DEAR EDITOR,
In this study, we sequenced the complete mitochondrial genome (mitogenome) of the Thai Red Junglefowl (RJF; Gallus gallus) using the next-generation sequencing (NGS) platform of the Ion Torrent PGM. Samples were taken from Mae Wang District, Chiang Mai Province, northern Thailand. Our data showed the complete mitogenome to be 16 785 bp in length, composed by 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. The genome nucleotide composition was 30.3% A, 23.7% T, 32.5% C, and 13.5% G, resulting in a high percentage of A+T (50.4%). Phylogenetic analysis revealed that the mitogenome belonged to haplogroup X, whereas those of all domestic chickens belong to haplogroups A to G. This newly released mitogenome sequence will advance further evolutionary and population genetics study of the RJF and domestic chicken. The availability of the G. gallus mitogenome will also contribute to further conservation genetics research of a unique species, listed as ‘data deficient’ in Thailand.
The Red Junglefowl (RJF; Gallus gallus) is a major wild ancestor of the domestic chicken (Darwin, 1875; Liu et al., 2006; Miao et al., 2013). Early studies based on mitochondrial DNA (mtDNA) revealed that the Thai RJF has a close relationship with the domestic chicken (Fumihito et al., 1994; Fumihito et al., 1996), implying that Thailand is likely a domestication center of the chicken. To the best of our knowledge, no complete mtDNA genome (i.e., mitogenome) sequence of the Thai RJF has been reported. In this study, we collected a RJF sample from the Mae Wang District of Chiang Mai Province in northern Thailand (permission provided by the Thai Institute of Animals for Scientific Purpose Development (No. U1-01205-2558)). The complete mitochondrial genome was submitted to GenBank (accession No.: MG605671).
Genomic DNA was extracted from whole blood using the HiPure Tissue DNA Micro Kit (Magen, China). The PCR amplification, library construction, and next-generation sequencing were in accordance with our earlier study (Chen et al., 2016). We used a de novo long fragment PCR and NGS strategy to obtain high quality mtDNA reads and exclude NTMT pseudogenes. We followed caveats for quality control in mtDNA genomic studies of domestic animals (Shi et al., 2014). The generated sequence was aligned against a reference sequence AP003321 (Nishibori et al., 2005), with all variants then output. Using the Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013), we checked the bam file exported by Torrent Suite 5.0.2 to confirm the scored variants.
Phylogenetic analysis was performed using complete mtDNA sequences of all major haplogroups and sub-haplogroups, as defined by Miao et al. (2013) and Peng et al. (2015). All mitogenomes were aligned by ClustalW, then analyzed by maximum parsimony (MP) in MEGA 7.0 with 1 000 bootstrap replicates (Tamura et al., 2011).
The complete mitogenome sequence of the Thai RJF (16 785 bp; GSA accession No.: PRJCA000287, GenBank accession No.: MG605671) had an overall base composition of 30.3% for A, 23.7% for T, 32.5% for C, and 13.5% for G, with high a A+T content of 54.0%. The mitogenome consisted of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a displacement loop (D-loop). Most mitogenome genes were encoded on the heavy strand, except for eight tRNA genes and one protein-coding gene (ND6), which were encoded on the light strand. Some protein-coding genes shared the start and stop codons; for instance, all 13 genes began with ATG, except for COX1, which started with GTG, and of the remaining 12 protein-coding genes, nine (ND1, COX2, ATPase8, ATPase6, ND3, ND4L, ND5, cyt b, and ND6) shared the stop codon TAA, two (COX3 and ND4) shard the stop codon “T– –”, and ND2 used TAG. The lengths of the 12S rRNA and 16S rRNA genes were 976 bp and 1 622 bp, respectively.
As the RJF is a wild type of Gallus gallus, it differed from all major domestic haplogroups (A to G). We calculated the differences between the Thai RJF and randomly selected individuals with haplogroups A to G (Table 1). Compared with the common domestic haplogroups, the RJF had 40 different base pairs on average (range: 29–53).
Table 1.
