ABSTRACT
Candida biofilms resist the effects of available antifungal therapies. Prior studies with Candida albicans biofilms show that an extracellular matrix mannan-glucan complex (MGCx) contributes to antifungal sequestration, leading to drug resistance. Here we implement biochemical, pharmacological, and genetic approaches to explore a similar mechanism of resistance for the three most common clinically encountered non-albicans Candida species (NAC). Our findings reveal that each Candida species biofilm synthesizes a mannan-glucan complex and that the antifungal-protective function of this complex is conserved. Structural similarities extended primarily to the polysaccharide backbone (α-1,6-mannan and β-1,6-glucan). Surprisingly, biochemical analysis uncovered stark differences in the branching side chains of the MGCx among the species. Consistent with the structural analysis, similarities in the genetic control of MGCx production for each Candida species also appeared limited to the synthesis of the polysaccharide backbone. Each species appears to employ a unique subset of modification enzymes for MGCx synthesis, likely accounting for the observed side chain diversity. Our results argue for the conservation of matrix function among Candida spp. While biogenesis is preserved at the level of the mannan-glucan complex backbone, divergence emerges for construction of branching side chains. Thus, the MGCx backbone represents an ideal drug target for effective pan-Candida species biofilm therapy.
KEYWORDS: biofilm, Candida, non-albicans, antifungal resistance, extracellular matrix
IMPORTANCE
Candida species, the most common fungal pathogens, frequently grow as a biofilm. These adherent communities tolerate extremely high concentrations of antifungal agents, due in large part, to a protective extracellular matrix. The present studies define the structural, functional, and genetic similarities and differences in the biofilm matrix from the four most common Candida species. Each species synthesizes an extracellular mannan-glucan complex (MGCx) which contributes to sequestration of antifungal drug, shielding the fungus from this external assault. Synthesis of a common polysaccharide backbone appears conserved. However, subtle structural differences in the branching side chains likely rely upon unique modification enzymes, which are species specific. Our findings identify MGCx backbone synthesis as a potential pan-Candida biofilm therapeutic target.
INTRODUCTION
Candida species are among the most common causes of fungal infection worldwide (1). More than a hundred Candida species have been identified, but fewer than two dozen have been implicated in human disease. Candida albicans is the predominant clinical species; however, other Candida species are increasingly encountered (1–5). Four species, Candida albicans, C. tropicalis, C. parapsilosis, and C. glabrata, account for nearly 95% of all infections. While the epidemiology of infection varies among these species, their disease manifestations are similar. Likewise, their general virulence attributes appear mostly conserved (6–17). Among these factors, the ability to live in the biofilm state is arguably responsible for a majority of invasive infections (18–24).
In a biofilm, microbes are protected from antimicrobial agents by an organism-produced extracellular matrix (25–30). Hence, biofilm-related infections are challenging to cure (18, 31–33). The structure, function, and genetic control of this process were recently defined for C. albicans (34–41). Biochemical investigation identified a unique mannan-glucan complex (MGCx) composed of three polysaccharide building blocks, α-1,6-mannan and β-1,6- and β-1,3-glucans (40). These polysaccharides assemble extracellularly to form a complex capable of impeding antifungal delivery through drug sequestration (39, 42). While recent studies have shed light on this process in C. albicans biofilms, only a few studies have begun to explore matrix production and function of other Candida spp., and there is a paucity of detailed structure-function knowledge for these emerging pathogens (27, 40, 43–47). Furthermore, the genetic pathways linked to matrix production or function in non-albicans Candida species remains unexplored. Elucidation of matrix biogenesis mechanisms in these emergent species thus addresses an intriguing biological question as well as a critical medical need.
Here, we present evidence of a conserved matrix mannan and glucan complex backbone (MGCx) which contributes to profound drug resistance exhibited by the four most prevalent Candida species. We show that select matrix synthesis C. albicans orthologs play similar roles across Candida species to synthesize a common polysaccharide backbone. However, structural and molecular divergence in matrix assembly is suggested by subtle differences in matrix branching and absence of the involvement of matrix modification enzyme orthologs. Our findings argue that broad-spectrum Candida biofilm drug discovery should target the level of MGCx backbone synthesis as opposed to side chain synthesis.
RESULTS
Comparison of Candida species biofilm growth and architecture.
We determined basic parameters of biofilm development for three non-albicans Candida species: C. parapsilosis, C. tropicalis, and the more distantly related C. glabrata. We also included wild-type C. albicans as a standard for comparison. Biofilm formation begins with adherence of cells to the substrate, and we found marked differences among the species in this property (Fig. 1A). Despite similar inocula, the yield of adherent cells was approximately fivefold lower for C. parapsilosis and C. glabrata than that measured for either C. albicans or C. tropicalis. Biofilm maturation over the next 24 h erased these differences in cell number estimates (Fig. 1B), thus indicating that all four species reach similar sessile equilibria under in vitro growth conditions. Scanning electron microscopy (SEM) of mature biofilms revealed that each of the species, with the exception of C. glabrata, demonstrated a preponderance of filamentous or elongated cell growth (Fig. 1C). In addition, abundant extracellular matrix material encased the cells within each biofilm (marked by white arrows). We extended our comparison to examine in vivo biofilm formed in the lumen of a rat vascular catheter. Basic architecture in this model recapitulated the biofilm characteristics observed in vitro: filamentous cells were present in C. albicans, C. parapsilosis, and C. tropicalis biofilms, and extracellular matrix material was abundant. In fact, extracellular matrix appeared more pronounced in vivo than in vitro, likely due to the contribution of host components (48). These results indicate that mature biofilms of the three non-albicans Candida species resemble those of C. albicans in terms of cellular content, matrix accumulation, and for C. parapsilosis and C. tropicalis, presence of filamentous cells.
FIG 1 .
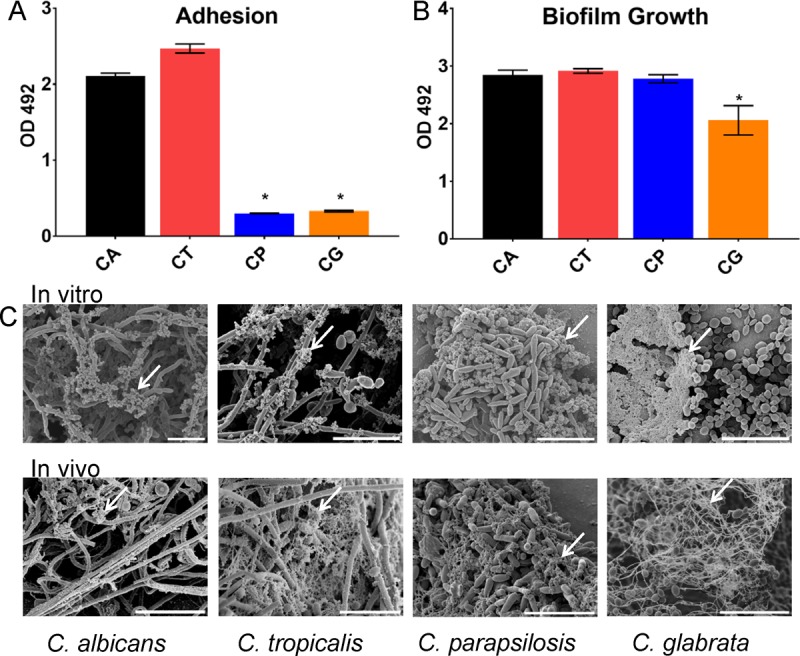
NAC form biofilms with variable characteristics. (A) Biofilm adhesion of reference strains for C. albicans (CA), C. tropicalis (CT), C. parapsilosis (CP), and C. glabrata (CG) was assessed using an XTT assay in a 96-well polystyrene plate after 1 h for adherence. The asterisks indicate statistically significant lower concentrations (P < 0.001) for C. parapsilosis and C. glabrata based upon ANOVA using the Holm-Sikak method for pairwise comparison. OD 492, optical density at 492 nm. (B) Mature biofilm formation for each of the four species was quantified in a 96-well format using an XTT endpoint after 24 h of incubation. The asterisk indicates a statistically significant lower concentration (P < 0.001) for C. glabrata based upon ANOVA using the Holm-Sikak method for pairwise comparison. (C) Mature biofilm architecture of wild-type biofilms from in vitro coverslips and the in vivo rat catheter model was assessed visually using SEM imaging after 24 h of incubation. The white arrows indicate extracellular matrix material. Bars, 20 µm.
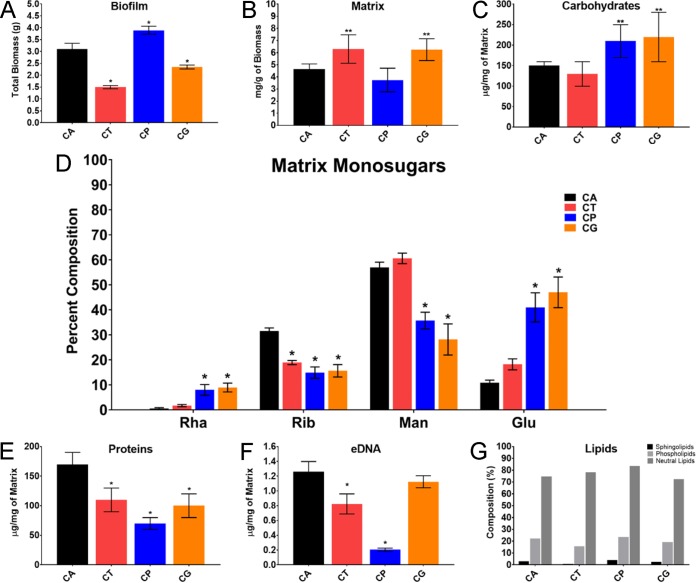
To determine the chemical nature of biofilm extracellular matrix material from each species, we used a large-scale, roller bottle apparatus for biofilm production and matrix isolation as described previously (49). Under these conditions, net biofilm biomass was comparable among the species (Fig. 2A), as it was in the small-scale experiments described above. The amounts of biofilm matrix mass (dry weight) in relation to the total biomass were relatively similar across species (Fig. 2B), but the relative abundance of each macromolecular matrix component varied among the species (Fig. 2C to G). Specifically, the protein component was greatest for C. albicans, while the carbohydrate component was greatest for C. parapsilosis and C. glabrata. The extracellular DNA (eDNA) component was comparable among the species (Fig. 2F). These results are consistent with those of a prior investigation (47) and indicate that macromolecular components vary slightly in their relative contributions to overall matrix composition.
FIG 2 .
