Abstract
Purpose
Group B Streptococcus (S. agalactiae, GBS) is a Gram-positive opportunistic pathogen that inhabits the respiratory, urogenital and gastrointestinal tracts of humans and animals. Maternal colonization of GBS is a risk factor for a spectrum of clinical diseases in humans and a principle cause of neonatal meningitis and septicaemia.
Methodology
We describe polymicrobial sepsis including GBS in two gravid adult female Long–Evans rats experiencing acute mortality from a colony of long-term breeding pairs. Fluorescent in situ hybridization confirmed GBS association with pathological changes in affected tissues, including the heart and uterus.
Results
Characterization of seven GBS strains obtained from clinically affected and non-affected animals indicated similar antibiotic resistance and susceptibility patterns to that of human strains of GBS. The rat strains have virulence factors known to contribute to pathogenicity, and shared serotypes with human invasive isolates. Phylogenetic analyses revealed that one rat-derived GBS strain was more closely related to human-derived strains than other rat-derived strains, strengthening the notion that interspecies transmission is possible.
Conclusions
To our knowledge, this is the first investigation of genotypic and phenotypic features of rat-derived GBS strains and their comparison to human- and other animal-derived GBS strains. Since GBS commonly colonizes commercially available rats, its exclusion as a potential pathogen for immunocompromised or stressed animals is recommended.
Keywords: group B Streptococcus, Streptococcus agalactiae, Long-Evans rats, genome, maternal, septicemia, meningitis, endocarditis
Introduction
Streptococcus agalactiae (group B Streptococcus; GBS) is a Gram-positive opportunistic bacterium that inhabits the gastrointestinal and urogenital tract of humans, and is estimated to colonize one in four humans of both sexes [1–3]. It is a major cause of neonatal septicaemia, bacteraemia and meningitis, with maternal vaginal colonization as the number one risk factor [1]. Strains of GBS possess one of 10 different serotypes (Ia, Ib, II–IX) based on the structure of the polysaccharide capsule, which has been associated with degree of virulence and patient prognosis [4]. Increasingly, invasive group B streptococcal disease has been reported to cause significant morbidity and mortality in pregnant women, the elderly and those with underlying health conditions such as diabetes mellitus [5, 6]. Overall, mortality due to GBS infection is higher in adults than in neonates and can manifest as skin, soft tissue and urinary tract infections, bacteraemia, pneumonia and endocarditis [7–9]. GBS can cause invasive disease in animals, including primates, dogs, fish and cattle as well as laboratory-reared mice [10–13]. Both rodents and primates have been used as experimental models for neonatal or adult GBS infections [14, 15]. Comparisons between human GBS isolates and isolates from fish, monkeys, dogs and cats have shown similar phenotypic relationships, that include serotype and antimicrobial resistance patterns [10, 16]. Despite these similarities, the nature of GBS as an agent transferable from humans to animals, or vice versa, remains a robust area of research [17–19]. There have been multiple reports of outbreaks of GBS infection in rats and mice; unfortunately due to limited colony size and research needs, further characterization of host and bacterial factors precipitating disease was not pursued [13, 20, 21].
In this study, we explored the pathogenesis of suspected sepsis in gravid Long–Evans rats, focusing on the possibility that GBS was playing a central role in the disease process. Given the anthropozoonotic potential of this organism and these cases’ similar clinical and pathological presentation to invasive disease reported in humans, the aim of our study was to screen and confirm GBS in affected tissues via fluorescent in situ hybridization; to further characterize whole-genome sequenced GBS strains isolated from both affected and non-affected rats [22]; and to compare these GBS strains phylogenetically to both human- and animal-derived GBS genomes.
Methods
Rats
Vendor-purchased adult male and female Long–Evans rats (Charles River, Wilmington, MA) were pair-housed in large stainless steel cages (70×46×46 cm) with clear polycarbonate front panels for neurobehavioural studies involving the resident– intruder paradigm under an IACUC-approved protocol [23]. Each pair was housed in cages with heat-treated pine shavings (P. J. Murphy Forest Products Corp., Montville, NJ) and provided with commercially available rodent chow (LabDiet, St. Louis, MO) and water ad libitum. The rats were maintained in cages in a temperature-controlled (20–21 °C) and humidity-controlled (40–50 %) vivarium under a reversed light cycle (12 h: 12 h, lights on between 8 : 00 P.M. and 8 : 00 A.M.). Cages were sanitized every 6 weeks, and bedding was changed weekly. Animals are observed at least once daily, with any exhibiting signs of declining health (e.g. reduced body condition, abnormal change in activity, hydration, appetite, urine or faecal production) directed to the attention of veterinary staff. Based on the animal’s condition, frequency of observation, diagnostics and treatment are performed under the discretion of a veterinarian. Sentinel serology was routinely negative for Mycoplasma pulmonis, rat parvovirus, Kilham rat virus, Toolan H1 virus, pneumonia virus of mice, sialodacryoadenitis virus, rat theilovirus and Sendai virus. Because it is not known to cause spontaneous disease in immunocompetent adult animals, no efforts were made to exclude GBS. All procedures were reviewed and approved by the Tufts University Institutional Animal Care and Use Committee, following the principles of the Eighth Edition of the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011).
Case history and identification
At our collaborating institution, from December 2014 to August 2015, there were 10 breeding female pair-housed rats that presented for necropsy after being found dead or moribund. Nine of 10 cases had histological signs of septicaemia and suppurative inflammation, including infections in the kidney, heart, meninges, liver, uterus and lungs (correspondence, L. Richey). Polymicrobial infections were documented in five rats, with one case that included GBS. This case was a 10-month-old multiparous female rat whose pathological findings included a right ventricular mural thrombus, placentitis and an autolysed fetus. Cultures obtained by our collaborating institution of both the ventricular thrombus and fetus were positive for GBS, Proteus mirabilis and E. coli (correspondence, L. Richey). Previous administration of antibiotics [Sulfatrim (trimethoprim/sulfamethoxazole) at 0.5 mg ml−1 for one month, Baytril (enrofloxacin) at 0.17 mg ml−1 in the drinking water of the animals for two weeks] had unknown efficacy. As part of the room depopulation, eight rats without clinical signs (including H15-11, H15-14, H15-16) were submitted to our institution for necropsy. After room depopulation and repopulation, the onset of two more cases of gravid female rats with invasive disease associated with GBS presented a unique opportunity to characterize and compare isolates from rat- and human-derived sources to understand the genetic basis of disease across these species. Two pair-housed gravid Long–Evans female rats presented with similar signs of lethargy, reduced body condition and bloody vaginal discharge. The first rat (case 1) was a 9- to 11-week-old primiparous gravid female and had been housed in the facility for 24 days. Due to worsening clinical signs, the rat was euthanized by CO2 inhalation and a diagnostic necropsy was performed. Case 2, a 22- to 25-week-old multiparous gravid Long–Evans rat, presented 4.5 months after arrival with signs similar to case 1. Baytril (enrofloxacin) in the water (0.17 mg ml−1) was provided to the rat for less than 24 h along with supportive nutrition. The next morning, the rat was found moribund but died just prior to transfer to our institution, where it was necropsied.
