Abstract
Extended hepatectomies may result in posthepatectomy liver failure, a condition with a high mortality. The main purpose of the present study was to investigate and compare the gene expression profiles in rats subjected to increasing size of partial hepatectomy (PH). Thirty Wistar rats were subjected to 30%, 70%, or 90% PH, sham operation, or no operation. Twenty-four hours following resection, liver tissue was harvested and genome-wide expression analysis was performed. Cluster analysis revealed two main groupings, one containing the PH(90%) and one containing the remaining groups [baseline, sham, PH(30%), and PH(70%)]. Categorization of specific affected molecular pathways in the PH(90%) group revealed a downregulation of cellular homeostatic function degradation and biosynthesis, whereas proliferation, cell growth, and cellular stress and injury were upregulated in the PH(90%) group. After PH(90%), the main upregulated pathways were mTOR and ILK. The main activated upstream regulators were hepatocyte growth factor and transforming growth factor. With decreasing size of the future liver remnant, the liver tended to prioritize expression of genes involved in cell proliferation and differentiation at the expense of genes involved in metabolism and body homeostasis. This prioritizing may be an essential molecular explanation for posthepatectomy liver failure.
Key words: Hepatectomy, Regeneration, Liver failure, Gene expression, Microarray analysis
INTRODUCTION
Partial hepatectomy (PH) remains the standard treatment with a curative intend for both primary and secondary liver malignancies, both of which are on the rise worldwide1–3. Because of the unique regenerative ability of the liver, PH of the healthy human liver can be performed, as long as the volume of the future liver remnant (FLR) is 20%–30% of the total initial liver volume4. Larger PHs may result in posthepatectomy liver failure (PHLF), which is associated with a high morbidity and mortality5. The corresponding minimal size of the FLR (MSFLR) in healthy rats is approximately 10%6–8.
The healthy liver’s ability to recover lost mass, without jeopardizing the viability of the entire organism, is unique. Liver regeneration is the result of hepatocyte hypertrophy and hyperplasia9. Regeneration terminates rather precisely when the normal liver-to-body weight ratio (LBWR) is restored. Initiation, progression, and termination of liver regeneration are very complex processes involving a large number of pathways10. The overall regulator of these complex processes is often termed the hepatostat11–13. The most widely used model to examine liver regeneration is the 70% PH14,15, in which the regenerative process is described from the early moments until termination of the process. The kinetics of liver regeneration seem to be different in the MSFLR model. In a recent study, we found a delay in hepatocyte proliferation, when comparing 90% PH to 70% PH16. This finding is also in agreement with other previous studies17,18. This suppression or delay of hepatocyte proliferation in the early phase of MSFLR regeneration could be crucial in understanding PHLF.
Genome-wide expression analysis can be used to uncover molecules and pathways affected by different sizes of the PH. The main purpose of the present study was to investigate and compare the gene expression profiles in rats subjected to the increasing size of the PH [PH(30%), PH(70%), and PH(90%)]. In addition, we aimed to determine which genes and gene networks are regulated in order to identify which pathways the liver prioritizes depending on the extent of PH and consequently the size of the FLR.
MATERIALS AND METHODS
Experimental Design
In total, 30 healthy, 10-week-old, male Wistar rats (M&B Taconic, Eiby, Denmark) with a mean weight of 206 g (range: 188–227) were randomized into blocks of 5 according to the size of the PH: PH(30%) (n = 6), PH(70%) (n = 6), PH(90%) (n = 6), sham (laparotomy without PH) (n = 6), and baseline (no surgery) (n = 6). All rats (except baseline) were euthanized 24 h postoperatively [postoperative day (POD) 1].
The experimental protocol was approved by the Danish Animal Research Committee, Copenhagen, Denmark (License No. 2012-15-2934-00591). Animals received care in accordance with the Guide for Care and Use of Laboratory Animals published by the US National Institutes of Health19. The animals were housed in standard animal laboratories with the temperature maintained at 23°C, with an artificial 12-h light–dark cycle, and with free access to food and water. The rats were kept in the animal facilities until the end of the experiment.
Surgical Procedure
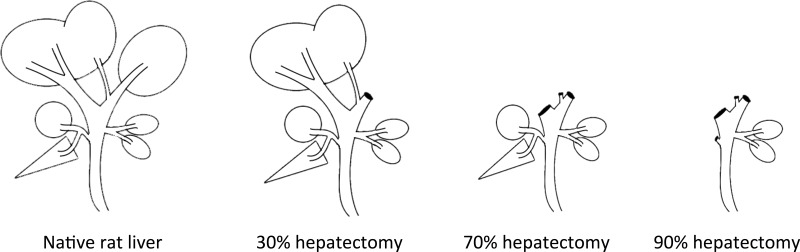
The surgical procedure was performed as described in a previous publication by this study group16. In brief, performing PH(30%), the left lateral lobe (LLL) was resected (Fig. 1). In PH(70%), the LLL and the median lobe (ML) were resected. In PH(90%), the right lobe (RL), ML, and LLL were all resected, leaving only the two caudate lobes [posterior caudate lobe (PCL) and anterior caudate lobe (ACL)] intact6.
Figure 1.
Surgical procedure.
After 24 h, the rats were reanesthetized, and a laparotomy was performed through the previous incision. Blood samples were collected from the heart by cannulation, immediately processed, and stored at −80°C until biochemical analyses. Animals were euthanized by cervical dislocation under anesthesia. The remnant liver was removed, and the ACL was snap frozen in liquid nitrogen and stored at −80°C until use.
Biochemical Analyses
Alanine aminotransferase (ALT), alkaline phosphatase (AP), haptoglobin (HG), and bilirubin (BR) levels were measured using Modular P (Roche Diagnostics, Mannheim, Germany).
The prothrombin/proconvertin ratio (PP) was measured using the automated coagulation analyzer Sysmex CS2100i (Sysmex©, Tokyo, Japan).