NAC form biofilm matrix of variable quantity and quality. (A) Total biofilm mass was assessed by measurements (dry weight) of in vitro biofilms grown in polystyrene roller bottles (three replicates of 20 bottles per species). The four species studied were C. albicans (CA), C. tropicalis (CT), C. parapsilosis (CP), and C. glabrata (CG). The single asterisks indicate statistically significant lower values (P < 0.001) for C. tropicalis and C. glabrata based upon ANOVA using the Holm-Sikak method for pairwise comparison. (B) Biofilm matrix biomass was quantified by the dry weight following matrix separation from biofilm cells. Biofilms were grown in polystyrene roller bottles (three replicates of five bottles per species). Two asterisks indicate statistically lower values for C. tropicalis (P = 0.003) and C. glabrata (P = 0.005) between strains based upon ANOVA using the Holm-Sikak method for pairwise comparison. (C) Biofilm matrix total carbohydrate concentration was assessed using the phenol-sulfuric acid assay. The results were normalized by matrix biomass. The single asterisks indicate statistically lower concentrations for C. parapsilosis (P = 0.009) and C. glabrata (P = 0.002) based upon ANOVA using the Holm-Sikak method for pairwise comparison. (D) Relative percent monosugar composition (Rha, rhamnose; Rib, ribose; Man, mannose; Glu, glucose) in the biofilm matrix of C. albicans, C. tropicalis, C parapsilosis, and C. glabrata. The single asterisks indicate statistically significant differences (P < 0.001) between strains based upon ANOVA. (E) Biofilm matrix total protein concentration was assessed using the BCA protein assay kit. The results were normalized by matrix biomass. The single asterisks indicate statistically lower concentrations for C. tropicalis, C. parapsilosis, and C. glabrata (P < 0.001) than for C. albicans based upon ANOVA using the Holm-Sikak method for pairwise comparison. (F) Biofilm matrix total eDNA. The results were normalized by matrix biomass. The single asterisks indicate statistically lower concentrations for C. tropicalis, C. parapsilosis, and C. glabrata (P < 0.001) than for C. albicans based upon ANOVA using the Holm-Sikak method for pairwise comparison. (G) Biofilm matrix total lipid concentration was assessed by gas chromatography. The results were normalized by matrix biomass.
Detection of matrix MGCx in non-albicans Candida species.
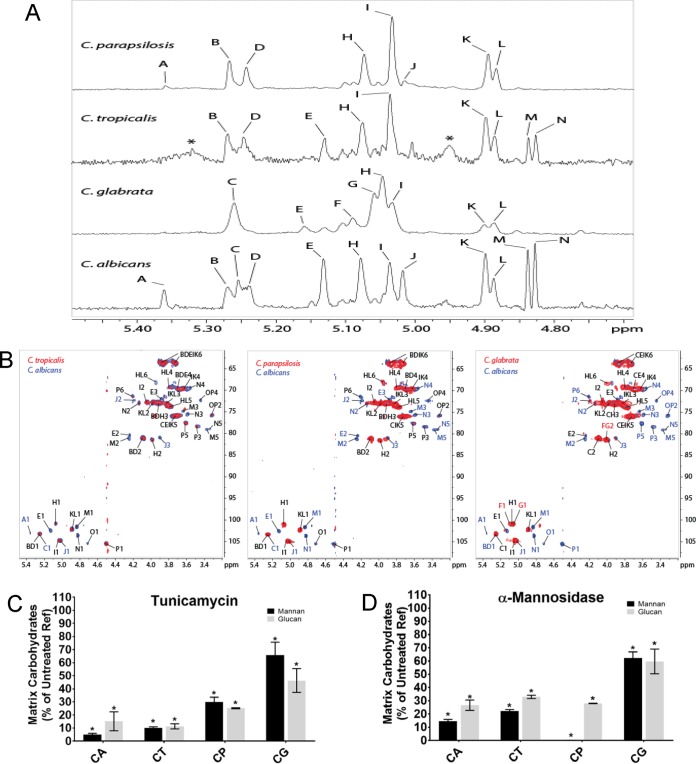
The mannan-glucan complex (MGCx) is a signature feature of the C. albicans biofilm (40). The MGCx is comprised of an α-1,6-mannan backbone and β-1,6-glucan (40). Based on our previous report, the MGCx constitute approximately 20% of the total carbohydrate pool in the C. albicans matrix. The MGCx isolated from the non-albicans Candida species (NAC) biofilm matrices constituted 13.5%, 34.8%, and 17.0%, in C. tropicalis, C. parapsilosis, and C. glabrata, respectively (Table 1). Further gas chromatography revealed the presence of both mannan and glucan in the matrix of each non-albicans Candida biofilm (Fig. 3A). The C. albicans MGCx has a mannan/glucan ratio of 89:11 (40). We found mannan/glucan ratios of 64:25 for C. tropicalis, 83:12 for C. parapsilosis, and 93:7 for C. glabrata (Fig. 3A and Table 1). These findings are consistent with the production of an MGCx by each non-albicans Candida species, though the MGCx of each species may have distinct structural features.
TABLE 1 .
Carbohydrate distribution in Candida species matrix following neutral sugar purification and fractionation
| Candida speciesa | Carbohydrate typeb |
||||
|---|---|---|---|---|---|
| Bound (%)c | Neutral |
||||
| %c | MWFd | Man/Glu ratio | Content (%)c | ||
| CT | 86.5 | 13.5 | HMWF | 72:28 | 2.6 |
| HMWF | 19:81 | 4.3 | |||
| LMWF | 73:27 | 0.9 | |||
| CP | 65.2 | 34.8 | HMWF | 13:87 | 20.9 |
| LMWF | 10:90 | 13.9 | |||
| CG | 83.0 | 17.0 | HMWF | 95:5 | 0.4 |
| HMWF | 82:18 | 1.0 | |||
| HMWF | 61:39 | 1.6 | |||
| HMWF | 52:48 | 3.4 | |||
| HMWF | 27:73 | 0.8 | |||
| LMWF | 80:20 | 1.1 | |||
| LMWF | 50:50 | 1.7 | |||
CT, C. tropicalis; CP, C. parapsilosis; CG, C. glabrata.
The carbohydrate type indicates whether carbohydrate is associated either with the uncharged neutral fraction or the charged bound fraction (glycoproteins).
Values are percentages of the listed fractions in the total matrix carbohydrate pool. Content (%) represents the percent content within the neutral pool for each species.
MWF is the molecular weight fraction for each isolated polymer within the neutral carbohydrate pool. HMWF and LMWF are the high-molecular-weight-reaction and low-molecular-weight fraction, respectively.
FIG 3 .
Comparative chromatographic fractionation and NMR analysis of carbohydrates from the Candida species biofilm extracellular matrix. (A) Comparison of the 500-MHz 1H NMR spectra of purified neutral matrix polysaccharides from C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata biofilm matrix. See Table 2. (B) Comparison of C. albicans NMR spectra spin systems to each of the NAC species (C. tropicalis, C. parapsilosis, and C. glabrata, respectively) as reflected by heteronuclear single quantum coherence (HSQC) and nuclear Overhauser effect spectroscopy (NOESY) data. (C and D) Carbohydrates in the matrix of wild-type biofilms treated with tunicamycin (C) and α-mannosidase (D) were quantified by gas chromatography. Data are presented as percentages of the reference strain (Ref), with means ± standard errors (SEs) (error bars) shown. All values were significantly lower than the reference value according to ANOVA as indicated by the single asterisks.
Nuclear magnetic resonance (NMR) analyses performed on total purified NAC MGCx revealed compelling structural similarities as well as striking differences among the neutral matrix polysaccharides of the species (Fig. 3B and C and Table 2; see Table S1 in the supplemental material). Control analysis of C. albicans samples revealed several α- and β-Man spin systems that resembled the high- and low-molecular-weight F2 and F17 glucomannans we reported previously (40). Of particular relevance, we found multiple signals specific for α-1→2-Manα-1→2 residues enclosed within branched regions, such as those found in the MGCx side chains. Several of these spin systems (Fig. 3A and B, peaks B, D, H, I, K, and L) were common to C. albicans, C. parapsilosis, and C. tropicalis. Fewer spin systems were common to C. albicans and C. glabrata (Fig. 3B, peaks C, I, K, and L), as may be expected from their greater phylogenetic distance. We also found peaks characteristic of β1,6-glucan in two-dimensional (2D) NMR analysis (Fig. 3B, peaks P and O). In addition to differences in the presence or absence of specific peaks, differences among the polysaccharide compositions were also evident from the quantitative differences among individual anomeric peaks (Fig. 3B and Table 2).
TABLE 2 .
Percentages of the different residues found in each sample following 1D 1H NMR
| No. | Residue | % of residue in Candida speciesa: |
% of MGCxb |
||||
|---|---|---|---|---|---|---|---|
| CA | CG | CT | CP | F2 | F17 | ||
| i | β-1-2-Manα-1-P | 1.8 | |||||
| ii | α-1-2-Manα-1-P | 1.8 | 0.4 | ||||
| A | α-1-2-Manα-1-3- | 3.7 | 2.1 | 0.9 | 1.1 | ||
| B | α-1-2-Manα-1-2- | 4.3 | 9.5 | 11.5 | 5.4 | ||
| C | α-1-2-Manα-1-2- | 7.2 | 21.8 | 7.1 | 14.1 | ||
| D | α-1-2-Manα-1-2- | 7.4 | 10.5 | 12.8 | 9.3 | 15.6 | |
| E | β-1-2-Manα-1-2- | 11.9 | 5.7 | 9.1 | 13 | 4.1 | |
| F | 2,6-Manα-1-6- (l)c | 5.3 | |||||
| G | 2,6-Manα-1-6- (l) | 15.1 | 1.6 | ||||
| H | 2,6-Manα-1-6- (b)d | 15.6 | 28.2 | 18.5 | 18.1 | 10.8 | 13.4 |
| I | Manα-1-2- | 13.5 | 14.1 | 23.7 | 29.1 | 10.6 | 20.9 |
| J | 3-Manα-1-2- | 7.2 | 4.4 | ||||
| K | Manα-1-6- | 10.6 | 6.4 | 13.5 | 15.6 | 16.1 | 15.8 |
| L | 6-Manα-1-6- | 6.4 | 3.5 | 7.2 | 6.3 | 2.4 | 6.6 |
| M | β-1-2-Manβ-1-2- | 5.4 | 3.7 | 10.3 | 3.2 | ||
| N | Manβ-1-2- | 6.7 | 4.4 | 10.3 | 3.2 | ||
CA, C. albicans; CG, C. glabrata; CT, C. tropicalis; CP, C. parapsilosis. Blank cells indicate that the residue was not found.
F2 and F17 refer to high- and low-molecular-weight MGCx previously reported in the biofilm matrix of C. albicans (40). Blank cells indicate that the MGCx was not found.
(l), residues within a linear region, i.e., residues whose neighbors are not branching residues.
(b), residues within a branched region, i.e., residues whose neighbors are branching residues.
