Abstract
Purpose
The microbiome from nursing home (NH) residents is marked by a loss in diversity that is associated with increased frailty. Our objective was to explore the associations of NH environment, frailty, nutritional status and residents’ age to microbiome composition and potential metabolic function.
Methodology
We conducted a prospective longitudinal cohort study of 23 residents, 65 years or older, from one NH that had four floors: two separate medical intensive floors and two floors with active elders. Residents were assessed using the mini nutritional assessment tool and clinical frailty scale. Bacterial composition and metabolic potential of residents' stool samples was determined by metagenomic sequencing. We performed traditional unsupervised correspondence analysis and linear mixed effect modelling regression to assess the bacteria and functional pathways significantly affected by these covariates.
Results/Key findings
NH resident microbiomes demonstrated temporal stability (PERMANOVA P=0.001) and differing dysbiotic associations with increasing age, frailty and malnutrition scores. As residents aged, the abundance of microbiota-encoded genes and pathways related to essential amino acid, nitrogenous base and vitamin B production declined. With increasing frailty, residents had lower abundances of butyrate-producing organisms, which are associated with increased health and higher abundances of known dysbiotic species. As residents became malnourished, butyrate-producing organisms declined and dysbiotic bacterial species increased. Finally, the microbiome of residents living in proximity shared similar species and, as demonstrated for Escherichia coli, similar strains.
Conclusion
These findings support the conclusion that a signature ‘NH’ microbiota may exist that is affected by the residents' age, frailty, nutritional status and physical location.
Keywords: intestinal microbiome, nursing home, elderly, frailty, malnutrition, physical location
Introduction
In the USA, nursing homes (NHs) provide custodial care to older adults who need both medical and non-medical assistance, such as support for daily living activities. These NH elders are grouped together and share a diet that is typically low in fibre with moderate to high fat content [1]. Residents living in NHs suffer from a high prevalence of Clostridium difficile and multi-drug resistant organisms (MDROs) that both colonize the intestinal tract and cause active disease. The prevalence of these pathogens colonizing the gut is so high that the NH is now considered a reservoir for introducing these pathogens into other healthcare settings [2, 3]. Unrelated individuals living together have more similar gut bacterial communities compared to individuals living in other households, suggesting a shared environment affects the similarity of the faecal microbiome [4]. Similarities of NH elders' microbiome have not previously been explored, warranting further research to better understand the factors leading to the development of the NH as a reservoir for MDROs for the community at large [5].
A healthy, diverse intestinal microbiome interacts positively with the host immune system and contributes to pathogen resistance [6]. Older adults that enter a NH experience an overall decline in intestinal microbiome function [7] and a significant loss in diversity when compared to community-dwelling elders [1]. Elders from NHs differ from community-dwelling counterparts in their microbiome composition, with higher proportions of the phylum Bacteroidetes compared to phylum Firmicutes and lower genus Coprococcus and Roseburia levels [1]. Most previous elder microbiome investigations have focused on comparing community-dwelling to NH elders. These studies have concluded that NH residents gain a NH-associated microbiome that is mostly a result of the NH-provided diet along with the increasing individual frailty associated with elders that need NH services [8]. Given the variability in diets of elders living in NHs or community-dwelling environments included in these studies, their conclusions have strongly highlighted the associations between diet and microbiome composition. This still leaves ambiguity as to what occurs to a NH resident microbiome as they age and their health status evolves with increasing frailty and malnutrition.
Elders are commonly grouped together in sections of the NH by medical or mental (i.e. dementia units) needs. Geographical location is known to influence the microbiome composition, however the influence of diet versus physical location is not well understood [9]. A better understanding of environmental influences on human microbial communities could be an important factor to consider when understanding disease etiologies, especially among vulnerable NH elders. Accordingly, the ‘NH microbiota’ remains poorly defined especially when it comes to other factors besides frailty and diet that may influence its composition such as increasing age, malnutrition or physical location. In addition, the complex interplay between patient-level and environmental (facility)-level factors and their influence on the microbiome in this vulnerable population is poorly understood.
Accordingly, we set out to follow a cohort of elders from one NH to investigate what associations age, frailty, malnutrition and physical location had on observed dysbiosis and the stability of the microbiome over time. We performed these observations among NH elders consuming the same diet, however they live in separate sections of the same NH facility with specific floors being isolated from others. Our findings contribute to the understanding of how patient-level factors, such as age, frailty and malnutrition level, influence the microbiota composition while adding novel discoveries as to the associations that facility-level factors (i.e. floor location) have with microbiome composition.
Methods
Study setting and population
This prospective cohort study was approved by the institutional review board at the University of Massachusetts Medical School. This cohort is of NH residents ≥65 years of age who lived in one NH facility that contained four different floors. The third floor was the facility’s locked dementia unit where residents were not allowed to leave the floor except for issues regarding medical care. The next floor (fourth floor), termed the medical care unit, housed residents with chronic medical issues requiring a higher level of nursing care, their food and care limited to this location. The fifth and sixth floors housed higher functioning long-term care residents who all ate and engaged in activities at one central location. They were also allowed to leave the floor and travel frequently into the community. We approached residents across all floors who had been living at that facility for ≥1 month and did not have any diarrheal illness or antimicrobial exposure within the preceding 4 weeks. All residents throughout the facility followed the same low-fibre diet across all floors prepared in one central kitchen that is typical for a NH diet. No patients suffered from dysphagia or had a feeding tube.