RNA Isolation
Each cryopreserved liver tissue was cut in a 2 × 2-mm piece and immediately transferred into 5-ml polypropylene tubes (BD, Franklin Lakes, NJ, USA) containing 200 μl of chilled homogenization solution and 4 μl of lysis buffer (Maxwell 16 LEV simply RNA; Promega, Fitchburg, MA, USA). While kept on ice, each tube was thoroughly mixed using a homogenizer before adding 200 μl of lysis buffer (Maxwell 16 LEV simply RNA; Promega) and vortexed at a maximum speed for 15 s before further processing according to the manufacturer’s protocol. The isolated RNA was stored at 80°C.
RNA quality was evaluated using the BioAnalyzer 21000 (Agilent Technologies, Santa Clara, CA, USA). Only samples with a RIN score >7 were used.
Microarray Analysis
Following the manufacturer’s instructions, the Ambion® WT Expression Kit (Thermo Fischer Scientific, Waltham, MA, USA) was used for labeling 100 ng of isolated RNA. Samples were hybridized to the Affymetrix GeneChip® Rat Gene 2.0 ST Array (Thermo Fischer Scientific) at 45°C and 60 rpm for 16 h and scanned with the Affymetrix GeneChip® Scanner 3000 7 G.
Quality control of the array data was performed in the Expression Console software (Affymetrix) with default RMA Gene analysis settings. Experimental data have been submitted to NBCI Gene Expression Omnibus under series record GSE97429 and will be available after March 31, 2018.
Gene Expression Analysis
Gene expression levels were analyzed by importing raw image files from the quantitative scanning into GeneSpring Version 13.0 (Agilent Technologies). Networks and functional analyses were generated through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA®; www.qiagen.com/ingenuity; QIAGEN; Redwood City, CA, USA).
Quantitative Real-Time-PCR Expression Analysis
Validation of microarray gene expression data was performed using quantitative real-time (qRT)-PCR expression analysis using LightCycler 480 Instrument 96-well (Roche Diagnostics, Hvidovre, Denmark). A total of 10 genes were selected for validation. Three of them were stably expressed housekeeping genes GAPDH, SDHA, and β-actin. Seven genes were selected because of different expression levels between groups (IL1rn, Lcn2, Tgfb1, Egf, Nr1I2, Gucy2c, and Por). Primer sequence details can be seen in Table 1. Analysis was performed using Multiwell Plate 96 Clear (Roche Diagnostics). All genes were analyzed in duplicates. From the isolated RNA, reverse transcription was performed in order to produce a suitable amount of cDNA. cDNA (2 μl) was mixed with 7.5 μl of SYBR Green l Master (Roche Diagnostics) and 3 μl of gene-specific primer mixture before PCR amplification according to the manufacturer’s protocol. The measured expression levels were normalized to the geometric mean of the three stable expressed genes.
Table 1.
Primer Sequence Design Used for PCR Analysis
| Interleukin-1rn | F: TGGTGCCTATTGACTTTCGGA |
| R: ACAGGGCTCTTTTGGTGTGT | |
| Lipocalin 2 | F: AGGCTTCTGGACCGAACG |
| R: TGTCTTTCTTTCTGGACCGCA | |
| Transforming growth factor-β1 | F: CGTCAGACATTCGGGAAGCA |
| R: CACTCAGGCGTATCAGTGGG | |
| Epidermal growth factor receptor 2 | F: GGAGGCCCCTTTGATCAGAT |
| R: GTCTGGTTTCTAGGCGCAGA | |
| Nuclear receptor subfamily 1 group I | F: GGCCGATGTGTCAACCTACA |
| R: GGTTCCTGTTTCCGTGTCGA | |
| Guanylate cyclase 2C | F: GAGGTGGTGGACATGCTGAAT |
| R: CGGTTGCCGTTTCTCATAGGC | |
| Cytochrome P450 oxidoreductase | F: AGGATTTCATCACGTGGAGG |
| R: CTCTTCAGACGGCCCATCTC |
A comparison of expression levels of the seven selected differently expressed genes is presented in Table 2. A concurrence was found between the two methods of measuring gene expression levels, even though the PCR measurements tended to vary more for some of the genes tested. Overall, the PCR data confirmed our microarray data.
Table 2.
Comparison of Expression Levels of the Seven Differently Expressed Genes
| Gene Name | Baseline (PCR/MA) | PH(30%) (PCR/MA) | PH(70%) (PCR/MA) | PH(90%) (PCR/MA) |
|---|---|---|---|---|
| Interleukin-1rn | −1.69/−1.72 | 1.00/1.29 | 1.36/1.26 | 2.50/2.44 |
| Lipocalin 2 | −60.63/−13.16 | 7.09/3.48 | 19.01/4.43 | 9.19/6.65 |
| Transforming growth factor-β1 | −1.01/−1.15 | 10.02/1.48 | 16.47/1.28 | −1.74/1.14 |
| Epidermal growth factor receptor | 1.05/−1.29 | 1.48/−1.28 | 1.38/−1.67 | 1.41/−3.43 |
| Nuclear receptor subfamily 1 group i member 2 | 1.02/−1.11 | −1.87/−1.41 | −2.00/−1.34 | −1.23/1.27 |
| Guanylate cyclase 2C | −7.89/1.12 | 17.03/1.03 | 33.74/1.45 | 81.00/8.23 |
| Cytochrome P450 oxidoreductase | −2.26/−1.69 | −1.15/1.05 | −1.39/1.05 | 1.29/1.85 |
Values indicate the mean fold change in expression levels compared to the sham group. PCR, polymerase chain reaction expression analysis; MA, microarray expression analysis.