2D HSQC NMR chemical shift assignment of the major spin systems found in C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis. Download TABLE S1, DOCX file, 0.02 MB (24.7KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We used a biochemical approach to test for a physical interaction among the matrix polysaccharide components. Specifically, we assessed the glycosyl composition of matrix polysaccharides following a series of neutral sugar purification and fractionation steps. For each species fraction, we identified both mannan and glucan in the high-molecular-weight (HMW) and low-molecular-weight (LMW) fractions. Comigration of both mannan and glucan components strongly suggests the presence of covalent bonds between those two polymers; however, their definite nature is not completely understood. This observation is consistent with coelution of each monosugar as a distinct mannan-glucan complex polysaccharide in the NAC biofilm matrix (Table 1). However, in keeping with analysis described above, the ratios of mannan and glucan varied somewhat across the species, suggesting structural differences in this matrix component among the NAC. In addition, there were several distinct mannan-glucan complexes of different sizes for C. glabrata. These cofractionation results support the model that each non-albicans Candida species produces a biofilm matrix MGCx.
We also used pharmacological and enzymatic approaches to test predictions of the MGCx model. Both polysaccharides are required for MGCx assembly in this model; hence, disruption of either mannan or glucan alone would be predicted to decrease matrix accumulation of the other (40). To inhibit matrix mannan production, we utilized tunicamycin, an inhibitor of N-glycosylation, and α-mannosidase, an enzyme that hydrolyzes and degrades mannan (39). Either tunicamycin or α-mannosidase treatment of biofilm reduced the matrix mannan content on average by 28% and 25%, respectively (Fig. 3C and D). Each treatment caused a concomitant decrease in the concentration of glucan that was nearly identical in magnitude (25% and 27%, respectively) (Fig. 3C and D). The finding that matrix glucan accumulation is dependent upon matrix mannan supports the model that polysaccharide interaction leading to an MGCx is conserved across these Candida species (39).
Mannan-glucan matrix function in NAC species.
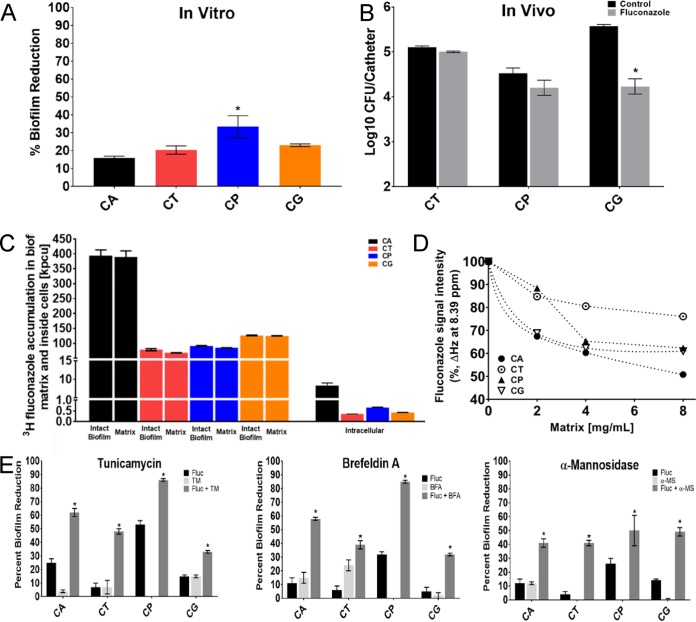
Prior studies have linked a majority of the C. albicans biofilm resistance phenotype to sequestration of antifungal drugs by the extracellular matrix (28, 34, 36, 37, 39, 50–52). The MGCx appears strongly linked to this drug sequestration phenomenon. Therefore, we examined the drug susceptibility of biofilms both in vitro and in vivo using a rat vascular catheter model for each of the Candida species (53, 54) (Fig. 4A and B). For C. albicans, C. tropicalis, and C. glabrata, the highest soluble concentration of the antifungal fluconazole (1,000 µg/ml, which is more than 1,000 times the planktonic MIC) did not appreciably impact biofilm cell burden in either model, consistent with prior biofilm antifungal testing for these species (55–58). For the strain of C. parapsilosis studied, treatment was more effective compared to the other species. However, the fluconazole concentration needed for effective treatment remained 100 times greater than that associated with planktonic efficacy. These results confirm that biofilms of these species are dramatically less sensitive to fluconazole than planktonic cells. To evaluate drug sequestration as a mechanism underlying the biofilm-associated drug resistance, we tracked radiolabeled fluconazole within the biofilm of each Candida species. Nearly all of the antifungal accumulated in the biofilm matrix for each species (Fig. 4C). Interestingly, despite the fact that each species was exposed to a similar concentration of fluconazole, the total sequestered drug concentrations were lower in the NAC biofilms. Because matrix levels are relatively similar among the species (Fig. 2B), we speculate that differences in matrix-drug binding affinity among the species account for differences in drug accumulation.
FIG 4 .
NAC biofilm drug resistance phenotype and mechanism. (A) Biofilm antifungal susceptibility (fluconazole [125 or 1,000 µg/ml] for 48 h) of wild-type C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata was assessed using an XTT assay in a 96-well polystyrene plate assay. The asterisk indicates a statistically significant difference (P < 0.001) between strains based upon ANOVA using the Holm-Sikak method for pairwise comparison. (B) Biofilm antifungal susceptibility (fluconazole [250 µg/ml] after 24-h exposure) of wild-type C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata was assessed using viable counts from the rat vascular catheter biofilm model. (C) Fluconazole sequestration and binding to the Candida species biofilm extracellular matrix. Sequestration of 3H-labeled fluconazole was assessed using in vitro intact biofilms as well as the extracellular matrix and intracellular components. (D) Fluconazole binding to the NAC biofilm extracellular matrices. Fluconazole interactions with the tested matrices studied by one-dimensional 1H NMR at 600 MHz were determined as decreases in the intensity of chemical shift peaks characteristic of protons present either in the heterocyclic azole rings or the aromatic ring of the drug. Spectra were recorded at the constant fluconazole concentration of 0.653 mM, and matrix concentrations ranged from 0 up to 8 mg/ml. (E) Biofilms were treated with pharmacological inhibitors of mannan or glucan or a mannan hydrolysis enzyme both with and without 1,000 μg/ml fluconazole for all species except C. parapsilosis for which we used 250 μg/ml. Efficacy was assessed in a 96-well plate format for quantification with the XTT assay. The asterisks indicate statistically significant differences (P < 0.001) between the combination and either treatment alone based upon ANOVA using the Holm-Sikak method for pairwise comparison. FLUC, fluconazole; TM, tunicamycin; BFA, brefeldin A; α-MS, α-mannosidase.
We further explored the capacity of NAC matrices to interact with fluconazole using one-dimensional 1H NMR. In our experiments, interactions of the antifungal drug with the matrix were evident by changes in the intensity of 1H peaks upon increasing concentrations of matrix material. Similarly to our previous report (40), the signals of both aromatic and azole protons decreased as a function of increasing concentration of matrices, suggesting that both the aromatic ring and the heterocyclic triazole rings were involved in interactions with matrix components. Interestingly, each NAC matrix had a distinct drug binding dynamics profile (Fig. 4D). The degree of interaction in this assay was greatest for C. albicans, consistent with differences in matrix affinity among the species.
If the MGCx were responsible for functional drug sequestration, then MGCx disruption should increase biofilm drug susceptibility. We tested this prediction through pharmacological and enzymatic treatments. Mannan accumulation was reduced by either tunicamycin or α-mannosidase treatment as shown above. In addition, both mannan and β-1,6-glucan accumulation were reduced by brefeldin A treatment (59). These treatments alone did not influence biofilm cell viability. However, each treatment augmented the activity of fluconazole, resulting in profound killing of biofilm cells (Fig. 4D). These effects were biofilm specific, as these agents did not enhance the activity of fluconazole against planktonic cells (Fig. S1). In sum, these results support the hypothesis that MGCx contributes to sequestration of antifungals to promote biofilm resistance across Candida species.
Planktonic cell growth following pharmacological and enzymatic treatments. Planktonic cells were grown in 96-well round-bottom plates for 24 h and quantified using XTT. For experiments with tunicamycin (TM) (at 1.0 μg/ml) and brefeldin A (BFA) (at 0.6 μg/ml), biofilms or cells were grown for 6 h and then treated for 24 h before quantification. Experiments with α-mannosidase (α-MS) (at 0.78 U/ml) used biofilms or cells first grown for 24 h before 24-h treatment. The means of three technical replicates and SEs are shown. Download FIG S1, TIF file, 2.2 MB (2.3MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic determinants of NAC matrix MGCx structure and function.
In order to identify genetic determinants of matrix production across Candida species, we used a candidate gene approach. Hypothesizing genetic conservation among species, we identified orthologs of 12 C. albicans genes shown to be involved in production or modification of matrix mannan or β-1,6-glucan (Table 3) (34, 39). We successfully constructed homozygous deletion mutations for the genes in each species with the exceptions of ALG11 and VRG4 in C. tropicalis, BIG1, KRE5, and VRG4 in C. parapsilosis, and KRE5 and VRG4 in C. glabrata. We speculate that these genes may be essential in the respective species. Most of the mutants formed biofilms similar to their reference strains. However, three mutants formed biofilms with reduced cell numbers (Fig. S2), including C. parapsilosis mnn11Δ/Δ, pmr1Δ/Δ, and phr1Δ/Δ mutants and C. glabrata mnn11Δ and big1Δ mutants.
TABLE 3 .
