ABSTRACT
Peptidoglycan is a sugar/amino acid polymer unique to bacteria and essential for division and cell shape maintenance. The d-amino acids that make up its cross-linked stem peptides are not abundant in nature and must be synthesized by bacteria de novo. d-Glutamate is present at the second position of the pentapeptide stem and is strictly conserved in all bacterial species. In Gram-negative bacteria, d-glutamate is generated via the racemization of l-glutamate by glutamate racemase (MurI). Chlamydia trachomatis is the leading cause of infectious blindness and sexually transmitted bacterial infections worldwide. While its genome encodes a majority of the enzymes involved in peptidoglycan synthesis, no murI homologue has ever been annotated. Recent studies have revealed the presence of peptidoglycan in C. trachomatis and confirmed that its pentapeptide includes d-glutamate. In this study, we show that C. trachomatis synthesizes d-glutamate by utilizing a novel, bifunctional homologue of diaminopimelate epimerase (DapF). DapF catalyzes the final step in the synthesis of meso-diaminopimelate, another amino acid unique to peptidoglycan. Genetic complementation of an Escherichia coli murI mutant demonstrated that Chlamydia DapF can generate d-glutamate. Biochemical analysis showed robust activity, but unlike canonical glutamate racemases, activity was dependent on the cofactor pyridoxal phosphate. Genetic complementation, enzymatic characterization, and bioinformatic analyses indicate that chlamydial DapF shares characteristics with other promiscuous/primordial enzymes, presenting a potential mechanism for d-glutamate synthesis not only in Chlamydia but also numerous other genera within the Planctomycetes-Verrucomicrobiae-Chlamydiae superphylum that lack recognized glutamate racemases.
KEYWORDS: Chlamydia, diaminopimelate epimerase, evolution, glutamate racemase, moonlighting enzyme, peptidoglycan, underground metabolism, primordial enzymes
IMPORTANCE
Here we describe one of the last remaining “missing” steps in peptidoglycan synthesis in pathogenic Chlamydia species, the synthesis of d-glutamate. We have determined that the diaminopimelate epimerase (DapF) encoded by Chlamydia trachomatis is capable of carrying out both the epimerization of DAP and the pyridoxal phosphate-dependent racemization of glutamate. Enzyme promiscuity is thought to be the hallmark of early microbial life on this planet, and there is currently an active debate as to whether “moonlighting enzymes” represent primordial evolutionary relics or are a product of more recent reductionist evolutionary pressures. Given the large number of Chlamydia species (as well as members of the Planctomycetes-Verrucomicrobiae-Chlamydiae superphylum) that possess DapF but lack homologues of MurI, it is likely that DapF is a primordial isomerase that functions as both racemase and epimerase in these organisms, suggesting that specialized d-glutamate racemase enzymes never evolved in these microbes.
INTRODUCTION
The members of the family Chlamydiaceae are obligate intracellular bacterial pathogens that cause ocular, sexually transmitted, and respiratory infections. They exhibit a unique biphasic developmental cycle, alternating between metabolically inert elementary bodies (EBs) and metabolically active reticulate bodies (RBs) (1). Infectious EBs attach to and invade host epithelial cells and subsequently reside within a specialized host cell phagosome termed an inclusion. After invasion of a host cell, EBs differentiate into RBs that begin replicating and result in slow expansion of the inclusion. As the inclusion matures, RBs secrete effector proteins via a type III secretion system, altering the host cell trafficking machinery and resulting in the redirection of nutrients to the growing inclusion. Late in the developmental cycle, RBs differentiate back into EBs and exit the cell either by extrusion of the inclusion into the extracellular environment or as a result of host cell lysis due to uncontrolled expansion of the inclusion.
Similar to nearly all other Gram-positive and Gram-negative bacteria, pathogenic chlamydiae are susceptible to antibiotics that target peptidoglycan (2), a heteropolymer essential for bacterial cell division and maintenance of hydrostatic pressure. Numerous studies over the last several decades have failed to definitively identify peptidoglycan within chlamydiae (3, 4), despite the presence of numerous peptidoglycan synthesis genes located in the genome (4–6), giving rise to the “chlamydial anomaly” (7). Only within the last few years, thanks in large part to advances in fluorescence labeling, cryoelectron/superresolution microscopy, and direct muropeptide analysis, has convincing evidence been presented establishing that this essential cell wall component is present in members of the Chlamydiae (7–9).
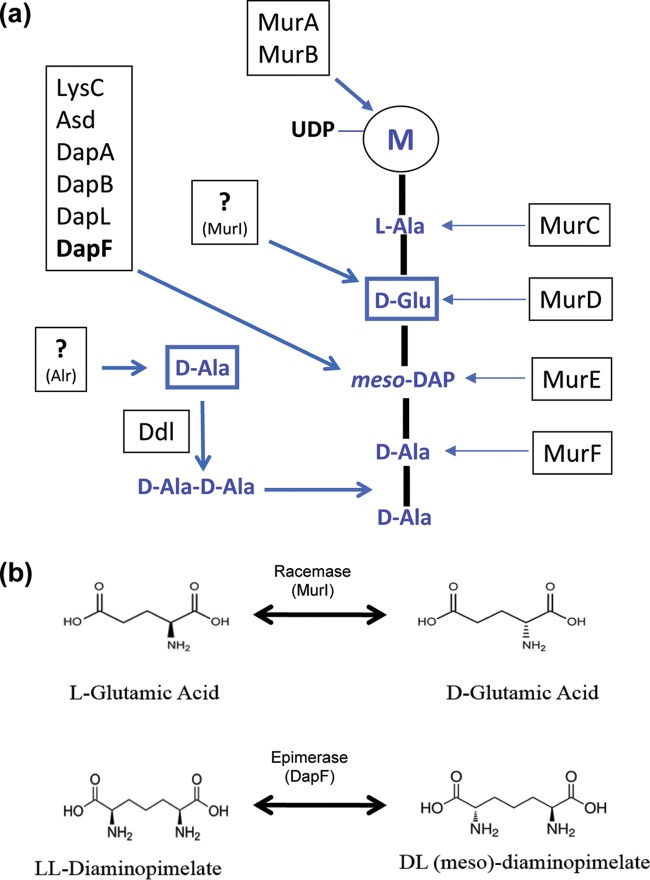
While a majority of the enzymes comprising the peptidoglycan biosynthetic pathway appear to be present in most members of the Chlamydiae (10), several key components are absent. No homologue of the gene for penicillin-binding protein 1 (Pbp1), encoding the glycosyltransferase essential for construction of the peptidoglycan glycan chains, has been identified in any of the Chlamydiae. Additionally, the biosynthesis pathway for generation of the Park nucleotide (the fundamental subunit of all bacterial peptidoglycan) in Chlamydia species is incomplete (Fig. 1a). Three noncanonical amino acids, d-alanine (d-Ala), d-glutamate (d-Glu), and meso-diaminopimelate (m-DAP), are essential for assembly of the Park nucleotide and, subsequently, peptidoglycan. As d-amino acids do not exist in abundance in nature, bacteria must either acquire them from an exogenous source or synthesize them de novo. The canonical pathway for the synthesis of d-amino acids involves the racemization of l-amino acids via racemase enzymes. Alternatively, specific d-amino acids can be synthesized from other d-amino acids via d-amino acid aminotransferase (Dat). Strikingly, the genomes of pathogenic Chlamydia species lack homologues of known racemases and d-amino acid aminotransferases (6). In most Gram-negative bacteria, d-Glu is generated via the glutamate racemase MurI (11) and is invariant in the second position of the peptidoglycan pentapeptide chain (12). All chlamydial genomes encode a UDP-N-acetylmuramoyl-l-alanine-d-glutamate ligase (13), which adds d-Glu to the pentapeptide chain in all of the bacteria examined to date. murI is an essential gene for the vast majority of bacterial pathogens (14), and while homologues are present in the genomes of various environmental chlamydiae (10), no murI homologue has been identified in any pathogenic Chlamydia species. Recent studies indicate that both pathogenic and environmental Chlamydia species contain m-DAP, d-Ala, and d-Glu in the stem peptides of their peptidoglycan (8, 9), indicating the presence of a potentially novel d-Glu synthesis pathway.