Statistical Analyses
Statistical analyses for gene expression data were performed using GeneSpring Version 13. By one-way ANOVA, unsupervised gene data were groupwise tested against each other and were considered significantly different with a value of p < 0.001 (Benjamini–Hochberg corrected), and fold change (FC) of group mean expression levels was >2.0. Also, in GeneSpring, unsupervised cluster analysis was performed.
In IPA, the Benjamini–Hochberg method was used to determine significant subset from our unsupervised gene data imported from GeneSpring. Values of p < 0.05 were considered significant. FC was set to be 1.0 as the data had already been selected with an FC > 2.0 from GeneSpring. Specifically for upstream regulators, the overlap p value was calculated by Fisher’s exact test, measuring whether there was a statistically significant overlap between the dataset genes and the genes regulated by a transcriptional regulator. Values of p < 0.001 were considered significant.
Statistical analyses for body weight- and liver-specific parameters were tested with regression analysis using Stata v.13.1 (StataCorp, College Station, TX, USA) and presented as mean values given with a 95% confidence interval (CI). Values of p < 0.05 were considered significant.
RESULTS
Animals and Circulating Markers
During the time period of our study, no animals died or were euthanized prior to evaluation. No observation was made preoperatively with regard to difference in bodyweight between groups (mean: 206 g, CI: 202–210). Bodyweight decreased significantly for all intervention groups in the first 24 h after surgery (mean: 199 g, CI: 194–204). Liver regeneration was calculated as the regeneration rate (RR), which we have previously described. Twenty-four hours after the liver resection was performed, the mean fold increase in RR was 1.93 PH(90%), 0.75 PH(70%), and 0.19 PH(30%)16.
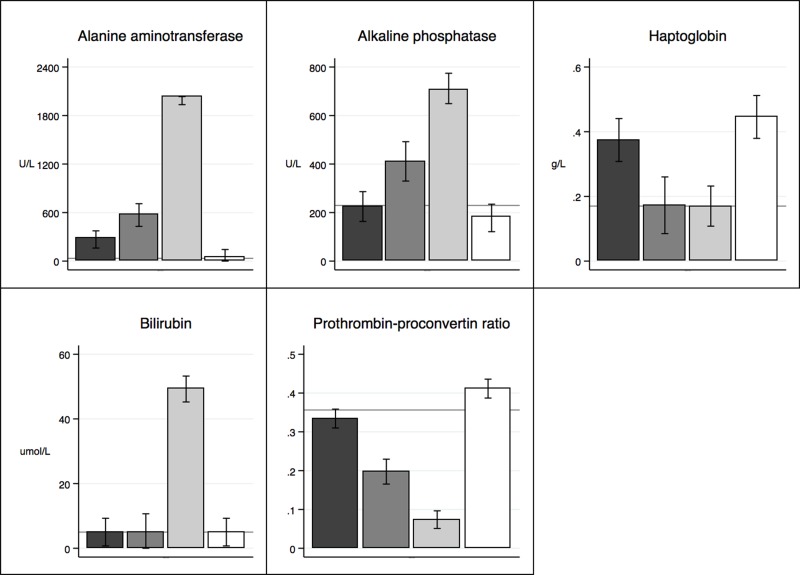
Liver-specific parameters are presented in Figure 2. In general, the group of PH(90%) showed significantly (p < 0.001) affected liver-specific parameters compared to all other groups with an increase in ALAT, AP, and BR and a decrease in PP. Compared with baseline, HG levels increased significantly for PH(30%) (p < 0.001) and sham (p < 0.001), while it remained steady for PH(70%) (p > 0.934) and PH(90%) (p ≥ 1.0).
Figure 2.
Liver-specific biochemistry: black bar, PH(30%); dark gray bar, PH(70%); light gray bar, PH(90%); white bar, sham; gray horizontal line, baseline.
Gene Expression
In total, microarray analysis revealed 29,489 genes. Data reduction of unsupervised gene data in GeneSpring (p < 0.001, FC > 2) reduced this number to 1,762 differentially expressed genes between groups, which were used for further analyses mentioned below.
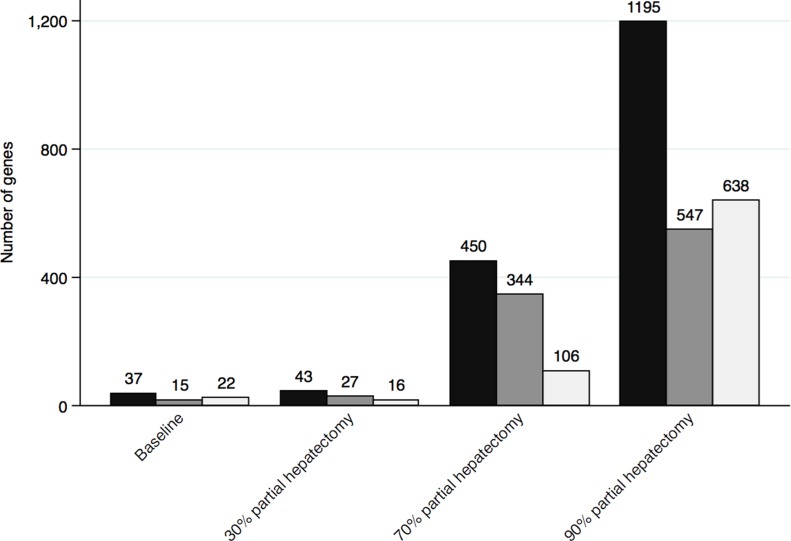
Comparing each intervention group to sham revealed a total of 1,471 varying genes and an increase in the number of differentially expressed genes with the extension of PH (Fig. 3).
Figure 3.
Number of genes significantly affected when comparing the sham group to all other groups. Black bar: total number of significantly affected genes; dark gray bar: number of genes significantly upregulated; light gray bar: number of genes significantly downregulated.