Data collection
We conducted baseline and end of study medical record abstraction for factors associated with key study outcomes. These factors included: age, nutritional status, comorbidities, use of proton pump inhibitors and frailty [1]. Prior history of hospitalizations, antibiotic exposures in the past year and histories of Clostridium difficile infection or urinary tract infections were collected from medical records. We obtained age, sex, race and length of NH stay from medical records. We categorized residents based on the continuous age variable into four age categories for analysis: (1) 65–74; (2) 75–84; (3) 85–94; and (4) ≥95 years of age. Frailty was categorized according to the validated and widely utilized Canadian Study of Health and Aging’s seven-point clinical frailty scale [10]. This has been previously validated in demonstrating signatures of frailty in the gut microbiota [11, 12]. We assessed nutritional status using the mini nutritional assessment (MNA) tool [13–15]. Residents were categorized as normal, at risk or malnourished based on the MNA survey administered to the residents by trained research staff or the nurse caring for the resident if mentally impaired. All residents were enrolled for a total of 5 months in which we monitored for any changes to their care.
Sample collection and processing
We collected four monthly stool samples from each resident. Additionally, from six residents we collected samples every 3 days for 2 weeks and then monthly for 4 months. DNA was extracted from samples using the PowerMagTM Soil DNA Isolation Kit on an epMotion 5075 TMX liquid handling workstation according to manufacture protocols (MO BIO Laboratories, #27100-4-EP). Sequencing libraries were constructed using the Nextera XT DNA Library Prep Kit (Illumina, #FC-131-1096) and sequenced on a NextSeq 500 Sequencing System as 2×150 base pair-end reads.
Sequence processing and analysis
Shotgun metagenomic reads were first trimmed and filtered to the host contamination using Trimmomatic [16] and Bowtie2 [17] as part of the KneadData pipeline (https://bitbucket.org/biobakery/kneaddata). Reads were then profiled for microbial species abundances using Metaphlan2 [18] and for abundance of Uniref genes, KEGG orthologues, and of the corresponding functional pathways (Metacyc pathways, KEGG pathways, and KEGG modules) using the software pipeline HUMAnN2 [19] and in-house written scripts (available upon request). Normalized taxonomic, gene and pathway abundances were then used for downstream statistical analysis in R (see below). Strain-level analysis of Escherichia coli present in metagenomic samples was performed using StrainPhlAn [20]. Reads were mapped against the MetaPhlan2 clade-specific marker gene database [18]. Reconstructed E.coli-specific consensus markers were derived from the mapping data. The reconstructed markers were then used to build a phylogeny of the strains. The tools cited, in turn, depend upon the following tools – for KneadData: Trimmomatic [16], Bowtie2 [17], SAMtools (https://github.com/samtools/); for StrainPhlan: MetaPhlan2 [18], muscle [21], RAxML [22], blastn [23]. The phylogenetic tree was visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/)
Data analysis
We performed traditional unsupervised correspondence analysis (NMDS and unsupervised hierarchical clustering) to first determine the similarity of samples with respect to the above covariate of interest. Permutation multivariate analysis of variance (PERMANOVA) was performed to evaluate inter- vs intra-individual variability in bacterial abundance. To determine the contribution of each covariate to changes in microbiome composition (including microbial and functional pathway abundances) we performed linear mixed effect modelling regression after arcsine square root transformation [24] using the R package lme4. P-values were calculated using a Kenward–Roger degrees of freedom approximation and the returned t-value from the regression modelling. Covariates with P<0.05 were retained and used for downstream visualization and data interpretation.
Results
Characteristics of the study subjects by floor location
Over a 6 month period we enrolled and followed 23 NH elders collecting monthly samples for a total of 4 months. Residents of the medical floor (fourth floor) and dementia unit (third floor) had higher frailty and malnutrition scores that those on the fifth and sixth floors (Table 1). Residents on the fourth floor were older with an average age of 96.4. There were no differences in how long the resident had been living on that floor (length of stay) nor with regards to Charlston comorbidity index score. No residents were exposed to antimicrobials or hospitalized during the study period. The last antimicrobial exposure occurred 3 months prior in only one subject.
Table 1. Characteristics of the NH cohort by floor of residence.
| Floor no. (n) | Third (3) | Fourth (4) | Fifth (7) | Sixth (8) | P-value |
|---|---|---|---|---|---|
| Age | 82 (2.0) | 96.4 (4.6) | 89.9 (7.1) | 86.3 (8.4) | 0.036 |
| Age category | 2 (0.0) | 3.6 (0.5) | 2.9 (0.9) | 2.5 (0.8) | 0.010 |
| Length of stay | 23 (18.2) | 14.6 (19.2) | 22.9 (32.3) | 17.9 (22.2) | 0.93 |
| CCI score | 2 (1.7) | 1 (1.7) | 1.3 (1.8) | 1.0 (1.4) | 0.82 |
| Malnutrition | 1 (0.0) | 1.6 (0.5) | 0.6 (0.5) | 0.4 (0.5) | 0.008 |
| Frailty | 6.7 (0.6) | 6.8 (0.8) | 4.9 (0.9) | 4.9 (0.6) | 0.053 |
Data presented as means (sd). CCI, Charlston comorbidity index; length of stay in months; age category: 1=65–74, 2=75–84, 3=85–94 and 4 =≥95.
