Abstract
We identify SMARCD2 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily D, member 2), also known as BAF60b (BRG1/Brahma-associated factor 60b), as a critical regulator of myeloid differentiation in humans, mice, and zebrafish. Studying patients from three unrelated pedigrees characterized by neutropenia, specific granule deficiency, myelodysplasia with excess of blast cells, and various developmental aberrations, we identified three homozygous loss-of-function mutations in SMARCD2. Using mice and zebrafish as model systems, we showed that SMARCD2 controls early steps in the differentiation of myeloid–erythroid progenitor cells. In vitro, SMARCD2 interacts with the transcription factor CEBPε and controls expression of neutrophil proteins stored in specific granules. Defective expression of SMARCD2 leads to transcriptional and chromatin changes in acute myeloid leukemia (AML) human promyelocytic cells. In summary, SMARCD2 is a key factor controlling myelopoiesis and is a potential tumor suppressor in leukemia.
Differentiation of hematopoietic stem cells (HSCs) follows a hierarchical program of transcription factor–regulated events1–3. Early myeloid cell differentiation is dependent on PU.1 and CEBPα (CCAAT/enhancer-binding protein α), and late myeloid cell differentiation is orchestrated by CEBPε (CCAAT/enhancer-binding protein ε)4. The influence of SWI/SNF (SWItch/Sucrose Non-Fermentable) chromatin-remodeling factors as novel master regulators of hematopoietic differentiation is only beginning to be explored3,5,6. PU.1, CEPBPα, CEBPε, and proteins in SWI/SNF complexes participate in transcription factor–mediated instructive events and less well-defined permissive events orchestrated by a variety of epigenetic modulators7,8. Dynamic chromatin remodeling adds another level of complexity. Embedding of promoter DNA into nucleosome landscapes restricts the accessibility of cognate binding sites for transcription factors and restricts gene expression9–11. The SWI/SNF complex is composed of multimeric units that use energy derived from ATP hydrolysis to unwrap or restructure nucleosomes12. SMARCD2 is a component of the SWI/SNF complex in HSCs and other hematopoietic cells6,13,14. The two paralogous proteins SMARCD1 (BAF60A) and SMARCD3 (BAF60C) control embryonic stem (ES) cell15 and heart muscle cell16 differentiation, respectively.
RESULTS
Clinical phenotype
Here we investigated three independent consanguineous pedigrees with four patients (for an explanation on kinship, see the Online Methods) who presented as neonates with delayed separation of umbilical cord and subsequently developed severe bacterial infections associated with neutropenia, parasitosis, or chronic diarrhea (Supplementary Table 1). Extrahematopoietic findings included mild-to-moderate developmental delay and dysmorphic features (Fig. 1, Supplementary Fig. 1, and Supplementary Table 1). The bone marrow of patients showed hypercellularity, paucity of neutrophil granulocytes, dysplastic features (Fig. 1), and progressive development of myelodysplasia (Fig. 2 and Supplementary Fig. 2). Neutrophil granulocytes were characterized by absence of granule proteins (Supplementary Fig. 3).
Figure 1.
Syndromic features in SMARCD2 deficiency. (a–e) The phenotype of patient AII.1 includes low-set ears, posteriorly rotated, with prominent concha, hypoplastic mandibula, saddle nose, midface hypoplasia, synophris, and asymmetric face (ear to ear) (a), misaligned, dysplastic teeth and incomplete amelogenesis imperfecta (filled arrowheads) (b), brachytelephalangy (unfilled arrowheads) and longitudinal ridges on finger nails (c), sandal gap/increased interdigital space D1–D2 (asterisk), brachymetatarsy D4 (filled arrowhead), and brittle nails (d), and severe osteopenia with relative constriction of diaphysis and flaring of metaphysis (Erlenmeyer deformity) (e). Images have been partially cropped; please compare to Supplementary Table 18. Patients or their parents gave informed consent for publication of their photographs.
Figure 2.
Bone marrow and peripheral blood cell analysis. (a–c) Healthy donor. (a) Regular maturation of hematopoietic lineages and no blast cell excess. Inset, magnification (bone marrow histology; hematoxylin and eosin). (b) Segmented neutrophil granulocytes (peripheral blood cytology; Giemsa). (c) Red and white blood cell maturation (bone marrow cytology; Giemsa). (d–f) AII.1. (d) Diffuse and compact blast cell infiltration with absence of megakaryocytes and erythroid islands. Inset, immature neutrophilic cells (bone marrow histology; hematoxylin and eosin). (e) Atypical neutrophilic cells with hypogranulated cytoplasm, hyposegmented nuclei, and pseudo-Pelger–Huët anomaly (PPHA) (black arrowhead) (peripheral blood cytology; Giemsa). (f) Left-shifted neutrophilic granulopoiesis, blast cells, and PPHA (black arrowheads) (under G-CSF) (peripheral blood cytology; Giemsa). (g–i) BII.1 and BII.2. (g) Hypercellularity with (sub)total adipocyte depletion and normal erythroid precursors. Diffuse infiltration by blast cells and starry sky pattern with disseminated activated macrophages (unfilled arrowheads). Inset, immature neutrophilic cells (bone marrow histology from BII.2; hematoxylin and eosin). (h) Circulating atypical neutrophil cells and PPHA (black arrowhead) (BII.1 peripheral blood cytology; Giemsa). (i) Left-shifted atypical neutrophilic granulopoiesis with increase of blast cells. PPHA (black arrowhead) and atypical neutrophils (unfilled arrowhead) (BII.1 bone marrow cytology; Giemsa). (j–l) CII.1. (j) Marked hypercellularity with (sub)total adipocyte depletion and normal erythrocytes. Diffuse and compact infiltration by blast cells and scattered activated macrophages (unfilled arrowheads). Inset, pleomorphic blast cells with round nuclei and small nucleoli (black arrowheads) (bone marrow histology; hematoxylin and eosin). (k) Glycophorin C staining shows erythropoietic islands (unfilled arrowheads) and blast infiltration (asterisks). (l) CD61 staining shows loosely scattered, small and immature megakaryocytes (micromegakaryocytes) (bone marrow histology; hematoxylin and eosin). Images have been cropped; see also Supplementary Table 18. Scale bars, approximately 20 µm.
Molecular genetics
In search of the underlying genetic defect, we performed homozygosity mapping and whole-exome sequencing, followed by Sanger sequencing of patients and family members (see the Online Methods and Supplementary Note for details). Homozygosity mapping identified an especially large perfect marker interval of over 50 Mb in family A on chromosome 17; within this interval, family B had two non-adjacent perfect intervals spanning 1.8 Mb and 0.5 Mb. The asymptotic logarithm of the odds (LOD) scores for these intervals are +4.2 (+1.8 for family A and +2.4 for family B), and peak observed LOD scores, with a more realistic disease haplotype frequency of 0.05, were 3.0 (+1.2 and +1.8). There were approximately 36 genes located in the two shared intervals, including SMARCD2.
We identified distinct segregating homozygous mutations in SMARCD2 in all three pedigrees (Fig. 3a–c). Mutations are described by their putative effect on transcript SMARCD2-001 (ENST00000448276; NM_001098426.1). Effects on hypothetical transcripts are shown in Supplementary Table 2. At the DNA level, the mutations in pedigrees A and C affected splice sites, while the mutation in pedigree B was a duplication of 25 bp, leading to a frameshift and premature termination (Supplementary Table 2). Immunoblot analyses showed an absence of SMARCD2 protein in patient-derived cells (Fig. 3d and Supplementary Data 1). To confirm that the SMARCD2 mutations lead to a loss of function, we sequenced reverse-transcribed mRNA from patient-derived cells (Fig. 3e) and determined their putatively encoded proteins. We then cloned (primers listed in Supplementary Table 3) two isoforms of patient AII.1 (AII.1a: p.Ile362Cysfs*3 and AII1b: p.Ser394Argfs*1), one isoform of patient BII.1 (BII.1: p.Gln147Glufs*5) and one isoform of patient CII.1 (CII.1: p.Arg73Valfs*8). FLAG-tagged expression vectors carrying mutated SMARCD2 versions and a red fluorescence protein gene separated by an internal ribosomal entry sequence (IRES.RFP) were transfected into 293T cells, and the encoded proteins were investigated for coimmunoprecipitation with native SWI/SNF core members. As shown in Figure 3f (Supplementary Data 2), only the wild-type version of SMARCD2 was able to co-precipitate with SMARCA4 (BRG1), SMARCC2 (BAF170), SMARCC1 (BAF155), and SMARCB1 (BAF47); none of the mutant versions were able to co-precipitate with any of these proteins, suggesting that the mutations constitute loss-of-function alleles.
Figure 3.