FIG 1 .
Missing enzymatic activities in the Park nucleotide biosynthesis pathway in pathogenic Chlamydia species. (a) Muramyl pentapeptide biosynthesis pathway in E. coli. Peptide synthesis is nonribosomal and instead carried out by a series of amino acid ligases (MurC, -D, -E, and -F) that serially add amino acids to newly forming peptide chains. Enzyme homologues not present in the C. trachomatis genome are also indicated (?). M, N-acetylmuramic acid. (b) Glutamate racemase versus DAP epimerase enzymatic reactions. Stereoisomers of glutamic acid and DAP. l,l-DAP, and d,l-m-DAP possess two amine groups, thus creating two centers of asymmetry, whereas l- and d-glutamic acids contain only a single center of asymmetry.
Chlamydiae are obligate intracellular organisms that grow within eukaryotic cells, and eukaryotic cells do not synthesize or maintain d-amino acids at appreciable levels (15). Therefore, in the absence of a murI (or dat) homolog, de novo synthesis appears to be the only method for the acquisition of d-Glu by pathogenic Chlamydia species. Obligate bacterial pathogens and symbionts frequently exhibit a reduced genome size due to gene loss (16, 17). Consequently, they are thought to be highly efficient in their remaining metabolic processes, although this phenomenon is not universal in all microbes (18). Chlamydia possesses a streamlined genome as a result of millions of years of coevolution with its vertebrate host(s), and this is indicative of a highly efficient metabolic network (19, 20). Often, “promiscuous enzymes” are relied upon by genetically reduced microbes to carry out various “moonlighting functions” otherwise performed by entirely separate proteins in other organisms (21, 22). Serine hydroxymethyltransferase (GlyA) from Escherichia coli is one such promiscuous enzyme in that it displays alanine racemase coactivity as a side reaction (23). Treponema denticola also possesses a glyA homologue that encodes a serine hydroxymethyltransferase with alanine racemase activity (12). A previous study indicated that Chlamydia pneumoniae is capable of producing d-Ala and that this activity is the direct result of its promiscuous GlyA enzyme (24).
We postulated that pathogenic chlamydiae possess a bifunctional enzyme capable of generating d-Glu for the synthesis of peptidoglycan. Racemases and epimerases both belong to the family of isomerases and are capable of altering the chirality of amino acids and their derivatives. While racemases catalyze the inversion of the configuration around an asymmetrical carbon in substrates having one center of asymmetry, epimerases perform the same function on substrates that have more than one center of asymmetry. Upon examination of the known epimerases encoded by genes in the C. trachomatis genome, we determined that chlamydial DapF, the DAP epimerase responsible for converting l,l-DAP to d,l-m-DAP (13), seemed a likely candidate for glutamate racemase activity. The structural similarity of l,l-DAP and d,l-m-DAP compared to l-glutamate (L-Glu) and d-Glu (Fig. 1b) led us to speculate that chlamydial DapF (DapFCT) may have evolved broad substrate specificity, allowing it to function as both a racemase and an epimerase in Chlamydia.
Here we demonstrate that DapFCT is capable of racemizing l-Glu to d-Glu (and vice versa) in the presence of the cofactor pyridoxal phosphate (PLP), thereby solving the mystery of how a microbe that lacks a canonical d-Glu racemase is capable of synthesizing d-Glu-containing peptidoglycan. Interestingly, DAP and glutamate appear to be competitive substrates, indicating that they share an active site despite the racemase reaction requiring the PLP cofactor. Comparison of the predicted structure of DapFCT to the solved structures of DAP epimerases encoded by other bacterial species revealed numerous amino acid changes in the enzyme’s substrate-binding site, offering a potential explanation for the relaxed substrate specificity of the chlamydial enzyme. We speculate that the reduced (i) growth rate and (ii) overall quantity of peptidoglycan synthesized by pathogenic Chlamydia species (as well as other members of the PVC [Planctomycetes-Verrucomicrobiae-Chlamydiae] superphylum) have allowed these microbes to successfully evolve for millions of years without canonical glutamate racemases. Given that the vast majority of bacteria belonging to the PVC group possess DapF homologues but lack MurI homologues, we believe that DapFCT is illustrative of a primordial isomerase capable of dual racemase and epimerase activities and whose existence likely predates the emergence of specialized glutamate racemases.
RESULTS
C. trachomatis dapF restores growth in an E. coli d-glutamate auxotroph.
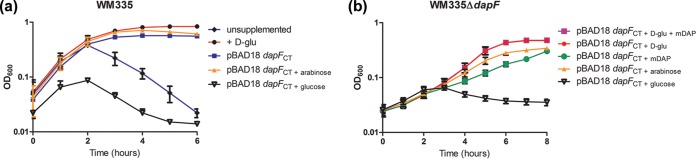
To assess whether chlamydial DAP epimerase is capable of generating sufficient d-Glu to restore growth in an E. coli d-Glu auxotroph, we inserted a copy of the C. trachomatis dapF gene into expression plasmid pBAD18, placing it under the control of an arabinose-inducible promoter. We subsequently transformed this plasmid, pBAD18::dapFCT, into E. coli strain WM335 (25), which contains a premature stop codon in murI (26), as well as an essential secondary mutation in gltS, a d-Glu transporter (27). WM335 requires exogenous d-Glu for sustained replication, and its growth in d-Glu-free medium is highly attenuated. Once internal d-Glu stores are depleted, this strain rapidly lyses as a result of its weakened peptidoglycan layer. Cell lysis is reflected by a drop in optical density at 600 nm (OD600) observed after 2 h of growth in medium without d-Glu after subculture from overnight cultures (Fig. 2a). Growth curves of the pBAD18::dapFCT transformant in the absence of exogenous d-Glu and in the presence of an inducer (arabinose) showed that expression of DapFCT restored WM335 growth and continued survival when stationary phase was reached (Fig. 2a). When expression of DapFCT was repressed by the addition of 0.25% glucose, no growth/survival complementation was observed.
FIG 2 .
Expression of C. trachomatis DapF restores growth in an E. coli d-glutamate auxotroph. (a) Overnight cultures of strains WM335 and WM335/dapFCT were subcultured to an OD600 of ~0.05 and grown in LB either unsupplemented or in the presence of d-Glu (200 µg/ml), arabinose (1%, vol/vol), or glucose (0.25%, vol/vol). OD600 was monitored every hour over a 6-h period. Mean values of three representative experiments are shown. (b) Overnight cultures of strain WM335ΔdapF/dapFCT were subcultured to an OD600 of ~0.05 and grown in LB with thymine (50 µg/ml) under the following conditions: d-Glu (200 µg/ml), m-DAP (100 µg/ml) (+ mDAP), with 1% (vol/vol) arabinose, or 0.25% (vol/vol) glucose. OD600 was monitored every hour over an 8-h period. Data represent mean values from technical replicates of three independent biological experiments, and error bars represent the standard deviation of the mean.
To determine if DapFCT can function as both a glutamate racemase and a DAP epimerase in a single background strain, we deleted the E. coli dapF allele from WM335 via lambda Red recombineering. The resulting mutant (WM335ΔdapF) was subsequently transformed with pBAD18::dapFCT and analyzed for growth as before. WM335ΔdapF transformed with pBAD18::dapFCT (WM335ΔdapF/dapFCT) exhibited slow but steady growth when induced with 1% arabinose (Fig. 2b). Upon supplementation with either d-Glu or m-DAP, growth complementation was achieved in the absence of the inducer. As DapFCT restored WM335ΔdapF growth, this indicates that this enzyme appears to be capable of both DAP epimerase and d-glut racemase activities.