C. tropicalis, C. parapsilosis, and C. glabrata mutant strains used in this study
| Species and gene | Systemic name | Genotype | Strain | Descriptiona | Homology (%)b |
|---|---|---|---|---|---|
| C. tropicalis | |||||
| MNN9 | CTRG_02261 | Δ/Δ | URZ565 | β-1,6-Mannosyltransferase | 85.6 |
| MNN11 | CTRG_04663 | Δ/Δ | URZ567 | α-1,6-Mannosyltransferase | 67.9 |
| VAN1 | CTRG_05614 | Δ/Δ | URZ570 | α-1,6-Mannosyltransferase | 94.4 |
| MNN4-4 | CTRG_05766 | Δ/Δ | URZ566 | Mannosylphosphate transferase | 55.2 |
| PMR1 | CTRG_04916 | Δ/Δ | URZ569 | Ca2+/Mn2+ ATPase | 88.2 |
| XOG1 | CTRG_04334 | Δ/Δ | URZ571 | β-1,3-Glucanase | 75.8 |
| BGL2 | CTRG_00169 | Δ/Δ | URZ562 | β-1,3-Glucosyltransferase | 61.5 |
| PHR1 | CTRG_03942 | Δ/Δ | URZ568 | β-1,3-Glucosyltransferase | 75.2 |
| BIG1 | CTRG_04070 | Δ/Δ | URZ563 | β-1,6-Glucan synthesis | 70.3 |
| KRE5 | CTRG_02572 | Δ/Δ | URZ564 | β-1,6-Glucan synthesis | 65.7 |
| C. parapsilosis | |||||
| ALG11 | CPAR2_601300 | Δ/Δ | EGD136 | α-1,2-Mannosyltransferase | 58.4 |
| MNN9 | CPAR2_806810 | Δ/Δ | EGD194 | α-1,6-Mannosyltransferase | 78.2 |
| MNN11 | CPAR2_106380 | Δ/Δ | EGD144 | α-1,6-Mannosyltransferase | 55.9 |
| VAN1 | CPAR2_807920 | Δ/Δ | EGD184 | α-1,6-Mannosyltransferase | 73.7 |
| MNN4-4 | CPAR2_106570 | Δ/Δ | EGD149 | Mannosylphosphate transferase | 36.0 |
| PMR1 | CPAR2_31360 | Δ/Δ | EGD141 | Ca2+/Mn2+ ATPase | 82.7 |
| XOG1 | CPAR2_106000 | Δ/Δ | EGD150 | β-1,3-Glucanase | 64.2 |
| BGL2 | CPAR2_401600 | Δ/Δ | EGD147 | β-1,3-Glucosyltransferase | 72.7 |
| PHR1 | CPAR2_302140 | Δ/Δ | EGD188 | β-1,3-Glucosyltransferase | 59.7 |
| C. glabrata | |||||
| ALG11 | CAGL0D01122g | Δ | EGD125 | α-1,2-Mannosyltransferase | 29.5 |
| MNN9 | CAGL0L12804g | Δ | EGD128 | α-1,6-Mannosyltransferase | 52.4 |
| MNN11 | CAGL0G07491g | Δ | EGD124 | α-1,6-Mannosyltransferase | 25.7 |
| VAN1 | CAGL0B02321g | Δ | EGD137 | α-1,6-Mannosyltransferase | 49.4 |
| MNN4-4 | CAGL0H01793g | Δ | EGD121 | Mannosylphosphate transferase | 26.4 |
| PMR1 | CAGL0J01870g | Δ | EGD143 | Ca2+/Mn2+ ATPase | 60.0 |
| XOG1 | CAGL0G09515g | Δ | EGD134 | β-1,3-Glucanase | 52.2 |
| BGL2 | CAGL0G00220g | Δ | EGD127 | β-1,3-Glucosyltransferase | 65.3 |
| PHR1 (GAS2) | CAGL0M13849g | Δ | EGD131 | β-1,3-Glucosyltransferase | 57.1 |
| BIG1 | CAGL0L11528g | Δ | EGD129 | β-1,6-Glucan synthesis | 23.2 |
Based on Candida or Saccharomyces Genome Database.
Protein sequence alignment to Candida albicans.
Biofilm formation capacity for non-albicans Candida mutants in the mannan and glucan pathways. Mature biofilm burden for each of the select deletion mutants was quantified in a 96-well format using an XTT endpoint after 24 h of incubation. Asterisks indicate statistically significant differences (P < 0.001) between strains based upon ANOVA. Download FIG S2, TIF file, 4.2 MB (4.3MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
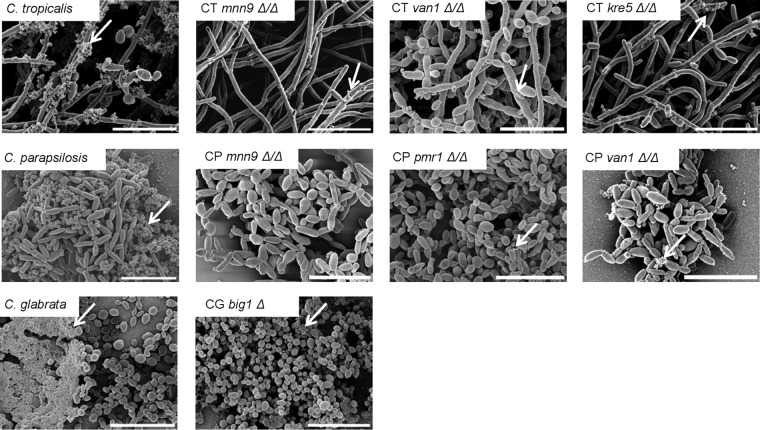
We examined each of the mutant biofilms by electron microscopy to visualize biofilm architecture and matrix deposition. Biofilm architecture seemed largely unaffected by the mutations, but there was a striking reduction of visible extracellular matrix in seven of the mutant biofilm strains (Fig. 5). This suggests that each of these genes is required for biofilm matrix production in the respective NAC species.
FIG 5 .
Genetic control of NAC biofilm extracellular matrix production. Mature biofilm architecture from in vitro coverslips was assessed visually using SEM imaging after 24 h of incubation. The white arrows indicate extracellular matrix material. Bars, 20 µm.
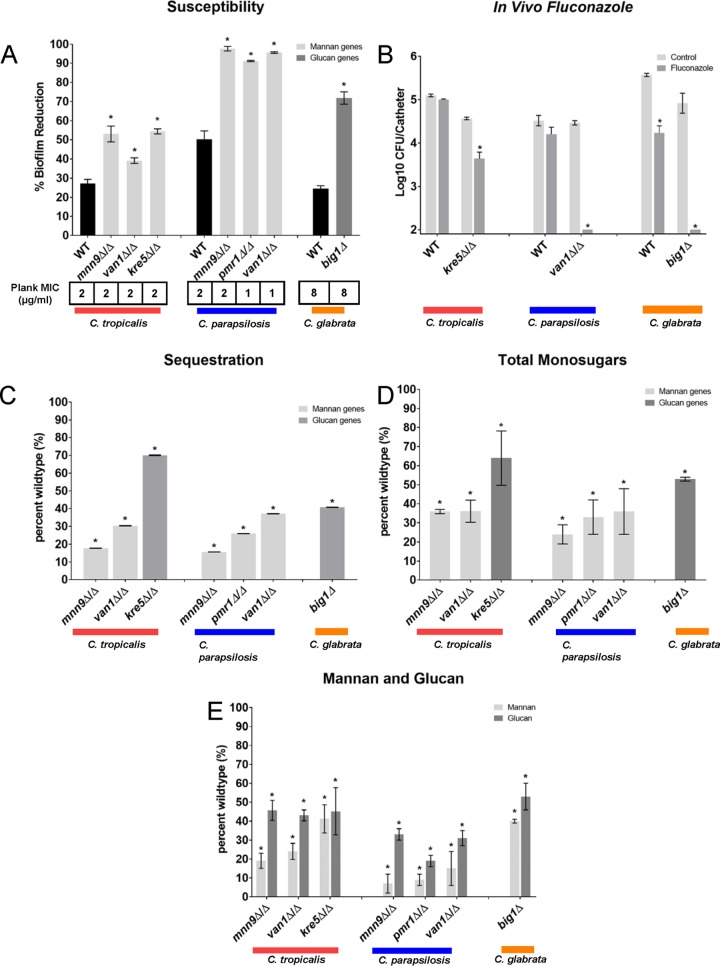
We next examined the influence of the genetic disruptions on the biofilm resistance phenotype in vitro (Fig. 5 and Fig. S3 and S4). Surprisingly, despite the importance of each of these genes for biofilm resistance in C. albicans, only a small subset impacted susceptibility in the NAC biofilms. Specifically, we found a total of 7 of 29 deletion mutants exhibiting enhanced biofilm drug susceptibility (Fig. 6A). For C. tropicalis, fluconazole-susceptible strains included mannan synthesis mutants, i.e., mnn9Δ/Δ and van1Δ/Δ mutants, and the β-1,6-glucan synthesis mutant, the kre5Δ/Δ mutant. Among the C. parapsilosis mutants screened, we identified three mannan synthesis mutants, mnn9Δ/Δ, van1Δ/Δ, and pmr1Δ/Δ mutants. Interestingly, two of the C. parapsilosis orthologs were also relevant for C. tropicalis (mnn9Δ/Δ and van1Δ/Δ). Only a single mutant from C. glabrata exhibited a biofilm susceptibility phenotype: a putative enzyme for β-1,6-glucan synthesis (big1Δ). The susceptibility to fluconazole appeared specific to the biofilm state, as planktonic MICs for all mutants were unchanged (Fig. 6A). The biofilm susceptibility phenotype was reversed for all strains in which we reintroduced a wild-type copy of the deleted gene (Fig. S5).
FIG 6 .
Genetic control of NAC biofilm drug resistance and extracellular matrix production. (A) The percentage of reduction in biofilm formation following 48-h treatment with fluconazole compared with untreated biofilms, as quantified using the 96-well XTT assay. The null mutant (Δ/Δ) is shown for each gene of interest. The graph shows data from three assay replicates of a representative example of three biological replicates. Asterisks indicate statistically significant difference (P < 0.001) between the reference (wild type [WT]) and mutant based upon ANOVA using the Holm-Sidak method for pairwise comparison. The MIC of fluconazole for planktonic cells of the Δ/Δ strains is shown below the bar graph by the Plank MIC values. (B) The NAC species reference strains, kre5Δ/Δ, van1Δ/Δ, and big1Δ/Δ mutants, were tested in vivo using a rat central venous catheter model, with the effects of fluconazole or saline treatment compared with the reference strains for C. tropicalis, C. parapsilosis, and C. glabrata, respectively. Biofilms were quantified using viable-cell counts following treatment. The values are means ± standard deviations (error bars) from three replicates. The asterisks indicate that the CFU values were significantly different from the CFU for the reference strain (P < 0.001) based upon ANOVA using the Holm-Sikak method for pairwise comparison. (C) Intact biofilms grown from the wild-type and mutant strains were exposed to [3H]fluconazole, washed, and harvested. Scintillation counting was performed in triplicate to determine the fluconazole content in the intact biofilms and the isolated matrix. Standard deviations are shown. (D) Mature in vitro biofilms from the wild-type strain and null mutants were assayed for matrix carbohydrate concentration using the phenol sulfuric acid method. The value for each mutant is presented as a percentage of the value for the wild-type strain. The graph shows data from three biological replicates and three assay replicates. The asterisks indicate that glucan measurements were significantly different (P < 0.0001) from the wild-type measurement based on ANOVA. (E) Mature in vitro biofilms from the wild-type strain and null mutants were assayed for matrix glucan and mannan concentrations by gas chromatography. The value for each mutant is presented as a percentage of the value for the wild-type strain. The graph shows data from three biological replicates and three assay replicates. The asterisks indicate that glucan measurements were significantly different (P < 0.0001) from the value for the wild-type strain based upon ANOVA.
