ABSTRACT
Antimicrobial resistance is a threat to public health globally and leads to an estimated 23,000 deaths annually in the United States alone. Here, we report the genomic characterization of an unusual Klebsiella pneumoniae, nonsusceptible to all 26 antibiotics tested, that was isolated from a U.S. patient. The isolate harbored four known beta-lactamase genes, including plasmid-mediated blaNDM-1 and blaCMY-6, as well as chromosomal blaCTX-M-15 and blaSHV-28, which accounted for resistance to all beta-lactams tested. In addition, sequence analysis identified mechanisms that could explain all other reported nonsusceptibility results, including nonsusceptibility to colistin, tigecycline, and chloramphenicol. Two plasmids, IncA/C2 and IncFIB, were closely related to mobile elements described previously and isolated from Gram-negative bacteria from China, Nepal, India, the United States, and Kenya, suggesting possible origins of the isolate and plasmids. This is one of the first K. pneumoniae isolates in the United States to have been reported to the Centers for Disease Control and Prevention (CDC) as nonsusceptible to all drugs tested, including all beta-lactams, colistin, and tigecycline.
KEYWORDS: carbapenems, colistin, Klebsiella pneumoniae, plasmid-mediated resistance, whole-genome sequencing
IMPORTANCE
Antimicrobial resistance is a major public health threat worldwide. Bacteria that are nonsusceptible or resistant to all antimicrobials available are of major concern to patients and the public because of lack of treatment options and potential for spread. A Klebsiella pneumoniae strain that was nonsusceptible to all tested antibiotics was isolated from a U.S. patient. Mechanisms that could explain all observed phenotypic antimicrobial resistance phenotypes, including resistance to colistin and beta-lactams, were identified through whole-genome sequencing. The large variety of resistance determinants identified demonstrates the usefulness of whole-genome sequencing for detecting these genes in an outbreak response. Sequencing of isolates with rare and unusual phenotypes can provide information on how these extremely resistant isolates develop, including whether resistance is acquired on mobile elements or accumulated through chromosomal mutations. Moreover, this provides further insight into not only detecting these highly resistant organisms but also preventing their spread.
OBSERVATION
Antimicrobial resistance is a major public health threat worldwide, leading to an estimated 23,000 deaths annually in the United States alone (1). Carbapenem-resistant Gram-negative bacteria are particularly concerning as they typically display multidrug-resistance and are associated with higher mortality rates (2). Carbapenem-resistant Enterobacteriaceae (CRE) can possess a variety of antimicrobial resistance mechanisms, both intrinsic and acquired, that render antimicrobials ineffective. Among these mechanisms, acquired carbapenemases, such as Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-beta-lactamase (NDM), are of greatest public health concern because of the potential for rapid dissemination of genes encoding these enzymes via mobile genetic elements, including plasmids and integrons (3).
Carbapenem resistance spurred the development of new antibiotics and combination drugs, including plazomicin, ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam (4) while also renewing use of older agents, such as colistin (5). However, resistance to these drugs also exists, as new beta-lactamase inhibitors (avibactam, vaborbactam, relebactam) do not inhibit metallo-beta-lactamases (4), plazomicin is inactivated by 16S rRNA methyltransferases (4), and mobile colistin resistance (mcr) has also been reported (6).
In August 2016, a female patient in her 70s in Nevada who had recently arrived from India was admitted to an acute care hospital with a diagnosis of systemic inflammatory response syndrome (7). In the 2 years preceding her U.S. hospitalization, the patient received health care multiple times in India for a right femur fracture and subsequent complications. Blood cultures during her last hospitalization were negative; however, a right lateral hip abscess culture revealed monomicrobial growth of Klebsiella pneumoniae that was nonsusceptible to all antimicrobial drugs tested. The patient developed septic shock in September 2016 and died with the CRE. Here, we report the genome sequence and genomic analysis of this isolate and elucidate contributing mechanisms leading to the observed phenotypic profile.