The individual NH microbiome demonstrates stability over short and long time-interval observations
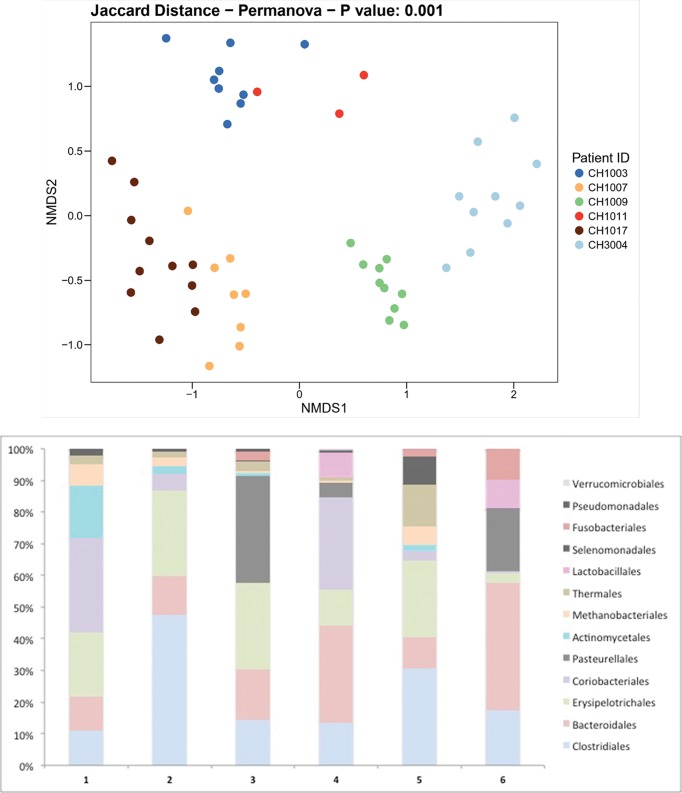
There were six residents for whom we collected samples every 3 days for 2 weeks and then monthly afterwards for 4 months, resulting in a total of 10 samples per resident. For one resident we were only able to obtain eight samples with two missed time points during the 2 week sampling time period. The average age of this group was 82.7 (9.2) years, all living on the fifth and sixth floors with an average frailty score of 5.2 (0.4) and malnutrition score of 0.2 (0.4). None of these residents had any changes in medications or healthcare exposure over the study period. Microbiota compositional differences were greater between individuals than within individuals demonstrating faecal microbiota stability in NH elders (PERMANOVA – Jaccard distance P=0.001; Fig. 1a). The microbiome composition between individuals varied as demonstrated at the order level in Fig. 1(b). These data indicate that, for long-stay NH residents, both 3 day and 30 day collection frequencies exhibit similar microbiome variation within an individual which remains stable over time as long as there are no changes to their diet or medication.
Fig. 1.
Individual variability over time. (a) Distance matrix of sample similarity from the presence or absence of bacterial species in each sample using the Jaccard binary index. Sample-to-sample distances are presented in a 2D principal component analysis plot. Samples from six NH residents are coloured by participant. No resident experienced any change in medications or a healthcare event over the sampling period. Each point represents a sample time point taken at 3 day and then 30 day intervals over a 4 month period. (b) The different community compositions among the six residents defined at the order level.
Distance measures demonstrate grouping patterns by age, frailty and malnutrition
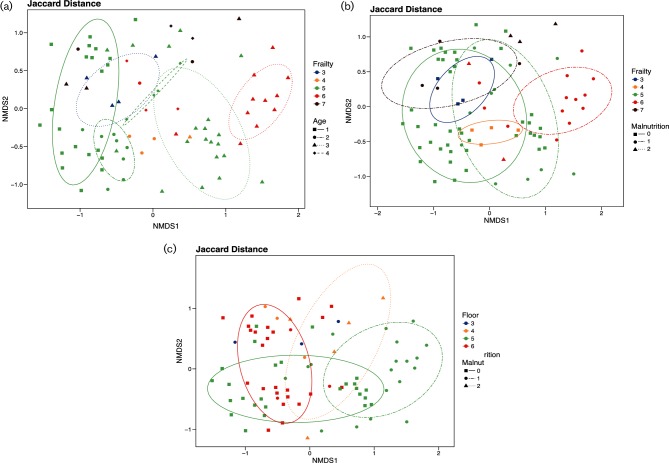
We explored beta-diversity by principal coordinate analysis (PCA) using Jaccard distances for a measure of community species dissimilarity among combinations of malnutrition scores, frailty scores, age categories and floor locations in Fig. 2. Microbiota compositional differences were greater between the covariates of malnutrition scores, frailty scores, age categories and floor location than within them with a PERMANOVA – Jaccard distance P<0.001 for each of the covariates. First, with regards to age and frailty, we note not only a pattern of composition shifting towards the right as the age category increases but also a clustering of groups by frailty score, Fig. 2(a). Comparing malnutrition and frailty, we see a similar pattern of shifting composition towards the top right as malnutrition scores increase, maintaining the clustering by frailty score, Fig. 2(b). However, when we explore PCA by floor location any discernable pattern seems to be mixed with floors three and four clustered towards the middle surrounded by resident samples from floors five and six, Fig. 2(c). We explored combinations of each of the variables represented in Fig. 2(a–c), with ellipses that represent 75 % confidence intervals. These ellipses demonstrate clustering of combinations of the grouping categorical variables. If there were less than four samples per grouping we omitted the ellipses. From these groupings certain patterns emerge. For example, in Fig. 2(a), residents in age category 3 shift in microbiome composition to the right as their frailty score increases from 3 to 6. From this analysis we noted clustering among the variables of malnutrition, frailty and age without any specific pattern to floor location, suggesting the overall microbiome composition did not correlate with location as it did to malnutrition, frailty and age.
Fig. 2.
PCO plots. (a) Jaccard binary index principal component analysis by age category and frailty score. Jaccard binary index compares similarity in the presence or absence of bacterial species in the microbiome between samples. Residents are categorized into different age groups represented by different symbols, where category 1 includes residents aged 65–74 years, category 2 is 75–84, category 3 is 85–94 and category 4 is ≥95 years of age. Frailty scores are each coloured differently. (b) Jaccard binary index principal component analysis by malnutrition and frailty score. Malnutrition scores are represented by different symbols where frailty scores are each coloured differently. (c) Jaccard binary index principal component analysis by malnutrition and floor location. Malnutrition scores are represented by different symbols where each floor is coloured differently. All panels display ellipses with a confidence interval of 75 %. Ellipses are drawn for each combination of the grouping variables (e.g. frailty 5 and malnutrition 0). Ellipses cannot be drawn for a group with less than four samples and are therefore omitted from the plots.