NAC biofilm drug susceptibility in mutants without an enhanced susceptibility phenotype. (A to C) Percentage of reduction in biofilm formation following 48-h treatment with fluconazole compared with untreated biofilms, as quantified using the 96-well XTT assay. The null mutant (Δ/Δ) is shown for each gene of interest. The figure shows data from three assay replicates of a representative example of three biological replicates. Asterisks indicate statistically significant differences (P < 0.001) between reference and mutant based upon ANOVA using the Holm-Sikak method for pairwise comparison. Download FIG S3, TIF file, 2.7 MB (2.8MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polyene and echinocandin biofilm activity against fluconazole-susceptible NAC mutants. After growth for 6 h, biofilms were treated with either 0.5 μg/ml amphotericin B or micafungin for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. All mutants were significantly more susceptible to treatment than the reference strain (*, P < 0.001; **, P < 0.05). The means of three technical replicates and SEs are shown. Download FIG S4, TIF file, 2 MB (2MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complemented C. tropical and C. glabrata glucan and mannan synthesis mutant biofilms are susceptible to fluconazole. After growth for 6 h, biofilms were treated with 1,000 μg/ml fluconazole for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. All mutants were significantly more susceptible to treatment than the reference strain (P < 0.001). Each of the complemented strains exhibited resistance similar to that of the reference strains. The means of three technical replicates and SEs are shown. Download FIG S5, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For three C. parapsilosis mutants (mnn9Δ/Δ, pmr1Δ/Δ, and van1Δ/Δ mutants), complementation was not successful; consequently, we examined multiple independent deletion mutant strains to verify the enhanced antifungal susceptibility phenotype (Fig. S6). Antifungal biofilm susceptibility was similarly assessed for two additional drugs, amphotericin B and micafungin (Fig. S4). Enhanced susceptibility was observed for the majority of mutants, consistent with a pan-antifungal mechanism. These results show that inferred defects in mannan or β-1,6-glucan production can cause increased biofilm drug susceptibility in NAC species.
Multiple independent transformants of noncomplemented mutant strains are susceptible to fluconazole. After growth for 6 h, biofilms were treated with 1,000 μg/ml fluconazole for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. Two or three independent transformants were tested for each homozygous deletion mutant. All mutants were significantly more susceptible to treatment than the reference strain (P < 0.001). A different biological replicate is presented here than in Fig. 5A. The means of three technical replicates and SEs are shown. Download FIG S6, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We explored the clinical relevance of these findings using the rat catheter in vivo biofilm model. Enhanced fluconazole efficacy was observed for representative mannan or β-1,6-glucan synthesis mutants from each species (Fig. 6B). These results support the hypothesis that the increased biofilm drug susceptibility that results from impaired mannan or β-1,6-glucan synthesis is relevant in a model infection environment.
To determine directly whether matrix mannan and glucan are required for drug sequestration in the NAC biofilm mutants, we conducted [3H]fluconazole binding assays. Compared to the respective wild-type strain, each of the mannan and glucan mutant strains had decreased matrix sequestration of radiolabeled drug (Fig. 6C). These findings strengthen the conclusion from our pharmacological and enzymatic matrix disruption treatments that mannan and glucan contribute to matrix drug sequestration. However, variability in the degree of sequestration relative to the change in susceptibility suggests the potential of additional mechanisms as previously described (28).
To identify matrix changes associated with increased biofilm susceptibility, we harvested matrix from biofilms of each mutant and assayed for total carbohydrate and specifically for mannan and glucan (Fig. 6D and 5E). We found that all seven mutants had significantly lower levels of the corresponding polysaccharide than the reference strain with remaining amounts less than 50% of the level in the wild type on average (Fig. 6E). Furthermore, genetic disruption of either a mannan or glucan synthase was associated with a lower content of both mannan and glucan. This observation is consistent with the model that the MGCx accounts for the drug resistance phenotype of the NAC biofilms. Unfortunately, matrix quantities did not allow for complementary NMR analysis. Because the synthesis pathways for the carbohydrates are distinct, inhibition of one pathway would not be expected to lead to a biochemical reduction in the nonimpacted pathway components. Our findings suggest that there is an extracellular physical interaction among the matrix components that is conserved among Candida species.
DISCUSSION
We theorized that the MGCx, recently found in C. albicans, is conserved and promotes the drug resistance phenotype across Candida species. In order to address this question, in this study, we first utilized large-scale biofilm growth and matrix composition analysis. We find that each of the major matrix polysaccharide constituents is required for assembly and function of matrix across Candida species. At the structural level, several complementary assays corroborate the presence of an MGCx in diverse Candida species. Specifically, we identified an α-1,6-mannan backbone with α-1,2 branches and β-1,6-glucan in the matrix. Analysis of the glycosyl composition of matrix after fractionation also revealed coelution of mannan and glucan components. Additionally, both pharmacological and genetic studies revealed codependence of accumulation of matrix mannan and glucan. Phenotypic studies documented the central role of this polysaccharide complex in matrix sequestration and biofilm-specific drug resistance under both in vitro and in vivo conditions.
However, the results of refined NMR analysis of the complex and phenotypic and genotypic assays suggested meaningful differences in the MGCx among the Candida species. Specifically, the relative ratios of the mannan and glucan components varied among the four species. Additionally, while NMR analysis revealed similar mannan and glucan backbone features, there were differences in branching pattern and length. The C. parapsilosis matrix mostly resembled low-molecular-weight glucomannans of C. albicans. The abundance of α-1→6-linked Man residues along with a substantially increased content of terminal Manα-1→2 residues indicated the presence of a similar α-1→6-linked Man backbone structure but with shorter side chains consisting of a blend of α-1→2- and α-1→3-linked Man residues. The lack of β-Man for C. parapsilosis matrix was unique, as β-Man was observed for the other species. The matrix of C. tropicalis exhibited characteristics similar to those of C. albicans and C. parapsilosis matrix. Like the latter, this pool contained amounts of α-1→2-Manα-1→2- and 1→6-linked Man as well as of β-Man residues that were comparable to those measured in C. albicans. On the other hand, C. tropicalis matrix neutral carbohydrates did not contain any 3-linked Man residues, whereas there was a larger terminal Manα-1→2 residue content, along with more 1→6-linked Man. This pattern suggests more branching but relatively shorter side chains. On the basis of measured distribution of 2,6-Manα-1→6 and α-1→2-Manα-1→2 residues, it appeared that the overall branching pattern is bit less diverse than that observed in C. albicans matrix. The analysis of C. glabrata matrix neutral carbohydrates revealed the most distinct highly branched structure, which also contained well-defined linear regions without any branching Man residues present. This observation suggested an uneven and infrequent distribution of side chains in this carbohydrate pool as reflected by the presence of only one type of α-1→2-Manα-1→2 signal, whereas the amount of the terminal Manα-1→2 residue indicated a side chain length distribution type resembling the distribution determined in C. albicans. Unlike the latter, C. glabrata matrix neutral carbohydrates did not contain 3-linked Man residues, while the content of β-Man was significantly reduced but still detected. Overall, the major Man residues were similar among all four tested Candida species. However, the relative proportion of the residues varied significantly, likely due to differences in the degree of branching, the length of side chains, and the presence of unique residues such as β-Man or 3-linked Man. We speculate that these structural differences in the MGCx and potentially other noncarbohydrate constituents determine the unique sequestration properties of the NAC.
Our genetic studies reveal both similarities and differences among the species with regard to the genetic pathways responsible for production of matrix. It was initially surprising that the only orthologous mutations in the NAC that impacted the antifungal resistance phenotype were those linked to component synthesis alone. Conversely, the mutations with putative glucan and mannan modification function based upon the C. albicans MGCx structure had no apparent impact on NAC matrix production. A plausible explanation for this observation is divergence in the genetic pathways responsible for the differences in the impact of C. albicans orthologous mutations in the NAC species. Researchers have often assumed that if a yeast species is related to another yeast species (especially within the same genus), the underlying molecular and cellular mechanisms must also be closely related. However, even within a Candida clade, the genetic relatedness between any two NAC species is often larger than the genetic distance between humans and reptiles (60). There is ample precedent for genetic rewiring among the Candida species, including for biofilm formation pathways (13, 15, 16, 61–63). While sequence identity is not a great predictor of function, the amino acid similarity across species for these C. albicans matrix resistance genes was only modest.
Another potential explanation for the lack of genetic conservation is the distinct differences in MGCx branching structure. We favor this model due to the biochemical differences that support a model of altered matrix branch chain structure across species. The latter model suggests that enzymes needed for production of the matrix backbone may be useful for pan-Candida biofilm development of useful therapies, while targeting modification enzymes would be predicted to be less effective across species. In future work, careful characterization of the remaining genetic components of the NAC biofilm synthesis pathway will help to elucidate additional therapeutic targets.
MATERIALS AND METHODS
Ethics statement.
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Wisconsin according to the guidelines of the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals (64), and Public Health Service policy. The approved animal protocol number is DA0031.
Media.
Strains were stored in 15% (vol/vol) glycerol stock at −80°C and maintained on yeast extract-peptone-dextrose (YPD) agar. Prior to biofilm experiments, all Candida strains were grown at 30°C in YPD, and biofilms were grown in RPMI 1640 buffered with morpholinepropanesulfonic acid (RPMI-MOPS).
Strains and strain construction.
Strains used for this study are listed in Table 3, and the genotypes of strains constructed in the present studies are shown in Table S2 in the supplemental material. The parent strains CAY3764, CPL2H1, and HTL were used to create homozygous deletion strains using fusion PCR disruption cassettes as previously described for C. tropicalis, C. parapsilosis, and C. glabrata, respectively (6, 65–67). Correct integration was confirmed by PCR. At least two independent mutants were created for each gene of interest. The primers used for strain construction and confirmation are listed in Table S3.
C. tropicalis, C. parapsilosis, and C. glabrata mutant strains developed in this study. A list of genes under study, strain names, and strain genotypes for the strains constructed and utilized in these studies is shown. Download TABLE S2, DOCX file, 0.02 MB (18.7KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for mutant strain creation and confirmation. The primers used for genetic modification of strains and the primers used to confirm correct modifications are listed. Download TABLE S3, DOCX file, 0.02 MB (24.5KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of C. tropicalis mutant strains was conducted using the background strain of CAY3764, which is auxotrophic for LEU2 and HIS1 (67). The primers used for strain construction and confirmation are listed in Table S3. For C. parapsilosis strain construction, we used the background strain of CPL2H1, which is auxotrophic for LEU2 and HIS1, as described by Holland et al. (6). The first allele was deleted by replacing one allele with HIS1 from C. dubliniensis, and the second with LEU2 from C. maltosa. C. glabrata transformations were conducted using auxotrophic background strain HTL (HIS3, LEU2, TRP1) (63). Candidate genes were deleted by replacing the allele with the gene conferring resistance to the antibiotic nourseothricin. All mutant strains were confirmed by PCR using primers inside nourseothricin and a primer outside the integration sites at both the 5′ and 3′ ends of the gene.
Complementation of C. tropicalis and C. parapsilosis mutant strains with a single wild-type gene copy used selection for nourseothricin resistance, whereas complementation of C. glabrata mutant strains used selection for hygromycin B resistance. A single copy of the gene was reintroduced to its endogenous location within the genome. Briefly, each open reading frame (ORF) (plus 1 kb upstream and downstream) was amplified by PCR, and using the PCR fusion method, a cassette was created with either hygromycin B or nourseothricin at the tail end of the PCR product. This fusion PCR product was then transformed into their respective species using the same method as previously described for mutant construction. Colony PCR was used to verify all genotypes; the primers are listed in Table S3.