Supernatants of WM335 expressing dapFCT provide evidence of d- to l-glutamate racemization.
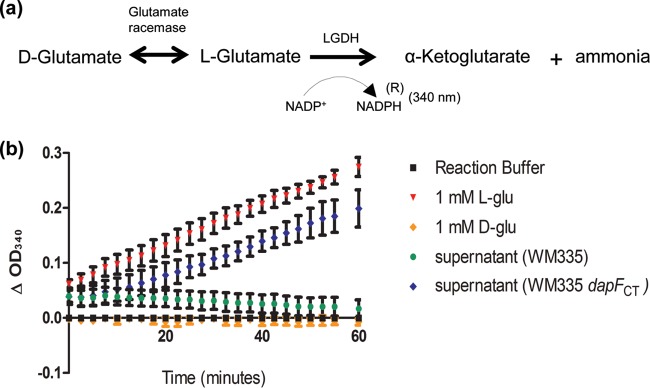
The standard biochemical approach for measurement of glutamate racemase activity is a coupled two-reaction assay: (i) racemization of d-Glu to l-Glu, followed by (ii) conversion of l-Glu to α-ketoglutarate subsequent to the addition of l-glutamic acid dehydrogenase (LGDH, Fig. 3a). The second reaction requires NADP as a cofactor. Conversion of l-Glu to α-ketoglutarate reduces NADP+ to NADPH, which is measured spectrophotometrically at 340 nm. As LGDH cannot utilize d-Glu as a substrate, production of α-ketoglutarate is indicative of glutamate racemase activity. Racemization of glutamate is reversible, and we postulated that if C. trachomatis DapF is capable of converting l-Glu to d-Glu, then alteration of the reaction stoichiometry should allow for the conversion of d-Glu to l-Glu in WM335 cells overexpressing DapFCT. To test this possibility, we assayed supernatants from overnight cultures of the parental WM335 strain and the complemented dapFCT mutant strain, each grown in the presence of exogenous d-Glu. We found that supernatants collected from WM335 expressing DapFCT contained l-Glu levels ~16-fold higher than those obtained from the parental strain (Fig. 3b), indicating that DapFCT is also capable of racemizing d-Glu to l-Glu.
FIG 3 .
Detection of high levels of l-Glu in supernatants from overnight cultures of E. coli strain WM335 expressing DapFCT grown with exogenous d-Glu. (a) Racemase activity was measured via a coupled biochemical reaction, i.e., the racemization of glutamate, followed by the dehydrogenation of l-Glu to α-ketoglutarate and ammonia. (b) Cultures of WM335 and WM335/pBAD18::dapFCT were grown overnight in the presence of arabinose and exogenous d-glutamate. Supernatants were measured for the presence of l-Glu by utilizing LGDH as described in Materials and Methods. Growth medium (Luria broth) supplemented with l- or d-Glu was used for a positive or negative control (red triangles and orange diamonds, respectively). Symbols represent mean values of two independent biological replicates, and error bars represent ranges.
C. trachomatis DapF possesses glutamate racemase activity that is cofactor PLP dependent.
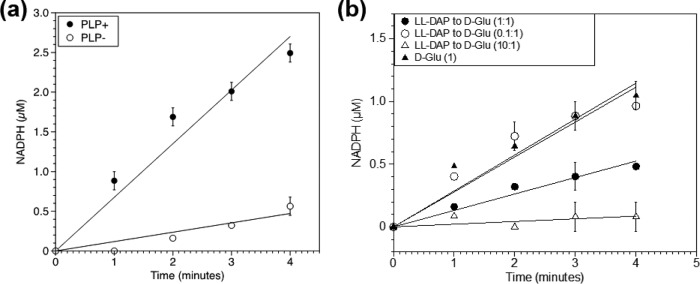
Alanine racemases require PLP as a cofactor, whereas canonical glutamate racemases and DAP epimerases are PLP-independent enzymes that utilize a two-cysteine-dependent acid-base catalytic mechanism (15, 28). Researchers recently discovered that the cystathionine beta-lyase isolated from Wolbachia and Thermotoga maritima exhibits PLP-dependent glutamate racemase activity (29), indicating that the dual functionality of these moonlighting enzymes may require PLP as a cofactor. To test this possibility, DapFCT-His was overexpressed and purified for in vitro characterization. Resolution of the purified DapFCT-His protein via SDS-PAGE yielded a single band corresponding to the expected molecular mass of ~32 kDa (see Fig. S1 in the supplemental material). Purified DapFCT-His was assayed for glutamate racemase activity with d-Glu as the substrate as described in Materials and Methods. In the in vitro assay, purified DapFCT-His showed an increase in absorbance at 340 nm while controls remained significantly lower than the experimental reaction (Fig. S2). d-Glu racemization to l-Glu was found to be PLP dependent (Fig. 4a).
FIG 4 .
Dependence of DapFCT on PLP for glutamate racemization. (a) Glutamate racemase activity of DapFCT in the presence (●) or absence (○) of PLP. The glutamate racemase activity of DapFCT was determined in a coupled assay as described in Materials and Methods. Error bars represent the standard deviation of two independent experiments. (b) Racemase activity of DapFCT on 10 mM d-Glu alone (▲) and in the presence of 1 mM (○), 10 mM (●), or 100 mM (△) l,l-DAP. Each symbol represents the average value of two independent biological replicates.
SDS-PAGE analysis of various eluted fractions of purified DapFCT-His expressed in E. coli BL21. Lanes: 1, molecular mass markers; 2 to 4, elution of DapFCT-His; 5, whole-cell lysate of E. coli BL21 expressing DapFCT-His. The protein corresponding to cloned DapFCT-His is boxed. Download FIG S1, TIF file, 11 MB (11.2MB, tif) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assay of DapFCT for glutamate racemase activity with the glutamate racemase-coupled assay. Reaction mixtures were prepared with 20 mM d-glutamate (●), without DapF (○), and without d-glutamate (△). An increase in NADPH formation is indicative of glutamate racemase activity of DapFCT. Error bars represent the standard deviations of two biological replicates. Download FIG S2, TIF file, 13.6 MB (13.9MB, tif) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As PLP-dependent and PLP-independent isomerases are thought to operate by dissimilar mechanisms, it was not entirely clear whether DapFCT utilized the same substrate-binding pocket to carry out both racemase and epimerase reactions. To assess whether l,l-DAP and l-Glu compete for the same substrate-binding/active sites on DapFCT, competition assays were performed. The addition of 10 and 100 mM l,l-DAP to the reaction mixture decreased glutamate racemase activity 47 and 95%, respectively, while 1 mM l,l-DAP did not appear to substantially inhibit the reaction (Fig. 4b). These data indicate that both substrates appear to have a binding site on DapFCT, that this site is occupied by either d-Glu or l,l-DAP in a concentration-dependent manner, and that the efficiency of glutamate racemization is inversely proportional to the amount of competing l,l-DAP present.
Structure of Chlamydia DAP epimerase exhibits significant remodeling in the substrate-binding pocket.