Mixed effect modelling demonstrated age, malnutrition, frailty and location affected bacterial species composition and functional pathways
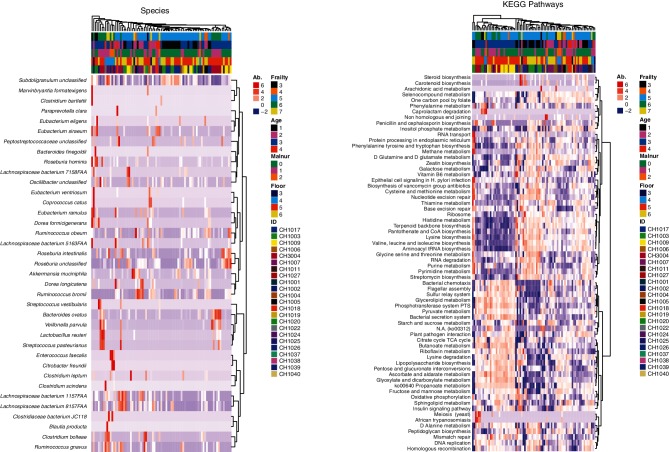
After unsupervised correspondence analysis, we performed linear mixed effect modelling regression to determine the bacterial species and contributed functional pathways that were significantly affected by the covariates of interest. Briefly, after arctangent-square root transformation of microbial species (and pathway) abundances, we performed linear mixed effect modelling regression. We fit the model abundance ~1+age+malnutrition+floor+frailty+(1|ID), where age, malnutrition, floor and frailty are the fixed effect and ID represents the random effect. Using this approach, we decouple the effect of each of the modelled covariates towards the microbial (or pathway) abundance and assess each covariate's statistical significance independently. Row-normalized abundances of significant species and KEGG pathways are displayed as hierarchical clustered heatmaps in Fig. 3(a, b). The species and KEGG pathways depicted in Fig. 3 are statististically significant for at least one of the model fixed effects.
Fig. 3.
Heatmap of the relative abundance of each gene type in each individual and hierarchical clustering, depicting (a) species and (b) pathways. (a) The heatmap depicts the relative abundances of gene sequences assigned to each bacterial genus (y-axis) across the 100 samples analysed (x-axis). The heatmap colours represent the relative abundances of the microbial genus assignments within each sample. Square colours shifted towards red to indicate higher abundance. The coloured bars across the top of the graph depict the frailty score, age category, malnutrition score, floor location and finally individual resident from top to bottom. (b) This heatmap differs by depicting the relative abundances of gene sequences assigned to each KEGG pathway (y-axis) across the 100 samples analysed (x-axis).
Dysbiosis with increasing age in both bacterial species and metabolic pathways
We observed the correlation of species and pathway abundances with increasing age. Akkermansia muciniphila, a mucin-degrading bacterium known to decrease in the elderly [25], was significantly decreased in residents in age category 3 (P=0.018) and Ruminococcus bromii, a keystone species in degradation of starch, was elevated in age category 2 (P=0.012) and then decreased in older age categories. The bacterial species Ruminococcus gnavus, which has been associated with a dysbiotic microbiota [26], was more abundant in age category 2 (0.003) and lower in older residents with the lowest abundances in age category 4 (0.015). Several bacterial species known to be butyrate producers [27] were elevated in older adults. Butyrate is known to contribute to the maintenance of the gut barrier functions, and has both immunomodulatory and anti-inflammatory properties [28]. Butyrate-producing Eubacterium siraeum was significantly elevated in age category 4 (P=0.004) and Roseburia intestinalis was elevated in age categories 2 (P=0.012), 3 (P=0.002) and 4 (P=0.016).
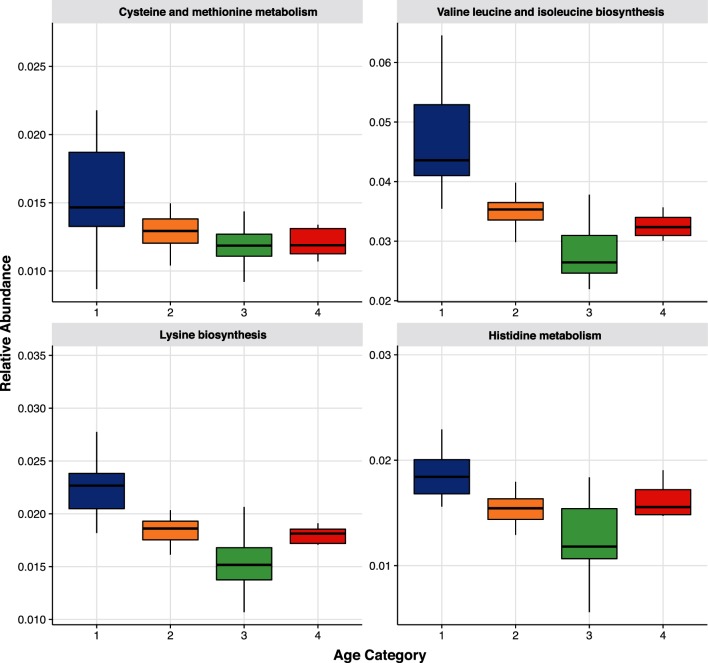
Metabolic pathways involved in essential amino acid biosynthesis and metabolism were higher in younger residents and decreased over the subsequent age categories, Fig. 4. This involved cysteine and methionine metabolism (ko00270; P=0.017), histidine metabolism (ko00340; P=0.020), valine, leucine and isoleucine biosynthesis (ko00290; P=0.005), and lysine biosynthesis (ko00300; P<0.001). In addition, nitrogenous base metabolism also decreased with increasing age as purine and pyrimidine metabolism decreased over age categories 2 (ko00230 and ko00240; P=0.004, 0.010) and 3 (0.028, 0.010). Finally, vitamin metabolism was also lower in older residents with vitamin B5, pantothenate and CoA biosynthesis, decreased in age categories 2 (ko00770; P=0.013) and 3 (P=0.004) and vitamin B1 thiamine metabolism decreased with increasing age and was lowest in age category 3 (ko00730; P=0.022). Combined with the species data, these results indicate that the dysbiosis associated with ageing included decreases in mucin and starch degradation, essential amino acid synthesis, and decreases in nitrogenous base and vitamin synthesis.