Identification of biallelic loss-of-function mutations in SMARCD2. (a–c) Pedigrees and Sanger sequencing chromatograms for patient (Pat) as compared to reference (Ref) sequences and specification of homozygous mutations (Mut). In a, the reverse read is shown for patient AII.1. (d) Immunoblot showing absence of SMARCD2 protein expression (molecular weight, 58.9 kDa; arrowhead) in fibroblasts (healthy donor 1 (HD1), healthy donor 2 (HD2), patients AII.1 and BII.1) and in Epstein–Barr virus (EBV)-transformed B cell lines (healthy donor (HD), patient CII.1). Images have been cropped; please compare to Supplementary Data 1. Replicates: 2. (e) SMARCD2 mRNA transcripts detected in patient-derived cells; ORFs are shown in black. Healthy donor (HD) transcript ENST00000448276; NM_001098426.1; CCDS45756 is shown in comparison to transcripts in patients AII.1 (a, p.Ile362Cysfs*2; b, p.Ser394Argfs*1; c, p.Ile362Valfs*85), BII.1 (p.Gln147Glufs*4), and CII.1 (p.Arg73Valfs*8). Replicates: 2. (f) Immunoprecipitation showing defective binding of patient-specific mutated SMARCD2 proteins to the SWI/SNF core complex components BRG1, BAF170, BAF155, and BAF47. FLAG-tagged SMARCD2 proteins (wild type and mutant), expressed in 293T cells, were immunoprecipitated using antibody to FLAG. Coimmunoprecipitation of endogenous SWI/SNF complex components was visualized by immunoblotting of input and immunoprecipitated (IP) samples. Exposure of the membrane analyzed for FLAG shows the presence of immunoprecipitated wild-type SMARCD2, SMARCD2-AII.1a, SMARCD2-AII.1b, SMARCD2-BII.1, and SMARCD2-CII.1 proteins. Images have been cropped; please compare to Supplementary Data 2. Replicates: 3 Please also see Supplementary Table 20 and the Supplementary Note.
Because all SMARCD2-deficient patients had either been subjected to allogeneic hematopoietic stem cell therapy (HSCT) or had died from their disease, primary SMARCD2-deficient HSCs were not available for further experiments. To further study the role of SMARCD2 in neutrophil differentiation, we established several in vivo and in vitro models.
smarcd2 regulates granulopoiesis in zebrafish
As a first model organism, we used zebrafish (Danio rerio), in which smarcd2 (XP_692749.2) is the ortholog of human SMARCD2. Using antisense morpholino oligonucleotides (MOs), we created Smarcd2-deficient zebrafish in two reporter strains with fluorescent neutrophil granulocytes: Tg(mpx:EGFP)i114 (Supplementary Fig. 4a–c and Supplementary Data 3a,b) and Tg(lyz:dsRed)nz50 (Fig. 4a)17–19. smarcd2 MOs were designed to block either translation initiation (label ATG) or splicing (labels SB1 and SB2, for MOs targeting splice donor and acceptor sites, respectively) of smarcd2. In both fish lines, there was a significant reduction in the number of neutrophil granulocytes in comparison to controls at 72 h post fertilization (h.p.f.) for the ATG and SB1 MOs (Fig. 4a and Supplementary Fig. 4c). MO SB2, which failed to disrupt smarcd2 splicing (Supplementary Fig. 4a), provided an additional negative control indicating specificity of the on-target smarcd2 MO effect to reduce neutrophil abundance. Using CRISPR/Cas9 genome editing in zebrafish, we also created a frameshift mutant smarcd2 allele smarcd21/1 (Supplementary Fig. 4d), which also showed reduced granulocyte abundance at 72 h.p.f. in comparison to wild-type controls (Fig. 4b,c). There were no marked effects of smarcd2 MO on zebrafish granulocyte morphology (Supplementary Fig. 5). There were no qualitative differences in O-dianisidine-stained hemoglobinized erythrocytes and no quantitative differences in numbers of Tg(mpeg1:mCherry)gl25xc264-marked macrophages and Tg(cd41:EGFP)la2-marked thrombocytes after smarcd2 MO knockdown (Supplementary Fig. 6). This underlines the lineage-specific effects of smarcd2. Collectively, these zebrafish models provide concordant evidence that a requirement for SMARCD2 in neutrophil granulocyte differentiation is evolutionarily conserved.
Figure 4.
Smarcd2 deficiency in zebrafish. (a) Neutrophil numbers in Tg(lyz:dsRed)nz50 zebrafish at 72 h.p.f. after injection with MOs (control (CRTL) versus translation-start-site blocker (ATG) and splice-site blocker (SB1 and SB2) MOs targeting smarcd2). Data represent the numbers of fluorescence-labeled neutrophils per individual fish embryo. Pooled data from two independent MO experiments are shown: CTRL n = 16, ATG n = 16, SB1 n = 16, SB2 n = 16 fish. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test. Replicates: 2. (b) Representative fluorescence images of zebrafish strain Tg(mpx:EGFP)i114: smarcd2wt/wt (wild type) and smarcd21/1 (knockout). Reduced numbers of GFP-expressing neutrophils are observed in smarcd21/1 mutant fish embryos. Acquired images: smarcd2wt/wt (n = 37 images) and smarcd21/1 (n = 10 images). (c) Enumeration of neutrophils in smarcd2wt/wt versus smarcd21/1 zebrafish. Numbers of fluorescence-labeled neutrophils were evaluated in caudal hematopoietic tissue for individual fish embryos. n = 38 smarcd2wt/wt and n = 10 smarcd21/1 fish were evaluated in two independent CRISPR/Cas9 experiments. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test. Replicates: 2. Please also see Supplementary Tables 19 and 20.
Knockout of Smarcd2 in mouse embryos
A second in vivo model was generated by injection of Smarcd2+/− mouse ES cells (KOMP repository) into blastocysts and transfer of these cells into pseudo-pregnant mice. Chimeric offspring were mated with wild-type mice, resulting in Smarcd2+/− mice, which were intercrossed (Supplementary Fig. 7a,b). We found that Smarcd2−/− embryos died late during fetal development (Supplementary Fig. 7c–e and Supplementary Data 3c–e) and were characterized by reduced size, pallor, and decreased temporal vascularization (Fig. 5a), suggestive of a compromised hematopoietic system. However, we did find Mendelian ratios of Smarcd2−/− embryos at 14.5 d post-coitum (d.p.c.) (Supplementary Fig. 7d). Smarcd2−/− embryos expressed SWI/SNF core members and the SMARCD1 and SMARCD3 paralog proteins (Supplementary Fig. 7f and Supplementary Data 4). Flow cytometry analysis of fetal liver single-cell suspensions showed comparable numbers of HSCs (Supplementary Fig. 7g), yet a striking reduction in granulocyte–macrophage progenitors (GMPs) and of CD11b+Gr1+ neutrophil granulocytes and CD11b+Ly6c+ monocytes in Smarcd2−/− embryos versus Smarcd2+/+ embryos (Fig. 5b,c).
Figure 5.
Defective hematopoiesis in Smarcd2−/− mouse embryos. (a) Morphology of Smarcd2+/+, Smarcd2+/−, and Smarcd2−/− littermates at 14.5 d.p.c. Images were acquired from four litters: wild type (+/+) n = 4, heterozygous (+/−) n = 10, knockout (−/−) n = 9. Replicates: 2. (b) FACS plots of CD11b, Gr1, and Ly6c expression in Smarcd2+/+, Smarcd2+/−, and Smarcd2−/− embryos. (c) Myeloid fetal liver cell quantification. Data were pooled from six litters: wild type n = 9, heterozygous n = 22, knockout n = 9. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test. Replicates: 3. (d) May–Grünwald and eosin staining of CFU cells derived from Smarcd2+/+, Smarcd2+/−, and Smarcd2−/− HSCs. Mature mouse neutrophils (with annular-shaped nuclei) are absent in Smarcd2−/− colonies. All images were acquired at 63× magnification. Replicates: 2. (e) CFUs derived from Smarcd2+/+, Smarcd2+/−, and Smarcd2−/− LSK cells upon differentiation with cytokines MethoCult M3434 contains SCF, IL-3, IL-6, and EPO. Data were pooled for LSK cells derived from five litters: wild type n = 4, heterozygous n = 5, knockout n = 5. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test. Replicates: 3. (f) Blood cytology for Smarcd2+/+ and Smarcd2−/− embryos assessed by May–Grünwald and eosin staining at 20× and 63× magnification, showing anisocytosis (unfilled arrowhead, 23×), increased mitosis (black arrowheads, 63×), and multinucleated cells (unfilled arrowheads, 63×) in Smarcd2−/− embryos at 14.5 d.p.c. Replicates: 2. (g,h) FACS analysis of erythropoietic progenitors derived from Smarcd2+/+, Smarcd2+/−, and Smarcd2−/− CFU GEMM colonies (myeloid colonies containing granulocytes, erythrocytes, monocytes, and megakaryocytes). (g) FACS scatterplots and pictogram showing the distribution of CD71/Ter119 staining and erythroid stages (S0–S5). (h) Percentage of cells in stages S0–S5; wild type n = 24, heterozygous n = 16, knockout n = 24. Center values, mean; error bars, s.e.m. P values were calculated by two-way ANOVA (shown for wild type versus knockout). Replicates: 2.