A K. pneumoniae strain was isolated from a hip abscess culture, and antimicrobial susceptibility testing was performed and interpreted using CLSI reference broth microdilution (M100, M07) (Table 1). The MIC for fosfomycin was determined by Etest (BioMérieux; Durham, NC) and the CLSI reference agar dilution using Mueller-Hinton agar II (BBL) with 25 µg/ml glucose 6-phosphate sodium salt (M100, M07). MICs of tigecycline and ceftazidime-avibactam were interpreted according to breakpoints established by the U.S. Food and Drug Administration (FDA; Silver Springs, MD). DNA was extracted according to methods previously described (8), and whole-genome sequencing data were generated using short-read (Illumina, San Diego, CA) and long-read (Pacific Biosciences, Menlo Park, CA) technology. PacBio long reads were assembled de novo using HGAP V3 and were subsequently polished with Quiver (9). Illumina short reads were then mapped to the polished long-read contigs for minor error correction using Pilon 1.21 (10). Acquired antimicrobial resistance genes were identified on high-quality contigs using the Resfinder repository (11) and SSTAR V1.1.01 (12). Finally, additional genes were detected with PGAP (13) and ISfinder (14) to identify genes contributing to displayed phenotypic resistance which could not be explained by acquired mechanisms present in the Resfinder collection alone.
TABLE 1 .
Antimicrobial susceptibility testing results using broth microdilution, agar dilution, and Etesta
| Class and antimicrobial(s) | MIC(s) (μg/ml) | Interpretation | Associated resistance gene(s) |
|---|---|---|---|
| Aminoglycoside | |||
| Amikacin | >64 | R | aacA4, rmtC |
| Gentamicin | >16 | R | aacA4, rmtC |
| Tobramycin | >16 | R | aacA4, rmtC |
| Beta-lactam | |||
| Ampicillin | >32 | R | blaCTX-M-15, blaSHV-28, blaCMY-6, blaNDM-1 |
| Aztreonam | >64 | R | blaCTX-M-15, blaCMY-6 |
| Cefazolin | >8 | R | blaCTX-M-15, blaSHV-28, blaCMY-6, blaNDM-1 |
| Cefepime | >32 | R | blaCTX-M-15, blaNDM-1 |
| Cefotaxime | >64 | R | blaCTX-M-15, blaCMY-6, blaNDM-1 |
| Cefotaxime-clavulanic acid | >32/4 | ND | blaCMY-6, blaNDM-1 |
| Cefoxitin | >16 | R | blaCMY-6, blaNDM-1 |
| Ceftazidime | >128 | R | blaCTX-M-15, blaCMY-6, blaNDM-1 |
| Ceftazidime-avibactamb | >16/4 | R | blaNDM-1 |
| Ceftazidime-clavulanic acid | >64/4 | ND | blaCMY-6, blaNDM-1 |
| Ceftriaxone | >32 | R | blaCTX-M-15, blaCMY-6, blaNDM-1 |
| Doripenem | >8 | R | blaNDM-1 |
| Ertapenem | >8 | R | blaNDM-1 |
| Imipenem | 32 | R | blaNDM-1 |
| Meropenem | >8 | R | blaNDM-1 |
| Piperacillin-tazobactam | >128/4 | R | blaNDM-1 |
| Chloramphenicol | |||
| Chloramphenicol | >16 | R | Truncated ramR |
| Fluoroquinolone | |||
| Ciprofloxacin | >8 | R | oqxA, oqxB, gyrA and parC mutations, truncated ramR |
| Levofloxacin | >8 | R | oqxA, oqxB, gyrA and parC mutations, truncated ramR |
| Fosfomycin | |||
| Fosfomycin | 32c, 16d | ND | fosA |
| Polymyxin | |||
| Colistin | >8 | NWT | Disrupted mgrB |
| Polymyxin-B | >8 | NWT | Disrupted mgrB |
| Sulfonamide | |||
| Trimethoprim-sulfamethoxazole | 8/152 | R | sul1 |
| Tetracycline | |||
| Tetracycline | >32 | R | tet(A), truncated ramR |
| Tigecyclineb | 4 | I | Truncated ramR |
| Macrolide | |||
| Not included in AST panel | Na | Na | mph(A) |
Interpretations were based on CLSI guidelines or FDA breakpoints. Cellular location for resistance genes blaCMY-6, blaNDM-1, aacA4, rmtC, and sul1 was plasmid IncA/C2; cellular location for resistance genes mph(A) and tet(A) was plasmid IncFIB(K). The cellular location for resistance genes blaCTX-M-15, blaSHV-28, fosA, oqxA, oqxB, mgrB, ramR, gyrA, and parC was chromosomal. R, resistant; I, intermediate; ND, not defined; NWT, non-wild type; Na, not applicable.