Fig. 4.
Essential amino acids by age. Relative abundances of essential amino acid pathways by age category: (1) 65–74; (2) 75–84; (3) 85–94; and (4) ≥95 years of age. Data presented as boxplots with the box being the first and third quartiles, the band inside the box is the median, and the whiskers represent the 95th percentiles. Each pathway depicted is as follows: (a) cystine and methionine metabolism; (b) valine, leucine and isoleucine biosynthesis; (c) lysine biosynthesis; and (d) histidine metabolism.
In conjunction with the observed butyrate-producing species being of higher abundance in older residents, we also noted that butyrate metabolism also rose with increasing age and was significantly elevated in age category 3 (ko00650; P=0.031). Pyruvate metabolism increased over the age categories and was highest in age category 3 (ko00620; P=0.019) while CoA biosynthesis increased in age category 2 (ko00770; P=0.013) and 3 (P=0.004). Butyrate is synthesized via pyruvate and acetyl-coenzyme A (CoA) [29]. Taken together this signifies that butyrate-producing organisms and butyrate production, a sign of intestinal health, increased with age.
Dysbiotic patterns with increasing frailty
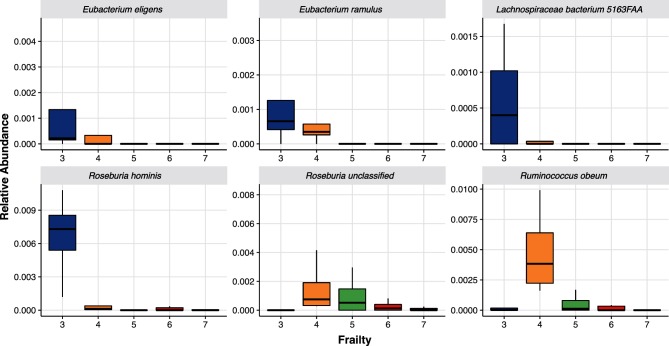
After adjusting for age, malnutrition and floor location, residents with lower frailty scores had higher abundances of butyrate-producing organisms in Fig. 5, notably members of the Clostridium cluster XIVa [30] as well as Lachnospiraceae bacterium 5_1_63FAA [31]. Conversely the bacterial species R. gnavus, which is associated with a dysbiotic microbiota [26], was higher in residents with higher frailty scores peaking at a clinical frailty score of 7 (P=0.009). From a metabolic potential standpoint, residents with higher clinical frailty scores had higher abundances of lipopolysaccharide (LPS) biosynthesis (ko00540; P=0.043), peptidoglycan (PGN) biosynthesis (ko00550; P=0.029) and sphingolipid metabolism (ko00600; P=0.014). The observed dysbiosis with increasing frailty included lower butyrate-producing organisms with increases in LPS and PGN biosynthesis as well as sphingolipid metabolism. Alterations in LPS, PGN, and sphingolipid synthesis and metabolism have all been linked to increased bowel inflammation [32, 33].
Fig. 5.
Butyrate-Producing Bacteria by Frailty. Relative abundances of butyrate-producing organisms by Clinical Frailty Score. Data presented as boxplots with the box being the first and third quartiles, the band inside the box is the median, and the wiskers represent the 95th percentiles. Each organism depicted is as follows: (a), Eubacterium eligens; (b), Eubacterium ramulus; (c), Lachnospiraceae bacterium; (d), Roseburia hominis; (e), Roseburia unclassified; (f), Ruminococcus obeum.
Malnutrition’s association with dysbiotic and opportunistic organisms
For residents who were either at risk of malnutrition or scored as malnourished, we noted trends of higher abundances of organisms associated with a dysbiotic microbiome. We found increased abundances of Citrobacter freundii in malnourished residents (P=0.020). These bacteria serve as opportunistic and super-infectious agents in immunocompetent and compromised patients [34]. Additionally, Enterococcus faecalis, which causes life-threatening hospital associated infections in humans such as sepsis, urinary tract infections and meningitis [35], was also elevated in malnourished residents (P=0.014). Similar to increasing frailty, the bacterial dysbiotic species R. gnavus, was lowest in non-malnourished residents (<0.001). Finally, the butyrate-producing organism R. intestinalis, was less abundant in both residents at risk of malnutrition (P<0.001) and those that were malnourished (P<0.001) as well as Subdoligranulum, a spore-forming butyrate producer [36], reduced in the malnourished (P=0.008). From a metabolic standpoint PGN biosynthesis was noted to be elevated in malnourished residents (ko00550; P=0.008). Malnutrition associated with opportunistic dysbiotic bacterial species as well as lower butyrate-producing organisms with increased inflammation associated PGN.