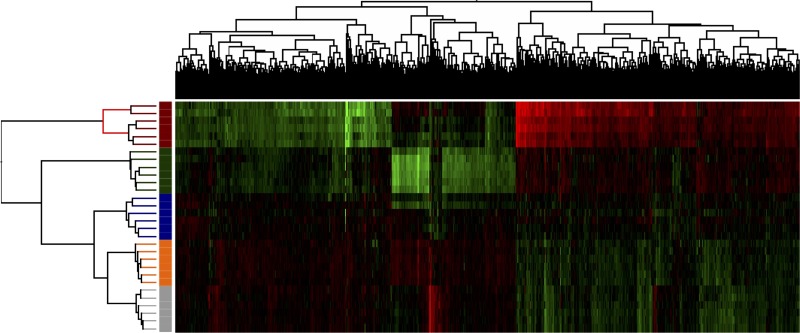
Figure 4 presents an unsupervised cluster analysis. From gene expression levels, groups were clustered as appearing in the study design most clearly distinguishing the PH(90%) group from the other groups.
Figure 4.
Cluster analysis (unsupervised) of the five groups: blue: PH(30%); green: PH(70%); red: PH(90%); orange: sham; gray: baseline. The range color of the analysis indicates the gene expression levels (red, decreased expression level; green, increased expression level).
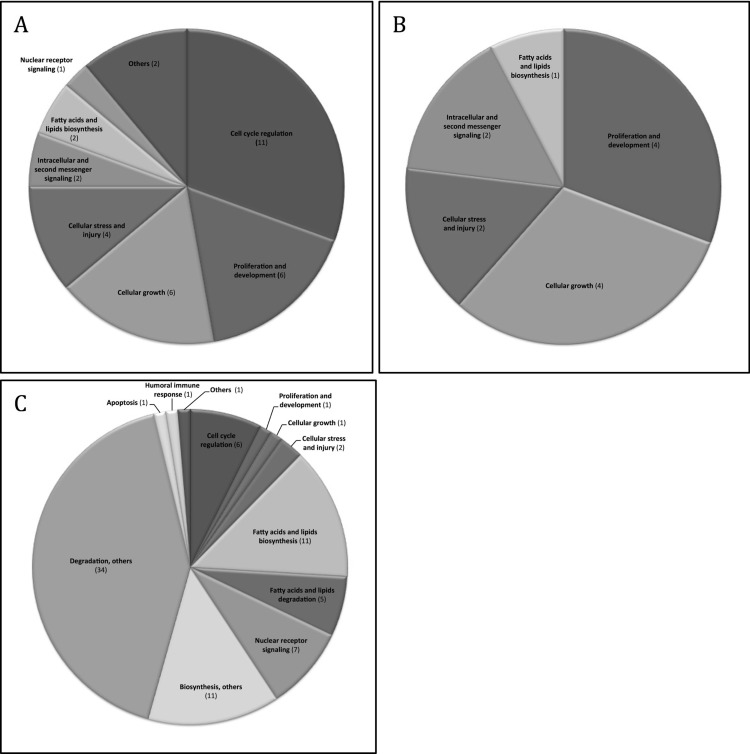
IPA was used to identify molecular pathways and upstream regulators within the differentially expressed genes. Analyzing the affected genes and comparing the PH(90%) group to the other groups, 20 molecular pathways were found to be upregulated. Of these, five pathways were significantly upregulated throughout the groups: eukaryotic initiation factor (eIFs)2 signaling, regulation of eIF4 and p70S6K, mammalian target of rapamycin (mTOR) signaling, integrin-linked kinase (ILK) signaling, and Acyl-CoA hydrolysis. Seventy-two molecular pathways were downregulated. Each pathway was associated with its signaling pathway category(ies) shown in the IPA report (Fig. 5A–C). Regarding the downregulated pathways and the categorization of these, no remarkable differences were found between groups. A pie chart of PH(90%) versus all groups is displayed in Figure 5C and is interpretable as a generalization for the overall downregulated pathway categories. Table 3 shows all the up- and downregulated pathways. Cascades of upstream transcriptional regulators explaining the observed gene expression changes and affected molecular pathways are presented in Table 4.
Figure 5.
Categorized Ingenuity Pathway Analysis (IPA) molecular pathways. The categorization was performed on the 20 upregulated pathways and 72 downregulated pathways found when comparing PH(90%) to all other groups. (A) Upregulated categorized pathways: PH(90%) versus all groups (n = 20). (B) Upregulated categorized pathways: PH(90%) versus PH(70%) (n = 5). (C) Downregulated categorized pathways: PH(90%) versus all groups (n = 72). The pie charts describe signaling pathway categories associated with the pathways found in IPA. The number in parentheses refers to the number of pathways clustered in the category. Several pathways are represented in more than one category. The IPA report (“Signaling Pathway Categories”) for each pathway was used for categorization. Some categories were accumulated as follows: Others: cancer, disease-specific pathway; Degradation, others: hormone degradation, amino acid degradation, xenobiotic metabolism, detoxification, degradation other categories; Biosynthesis, others: hormone biosynthesis, vitamin biosynthesis, biosynthesis other categories.
Table 3.