Culture and isolation
At necropsy, culture samples from any grossly abnormal organs from clinically ill rats (cases 1 and 2) and nares samples of rats without clinical signs prior to room depopulation (8 rats including H15-11, H15-14, H15-16) were collected directly with either sterile instruments or Calgiswabs (Puritan Medical Products, Guilford, ME) and submerged directly into Brucella broth [Becton Dickinson (BD), Franklin Lakes, NJ] containing 20 % glycerol [Macron Chemicals (Avantor Performance Materials, Inc.), Center Valley, PA] (Table 1). A sterile swab was inserted into the glycerol-based broth and inoculated onto two plates: a split plate of Trypticase soy agar with 5 % sheep’s blood and MacConkey agar, and a plate of chocolate agar [Becton Dickinson (BD), Sparks, MD]. Media were incubated at 37 °C for 24 h. The swab was enriched overnight in a Trypticase soy broth in case GBS was not isolated after the first 24 h. Colonies were isolated on blood agar plates and identified as GBS with the use of conventional methods such as colony morphology, beta-haemolysis and verification using the API-20 STREP identification product (Biomérieux, France).
Table 1. Summary of gross necropsy, histological and microbiological findings from cases 1 and 2, and archived case.
| Case | Gross findings | Histopathologic diagnoses | Culture site results |
|---|---|---|---|
| 1* | Focal pale tan discoloration at dorsal base of the heart; pale circumferential dark circular lesions on splenic surface; mottled and tan serosal surface of kidney; cortical pitting on surface of left kidney; 3 implantation sites in left horn of uterus | Streptococcal septicaemia Severe multifocal acute necrotizing suppurative myocarditis, epicarditis and valvular endocarditis with intra-lesional cocci Moderate multifocal acute suppurative necrotizing splenitis with intra-lesional cocci Moderate multifocal neutrophilic and histiocytic interstitial pneumonia with vasculitis and fibrin thrombi admixed with bacteria Moderate multifocal acute suppurative necrotizing nephritis with intra-lesional cocci Mild multifocal acute suppurative meningoencephalitis Severe multifocal acute necrotizing suppurative metritis with intra-lesional cocci |
Blood – Streptococcus agalactiae Cardiac lesion – S. agalactiae - 3 isolates Spleen – S. agalactiae, Enterobacter aerogenes, Staphylococcus sciuri, Enterococcus faecalis Vaginal swab – S. agalactiae, S. sciuri, E. aerogenes, Proteus mirabilis, E. coli, E. faecalis Cervical contents – S. agalactiae, E. faecalis - 2 isolates, E. aerogenes, P. mirabilis, Enterococcus avium |
| 2* | 3–4 tan to pale yellow plaque-like lesions on the pericardium extending into the parenchyma; two pinpoint dark red to haemorrhagic lesions on the lung surface; left kidney with pinpoint tan-white focal lesions extending 1.5 mm into the parenchyma; mildly distended uterus with 1 haemorrhagic and autolysed fetus in each horn | Severe multifocal acute necrotizing myocarditis, epicarditis and endocarditis with atrial and ventricular thrombi admixed with intra-lesional cocci Moderate multifocal subacute neutrophilic and histiocytic interstitial pneumonia Severe multifocal acute necrotizing suppurative metritis with intra-lesional cocci Mild multifocal subacute lymphocytic pancreatitis Streptococcal septicaemia |
Nares wash – S. agalactiae, E. faecalis, Streptococcus mitis, S. sciuri, E. aerogenes - S. agalactiae, E. faecalis, Streptococcus mitis, S. sciuri, E. aerogenes Cardiac lesion – S. agalactiae - S. agalactiae Lung -–S. agalactiae, E. faecalis, E. avium, E. coli - S. agalactiae, E. faecalis, E. avium, E. coli Kidney – S. agalactiae - S. agalactiae Brain – S. agalactiae, E. faecalis, Streptococcus oralis - S. agalactiae, E. faecalis, Streptococcus oralis Vaginal swab – S. agalactiae, E. faecalis, E. avium, E. coli, Morganella morganii, P. mirabilis - S. agalactiae, E. faecalis, E. avium, E. coli, Morganella morganii, P. mirabilis Uterine swab – S. agalactiae, E. faecalis, E. avium, E. coli Fetus – S. agalactiae, E. faecalis - 2 isolates, E. coli |
| Archived case† | Ventricular thrombus; autolysed fetus‡ | Septic right ventricular thrombosis with Gram-positive and -negative bacteria Grossly fibrinopurulent inflammation of the placenta, macerated fetus Renal abscess with Gram-positive bacteria Severe submassive and centrilobular hepatic necrosis Pleural effusion Extramedullary splenic myelopoiesis |
Cardiac thrombus – S. agalactiae, P. mirabilis, E. coli Fetus – S. agalactiae, P. mirabilis, E. coli |
*Histopathology reviewed by V. B.
†Histopathology reviewed by L. R.
‡Notes from necropsy of archived case with GBS-related pathology not further detailed.
Histopathology
For all rats necropsied, tissues (heart, uterus, brain, lungs, spleen, kidneys, liver, salivary gland, trachea, oesophagus, pituitary gland, thyroid, parathyroid, mesentery, pancreas, mesenteric lymph node, gastrointestinal tract, urogenital tract) were fixed in 10 % formalin, embedded in paraffin, sectioned at ~4 µm and stained with haematoxylin and eosin (H and E). Additional slides (heart, brain) were stained with Brown and Brenn Gram stain. Slides were reviewed by a board-certified veterinary pathologist (V. B.).
Fluorescent in situ hybridization (FISH)
Oligonucleotide probes Saga and EUB338, which were synthesized and 5′-labelled (IDT Technologies, Coralville, IA) were used. The probe Saga (5′-GTA AAC ACC AAA CMT CAG CG −3′), which specifically targets and hybridizes to a 16S rRNA region of S. agalactiae, was used for the identification of this bacterium [24]. The 5′ end of Saga was labelled with fluorochrome Cy3, which emits a red fluorescent signal. The probe EUB338 (5′-GCT GCC TCC CGT AGG AGT-3′), which hybridizes the 16S rRNA of almost all bacteria [25], was 5′-labelled with fluorochrome Fluo, which shows a green fluorescent signal. Before use on paraffin formalin-fixed slides, FISH was performed on separate smeared slide preparations of E. coli, Enterococcus faecalis and Streptococcus agalactiae cultured from the rats to verify expected fluorescence signals on observation with microscopy with non-target and target species. FISH was performed using a modified protocol described previously in use for human GBS isolates [26]. Briefly, tissue sections of organs from the cases were fixed in 10 % formalin, embedded in paraffin and sectioned at ~4 µm. These unstained slides were then deparaffinated and rehydrated in an ascending ethanol series. Slides were left to air-dry and, for hybridization, slides were covered with a hybridization buffer [0.9M NaCl, 20 mM Tris-HCL (pH 8), 0.01 % SDS, 20 % formamide] containing both the probes Saga-Cy3 and EUB338-Fluo at 10 ng µl–1, which had been preheated at 74.5 °C for 10 min. Slides were then incubated at 46 °C overnight in a humid chamber in the dark. The hybridized slides were then incubated in a pre-warmed washing buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.2, 0.01 % SDS) in the dark for 15 min at 48 °C. The slides were subjected to a second washing buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.2) in the dark for 15 min at 48 °C. After the washing steps, slides were rinsed in double-distilled H2O and air-dried. DNA was stained with Vectashield with 4′, 6-diamidino-2-phenylindole (DAPI; Vector Laboratories) and cover-slipped for viewing. Slides were observed and imaged with a Zeiss Axioshop 2 plus microscope and QImaging-QI click camera (QImaging, Surrey, BC, Canada).