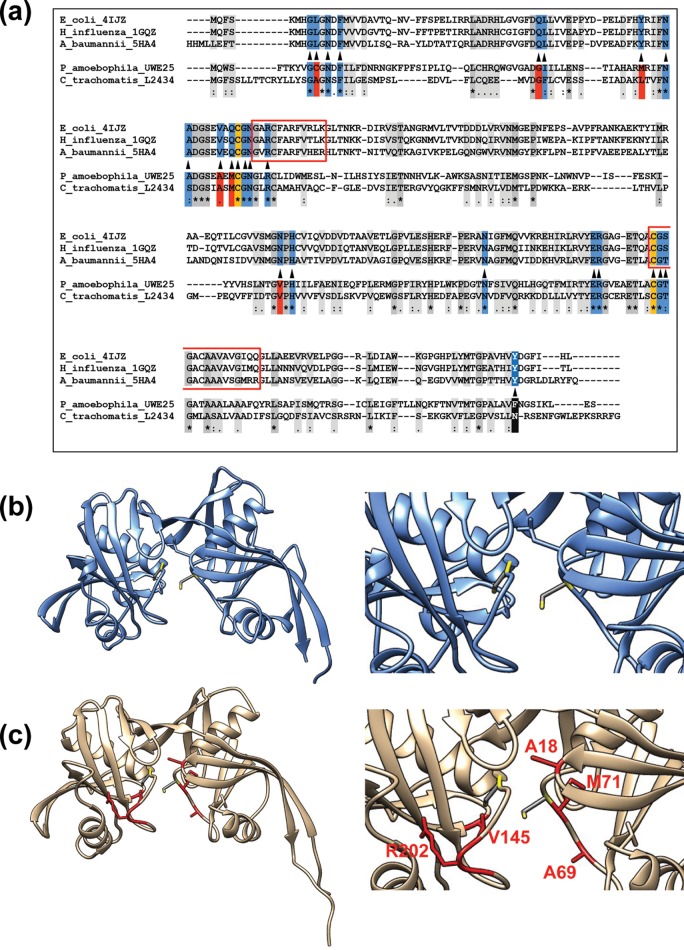
As DapFCT appears to make use of the same substrate-binding pocket when carrying out both epimerase and racemase activities, we reasoned that the enzyme’s active site likely possesses unique amino acid substitutions compared to the more specialized DAP epimerases encoded by other bacterial species. In a majority of the DAP epimerases whose crystal structures have been resolved, the amino acids lining the active-site cavity are highly conserved (15, 30, 31) (Fig. 5a). In the DAP epimerases encoded by Haemophilus influenzae and Bacillus anthracis, the active-site residues are flanked by two internal α-helixes cradled by β-sheets. These helices are positioned end-on toward each other (Fig. 5b), and their orientation and lengths are crucial to the relative proximity of two active-site cysteine residues (30, 31). One active-site cysteine thiolate acts as a base to deprotonate the α-carbon, while a second cysteine thiol acts as an acid to reprotonate the resulting planar carbanionic intermediate from the opposite face (13). To access the level of conservation of the DapFCT active site, we sorted proteins present in the Protein Data Bank (PDB) for enzymes that were most similar to DapFCT on the basis of structural alignments of amino acid sequences (paired with solved PDB structures). The DAP epimerase encoded by B. anthracis (2OTN) was the most structurally similar to the Chlamydia enzyme (as determined by RaptorX and I-TASSER structural prediction web-based applications) and was used as a template for modeling of a structural approximation of DapFCT (Fig. 5b and c). Both active-site cysteines (C72 and C207) are conserved in the chlamydial enzyme, as are the histidine and glutamic acid residues (H147 and E197) that are essential for epimerase catalytic activity (30). Of the 18 remaining amino acids that reside within the enzyme active site, only 9 (G17, N20, F22, N63, G73, N74, N179, R198, and G208) are well conserved in the C. trachomatis protein (Fig. 5a). Of the remaining nine amino acids, three are replaced with similar amino acids (F46, S64, T209) and six represent dissimilar amino acid substitutions (A18, G45, L59, V83, Q85, V145). We hypothesize that amino acid alterations within the binding pocket (A18, A69, M71, V145, and R202) likely influence the accessibility of substrates to the chlamydial enzyme’s active-site cysteine residues (C72 and C207).
FIG 5 .
C. trachomatis DAP epimerase exhibits significant remodeling of the enzyme substrate-binding pocket. (a) Amino acid structural alignments indicate that of the 18 to 20 generally well-conserved amino acid residues lining the active site within the enzyme substrate-binding pocket (blue), 6 are highly dissimilar in chlamydial DapF (red). Boxed residues indicate alpha-helical domains critical for determination of the distance between active-site cysteine residues C72 and C207 (gold). DAP epimerase encoded by B. anthracis (b) is the enzyme most structurally similar to the DAP epimerase encoded by C. trachomatis present in the PDB (2OTN) and was used as the template for the modeling of a structural approximation of DapFCT (c). Active-site cysteine residues (yellow) are conserved, as is their orientation within the active site, because of the presence of similar alpha-helical domains (a, boxed in red). Amino acid alterations within the binding pocket (A18, A69, M71, V145, and R202) are red, and the chlamydial enzyme’s active-site cysteine residues (C72 and C207) are yellow.
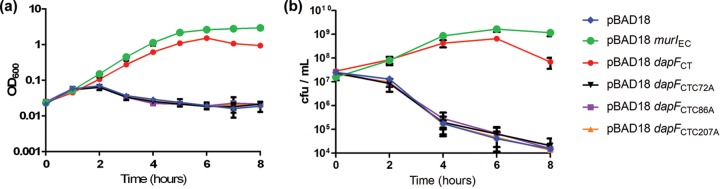
Chlamydial DapF racemase activity requires active-site cysteines.
As mentioned previously, both canonical d-Glu racemase and DAP epimerase are PLP-independent enzymes and utilize a two-cysteine-dependent acid-base catalytic mechanism (32). Sequence alignments revealed that the two active-site cysteines for epimerase function are conserved in DapFCT (Fig. 5a). As our enzymatic assays established that the glutamate racemase activity of DapFCT is PLP dependent, we questioned whether these active-site cysteines play any role in this activity. We mutated each of the two putative active-site cysteines in DapFCT and assessed the mutated enzyme’s ability to complement the d-Glu auxotrophy of the WM335 parental strain. Interestingly, neither of these mutated alleles (C72A or C207A) was capable of restoring WM335 growth in the absence of exogenous d-Glu (Fig. 6), indicating that, despite being a PLP-dependent reaction, the racemase activity of DapFCT appears to be dependent on both active-site cysteine residues. DapFCT possesses an additional cysteine (C86) that is not highly conserved in other bacterial species (Fig. 5a). This cysteine sits at the terminus of one of the two predicted alpha-helical domains (opposite C72), which together determine the distance between the enzyme’s active-site cysteines and thus the functionality of the enzyme. A C86A substitution also resulted in loss of the ability of DapFCT to restore growth (Fig. 6), suggesting that despite requiring PLP as a cofactor for the racemization of glutamate, DapFCT requires the epimerase active-site cysteines for glutamate racemase activity.
FIG 6 .
Chlamydial DapF requires the epimerase active-site cysteines for racemase activity. Mutagenesis of cysteine resides within the predicted active site of chlamydial DapF results in loss of growth/survival complementation of WM335. Cultures were grown overnight in LB supplemented with ampicillin, thymine, and d-Glu. Overnight cultures were subcultured to an OD600 of 0.025 in LB supplemented with ampicillin, thymine, and arabinose (1%, vol/vol). Cultures were then grown with shaking at 37°C, and the OD600 (a) was recorded hourly for 8 h. To determine the number of CFU/ml (b), bacterial samples taken every 2 h were plated on LB agar containing ampicillin, thymine, and d-Glu. Complementation of WM335 with E. coli murI was included as a positive control (green). The data are mean values of technical replicates of three independent biological experiments, and error bars represent the standard deviation of the mean.
Phylogenetic analysis of Chlamydia DapF shows high similarity to DapF in other members of the PVC group superphylum.