Upregulated and Downregulated Pathways of Differentially Expressed Genes
| Upregulated IPA Pathways: PH(90%) Versus All Other Groups (n = 20) |
| 1. EIF2 signaling |
| 2. Cell cycle: G2/M DNA damage checkpoint regulation |
| 3. GADD45 signaling |
| 4. Mitotic roles of Polo-like kinase |
| 5. Regulation of eIF4 and p70S6K |
| 6. Cell cycle control of chromosomal replication |
| 7. mTOR signaling |
| 8. Estrogen-mediated S-phase entry |
| 9. Cyclins and cell cycle regulation |
| 10. Role of CHK proteins in cell cycle checkpoint control |
| 11. Hereditary breast cancer signaling |
| 12. RAN signaling |
| 13. ILK signaling |
| 14. ATM signaling |
| 15. DNA damage-induced 14-3-3o signaling |
| 16. Cell cycle: G1/S checkpoint regulation |
| 17. Stearate biosynthesis I |
| 18. Acyl-CoA hydrolysis |
| 19. Cell cycle regulation by BTG family proteins |
| 20. Breast cancer regulation by stathmin 1 |
| Downregulated IPA Pathways: PH(90%) Versus All Other Groups (n = 72) |
| 1. Acetone degradation I (to methylglyoxal) |
| 2. Adenosine nucleotide degradation II |
| 3. Androgen biosynthesis |
| 4. Aryl hydrocarbon signaling |
| 5. Bile acid biosynthesis, neutral pathway |
| 6. Bupropion degradation |
| 7. Cell cycle control of chromosomal replication |
| 8. Cell cycle: G2/M DNA damage checkpoint regulation |
| 9. Cholesterol biosynthesis I |
| 10. Cholesterol biosynthesis II (via 24,25-dihydrolanosterol) |
| 11. Cholesterol biosynthesis III (via desmosterol) |
| 12. Citrulline biosynthesis |
| 13. Complement system |
| 14. DNA damage-induced 14-3-3o signaling |
| 15. Dopamine degradation |
| 16. Estrogen biosynthesis |
| 17. Estrogen-mediated S-phase entry |
| 18. Ethanol degradation II |
| 19. Ethanol degradation IV |
| 20. Fatty acid activation |
| 21. Fatty acid α-oxidation |
| 22. Fatty acid β-oxidation I |
| 23. FXR/RXR activation |
| 24. γ-Glutamyl cycle |
| 25. γ-Linolenate biosynthesis II |
| 26. Glucocorticoid biosynthesis |
| 27. Glutaryl-CoA degradation |
| 28. Glutathione-mediated detoxification |
| 29. Glycine betaine degradation |
| 30. Guanosine nucleotides degradation III |
| 31. Histamine degradation |
| 32. Leucine degradation |
| 33. LPS/IL-1-mediated inhibition of RXR function |
| 34. LXR/RXR activation |
| 35. Melatonin degradation I |
| 36. Methylglyoxal degradation III |
| 37. Mineralocorticoid biosynthesis |
| 38. Mitochondrial l-carnitine shuttle pathway |
| 39. Mitotic roles of Polo-like kinase |
| 40. Molybdenum cofactor biosynthesis |
| 41. NAD biosynthesis II (from tryptophan) |
| 42. Nicotine degradation II |
| 43. Nicotine degradation III |
| 44. Noradrenaline and adrenaline degradation |
| 45. NRF2-mediated oxidative stress response |
| 46. Oleate biosynthesis II |
| 47. Oxidative ethanol degradation III |
| 48. Proline degradation |
| 49. Putrescine degradation III |
| 50. PXR/RXR activation |
| 51. Retinol biosynthesis |
| 52. Role of BRCA1 in DNA damage response |
| 53. Serotonin degradation |
| 54. Stearate biosynthesis I |
| 55. Sucrose degradation |
| 56. Superoxide radicals degradation |
| 57. Superpathway of cholesterol biosynthesis |
| 58. Superpathway of melatonin degradation |
| 59. Superpathway of methionine degradation |
| 60. Taurine biosynthesis |
| 61. The visual cycle |
| 62. Thymine degradation |
| 63. Thyroid hormone metabolism II (via conjugation and/or degradation) |
| 64. TR/RXR activation |
| 65. Triacylglycerol degradation |
| 66. Tryptophan degradation III |
| 67. Tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde |
| 68. Tryptophan degradation X (mammalian, via tryptamine) |
| 69. Uracil degradation II (reductive) |
| 70. Urate biosynthesis/inosine 5*-phosphate degradation |
| 71. Xenobiotic metabolism signaling |
| 72. Zymosterol biosynthesis |
Table 4.
Cascades of Upstream Transcriptional Regulators Explaining the Observed Gene Expression Changes and Affected Molecular Pathways
| IPA Top Upstream Regulators | p Value | Name | Molecule Type (IPA) | Role in Cell (IPA Report) | |||
|---|---|---|---|---|---|---|---|
| 30% PH | 70% PH | Sham | Baseline | ||||
| Activated | |||||||
| MYC | S | S | S | S | V-Myc avian myelocytomatosis viral oncogene homolog | Transcription regulator | Cell cycle progression; apoptosis; cellular transformation |
| MYCN | S | S | S | S | V-Myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog | Transcription regulator | Tissue development |
| HGF | S | S | S | S | Hepatocyte growth factor | Growth factor | Organ regeneration; wound healing |
| FOS | S | S | S | S | FOS proto-oncogene | Transcription regulator | Cell proliferation; differentiation; apoptosis; transformation |
| TGFB1 | S | NSD | S | S | Transforming growth factor-β1 | Kinase | Cell growth; cell differentiation; apoptosis |
| F2 | S | S | S | S | Coagulation factor II; thrombin | Peptidase | Coagulation |
| POR | S | S | NSD | NSD | Cytochrome p450 oxidoreductase | Enzyme | – |
| Inhibited | |||||||
| PPARA | S | S | S | S | Peroxisome proliferator-activated receptor α | Ligand-dependent nuclear receptor | Proliferation; abnormal morphology |
| NR1I2 | S | S | S | S | Nuclear receptor subfamily 1 group I member 1 | Ligand-dependent nuclear receptor | Cell death; morphology; phosphorylation; migration; proliferation; contact growth inhibitor; apoptosis |
| HNF1A | S | S | S | S | Hepatocyte nuclear factor 1 α | Transcription regulator | Cell growth; cell differentiation |
| RXRA | S | S | S | S | Retinoid X receptor α | Ligand-dependent nuclear receptor | – |
| SCAP | NSD | NSD | S | S | Sterol regulatory element-binding protein (SREBF) chaperone | Other | – |
The goal of the IPA Upstream Regulator analytic is to identify the cascade of upstream transcriptional regulators that can explain the observed gene expression changes in a user’s dataset, which can help illuminate the biological activities occurring in the tissues or cells being studied (http://pages.ingenuity.com/rs/ingenuity/images/0812%20upstream_regulator_analysis_whitepaper.pdf).
