Abstract
Autosomal recessive primary microcephaly (microcephaly primary hereditary, MCPH) is a genetically heterogeneous rare developmental disorder that is characterized by prenatal onset of abnormal brain growth, which leads to intellectual disability of variable severity. We report a 5-year-old male who presented with a severe form of primary microcephaly. Targeted panel sequencing revealed compound heterozygous truncating mutations of the abnormal spindle-like microcephaly-associated (ASPM) gene, which confirmed the MCPH5 diagnosis. A novel NM_018136.4: c.9742_9745del (p.Lys3248Serfs*13) deletion mutation was identified.
Autosomal recessive primary microcephaly (microcephaly primary hereditary, MCPH, MIM#251200) is a genetically heterogeneous developmental disorder that is characterized by prenatal onset of abnormal brain growth, resulting in a reduced brain volume, which is indirectly measured as an occipitofrontal circumference (OFC) that is equal to or greater than 2 standard deviations (SDs) and 3 SDs below the age- and sex-matched means at birth and 6 months, respectively. These physical abnormalities lead to varying degrees of non-progressive intellectual disability in the absence of any significant neurological deficit.1 Eighteen MCPH loci are implicated in MCPH (MCPH1-18). Mutations of abnormal spindle-like microcephaly-associated (ASPM) gene (MIM#605481) are the most common cause of MCPH accounting for up to 40% in consanguineous and non-consanguineous families,2 followed in frequency by mutations in WDR62. The phenotypic similarity between patients with MCPH-causing genes is remarkable.3,4 Therefore, whole-exome sequencing or targeted panel sequencing (TPS) as a genetic screening technique is mandatory to delineate the etiology and obtain the proper prenatal diagnosis. We report a severe form of primary microcephaly exhibiting compound heterozygous ASPM mutations that included a novel deletion mutation.
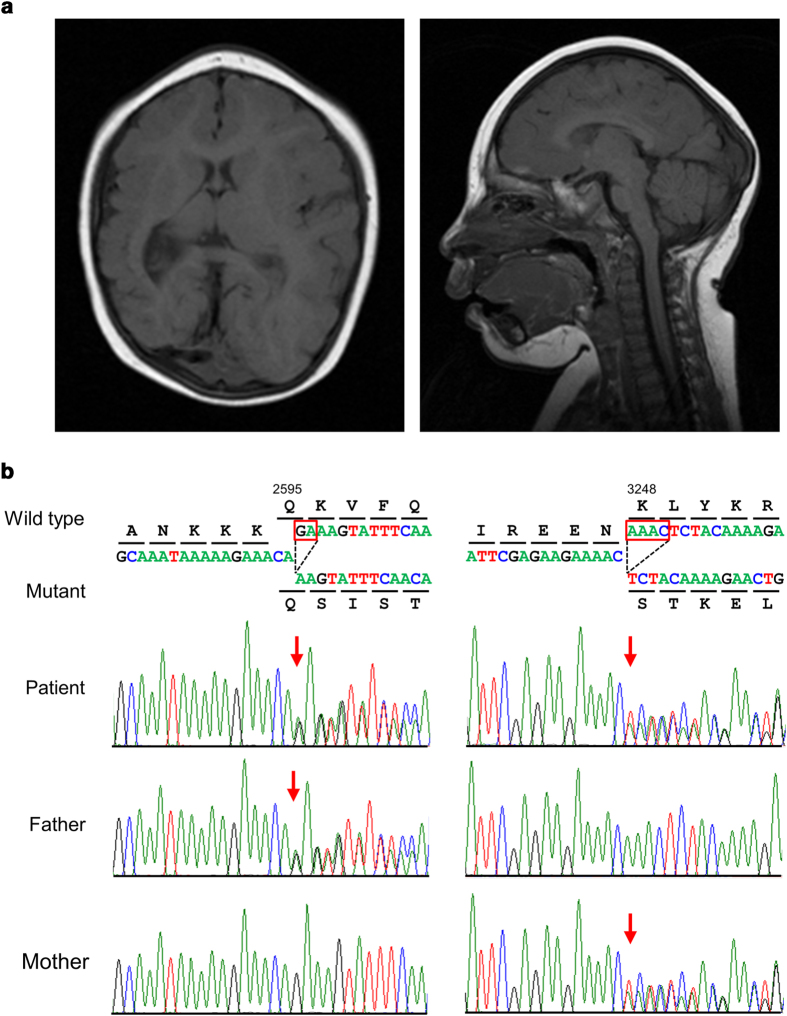
The patient was a 5-year-old male who was the first child of Japanese non-consanguineous parents. Microcephaly was noted prenatally on ultrasonography. He was delivered at 38 weeks of gestation, with a birth weight of 2058 g (−2.3 SD), body length of 43 cm (−2.3 SD) and OFC of 27 cm (−4.2 SD). He exhibited hypertonicity, opisthotonic posture and sleep disturbance. Trembling was noted in his body and neck. Brain magnetic resonance imaging (MRI) revealed a global reduction in the size of his cerebral structures, including the cortex and white matter, simplified gyral pattern and enlarged lateral ventricle with clear laterality (Figure 1a). Delayed motor development was marked from the infantile period. The patient sat with support at 1 year of age and began to walk independently at 3 years of age. He spoke incohesive words and exhibited a severe intellectual disability. His recognition and ability to communicate with others developed slowly. Routine laboratory investigations and metabolic surveys revealed normal results. Standard karyotyping and array-based comparative genomic hybridization using peripheral blood revealed no abnormalities. His body weight at 8 years of age was 12.5 kg (−1.8 SD), height was 90 cm (−3.0 SD) and OFC was 41.5 cm (−5.2 SD). Craniofacial dysmorphic features, including up-slanting palpebral fissures, epicanthus, high-arched palate and micrognathism, were noted. The clinical picture was typical of primary microcephaly, particularly MCPH. However, TPS with a panel of multiple potential disease-causing genes was used as the genetic screening test because of the absence of parental consanguinity.
