b;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia MA, Wang B, Feig M, Hausinger RP. Mutational and Computational Evidence That a Nickel-Transfer Tunnel in UreD Is Used for Activation of Klebsiella aerogenes Urease. Biochemistry. 2015;54:6392–6401. doi: 10.1021/acs.biochem.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YH, Wong HC, Yuen MH, Lau PH, Chen YW, Wong KB. Structure of UreG/UreF/UreH Complex Reveals How Urease Accessory Proteins Facilitate Maturation of Helicobacter pylori Urease. PLoS Biology. 2013;11:e1001678. doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Mol Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- Hall Z, Politis A, Robinson CV. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility-mass spectrometry. Structure (London, England: 1993) 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Hausinger RP. Urease Activation. In: Johnson MK, Scott RA, editors. Metalloprotein Active Site Assembly. Chichester, United Kingdom: John Wiley and Sons, Ltd; 2017. [Google Scholar]
- Ligabue-Braun R, Real-Guerra R, Carlini CR, Verli H. Evidence-based docking of the urease activation complex. Journal of Biomolecular Structure and Dynamics. 2013;31:854–861. doi: 10.1080/07391102.2012.713782. [DOI] [PubMed] [Google Scholar]
- Marklund EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JL. Collision cross sections for structural proteomics. Structure. 2015;23:791–9. doi: 10.1016/j.str.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Teichmann SA. Structure, dynamics, assembly, and evolution of protein complexes. Annual Review of Biochemistry. 2015 doi: 10.1146/annurev-biochem-060614-034142. [DOI] [PubMed] [Google Scholar]
- Mazzei L, Musiani F, Ciurli S. The Biological Chemistry of Nickel. The Royal Society of Chemistry; 2017. CHAPTER 5 Urease. [Google Scholar]
- Mckay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J Am Chem Soc. 2006;128:11433–42. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiological Reviews. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief MB, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. Journal of Bacteriology. 1997;179:4081–6. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IS, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proceedings of the National Academy of Sciences. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MA, Michel LO, Hausinger RP, Karplus PA. Structures of Cys319 Variants and Acetohydroxamate-Inhibited Klebsiella aerogenes Urease. Biochemistry. 1997;36:8164–8172. doi: 10.1021/bi970514j. [DOI] [PubMed] [Google Scholar]
- Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient Protein-Protein Interactions: Structural, Functional, and Network Properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Politis A, Park A, Hyung S-J, Barsky D, Ruotolo BT, Robinson CV. Integrating Ion Mobility Mass Spectrometry with Molecular Modelling to Determine the Architecture of Multiprotein Complexes. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz-Valenzuela S, Sukuru SCK, Hausinger RP, Kuhn LA, Heller WT. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Archives of Biochemistry and Biophysics. 2008;480:51–57. doi: 10.1016/j.abb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility–mass spectrometry analysis of large protein complexes. Nature protocols. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the Pieces Together: Integrative Modeling Platform Software for Structure Determination of Macromolecular Assemblies. PLoS Biology. 2012:10. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Levy Emmanuel D, Teichmann Sarah A. Homomeric protein complexes: evolution and assembly. Biochemical Society Transactions. 2010;38:879–882. doi: 10.1042/BST0380879. [DOI] [PubMed] [Google Scholar]
- Winkler R. ESIprot: a universal tool for charge state determination and molecular weight calculation of proteins from electrospray ionization mass spectrometry data. Rapid Communications in Mass Spectrometry. 2010;24:285–294. doi: 10.1002/rcm.4384. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Hyung SJ, Ruotolo BT. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst. 2011;136:3534–3541. doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- Yang X, Li H, Lai TP, Sun H. UreE-UreG Complex Facilitates Nickel Transfer and Preactivates GTPase of UreG in Helicobacter pylori. Journal of Biological Chemistry. 2015;290:12474–12485. doi: 10.1074/jbc.M114.632364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.