Key Points
Question
Does continuous blockade of EGFR (epidermal growth factor receptor) by Sym004 (a mixture of futuximab and modotuximab) lead to a survival benefit in patients with anti-EGFR refractory metastatic colorectal cancer?
Findings
In this randomized clinical trial that included 254 patients, Sym004 did not improve survival compared with investigators’ treatments of choice in the intent-to-treat population. Preplanned circulating tumor DNA biomarker profiling captured high intrapatient heterogeneity and identified a Sym004-sensitive subpopulation with clinically meaningful improvement of overall survival.
Meaning
These findings provide the rationale for a prospective clinical validation of Sym004 efficacy in a molecularly defined subgroup of patients with acquired resistance to anti-EGFR therapy.
Abstract
Importance
Acquired resistance to anti-EGFR therapy (epidermal growth factor receptor) is frequently due to RAS and EGFR extracellular domain (ECD) mutations in metastatic colorectal cancer (mCRC). Some anti-EGFR–refractory patients retain tumor EGFR dependency potentially targetable by agents such as Sym004, which is a mixture of 2 nonoverlapping monoclonal antibodies targeting EGFR.
Objective
To determine if continuous blockade of EGFR by Sym004 has survival benefit.
Design, Setting, and Participants
Multicenter, phase 2, randomized, clinical trial comparing 2 regimens of Sym004 with investigator’s choice from March 6, 2014, through October 15, 2015. Circulating tumor DNA (ctDNA) was analyzed for biomarker and tracking clonal dynamics during treatment. Participants had wild-type KRAS exon 2 mCRC refractory to standard chemotherapy and acquired resistance to anti-EGFR monoclonal antibodies.
Interventions
Participants were randomly assigned in a 1:1:1 ratio to Sym004, 12 mg/kg/wk (arm A), Sym004, 9 mg/kg loading dose followed by 6 mg/kg/wk (arm B), or investigator’s choice of treatment (arm C).
Main Outcomes and Measures
Overall survival (OS). Secondary end points included preplanned exploratory biomarker analysis in ctDNA.
Results
A total of 254 patients were randomized (intent-to-treat [ITT] population) (median age, 63 [range, 34-91] years; 63% male; n = 160). Median OS in the ITT population was 7.9 months (95% CI, 6.5-9.9 months), 10.3 months (95% CI, 9.0-12.9 months), and 9.6 months (95% CI, 8.3-12.2 months) for arms A, B, and C, respectively (hazard ratio [HR], 1.31; 95% CI, 0.92-1.87 for A vs C; and HR, 0.97; 95% CI, 0.68-1.40 for B vs C). The ctDNA revealed high intrapatient genomic heterogeneity following anti-EGFR therapy. Sym004 effectively targeted EGFR ECD-mutated cancer cells, and a decrease in EGFR ECD ctDNA occurred in Sym004-treated patients. However, this did not translate into clinical benefit in patients with EGFR ECD mutations, likely owing to co-occurring resistance mechanisms. A subgroup of patients was defined by ctDNA (RAS/BRAF/EGFR ECD-mutation negative) associated with improved OS in Sym004-treated patients in arm B compared with arm C (median OS, 12.8 and 7.3 months, respectively).
Conclusions and Relevance
Sym004 did not improve OS in an unselected population of patients with mCRC and acquired anti-EGFR resistance. A prospective clinical validation of Sym004 efficacy in a ctDNA molecularly defined subgroup of patients with refractory mCRC is warranted.
Trial Registration
clinicaltrialsregister.eu Identifier: 2013-003829-29
This phase 2 randomized clinical trial evaluates the efficacy and safety of Sym004 (a mixture of futuximab and modotuximab) for the treatment of refractory metastatic colorectal cancer with acquired resistance to anti-EGFR therapy.
Introduction
Panitumumab and cetuximab are 2 anti-EGFR (epidermal growth factor receptor) monoclonal antibodies (mAbs) approved for treatment of RAS wild-type (WT) metastatic colorectal cancer (mCRC). However, response is transient due to the emergence of acquired resistance. Our research group and others previously elucidated the molecular mechanisms responsible for treatment progression to anti-EGFR mAbs. Alterations in components of the RAS signaling pathway, together with mutations in the extracellular domain (ECD) of the EGFR gene (OMIM 131550), are the most common mechanisms of acquired resistance to EGFR blockade in mCRC. In addition, a specific mutation of the BRAF gene at amino acid position 600 (BRAF V600E) has been documented to be associated with poor prognosis in patients with mCRC.
A potential approach to overcome acquired resistance to approved anti-EGFR mAbs is treatment with mixtures of mAbs targeting nonoverlapping epitopes of EGFR. Sym004 is a mixture of 2 mAbs, futuximab and modotuximab, that bind to nonoverlapping epitopes in EGFR ECD III. Sym004 has been extensively evaluated in preclinical models and in early clinical development. Binding of the Sym004 mAbs to EGFR leads to highly efficient receptor internalization and degradation, which in turn leads to profound inhibition of cancer cell growth. This novel synergistic mechanism of EGFR elimination results in more effective blockade of EGFR signaling pathways and higher antitumor activity than that observed with single mAbs.