To assess the differentiation capacity of HSCs, we next purified CD45.2+Lin–Mac+/lo Sca1+c-Kit+ (LSK) cells from wild-type, heterozygous, and homozygous fetal livers and performed colony-forming unit (CFU) assays in vitro. In comparison to CFU colonies derived from wild-type or heterozygous fetal liver LSK cells, knockout CFU colonies showed a marked reduction in size and numbers (data not shown and Supplementary Fig. 8a) and maturation arrest (Fig. 5d). Smarcd2−/− myeloid CFU colonies, generated in the presence of myeloid cytokine cocktail, were deficient in cell surface expression of CD11b, Gr1, and Ly6c (Supplementary Fig. 8b). A block in myeloid differentiation was also seen when LSK cells (native) were exposed to any of GM-CSF, M-CSF, or G-CSF, suggesting that none of the corresponding cytokine receptors were able to induce myeloid cell growth (Fig. 5e).
Aberrant hematopoiesis was not restricted to the myeloid compartment in Smarcd2−/− embryos but also affected erythroid differentiation. Fetal/umbilical cord blood cytology at 14.5 d.p.c. showed marked dysplastic changes in Smarcd2−/− erythropoiesis: In contrast to wild-type embryos, characterized by normochromic, orthochromatic erythrocytes and the presence of few nucleated erythrocytes, Smarcd2−/− embryos showed extensive anisocytosis of erythrocytes, multinucleated cells, perturbed mitosis, and increased apoptosis (Fig. 5f). Furthermore, in vitro erythroid differentiation of LSK cells in the presence of recombinant mouse SCF, recombinant mouse IL-3, recombinant human IL-6, and recombinant human EPO hints at a partial differentiation block or delay at the immature S1 stage, as determined by CD71/Ter119 expression20 in Smarcd2−/− GEMM colonies (Fig. 5g,h). Taken together, mouse SMARCD2-deficient hematopoietic cell differentiation is characterized by a maturation arrest in myeloid and erythroid cells in vitro and in vivo, reminiscent of the hematological phenotype in SMARCD2−/− patients.
Various previous studies found that SWI/SNF complex members increase or decrease primitive or definite hematopoiesis6. Hence, we hypothesize that (i) the functional effects of SMARCD2 deficiency on granulopoiesis are due to its absence from SWI/SNF complexes, (ii) SWI/SNF complexes that contain SMARCD2 have a specific role in granulopoiesis, and (iii) mechanistically, SMARCD2 governs granulopoiesis via chromatin accessibility and interaction with CEBPε.
SMARCD2 takes stage-specific roles in granulopoiesis
We continued our studies in mouse cells, taking a systems biology approach. To identify alterations in transcriptional networks controlling differentiation of fetal liver HSCs, we isolated LSK and myeloid progenitor cells (Fig. 6a). We profiled the transcriptome by RNA–seq of LSK cells from five Smarcd2+/+ and nine Smarcd2−/− fetal livers. Among a total of 12,362 detected genes, we found 4,290 to be differentially expressed at a false discovery rate (Online Methods) lower than 10%. As expected, Smarcd2 showed the largest fold change in expression of all genes (Fig. 6b and Supplementary Table 4). Interestingly, the majority (79%) of the 605 genes with a relatively large difference (fold change > 1.4, FDR < 1%) were upregulated and not downregulated. This had also been reported for embryonic fibroblasts deficient for SMARCB1 (Snf5) and SMARCA4 (Brg1), two other members of the SWI/SNF complex11.
Figure 6.
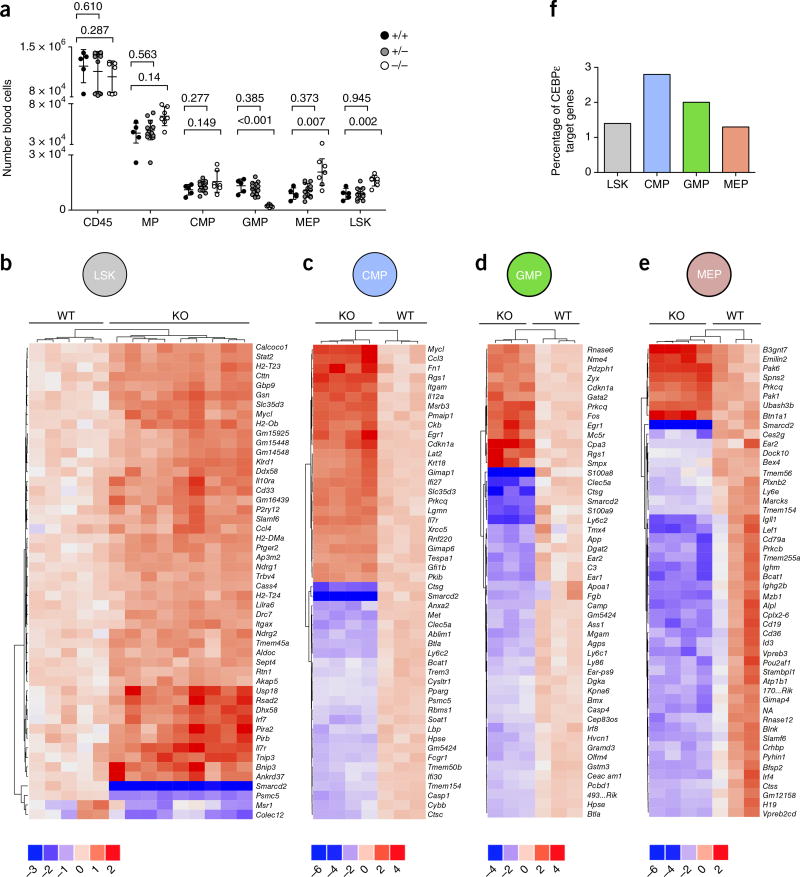
SMARCD2 regulates transcriptional networks in hematopoietic progenitor cells. (a) Quantification of fetal myeloid blood cells (CD45+), progenitors (MPs, CMPs, GMPs, MEPs), and LSK stem cells at 14.5 d.p.c. Data were pooled from four litters: wild type (+/+) n = 4, heterozygous (+/−) n = 14, knockout (−/−) n = 7 embryos. Blood cell numbers per embryo are shown. Smarcd2−/− fetal hematopoiesis shows more cells in the LSK, MP, CMP, and MEP compartments and fewer cells in the GMP compartment. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test with Welsh correction: GMP wild type versus knockout, P = 0,003. Replicates: 2. (b–e) RNA–seq analysis of Smarcd2+/+ and Smarcd2−/− fetal liver hematopoietic cell samples at 14–15 d.p.c. Shown are heat maps of the 50 genes with the lowest P values. Each column represents a fetal liver sample from one embryo. The color keys below the heat maps show the range of log2-transformed fold change in expression. (b) RNA–seq analysis of Smarcd2+/+ (n = 5) and Smarcd2−/− (n = 9) fetal liver LSK cell samples at 14–15 d.p.c. (c) RNA–seq analysis of Smarcd2+/+ (n = 3) and Smarcd2−/− (n = 4) fetal liver CMP cell samples at 14–15 d.p.c. (d) RNA–seq analysis of Smarcd2+/+ (n = 3) and Smarcd2−/− (n = 3) fetal liver GMP cell samples at 14–15 d.p.c.; 4930523C07Rik is abbreviated as 493...Rik. (e) RNA–seq analysis of Smarcd2+/+ (n = 3) and Smarcd2−/− (n = 4) fetal liver MEP cell samples at 14–15 d.p.c.; 700048O20Rik is abbreviated as 170...Rik; and NA is a gene without a name, described as ENSMUSG00000099065. Replicates: 1. (f) Percentage of CEBPε target genes among the differentially expressed genes for the different subpopulations (LSK, CMP, GMP, and MEP); see Supplementary Table 20.
The upregulated genes were most enriched in categories related to membrane proteins, including major histocompatibility complex (MHC) proteins, immunoglobulin domains, and G-protein-coupled receptors that included signaling pathways related to immunodeficiency and host defense (Supplementary Fig. 9a and Supplementary Tables 5 and 6a). A subset of CEBPε-dependent genes (Supplementary Table 6b) was also deregulated in Smarcd2−/− mouse LSK cells (Supplementary Fig. 9b,c and Supplementary Table 6c). Consistent with the finding that CpG island (CGI) promoters can facilitate promiscuous induction without a requirement for SWI/SNF21, we found that genes containing CGI promoters were significantly under-represented within the group of differentially expressed genes (Fisher’s exact test, P = 0.004; odds ratio = 0.71). Thus, a considerable fraction of the genes that were found to be differentially expressed are directly dependent on SWI/SNF and/or transcription factors.