FDA breakpoints.
Agar dilution.
Etest.
The sequenced K. pneumoniae isolate contained a 5.8-Mb genome, including a 5.4-Mb chromosome and three plasmids, and belonged to multilocus sequence typing (MLST) strain 15 (ST15). K. pneumoniae ST15 has spread globally and is responsible for the high prevalence of CTX-M-15 in Asia and Europe (15). This sequence type often displays resistance to beta-lactams and fluoroquinolones and was recently associated with transferable mcr-1 colistin resistance (16). Our isolate harbored three different plasmids of incompatibility groups A/C2, FIB(pKPHS1), and FIB(K).
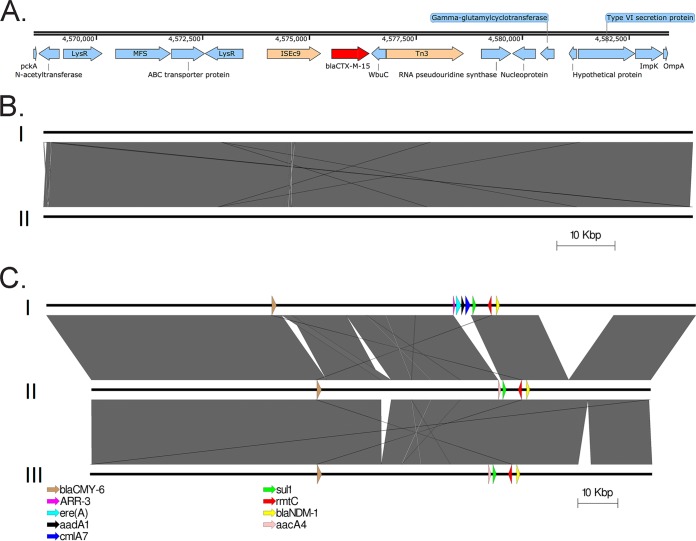
Details for mechanisms of resistance are described in Table 1. The conjugative IncA/C2 plasmid carried blaNDM-1 carbapenemase and blaCMY-6 class C beta-lactamase genes, both of which are commonly inserted into broad-host-range plasmids, such as IncA/C2 (17). In addition to plasmid-mediated beta-lactamase genes, we identified two chromosomally located beta-lactamase genes, blaCTX-M-15 and blaSHV-28. The extended-spectrum-beta-lactamase (ESBL) blaCTX-M-15 gene likely inserted itself into the chromosome via an ISEc9 (also called ISEcp1) insertion sequence (Fig. 1A), a previously described resistance gene mobile element association (18). The presence of these four beta-lactamases accounted for all displayed phenotypic resistance to beta-lactams, including the new combination drug ceftazidime-avibactam (Table 1). Also of note, this K. pneumoniae isolate would likely be resistant to the newer agents meropenem-vaborbactam and imipenem-relebactam. Although not tested, the beta-lactamase inhibitors in these agents do not inactivate metallo-beta-lactamases, such as NDM. Carbapenem resistance can also originate from intrinsic mechanisms due to mutations in outer membrane porins (OMPs). Although we identified a previously described OmpK36 variant that is associated with carbapenem nonsusceptibility likely resulting from reduced outer membrane permeability (GenBank accession number ACM07443) (19), we did not observe any premature stop codons leading to truncations in OmpK35, OmpK36, or OmpK37 previously associated with reduced outer membrane permeability.