In vitro biofilm models.
Biofilms were grown in one of four models: 96-well or 6-well polystyrene plate, polystyrene roller bottle, or glass coverslip. Ninety-six-well flat-bottom polystyrene plates were used to assess biofilm adherence, maturation, and treatment effect as previously described (68, 69). The Candida species inocula (106 cells/ml) were prepared by growth in YPD with uridine overnight at 30°C, followed by dilution in RPMI-MOPS based on hemocytometer counts. The six-well plate assay was used to assess matrix composition. For this assay, 1 ml of culture was inoculated in each well. After a 60-min adherence period at 30°C, the nonadherent inoculum was removed and 1 ml of fresh medium (RPMI-MOPS) was applied to each well. The plates were incubated at 37°C for 48 h on an orbital shaker set at 50 rpm. The medium was removed, and fresh medium was added midway through the incubation period. The coverslip assay was used for in vitro biofilm imaging. Briefly, in vitro biofilms were grown on sterile coverslips (Thermanox) in sterile 12-well plates that had previously been coated with 10 µl of human NaEDTA plasma each and allowed to dry at 30°C. Forty microliters of yeast in RPMI was counted and diluted as in the biofilm models described above and added to each coverslip for 60 min at 30°C. The initial inoculum was then removed, 1 ml of RPMI-MOPS plus 5% NaEDTA human plasma was added to each well, and the plates were incubated for 20 h at 37°C and 50 rpm on an orbital shaker for an additional 24 h. A rolling bottle system was used to generate matrix for analyses (49). Briefly, aliquots of C. albicans grown in RPMI-MOPS were used to inoculate a polystyrene roller. Bottles were placed on a roller apparatus (Wheaton Science Products, Millville, NJ), rolling at the rate of 20 rpm at 37°C. After 24 h, the biofilm culture medium was replaced with fresh medium, and the bottles were incubated for another 24 h. At least three biological replicates were performed for each assay.
In vitro biofilm and planktonic antifungal susceptibility testing.
A tetrazolium salt XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt] reduction assay was used to measure in vitro biofilm drug susceptibility (70, 71). Biofilms were formed in the wells of 96-well microtiter plates as described above. After a 6-h biofilm formation period, the biofilms were washed with phosphate-buffered saline (PBS) twice to remove nonadherent cells. Fresh RPMI-MOPS and drug dilutions were added, followed by additional periods of incubation (48 h). The antifungal studies included fluconazole at 4 to 1,000 mg/ml, amphotericin B at 125 to 0.5 µg/ml, and micafungin at 125 to 0.5 µg/ml. For experiments with tunicamycin (1.0 µg/ml) and brefeldin A (0.6 µg/ml), biofilms were treated alone or in combination with fluconazole after an initial 6-h growth phase. Biofilms treated with α-mannosidase (0.78 U/ml; jack bean; Sigma) were grown for 24 h before a 24-h dose either alone or in combination with fluconazole. Drugs were reapplied after 24 h, and the plates were incubated for an additional 24 h. Following treatment with 90 µl XTT (0.75 mg/ml) and 10 µl phenazine methosulfate (3.20 mg/ml) for 30 min, absorbance at 492 nm was measured using an automated plate reader. The percent reduction in biofilm growth was calculated using the reduction in absorbance compared to that of controls with no antifungal treatment. Assays were performed in triplicate, and significant differences were measured by analysis of variance (ANOVA) with pairwise comparisons using the Holm-Sidak method. The CLSI M27 A3 broth microdilution susceptibility method was used to examine the activities of fluconazole against planktonic Candida. sp. (72). Endpoints were assessed after 24 h by visible turbidity in triplicate assays.
Biofilm SEM.
In vitro biofilms from sterile coverslips were grown as described above. Following a 24-h incubation period, the medium was replaced with 1 ml of fixative (4% formaldehyde, 1% glutaraldehyde in PBS), and the coverslips were incubated at 4°C for 24 h. The coverslips were then washed with PBS and treated with 1% osmium tetroxide for 30 min at ambient temperature. After a series of alcohol washes (30 to 100%), final desiccation was performed by critical point drying. Coverslips were mounted, palladium-gold coated, and imaged in a scanning electron microscope (SEM) (LEO 1530) at 3 kV. The images were assembled using Adobe Photoshop 7.0.1.
Matrix isolation from roller bottle and six-well biofilms.
A rolling bottle system was used to generate matrix for composition analyses (49). After incubation for 48 h, the medium was removed, and the Candida biofilms were dislodged with a spatula and gently sonicated to avoid cell wall disruption (sonication with a 6-mm microtip at 20 kHz with an amplitude of 30% for 8 min). The aggregate biofilm was then centrifuged to separate fungal cells and matrix. The supernatant-containing matrix was then collected and lyophilized. The sample was resuspended in water and dialyzed (molecular size cutoff of 3.5 kDa) for 5 days and again lyophilized, yielding the “crude” biofilm matrix. Overall, a total of 400 bottles of the matrix corresponding to the biofilm area of 59.5 m2 were collected for analysis.
Matrix was similarly collected from six-well plates as described previously (34). Following incubation, biofilms were harvested by removing and discarding the medium, washing each well with 1 ml of double-distilled water (ddH2O), and then removing the biofilms using a spatula. Biofilms were collected in 1 ml of ddH2O per well and then sonicated for 20 min. The soluble matrix was then separated from the cells by centrifuging the samples at 2,880 × g for 20 min at 4°C.
Biofilm matrix analysis. (i) Dry weight.
Following biofilm matrix collection described above, matrix was lyophilized, and weighed to obtain total biomass for each biofilm.
(ii) Carbohydrate analysis.
The carbohydrate concentration of crude matrix was determined colorimetrically (492 nm) using the phenol-sulfuric acid method (73). Structural analysis was performed after a series of purification and fractionation steps, including size exclusion chromatography.
(iii) Protein analysis.
The protein concentration of the crude matrix sample was assessed colorimetrically at 562 nm using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL) (74).
(iv) Nucleic acid analysis.
Nucleic acid concentrations were measured spectrophotometrically (260 nm) (75).
(v) Monosaccharide analysis.
Sugars were detected and quantified by gas-liquid chromatography with a flame ionization detector (GLC-FID) on a Shimadzu GC-2010 system after conversion to alditol acetate derivatives as previously described (76). A 50% cyanopropylmethyl–50% phenylmethyl polysiloxane column was used (Restek) under the GLC conditions previously described (77). Data for each monosaccharide were calculated and presented as a percentage of the total detected sugars.
(vi) Lipid analysis.
Lipids were extracted from the desalted lyophilized matrix powder with a mixture of CHCl3 and methanol (MeOH) (2:1, by volume) as described elsewhere (78). Methylation of fatty acids was performed using 0.5 ml of 14% boron trifluoride (BF3) in MeOH, and methyl esters were recovered with hexane. Fatty acid methyl esters were analyzed by gas chromatography using a Hewlett-Packard 5890 instrument (Hewlett Packard, Palo Alto, CA).
(vii) eDNA analysis.
Extracellular DNA (eDNA) was isolated from non-albicans Candida species (NAC) matrices using the MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI). Nucleic acid concentrations were measured spectrophotometrically with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, San Jose, CA). The average measured ratio of absorbance at 260/280 nm was about ~1.8, which indicated that the DNA was pure and free of contaminants.
NMR of neutral carbohydrates.
After isolation, matrix samples were resuspended in 1 ml of 20 mM bis-Tris HCl (pH 6.5) loading buffer and fractionated on a HiPrep 26/10 desalting column prepacked with Sephadex G-25 Fine (GE Healthcare Life Sciences, Uppsala, Sweden). Column-dialyzed fractions were then separated on an anion exchanger HiPrep 16/10 DEAE FF column (GE Healthcare Life Sciences) equilibrated with 20 mM bis-Tris HCl (pH 6.5). Elution was performed in a 20 mM bis-Tris HCl (pH 6.4)–0.5 M NaCl buffer system at a flow rate of 1 ml/min in a linear gradient of salt from 0 to 100% in 20 column volumes. Neutral free carbohydrates were detected in flowthrough fractions, which were then pooled, lyophilized, resuspended in 2 ml of 150 mM NH4HCO3, and applied to gel filtration on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences). Matrix components were eluted at a flow rate of 0.5 ml/min, and 1-ml fractions were collected. All chromatographic separation steps were performed at room temperature on the high-performance liquid chromatography Äkta-Purifier 10 system (GE Healthcare Life Sciences). All buffers used were filtered through 0.2-μm nylon membrane filters (Nalgene, Rochester, NY) and degassed prior to use. Isolated polysugar fractions were lyophilized, resuspended in a small volume of water, and incubated at 55°C overnight in order to decompose and remove any remaining ammonium bicarbonate. These steps were repeated until all of the salt was removed and the isolated sugars appeared as an anamorphous cotton-like material after the final lyophilization. The molecular sizes of biofilm matrix neutral carbohydrates were estimated using size exclusion column calibration with a set of Leuconostoc species dextran standards, which included 100-kDa, 70-kDa, 40-kDa, 25-kDa, and 6-kDa polymers.
To elucidate the structure of the isolated carbohydrates, we utilized a combination of one-dimensional (1D) 1H nuclear magnetic resonance (NMR) and 2D heteronuclear single quantum coherence (HSQC) NMR experiments as well as known chemical shift assignments characteristic of individual mannosyl and glucosyl motifs in mannan and glucan structures based on previously published studies (79–83). All data were collected at 70°C on a Bruker Biospin Avance III 500-MHz NMR spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) equipped with a 5-mm triple resonance, cryogenic probe, CPTXI 500 H-C/N-D. One-dimensional spectra were collected with 32 acquisitions using a standard one-pulse experiment. The spectral width was 10 ppm centered at 4.7 ppm. The relaxation delay time was 2 s with an acquisition time of 3.3 s (32,768 data points). Thirty-two acquisitions were collected. Multiplicity-edited, phase-sensitive, echo-antiecho 1H HSQC spectra were obtained using four acquisitions per indirect time point with 1H decoupling during acquisition (84). Matched swept adiabatic 13C inversion pulses were used. The raw data matrix size was 2,048 × 128 blocks. Spectra were collected with a relaxation time delay of 2 s and an acquisition time of 0.2 s with sweep widths of 10 ppm (1H) and 65 ppm (13C), respectively. The centers of the spectra were 4.7 ppm for 1H and 82 ppm for 13C.
Matrix carbohydrate fractionation and analysis for an MGCx.