As DapFCT has glutamate racemase activity, we deduced that DAP epimerases might function similarly in other bacterial species closely related to C. trachomatis. DAP epimerases encoded by pathogenic Chlamydia species all share moderate sequence similarity (>48% identity, >65% similarity) and all lack murI homologues, suggesting that their DapF enzymes are also capable of racemizing glutamate. We questioned whether similarity to the C. trachomatis DapF enzyme significantly correlated with the absence of homologues of murI in other members of the PVC superphylum. Using protein sequences from Parachlamydia acanthamoebae, a chlamydia-like endosymbiont of amoebae that possesses both DapF and MurI homologues, we screened members of the PVC group for the presence/absence of these two enzymes. For the PVC group members on the NCBI database at the time of analysis (PVC group: taxid: 1783257, 5 December 2017), we performed BLAST searches for homologues of P. acanthamoebae 50S ribosomal protein L3, MurA (initiates the first committed step in peptidoglycan synthesis), DapF (DAP epimerase), Alr (alanine racemase), and MurI (glutamate racemase). The analysis produced affirmative alignment scores (>e−4) for 396, 367, 270, and 174 enzyme homologues, respectively. These numbers did not vary significantly when the enzyme sequences of other representative species were BLAST searched against the PVC group taxonomic identity (Table S1), minimizing the possibility that potential MurI homologues were missed in the initial BLAST search because of divergence from the P. acanthamoebae homologue. Additionally, the 174 examples of PVC group members that possess a MurI enzyme is likely an overestimate of potential d-Glu racemase homologues; several of these “positive” hits were assigned to C. trachomatis and C. abortus isolates. Upon closer inspection, the MurI homologues assigned to these isolates have low sequence similarity to each other (between species) and high sequence similarity to commensal/pathogenic microbes known to share the mammalian reproductive/vaginal niche (Table S2). This analysis further indicates that the absence of a specialized glutamate racemase is highly prevalent among members of the PVC group (Fig. 7).
FIG 7 .
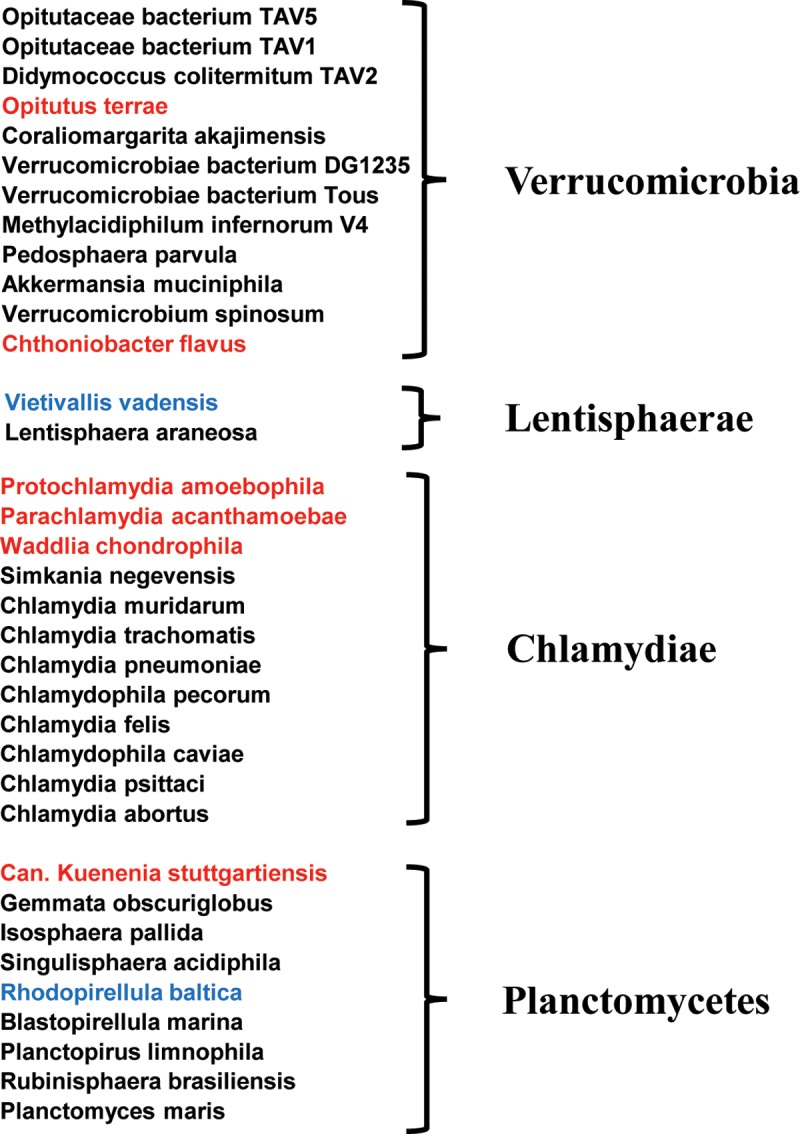
Absence of MurI homologues is prevalent in microbes of the PVC group superphylum. Shown is a list of representative species comprising the PVC group superphylum. Species possessing homologues of the DAP epimerase encoded by P. acanthamoebae (DapFPA) but lacking homologues of the glutamate racemase (MurIPA) are black. Species lacking both DapFPA and MurIPA homologues are blue. Species encoding homologues of both DapFPA and MurIPA are red. More comprehensive lists of PVC group members arranged by DAP epimerase/glutamate racemase status are provided in Tables S3 and S4.
Glutamate racemase homologues of enzymes encoded by different PVC group members resulting from pBLAST searches. Download TABLE S1, XLSX file, 0.01 MB (8.9KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of “glutamate racemase” homologues assigned to pathogenic Chlamydia species. Download TABLE S2, XLSX file, 0.01 MB (11.4KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PVC group members possessing DAP epimerase (DapF) and lacking d-glutamate racemase (MurI) homologues. Download TABLE S3, XLSX file, 0.02 MB (23KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PVC group members possessing both DAP epimerase (DapF) and d-glutamate racemase (MurI) homologues. Download TABLE S4, XLSX file, 0.02 MB (19.4KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Every chlamydial genome sequenced to date encodes a nearly complete peptidoglycan synthesis pathway, with the exception of two key genes, murI and alr, which encode glutamate and alanine racemases, respectively (6). We hypothesized that the structural similarity between l-Glu and l,l-DAP makes DapF a good candidate for Chlamydia’s “missing” glutamate racemase. Our data indicate that C. trachomatis DapF can complement the d-Glu auxotrophy of an E. coli mutant that expresses a nonfunctional glutamate racemase. Biochemical assays confirmed that chlamydial DapF possesses glutamate racemase activity.
These findings, while solving one long-standing mystery of chlamydial cell wall biology, also raise several questions regarding the functionality of this enzyme and its potential regulation by substrate availability. Glutamate racemization is a PLP-independent mechanism in most model organisms; however, another example of PLP-dependent glutamate racemase activity has recently been reported (29). While this study is the first to demonstrate glutamate racemase activity of a DapF protein, surprisingly, this activity is PLP dependent. Paradoxically, the two active-site cysteine residues are not thought to play any role in PLP-dependent racemase activity (29), and yet our data suggest that they are essential for DapFCT to complement growth in an E. coli d-Glu auxotroph. While epimerase active-site cysteines have been replaced in other systems without significantly affecting protein expression/integrity (33, 34), we cannot rule out the possibility that, in addition to being essential for epimerase function, these cysteine residues may also be indispensable for the overall structural stability of the Chlamydia enzyme. Regardless, as both substrates likely compete for the same substrate-binding pocket, it appears that Chlamydia has sacrificed enhanced efficiency to maintain the dual functionality of this enzyme. DapFCT is capable of racemizing glutamate while maintaining enzyme kinetics similar to those of MurIEC; however, this high rate of activity is only possible at low levels of exogenous l,l-DAP (Fig. 4b). It is possible that the presence of competing substrates or availability of the PLP cofactor plays a role in the favoring of one reaction over the other in vivo. Further studies are needed to assess if and how these competing reactions are regulated, how Chlamydia maintains the balance between competing substrates, and how reliance on this single enzyme for the construction of two essential building blocks of the bacterial cell wall affects Chlamydia growth and development.