We next compared LSK cells with myeloid progenitor cells. During their maturation, neutrophil granulocytes pass through various stages of development. These stages include LSK cells (CD45+Lin–Sca-1+c-Kit+), common myeloid progenitors (CMPs) defined as CD45+Lin–Sca-1–c-Kit+CD34+CD16/32 (FCGR)int, and GMPs defined as CD45+Lin–Sca-1–c-Kit+CD34+CD16/32 (FCGR)high or megakaryocyte–erythroid progenitors (MEPs) defined as CD45+Lin–Sca-1–c-Kit+CD34–CD16/32 (FCGR)low (Fig. 6a and Supplementary Fig. 10a–c). In contrast to the LSK, CMP, or MEP compartments, GMP cells were almost absent in Smarcd2−/− fetal livers (compare Supplementary Fig. 10a and Supplementary Fig. 10b). The distinct subpopulations were FACS sorted and analyzed by RNA–seq. As shown in Figure 6b–e, SMARCD2 is a transcriptional suppressor in immature cells (LSK and CMP cells), while it adopts the role of a transcriptional activator in further differentiated stages (MEP and GMP cells). Among a total of 15,465 detected genes in CMP cells, we found 852 to be differentially expressed at an FDR lower than 10%; the majority (59%) of the 170 genes with a relatively large difference in expression (fold change > 0.8, FDR < 1%) were upregulated and not downregulated (Fig. 6c). In contrast, among a total of 26,595 detected genes in GMP cells, we found 136 to be differentially expressed at an FDR lower than 10%; the majority (70%) of the 56 genes with a relatively large difference in expression (fold change > 1.5, FDR < 1%) were downregulated and not upregulated (Fig. 6d). A similar pattern was observed in MEP cells. Among a total of 13,049 detected genes in MEP cells, we found 150 to be differentially expressed at an FDR lower than 10%; again, the majority (84%) of the 37 genes with a relatively large difference in expression (fold change > 1.5, FDR < 1%) were downregulated and not upregulated (Fig. 6e). For differentially expressed genes, see Supplementary Tables 4 and 7–10; for enriched categories, see results in Supplementary Tables 5 and 11–14. A relatively large proportion of CEBPε-dependent genes were deregulated in Smarcd2−/− mouse CMP cells (Fig. 6f).
SMARCD2 regulates human granule gene expression
The mouse experiments described in the previous subsection suggest that SMARCD2 orchestrates transcriptional networks in early HSCs, but they do not directly explain the striking absence of neutrophil granules and perturbed differentiation of mature neutrophils seen in SMARCD2-deficient human individuals. To shed light on the mechanisms of SMARCD2 in late human neutrophil maturation, we set out to establish a human in vitro system to further study the function of SMARCD2. We chose the promyelocytic cell line NB4 that is responsive to retinoic acid signaling and can be differentiated toward mature neutrophil granulocytes in vitro. Because our attempts to generate SMARCD2-deficient NB4 cells using CRISPR/Cas9 tools were unsuccessful, we decided to make use of RNA interference to establish cell lines characterized by lower SMARCD2 protein expression. We designed lentiviral short hairpin RNA (shRNA) constructs expressing a SMARCD2-specific shRNA and the marker gene GFP, and we transduced and flow sorted NB4 cells for further analysis NB4 cells represent a human promyelocytic leukemia cell line, derived from a patient with a T15/17 translocation.
NB4 cells express SMARCD1, SMARCD2, SMARCD3, and CEBPE mRNA/cDNA at detectable levels (Fig. 7a and ref. 22). RNA expression of SMARCD2, but not of the family members SMARCD1 and SMARCD3, was significantly reduced upon lentiviral expression of shRNA directed against SMARCD2 (Fig. 7a). The expression of CEBPE was not affected by SMARCD2 knockdown and increased after differentiation with all-trans retinoic acid (ATRA) (data not shown), as previously described (for example, see ref. 23). Next, we systematically analyzed the RNA expression of genes encoding proteins that are expressed and stored in primary and specific granules in neutrophil granulocytes (Fig. 7a). Interestingly, during differentiation with ATRA, transcript levels of the primary granule proteins cathelicidin (CAMP) and α1-antitrypsin (AAT), also known as SERPIN A1, as well as the specific granule proteins matrix metalloproteinase 8 (MMP8), transcobalamin 1 (TCN1), and lactoferrin (LTF), were reduced in SMARCD2-deficient cells.
Figure 7.
SMARCD2, granule formation, and transcriptional regulation. (a) The relative mRNA expression of SMARCD genes, primary granule genes (CAMP, AAT), and secondary granule genes (MMP8, TCN1, LTF) is shown in human NB4 AML cells upon shRNA-mediated knockdown of SMARCD2. Data points show relative expression in cells treated with shRNA 1 or shRNA 2 versus control (CTRL) in three independent experiments for SMARCD1, SMARCD3, CAMP, AAT, and MMP8 and in four independent experiments for SMARCD2 and LTF. The expression levels of SMARCD1, SMARCD2, and SMARCD3 were determined in undifferentiated cells, and granule gene expression was measured in ATRA-differentiated NB4 cells. Center values, mean, error bars, s.d. Replicates: 3 or 4. (b) Chromatin immunoprecipitation (ChIP) in ATRA-differentiated NB4 cells. Shown is the percent input to describe the enrichment of CEBPε and BRG1 at the LTF promoter (Online Methods and Supplementary Note). CEBPε binds to the LTF promoter, and binding is significantly reduced in cells transduced with shRNA 1) or shRNA 2. BRG1 binds to the LTF promoter, and binding is significantly reduced in cells transduced with shRNA 1 or shRNA 2. Data from two experiments with a total of n = 5 independent NB4 cell cultures are shown; in total, three experiments were performed. Center values, mean; error bars, s.d. P values were calculated by two-tailed unpaired t test. Replicates: 3.
SMARCD2 mediates transcription via CEBPε
Mice with targeted mutations in Cebpe24 and human patients with rare mutations in CEBPE25 are characterized by specific granule deficiency and susceptibility to bacterial infections. In view of these phenotypic similarities, we asked whether SMARCD2 controls the effects of CEBPε. RNA expression of CEBPE was not directly affected in SMARCD2-deficient cells. As an alternative, we hypothesized that SMARCD2 might be relevant for recruiting CEBPε to transcription start sites or open chromatin and thus facilitating expression of CEBPε-dependent genes.
CEBPε binds to the promoters of primary granule genes of CAMP and SERPINA1 (AAT) as well as to the promoters of specific granule genes LTF (lactoferrin) and MMP8 (matrix metalloproteinase 8/neutrophil collagenase) (Fig. 7b and Supplementary Fig. 11a–c). shRNA-mediated knockdown of SMARCD2 significantly impaired binding of CEBPε (CTRL versus shRNA 1, P = 0.004; CTRL versus shRNA 2, P = 0.011, two-tailed unpaired t tests) and BRG1 (CTRL versus shRNA 1, P = 0.036; CTRL versus shRNA 2, P = 0.031, two-tailed unpaired t tests) to the LTF promoter (Fig. 7b).
To address the question of whether SMARCD2 interacts directly with CEBPε, we performed immunoprecipitation studies in 293T cells engineered to express HA-tagged CEBPε and FLAG-tagged SMARCD2. As shown in Figure 8a,b (Supplementary Data 5), immunoprecipitation studies confirmed a physical interaction between both proteins. The interaction of the endogenous proteins was demonstrated by immunoprecipitation with antibody to CEBPε and co-precipitation of SMARCD2 (Supplementary Fig. 12a–j).
Figure 8.
SMARCD2 transcriptional regulation. (a,b) Coimmunoprecipitation of FLAG-tagged SMARCD2 and HA-tagged CEBPε in 293T cells. (a) Immunoblot detection of immunoprecipitated FLAG-tagged SMARCD2 and coimmunoprecipitated HA-tagged CEBPε. (b) Immunoblot detection of immunoprecipitated HA-tagged CEBPε and coimmunoprecipitated FLAG-tagged SMARCD2. Images have been cropped; please compare to Supplementary Data 5. GAPDH is probed as a control. Replicates: 3. (c) Venn diagram showing the intersection of differentially expressed genes in undifferentiated NB4 cells (UD) and ATRA-differentiated NB4 cells (ATRA) with and without SMARCD2 knockdown in comparison to CEBPε target genes. For a list of intersections, see Supplementary Table 4. (d–g) Representation of differentially expressed genes in SMARCD2-knockdown versus control NB4 cells. (d) Undifferentiated NB4 cells (SMARCD2 knockdown versus control) analyzed by ATAC–seq and RNA–seq. An overlapping set of 12 genes was deregulated in both assays. (e) Heat map showing the relative expression (log2-transformed fold change) of the genes identified in undifferentiated cells in both assays. The heat map legend is the same as in g. NB4 cells were transduced with shRNA 1, shRNA 2, or CTRL, mock treated with DMSO, and sequenced twice (twice for RNA–seq and twice for ATAC–seq). Technical replicates: 2. (f) ATRA-differentiated NB4 cells (SMARCD2 knockdown versus control) analyzed by ATAC–seq and RNA–seq. A total of 16 genes were deregulated in both assays. (g) Heat map showing the relative expression (log2-transformed fold change) of the genes identified in ATRA-differentiated cells in both assays. NB4 cells were transduced with shRNA 1, shRNA 2, or CTRL, treated with ATRA, and sequenced twice (twice for RNA–seq and twice for ATAC–seq). Technical replicates: 2.
A functional link between SMARCD2 and CEBPε is further supported by our finding, that documented CEPBε-dependent genes are deregulated in the absence of SMARCD2 in human (Fig. 8c, Supplementary Fig. 13, and Supplementary Table 6d) and mouse (Fig. 6f and Supplementary Fig. 9b,c) hematopoietic cells. The proportion of CEBPε-dependent genes affected by Smarcd2−/− was highest in the CMP compartment than in LSK, GMP, or MEP cells (Fig. 6f). This is in keeping with an established role for CEBPε in intermediate stages of differentiation26.