Residents from the same location share similar bacterial organisms
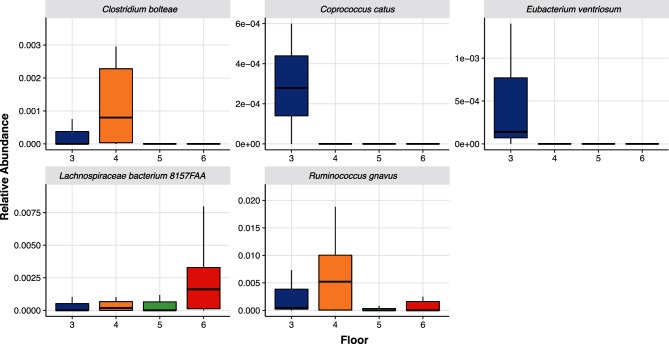
Specific bacterial species were found to be more abundant among residents located on different floors, Fig. 6(a–e). Residents living on the medical floor (4th floor) had higher abundances of the dysbiotic bacterial species, R. gnavus (P<0.001), and organisms that can cause opportunistic infections such as Clostridium bolteae [37, 38] (P=0.038). Other bacterial species, such as Coprococcus catus and Eubacterium ventriosum were of higher abundances in residents on the dementia unit (3rd floor) in comparison to the residents on other floors. Not much is known of these two bacterial species. The anti-inflammatory Lachnospiraceae bacterium 8_1_57FAA [39] was of greater abundance in residents living on floors with higher functioning elders (5th floor; P=0.015 and 6th floor; P=0.006). Lactobacillus reuteri, and anti-inflammaroty bacterial species [40], was also higher in residents on the 5th and 6th floors (P=0.003 and P=0.006). Taking all of these associations together, it points towards elderly residents that live together share specific bacterial species.
Fig. 6.
Bacterial Species by floor. Relative abundances of organisms by floor in the NH. Data presented as boxplots with the box being the first and third quartiles, the band inside the box is the median, and the wiskers represent the 95th percentiles. The dots represent the outliers. Each organism depicted is as follows: (a), Clostridium bolteae; (b), Coprococcus catus; (c), Eubacterium ventriosum; (d), Lachnospiraceae bacterium; (e), Ruminococcus gnavus.
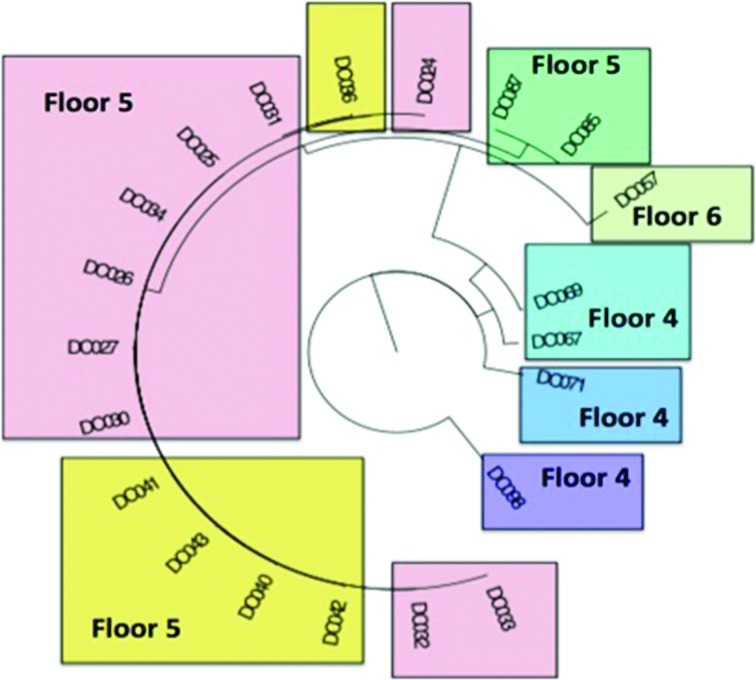
Finally, we wondered if residents shared genetically similar strains of bacteria. Observing that strains are shared between NH residents could have implications with respect to transmission of infections, we constructed the genetic relationship of E. coli carried by seven individual residents from metagenomic sequence data, Fig. 7. E. coli was chosen for strain-level analysis due to it being both a common bacterial species that colonizes the intestines and being a species known to cause disease (e.g. urinary tract infections). We also performed strain-level analysis on other common commensal bacterial species but did not note any such floor association patterns. The phylogenic tree demonstrates that ambulatory residents on the fifth and sixth floors had E. coli strains sharing more similar phylogenetic relationships than patients on the fourth floor (medical floor). Interestingly, two of the resident E. coli phylogeny intermingle (yellow and pink, Fig. 7), suggesting a high degree of strain similarity. This data suggests that residents who share common living areas had genetically related E. coli strains compared to other residents living in a separate area of the same facility who did not intermingle.
Fig. 7.
Phylogenetic tree of E. coli by individual resident and floor. The phylogenic tree of E. coli species from seven of the individual residents for whom sequence data was abundant enough to map. The identification numbers listed represent a unique sample time-point. Each individual resident is colour-coded and their floor location noted on the graph. The ambulatory residents on the fifth and sixth floors (pink/yellow/green coloured) had more similar E. coli phylogeny than patients on the fourth floor (blue/purple coloured).
Discussion
NH residents demonstrated different dysbiotic associations with increasing age, frailty and malnutrition scores. As residents aged, the abundance of microbiota-encoded genes and pathways related to essential amino acid, nitrogenous base and vitamin B production declined. With increasing frailty, residents had lower abundances of butyrate-producing organisms, higher abundances of known dysbiotic species, and higher LPS and PGN biosynthesis, and higher sphingolipid metabolism. Alterations in LPS, PGN and sphingolipid biosynthesis and metabolism have all been linked to increased bowel inflammation [32, 33]. As residents became at risk of becoming or were malnourished, butyrate-producing organisms declined and opportunistic and dysbiotic bacterial species increased along with PGN biosynthesis. Interestingly, when looking at physical location within the NH, residents living together shared similar bacterial species and had similar E. coli phylogeny. Taken together, this supports the conclusion that the ‘NH’ microbiota is influenced by resident age, frailty, nutritional status and physical location.