Further characterization of rat GBS whole-genome sequences
Seven GBS genomes obtained from both affected (cases 1 and 2) and non-affected rats (H15-11, H15-14, H15-16) in the colony were characterized by serotype, multilocus sequence type (MLST), identification of antimicrobial resistance genes and virulence factors, and antimicrobial susceptibility testing [22]. Here, we additionally report fluoroquinolone resistance mutations, susceptibility to the antimicrobial trimethoprim-sulfamethoxazole (TMS) using the E-test and elaborate on the significance of the genotypic and phenotypic profiles of these isolates. The seven published genomes are available in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under the following accession numbers: MJHP0000000, MJHQ00000000, MJHR00000000, MJHS00000000, MJHT00000000, MJHU00000000, MJHV00000000 [22].
Antimicrobial susceptibility testing
Initial antimicrobial susceptibility testing results using the broth microdilution method for the seven GBS isolates were previously published, for the antimicrobials ampicillin, ceftriaxone, meropenem, vancomycin, erythromycin, tetracycline, enrofloxacin and clindamycin [22]. Antibiotics that are commonly used in the treatment of rodent and human bacterial infection include: penicillin or ampicillin, ceftriaxone, meropenem, vancomycin, erythromycin, tetracycline, enrofloxacin, clindamycin and TMS. Here, the E-test (Biomérieux, France) was used to determine the minimum inhibitory concentrations (MICs) for TMS according to the manufacturer’s instructions. Streptococcus pneumoniae ATCC 49619 was used as the quality-control strain to ensure the accuracy of the results. Bacteria were grown on blood agar supplemented with 5 % sheep’s blood and resuspended in sterile saline to a 0.5 McFarland standard. The bacteria were inoculated onto a Mueller–Hinton agar plate by streaking evenly with a swab over the entire surface to obtain a uniform lawn of growth. Plates were allowed to dry for 5 min and the antibiotic strips were placed. Plates were incubated in an aerobic incubator at 35 °C for 20–24 h. The zone sizes were recorded and interpreted according to the CLSI interpretative criteria for streptococci [27].
Detection of fluoroquinolone resistance mutations
Fluoroquinolone resistance can be due to amino acid substitutions in a region of the GyrA, GyrB, ParC or ParE subunit that is known as the quinolone resistance determining region (QRDR) [28]. QRDR mutations involving the gyrA, gyrB, parC and parE genes were investigated using multiple sequence alignment using the Pathosystems Resource Integration Center (PATRIC) in comparison to S. agalactiae reference strain ATCC 13813 (GenBank accession no. AEQQ00000000) [29].
Single nucleotide polymorphism (SNP) analysis of human- and rat-derived GBS isolates
We next sought to define the phylogenetic relationship between rat-derived and human invasive GBS whole genomes to explore the possibility of interspecies transmission. Staff at the Streptococcus Laboratory of the CDC constructed a phylogenetic tree of S. agalactiae strains of human invasive isolates obtained from the Active Bacterial Core surveillance (ABCs) archive from 2015 (B. J. Metcalf, S. Chochua and B. W. Beall, unpublished data) and rat isolates with the same serotype and sequence type (V, ST1 and Ib, ST 12) using the SNP identification program kSNP 3.0 [30]. In addition, to determine genetic variation among the GBS strains isolated from each of the five rats, the number of SNPs was compared between isolates. There were 9228 core SNPs, 17 345 non-core SNPs and 14 226 SNPs in at least a fraction 0.5 of the genomes analysed; estimated core genome size was ~1 500 000 base pairs (correspondence, B. J. Metcalf).
Phylogram of human- and animal-derived GBS genomes
To ascertain the phylogenetic relationship between not only human invasive GBS genomes, but also other animal-derived GBS genomes, selected streptococcal genome groups, including rat-derived GBS genomes, were submitted to Pathosystems Resource Integration Center (PATRIC) staff for phylogenetic analysis. The PATRIC database was used to access publicly available annotated bacterial genomes of GBS from GenBank [29]. The accession numbers of sequences derived from the GenBank database are as follows: S. agalactiae GD201008-001 (SAMN02603155), S. agalactiae SA20-06 (SAMN02603506), S. agalactiae ZQ0910 (SAMN02469855), S. agalactiae STIR-CD-17 (SAMN02470563), S. agalactiae A909 (SAMN02604011), S. agalactiae NEM316 (SAMEA 3138330), S. agalactiae 2603 V/R (SAMN02604013), S. agalactiae 18RS21 (SAMN02435897), S. agalactiae 515 (SAM N02435850), S. agalactiae CJB111 (SAMN02435851), S. agalactiae COH1 (SAMEA3139000), S. agalactiae GB00112 (SAMN01084069), S. agalactiae H36B (SAMN02435895), S. agalactiae ATCC 13813 (SAMN00217013), S. agalactiae FSL S3-026 (SAMN02428925), S. gallolyticus subsp. gallolyticus ATCC 43143 (AP012053), S. gallolyticus subsp. gallolyticus ATCC BAA-2069 (SAMEA2272408), S. pasteurianus ATCC43144 (AP012054). With the selected genomes, an order-level phylogenetic tree was created using Phylogenomic Estimation with Progressive Refinement (PEPR), that builds trees based on shared protein families [31–33]. Briefly, to compare similar protein sequences between all genomes, including the outgroup taxa, blast searches were performed. To predict initial homologous protein sets, Markov Clustering (MCL) was used to cluster blast results into protein families [34]. These were then refined using hidden Markov models as previously described [35]. The families included only those with membership in more than 80 % of the genomes analysed. Multiple sequence alignment of each protein family was performed using muscle (at default parameters) [36]. These protein families were then filtered to include only those with membership in more than 80 % of the genomes analysed. Poorly aligned (length heterogeneous) regions were eliminated using Gblocks [37]. Modified alignments were then concatenated into a single data set for phylogeny estimation. In post-analysis, RAxML was used for preliminary phylogenetic tree-building [38]. Bootstrap analysis was used to estimate support values for generated lineages, with 100 pseudo-replications sampling only half of aligned protein sets per replication. Support trees inferring maximum-likelihood were constructed using FastTree [39]. When highly supported nodes had subnodes with low support, local refinements to tree topology were made by running the entire pipeline using only those genomes represented by the node being refined (with additional sister taxa for rooting purposes). The refined subtree was then spliced back into the full tree. Information was compiled in Newick format to create a phylogenetic tree using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Bacterial isolates/culture results
The microbiological findings are shown in Table 1. For cases 1 and 2, the tissue samples submitted for microbiological examination from necropsy tested positive for GBS, with pure cultures obtained from the blood, cardiac tissue and lung of case 1, and cardiac tissue and kidney of case 2. Other species (e.g. E. coli, Enterococcus faecalis) mixed with GBS were isolated from other organs of case 1 and case 2 (Table 1). One of 2 female rats and 6 of 7 male rats (including H15-11, H15-14, H15-16) without clinical signs prior to room depopulation had GBS cultured from the nares. Of these animals, 5 of 6 rats previously treated with enrofloxacin (0.17 mg ml−1) in the water, and 1 of 2 untreated rats, cultured positive for GBS.