Figure 1.
(a) Brain MRI in the proband at 5 years of age. Axial (left) and sagittal (right) T1-weighted images reveal a global reduction in the size of the cerebral structures, including the cortex and white matter, simplified gyral pattern, enlarged lateral ventricle and fully formed but diffusely thin corpus callosum. Notably, the right side is more remarkably affected than the left side. The sloping of the forehead reflects a severe decrease in the cranial-to-facial proportions. (b) Partial sequence chromatograms around codons 2595 and 3248 on exons 18 and 24, respectively, of ASPM in the patient and his parents. Red boxes denote deleted bases, and red arrows denote the start sites of heterozygous sequences due to these deletions. DNA and corresponding amino acid sequences of the wild-type and mutant ASPM alleles are also shown.
Informed consent was obtained from the parents, and molecular diagnosis was performed using genomic DNA extracted from the patient’s blood. The ethics committees of Tokushima University and Osaka Medical Center and Research Institute for Maternal and Child Health approved the study. The TruSight One Sequencing Panel (Illumina, San Diego, CA, USA) was used with an MiSeq sequencer (Illumina), followed by analysis using our pipeline for NGS data as described previously5 with a minor modification of a software update specific to the bioinformatics pipeline.6 Sequence variants with higher allele frequencies (i.e., >0.01) in the following databases were excluded to identify presumably pathogenic single nucleotide variants: 1000 Genomes Project database (http://www.1000genomes.org), National Heart, Lung, and Blood Institute Grand Opportunity (NHLBI GO) Exome Sequencing Project (ESP6500, http://evs.gs.washington.edu/EVS), Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB) and integrative Japanese Genome Variation Database (iJGVD, https://ijgvd.megabank.tohoku.ac.jp). Copy-number variation analysis using TPS data was also performed as described previously.6,7 These analyses detected two different 2- and 4-nucleotide deletions in exons 18 and 24 of the ASPM gene, namely, [NM_018136.4(ASPM_v001)], c.7782_7783del and c.9742_9745del, respectively, which caused reading frameshifts that ended with premature stop codons, p.Lys2595Serfs*6 and p.Lys3248Serfs*13, respectively. Sanger sequencing confirmed these deletions. The patient’s father and mother were heterozygous carriers of the c.7782_7783del and c.9742_9745del variants, respectively, which suggested the presence of these variations in the patient in a compound heterozygous state (Figure 1b). c.7782_7783del was present in the Human Gene Mutation Database (HGMD, Professional 2017.4, http://www.hgmd.org/) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) as a pathogenic variant, but c.9742_9745del was not present in any database. No other possibly pathogenic variants or gross deletions were detected in the coding regions of other MCPH-related genes in the panel for TPS (data not shown). The patient was diagnosed with MCPH5 caused by one known and one novel deletion of ASPM in a compound heterozygous state based on the results of this molecular diagnosis and re-evaluation of the affected patient’s clinical features.