Additional structural analysis was performed after a series of purification and fractionation steps to further ascertain the presence of a mannan-glucan complex. These steps include size exclusion chromatography followed by separation on an anion exchanger HiPrep 16/10 DEAE FF column (GE Healthcare Life Sciences). Neutral free carbohydrates were collected in flowthrough fractions, which were pooled and applied to gel filtration on a HighPrep 16/60 Sephacryl S-300 HR column (GE Healthcare Life Sciences), yielding 22 individual polysaccharide peaks, F1 to F22. The molecular weight of biofilm matrix neutral carbohydrates was determined using size exclusion column calibration with a set of Leuconostoc species dextran standards (polymers with molecular weights [in thousands] of 100, 70, 40, 25, and 6). Both high-molecular-weight (HMW) and low-molecular-weight (LMW) fractions were examined by GC for monosugar analysis. Matrix monosugar composition and quantification was performed on alditol acetate derivatives by GLC-FID (Shimadzu GC-2010 system; Shimadzu, Kyoto, Japan).
In vivo Candida venous catheter biofilm model.
A jugular vein rat central venous catheter infection model was used for in vivo biofilm studies (53). Candida strains were grown to logarithmic phase in YPD at 30°C. After a 24-h conditioning period after catheter placement, infection was achieved by intraluminal instillation of 500 ml of C. albicans (106 cells/ml). After an adherence period of 6 h, the 500-ml volume was withdrawn, and the catheter was flushed with heparinized saline. Following a 24-h incubation period, the catheters were removed, and the animals were prepared for SEM imaging as described above. The images were assembled using Adobe Photoshop 7.0.1.as described above. For drug treatment experiments, fluconazole (250 µg/ml) was instilled in the catheter after 24 h of biofilm growth. After a 24-h drug treatment period, the posttreatment viable burden of Candida biofilm on the catheter surface was measured by viable plate counts on Sabouraud’s dextrose agar (SDA) following removal of the biofilm by sonication and vortexing. We utilized three replicates for each condition.
Sequestration of [3H]fluconazole in biofilms.
Radiolabeled fluconazole was used in an assay to assess drug retention in biofilms formed in six-well plates (85). Biofilms were grown for 48 h in six-well polystyrene plates as described above, washed, and then incubated with 8.48 × 105 cpm of [3H]fluconazole (Moravek Biochemicals; 50 µM, 0.001 mCi/ml in ethanol) in RPMI-MOPS for 30 min at 37°C with orbital shaking at 50 rpm. Unlabeled fluconazole (20 µM) in RPMI-MOPS was added for an additional 15-min incubation period. After washing, biofilms and matrix were collected and isolated as described above. For a subset of biofilm cells, cells were disrupted by bead beating to yield cell wall and intracellular portions. Samples were added to a Tri-Carb 2100 TR liquid scintillation analyzer after adding ScintiSafe 30% liquid scintillation counting reagent (LSC) mixture to each sample fraction. The values for three technical replicates were averaged, the standard errors (SEs) were calculated, with values compared to the reference strain using pairwise comparisons with ANOVA by the Holm-Sidak method.
Fluconazole-matrix interaction determination.
Interactions between the biofilm matrix and fluconazole were also probed using one-dimensional 1H NMR. Data were collected at 37°C on a Bruker Biospin Avance III 600-MHz NMR spectrometer (Bruker BioSpin GmbH) equipped with a 1.7-mm triple resonance, cryogenic probe, CPTXI 500 H-C/N-D. One-dimensional spectra were collected with 512 acquisitions using a one-pulse sequence experiment with water suppression and excitation sculpting with gradients (zgesgp). The spectral width was 16 ppm centered at 4.7 ppm. The relaxation delay time was 2 s. The approach was based on monitoring chemical shifts of fluconazole-specific protons in the presence and absence of the biofilm matrix under different pH conditions. In this study, all tested reactions were prepared in PBS (pH 7.2) and fluconazole was used at a constant concentration of 0.653 mM. In this drug-matrix system, interactions were represented by decreasing in signal intensities of the chemical shift peaks of protons present in fluconazole.
ACKNOWLEDGMENTS
This study made use of the National Magnetic Resonance Facility at the University of Wisconsin—Madison, which is supported by NIH grant P41GM103399 (NIGMS) (old number, P41RR002301). Equipment was purchased with funds from the University of Wisconsin—Madison, NIH (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, and S10RR029220), NSF (DMB-8415048, OIA-9977486, and BIR-9214394), and USDA.
Footnotes
Citation Dominguez E, Zarnowski R, Sanchez H, Covelli AS, Westler WM, Azadi P, Nett J, Mitchell AP, Andes DR. 2018. Conservation and divergence in the Candida species biofilm matrix mannan-glucan complex structure, function, and genetic control. mBio 9:e00451-18. https://doi.org/10.1128/mBio.00451-18.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C, Franks B, Azie NE. 2014. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004-2008. PLoS One 9:e101510. doi: 10.1371/journal.pone.0101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland AA, Farley MM, Harrison LH, Stein B, Hollick R, Lockhart SR, Magill SS, Derado G, Park BJ, Chiller TM. 2012. Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008-2011. Clin Infect Dis 55:1352–1361. doi: 10.1093/cid/cis697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, Baughman W, Stein B, Hollick R, Park BJ, Chiller T. 2012. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andes DR, Safdar N, Baddley JW, Alexander B, Brumble L, Freifeld A, Hadley S, Herwaldt L, Kauffman C, Lyon GM, Morrison V, Patterson T, Perl T, Walker R, Hess T, Chiller T, Pappas PG, The TRANSNET Investigators . 2016. The epidemiology and outcomes of invasive Candida infections among organ transplant recipients in the United States: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Transpl Infect Dis 18:921–931. doi: 10.1111/tid.12613. [DOI] [PubMed] [Google Scholar]
- 6.Holland LM, Schröder MS, Turner SA, Taff H, Andes D, Grózer Z, Gácser A, Ames L, Haynes K, Higgins DG, Butler G. 2014. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog 10:e1004365. doi: 10.1371/journal.ppat.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porman AM, Alby K, Hirakawa MP, Bennett RJ. 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A 108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran GP, Coleman DC, Sullivan DJ. 2011. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryot Cell 10:34–42. doi: 10.1128/EC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson DS, Carlisle PL, Kadosh D. 2011. Coevolution of morphology and virulence in Candida species. Eukaryot Cell 10:1173–1182. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maestre-Reyna M, Diderrich R, Veelders MS, Eulenburg G, Kalugin V, Brückner S, Keller P, Rupp S, Mösch HU, Essen LO. 2012. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc Natl Acad Sci U S A 109:16864–16869. doi: 10.1073/pnas.1207653109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trofa D, Gácser A, Nosanchuk JD. 2008. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 21:606–625. doi: 10.1128/CMR.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lackey E, Vipulanandan G, Childers DS, Kadosh D. 2013. Comparative evolution of morphological regulatory functions in Candida species. Eukaryot Cell 12:1356–1368. doi: 10.1128/EC.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papon N, Courdavault V, Clastre M, Bennett RJ. 2013. Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog 9:e1003550. doi: 10.1371/journal.ppat.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priest SJ, Lorenz MC. 2015. Characterization of virulence-related phenotypes in Candida species of the CUG clade. Eukaryot Cell 14:931–940. doi: 10.1128/EC.00062-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YL, Konieczka JH, Springer DJ, Bowen SE, Zhang J, Silao FG, Bungay AA, Bigol UG, Nicolas MG, Abraham SN, Thompson DA, Regev A, Heitman J. 2012. Convergent evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. G3 2:675–691. doi: 10.1534/g3.112.002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur R, Domergue R, Zupancic ML, Cormack BP. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol 8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uppuluri P, Pierce CG, López-Ribot JL. 2009. Candida albicans biofilm formation and its clinical consequences. Future Microbiol 4:1235–1237. doi: 10.2217/fmb.09.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramage G, Saville SP, Thomas DP, López-Ribot JL. 2005. Candida biofilms: an update. Eukaryot Cell 4:633–638. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai JV, Mitchell AP, Andes DR. 2014. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med 4:a019729. doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blankenship JR, Mitchell AP. 2006. How to build a biofilm: a fungal perspective. Curr Opin Microbiol 9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Andes DR, Safdar N, Baddley JW, Playford G, Reboli AC, Rex JH, Sobel JD, Pappas PG, Kullberg BJ, Mycoses Study Group . 2012. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis 54:1110–1122. doi: 10.1093/cid/cis021. [DOI] [PubMed] [Google Scholar]
- 24.Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin Microbiol Rev 17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 27.Baillie GS, Douglas LJ. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J Antimicrob Chemother 46:397–403. doi: 10.1093/jac/46.3.397. [DOI] [PubMed] [Google Scholar]
- 28.Taff HT, Mitchell KF, Edward JA, Andes DR. 2013. Mechanisms of Candida biofilm drug resistance. Future Microbiol 8:1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vediyappan G, Rossignol T, d’Enfert C. 2010. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother 54:2096–2111. doi: 10.1128/AAC.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katragkou A, Chatzimoschou A, Simitsopoulou M, Dalakiouridou M, Diza-Mataftsi E, Tsantali C, Roilides E. 2008. Differential activities of newer antifungal agents against Candida albicans and Candida parapsilosis biofilms. Antimicrob Agents Chemother 52:357–360. doi: 10.1128/AAC.00856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 32.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 33.Mah TF, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 34.Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother 54:3505–3508. doi: 10.1128/AAC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nett JE, Sanchez H, Cain MT, Andes DR. 2010. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis 202:171–175. doi: 10.1086/651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell KF, Zarnowski R, Sanchez H, Edward JA, Reinicke EL, Nett JE, Mitchell AP, Andes DR. 2015. Community participation in biofilm matrix assembly and function. Proc Natl Acad Sci U S A 112:4092–4097. doi: 10.1073/pnas.1421437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, Lounes-Hadj Sahraoui A, Fontaine J, Sanchez H, Hatfield RD, Ntambi JM, Nett JE, Mitchell AP, Andes DR. 2014. Novel entries in a fungal biofilm matrix encyclopedia. mBio 5:e01333-14. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Ribot JL. 2014. Large-scale biochemical profiling of the Candida albicans biofilm matrix: new compositional, structural, and functional insights. mBio 5:e01781-14. doi: 10.1128/mBio.01781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell KF, Zarnowski R, Andes DR. 2016. Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog 12:e1005828. doi: 10.1371/journal.ppat.1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajendran R, Sherry L, Lappin DF, Nile CJ, Smith K, Williams C, Munro CA, Ramage G. 2014. Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation. BMC Microbiol 14:303. doi: 10.1186/s12866-014-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169:323–331. doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol 55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- 46.Fernandes T, Silva S, Henriques M. 2015. Candida tropicalis biofilm’s matrix—involvement on its resistance to amphotericin B. Diagn Microbiol Infect Dis 83:165–169. doi: 10.1016/j.diagmicrobio.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Silva S, Henriques M, Martins A, Oliveira R, Williams D, Azeredo J. 2009. Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med Mycol 47:681–689. doi: 10.3109/13693780802549594. [DOI] [PubMed] [Google Scholar]