The frequency of promiscuous enzymes is generally thought to be a function of genome size and the underlying redundancy often seen in larger bacterial genomes (35). Mutations in extra copies of a given allele give rise to novel enzymes with new substrates and activities (36). This is exemplified in the patchwork model of enzyme evolution in which enzymatic specialization increases over time as a result of gene duplication and divergence (35). This model, currently favored for modern enzyme evolution, is based on the underlying assumptions that increased enzymatic efficiency is selectively advantageous and that this pressure for faster reactions results in a narrowing of enzyme function and substrate specificity. While this may be the case for a majority of the bacterial species studied to date, these efficiencies may confer less of a benefit, or may even be detrimental, in slow-growing microbes that live in relatively stable, nutrient-rich environments (such as pathogenic Chlamydia species and potentially other microbes comprising the PVC superphylum).
D’Ari and Casadesus coined the term “underground metabolism” to describe reactions that occur when enzymes act on substrate analogues that are also endogenous metabolites utilized by an organism (37). They argued that a certain level of metabolic inaccuracy is actually energetically efficient and that the resulting metabolic plasticity may provide an evolutionary advantage in some environments. In the case of DapFCT, substrate diversity appears to have been selected for as both substrates (d-Glu and m-DAP) are useful to the cell. DapF is not the only enzyme in the Chlamydia peptidoglycan biosynthetic pathway that exhibits substrate promiscuity; C. trachomatis UDP-N-acetylmuramate-l-alanine ligase (MurC) incorporates glycine, as well as l-alanine, into the first position on the peptidoglycan stem peptide (9, 38, 39). It is likely that in environments where enhanced metabolic rates are not required (or are potentially even selected against), the pressure for enzymatic efficiency is relaxed, and the activities of promiscuous enzymes can be maintained or returned to a more primordial state. Given the number of PVC group members lacking MurI homologues and encoding DapF enzymes with high similarity to DapFCT, it is likely that in this case a primordial isomerase with dual substrate specificity was maintained by numerous species within this superphylum. Promiscuous enzymes have recently been identified in a variety of other bacteria with reduced genomes (29) and likely play an essential role in the adaptive strategies associated with genomic/metabolic streamlining.
Numerous other bacteria outside the PVC superphylum, including Rickettsia, Xanthomonas, Xylella, and Bordetella species, also contain a nearly complete peptidoglycan biosynthesis pathway, lack a MurI homologue, and retain a DAP epimerase. It is tempting to speculate that the DAP epimerases encoded by these diverse species also possess glutamate racemase activity. Furthermore, the genomes of chlamydiae contain a high number of plant-like genes whose products are targeted to the chloroplast, suggesting an evolutionary relationship between chloroplasts and chlamydiae (40). The DAP synthesis pathway is unique in pathogenic Chlamydia species; these microbes lack dapD, dapC, and dapE homologues in their genomes and instead produce DAP by an aminotransferase thought previously to be utilized only by plants and cyanobacteria (13). This aminotransferase enzyme (DapL) also exhibits substrate promiscuity; however, its active-site resides are largely conserved with those of homologous enzymes in other bacteria (41). The genome of the nonseed moss Physcomitrella patens contains 10 genes involved in peptidoglycan synthesis, with several of those genes, including murE and pbps, encoding functional proteins. Moreover, P. patens is sensitive to β-lactam antibiotics, fosfomycin, and d-cycloserine (42–44) and incorporates d-amino acids (45), indicating that the plastid contains peptidoglycan. While a murI homologue has not been identified in the P. patents genome, it does encode a homologue of DAP epimerase. Should this enzyme also function as a glutamate racemase, it would further the evolutionary link between Chlamydia and chloroplasts and provide insight into how primordial microbial systems met their fundamental growth requirements with genomic efficiency and enzymatic versatility. Additionally, Chlamydia species lack the final enzyme in the lysine biosynthesis pathway (DAP decarboxylase, LysA), indicating that the sole purpose of m-DAP production by the microbe is peptidoglycan biosynthesis. It is worth noting that Chlamydia possesses numerous other examples of metabolic pathways apparently lacking critical enzymes essential for their ultimate functionality (22). More examples of promiscuous, primordial enzymes likely remain hidden within its efficiently streamlined genome, waiting to be discovered.
MATERIALS AND METHODS
Bacterial strains, medium, and growth conditions.
The E. coli WM335 dapF double mutant was constructed by lambda Red recombination (46) with WM335 (25) as the parent strain. All strains were grown in LB medium with aeration or on agar plates. The medium was supplemented with ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), thymine (50 μg/ml), d-glutamic acid (200 μg/ml), arabinose (1%), glucose (0.25%), and DAP (racemic mixture; 100 μg/ml; Sigma, St. Louis, MO) as needed, and cultures were incubated at 37°C.
Cloning, in vivo complementation, and recombinant expression of DapF from C. trachomatis.
dapF of C. trachomatis and murI of E. coli were cloned into expression vector pBAD for complementation of the murI mutant of E. coli. To express and purify DapFCT, dapFCT was cloned into pTBSG (47) with a 6-His tag placed at the 5′ end of dapFCT. pTBSG::dapFCTL2 was graciously provided by Scott Hefty (University of Kansas). This plasmid was expressed in E. coli BL21(DE3) at small and large scales. Initially, this strain was grown at 37°C and slower induction at 16°C was done at an OD600 of 0.45 with a final isopropyl-β-d-thiogalactopyranoside (IPTG) concentration of 0.5 mM. Cells were harvested after 12 h of growth and resuspended in 20 mM Tris-Cl (pH 8.0)–500 mM NaCl–20 mM imidazole. Cells were lysed by incubation with lysozyme for 30 min on ice and then sonicated with an Ultrasonic Processor. A cell extract was obtained after separation of cell debris by centrifugation (10,000 × g for 15 min at 4°C). The soluble lysate was applied to a Ni-nitrilotriacetic acid column. After the column was washed with washing buffer (20 mM Tris-Cl [pH 8.0], 500 mM NaCl, 20 mM imidazole), the His-tagged DapF protein was eluted from the column with elution buffer (20 mM Tris-Cl [pH 8.0], 500 mM NaCl, 300 mM imidazole). All protein fractions were analyzed by SDS-PAGE. Fractions containing the greatest abundance of His-tagged DapFCT were pooled and dialyzed with a dialysis tube with a 10-kDa molecular mass cutoff (Pierce Biotechnology, Waltham, MA) against storage buffer that contained 20 mM Tris-Cl (pH 7.7) and 150 mM NaCl. The His tag was not removed, as the His-tagged version of DapF complemented the murI mutant and the purified enzyme with or without the tag showed no significant difference in enzymatic activity (data not shown).
Site-directed mutagenesis.
Overlapping oligonucleotides were used to introduce the desired site-directed mutations via PCR. NEA292 (5′-CAATGGCGCACGTAGCCCAGGCCTTTGGACTTGAAGATGTTTC-3′) and NEA293 (5′-GAAACATCTTCAAGTCCAAAGGCCTGGGCTACGTGCGCCATTG-3′) or NEA294 (5′-GTGAGCGAGAAACCTTATCTGCTGGGACAGGGATGTTGGCAAG-3′) and NEA295 (5′-CTTGCCAACATCCCTGTCCCAGCAGATAAGGTTTCTCGCTCAC-3′) were used to amplify dapFCT with a C86A or C207A mutation, respectively, by using PfuUltra HF polymerase (Stratagene; La Jolla, CA). The boldface base pairs are the changed nucleotides. The PCR was treated with DpnI and transformed into DH5α competent cells. Plasmids from the resulting ampicillin-resistant colonies were sequence verified for the desired site-directed mutations.
Growth curves.