SMARCD2 modulates chromatin accessibility
The consequences of defective nucleosome positioning due to dysfunctional SWI/SNF molecules may be complex. We attempted to interrogate the effects of SMARCD2 deficiency on global chromatin accessibility using the assay for transposase accessible chromatin with high-throughput sequencing (ATAC–seq) (Fig. 8d–g, Supplementary Fig. 14, and Supplementary Tables 15 and 16). We compared all genes that showed differential chromatin accessibility in SMARCD2-knockdown cells (Supplementary Fig. 14) with differentially expressed genes determined by RNA–seq studies both in undifferentiated (Fig. 8d,e) and ATRA-differentiated (Fig. 8f,g) promyelocytic leukemia cell line NB4. A specific subset of genes was found to be deregulated in both assays, ATAC–seq and RNA–seq. These genes are involved in vesicular trafficking, migration, and signaling. The specificity of this observation is in line with findings in mouse embryonal fibroblasts11 and yeast27, which show only a moderate correlation of SWI/SNF-governed chromatin accessibility and transcription in response to knockout of single SWI/SNF units. To examine the role of the discovered gene sets, further studies are needed. Differentially expressed genes in both the mouse transcriptome (Supplementary Tables 4 and 7– 10) and the human transcriptome (Supplementary Tables 15 and 16) clustered significantly in signaling pathways relevant to immune system functions (Supplementary Fig. 9a and Supplementary Table 6a (mouse); Supplementary Fig. 13a,b and Supplementary Table 6e,f (human)). Taken together, DNA accessibility studies, transcriptome studies, and protein–protein interaction studies suggest that SMARCD2 has a direct role to remodel the chromatin and to mediate downstream effects partly by interaction with the myeloid transcription factor CEBPε.
DISCUSSION
In this study, we identify patients with SMARCD2 deficiency, characterized by hematopoietic defects and developmental aberrations. On the basis of our comparative studies, also involving zebrafish and mice, we conclude that SMARCD2 orchestrates HSC differentiation.
Clinically, the phenotype of SMARCD2-deficient neutrophil granulocytes is reminiscent of the phenotype of specific granule deficiency caused by mutations in CEBPE28. CEBPE-deficient neutrophil granulocytes show bilobed nuclei29,30 with nuclear blebs and pockets31 or pseudo-Pelger–Huët-type bilobed nuclei32 in conjunction with a lack of specific granules30.
Both SMARCD2 deficiency and CEBPε deficiency may be associated with decreased counts of peripheral neutrophil granulocytes, yet the capacity to mobilize neutrophils from the bone marrow remains intact.
All patients with SMARCD2 deficiency had evidence of myelodysplasia and blast excess, a feature not typically seen in CEBPε deficiency. However, allelic loss of CEBPE has been detected in 4 of 20 cases of evolving myelodysplastic syndromes (MDS)33, and myelodysplasia is also a feature in Cebpe−/− mice34,35. CEPBε controls terminal differentiation of neutrophil granulocytes, whereas SMARCD2 appears to also control early stages of HSC differentiation, as documented by imbalances in the transcriptome in hematopoietic progenitor cells. Even though the effects of SMARCD2 deficiency on late neutrophil granulocytes are mediated, at least in part, by CEBPε, there are other less well-defined modes of action of SMARCD2.
The role of the SWI/SNF complex and SMARCD2 in leukemogenesis is intriguing. Previous studies had shown that the SWI/SNF complex controls maintenance of myeloid leukemia cells13,36. Specifically, knockdown of SMARCD2 in human MLL-rearranged leukemia cells resulted in reduced self-renewal capacity37. Furthermore, mutational analysis of acute promyelocyte leukemia samples identified somatic loss-of-function mutations in ARID1B and ARID1A, encoding two components of the SWI/SNF complex38.
The companion paper by Priam et al.39 documents a specific and non-redundant role for mouse SMARCD2 in controlling granulocytopoiesis. Similar to our patients with SMARCD2 deficiency, adult mice with SMARCD2-deficient hematopoietic cells develop myelodysplasia and blast excess. It remains to be determined whether this proliferative disorder is indeed clonal.
Interestingly, our transcriptome studies in defined subpopulations of hematopoietic progenitor cells showed an evolving pattern, consistent with the concept that SMARCD2 acts as a transcriptional suppressor in early HSCs and as a transcriptional activator in later stages. We interrogated the genomic landscape in SMARCD2-deficient leukemia cells but could only identify a relatively small number of overlapping genes in ATAC–seq and corresponding RNA transcripts. This may be due to intrinsic limitations of the model system.
The complexity of SMARCD2-dependent transcriptional regulation in early and late hematopoiesis prohibits a simple mechanistic explanation of the leukemogenic disposition. Our studies in human cells and studies in mouse cells39 documented that SMARCD2 is essential for CEBPε and SWI/SNF recruitment to the promoter of neutrophilic granule genes. Further studies are however needed to shed light on the critical role of SMARCD2 in early stages of HSC differentiation.
Systematic studies in patients with rare disorders may highlight the critical role of defined genes and pathways controlling differentiation and function of the blood and immune system. Our clinical and molecular studies in SMARCD2-deficient patients provide an example and reveal SMARCD2 as a key factor controlling transcriptional networks governing stem cell differentiation and lineage specification in the hematopoietic system.
URLs
CHOPCHOP tool, https://chopchop.rc.fas.harvard.edu/; Cytoscape, http://wiki.reactome.org/index.php?title=ReactomeFIViz&oldid=7168; Venny, http://bioinfogp.cnb.csic.es/tools/venny/index.html; KOMP repository, http://www.komp.org/.
ONLINE METHODS
The Online Methods and Supplementary Note include statistical methods to analyze different types of data. Available blood counts for patients (Supplementary Table 17), patient materials used in this study (Supplementary Table 18), raw data and statistical test results (Supplementary Table 19), and an overview of the replicate experiments performed (Supplementary Table 20) have been provided.
Patients
Patients were referred by A.S.-P., P.D.A., and M.R.A. for genetic assessment of congenital neutrophil deficiencies. The study was approved by the ethics committees of the University Medical School of Hannover and the Faculty of Medicine at LMU, Munich. Patient recruitment, genetic analysis, and data handling were carried out in accordance with the tenets of the Declaration of Helsinki. Patients or their parents gave informed consent for the genetic and functional studies and for publication of their photographs; this explicitly includes the publication of photographs showing faces without black bars.
Hematology, biochemistry, and pathological bone marrow studies
Clinical laboratory–based assays, such as blood cell counting, were performed by referring centers according to good clinical practices. Bone marrow histological studies were performed on paraffin-embedded samples provided by the referring clinical immunology centers. Following standard histopathological procedures, specimens were sectioned with a microtome (Leica) and stained with a SAKURA Tissue-Tek Prisma and Film Automated Slide Stainer (hematoxylin–eosin) or a BenchMark XT fully automated IHC/ISH staining instrument (immunohistochemistry). In addition to antibody against lactoferrin (ab15811, Abcam; dilution 1:100), antibodies against myeloperoxidase (A0398, Dako; dilution 1:4,000), CD15 (PNIM1921, Beckman Coulter; dilution 1:100), glycophorin C (M0820, Dako; dilution 1:200), and CD61 (760-4249, Ventana/Roche; ready to use/undiluted) were routinely used.
Homozygosity mapping and next-generation sequencing
Patient AII.1 served as the index case. Patient BII.1, previously described in a clinical case report40, and patient BII.2 (not previously described) were analyzed by homozygosity mapping using the Affymetrix 6.0 chip, as in ref. 41. We searched for perfectly segregating intervals in the SNP data using the software findhomoz42. To compute LOD scores, we assumed that the parents of the affected individuals were second cousins as in refs. 43,44 because they are known not to be first cousins and, if they are more distantly related than second cousins, the LOD scores would be higher. Indeed, in the initial case report, family B was erroneously described as “non-consanguineous” (ref. 40).
Genomic DNA from the two parents and two affected children in family B was enriched for all coding exons using Agilent’s SureSelect Human All-Exon kit V3-50MB (Agilent Technologies) according to the manufacturer’s protocol and subjected to sequencing on an Illumina Genome Analyzer II. Short sequence reads were mapped to human reference genome GRCh37 with Novoalign, and variants were detected as previously described45–47. For each possible mutation found in family B, we designed a sequencing assay to test the affected individual in family A (our index patient) for that mutation. As this approach failed, we performed high-throughput sequencing in family A and identified a likely pathological variant in SMARCD2: c.1181+1G>A (ENST00000448276; NM_001098426.1) confirmed by Sanger sequencing. Sanger sequencing of SMARCD2 in family B identified a large homozygous insertion in patients BII.1 and BII.2 (c.414_438dup), segregating in family B.