In a recent phase 1 study, Sym004 was found to be well tolerated at doses up to 12 mg/kg/wk, with grade 3 skin toxic effects and hypomagnesemia as mechanism-based dose-limiting toxic effects. Notably, Sym004 showed early signs of clinical activity in an expansion cohort of 39 anti-EGFR antibody pretreated patients with mCRC. Tumor shrinkage was observed in 44% of patients, and 13% achieved partial responses, providing a clear rationale for further exploring the activity of Sym004 in mCRC.
In the present study, we report safety and efficacy data from treatment with 2 dose regimens of Sym004 or with investigator’s choice (IC) of chemotherapy or best supportive care (BSC) in a randomized phase 2 clinical trial in chemorefractory patients with mCRC and acquired resistance to approved anti-EGFR mAbs. As part of the planned biomarker analysis, tumor circulating tumor DNA (ctDNA) assessment was used to identify a biomarker-defined patient population that could obtain significant clinical benefit from Sym004 treatment.
Methods
Study Design and Participants
Sym004-05 was a multicenter, phase 2, randomized clinical trial in patients with treatment-refractory mCRC and acquired resistance to therapy with anti-EGFR mAbs. The trial protocol is available at Supplement 1. The study was reviewed and approved by the ethics committee of all participating institutions. Informed consent was obtained from all study participants.
Patients with histologically or cytologically confirmed mCRC that was exon 2 KRAS WT at the time of initial diagnosis and who gave written informed consent were screened for enrollment to this trial. Included patients were required to have prior intolerance to or failure of standard chemotherapy regimens including fluorouracil, oxaliplatin, and irinotecan, and were allowed to have received bevacizumab or ziv-aflibercept. Prior therapy with regorafenib was not permitted. In addition, all included patients had acquired resistance to prior therapy with a marketed anti-EGFR mAb, as defined by having achieved either an objective partial response (PR), complete response (CR), or stable disease for more than 16 weeks followed by documented progressive disease (PD) during or within 6 months of completion of this therapy. Included patients were required to have measurable disease according to Response Evaluation Criteria in Solid Tumors (RECIST), and an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1.
Patients were randomly assigned in a 1:1:1 ratio to 1 of 3 treatment arms, 1 of 2 dose regimens of Sym004, 12 mg/kg/wk (arm A) or 9 mg/kg loading dose followed by 6 mg/kg/wk (arm B) compared with a control group, which included IC of capecitabine, fluorouracil, or BSC (arm C). Treatment continued until radiographically confirmed disease progression on standard imaging, unacceptable toxic effects, death, or the patient or physician decided to stop. Doses could be modified to manage treatment-related toxic effects.
Overall survival (OS), defined as time from randomization to date of death or censored at last day of contact, was the primary efficacy end point. Secondary end points included safety; progression-free survival (PFS), defined as time from randomization to date of disease progression or death from any cause; response rate; and exploratory biomarker analysis.
To establish a robust estimate of Sym004 survival benefit, a minimum of 240 patients were to be randomized based on 80% statistical power to differentiate assumed median OS of 6 months for arm C group and 9.2 months for arm A or B, with a 2-sided level of significance of P = .121. The primary analysis was to be performed when at least 181 events (deaths) had been reported or 12 months after the last patient was randomized in the trial, whichever occurred later. The data cutoff was October 24, 2016. We used the Kaplan-Meier method to generate curves for OS and PFS. Hazard ratios (HRs) for treatment effects were estimated with an unstratified Cox proportional hazards model.
Baseline ctDNA profiles (Guardant360, version 2.9; Guardant Health) were obtained from blood samples collected from patients in the trial (eFigure 1 in Supplement 2). Serial samples obtained prior to and after 3 weeks of treatment were also analyzed for EGFR ECD mutation dynamics. The ctDNA was amplified using droplet digital polymerase chain reaction (ddPCR) Supermix for Probes with EGFR ECD mutation assays (Bio-Rad). This ddPCR was then performed according to the manufacturer’s protocol. We performed 3 independent ddPCR experiments for each of the point mutations assessed and for each longitudinal time point. Fractional abundances (%) were calculated as follows: fractional abundance = (number of mutations events/[number of mutation + WT events]) × 100 and were Poisson corrected by QuantaSoft analysis software (BioRad).