The ASPM protein plays a crucial role in the maintenance of the cleavage plane orientation, which allows for the symmetric proliferative division of neuroepithelial cells during brain development.8 A total of 167 ASPM pathogenic variants have been reported in cases with microcephaly in HGMD professional 2017.4. Most of these mutations are nonsense (69 variants), frameshift (small deletions and insertions, 79 variants) or splice-site mutations (15 variants), which are predicted to result in unstable RNA that is degraded by nonsense-mediated RNA decay (NMD) or truncated protein synthesis.1–4 Therefore, most of the pathogenic variations of ASPM that are responsible for microcephaly reduce protein synthesis or produce partially or wholly non-functional short proteins. However, few experiments have verified this hypothesis.4 The mutations were spread throughout the coding sequence of the ASPM gene, with no apparent hotspots, and there were no clear cut genotype–phenotype correlations between the head size centile, degree of intellectual disability or presence of epileptic fits and mutation type or position within the gene,2 which suggests that NMD is the major mechanism through which ASPM gene mutations cause reduced fetal brain growth and result in MCPH5.9,10 However, mice homozygously expressing truncated forms of ASPM similar to ASPM mutants that cause microcephaly in humans exhibit mild microcephaly associated with a reduced relative brain weight and cortical thickness, likely due to a reduced number of neural stem cells.11 Therefore, the precise mechanisms that underlie the microcephaly caused by ASPM mutations in humans are not known.4
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank the patient and his family for their participation in this study. Portions of this work were performed at the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. This work was supported by the Japan Society for the Promotion of Science (15K19620, 16K15618) and Japan Agency for Medical Research and Development (JP16ek0109151, JP16kk0205012).
Footnotes
The authors declare no conflict of interest.
References
- Mahmood S, Wasim Ahmad W, Hassan MJ. Autosomal recessive primary microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J Rare Dis 2011; 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K et al. The molecular landscape of ASPM mutations in primary microcephaly. J Med Genet 2009; 46: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hamid MS, Ismail MF, Darwish HA, Effat LK, Zaki MS, Abdel-Salam GMH. Molecular and phenotypic spectrum of ASPM-related primary microcephaly: identification of eight novel mutations. Am J Med Genet A 2016; 170A: 2133–2140. [DOI] [PubMed] [Google Scholar]
- Létard P, Drunat S, Vial Y, Duerinckx S, Ernault A, Amram D et al. Autosomal recessive primary microcephaly due to ASPM mutations: an update. Hum Mutat 2018; 39: 319–332. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Naruto T, Kohmoto T, Komori T, Imoto I.. A novel PTCH1 mutation in a patient with Gorlin syndrome. Hum Genome Var 2014; 1: 14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Nakagawa R, Naruto T, Kohmoto T, Suga K, Goji A et al. A novel missense mutation of COL5A2 in a patient with Ehlers–Danlos syndrome. Hum Genome Var 2016; 3: 16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hayabuchi Y, Ono A, Naruto T, Horikawa H, Kohmoto T et al. Detection of 1p36 deletion by clinical exome-first diagnostic approach. Hum Genome Var 2016; 3: 16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB.. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA 2006; 103: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Pattison L, Scott S et al. ASPM is a major determinant of cerebral cortical size. Nat Genet 2002; 32: 316–320. [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet 2003; 73: 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Bräuninger M, Arai Y, Schreier D et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci USA 2010; 107: 16595–16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Citations
- Imoto Issei.HGV Database. 2018. 10.6084/m9.figshare.hgv.1929. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Imoto Issei.HGV Database. 2018. 10.6084/m9.figshare.hgv.1929. [DOI]