Within our cohort of approximately 250 families including a total of 400 patients with SCN, in patient CII.1, a homozygous mutation in SMARCD2 (c.401+2T>C) was identified by whole-exome sequencing with a SureSelect XT Human All-Exon V3 + UTRs kit according to the manufacturer’s instructions (Agilent Technologies) using a SOLiD 5500 next-generation sequencing platform (Life Technologies) to obtain an average coverage depth of 100× (75-bp forward and 35-bp reverse paired-end reads). Segregation of this variant in family C was confirmed by Sanger sequencing. In all three families (A, B, and C), CEBPE and several other candidate genes were excluded (shown not to contain germline biallelic mutations) by Sanger sequencing or whole-exome sequencing (ref. 40 and new data, data not shown). The Exome Aggregation Consortium (EXAC)48 describes seven loss-of-function variants, each occurring once in the heterozygous state among a cohort of 60,000 individuals. This suggests a frequency for homozygous SMARCD2 loss of function in the general population in the range of 1 in 14,400,000,000 to approximately 1 in 2,000,000,000. If multiple loss-of-function variants occur within the same subpopulation, the probability of compound-heterozygous individuals would be higher. See the Supplementary Note for statistics.
Sanger sequencing of SMARCD2
Human SMARCD2 isoform SMARCD2-001 (ENST00000448276; NM_001098426.1) is consistently annotated (CCDS45756) and was used as the reference sequence for specific sequence-based experiments. Targeted sequencing included all 13 exons of ENST00000448276; NM_001098426.1, as well as one potential alternative exon 1 derived from isoform SMARCD2-003 (ENST00000323347; no RefSeq ID). Throughout the text, mutations are described by their putative effect on transcript SMARCD2-001 only. Effects on other transcripts are shown in Supplementary Table 2. See also the Supplementary Note.
Mouse model
Generation of the Smarcd2-deficient mouse model
C57BL/6 ES cell clone 11930A-F4 carrying a mutant Smarcd2 allele was generated by Regeneron Pharmaceuticals and obtained from the KOMP repository (see URLs). To generate Smarcd2-deficient mice, clonal ES cells were injected into C57BL/6BrdCrHsd-Tyrc (albino) blastocysts and these were transferred to pseudo-pregnant NMRI foster mothers. The resulting chimeras were crossed to C57BL/6 albino mice to identify germline transmission of the targeted allele and to produce mice heterozygous for the mutation. F1 intercrosses of heterozygous mice resulted in smarcd2+/+, smarcd2+/−, and smarcd2−/− embryos, which were genotyped using standard PCR reaction conditions (Supplementary Fig. 7 and Supplementary Table 3).
Animals were maintained under specific-pathogen-free conditions at 23 °C, 65% humidity with a 12-h light/12-h dark cycle and had free access to a standard rodent diet (V1534, Ssniff) and water. All animal experiments were carried out in accordance with the German Animal Welfare Act with permission from the responsible veterinary authority.
Flow cytometry (FACS)
For FACS analysis of fetal liver hematopoietic cells, single-cell suspensions were prepared by homogenization of fetal liver tissue with a 1-ml Eppendorf pipette and Hank’s buffered salt solution (HBSS) containing 3% FCS. Fetal liver cells were kept on ice until genotyping. For FACS analysis of CFU-derived hematopoietic cells, CFU colonies were picked after evaluation by light microscopy and washed once in HBSS containing 3% FCS.
Fetal liver hematopoietic cells and CFU-derived cells were stained with the following fluorochrome- or biotin-conjugated monoclonal antibodies for 20 min on ice: anti-B220–Alexa Fluor 780 (eBioscience, RA3-6B2; 1:200), anti-CD3-FITC (eBioscience, 145-2C11; 1:200), anti-CD19-PeCy7 (eBioscience, eBio1D3; 1:200), anti-Ter119-PE (BD Pharmigen, TER-119; 1:200), anti-Gr1-FITC (BD Pharmigen, RB6-8C5; 1:200), anti-Ly6c-PerCP-Cy5.5 (eBioscience, Hk1.4; 1:200), anti-Mac1/CD11b-biotin/-eFluor 450 (eBioscience, M1/70; 1:200), and anti-CD71-FITC (BD Pharmigen, C2; 1:200). Cells stained with biotinylated monoclonal antibodies were washed and incubated with streptavidin-APC or PerCP (eBioscience, 17-4317, 1:200 or 45-4317, 1:100). Samples were acquired on either a FACSCanto or LSR II flow cytometer (BD), and data were analyzed using FlowJo software (Tree Star). Fluorescence intensity plots are shown on a log10 scale. Relative abundance (percentage of the parental gate) was analyzed with Prism software (GraphPad); center values, mean; error bars, s.e.m. P values were calculated by two-tailed unpaired t test. See the Supplementary Note for further information on statistics.
Mouse fetal blood cytology
Fetal blood was recovered from sacrificed embryos and washed in HBSS with 3% FCS. Cytological assessment of equal numbers of nucleated cells was performed by cytospins (Shandon Cytofunnel Thermo) and May–Grünwald–Giemsa staining. Blood cells were morphologically assessed using an inverted microscope (Axiovert-II, Zeiss) and photographed.
Zebrafish experiments
Zebrafish
Tg(mpx:EGFP)i114 (ref. 19), Tg(lyz:dsRed)nz50 (ref. 17), Tg(mpeg1:Gal4FF)gl25/(UAS-E1b:Eco.NfsBmCherry)c264 (ref. 49), and Tg(itga2b:eGFP)la2 (ref. 50) strains were used. Fish were held in the FishCore (Monash University) using standard practices. Embryos were held in egg water (0.06 g/l salt (Red Sea)) or E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, equilibrated to pH 7.0); from 12 h.p.f., 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) was added to inhibit pigmentation. Embryos were held at 28 °C in an incubator (Thermoline Scientific) upon collection. Animal experiments followed NHMRC guidelines (Australian Code of the Care and Use of Animals for Scientific Purposes, 8th edition, NHMRC, 2013) and were approved by the Monash University Animal Ethics Committees.
CRISPR/Cas9 mutagenesis in zebrafish model: single-guide RNA synthesis for the CRISPR mutagenesis model
The zebrafish smarcd2 gene was mutated by CRISPR/Cas9 technology using the method of Gagnon et al.51. The web tool CHOPCHOP51 was used to design gene-specific spacer sequences to contribute to two single-guide RNAs (sgRNAs) for smarcd2 targeting (named S1 and S2 in Supplementary Table 3). All CHOPCHOP results were checked against the zebrafish genome database using the Ensembl genome browser. DNA templates for sgRNA synthesis were generated by annealing of two single-stranded DNA oligonucleotides (Sigma-Aldrich) followed by T4 DNA polymerase (New England BioLabs) fill-in, to make a full double-stranded DNA oligonucleotide. For each sgRNA DNA template, one oligonucleotide provided the site-specific sequence (incorporating either S1 or S2) and the second ‘constant’ oligonucleotide supplied the binding site for the Cas9 enzyme. The sgRNAs were generated by in vitro transcription (mMESSAGE mMACHINE SP6 or T7 Transcription Kit, Thermo Fisher Scientific). Transcribed sgRNA was cleaned (Sephadex G-50 spin columns, Roche Diagnostics), and its integrity was checked on 1% agarose TBE gels (Bioline, BIO-41025). See the Supplementary Note for details on statistics.
sgRNA microinjection
Individual sgRNAs (50–200 ng/µl) mixed with 20 µM Cas9 nuclease (New England BioLabs) at a 1:1 ratio were microinjected (500–1,000 pg) into the cytoplasm of one-cell-stage Tg(mpx:EGFP) embryos.
Genotyping of zebrafish
smard2 locus genotyping was performed by DNA sequencing. DNA samples were extracted from single embryos or fin clips of adult fish using the HotSHOT protocol52 and amplified by PCR (for primers and PCR conditions, see Supplementary Table 3). Following gel electrophoresis, excised bands (AccuPrep Gel Purification Kit, BIONEER) were sequenced in the Micromon sequencing facility (Monash University) using an Applied Biosystems 3730s Genetic Analyzer. F0 genotyping documented sgRNA activity. F1 genotyping was used to identify founders carrying mutated alleles. F2 genotyping assisted colony management and confirmed the genotypes of all embryos contributing to the phenotype comparison. The CRISPR/Cas9-induced smarcd2 c.66dup allele has been designated smarcd2gl35.
Sequence analysis of zebrafish results
Sequencing traces were analyzed in the DNASTAR navigator (version 2.2.1.1) and ApE (A Plasmid Editor, v.2.0.47; ref. 53). Analysis of complex, compound CRISPR/Cas9 genotypes required manual curation and interpretation of sequence chromatograms.
Mouse LSK transcriptome: RNA–seq
Mouse cell populations were sorted into RLT lysis buffer or buffer composed of 0.2% Triton X-100 (Sigma) and 2 U/µl of RNase Inhibitor (Promega). ERCC spike-in controls (Life Technologies) were added to the cell lysis mix at a 1:1,000 dilution. RNA was cleaned from the crude lysate with Agencourt RNAclean XP SPRI beads (Beckman Coulter). cDNA was synthesized and pre-amplified from 5 µl of lysate as described elsewhere54, and 0.7 ng of pre-amplified cDNA was used as input for tagmentation with the Nextera XT Sample Preparation kit (Illumina), where a second amplification round was performed for 12 cycles. For each sample, 5 ng of the final library was pooled, and 10 pmol of the library pool was sequenced with 1 × 50 base sequencing on an Illumina HiSeq 1500.