Results
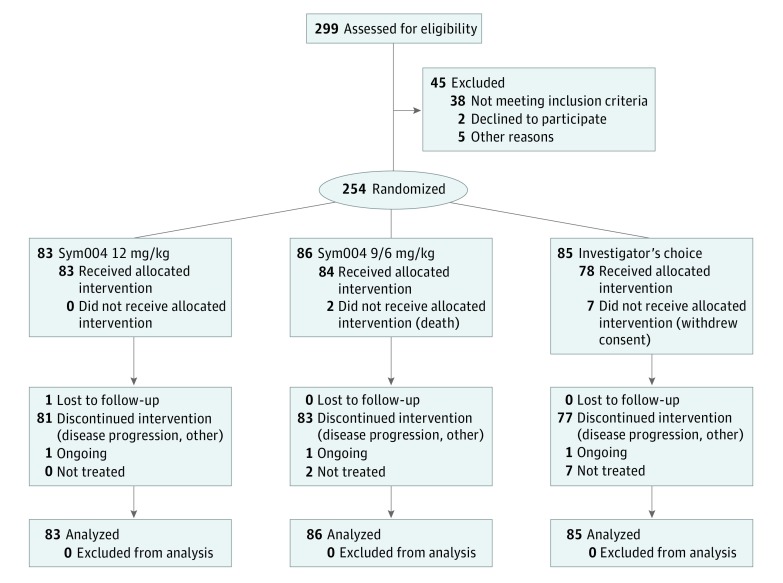
Between March 6, 2014, and October 15, 2015, of 299 patients screened, 254 were randomly assigned, and these made up the intent-to-treat (ITT) population (Figure 1). The study population was balanced at baseline (Table 1). Patients were heavily pretreated: 72%, 78%, and 72% in arms A, B, and C, respectively, had received at least 3 previous regimens.
Figure 1. Study Enrollment Flow Diagram for Patients With Metastatic Colorectal Cancer Included in the Sym004-05 Study.
Table 1. Baseline Characteristics for the Intent-to-Treat Study Population.
| Characteristic | Arm A, Sym004 12 mg/kg (n = 83) | Arm B, Sym004 9/6 mg/kg (n = 86) | Arm C, Investigator’s Choice (n = 85) |
|---|---|---|---|
| Age, mean (SD)a, y | 62 (10) | 64 (10) | 61 (11) |
| Sex, No. (%) | |||
| Male | 52 (63) | 54 (63) | 54 (64) |
| Female | 31 (37) | 32 (37) | 31 (37) |
| Race, No. (%) | |||
| White | 72 (88) | 75 (87) | 73 (86) |
| Other or NA | 11 (13) | 11 (13) | 12 (14) |
| ECOG performance status, No. (%) | |||
| 0 | 33 (40) | 35 (41) | 35 (41) |
| 1 | 50 (60) | 50 (58) | 50 (59) |
| 2 | 0 | 1 (1)a | 0 |
| Tumor site, No. (%) | |||
| Right colon | 12 (15) | 10 (12) | 9 (11) |
| Left colon/rectum | 67 (81) | 72 (84) | 70 (82) |
| Prior mCRC treatments, No. (%)b | |||
| 2 | 23 (28) | 19 (22) | 24 (28) |
| 3 | 27 (33) | 24 (28) | 29 (34) |
| ≥4 | 33 (40) | 43 (50) | 32 (38) |
| Prior anti-EGFR mAb therapies, No. (%) | |||
| Cetuximab only | 55 (66) | 54 (63) | 53 (62) |
| Cetuximab and panitumumab | 12 (15) | 14 (16) | 14 (17) |
| Panitumumab only | 16 (19.3) | 18 (21) | 18 (21.2) |
| Time since last anti-EGFR mAb therapy, mean (SD), d | 78 (48) | 80 (51) | 72 (46) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; EGFR, epidermal growth factor receptor; mAb, monoclonal antibody; mCRC, metastatic colorectal cancer; NA, not available.
Patient had an ECOG performance status of 1 at screening and therefore met eligibility criteria.
All patients received at least 1 prior anti-EGFR mAb cancer therapy.
Median OS in the ITT population was 7.9 (95% CI, 6.5-9.9) months, 10.3 (95% CI, 9.0-12.9) months, and 9.6 (95% CI, 8.3-12.2) months for arms A, B, and C, respectively (HR, 1.31; 95% CI, 0.92-1.87 for A vs C; and HR, 0.97; 95% CI, 0.68-1.4 for B vs C) (Table 2 and eFigure 2 in Supplement 2). Median PFS in the ITT population was 2.8 (95% CI, 1.8-3.2) months, 2.7 (95% CI, 2.6-3.3) months, and 2.6 (95% CI, 1.4-3.1) months for arms A, B, and C, respectively. Response rates for evaluable patients in the ITT population were 11 PRs (14.1%), 8 PRs (9.6%), and 1 CR and 1 PR (2.9%) in arms A, B, and C, respectively (eTable 1 in Supplement 2). The unexpectedly long median OS of 9.6 months in the IC arm led to evaluation of potential factors that might explain this finding. It became evident that a subgroup of patients (n = 30) had been subject to nonstandard medical practice in first- or second-line therapies that had an impact on the efficacy of rescue chemotherapy in the anti-EGFR refractory setting as well as in the molecular characterization of these patients (details provided in eMethods and eTable 2 in Supplement 2). This population was excluded from the genomic analysis.