FIG 1 .
(A) Chromosomal integration of blaCTX-M-15 by an ISEc9 insertion sequence. The blaCTX-M-15 gene is highlighted in red, mobile elements are peach, and all other genes are blue. Numbers indicate chromosomal locations. (B) The nonconjugative IncFIB(pKPHS1) p1605752FIB_2 plasmid carried by our isolate (I) was highly similar to a plasmid previously isolated in Nepal from K. pneumoniae strain ST15 (II). Gray areas indicate regions of similarity; white areas represent nonhomologous regions. No known antimicrobial resistance genes were present on either plasmid. (C) Conjugative IncA/C2 p1605752AC2 plasmid of our isolate (II) compared to the two most closely related IncA/C2 plasmids, pNDM-KN (I) and pNDM-US (III). Gray areas indicate regions of similarity; white areas represent nonhomologous regions. Known antimicrobial resistance genes are indicated with colored arrows.
Although transferable mcr has been documented in ST15 strains, this isolate did not harbor any mcr genes. Colistin resistance was likely explained by the insertional inactivation of the mgrB regulator gene by an IS1 element (20). The MgrB protein acts as a negative-feedback regulator controlling the PhoP/PhoQ signaling system. Upregulation of this system due to mgrB inactivation activates the lipopolysaccharide modification system of the cell, which subsequently leads to polymyxin resistance (20). No amino acid changes in either PhoP/PhoQ or PmrA/PmrB, previously associated with intrinsic resistance to colistin, were identified.
Using K. pneumoniae strain ATCC 43816 as a reference, mutations leading to amino acid changes that render fluoroquinolones inactive were identified in two chromosomal genes, gyrA (S83F and D87A) and parC (S80I) (21). We also identified a frameshift mutation resulting in internal stop codons in the ramR transcriptional repressor gene which likely rendered it nonfunctional, leading to overexpression of the AcrAB efflux pump and to resistance to tigecycline (22), tetracycline, fluoroquinolones, and chloramphenicol (23).
To further confirm efflux pump contributions to the displayed MIC values, an efflux inhibitor assay was performed, as previously described (24). Antibiotic susceptibility testing was performed with and without 43.75 µg/ml of PAβN, a known efflux pump inhibitor. MIC values were lower in the presence of PAβN for chloramphenicol (4 μg/ml versus >16 μg/ml), minocycline (<4 μg/ml versus >16 μg/ml), and tigecycline (2 μg/ml versus 4 μg/ml). MIC values for both fluoroquinolones tested did not differ in the presence or absence of PAβN, likely due to mutations in gyrA and parC.
Although the isolate harbored a fosA gene and susceptibility testing was performed, currently, no CLSI MIC breakpoints exist for fosfomycin susceptibility in K. pneumoniae. Finally, we identified one gene, mph(A), which confers resistance to macrolides and has the potential to spread due to its location on a plasmid. However, phenotypic resistance was not documented, as macrolides are not included in our antimicrobial susceptibility test panel.