With increasing resident age, we found that bacterial species previously observed to decline with age were reduced in our older residents. Specifically, A. muciniphila and R. bromii decline as a likely result of changes in the diet of older adults. This dietary change favours growth of bacteria that are able to degrade mucins [25, 41] and metabolizing dietary plant polysaccharides [42]. Additionally, R. gnavus, a species known to decrease with age [43], was higher in residents aged 65–74 and exhibited lowest abundances in those 95 and older. In older NH residents, we also observed a disbiotic decrease of metabolic pathways involved in essential amino acid, nucleotide and vitamin B biosynthesis. Ageing has been associated with a progressive loss of muscle mass (sarcopenia) which is linked to lower availability of essential amino acid [44, 45]. Purine and pyrimidine nucleotide biosynthesis modules are known to be globally decreased in inflammatory bowel disease (IBD) patients [46] as well as the vitamin B complex [47]. Taken together, these metabolic pathways reflect an age-specific dysbiosis specific to chronic intestinal inflammation seen in the elderly.
With increasing frailty or malnourishment, we noted increased abundance of R. gnavus and decreased Lachnospiracae and Ruminococcaceae families. The abundance of butyrate-producing organisms also declined with increasing frailty and malnutrition. Similar dysbiotic patterns have been observed in the disease states of IBDs [26, 27, 48, 49] as well as in systemic inflammatory disorders such as multiple sclerosis [50]. Butyrate is an essential metabolite in the human colon. It is the preferred energy source for the colonic epithelial cells and it contributes to gut barrier maintenance as well as having both immunomodulatory and anti-inflammatory properties [28]. Our finding adds to the growing evidence that a dysbiotic gut microbiota, with reduced butyrate production, is linked to medical disorders and may be a target of dietary and probiotic interventions.
LPS and PGN biosynthesis was increased in frail or malnourished residents. Both of these gut-microbiota-derived molecules stimulate specific systemic inflammatory pathways that result in low-grade systemic inflammation [33]. LPS- and PGN-associated inflammation has been linked to central nervous system disorders (e.g. chronic fatigue syndrome and complex regional pain syndrome) [51], as well as colorectal carcinoma tumour progression [52], and obesity and obesity-related pathologies [53]. Additionally, patients who have had a stroke or cardiovascular disease have had a greater inflammatory gut profile with an increase PGN-producing enzymes [54]. Besides LPS and PGN biosynthesis, residents with higher frailty scores also were enriched in genes for sphingolipid metabolism. Alterations of sphingolipid metabolisms resulting in dysbiotic bioactive sphingoid level have been associated with IBDs [47] and nonalcoholic fatty liver disease [55]. Changes in sphingolipid levels result in a variety of effects on the epithelial barrier integrity, immune cell targeting and signalling, and innate/adaptive immune responses [32]. Taken together, elderly NH residents with higher frailty scores associated with a dysbiotic microbiota resembling an inflammatory gut profile.
The NH had two floors where the residents did not leave that location and two floors of higher functioning elders that intermingled and used common socialization areas. Importantly, all residents were provided with the same diet. From the linear mixed effect modelling, we noted that several bacterial species uniquely associated with specific floors irrespective of age, frailty and malnutrition. The dementia floor had greater abundances of C. catus and E. ventriosum, whereas the dementia and medical floors both had higher abundances of dysbiotic bacterial species. C. bolteae, an organism that causes opportunistic infections [37, 38], as well as R. gnavus, which characterizes the dysbiosis seen in IBD patients [38, 49] and also causes opportunistic infections [56], were both present in residents living on the third and fouth floors. Of note, residents from these floors had more frequent contact with the hospital setting. The healthier residents of the fifth and sixth floors had higher abundances of L. bacterium, a butyrate-producing organism [30, 31], as well as L. reuteri, a species used as a probiotic for its anti-inflammatory effects [40]. When we performed the phylogenetic tree analysis of E. coli, resident sequences from the fifth and sixth floors intermingled while the fourth floor residents were genetically separate. Taken together, the location of the resident in the NH had associations with both specific enriched bacteria organisms and genetically similar E. coli phylogeny.
This study had several notable strengths and limitations. One limitation of was that it reports data from only a single site, however this is balanced by the fact that all study participants had the same diet, effectively removing this important covariate. This study is also limited in the number of residents enrolled. Other potentially confounding variables such as polypharmacy and specific classes of medications the residents were exposed to were not evaluated in this cohort. Finally, the physical location microbiome association findings may have been biased by the clustering of residents with similar medical conditions onto the same floor. Following up this investigation with a multi-centre cohort study would strengthen the findings and further explore the dysbiosis associations with age and frailty but we report new findings with regards to malnutrition and physical location within a unit of other elders. Confirming our study’s findings in multiple NH facilities and addressing how these dysbiotic patterns associate to C. difficile or MDRO colonization would be key to improving the health and wellbeing of elders living in a NH setting.
In conclusion, the dysbiosis of the NH elderly gut microbiome not only differs with increasing age, frailty and nutrition, but also physical location within the NH. This supports the conclusion that a signature ‘NH’ microbiota that is influenced by each of these variables may exist. Further work is needed to see if introducing dietary changes or probiotics could affect these relationships with a goal of moving the microbiome away from being dysbiotic and supporting inflammation towards a healthier one that could help prevent disease.
Funding information
J.P.H. was supported by an intradepartmental grant through the Department of Emergency Medicine, a grant from the Center for Microbiome Research at the University of Massachusetts Medical School and from a National Institute on Aging award 1R03AG056356-01. V.B. was supported by an NIH award 1R15AI112985-01A1.
Acknowledgements
We would like to thank the administration, staff and residents living at Bethany Health Care Center for their support in this project. We would especially like to thank Sister Jacquelyn McCarthy and Dr Rohit Jangi for their administration help and Barbara Galluzzo for the day-to-day logistical help.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the University of Massachusetts Medical School’s Institutional Review Board (IRB docket H00010892). All subjects, or the subject’s legally authorized representative, gave informed consent to participate in this study.