Histopathology
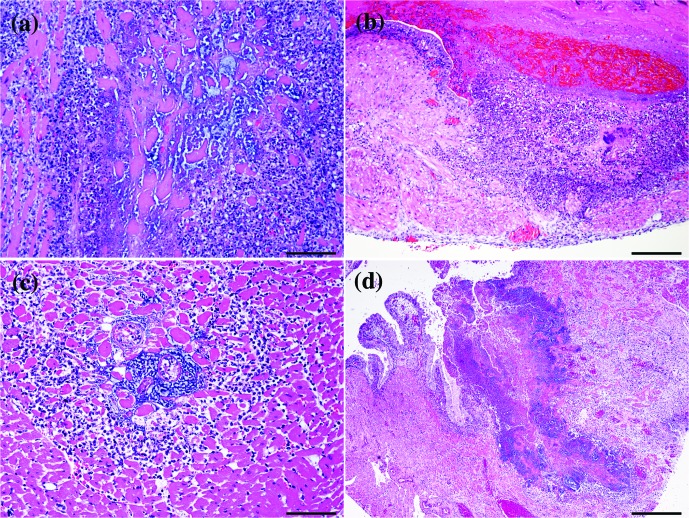
Final histopathological diagnoses for case 1 included a necrotizing suppurative myocarditis, epicarditis and valvular endocarditis (Fig. 1a), necrotizing suppurative metritis with intra-lesional cocci (Fig. 1b), moderate neutrophilic and histiocytic pneumonia with vasculitis and intra-lesional bacteria, suppurative necrotizing splenitis with intra-lesional cocci, suppurative necrotizing nephritis with intra-lesional cocci, and a mild suppurative meningo-encephalitis. Case 2 also had a severe necrotizing suppurative myocarditis, epicarditis and endocarditis, that included atrial and ventricular thrombi admixed with intra-lesional cocci (Fig. 1c). A severe necrotizing suppurative metritis with intra-lesional cocci was observed similar to case 1 (Fig. 1d). Other significant findings included a moderate multifocal, subacute neutrophilic and histiocytic pneumonia. Based on these findings, the presumptive cause of death was consistent with streptococcal septicaemia. For cases 1 and 2, the gross, histological and microbiological findings are summarized in Table 1. There were no significant findings for the eight rats (including H15-11, H15-14, H15-16) necropsied prior to room depopulation.
Fig. 1.
Histopathology. (a) Heart, case 1. Diffuse necrotizing suppurative endocarditis with large colonies of cocci. Haematoxylin and eosin stain; 20×, bar: 100 µm. (b) Uterus, case 1. Multifocal and transmural necrotizing suppurative metritis. The lumen is expanded by cellular debris and hyalinosis with abundant cocci. Haematoxylin and eosin stain; 10×, bar: 200 µm. (c) Heart, case 1. Necrotizing suppurative endocarditis with moderate amounts of cellular debris, large colonies of cocci, haemorrhage and fibrin; 20×; bar: 100 µm. (d) Uterus, case 2. Inflammation and necrosis effacing the endometrium into the myometrium. Affected areas have moderate amounts of cellular debris, fibrin and haemorrhage admixed with cocci; 4×; bar: 500 µm.
FISH
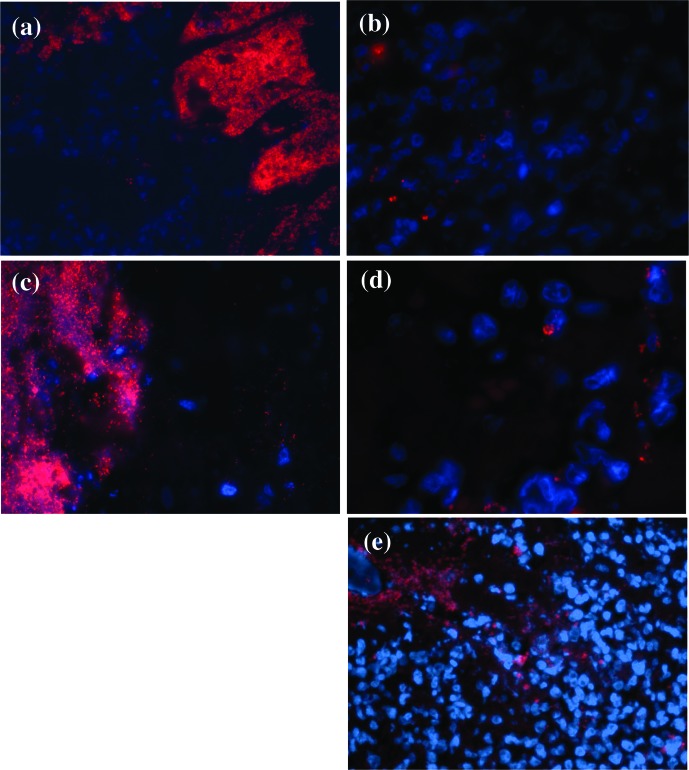
On prepared tissue slides, using Saga, a probe specific to a 16S RNA region of GBS, and EUB338, a probe specific to almost all bacteria, the bacteria involved in the metritis, myocarditis, nephritis, splenitis, meningitis and pneumonia of case 1, and metritis and myocarditis of case 2, hybridized with the oligonucleotide probes. Prepared slides of the cardiac lesion from the archived case, involving a 10-month-old multiparous female rat that presented with polymicrobial sepsis including GBS, also showed bacteria that hybridized with Saga and EUB338. Other commensal bacteria did not hybridize to the Saga probe and were not affiliated with the lesions. Fig. 2 shows representative images of Saga hybridization of bacteria intimately related with pathology in the heart and uterus of cases 1 and 2, and the archived case.
Fig. 2.
Fluorescent in situ hybridization (FISH) of sections of paraffin-fixed slides from cases 1 and 2, and archived case. The sections were simultaneously hybridized with nuclear stain DAPI (blue) and Saga probe (specific to GBS) (red). Case 1’s affected heart (a; 63×) and uterus (b; 100×), Case 2’s affected heart (c; 63×) and uterus (d; 100×) and the archived case’s affected heart (e; 63×) are shown.