Table 2. Efficacy Data for the ITT Study Population and Population With Biomarker Data.
| Study Group | OS (95% CI), mo | 1-Year Survival, No. (95% CI) | HR (95% CI) |
|---|---|---|---|
| ITT population (n = 254) | |||
| Sym004 12 mg/kg (n = 83) | 7.9 (6.5-9.9) | 37 (26-47) | 1.31 (0.92-1.87) |
| Sym004 9/6 mg/kg (n = 86) | 10.3 (9.0-12.9) | 44 (33-54) | 0.97 (0.68-1.40) |
| Investigator’s choice | 9.6 (8.3-12.2) | 40 (29-51) | 1 [Reference] |
| Population with biomarker data (n = 193)a | |||
| Sym004 12 mg/kg (n = 70) | 7.7 (5.5-11.3) | 38 (26-49) | 1.03 (0.69-1.54) |
| Sym004 9/6 mg/kg (n = 67) | 9.9 (7.1-12.9) | 44 (32-56) | 0.79 (0.52-1.20) |
| Investigator’s choice (n = 56) | 8.5 (6.4-9.9) | 27 (16-41) | 1 [Reference] |
Abbreviations: ITT, intent-to-treat; HR, hazard ratio; OS, overall survival.
Sixty-one patients are excluded from this population, 31 due to lack of biomarker data and 30 due to treatment and OS data inconsistent with the study population (see Supplement 2 for more information).
Treatment with both regimens of Sym004 led to a higher frequency and severity of adverse events (AEs) than treatment administered to patients on the IC arm, and the Sym004 regimen of 12 mg/kg/wk (arm A) was more poorly tolerated than the Sym004 9 mg/kg loading dose followed by 6 mg/kg/wk (arm B) dosing schedule (eTable 3 in Supplement 2). The Sym004 AE profile was consistent with other anti-EGFR mAbs, although the frequency and severity of both dermatologic AEs (94.0% and 92.9% for arms A and B, respectively, compared with 10.3% in the IC arm) and hypomagnesemia (68.7% and 56.0% for arms A and B, respectively, compared with 7.7% in the IC arm) were higher than those found with other approved anti-EGFR mAb therapies. In contrast, the frequency of gastrointestinal AEs appeared to be lower than has been reported for other anti-EGFR mAbs (51.8% and 48.8% for arms A and B, respectively, compared with 47.4% in IC arm). The frequency of treatment-emergent AEs leading to study treatment discontinuation (6.0% in arm B vs 7.7% in arm C) and related treatment-emergent AEs leading to study treatment discontinuation (2.4% in arm B vs 3.8% in arm C) was not different in patients treated with the lower Sym004 dose and IC (eTable 3 in Supplement 2).
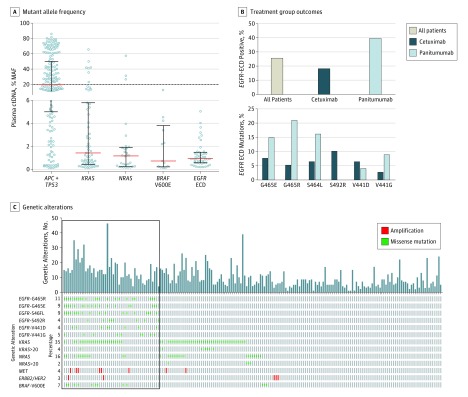
Baseline ctDNA profiles of alterations in 70 genes (eFigure 1 in Supplement 2) were obtained from blood samples collected from 193 patients in the trial. Genotyping of baseline ctDNA captured high intrapatient genomic heterogeneity and confirmed previously reported mechanisms of acquired resistance to cetuximab and panitumumab, including mutations in RAS (29.5% of patients), EGFR ECD (25% of patients), and BRAF V600E (6.7% of patients), as well as amplification of ERBB2/HER2 and MET (eFigures 3 and 4 in Supplement 2). Inactivation of APC and/or TP53 is an early event in the development of CRC and the APC/TP53 highest mutant allele frequency (MAF) alteration in a patient’s ctDNA can therefore serve as an arbitrary marker for clonal mutations (present in all tumor cells). The median MAF for the most prevalent TP53/APC alterations was close to 20%. The median MAFs for KRAS, NRAS, EGFR ECD and BRAF were much lower than 20%, indicating that these mutations are primarily subclonal, although a subset of 10 patients harbored RAS mutations at allele frequencies above 20% (Figure 2A). Six EGFR ECD mutations (V441D, V441G, S464L, G465E, G465R, and S492R) were the most frequent (each present in ≥5% of patients, totally detected in 25% of patients; Figure 2C and eFigure 5 in Supplement 2). Patients with EGFR ECD mutations had more genetic alterations (median number of alterations per patient, 14; interquartile range [IQR], 10.0-18.5) compared with the full biomarker patient population (median, 9; IQR, 5-14) (Figure 2C). The frequency and type of EGFR ECD mutations varied depending on previous treatment with cetuximab or panitumumab (Figure 2B).
Figure 2. Mutant Allele Frequency, Genetic Alterations, and Treatment Group Outcomes for Patients Included in the Sym004-05 Study.