The p value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcriptional regulator. The purpose of the overlap p value is to identify transcriptional regulators that are able to explain observed gene expression changes. The overlap p value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcriptional regulator (TR). It is calculated using Fisher’s exact test, and significance is generally attributed to p < 0.01 (http://pages.ingenuity.com/rs/ingenuity/images/0812%20upstream_regulator_analysis_whitepaper.pdf). S, significant difference, p < 0.001; NSD, no significant difference, p > 0.001.
DISCUSSION
In the present study, we investigated and compared the gene expression profiles in rats subjected to increasing size of PH approaching the lower limit of the FLR. We demonstrated that the gene expression profile was significantly affected by increasing PH. Overall, we found that hepatocytes tended to express genes involved in cell proliferation and differentiation at the expense of genes involved in metabolism and thereby body homeostasis with decreasing FLR.
The bodyweight for all intervention groups was significantly lower at euthanasia compared to the bodyweight preoperatively, with a significant decrease in the resected compared to the sham group. As a change in bodyweight is a reliable marker of acute stress in rats20, this finding indicates that PH induces a higher level of surgical stress than the sham operation.
We observed a significant increase in ALT, AP, and BR being proportional to the amount of liver parenchyma resected. As no ischemic liver parenchyma could explain this rise in liver parameters, a plausible explanation could be that the increase in portal blood flow to the FLR was proportional to the amount of liver tissue resected. This portal hyperperfusion will cause intravascular shear stress, and an associated HABR (hepatic arterial buffer response) induces constriction of the hepatic artery21. Enzyme leakage and PHLF seems to be linked to this phenomenon. PP was as expected, adversely affected by increasing size of the PH. We interpret this as a decrease in the FLR’s synthetic capacity being inversely related to its size. The same pattern was found for HG, which was elevated in rats sustained to PH(30%) and sham operation compared to baseline, PH(70%), and PH(90%). HG is an acute phase reactant that is synthesized by hepatocytes and plays a key role in the innate immune system22. This finding is probably a consequence of an initially nonresponsive synthetic capacity of the FLR.
Fausto divided the gene expression after PH in rats into four phases depending on time after surgery (immediate-early genes: 0–4 h post-PH, delayed-early genes: 4–8 h post-PH, cell cycle genes: 8–20 h post-PH, and DNA replication and mitosis: 20–48 h post-PH)23. We analyzed the rat liver tissue at POD1 (24 h), and the gene expression profiles found are compatible with the category of cell cycle genes and genes of DNA replication and mitosis.
A comparative analysis with the sham group as a reference showed a significant increase in the number of genes significantly affected with increasing extent of PH (Fig. 3). A subsequent cluster analysis (Fig. 4) divided these genes into two main clusters: one containing the PH(90%) group and one containing all the other groups. This leads us to conclude that extended PH(90%) affects genes significantly different than minor PHs. Also, the cluster analysis revealed a subdivision of the main cluster analysis group containing all other groups than PH(90%) with a differentiation into groups based on the type of intervention. Interestingly, the influence of PH(70%) on gene expression seems to be more similar to PH(30%), sham, and baseline than the extended PH(90%). We have previously shown that the extended PH(90%) in rats leads to higher mortality than minor PHs16, and hypothesized that the early postoperative period after PH(90%) is critical with regard to PHLF and survival. In this study, we show that there are observable and significant differences in the genetic expression between the extended PH(90%) and minor PHs in this period. The key to the sufficient regeneration of the MSFLR parallel to the continuous maintenance of liver homeostasis and survival could be a consequence in these genetic differences.
These observations prompted us to identify pathways that the liver prioritizes depending on the extent of PH. Our frame of reference was the PH(90%) group. Seventy-two pathways were downregulated in the PH(90%) group compared to all the other groups. Degradation and biosynthesis were the pathway categories mainly found to be downregulated, thus indicating how the hepatostat prioritizes when the FLR approaches the lower limit.
Twenty pathways were significantly upregulated comparing PH(90%) with all other groups. The main pathway categories were found to be cell cycle regulation, proliferation and development, cellular growth, and cellular stress and injury (Fig. 5A). When comparing PH(90%) only to PH(70%), proliferation and development, cellular growth, and cellular stress and injury still appeared as significantly upregulated pathway categories, whereas cell cycle regulation is no longer significantly differently affected (Fig. 5B).
When cell cycle is active, the regulation may be independent of the size of PH. Only when PH exceeds 30% is regeneration achieved by proliferation; until then, the regeneration depends on hypertrophy only9,16, and no cell cycle regulation is needed. On the contrary, proliferation and cellular growth seem to be alterable with the size of PH. As expected, cellular stress and injury are genetically largely expressed in the extended PH(90%) group, indicating that this extended surgical procedure is followed by a very critical phase determining survival or PHLF.
The main upregulated pathways in the PH(90%) group compared to all the other groups were eIF2 signaling, regulation of eIF4 and p70S6K, mTOR signaling, ILK signaling, and Acyl-CoA hydrolysis. The mTOR kinase is the central modulator of proliferative signal transduction. Its activation triggers a phosphorylation cascade inducing cell growth and proliferation24. Included in the mTOR axis are the eIFs and the p70s6K24,25; they are also upregulated in the PH(90%) group. Contrary to our present and previous16 results, Zhang et al.17 performed the extended PH(90%) and found suppressed regeneration of the FLR, mTOR downregulation, and high mortality. This could indicate that the expression of mTOR might be crucial to the balancing of regeneration and survival or PHLF and death.