Statistics: RNA–seq data analysis
We aimed for a sample size of n = 5 according to recommendations for power in RNA–seq55, which was reached in LSK samples. For CMP, GMP, and MEP samples, a minimum sample size of n = 3 was accepted. All sorted mouse samples were processed if at least two genotypes per litter were available. The mouse fetal liver samples/cell lysates were randomized for RNA-seq library preparation by assigning a random sample number. During analysis, samples had to be unblinded. Sequencing reads were demultiplexed from the Nextera (i5 and i7) indices. Demultiplexed reads were aligned to the mouse genome (mm10) and ERCC reference using NextGenMap56. Count data were generated from mapped reads using feature-Counts57 on Ensembl gene models (GRCm38.74). To remove noise from genes with low expression levels, count data sets were subjected to data-driven gene filtering using the HTSFilter R package58.
Differential expression analysis was performed in the DESeq2 R package59. For each cell type, the full set of detected genes, their estimated log2-transformed fold change in expression, and the adjusted P values (Padj) from the Wald test are given in Supplementary Tables 4 and 7–10. For Figure 6b–e, we used the 50 genes showing the largest difference (based on P values) between the two groups and applied hierarchical clustering gene-wise and sample-wise with complete linkage based on Euclidian distances of variance-stabilized counts for differentially expressed genes. We display the 2D hierarchical cluster results as a heat map. The reference expression value is the expression average for wild-type cells. To test enrichment of functional categories, we used upregulated (log2(fold change) > 0.5) and downregulated (log2 (fold change) < −0.5) genes as the input list and all detected genes as the background list for gene set enrichment analysis using TopGO. Results obtained using the ‘elim’ algorithm are shown with the Fisher score in Supplementary Table 5. Of note, LSK sets were analyzed twice; test results and GO terms are given for both experiments in Supplementary Tables 4 and 5 (first experiment) and Supplementary Tables 7 and 13 (second experiment). Because of low sample size in the second experiment, only the heat map for the first experiment is shown (Fig. 6b).
Expression of neutrophil-specific granule genes in NB4 cells
NB4 AML cells were transduced with specific shRNAs against SMARCD2 (clone 1, V3LHS_300463; clone 2, V3LHS_400374) or non-silencing control (RHS4531) and grown in complete RPMI medium with 1 µM ATRA (dissolved in DMSO). The medium was replaced with RPMI medium supplemented with ATRA after 3 d. Cells were analyzed on days 3 and 6. RNA was extracted, cDNA was transcribed, and the expression levels of SMARCD1, SMARCD2, and SMARCD3 as well as granule genes LTF, MMP8, TCN1, CAMP, and AAT (SERPIN A1) normalized to GAPDH were detected by SYBR Green–based qPCR on an ABI Step One plus cycler. NB4 cells (obtained from DSMZ–German Collection of Microorganisms and Cell Cultures) have been regularly tested for mycoplasma (according to laboratory routine) and were found negative.
Statistics: granule gene expression
Differential expression of genes was calculated by the ΔΔCT method. Data points represent the relative fold change in cells transduced with shRNA clone 1 or 2 versus non-silencing control and individual repeat differentiation experiments. Descriptive and analytical statistics were prepared in Prism 5.0 (GraphPad), and P values are from two-tailed unpaired t tests. Center values, mean; error bars, s.d.
Chromatin immunoprecipitation
ChIP was performed as previously described60–63. Details are provided in the Supplementary Note.
RNA–seq in differentiated NB4 AML cells
NB4 AML cells transduced with specific shRNAs against SMARCD2 (clone 1, V3LHS_300463; clone 2, V3LHS_400374) or non-silencing control (RHS4531) were grown in complete RPMI medium containing 1 µM ATRA (dissolved in DMSO) or DMSO only (control) for 3 d. RNA was extracted from 1 million NB4 cells (shRNA-treated and control cells) with or without ATRA-induced differentiation. RNA was extracted using the GeneJET RNA Purification kit (Thermo Fisher Scientific), and RNA–seq library preparation was performed with the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530 S, New England BioLabs) according to the manufacturer’s instructions and sequenced on an Illumina NextSeq 500 instrument at the Dr. von Hauner Children’s Hospital NGS facility. The six libraries were sequenced together using a Mid output cartridge (FC-404-2001, 150 cycles, paired-end sequencing) reaching approximately 2 × 5 Gb per sample.
ATAC–seq in differentiated NB4 AML cells
ATAC–seq was performed as described previously64. NB4 cells (ACC207) were cultured as described above. DMSO (Sigma-Aldrich) was used as a carrier for ATRA. Cells were kept in logarithmic growth and stimulated with 1 µM ATRA or DMSO as a control. After 72 h, 50,000 cells per condition were collected and nucleus preparation was performed as described64. Isolated nuclei were treated with Tn5 transposase from the Nextera DNA Library Preparation kit (Illumina, FC-140-1089) for 30 min at 300 r.p.m. in a Thermomixer. Transposed DNA was purified with the Qiagen MinElute Reaction Cleanup kit (Qiagen, 28204) and amplified with Illumina Tn5-compatible barcoding primers (Supplementary Table 3; NEBNext Multiplex Oligos for Illumina, New England BioLabs). We ran a qPCR side reaction with 5 µl of the previously amplified library to determine the minimum number of additional PCR cycles needed (Supplementary Table 3). Minimally PCR-amplified libraries were again purified with the Qiagen MinElute Reaction Cleanup kit. Libraries were analyzed on an Agilent Bioanalyzer 2100 (High-Sensitivity DNA Chip), and size selection for the fragments was performed using AMPure beads. The 16 ATAC libraries were pooled and sequenced using a Mid output cartridge (FC-404-2001, 150 cycles, paired-end sequencing) on an Illumina NextSeq 500 instrument reaching approximately 2 × 1.8 Gb per sample.
Human RNA–seq and ATAC–seq data analysis
Demultiplexed FASTQ files were generated using bcl2fastq v2.17 (BCL2FASTQ Conversion Software 2.17, Illumina). ATAC–seq reads were mapped with BWA-MEM65, using default parameters, to the human genome (GRCh37.p13). RNA–seq reads were mapped with STAR (v2.5.0a) to the same genome in combination with the gene model annotation of GENCODE 19 (ref. 66). The R/Bioconductor67 package GenomicAlignments was used to generate gene-level count data for the RNA–seq data. The data analysis methods are described in the Supplementary Note.
Statistics: pathway analysis
Pathway analysis of human and mouse transcriptomes was carried out using Cytoscape 3.3.0 (ref. 68) together with the Reactome Functional Interaction plugin69,70. Differentially expressed mouse genes (log2 (fold change) −0.5 < x > 0.5, Padj < 0.1) or differentially expressed human genes (Padj <0.1) or intersections thereof with CEBPε target genes were loaded into the gene set–mutation analysis interface. Networks were generated with or without linker genes, as indicated. Spectral-partition-based network clustering according to ref. 71 was performed, and individual spectral clusters were analyzed by Reactome Pathway Enrichment (see URLs). Abstraction of spectral clusters (Supplementary Figs. 9 and 13) and lists of significantly enriched pathways (Supplementary Table 4) are provided. For intersection of gene lists, the Venny Venn online tool (see URLs) was used.
For further information on cell lines, plasmids, and molecular cloning; immunoprecipitation experiments and immunoblotting; flow cytometry sorting; colony-forming unit assays; morpholino knockdown experiments; phenotype analysis of zebrafish; and statistics for zebrafish morpholino experiments, see the Supplementary Note.
Data availability
The human mutation data and phenotypic descriptions that support the findings of this study have been deposited in ClinVar under accession 506022. Owing to institutional IRB restrictions and international laws, the SNP-based homozygosity data and whole-exome sequencing data for patients must not be disclosed. Mouse transcriptome data (RNA–seq) that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession GSE84703. Human NB4 cell line transcriptome data (RNA–seq) and chromatin accessibility data (ATAC–seq) that support the findings of this study have been deposited in GEO under accessions GSE93876 and GSE93877.
All other data that support the findings of this study are either available within the paper and its supplementary information files or are available from the corresponding author (C.K.) upon reasonable request.
Supplementary Material
Acknowledgments
We thank all medical and laboratory staff members involved in taking care of patients and performing scientific experiments, in particular R. Conca (FACS sorting), J. Hinke (genomic facility), and P. Robinson and S. Mundlos for next-generation sequencing expertise. We thank S. Hollizeck, D. Kotlarz, M. Lyszkiewicz, and N. Zietara for critical scientific discussion. We thank B. Zeller, R. Abdennour, and H. Nordgarden for clinical care of patients and A. Tierens for initial FACS and histological workup. We thank J. Lessard (IRIC, Université de Montréal) for providing antibodies to SMARCD1, SMARCD2, and SMARCD3 and for critical discussion.
The study has been supported by the European Research Council (ERC Advanced Grant ‘Explore’), the Else Kröner-Fresenius-Stiftung, the DZIF (German Center for Infection Research), the Deutsche Forschungsgemeinschaft (Gottfried Wilhelm Leibniz Program), the German PID-NET (BMBF), and the Care-for-Rare Foundation.