Growth curves were determined by diluting overnight cultures into Difco LB Miller (Luria-Bertani) broth base (Becton, Dickinson and Company, NJ) with appropriate antibiotics and supplements as required to an initial OD600 of either 0.025 or 0.05. Cultures were then grown with shaking at 37°C for 6 to 8 h with OD600 readings taken every hour. To ensure that changes in OD600 values correlated with bacterial numbers, samples were taken every 2 h, bacteria were serially diluted and plated on LB agar, and CFU counts were determined.
Enzyme assays.
The generation of l-Glu from d-Glu in E. coli expressing chlamydial DapF was measured by an enzyme-coupled reaction. Supernatants from E. coli grown overnight in the presence of exogenous d-Glu (200 μg/ml) were passed through a 0.22-μm filter. Ten microliters was then added to assay buffer (2.5 mM NADP+) containing 3 μl of l-glutamate dehydrogenase (LGDH) from bovine liver (Sigma), which catalyzes the conversion of l-Glu to α-ketoglutarate. The increase in absorbance at 340 nm (which indicates the reduction of NADP+ to NADPH) was measured in a BioTek Synergy 2 plate reader at 25°C, and measurements were taken every 2.5 min for an hour. For the purified, His-tagged DapFCT protein, the reaction mixture with a final volume of 0.1 ml contained 20 mM Tris-Cl (pH 8.0), 20 mM d-Glu, 10-μM PLP, 10 mM MgCl2, and 20 μg of purified, His-tagged DapFCT. Reaction mixtures were incubated at 37°C for 1 h and then heat inactivated at 95°C prior to the l-Glu assay. The l-Glu assay reaction mixture with a final volume of 0.1 ml contained 20 mM Tris-Cl (pH 8.0), 2.5 mM NADP+, 10 mM MgCl2, and 50 μl of the first reaction mixture (d-Glu→l-Glu). The OD340 was noted immediately after the addition of 2.5 μl of LGDH. Each reaction was performed in biological replicates. To test the substrate affinity of DapFCT and assess the effects of the competing substrate on the glutamate racemization reaction, various concentrations of l,l-DAP (1 to 100 mM) were added to the reaction mixture while the concentration of d-Glu was kept constant (10 mM). The resulting glutamate racemase activity (in the presence or absence of l,l-DAP) was used to describe the relationships between competing substrates (d-Glu and l,l-DAP).
Bioinformatics.
Sequence alignments and protein structural prediction were conducted with RaptorX (48) and I-TASSER (49) web-based applications. Mapping of amino acids and visualization of DapFCT structural predictions were accomplished with the UCSF Chimera imaging, analysis, and visualization suite (50).
ACKNOWLEDGMENTS
We thank Scott Hefty (University of Kansas) for providing pTBSG::dapFCTL2 for expression of His-tagged DapFCT. We thank Josephine Clark-Curtiss and Hedwin Kitdorlang Dkhar for assistance with protein purification. We also thank Rhoel Dinglasan and Timothy Hamerly for help in developing analytic methods for the detection of glutamate isomers.
Molecular graphics and analyses were performed with the UCSF Chimera package developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). This work was supported by NIH grants to Anthony Maurelli (AI044033) and George Liechti (F32AI112209) in addition to a USU faculty start up award (HP73LIEC17) to George Liechti. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The opinions or assertions contained herein are ours and are not to be construed as official or as reflecting the views of the Department of Defense or the Uniformed Services University.
Footnotes
Citation Liechti G, Singh R, Rossi PL, Gray MD, Adams NE, Maurelli AT. 2018. Chlamydia trachomatis dapF encodes a bifunctional enzyme capable of both d-glutamate racemase and diaminopimelate epimerase activities. mBio 9:e00204-18. https://doi.org/10.1128/mBio.00204-18.
REFERENCES
- 1.Wyrick PB. 2000. Intracellular survival by Chlamydia. Cell Microbiol 2:275–282. doi: 10.1046/j.1462-5822.2000.00059.x. [DOI] [PubMed] [Google Scholar]
- 2.Heilman FR, Herrell WE. 1944. Penicillin in the treatment of experimental psittacosis. Proe Staff Meet Mayo Clin 19:204–207. [Google Scholar]
- 3.Fox A, Rogers JC, Gilbart J, Morgan S, Davis CH, Knight S, Wyrick PB. 1990. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun 58:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopra I, Storey C, Falla TJ, Pearce JH. 1998. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology 144:2673–2678. doi: 10.1099/00221287-144-10-2673. [DOI] [PubMed] [Google Scholar]
- 5.Moulder JW. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis 2:87–99. [PubMed] [Google Scholar]
- 6.McCoy AJ, Maurelli AT. 2006. Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol 14:70–77. doi: 10.1016/j.tim.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packiam M, Weinrick B, Jacobs WR Jr, Maurelli AT. 2015. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly.” Proc Natl Acad Sci U S A 112:11660–11665 doi: 10.1073/pnas.1514026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquier N, Viollier PH, Greub G. 2015. The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly. FEMS Microbiol Rev 39:262–275. doi: 10.1093/femsre/fuv001. [DOI] [PubMed] [Google Scholar]
- 11.Schell MJ. 2004. The N-methyl d-aspartate receptor glycine site and d-serine metabolism: an evolutionary perspective. Philos Trans R Soc Lond B Biol Sci 359:943–964. doi: 10.1098/rstb.2003.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertoldi M, Cellini B, Paiardini A, Di Salvo M, Borri Voltattorni C. 2003. Treponema denticola cystalysin exhibits significant alanine racemase activity accompanied by transamination: mechanistic implications. Biochem J 371:473–483. doi: 10.1042/BJ20020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT. 2006. l,l-Diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci U S A 103:17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundqvist T, Fisher SL, Kern G, Folmer RH, Xue Y, Newton DT, Keating TA, Alm RA, de Jonge BL. 2007. Exploitation of structural and regulatory diversity in glutamate racemases. Nature 447:817–822. doi: 10.1038/nature05689. [DOI] [PubMed] [Google Scholar]
- 15.Pillai B, Cherney MM, Diaper CM, Sutherland A, Blanchard JS, Vederas JC, James MN. 2006. Structural insights into stereochemical inversion by diaminopimelate epimerase: an antibacterial drug target. Proc Natl Acad Sci U S A 103:8668–8673. doi: 10.1073/pnas.0602537103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. doi: 10.1016/S0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 17.Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet 17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 18.Juárez-Vázquez AL, Edirisinghe JN, Verduzco-Castro EA, Michalska K, Wu C, Noda-García L, Babnigg G, Endres M, Medina-Ruíz S, Santoyo-Flores J, Carrillo-Tripp M, Ton-That H, Joachimiak A, Henry CS, Barona-Gómez F. 2017. Evolution of substrate specificity in a retained enzyme driven by gene loss. Elife 6:e22679. doi: 10.7554/eLife.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 20.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 21.Price DR, Wilson AC. 2014. A substrate ambiguous enzyme facilitates genome reduction in an intracellular symbiont. BMC Biol 12:110. doi: 10.1186/s12915-014-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams NE, Thiaville JJ, Proestos J, Juárez-Vázquez AL, McCoy AJ, Barona-Gómez F, Iwata-Reuyl D, de Crécy-Lagard V, Maurelli AT. 2014. Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. mBio 5:e01378-14. doi: 10.1128/mBio.01378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shostak K, Schirch V. 1988. Serine hydroxymethyltransferase: mechanism of the racemization and transamination of d- and l-alanine. Biochemistry 27:8007–8014. doi: 10.1021/bi00421a006. [DOI] [PubMed] [Google Scholar]