ILK is involved in the transmission of extracellular matrix signals through integrins and is thought to have a major influence on the hepatostat10. If ILK is eliminated, the FLR exceeds the original liver size, indicating a defect in the termination process26. Findings show that ILK is a suppressor of hepatocyte growth, and the expression gradually increases toward the end of regeneration26. At first glance, it seems to be a paradox that ILK is significantly upregulated in the PH(90%) group compared to all the other groups. An explanation may be that the hepatostat dampens excess proliferation, which may take place at the cost of metabolism and homeostasis in an attempt to avoid PHLF. This theory is supported by a previous study we carried out on rats subjected to PH(30%), PH(70%), and PH(90%)16. In that study, we demonstrated an initially very low number of proliferating hepatocytes in the PH(90%) group compared to the PH(70%) group. However, this is followed by a burst in hepatocyte proliferation in the PH(90%) group, surpassing the levels seen after PH(70%) at any time point.
Based on our genetic dataset, we identified cascades of upstream transcriptional regulators (Table 4). These regulators indicate what may have been happening during the regenerative process prior to the genetic expression levels found in our dataset. Again, we chose our frame of reference to be PH(90%). Six of the seven upregulated activated genes were involved in cell proliferation, cell differentiation, and tissue development. The upregulated activation of hepatocyte growth factor (HGF) and transforming growth factor-β (TGF-β) was of special interest. The expression of immediate and delayed early genes in liver regeneration does not lead to DNA replication unless the cells can progress through the cell cycle. This progression is accomplished by growth factors such as HGF and TGF23. Collectively, HGF and epidermal growth factor (EGF) have a stronger proliferative effect on hepatocytes in culture than any other mitogen10. TGF-β was significantly activated in the PH(90%) group compared to the baseline, sham, and the PH(30%) groups, but not compared to the PH(70%) group. TGF-β has been shown to play an important role in the formation of extracellular matrix and vessels in the regenerating liver, including the sinusoids27,28.
Of the five inhibited upstream regulators, hepatocyte nuclear factor 1-α (HFN1-α) is of special interest. HNF1-α regulates the transcription of genes into proteins that among other things are involved in glucose, cholesterol, and fatty acid transport, as well as metabolism29,30. Our IPA analysis further supports the theory that livers sustained to extended PH, close to the lower limit of the FLR, prioritize genes involved in cell proliferation and differentiation at the expense of genes involved in cellular homeostasis.
PH of the rat liver is a widely used model investigating liver regeneration. Despite obvious differences in rat liver size and anatomy7 compared to the human liver, and the fact that rat liver metabolism is more rapid31, it has been shown that the regulation of liver regeneration in rats and humans underlies the same principles32,33. Using the rat for experimental studies makes it possible to include a higher number of homogenous animals to the experimental groups contributing to significant data.
In previous studies, we have demonstrated that the FLR after PH(70%)16,34 and PH(90%)16 is almost fully regenerated (by measure of LBWR and hepatocyte proliferation) at POD5. However, with a delay in the initiation and peak of hepatocyte proliferation in the PH(90%) group at POD1, this led us to evaluate the gene expression on POD1.
Hepatocytes are the major cells in the liver constituting approximately 80% of its mass35. Other cells are cholangiocytes, Kupffer cells, stellate cells, blood cells, and liver sinusoidal cells. In the present study, gene analysis was performed on whole-liver biopsies, and as such, all other cells in the liver other than hepatocytes have contributed to the gene expression profiles we present. We believe that the contribution from other cells has been minor; regardless it was impossible for us to singularly isolate hepatocytes for analysis.
In conclusion, we demonstrated that the hepatostat’s influence on gene expression was pronouncedly affected by the size of the PH and thereby the size of the FLR. With decreasing size of the FLR, the hepatosat tended to prioritize the expression of genes involved in cell proliferation and differentiation at the expense of genes involved in metabolism and body homeostasis. This prioritization may be a two-edged sword (i.e., cell proliferation and differentiation seem to be rational after extended PHs). However, should this take place at the expense of metabolism and body homeostasis, it may be the genetic explanation for PHLF. Future research should focus on ways to ensure that hepatocytes regenerate while at the same time preserving their metabolic and homeostatic functions to decrease the risk of PHLF.
ACKNOWLEDGMENTS
The authors thank the Department of Molecular Medicine at Aarhus University Hospital, Kasper Thorsen, and Klaus Nielsen for their excellent technical assistance. The “Brødrene Hartmann Foundation” and “Tømrermester Jørgen Holm og hustru Elisa F. Hansen’s Memorial Fund” are thankfully acknowledged for their financial support. Author contributions: M.M. (study design, surgical procedures, data collection, data analysis, and manuscript writing), A.R.K. (study design, surgical procedure consultancy, data analysis, and manuscript writing), N.C.B. (study design, data analysis, and manuscript writing), K.J.A. (study design, surgical procedure consultancy, and drafting of manuscript), U.B.J. (data analysis and genetic consultancy), F.V.M. (study design, data analysis, and manuscript writing). All the authors have read, commented, and approved the final manuscript. The authors declare no conflicts of interest.