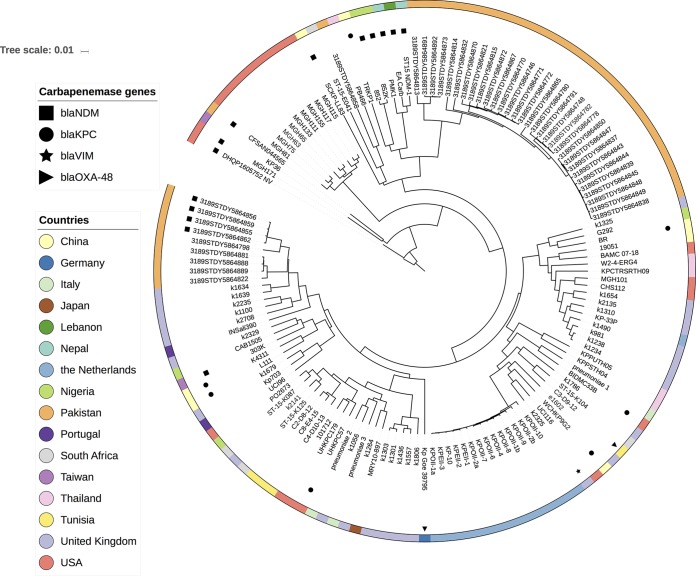
To estimate the origin of our isolate, we screened the whole-genome sequence against publicly available K. pneumoniae isolates in the genome database at NCBI (n = 2,919) using a combination of the Genome Tree Report (https://www.ncbi.nlm.nih.gov/genome/tree) and LYVE-SET 1.1.4f (https://github.com/lskatz/lyve-SET). Our isolate grouped with 140 K. pneumoniae isolates with a single nucleotide polymorphism (SNP) range of 89 to 3,248, and the cluster was dominated by isolates from Europe, South Asia, and the United States (Fig. 2). Our isolate was most closely related to NDM-producing ST15 K. pneumoniae MGH171 (GenBank accession number NGTL00000000.1) from Massachusetts General Hospital, Boston, MA, USA (89 SNPs over a 5.4-Mb core genome, or 93% of the genome). The blaNDM gene identified on the MGH171 genome assembly was partial, possibly due to an assembly artifact, and was located on a contig that also harbored the IncA/C2 replicon gene and blaCMY-6, increasing the likelihood of the presence of a true plasmid-mediated NDM in MGH171. Additionally, blaNDM presence was observed in 12 other K. pneumoniae ST15 isolates, and the phylogenetic cluster also included ST15 specimens harboring carbapenemase genes blaKPC-1 (n = 7), blaOXA-48 (n = 2), and blaVIM-1 (n = 1) (Fig. 2).
FIG 2 .
Our isolate, DHQP1605752_NV, clustered with 140 publicly available Klebsiella pneumoniae genome sequences. Presence or absence of known carbapenamase genes is listed for all isolates.
The nonconjugative IncFIB(pKPHS1) plasmid (p1605752FIB_2) carried by our isolate, 111,692 bp in length, was highly similar to a 111,693-bp plasmid previously isolated in Nepal from a K. pneumoniae ST15 strain (25) (100% query coverage, 99% sequence similarity) (GenBank accession number CP008931) (Fig. 1B). This isolate (GenBank accession number GCA_000764615.1) was also part of the cluster of 140 K. pneumoniae isolates described above and differed from our isolate by 354 SNPs over a core genome of 5.2 Mb, or 89% of the genome.
The sequence of the conjugative IncA/C2 plasmid (p1605752AC2) carrying blaNDM-1 was 140,133 bp in size and was similar to 13 other publically available plasmid sequences (>98% BLAST query coverage, >99% sequence identity), including pNDM-US from a K. pneumoniae ST11 strain isolated in the United States from a patient with a history of travel in India (GenBank accession number CP006661) and pNDM-KN from Kenya (GenBank accession number JN157804) (Fig. 1C). Lastly, conjugative plasmid IncFIB(K) (p1605752FIB), which carried antimicrobial resistance genes mph(A) and tet(A), did not share close homology with any known plasmids.