V.P. was supported by a Monash International Postgraduate Research Scholarship (MIPRS) and a Monash Graduate Scholarship (MGS). G.L. was supported by the NHMRC (1069284, 1044754). The Australian Regenerative Medicine Institute (ARMI) is supported by grants from the State Government of Victoria and the Australian Government. This research was supported in part by the Intramural Research Program of the US National Institutes of Health, NLM. W.E. and C.Z. were supported by the Deutsche Forschungsgemeinschaft (DFG) through LMUexcellent and SFB1243 (subproject A14). J.G. was supported by the Bundesministerium für Bildung und Forschung, Juniorverbund in der Sytemmedizin ‘mitOmics’ grant FKZ 01ZX1405A, and C.M. is supported by EU Horizon2020 Collaborative Research Project SOUND (633974).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.W. designed, performed, and interpreted experiments and wrote and edited the manuscript. D.P. performed ATAC–seq and RNA–seq, Y.F., E.B., T.R., and M.R. were involved in genomic and biochemical analyses, J.P. led the computational biology efforts, C.M. and J.G. analyzed ATAC–seq and RNA–seq data, and C.Z. and W.E. performed mouse RNA–seq and digital gene expression analysis. A.S.-P., P.D.A., and M.R.A. provided clinical care for patients, V.P. and G.J.L. generated and analyzed zebrafish models, and P.M.K. analyzed whole-exome sequencing in initial patients. M.D., M.R.S., and E.W. generated mice. H.-P.H. performed immunohistochemistry analysis of bone marrow biopsies, H.S. provided expert clinical genetic consulting, and A.A.S. guided bioinformatics studies and helped write and edit the manuscript. C.K. designed and guided the study, supervised M.W., provided laboratory resources, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Krumsiek J, Marr C, Schroeder T, Theis FJ. Hierarchical differentiation of myeloid progenitors is encoded in the transcription factor network. PLoS One. 2011;6:e22649. doi: 10.1371/journal.pone.0022649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krosl J, et al. A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood. 2010;116:1678–1684. doi: 10.1182/blood-2010-03-273862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 5.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang HT, et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat. Cell Biol. 2013;15:1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Álvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 2015;15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 8.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 9.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 10.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 11.Tolstorukov MY, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. USA. 2013;110:10165–10170. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 13.Buscarlet M, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jojic V, et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alajem A, et al. Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep. 2015;10:2019–2031. doi: 10.1016/j.celrep.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 16.Lickert H, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 17.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liongue C, Hall CJ, O’Connell BA, Crosier P, Ward AC. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood. 2009;113:2535–2546. doi: 10.1182/blood-2008-07-171967. [DOI] [PubMed] [Google Scholar]
- 19.Renshaw SA, et al. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 20.Koulnis M, et al. Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J. Vis. Exp. 2011;5:2809. doi: 10.3791/2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iriyama N, et al. Enhancement of differentiation induction and upregulation of CCAAT/enhancer-binding proteins and PU.1 in NB4 cells treated with combination of ATRA and valproic acid. Int. J. Oncol. 2014;44:865–873. doi: 10.3892/ijo.2013.2236. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka M, Gombart AF, Koeffler HP, Shiohara M. Expression of bactericidal/permeability-increasing protein requires C/EBPε. Int. J. Hematol. 2007;85:304–311. doi: 10.1532/IJH97.05162. [DOI] [PubMed] [Google Scholar]
- 24.Lekstrom-Himes J, Xanthopoulos KG. CCAAT/enhancer binding protein ε is critical for effective neutrophil-mediated response to inflammatory challenge. Blood. 1999;93:3096–3105. [PubMed] [Google Scholar]
- 25.Lekstrom-Himes JA, Dorman SE, Kopar P, Holland SM, Gallin JI. Neutrophil-specific granule deficiency results from a novel mutation with loss of function of the transcription factor CCAAT/enhancer binding protein ε. J. Exp. Med. 1999;189:1847–1852. doi: 10.1084/jem.189.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu H, et al. Maturity-dependent fractionation of neutrophil progenitors: a new method to examine in vivo expression profiles of differentiation-regulating genes. Exp. Hematol. 2012;40:675–681. doi: 10.1016/j.exphem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Gkikopoulos T, et al. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada T, et al. A novel in-frame deletion in the leucine zipper domain of C/EBPε leads to neutrophil-specific granule deficiency. J. Immunol. 2015;195:80–86. doi: 10.4049/jimmunol.1402222. [DOI] [PubMed] [Google Scholar]
- 29.Gallin JI, et al. Human neutrophil-specific granule deficiency: a model to assess the role of neutrophil-specific granules in the evolution of the inflammatory response. Blood. 1982;59:1317–1329. [PubMed] [Google Scholar]
- 30.Breton-Gorius J, Mason DY, Buriot D, Vilde JL, Griscelli C. Lactoferrin deficiency as a consequence of a lack of specific granules in neutrophils from a patient with recurrent infections. Detection by immunoperoxidase staining for lactoferrin and cytochemical electron microscopy. Am. J. Pathol. 1980;99:413–428. [PMC free article] [PubMed] [Google Scholar]
- 31.Komiyama A, Morosawa H, Nakahata T, Miyagawa Y, Akabane T. Abnormal neutrophil maturation in a neutrophil defect with morphologic abnormality and impaired function. J. Pediatr. 1979;94:19–25. doi: 10.1016/s0022-3476(79)80343-1. [DOI] [PubMed] [Google Scholar]
- 32.Khanna-Gupta A, et al. Growth factor independence-1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene-inactivating mutation in the C/EBPε gene. Blood. 2007;109:4181–4190. doi: 10.1182/blood-2005-05-022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike M, et al. C/EBP-ε: chromosomal mapping and mutational analysis of the gene in leukemia and preleukemia. Leuk. Res. 1997;21:833–839. doi: 10.1016/s0145-2126(97)00072-6. [DOI] [PubMed] [Google Scholar]
- 34.Verbeek W, Wachter M, Lekstrom-Himes J, Koeffler HP. C/EBPε−/− mice: increased rate of myeloid proliferation and apoptosis. Leukemia. 2001;15:103–111. doi: 10.1038/sj.leu.2401995. [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka R, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein ε–deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruickshank VA, et al. SWI/SNF subunits SMARCA4, SMARCD2 and DPF2 collaborate in MLL-rearranged leukaemia maintenance. PLoS One. 2015;10:e0142806. doi: 10.1371/journal.pone.0142806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madan V, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia. 2016;30:1672–1681. doi: 10.1038/leu.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priam P, et al. SMARCD2 subunit of the SWI/SNF chromatin-remodeling complex mediates granulopoiesis through a CEBPε- dependent mechanism. Nat. Genet. 2017 doi: 10.1038/ng.3812. http://dx.doi.org/10.1038/ng.3812. [DOI] [PubMed]
- 40.Wynn RF, et al. Intractable diarrhoea of infancy caused by neutrophil specific granule deficiency and cured by stem cell transplantation. Gut. 2006;55:292–293. doi: 10.1136/gut.2005.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotlarz D, et al. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J. Exp. Med. 2013;210:433–443. doi: 10.1084/jem.20111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohn G, et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat. Med. 2007;13:38–45. doi: 10.1038/nm1528. [DOI] [PubMed] [Google Scholar]
- 44.Hamada T, et al. Lipoid proteinosis maps to 1q21 and is caused by mutations in the extracellular matrix protein 1 gene (ECM1) Hum. Mol. Genet. 2002;11:833–840. doi: 10.1093/hmg/11.7.833. [DOI] [PubMed] [Google Scholar]
- 45.Krawitz P, et al. Microindel detection in short-read sequence data. Bioinformatics. 2010;26:722–729. doi: 10.1093/bioinformatics/btq027. [DOI] [PubMed] [Google Scholar]
- 46.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin HF, et al. Analysis of thrombocyte development in CD41–GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 2007;43:610, 612, 614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 53.Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a Bioconductor package to identify target-specific guide RNAs for CRISPR–Cas9 genome-editing systems. PLoS One. 2014;9:e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Zhou J, White KP. RNA–seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedlazeck FJ, Rescheneder P, von Haeseler A. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 2013;29:2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 58.Rau A, Gallopin M, Celeux G, Jaffrezic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29:2146–2152. doi: 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA–seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP–seq. Nat. Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cernilogar FM, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat. Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 63.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC–seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milacic M, et al. Annotating cancer variants and anti-cancer therapeutics in Reactome. Cancers (Basel) 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newman ME. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human mutation data and phenotypic descriptions that support the findings of this study have been deposited in ClinVar under accession 506022. Owing to institutional IRB restrictions and international laws, the SNP-based homozygosity data and whole-exome sequencing data for patients must not be disclosed. Mouse transcriptome data (RNA–seq) that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession GSE84703. Human NB4 cell line transcriptome data (RNA–seq) and chromatin accessibility data (ATAC–seq) that support the findings of this study have been deposited in GEO under accessions GSE93876 and GSE93877.
All other data that support the findings of this study are either available within the paper and its supplementary information files or are available from the corresponding author (C.K.) upon reasonable request.