A, Mutant allele frequency (MAF) of APC+TP53, KRAS exons 2, 3, and 4, NRAS exons 2, 3, and 4, BRAF V600E, and the EGFR extracellular domain (ECD) mutations V441D, V441G, S464L, G465E, G465R, and S492R in the 193 patients analyzed. For patients with more than 1 alteration in the same gene, only the highest MAF alteration for each gene is shown. Red lines denote the medians, black bars denote the interquartile ranges, and the dotted line depicts MAF = 20%. B, Top, fraction of EGFR-ECD positive patients (including the 6 most frequent EGFR ECD mutations) grouped by last anti-EGFR treatment received prior to enrolment. Bottom, number of patients with each of the 6 EGFR ECD mutations who were treated with either cetuximab or panitumumab as the last anti-EGFR treatment prior to study enrollment. C, Bar graph depicting number of genetic alterations for each genetically profiled patient, grouped by patients with EGFR ECD mutations to the left (box). The oncoprint denotes the 6 individual most frequent EGFR ECD mutations, KRAS mutations in exon 2, 3, or 4 at all MAFs (KRAS) and at MAF greater than 20% (KRAS >20), NRAS mutations in exon 2, 3, or 4 at all MAFs (NRAS) and at MAF greater than 20% (NRAS >20), MET and ERBB2/HER2 gene amplifications (defined as copy number >5), and BRAF V600E. Percentages denote the percentages of patients harboring the defined alteration.
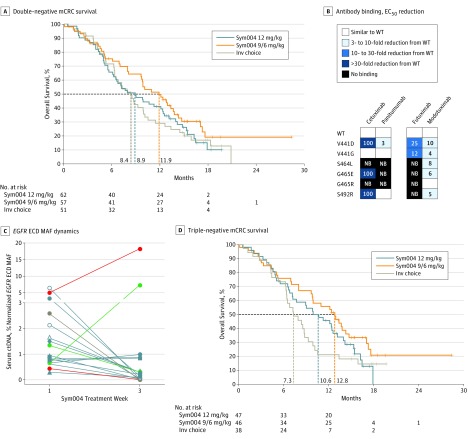
As a predefined exploratory secondary objective of the study, we next aimed to investigate ctDNA-defined molecular subgroups that would predict Sym004 efficacy. A 20% RAS MAF cutoff accurately selected a subgroup of 10 patients in which RAS ctDNA mutations and other mutations of acquired resistance were virtually mutually exclusive, suggesting RAS clonality (Figure 2C and eFigure 10 in Supplement 2). In these patients, RAS mutations potentially existed at the time of diagnosis, before the patient received cetuximab or panitumumab therapy. In preclinical patient-derived xenograft CRC models with RAS and BRAF V600E mutations, poor or limited Sym004 activity was observed, indicating that clonal mutations in these genes caused resistance to Sym004, as is the case for cetuximab and panitumumab (eFigure 11 in Supplement 2). Based on these data, we performed an exploratory analysis of efficacy in a genomically defined subpopulation, which excluded patients with clonal RAS (MAF >20%) and BRAF V600E mutation (named double-negative mCRC; 170 patients; eTable 3 in Supplement 2). Results showed an increase in OS for the Sym004 9/6 group (arm B) (11.9 vs 9.9 months) and a smaller increase in the Sym004 12 group (arm A) (8.9 vs 7.7 months) compared with the subpopulation of patients with biomarker data. Overall survival in the IC population (arm C) was unchanged (8.4 and 8.5 months in the subpopulation and double-negative mCRC subgroup, respectively). Thus, this exploratory biomarker-defined analysis showed an increase in median OS of 3.5 months in the double-negative mCRC population of patients treated with Sym004 9/6 (arm B; 11.9 months) compared with patients randomized to IC (arm 3; 8.4 months) (Figure 3A).
Figure 3. Survival and Other Characteristics of Molecularly Defined Groups of Study Patients With Metastatic Colorectal Cancer (mCRC).
A, Kaplan-Meier survival estimates for overall survival in patients with double-negative mCRC, as defined in the Results section. B, Heat map showing how the binding of different antibodies is affected by the different EGFR extracellular domain (ECD) mutations; the effect on binding was determined as the fold reduction in half maximal effective drug concentration (EC50) relative to wild-type (WT) EGFR. Inv indicates investigator’s’ NB, no binding. C, Dynamics of mutant allele frequencies (MAFs) of EGFR ECD mutations detected in serum samples obtained prior to treatment (week 1) and at week 3 of treatment with Sym004; EGFR ECD mutation dynamics are highlighted for 2 patients (green and red), demonstrating an increase in MAF for 1 EGFR ECD mutation and a decreased MAF for 1 or more other EGFR ECD mutations from week 1 to week 3. For each EGFR ECD mutation, the EGFR ECD MAF was normalized to the TP53 MAF; ctDNA indicates circulating tumor DNA. D, Kaplan-Meier survival estimates for overall survival in patients with triple-negative mCRC, as defined in the Results section.