- 48.Nett JE, Zarnowski R, Cabezas-Olcoz J, Brooks EG, Bernhardt J, Marchillo K, Mosher DF, Andes DR. 2015. Host contributions to construction of three device-associated Candida albicans biofilms. Infect Immun 83:4630–4638. doi: 10.1128/IAI.00931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zarnowski R, Sanchez H, Andes DR. 2016. Large-scale production and isolation of Candida biofilm extracellular matrix. Nat Protoc 11:2320–2327. doi: 10.1038/nprot.2016.132. [DOI] [PubMed] [Google Scholar]
- 50.Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, Andes D, Cowen LE. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog 7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramage G, Bachmann S, Patterson TF, Wickes BL, López-Ribot JL. 2002. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother 49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- 53.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun 72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce CG, Uppuluri P, Tristan AR, Wormley FL Jr, Mowat E, Ramage G, Lopez-Ribot JL. 2008. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc 3:1494–1500. doi: 10.1038/nprot.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maiolo EM, Oliva A, Furustrand Tafin U, Perrotet N, Borens O, Trampuz A. 2016. Antifungal activity against planktonic and biofilm Candida albicans in an experimental model of foreign-body infection. J Infect 72:386–392. doi: 10.1016/j.jinf.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Silva S, Henriques M, Oliveira R, Williams D, Azeredo J. 2010. In vitro biofilm activity of non-Candida albicans Candida species. Curr Microbiol 61:534–540. doi: 10.1007/s00284-010-9649-7. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun 70:878–888. doi: 10.1128/IAI.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maiolo EM, Furustrand Tafin U, Borens O, Trampuz A. 2014. Activities of fluconazole, caspofungin, anidulafungin, and amphotericin B on planktonic and biofilm Candida species determined by microcalorimetry. Antimicrob Agents Chemother 58:2709–2717. doi: 10.1128/AAC.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nebenführ A, Ritzenthaler C, Robinson DG. 2002. Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130:1102–1108. doi: 10.1104/pp.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldauf SL, Palmer JD. 1993. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci U S A 90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalal CK, Zuleta IA, Mitchell KF, Andes DR, El-Samad H, Johnson AD. 2016. Transcriptional rewiring over evolutionary timescales changes quantitative and qualitative properties of gene expression. Elife 5:e18981. doi: 10.7554/eLife.18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding C, Vidanes GM, Maguire SL, Guida A, Synnott JM, Andes DR, Butler G. 2011. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS One 6:e28151. doi: 10.1371/journal.pone.0028151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner I, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldón T, Hube B, d’Enfert C, Rupp S, Cormack B, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e1004211. doi: 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 65.Chang DC, Grant GB, O’Donnell K, Wannemuehler KA, Noble-Wang J, Rao CY, Jacobson LM, Crowell CS, Sneed RS, Lewis FM, Schaffzin JK, Kainer MA, Genese CA, Alfonso EC, Jones DB, Srinivasan A, Fridkin SK, Park BJ, Fusarium Keratitis Investigation Team . 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 66.Istel F, Schwarzmüller T, Tscherner M, Kuchler K. 2015. Genetic transformation of Candida glabrata by electroporation. Bio Protoc 5:e1528. doi: 10.21769/BioProtoc.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mancera E, Porman AM, Cuomo CA, Bennett RJ, Johnson AD. 2015. Finding a missing gene: EFG1 regulates morphogenesis in Candida tropicalis. G3 5:849–856. doi: 10.1534/g3.115.017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramage G, Vande Walle K, Wickes BL, López-Ribot JL. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother 45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taff HT, Nett JE, Andes DR. 2012. Comparative analysis of Candida biofilm quantitation assays. Med Mycol 50:214–218. doi: 10.3109/13693786.2011.580016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nett JE, Cain MT, Crawford K, Andes DR. 2011. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J Clin Microbiol 49:1426–1433. doi: 10.1128/JCM.02273-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramage G, López-Ribot JL. 2005. Techniques for antifungal susceptibility testing of Candida albicans biofilms. Methods Mol Med 118:71–79. doi: 10.1385/1-59259-943-5:071. [DOI] [PubMed] [Google Scholar]
- 72.Clinical and Laboratory Standards Institute 2007. Reference method for broth microdilution antifungal susceptibility testing of yeasts, 3rd ed Approved standard M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 73.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 74.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 75.Sandhu LC, Warters RL, Dethlefsen LA. 1985. Fluorescence studies of Hoechst 33342 with supercoiled and relaxed plasmid pBR322 DNA. Cytometry 6:191–194. doi: 10.1002/cyto.990060304. [DOI] [PubMed] [Google Scholar]
- 76.Henry RJ, Blakeney AB, Harris PJ, Stone BA. 1983. Detection of neutral and aminosugars from glycoproteins and polysaccharides as their alditol acetates. J Chromatogr 256:419–427. doi: 10.1016/S0021-9673(01)88259-5. [DOI] [PubMed] [Google Scholar]
- 77.Sassaki GL, Gorin PA, Souza LM, Czelusniak PA, Iacomini M. 2005. Rapid synthesis of partially O-methylated alditol acetate standards for GC-MS: some relative activities of hydroxyl groups of methyl glycopyranosides on Purdie methylation. Carbohydr Res 340:731–739. doi: 10.1016/j.carres.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 78.Zarnowski R, Jaromin A, Certik M, Czabany T, Fontaine J, Jakubik T, Iqbal MC, Grandmougin-Ferjani A, Kozubek A, Pietr SJ. 2004. The oil of Adenanthera pavonina L. seeds and its emulsions. Z Naturforsch C 59:321–326. doi: 10.1515/znc-2004-5-605. [DOI] [PubMed] [Google Scholar]
- 79.Shibata N, Kobayashi H, Okawa Y, Suzuki S. 2003. Existence of novel beta-1,2 linkage-containing side chain in the mannan of Candida lusitaniae, antigenically related to Candida albicans serotype A. Eur J Biochem 270:2565–2575. doi: 10.1046/j.1432-1033.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 80.Shibata N, Suzuki A, Kobayashi H, Okawa Y. 2007. Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem J 404:365–372. doi: 10.1042/BJ20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shibata N, Okawa Y. 2010. Enzymatic synthesis of new oligosaccharides using mannosyltransferases from Candida species and their NMR assignments. Biol Pharm Bull 33:895–899. doi: 10.1248/bpb.33.895. [DOI] [PubMed] [Google Scholar]
- 82.Lowman DW, Ensley HE, Greene RR, Knagge KJ, Williams DL, Kruppa MD. 2011. Mannan structural complexity is decreased when Candida albicans is cultivated in blood or serum at physiological temperature. Carbohydr Res 346:2752–2759. doi: 10.1016/j.carres.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lowman DW, West LJ, Bearden DW, Wempe MF, Power TD, Ensley HE, Haynes K, Williams DL, Kruppa MD. 2011. New insights into the structure of (1→3,1→6)-beta-d-glucan side chains in the Candida glabrata cell wall. PLoS One 6:e27614. doi: 10.1371/journal.pone.0027614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boyer RD, Johnson R, Krishnamurthy K. 2003. Compensation of refocusing inefficiency with synchronized inversion sweep (CRISIS) in multiplicity-edited HSQC. J Magn Reson 165:253–259. doi: 10.1016/j.jmr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 85.Downs JS, Arslanian S, de Bruin WB, Copeland VC, Doswell W, Herman W, Lain K, Mansfield J, Murray PJ, White N, Charron-Prochownik D. 2010. Implications of type 2 diabetes on adolescent reproductive health risk: an expert model. Diabetes Educ 36:911–919. doi: 10.1177/0145721710383586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2D HSQC NMR chemical shift assignment of the major spin systems found in C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis. Download TABLE S1, DOCX file, 0.02 MB (24.7KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Planktonic cell growth following pharmacological and enzymatic treatments. Planktonic cells were grown in 96-well round-bottom plates for 24 h and quantified using XTT. For experiments with tunicamycin (TM) (at 1.0 μg/ml) and brefeldin A (BFA) (at 0.6 μg/ml), biofilms or cells were grown for 6 h and then treated for 24 h before quantification. Experiments with α-mannosidase (α-MS) (at 0.78 U/ml) used biofilms or cells first grown for 24 h before 24-h treatment. The means of three technical replicates and SEs are shown. Download FIG S1, TIF file, 2.2 MB (2.3MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biofilm formation capacity for non-albicans Candida mutants in the mannan and glucan pathways. Mature biofilm burden for each of the select deletion mutants was quantified in a 96-well format using an XTT endpoint after 24 h of incubation. Asterisks indicate statistically significant differences (P < 0.001) between strains based upon ANOVA. Download FIG S2, TIF file, 4.2 MB (4.3MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NAC biofilm drug susceptibility in mutants without an enhanced susceptibility phenotype. (A to C) Percentage of reduction in biofilm formation following 48-h treatment with fluconazole compared with untreated biofilms, as quantified using the 96-well XTT assay. The null mutant (Δ/Δ) is shown for each gene of interest. The figure shows data from three assay replicates of a representative example of three biological replicates. Asterisks indicate statistically significant differences (P < 0.001) between reference and mutant based upon ANOVA using the Holm-Sikak method for pairwise comparison. Download FIG S3, TIF file, 2.7 MB (2.8MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polyene and echinocandin biofilm activity against fluconazole-susceptible NAC mutants. After growth for 6 h, biofilms were treated with either 0.5 μg/ml amphotericin B or micafungin for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. All mutants were significantly more susceptible to treatment than the reference strain (*, P < 0.001; **, P < 0.05). The means of three technical replicates and SEs are shown. Download FIG S4, TIF file, 2 MB (2MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complemented C. tropical and C. glabrata glucan and mannan synthesis mutant biofilms are susceptible to fluconazole. After growth for 6 h, biofilms were treated with 1,000 μg/ml fluconazole for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. All mutants were significantly more susceptible to treatment than the reference strain (P < 0.001). Each of the complemented strains exhibited resistance similar to that of the reference strains. The means of three technical replicates and SEs are shown. Download FIG S5, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple independent transformants of noncomplemented mutant strains are susceptible to fluconazole. After growth for 6 h, biofilms were treated with 1,000 μg/ml fluconazole for 48 h. Biofilms were quantified using the 96-well XTT assay, and reduction was determined by comparing treated and untreated biofilms. Two or three independent transformants were tested for each homozygous deletion mutant. All mutants were significantly more susceptible to treatment than the reference strain (P < 0.001). A different biological replicate is presented here than in Fig. 5A. The means of three technical replicates and SEs are shown. Download FIG S6, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. tropicalis, C. parapsilosis, and C. glabrata mutant strains developed in this study. A list of genes under study, strain names, and strain genotypes for the strains constructed and utilized in these studies is shown. Download TABLE S2, DOCX file, 0.02 MB (18.7KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for mutant strain creation and confirmation. The primers used for genetic modification of strains and the primers used to confirm correct modifications are listed. Download TABLE S3, DOCX file, 0.02 MB (24.5KB, docx) .
Copyright © 2018 Dominguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.