Antimicrobial susceptibility to TMS
As noted previously, isolates were susceptible to ampicillin (<0.25 µg ml−1), ceftriaxone (<0.5 µg ml−1), erythromycin (<0.25 µg ml−1) and clindamycin (<0.25 µg ml−1), but resistant to tetracycline (>8 µg ml−1) and intermediate-resistant to enrofloxacin (<0.5 µg ml−1) [22]. The control strain S. pneumoniae ATCC49619 had an MIC of 0.25 µg for TMS. All seven rat GBS strains (two from case 1, two from case 2, one from H15-11, one from H15-14 and one from H15-16) had an MIC of <0.25 µg ml−1, indicating that the strains were susceptible to TMS.
Genotypic profiles of GBS rat isolates
Table 2 presents a selected genotypic profile, including multi-locus sequence type, serotype, antibiotic resistance genes and virulence-associated genes of each GBS rat isolate sequenced that was previously described [22], in addition to current quinolone resistance determining region (QRDR) mutation findings. No previously reported mutations in the quinolone resistance determining region (QRDR) codons were observed in the gyrA, gyrB, parC and parE genes of all seven isolates when compared to the reference strain ATCC 13813. Novel polymorphisms were found at eight different codons and comprised gyrA (Asp307Ala and Glu371Gly), in parC (Ser638Asn, Glu641Asp) and parE (Ile495Leu) (n=7). The six ST12 rat-derived isolates had alterations in parC (Asp810Gly) and parE (Glu360Ser). The one ST1 rat-derived isolate had a polymorphism in parC (Glu812Gly). No amino acid substitutions were observed in gyrB.
Table 2. Clinical associations and genotypic profiles of Long–Evans rat GBS isolates.
| Case ID | Sex | Enrofloxacin in drinking water | Source | Serotype† | ST† | Resistance genes*† | Genes for Virulence Factors*†‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Haemolysin/cytolysin (cylE) | Hyaluronate lyase (hylB) | CAMP factor (cfa/cfb) | Laminin-binding protein (lmb) | C5a peptidase (scpB) | Pili [PI-1, PI-2a (pilA, pilB, pilC, srtC3, srtC4)] | Fg-binding surface proteins A and B (fbsA and B) | |||||||
| Case 1 | Female | Untreated | Cardiac | 1b | 12 | tet(M) | X | X | X | X | X | X | |
| Blood | 1b | 12 | tet(M) | X | X | X | X | X | X | ||||
| Case 2 | Female | Treated (1 day) | Cardiac | 1b | 12 | tet(M) | X | X | X | X | X | X | |
| Fetus | 1b | 12 | tet(M) | X | X | X | X | X | X | ||||
| Carrier H15-14 | Male | Untreated | Nares wash | V | 1 | tet(M), ermA | X | X | X | X | X | X | X |
| Carrier H15-11 | Male | Treated | Nares wash | 1b | 12 | tet(M) | X | X | X | X | X | X | |
| Carrier H15-16 | Male | Treated | Nares wash | 1b | 12 | tet(M) | X | X | X | X | X | X | |
ST, Sequence Type.
*Data from table obtained using ResFinder and Comprehensive Antimicrobial Resistance Database (CARD).
†Preliminary data described in [22].
‡Virulence Factor Database (VFDB).
Phylogenetic comparison of rat and human invasive GBS isolates
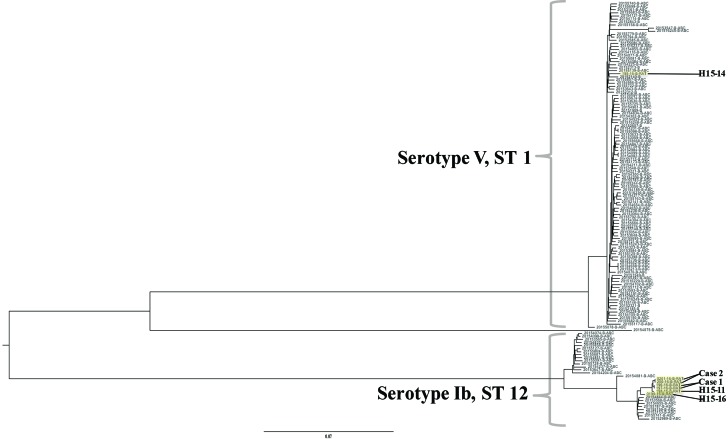
Phylogenetic analysis of human invasive GBS isolates from the CDC’s ABCs from the year 2015 and the seven rat isolates of the same MLST and serotype distribution was performed using kSNP 3.0 (Fig. 3) [30]. Rat GBS genomes are highlighted in yellow. Serotype and sequence type are designated for each genome. The six serotype Ib-ST12 isolates were phylogenetically similar to human invasive GBS isolates of the same serotype and sequence type. Similarly, the single serotype V-ST1 isolate belonged to the same evolutionary clade as human invasive isolates of the same serotype and sequence type. As expected, the two isolates collected from case 1 (196-16-B-RAT from cardiac tissue, 197-16-B-RAT from blood) had no difference in SNPs, and thus were identical. The two isolates from case 2 (200-16-B-RAT from cardiac tissue, 201-16-B-RAT from fetus) also had no difference in SNPs (Table S1, available in the online version of this article). The remaining GBS genomes isolated were all distinct and thus had a varying number of SNPs. The single serotype V-ST1 isolate (195-16-B-RAT) had the greatest number of SNPs difference between rat isolates, with 3737 SNPs distance between 199-16-B-RAT, and 3793 SNPs distance from the remainder of the rat GBS genomes. These genomes had less difference in SNPs compared to each other, with the highest difference being 88 SNPs between 199-16-B-RAT and 200-16-B-RAT and 201-16-B-RAT.
Fig. 3.
Phylogenetic tree of rat-derived and human GBS invasive genomes. Whole-genome data was collected from the Active Bacterial Core surveillance (ABCs) archive from 2015 of human GBS invasive genomes and collected rat GBS genomes (highlighted in yellow). Serotype and sequence type are designated for each branch in the tree.
Phylogram of GBS and other streptococcal genomes
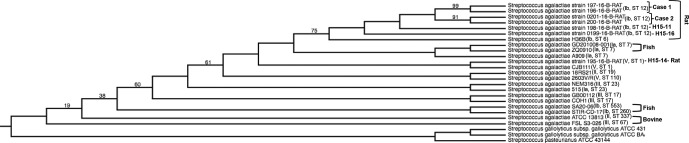
Phylogenetic analyses of GBS genomes isolated from different hosts (rat, bovine, human, fish), including the seven rat isolate genomes (195-16-B-RAT, 196-16-B-RAT, 197-16-B-RAT, 198-16-B-RAT, 199-16-B-RAT, 200-16-RAT, 0201-16-B-RAT), are represented in Fig. 4. For all GBS genomes, species source, serotype and ST are provided. The six rat-derived serotype Ib-ST12 isolates were most phylogenetically similar to each other compared to other GBS genomes, grouped into the same evolutionary branch. The next closest relation is human strain H36B of the same serotype (Ib). The single rat-derived serotype V-ST1 isolate was most closely related to a human invasive strain of the same serotype and sequence type, CJB111, and was more related to the fish strains GD201008-001 and ZQ0910 and human strain A909 than the other rat-derived genomes.
Fig. 4.