We then investigated the role of EGFR ECD mutations as a biomarker of Sym004 activity. Preclinical studies showed that EGFR ECD mutations negatively affected the binding and activity of cetuximab, panitumumab, and futuximab, all of which bind to surface-exposed amino acids in the V441-S492 region of domain III of EGFR. The inhibitory activity of Sym004, however, was partially rescued by the modotuximab component of Sym004, which binds to a different region and retains full binding and activity toward the most frequent EGFR ECD mutations (Figure 3B and eFigures 6, 7, 8, and 9 in Supplement 2). Unexpectedly, the presence of EGFR ECD mutations in the ctDNA of patients was not linked to clinical benefit of Sym004 (eFigure 12 in Supplement 2). The most plausible explanation was the subclonal nature of the EGFR ECD mutations in the patients and intrapatient heterogeneity (Figure 2C and eFigure 3 in Supplement 2). To study the molecular basis of these findings in more detail, we sought to analyze the dynamics of EGFR ECD mutations in blood from patients during Sym004 treatment. The percentage of EGFR ECD mutations decreased in the majority of patients treated with Sym004 (Figure 3C), suggesting that subclones carrying EGFR ECD mutations might be targeted by the modotuximab component of Sym004, as shown in the preclinical studies (Figure 3B and eFigures 7, 8, and 9 in Supplement 2). However, this retained activity did not translate into a clinically meaningful OS benefit, likely owing to other co-occurring resistance mechanisms (Figure 2C and eFigures 13, 14, and 15 in Supplement 2) and subclonality of EGFR ECD mutations (eFigure 3 in Supplement 2). Of note, in the 2 patients who experienced an increase in percentage of 1 EGFR ECD mutation in ctDNA during Sym004 therapy, other EGFR ECD mutations that were concomitantly detected in the same sample declined following Sym004 therapy (Figure 3C).
Most notably, we found that the molecular heterogeneity related to known resistance markers (which we defined as the number of resistance alterations, including RAS, BRAF, and EGFR ECD mutations and ERBB2/HER2 and MET amplifications) was associated with worse OS (eFigure 12 in Supplement 2). This led us to postulate that the occurrence of heterogeneous resistance mutations (including EGFR ECD mutations) might pinpoint a subset of patients in which high genomic complexity due to previous chemotherapy and EGFR blockade impaired effectiveness of Sym004 treatment. To test this hypothesis, we assessed outcomes in patients with RAS less than 20% MAF, BRAF WT, and EGFR ECD WT, which we named triple-negative mCRC (eTable 4 in Supplement 2). We found that the triple-negative mCRC population had markedly prolonged OS for Sym004 treatment arms: 12.8 (95% CI, 9.7-14.7) months in the Sym004 9/6 group (arm B; n = 46) and 10.6 (95% CI, 6.8-13.3) months in the Sym004 12 group (arm A; n = 47), compared with 7.3 (95% CI, 6.3-8.8) months in the IC group (arm C; n = 38) (Figure 3D and eTable 4 in Supplement 2).
Discussion
Sym004 is a mixture of 2 mAbs that target nonoverlapping epitopes on domain III with more efficient mediated down-modulation and subsequent degradation of cell-surface EGFR than the clinically approved antibodies cetuximab and panitumumab, leading to more complete and durable pathway inhibition. Here, we report safety and efficacy data from a randomized, multicenter, phase 2 clinical trial of Sym004 investigating 2 Sym004 dose regimens vs IC (capecitabine, fluorouracil, or BSC) in patients with refractory mCRC and with acquired resistance to anti-EGFR therapy.
In the ITT population, OS was similar for all arms of treatment. Based on historical data for last-line mCRC, the OS of 9.6 months in the IC arm is noteworthy. Currently, trifluridine/tipiracil (TAS-102) and regorafenib are the only treatment options for last-line mCRC, based on an increase in median OS in randomized clinical trials. While patients in the control arms in these 2 earlier studies received placebo, those in the present Sym004 study had a potentially active control arm, in which patients were able to receive capecitabine (68 patients), fluorouracil (13 patients), or BSC (4 patients). It is therefore plausible that a potential benefit of Sym004 in the ITT population was masked by a control arm in which most patients received an active treatment.
Recently, EGFR ECD mutations have emerged as a potential novel mechanism of acquired resistance to cetuximab and panitumumab in mCRC. In the present study, EGFR ECD mutations were detected in approximately 25% of patients, and 6 ECD mutations were particularly abundant. A significantly higher number of EGFR ECD mutations was found in patients treated with panitumumab than in those treated with cetuximab. The biology behind this apparent increased frequency of EGFR ECD mutations in panitumumab-treated patients is currently unknown, but lack of secondary effector functions such as antibody-dependent, cell-mediated cytotoxic effects and the preference for monovalent binding of panitumumab could be part of the explanation.
While preclinical studies showed Sym004 efficacy in EGFR ECD-mutated cells, and dynamic ctDNA analysis showed a decrease in EGFR ECD MAF following Sym004 treatment, the presence of these mutations was not linked to clinical benefit of Sym004 in the present trial. The EGFR ECD mutations were always subclonal and coexisted with other genetic alterations related to anti-EGFR resistance, which suggests that although the EGFR ECD-mutated cells are targeted by Sym004, Sym004-resistant clonal cell populations that fail to respond to Sym004 exist within the tumor. This finding provides a plausible explanation for the lack of clinical benefit of Sym004 in patients with EGFR ECD mutations. Our data thus suggest that, in a subset of patients, treatment with the anti-EGFR antibodies cetuximab and panitumumab results in the emergence of extremely heterogenous molecular landscapes in which subclones with distinct mechanisms of resistance coexist. These results are in agreement with evidence that genomic complexity following treatment pressure might be a poor prognostic factor and severely limit the impact of subsequent lines of treatment.