REFERENCES
- 1. Abdalla EK, Bauer TW, Chun YS, D’Angelica M, Kooby DA, Jarnagin WR. Locoregional surgical and interventional therapies for advanced colorectal cancer liver metastases: Expert consensus statements. HPB (Oxford) 2013;15(2):119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tejeda-Maldonado J, García-Juárez I, Aguirre-Valadez J, González-Aguirre A, Vilatobá-Chapa M, Armengol-Alonso A, Escobar-Penagos F, Torre A, Sánchez-Ávila JF, Carrillo-Pérez DL. Diagnosis and treatment of hepatocellular carcinoma: An update. World J Hepatol. 2015;7(3):362–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Greef K, Rolfo C, Russo A, Chapelle T, Bronte G, Passiglia F, Coelho A, Papadimitriou K, Peeters M. Multisciplinary management of patients with liver metastasis from colorectal cancer. World J Hepatol. 2016;22(32):7215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guglielmi A, Ruzzenente A, Conci S, Valdegamberi A, Iacono C. How much remnant is enough in liver resection? Dig Surg. 2012;29(1):6–17. [DOI] [PubMed] [Google Scholar]
- 5. Kauffmann R, Fong Y. Post-hepatectomy liver failure. Hepatobiliary Surg Nutr. 2014;3(5):238–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Madrahimov N, Dirsch O, Broelsch C, Dahmen U. Marginal hepatectomy in the rat. Ann Surg. 2006;244(1):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martins PNA, Theruvath TP, Neuhaus P. Rodent models of partial hepatectomies. Liver Int. 2008;28(1):3–11. [DOI] [PubMed] [Google Scholar]
- 8. Kubota T, Takabe K, Yang M, Sekido H, Endo I, Ichikawa Y. Minimum sizes for remnant and transplanted livers in rats. J Hepatobiliary Pancreat Surg. 1997;4:398–404. [Google Scholar]
- 9. Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22(13):1166–75. [DOI] [PubMed] [Google Scholar]
- 10. Michalopoulos GK. Advances in liver regeneration. Expert Rev Gastroenterol Hepatol. 2014;8(8):897–907. [DOI] [PubMed] [Google Scholar]
- 11. Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michalopoulos GK. Liver regeneration after partial hepatectomy: Critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3(1):485–513. [DOI] [PubMed] [Google Scholar]
- 14. Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–92. [Google Scholar]
- 15. Forbes SJ, Newsome PN. Liver regeneration—Mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol. 2016;13(8):473–85. [DOI] [PubMed] [Google Scholar]
- 16. Meier M, Andersen KJ, Knudsen AR, Nyengaard JR, Hamilton-Dutoit S, Mortensen FV. Liver regeneration is dependent on the extent of hepatectomy. J Surg Res. 2016;205(1):76–84. [DOI] [PubMed] [Google Scholar]
- 17. Zhang DX, Li CH, Zhang AQ, Jiang S, Lai YH, Ge XL, Pan K, Dong JH. mTOR-dependent suppression of remnant liver regeneration in liver failure after massive liver resection in rats. Dig Dis Sci. 2015;60(9):2718–29. [DOI] [PubMed] [Google Scholar]
- 18. Sakaguchi K, Takeuchi E, Suzuki M, Oda K, Nagino M, Nimura Y, Yoshida S. DNA polymerases and Ki-67 nuclear antigen are induced in correlation with the resected mass of rat liver up to 90%. Langenbecks Arch Surg. 2000;385(2):135–42. [DOI] [PubMed] [Google Scholar]
- 19. National Research Council. Guide for the care and use of laboratory animals, 8th ed. Washington (DC): National Academies Press; 2010. [Google Scholar]
- 20. Martí O, Martí J, Armario A. Effects of chronic stress on food intake in rats: Influence of stressor intensity and duration of daily exposure. Physiol Behav. 1994;55(4):747–53. [DOI] [PubMed] [Google Scholar]
- 21. Eipel C. Regulation of hepatic blood flow: The hepatic arterial buffer response revisited. World J Gastroenterol. 2010;16(48):6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cray C, Zaias J, Altman NH. Acute phase response in animals: A review. Comp Med. 2009;59(6):517–26. [PMC free article] [PubMed] [Google Scholar]
- 23. Fausto N. Liver regeneration. J Hepatobiliary Pancreat Surg. 2000;32(1 Suppl):19–31. [DOI] [PubMed] [Google Scholar]
- 24. Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(20):3589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. [DOI] [PubMed] [Google Scholar]
- 26. Apte U, Gkretsi V, Bowen WC, Mars WM, Luo J-H, Donthamsetty S, Orr A, Monga SPS, Wu C, Michalopoulos GK. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology 2009;50(3):844–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pepper MS, Vassalli JD, Orci L, Montesano R. Biphasic effect of transforming growth factor-beta 1 on in vitro angiogenesis. Exp Cell Res. 1993;204(2):356–63. [DOI] [PubMed] [Google Scholar]
- 28. Roberts AB, McCune BK, Sporn MB. TGF-beta: Regulation of extracellular matrix. Kidney Int. 1992;41:557–9. [DOI] [PubMed] [Google Scholar]
- 29. Costa RH, Kalinichenko VV, Holterman A-XL, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology 2003;38(6):1331–47. [DOI] [PubMed] [Google Scholar]
- 30. Mitchell SMS, Frayling TM. The role of transcription factors in maturity-onset diabetes of the young. Mol Genet Metab. 2002;77(1–2):35–43. [DOI] [PubMed] [Google Scholar]
- 31. Martignoni M, Groothuis GMM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opinion Drug Metab Toxicol. 2006;2(6):875–94. [DOI] [PubMed] [Google Scholar]
- 32. Helling TS. Liver failure following partial hepatectomy. HPB (Oxford) 2006;8(3):165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fausto N. Liver regeneration. In The liver: Biology and pathology, 4th ed. Philadelphia (PA): Lippincott, Williams & Wilkins; 2001. p. 591–610. [Google Scholar]
- 34. Andersen KJ, Knudsen AR, Kannerup A-S, Sasanuma H, Nyengaard JR, Hamilton-Dutoit S, Erlandsen EJ, Jørgensen B, Mortensen FV. The natural history of liver regeneration in rats: Description of an animal model for liver regeneration studies. Int J Surg. 2013;11(9):903–8. [DOI] [PubMed] [Google Scholar]
- 35. Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120(1):117–30. [DOI] [PubMed] [Google Scholar]