Whole-genome phylogeny comparing different species-derived GBS strains. The phylogenetic tree was generated using rat-derived GBS isolates (labelled as 'Case 1', 'Case 2', 'H15-11', H15'14 and 'H15-16') and streptococcal genomes derived from different species. Serotype and sequence type (ST) are labelled next to each individual genome. Species derivation is indicated next to rat, fish and bovine isolates. The remainder of unmarked S. agalactiae isolates are human-derived genomes. The align and tree were constructed using the pipeline Phylogenomic Estimation with Progressive Refinement (PEPR). Numbers indicate branch support values.
Colony follow-up
Without criteria for exclusion of rats colonized with GBS obtained from the commercial vendor, depopulation and repopulation did not prevent development of GBS invasive disease in selected rats in the colony. However, prophylactic measures that included stringent personal protective equipment practices and uterine tubal ligation have decreased and virtually eliminated the incidence of GBS-related clinical disease. There have been no cases of GBS-related morbidity or mortality in the past year.
Discussion
In this study, we characterized GBS isolates from cases of polymicrobial sepsis associated with spontaneous GBS disease in laboratory-reared adult gravid Long–Evans rats and clinically unaffected rats. The sudden onset of clinical signs and pathology observed in the affected cases that included the presence of myocarditis, metritis, meningitis and septicaemia closely recapitulates the presentations increasingly seen in human disease [5, 8]. The pathology seen in gravid female rats in the colony also parallels invasive disease described in well-established mouse models of GBS infection, as pregnant mice are more sensitive to GBS infection compared to their non-pregnant conspecifics [40, 41]. In experimental inoculation models, pregnant rats had higher carriage rates of GBS compared to virgin animals [42]. It is possible that due to gestational immunosuppression, the gravid female rats were more susceptible to GBS infection than the non-pregnant and male rats in the colony. Due to rising concerns for antimicrobial resistance and GBS invasive disease in both neonates and adults, and anthropozoonotic potential, rat GBS isolates were submitted to the Streptococcus Laboratory at the CDC for whole-genome sequencing as part of their Active Bacterial Core surveillance (ABCs) [22]. The rat GBS genomes were further compared to both reference human GBS genomes and GBS genomes from other animals using the bioinformatics database PATRIC. By working with the CDC and utilizing the expanding database of shared genomic information, characterization and comparison of GBS isolates within and across species provides critical information for public health epidemiology, surveillance and prevention of GBS infections.
Due to widespread use of intrapartum antibiotic prophylaxis to prevent GBS disease in the early neonatal period in humans, there has been concern for the development of antimicrobial resistance. Fortunately, reports of antimicrobial resistance of GBS to beta-lactams and cephalosporin antibiotics remain infrequent [1]. Previous MIC testing of the rat GBS isolates showed that all isolates were susceptible to many of the antimicrobial agents used in humans, including ampicillin and penicillin, the first-line antibiotics recommended by the CDC [22]. They were also susceptible to erythromycin and clindamycin, antibiotics that are advocated for use in cases of beta-lactam hypersensitivity [1]. Isolates were susceptible to TMS, an antibiotic that was administered to the animals before room depopulation. This finding supports the use of TMS to treat GBS in the colony, though it is unclear whether previous administration had any effect on morbidity and mortality. This antibiotic was clinically effective in treating affected rats in a previous GBS enzootic, and preferred over penicillin due to oral formulation [20]. There was widespread resistance (100 %) of the rat GBS isolates to tetracycline, consistent with the presence of the tet(M) resistance gene in these isolates. GBS resistance to tetracycline is fairly common, and has been attributed to its widespread use since the 1950s and subsequent selection for and spread of a few resistant clones [43]. Despite the isolates’ susceptibility to erythromycin and clindamycin, the one serotype V-ST 1 isolate had a gene for macrolide and lincosamide resistance, ermA. This may be due to inducible, non-constitutive expression of the resistance-causing methylase enzyme, which requires further testing to be confirmed [44]. No previously reported mutations were observed in the QRDR in the gyrA, gyrB, parC and parE genes in all seven isolates to explain the intermediate resistance to enrofloxacin. Further study is needed to understand the significance of the amino acid substitutions in gyrA, parC and parE to the fluoroquinolone resistance type. Other possible mechanisms for resistance include undetected upregulated efflux pumps or altered permeability [28, 45]. These findings also emphasize the importance of judicious determination of a broad-spectrum antibiotic protocol. Unfortunately the aetiological agent of the endemic was initially unclear based on polymicrobial culture results and histopathology. Even though GBS isolates were sensitive to TMS, and intermediate sensitivity to enrofloxacin, this oral antibiotic treatment was not successful in preventing GBS-associated sepsis. The oral route of antibiotic administration may also be a limiting factor, given that oral antibiotics alone were inadequate for GBS prophylaxis in humans [46]. Furthermore, clinically ill rats may restrict their water intake. Overall, these findings suggest that rat-derived GBS isolates have similar antimicrobial resistance profiles to most human GBS isolates, and that the use of beta-lactam antibiotics, and TMS, is appropriate for prophylaxis and treatment against GBS.
To examine the nature and incidence of GBS-related disease in the colony, a review of pathology reports of gravid female rats from December 2014 to August 2015 was performed. Notably, polymicrobial cultures, not just GBS, were obtained from many samples of animals that had experienced acute mortality in the colony. An important differential diagnosis experienced by some of the clinically affected female rats is maternal sepsis due to chorioamnionitis, a syndrome of ascending polymicrobial infection involving rupture of the fetal membranes, which can present with diverse clinical presentations such as nephritis, endometritis and septic abortion [47, 48]. Cultures obtained from women experiencing polymicrobial sepsis often grow more than one organism, including the genital mycoplasma Ureaplasma urealyticum, along with E. coli, GBS and group A Streptococcus [48]. Certainly, mixed bacterial cultures from vaginal and rectal swabs from women makes initial suspicion of the aetiology complicated [49]. Despite mixed bacterial cultures obtained in some samples, FISH confirmed GBS involvement with pathological tissue lesions for cases 1 and 2. FISH was used to confirm GBS-related pathology in the heart from a gravid female that had polymicrobial sepsis before the presentation of cases 1 and 2, supporting its use as a screening tool for GBS pathology in septic rats. Indeed, use of FISH as a quick and sensitive screening method of GBS in pregnant women at time of delivery has been developed [26]. Unfortunately the GBS isolate from the archived case was not available for further characterization.