- 24.De Benedetti S, Bühl H, Gaballah A, Klöckner A, Otten C, Schneider T, Sahl HG, Henrichfreise B. 2014. Characterization of serine hydroxymethyltransferase GlyA as a potential source of d-alanine in Chlamydia pneumoniae. Front Cell Infect Microbiol 4:19. doi: 10.3389/fcimb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lugtenberg EJ, Wijsman HJ, van Zaane D. 1973. Properties of a d-glutamic acid-requiring mutant of Escherichia coli. J Bacteriol 114:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doublet P, van Heijenoort J, Mengin-Lecreulx D. 1992. Identification of the Escherichia coli murI gene, which is required for the biosynthesis of d-glutamic acid, a specific component of bacterial peptidoglycan. J Bacteriol 174:5772–5779. doi: 10.1128/jb.174.18.5772-5779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dougherty TJ, Thanassi JA, Pucci MJ. 1993. The Escherichia coli mutant requiring d-glutamic acid is the result of mutations in two distinct genetic loci. J Bacteriol 175:111–116. doi: 10.1128/jb.175.1.111-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams G, Maziarz EP III, Amyes TL, Wood TD, Richard JP. 2003. Formation and stability of the enolates of N-protonated proline methyl ester and proline zwitterion in aqueous solution: a nonenzymatic model for the first step in the racemization of proline catalyzed by proline racemase. Biochemistry 42:8354–8361. doi: 10.1021/bi0345992. [DOI] [PubMed] [Google Scholar]
- 29.Ferla MP, Brewster JL, Hall KR, Evans GB, Patrick WM. 2017. Primordial-like enzymes from bacteria with reduced genomes. Mol Microbiol 105:508–524. doi: 10.1111/mmi.13737. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd AJ, Huyton T, Turkenburg J, Roper DI. 2004. Refinement of Haemophilus influenzae diaminopimelic acid epimerase (DapF) at 1.75 A resolution suggests a mechanism for stereocontrol during catalysis. Acta Crystallogr D Biol Crystallogr 60:397–400. doi: 10.1107/S0907444903027999. [DOI] [PubMed] [Google Scholar]
- 31.Matho M, Fakuda M, Santelli E, Jaroszewski L, Liddington RC, Roper D. 2009. Crystal structure and inhibition of a catalytically active form of diaminopimelate epimerase (DapF) from Bacillus anthracis. J Mol Biol 385:580–594. doi: 10.1016/j.jmb.2008.10.072. [DOI] [PubMed] [Google Scholar]
- 32.Tanner ME. 2002. Understanding nature’s strategies for enzyme-catalyzed racemization and epimerization. Acc Chem Res 35:237–246. doi: 10.1021/ar000056y. [DOI] [PubMed] [Google Scholar]
- 33.Usha V, Dover LG, Roper DL, Besra GS. 2008. Characterization of Mycobacterium tuberculosis diaminopimelic acid epimerase: paired cysteine residues are crucial for racemization. FEMS Microbiol Lett 280:57–63. doi: 10.1111/j.1574-6968.2007.01049.x. [DOI] [PubMed] [Google Scholar]
- 34.Koo CW, Blanchard JS. 1999. Chemical mechanism of Haemophilus influenzae diaminopimelate epimerase. Biochemistry 38:4416–4422. doi: 10.1021/bi982911f. [DOI] [PubMed] [Google Scholar]
- 35.Jensen RA. 1976. Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Núñez MA, Rodríguez-Vázquez K, Pérez-Rueda E. 2015. The lifestyle of prokaryotic organisms influences the repertoire of promiscuous enzymes. Proteins 83:1625–1631. doi: 10.1002/prot.24847. [DOI] [PubMed] [Google Scholar]
- 37.D’Ari R, Casadesús J. 1998. Underground metabolism. Bioessays 20:181–186. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Hesse L, Bostock J, Dementin S, Blanot D, Mengin-Lecreulx D, Chopra I. 2003. Functional and biochemical analysis of Chlamydia trachomatis MurC, an enzyme displaying UDP-N-acetylmuramate:amino acid ligase activity. J Bacteriol 185:6507–6512. doi: 10.1128/JB.185.22.6507-6512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Maurelli AT. 2005. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting d-alanyl-d-alanine ligase activity involved in peptidoglycan synthesis and d-cycloserine sensitivity. Mol Microbiol 57:41–52. doi: 10.1111/j.1365-2958.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- 40.Brinkman FS, Blanchard JL, Cherkasov A, Av-Gay Y, Brunham RC, Fernandez RC, Finlay BB, Otto SP, Ouellette BF, Keeling PJ, Rose AM, Hancock RE, Jones SJ, Greberg H. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res 12:1159–1167. doi: 10.1101/gr.341802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe N, Clay MD, van Belkum MJ, Fan C, Vederas JC, James MN. 2011. The structure of ll-diaminopimelate aminotransferase from Chlamydia trachomatis: implications for its broad substrate specificity. J Mol Biol 411:649–660. doi: 10.1016/j.jmb.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Kasten B, Reski R. 1997. β-Lactam antibiotics inhibit chloroplast division in a moss (Physcomitrella patens) but not in tomato (Lycopersicon esculentum). J Plant Physiol 150:137–140. doi: 10.1016/S0176-1617(97)80193-9. [DOI] [Google Scholar]
- 43.Katayama N, Takano H, Sugiyama M, Takio S, Sakai A, Tanaka K, Kuroiwa H, Ono K. 2003. Effects of antibiotics that inhibit the bacterial peptidoglycan synthesis pathway on moss chloroplast division. Plant Cell Physiol 44:776–781. doi: 10.1093/pcp/pcg096. [DOI] [PubMed] [Google Scholar]
- 44.Takano H, Takechi K. 2010. Plastid peptidoglycan. Biochim Biophys Acta 1800:144–151. doi: 10.1016/j.bbagen.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 45.Hirano T, Tanidokoro K, Shimizu Y, Kawarabayasi Y, Ohshima T, Sato M, Tadano S, Ishikawa H, Takio S, Takechi K, Takano H. 2016. Moss chloroplasts are surrounded by a peptidoglycan wall containing d-amino acids. Plant Cell 28:1521–1532. doi: 10.1105/tpc.16.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin H, Hu J, Hua Y, Challa SV, Cross TA, Gao FP. 2008. Construction of a series of vectors for high throughput cloning and expression screening of membrane proteins from Mycobacterium tuberculosis. BMC Biotechnol 8:51. doi: 10.1186/1472-6750-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng J, Xu J. 2011. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins 79(Suppl 10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS-PAGE analysis of various eluted fractions of purified DapFCT-His expressed in E. coli BL21. Lanes: 1, molecular mass markers; 2 to 4, elution of DapFCT-His; 5, whole-cell lysate of E. coli BL21 expressing DapFCT-His. The protein corresponding to cloned DapFCT-His is boxed. Download FIG S1, TIF file, 11 MB (11.2MB, tif) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assay of DapFCT for glutamate racemase activity with the glutamate racemase-coupled assay. Reaction mixtures were prepared with 20 mM d-glutamate (●), without DapF (○), and without d-glutamate (△). An increase in NADPH formation is indicative of glutamate racemase activity of DapFCT. Error bars represent the standard deviations of two biological replicates. Download FIG S2, TIF file, 13.6 MB (13.9MB, tif) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Glutamate racemase homologues of enzymes encoded by different PVC group members resulting from pBLAST searches. Download TABLE S1, XLSX file, 0.01 MB (8.9KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of “glutamate racemase” homologues assigned to pathogenic Chlamydia species. Download TABLE S2, XLSX file, 0.01 MB (11.4KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PVC group members possessing DAP epimerase (DapF) and lacking d-glutamate racemase (MurI) homologues. Download TABLE S3, XLSX file, 0.02 MB (23KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PVC group members possessing both DAP epimerase (DapF) and d-glutamate racemase (MurI) homologues. Download TABLE S4, XLSX file, 0.02 MB (19.4KB, xlsx) .
Copyright © 2018 Liechti et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.