Evaluation of OS in the triple-negative mCRC) population (patients with double-negative mCRC who are also without EGFR ECD mutations) showed clinically meaningful increases in median OS in both Sym004 treatment arms. In this exploratory analysis, the definition of triple-negative mCRC appeared to be an effective method to enrich the patient population for responsiveness to Sym004.
Limitations
A potential limitation of the study is that the treatment in the control arm was by investigator’s choice, and 96% of patients in the control arm received chemotherapy. It is therefore plausible that a potential benefit of Sym004 in the ITT population was masked by a control arm in which most patients received an active treatment. Second, the study was designed to assess OS in chemorefractory disease. However, patients were allowed to have discontinued previous lines of therapy because of intolerance without failure of that chemotherapy, and this potentially allowed for the recruitment of nonrefractory patients who could have benefited from chemotherapy in the control arm of the trial. Finally, although the increase in survival with Sym004 in the triple-negative population is promising, this is a retrospective hypothesis-generating analysis that has to be confirmed in a randomized clinical trial.
Conclusions
The results of this study showed that Sym 004 does not improve OS or PFS when used in an unselected group of patients with mCRC and acquired EGFR resistance. In a hypothesis-generating analysis, ctDNA profiling identified a subset of patients (clonal RAS, BRAF, and EGFR ECD WT) that gained clinically meaningful benefit from therapy with Sym004. These findings provide the rationale for a prospective clinical validation of Sym004 efficacy in a molecularly defined subgroup of patients for whom prior anti-EGFR therapy had failed. Moreover, these data support the use of liquid biomarker genomic profiling to guide treatment of patients with mCRC and to track cancer evolution.
Trial Protocol
eMethods.
eFigure 1. Overview of the 70 genes included in the Guardant360 version 2.9 panel
eFigure 2. Kaplan-Meier survival estimates for Overall Survival in ITT population
eFigure 3. Baseline genotyping for genomic biomarker analysis for patients included in the Sym004-05 study
eFigure 4. Number of genomic alterations (single nucleotide variants, copy number variants, indels, and fusions) identified in circulating tumor DNA from patients (N=193), listed by gene.
eFigure 5. Patient distribution and frequency of EGFR single nucleotide variant missense mutations
eFigure 6. Structural modeling of EGFR and mapping of EGFR ECD mutations identified in the patient cohort
eFigure 7. Dose-response curves showing the effect of the indicated antibodies on cell viability in NIH-3T3 cells stably overexpressing WT or mutant EGF
eFigure 8. Ability of Sym004 to block ligand induced phosphorylation of EGFR in NIH-3T3 cells transfected with either WT or mutant EGFR
eFigure 9. Total EGFR levels after 48 hours of treatment with the indicated antibodies, as determined by Simple Western analysis
eFigure 10. Venn diagrams depicting the number (fraction of all profiled patients in parentheses) of patients harboring concurrent mutations in the EGFR ECD (G465E, G465R, S464L, S492R, V441D, and V441G) and KRAS/NRAS exons 2, 3, and 4 (RAS), as well as BRAF V600E, at various mutant allele frequencies (MAFs)
eFigure 11. Examples of tumor growth curves in PDX models
eFigure 12. Bar graphs depicting overall survival (OS) for each genetically profiled patient
eFigure 13, 14 and 15. Oncoprints depicting the full ctDNA profiles of patients treated with Sym004 12 mg/kg (eFigure 12), Sym004 9/6 mg/kg (eFigure 13), or investigator’s choice (eFigure 14)
eFigure 16. Number of genetic alterations in all ctDNA profiled patients compared to patients harboring EGFR ECD mutations
eTable 1. Response in ITT population (evaluable patients)
eTable 2. Overall survival subsets analysis
eTable 3. Incidence of treatment emergent adverse effects (TEAE)
eTable 4. Baseline characteristics of DNmCRC and TNmCRC populations
References
- 1.Van Cutsem E, Köhne C-H, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011-2019. [DOI] [PubMed] [Google Scholar]
- 2.Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22(7):1535-1546. [DOI] [PubMed] [Google Scholar]
- 3.Peeters M, Oliner KS, Price TJ, et al. Analysis of KRAS/NRAS mutations in a phase III study of panitumumab with FOLFIRI compared with FOLFIRI alone as second-line treatment for metastatic colorectal cancer. Clin Cancer Res. 2015;21(24):5469-5479. [DOI] [PubMed] [Google Scholar]
- 4.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18(2):221-223. [DOI] [PubMed] [Google Scholar]
- 7.Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21(9):2157-2166. [DOI] [PubMed] [Google Scholar]
- 8.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4(11):1269-1280. [DOI] [PubMed] [Google Scholar]
- 9.Dienstmann R, Salazar R, Tabernero J. Overcoming resistance to anti-EGFR therapy in colorectal cancer. Am Soc Clin Oncol Educ Book. 2015;e149-e156. [DOI] [PubMed] [Google Scholar]
- 10.Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51(5):587-594. [DOI] [PubMed] [Google Scholar]