Footnotes
Abbreviations: IBD, inflammatory bowel disease; LPS, lipopolysaccharide; MDRO, multi-drug resistant organism; MNA, mini nutritional assessment; NH, nursing home; PCA, principal coordinate analysis.
References
- 1.Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 2.Lee DC, Barlas D, Ryan JG, Ward MF, Sama AE, et al. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: prevalence and predictors of colonization in patients presenting to the emergency department from nursing homes. J Am Geriatr Soc. 2002;50:1463–1465. doi: 10.1046/j.1532-5415.2002.50377.x. [DOI] [PubMed] [Google Scholar]
- 3.Pop-Vicas A, Tacconelli E, Gravenstein S, Lu B, D'Agata EM. Influx of multidrug-resistant, gram-negative bacteria in the hospital setting and the role of elderly patients with bacterial bloodstream infection. Infect Control Hosp Epidemiol. 2009;30:325–331. doi: 10.1086/596608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassone M, Mody L. Colonization with multi-drug resistant organisms in nursing homes: scope, importance, and management. Curr Geriatr Rep. 2015;4:87–95. doi: 10.1007/s13670-015-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampelli S, Candela M, Turroni S, Biagi E, Collino S, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging. 2013;5:902–912. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 9.Rehman A, Rausch P, Wang J, Skieceviciene J, Kiudelis G, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65:238–248. doi: 10.1136/gutjnl-2014-308341. [DOI] [PubMed] [Google Scholar]
- 10.Rockwood K, Song X, Macknight C, Bergman H, Hogan DB, et al. A global clinical measure of fitness and frailty in elderly people. CMAJ. 2005;173:489–495. doi: 10.1503/cmaj.050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson MA, Jackson M, Jeffery IB, Beaumont M, Bell JT, et al. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016;6:25945. doi: 10.1038/srep25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubenstein LZ, Harker JO, Salvà A, Guigoz Y, Vellas B. Screening for undernutrition in geriatric practice: developing the short-form mini-nutritional assessment (MNA-SF) J Gerontol A Biol Sci Med Sci. 2001;56:M366–M372. doi: 10.1093/gerona/56.6.M366. [DOI] [PubMed] [Google Scholar]
- 14.Saarela RK, Lindroos E, Soini H, Hiltunen K, Muurinen S, et al. Dentition, nutritional status and adequacy of dietary intake among older residents in assisted living facilities. Gerodontology. 2016;33:225–232. doi: 10.1111/ger.12144. [DOI] [PubMed] [Google Scholar]
- 15.Guigoz Y. The mini nutritional assessment (MNA) review of the literature–what does it tell us? J Nutr Health Aging. 2006;10:485–487. [PubMed] [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:1–3. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 19.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:2–18. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in crohn's disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 28.Rivière A, Selak M, Lantin D, Leroy F, de Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, de Vos WM, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. Isme J. 2013;7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdel Hadi L, di Vito C, Riboni L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Mediators Inflamm. 2016;2016:1–13. doi: 10.1155/2016/3827684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leclercq S, de Saeger C, Delzenne N, de Timary P, Stärkel P. Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol Psychiatry. 2014;76:725–733. doi: 10.1016/j.biopsych.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Whalen JG. Spontaneous Citrobacter freundii infection in an immunocompetent patient. Arch Dermatol. 2007;143:115–125. doi: 10.1001/archderm.143.1.124. [DOI] [PubMed] [Google Scholar]
- 35.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 36.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, et al. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. 2015;82:1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finegold SM, Song Y, Liu C, Hecht DW, Summanen P, et al. Clostridium clostridioforme: a mixture of three clinically important species. Eur J Clin Microbiol Infect Dis. 2005;24:319–324. doi: 10.1007/s10096-005-1334-6. [DOI] [PubMed] [Google Scholar]
- 38.Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, et al. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 2012;22:1974–1984. doi: 10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:e1128. doi: 10.1016/j.cell.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 40.Lin YP, Thibodeaux CH, Peña JA, Ferry GD, Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. 2008;14:1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- 41.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar M, Babaei P, Ji B, Nielsen J. Human gut microbiota and healthy aging: recent developments and future prospective. Nutr Healthy Aging. 2016;4:3–16. doi: 10.3233/NHA-150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biagi E, Candela M, Turroni S, Garagnani P, Franceschi C, et al. Ageing and gut microbes: perspectives for health maintenance and longevity. Pharmacol Res. 2013;69:11–20. doi: 10.1016/j.phrs.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Fujita S, Volpi E. Amino acids and muscle loss with aging. J Nutr. 2006;136:277S–280. doi: 10.1093/jn/136.1.277S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volpi E, Kobayashi H, Sheffield-Moore M, Mittendorfer B, Wolfe RR. Essential amino acids are primarily responsible for the amino acid stimulation of muscle protein anabolism in healthy elderly adults. Am J Clin Nutr. 2011;78:250–258. doi: 10.1093/ajcn/78.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basson A, Trotter A, Rodriguez-Palacios A, Cominelli F. Mucosal interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front Immunol. 2016;7:290. doi: 10.3389/fimmu.2016.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joossens M, Huys G, Cnockaert M, de Preter V, Verbeke K, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 50.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galland L. The gut microbiome and the brain. J Med Food. 2014;17:1261–1272. doi: 10.1089/jmf.2014.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Zhao F, Wang Y, Chen J, Tao J, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. doi: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 55.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen SG, Skov MN, Justesen US. Two cases of Ruminococcus gnavus bacteremia associated with diverticulitis. J Clin Microbiol. 2013;51:1334–1336. doi: 10.1128/JCM.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]