To our knowledge, this is the first comparative genomic analysis of rat-derived and other GBS genomes. Systematic comparison of isolates can be used to determine how closely related the populations are to each other, and may also infer interspecies GBS transmission. A previous study comparing bovine-derived isolates to isolates obtained from human workers and their families indicated close genotypic relationships suggesting the potential for GBS colonization and infection between species, highlighting potential public health consequences [50]. Indeed, WGS is increasingly being incorporated as a tool to relate strains’ genotypes and phylogeny for molecular epidemiology, screening guidelines, microbiological diagnosis and clinical management of GBS cases [51]. SNP analysis of human invasive strains from the CDC and the seven rat isolates revealed close phylogenetic relationships, with those of the same serotype and sequence type belonging to the same evolutionary clade. Comparison of accessory genetic elements among the human- and rat-derived isolates, such as virulence- and antimicrobial resistance-associated genes, that are not part of the SNP analysis, could further substantiate these results. The six rat genomes in the same clade of serotype Ib, ST12, had no more than 88 SNPs separating them, which is consistent with previously reported comparisons between strains of the same sequence type [52]. As anticipated, the rat GBS strain 195-16-B-RAT of serotype V-ST1 was the least related, with 3793 and 3737 SNPs separation from the other rat genomes. Based on the phylogenetic work and proximity in housing, the isolates from the same clade (from cases 1, 2, H15-11 and H15-16) are likely clonally related, and the single V-ST 1 isolate (from case H15-14) is distinct. One limitation of this study was that personnel manipulating these rats were not tested for GBS carrier status; this would have assisted in determining interspecies transmission. It is possible that the serotype Ib-ST 12 isolates were transmitted from animal to animal when shipping or that there was one point source (animal or human) of transmission, either at the facility or vendor. It is unclear where the distinct serotype V-ST 1 isolate originated, but an animal or human source is suspected.
Further comparison between human-, rat- and other animal-derived genomes showed GBS isolates from the same species were not necessarily closely related – for example, the rat-derived V-ST1 isolate was more closely related to the human invasive S. agalactiae strain CJB111 than the other rat-derived 1b-ST12 isolates. This complements previous findings of invasive GBS strains cultured from tilapia being closely related to the human GBS strain A909 [12]. Though still undocumented, previous reports and our current findings strengthen the notion that transmission across species is possible, given they are not distinguishable from human and other animal GBS strains, and thus could present as an anthropozoonotic hazard. Importantly, depending on the source, rats are colonized with GBS on arrival to the facility, since a particular vendor may not exclude GBS from their respective rat colony. It is likely that these rats may have originally been infected by human handlers, since approximately 25 % of humans are GBS carriers [1]. In this study, based on the vendor’s health monitoring records, commercially available rats’ GBS status varied by the barrier where the rats were housed, with one barrier having no rats culture positive for GBS while another had 21/44 (47 %) of the rats culture positive for GBS in the 18 months prior to September 2015. Due to the potential for transient colonization, test-and-cull strategies would be difficult and likely of limited efficacy in eliminating GBS from infected colonies [49]. Thus, within an animal facility, GBS may represent an occupational health risk when handling colonized rats. Conversely, to avoid transmission from human handlers, rats should be handled with appropriate personal protective equipment. Other preventative measures, including training on proper animal handling, along with providing salient information about GBS to veterinary staff, technical personnel and researchers, are recommended. Furthermore, thorough review of health monitoring reports and requests of pre-arrival health screening of animals are recommended for prevention of GBS introduction into the rodent colony.
In conclusion, this study presents the first known incidence of naturally occurring invasive GBS infection in adult gravid female rats with a similar presentation to disease in humans. This study also presents a genetic comparison of whole-genome sequenced human- and animal-derived invasive GBS isolates to those from both clinically affected rats and rats colonized with GBS, but without clinical signs. In collaboration with the CDC, use of bioinformatics tools allowed characterization of these isolates. These data will hopefully influence approaches to treating clinical disease and help determine the risk of GBS-associated disease in both human and animal populations. Further investigations are needed to understand the dynamics of host and environmental factors during GBS infection, as well as other mechanisms of GBS-associated virulence that precipitate disease. Rodents represent a useful research model to study human GBS invasive infections due to similarities in disease presentation and close genetic relationships of GBS strains isolated from both rats and humans.
Funding information
This work was supported by NIH Grants T32-OD010978 and P30-ES002109 (Principal Investigator, J. G. Fox).
Acknowledgements
We thank Bernard W. Beall, Benjamin J. Metcalf and Sopio Chochua at the CDC Streptococcus Laboratory for whole-genome sequencing of rat GBS isolates and expertise in bioinformatics. We thank Benjamin J. Metcalf for phylogenetic analysis. We thank Carolyn Madden and Ellen Buckley Jordan for culture, isolation and identification of bacteria; Zeli Shen for DNA extraction, PCR extraction and sequencing; Anthony Mannion and Mia Lieberman for technical support; and Alyssa Pappa for her excellent support and assistance.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All procedures regarding the care and use of animals were reviewed and approved by the Tufts University Institutional Animal Care and Use Committee, following the principles of the Eighth Edition of the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011), and in accordance with state and local regulations.
Supplementary Data
Footnotes
Abbreviations: GBS, group B Streptococcus; TMS, trimethoprim-sulfa.
'The GenBank accession numbers for the S. agalactiae strains referenced are as follows: ‘195-16-B-RAT’ : MJHP00000000, ‘196-16-B-RAT’ : MJHQ00000000, ‘197-16-B-RAT’ : MJHR00000000, ‘198-16-B-RAT’ : MJHS00000000, ‘199-16-B-RAT’ : MJHT00000000, ‘200-16-B-RAT’ : MJHU00000000, ‘201-16-B-RAT’ : MJHV00000000.
One supplementary table is available with the online version of this article.