- 12.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arena S, Siravegna G, Mussolin B, et al. MM-151 overcomes acquired resistance to cetuximab and panitumumab in colorectal cancers harboring EGFR extracellular domain mutations. Sci Transl Med. 2016;8(324):324ra14. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-Martín FJ, Bellosillo B, Gelabert-Baldrich M, et al. The first-in-class anti-EGFR antibody mixture Sym004 overcomes cetuximab resistance mediated by EGFR extracellular domain mutations in colorectal cancer. Clin Cancer Res. 2016;22(13):3260-3267. [DOI] [PubMed] [Google Scholar]
- 15.Dienstmann R, Patnaik A, Garcia-Carbonero R, et al. Safety and activity of the first-in-class Sym004 anti-EGFR antibody mixture in patients with refractory colorectal cancer. Cancer Discov. 2015;5(6):598-609. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen MW, Jacobsen HJ, Koefoed K, et al. Sym004: a novel synergistic anti-epidermal growth factor receptor antibody mixture with superior anticancer efficacy. Cancer Res. 2010;70(2):588-597. [DOI] [PubMed] [Google Scholar]
- 17.Koefoed K, Steinaa L, Søderberg JN, et al. Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor. MAbs. 2011;3(6):584-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iida M, Brand TM, Starr MM, et al. Sym004, a novel EGFR antibody mixture, can overcome acquired resistance to cetuximab. Neoplasia. 2013;15(10):1196-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendell JC, Ervin TJ, Senzer NN, et al. Results of the X-PECT study: a phase III randomized double-blind, placebo-controlled study of perifosine plus capecitabine (P-CAP) versus placebo plus capecitabine (CAP) in patients (pts) with refractory metastatic colorectal cancer (mCRC). J Clin Oncol. 2012;30:LBA3501. [Google Scholar]
- 20.Newhall K, Price T, Peeters M, et al. O-0011 * frequency of S492R mutations in the epidermal growth factor receptor: analysis of plasma DNA from metastatic colorectal cancer patients treated with panitumumab or cetuximab monotherapy. Ann Oncol. 2014;25:ii109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1(6):508-523. [DOI] [PubMed] [Google Scholar]
- 23.Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3(6):658-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grothey A, Van Cutsem E, Sobrero A, et al. ; CORRECT Study Group . Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381(9863):303-312. [DOI] [PubMed] [Google Scholar]
- 25.Mayer RJ, Van Cutsem E, Falcone A, et al. ; RECOURSE Study Group . Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372(20):1909-1919. [DOI] [PubMed] [Google Scholar]
- 26.Voigt M, Braig F, Göthel M, et al. Functional dissection of the epidermal growth factor receptor epitopes targeted by panitumumab and cetuximab. Neoplasia. 2012;14(11):1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sickmier EA, Kurzeja RJM, Michelsen K, Vazir M, Yang E, Tasker AS. The panitumumab EGFR complex reveals a binding mechanism that overcomes cetuximab induced resistance. PLoS One. 2016;11(9):e0163366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbosh C, Birkbak NJ, Wilson GA, et al. ; TRACERx consortium; PEACE consortium . Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. ; TRACERx Consortium . Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109-2121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial Protocol
eMethods.
eFigure 1. Overview of the 70 genes included in the Guardant360 version 2.9 panel
eFigure 2. Kaplan-Meier survival estimates for Overall Survival in ITT population
eFigure 3. Baseline genotyping for genomic biomarker analysis for patients included in the Sym004-05 study
eFigure 4. Number of genomic alterations (single nucleotide variants, copy number variants, indels, and fusions) identified in circulating tumor DNA from patients (N=193), listed by gene.
eFigure 5. Patient distribution and frequency of EGFR single nucleotide variant missense mutations
eFigure 6. Structural modeling of EGFR and mapping of EGFR ECD mutations identified in the patient cohort
eFigure 7. Dose-response curves showing the effect of the indicated antibodies on cell viability in NIH-3T3 cells stably overexpressing WT or mutant EGF
eFigure 8. Ability of Sym004 to block ligand induced phosphorylation of EGFR in NIH-3T3 cells transfected with either WT or mutant EGFR
eFigure 9. Total EGFR levels after 48 hours of treatment with the indicated antibodies, as determined by Simple Western analysis
eFigure 10. Venn diagrams depicting the number (fraction of all profiled patients in parentheses) of patients harboring concurrent mutations in the EGFR ECD (G465E, G465R, S464L, S492R, V441D, and V441G) and KRAS/NRAS exons 2, 3, and 4 (RAS), as well as BRAF V600E, at various mutant allele frequencies (MAFs)
eFigure 11. Examples of tumor growth curves in PDX models
eFigure 12. Bar graphs depicting overall survival (OS) for each genetically profiled patient
eFigure 13, 14 and 15. Oncoprints depicting the full ctDNA profiles of patients treated with Sym004 12 mg/kg (eFigure 12), Sym004 9/6 mg/kg (eFigure 13), or investigator’s choice (eFigure 14)
eFigure 16. Number of genetic alterations in all ctDNA profiled patients compared to patients harboring EGFR ECD mutations
eTable 1. Response in ITT population (evaluable patients)
eTable 2. Overall survival subsets analysis
eTable 3. Incidence of treatment emergent adverse effects (TEAE)
eTable 4. Baseline characteristics of DNmCRC and TNmCRC populations