We report the isolation and characterization of a K. pneumoniae ST15 strain that was nonsusceptible to all antimicrobials tested and that likely shares a common ancestor with 140 globally disseminated K. pneumoniae strains, which all have publicly available genome sequences. We detected resistance determinants to explain nonsusceptibility to all 26 antimicrobials tested. All identified determinants were described previously in other Gram-negative bacteria and included both intrinsic and acquired mechanisms.
Evaluation of multidrug-resistant organisms using advanced molecular detection, including whole-genome sequencing, is needed to better understand the origins, acquisition, and spectrum of antimicrobial resistance mechanisms and their combination within one bacterial host. In addition, sequencing provides a broader context regarding the potential mobility of resistance determinants. Consequently, the additional information provided by sequencing may aid in identifying predecessor genotypes that could evolve to pan-resistance and in determining their global dissemination to inform public health response and containment.
Accession number(s).
Raw sequencing reads, genome assemblies, and MIC data were placed under BioProject PRJNA391323. The chromosome, IncA/C2, IncFIB(pKPHS1), and IncFIB(K) genome sequences were deposited under GenBank accession numbers CP022127, CP022126, CP022128, and CP022125, respectively.
ACKNOWLEDGMENTS
We thank the CDC Biotechnology Core (PacBio) and Jonathan Daniels (Illumina) for their assistance with sequencing data generation. We also thank Natashia Reese for her assistance with drug susceptibility testing.
This work was made possible, in part, through support from CDC’s investments to combat antibiotic resistance and the Advanced Molecular Detection (AMD) program at CDC.
The findings and conclusions in this report are solely ours and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Citation de Man TJB, Lutgring JD, Lonsway DR, Anderson KF, Kiehlbauch JA, Chen L, Walters MS, Sjölund-Karlsson M, Rasheed JK, Kallen A, Halpin AL. 2018. Genomic analysis of a pan-resistant isolate of Klebsiella pneumoniae, United States 2016. mBio 9:e00440-18. https://doi.org/10.1128/mBio.00440-18.
REFERENCES
- 1.CDC 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA: http://www.cdc.gov. [Google Scholar]
- 2.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. 2008. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 29:1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 3.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 4.Wright H, Bonomo RA, Paterson DL. 2017. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect 23:704–712. doi: 10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Todd R, Kiehlbauch J, Walters M, Kallen A. 2017. Notes from the field: pan-resistant New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae—Washoe County, Nevada, 2016. Morb Mortal Wkly Rep 66:33. doi: 10.15585/mmwr.mm6601a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Man TJB, Perry KA, Lawsin A, Coulliette AD, Jensen B, Toney NC, Limbago BM, Noble-Wang J. 2016. Draft genome sequence of Mycobacterium wolinskyi, a rapid-growing species of nontuberculous mycobacteria. Genome Announc 4:e00138-16. doi: 10.1128/genomeA.00138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 10.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Man TJB, Limbago BM. 2016. SSTAR, a stand-alone easy-to-use antimicrobial resistance gene predictor. mSphere 1:e00050-15. doi: 10.1128/mSphere.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. 2011. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents 38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Caspar Y, Maillet M, Pavese P, Francony G, Brion JP, Mallaret MR, Bonnet R, Robin F, Beyrouthy R, Maurin M. 2017. mcr-1 colistin resistance in ESBL-producing Klebsiella pneumoniae, France. Emerg Infect Dis 23:874–876. doi: 10.3201/eid2305.161942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 19.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol 58:1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM; COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. 1997. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 41:699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng ZK, Hu F, Wang W, Guo Q, Chen Z, Xu X, Zhu D, Wang M. 2014. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 58:6982–6985. doi: 10.1128/AAC.03808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers RP, Cavallari JF, Burrows LL. 2013. The efflux inhibitor phenylalanine–arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One 8:e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, Didelot X, Bashir A, Sebra R, Kasarskis A, Sthapit B, Shakya M, Kelly D, Pollard AJ, Peto TEA, Crook DW, Donnelly P, Thorson S, Amatya P, Joshi S. 2014. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]