References
- 1.Verani JR, McGee L, Schrag SJ. Prevention of perinatal group B streptococcal disease–revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59:1–36. [PubMed] [Google Scholar]
- 2.Marió MJ, Valenzuela I, Vásquez AE, Illanes SE. Prevention of early-onset neonatal group B streptococcal disease. Rev Obstet Gynecol. 2013;6:63–68. [PMC free article] [PubMed] [Google Scholar]
- 3.Bliss SJ, Manning SD, Tallman P, Baker CJ, Pearlman MD, et al. Group B Streptococcus colonization in male and nonpregnant female university students: a cross-sectional prevalence study. Clin Infect Dis. 2002;34:184–190. doi: 10.1086/338258. [DOI] [PubMed] [Google Scholar]
- 4.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, et al. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect Immun. 2005;73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skoff TH, Farley MM, Petit S, Craig AS, Schaffner W, et al. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990–2007. Clin Infect Dis. 2009;49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 6.Wang YH, Cc L, Chiu CH, Wang MH, Yang TH, et al. Genetically diverse serotypes III and VI substitute major clonal disseminated serotypes Ib and V as prevalent serotypes of Streptococcus agalactiae from 2007 to 2012. J Microbiol Immunol Infect. 2015:52–57. doi: 10.1016/j.jmii.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA. 2008;299:2056. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 8.Farley MM. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis. 2001;33:556–561. doi: 10.1086/322696. [DOI] [PubMed] [Google Scholar]
- 9.Edwards MS, Baker CJ. Group B streptococcal infections in elderly adults. Clin Infect Dis. 2005;41:839–847. doi: 10.1086/432804. [DOI] [PubMed] [Google Scholar]
- 10.Lämmler C, Abdulmawjood A, Weiss R. Properties of serological group B streptococci of dog, cat and monkey origin. Zentralbl Veterinarmed B. 1998;45:561–566. doi: 10.1111/j.1439-0450.1998.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 11.Keefe GP. Streptococcus agalactiae mastitis: a review. Can Vet J. 1997;38:429–437. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Zhang W, Lu C. Comparative genomics analysis of Streptococcus agalactiae reveals that isolates from cultured tilapia in China are closely related to the human strain A909. BMC Genomics. 2013;14:775. doi: 10.1186/1471-2164-14-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schenkman DI, Rahija RJ, Klingenberger KL, Elliott JA, Richter CB. Outbreak of group B streptococcal meningoencephalitis in athymic mice. Lab Anim Sci. 1994;44:639–641. [PubMed] [Google Scholar]
- 14.Gravett MG, Witkin SS, Haluska GJ, Edwards JL, Cook MJ, et al. An experimental model for intraamniotic infection and preterm labor in rhesus monkeys. Am J Obstet Gynecol. 1994;171:1660–1667. doi: 10.1016/0002-9378(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Surve MV, Anil A, Kamath KG, Bhutda S, Sthanam LK, et al. Membrane vesicles of group B streptococcus disrupt feto-maternal barrier leading to preterm birth. PLoS Pathog. 2016;12:e1005816. doi: 10.1371/journal.ppat.1005816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delannoy CM, Crumlish M, Fontaine MC, Pollock J, Foster G, et al. Human Streptococcus agalactiae strains in aquatic mammals and fish. BMC Microbiol. 2013;13:41. doi: 10.1186/1471-2180-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalimuddin S, Chen SL, Lim CTK, Koh TH, Tan TY, et al. 2015 Epidemic of severe Streptococcus agalactiae sequence type 283 infections in singapore associated with the consumption of raw freshwater fish: a detailed analysis of clinical, epidemiological, and bacterial sequencing data. Clin Infect Dis. 2017;64:S145–S152. doi: 10.1093/cid/cix021. [DOI] [PubMed] [Google Scholar]
- 18.Dogan B, Schukken YH, Santisteban C, Boor KJ. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J Clin Microbiol. 2005;43:5899–5906. doi: 10.1128/JCM.43.12.5899-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyhs U, Kulkas L, Katholm J, Waller KP, Saha K, et al. Streptococcus agalactiae serotype IV in humans and Cattle, Northern Europe. Emerg Infect Dis. 2016;22:2097–2103. doi: 10.3201/eid2212.151447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuster KA, Hish GA, Selles LA, Chowdhury MA, Wiggins RC, et al. Naturally occurring disseminated group B streptococcus infections in postnatal rats. Comp Med. 2013;63:55–61. [PMC free article] [PubMed] [Google Scholar]
- 21.Geistfeld JG, Weisbroth SH, Jansen EA, Kumpfmiller D. Epizootic of group B Streptococcus agalactiae serotype V in DBA/2 mice. Lab Anim Sci. 1998;48:29–33. [PubMed] [Google Scholar]
- 22.Bodi Winn C, Dzink-Fox J, Feng Y, Shen Z, Bakthavatchalu V, et al. Whole-genome sequences and classification of Streptococcus agalactiae strains isolated from laboratory-reared long-evans rats (Rattus norvegicus) Genome Announc. 2017;5:e01435-16. doi: 10.1128/genomeA.01435-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Covington HE, Kikusui T, Goodhue J, Nikulina EM, Hammer RP, et al. Brief social defeat stress: long lasting effects on cocaine taking during a binge and zif268 mRNA expression in the amygdala and prefrontal cortex. Neuropsychopharmacology. 2005;30:310–321. doi: 10.1038/sj.npp.1300587. [DOI] [PubMed] [Google Scholar]
- 24.Trebesius K, Leitritz L, Adler K, Schubert S, Autenrieth IB, et al. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med Microbiol Immunol. 2000;188:169–175. doi: 10.1007/s004300000035. [DOI] [PubMed] [Google Scholar]
- 25.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajbakhsh S, Norouzi Esfahani M, Emaneini M, Motamed N, Rahmani E, et al. Identification of Streptococcus agalactiae by fluorescent in situ hybridization compared to culturing and the determination of prevalence of Streptococcus agalactiae colonization among pregnant women in Bushehr, Iran. BMC Infect Dis. 2013;13:420. doi: 10.1186/1471-2334-13-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100–S23, ed. Wayne, PA: CLSI; 2013. [Google Scholar]
- 28.Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci. 2015;1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 31.Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, et al. Phylogeny of gammaproteobacteria. J Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol. 2013;5:621–645. doi: 10.1093/gbe/evt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dongen S. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl. 2008;30:121–141. doi: 10.1137/040608635. [DOI] [Google Scholar]
- 35.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis : Probabalistic Models of Proteins and Nucleic Acids. 1st ed. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 36.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poutrel B, Dore J. Virulence of human and bovine isolates of group B streptococci (types Ia and III) in experimental pregnant mouse models. Infect Immun. 1985;47:94–97. doi: 10.1128/iai.47.1.94-97.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coid CR, O'Sullivan AM, Rossiter CE. The influence of different stages of pregnancy on group B streptococcal infection in mice. Placenta. 1985;6:65–68. doi: 10.1016/S0143-4004(85)80033-3. [DOI] [PubMed] [Google Scholar]
- 42.Ancona RJ, Ferrieri P. Experimental vaginal colonization and mother-infant transmission of group B streptococci in rats. Infect Immun. 1979;26:599–603. doi: 10.1128/iai.26.2.599-603.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, et al. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun. 2014;5:4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 45.Varaldo PE, Montanari MP, Giovanetti E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother. 2009;53:343–353. doi: 10.1128/AAC.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baecher L, Grobman W, Schrag SJ, Gorwitz R, Fultz-Butts K. Prenatal antibiotic treatment does not decrease group B streptococcus colonization at delivery. Int J Gynaecol Obstet. 2008;101:125–128. doi: 10.1016/j.ijgo.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Cape A, Tuomala RE, Taylor C, Puopolo KM. Peripartum bacteremia in the era of group B streptococcus prophylaxis. Obstet Gynecol. 2013;121:812–818. doi: 10.1097/AOG.0b013e3182888032. [DOI] [PubMed] [Google Scholar]
- 48.Tita AT, Andrews WW. Diagnosis and management of clinical chorioamnionitis. Clin Perinatol. 2010;37:339–354. doi: 10.1016/j.clp.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayó M, Berlanga M, Agut M. Vaginal microbiota in healthy pregnant women and prenatal screening of group B streptococci (GBS) Int Microbiol. 2002;5:87–90. doi: 10.1007/s10123-002-0064-1. [DOI] [PubMed] [Google Scholar]
- 50.Manning SD, Springman AC, Million AD, Milton NR, McNamara SE, et al. Association of Group B Streptococcus colonization and bovine exposure: a prospective cross-sectional cohort study. PLoS One. 2010;5:e8795. doi: 10.1371/journal.pone.0008795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almeida A, Villain A, Joubrel C, Touak G, Sauvage E, et al. Whole-genome comparison uncovers genomic mutations between group B streptococci sampled from infected newborns and their mothers. J Bacteriol. 2015;197:3354–3366. doi: 10.1128/JB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flores AR, Galloway-Peña J, Sahasrabhojane P, Saldaña M, Yao H, et al. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc Natl Acad Sci USA. 2015;112:6431–6436. doi: 10.1073/pnas.1504725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.