Mitochondrial genome differences vs. Thai Red Junglefowl (PRJA000287)
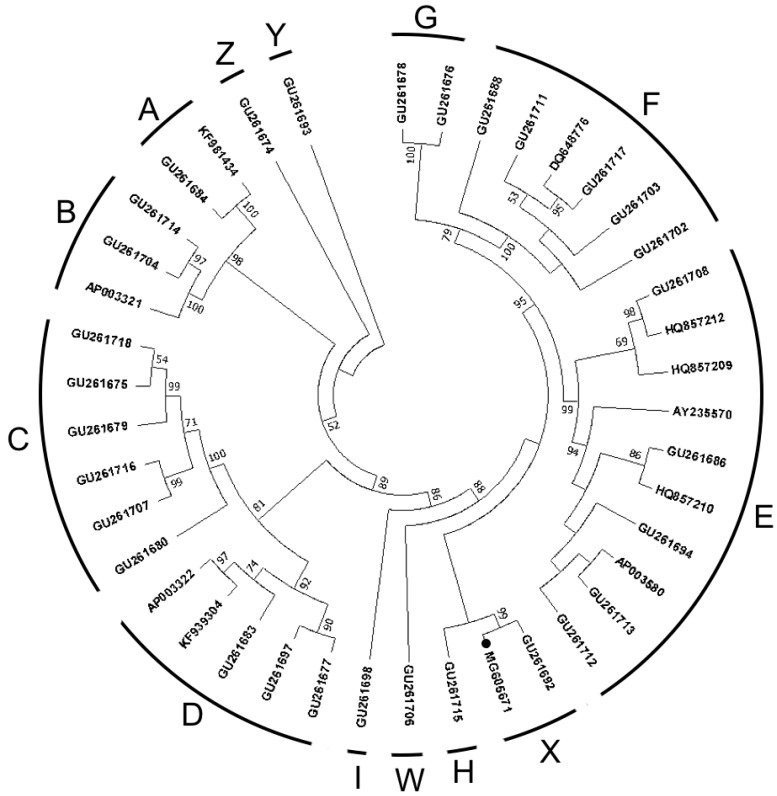
A phylogenetic tree (Figure 1) of all known Gallus gallus mitogenomes was constructed and tested with 1 000 bootstrap replications using the MEGA 7 software package (Tamura et al., 2013). The tree showed that the Thai RJF mitogenome clustered with GU261692 from Yunnan (Miao et al., 2013) into haplogroup X, as defined by Miao et al. (2013) and Peng et al. (2015). To the best of our knowledge, all domestic chickens are distributed in haplogroups A to G. However, the arrangement of the RJF was identical to haplogroup X, which is only found in wild chickens. In the MP tree, all haplogroups had very high bootstrap values, all larger than 90%. The root of the MP tree was in haplogroup Y, according to the Chicken Reference Tree in dometree.org (Peng et al., 2015). Thus, our study strongly supported the previously defined reference tree.
Figure 1.
Phylogenetic tree of 41 complete mitochondrial genomes of Gallus gallus constructed with maximum parsimony.
Black dot represents the new sample in this study. Bootstrap support values (>50) are shown at the nodes.
Here, we assembled the first complete mitogenome of the Thai RJF. This study will provide useful information for future evolutionary and population genetics analyses of the RJF and domestic chicken.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
The authors alone are responsible for the content and writing of this paper.
Funding Statement
This study was supported by the National Key Research and Development Program (2016YFC1200705), Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01: 2015CASEABRI002), Ministry of Science and Technology of China (2012FY110800), Chinese Academy of Sciences (KFZD-SW-208), and Yunnan Provincial Science and Technology Department
REFERENCES
- Chen X., Ni G., He K., Ding Z.L., Li G.M., Adeola A.C., Murphy R.W., Wang W.Z., Zhang Y.P. An improved de novo pipeline for enrichment of high diversity mitochondrial genomes from Amphibia to high-throughput sequencing. bioRxiv. 2016:080689. [Google Scholar]
- Darwin C. The variation of animals and plants under domestication. In: Barrett P.H., Freeman R.B., editors. New York University Press; New York: 1875. [Google Scholar]
- Fumihito A., Miyake T., Sumi S.I., Takada M., Ohno S., Kondo N. One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12505–12509. doi: 10.1073/pnas.91.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumihito A., Miyake T., Takada M., Shingu R., Endo T., Gojobor T., Kondo N., Ohno S. Monophyletic origin and unique dispersal patterns of domestic fowls. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P., Wu G.S., Yao Y.G., Miao Y.W., Luikart G., Baig M., Beja-Pereira A., Ding Z.L., Palanichamy M.G., Zhang Y.P. Multiple maternal origins of chickens: out of the Asian jungles. Molecular Phylogenetics and Evolution. 2006;38(1):12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Miao Y.W., Peng M.S., Wu G.S., Ouyang Y.N., Yang Z.Y., Yu N., Liang J.P., Pianchou G., Beja-Pereira A., Mitra B., Palanichamy M.G., Baig M., Chaudhuri T.K., Shen Y.Y., Kong Q.P., Murphy R.W., Yao Y.G., Zhang Y.P. Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity. 2013;110(3):277–282. doi: 10.1038/hdy.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibori M., Shimogiri T., Hayashi T., Yasue H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Animal Genetics. 2005;36:367–375. doi: 10.1111/j.1365-2052.2005.01318.x. [DOI] [PubMed] [Google Scholar]
- Peng M.S., Fan L., Shi N.N., Ning T., Yao Y.G., Murphy R.W., Wang W.Z., Zhang Y.P. DomeTree: a canonical toolkit for mitochondrial DNA analyses in domesticated animals. Molecular Ecology Resouces. 2015;15:1238–1242. doi: 10.1111/1755-0998.12386. [DOI] [PubMed] [Google Scholar]
- Shi N.N., Fan L., Yao Y.G., Peng M.S., Zhang Y.P. Mitochondrial genomes of domestic animals need scrutiny. Molecular Ecology. 2014;23:5393–5397. doi: 10.1111/mec.12955. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Q., Song F.H., Zhu J.W., Zhang S.S., Yang Y.D., Chen T.T., Tang B.X., Dong L.L., Ding N., Zhang Q., Bai Z.X., Dong X.N., Chen H.X., Sun M.Y., Zhai S., Sun Y.B., Yu L., Lan L., Xiao J.F., Fang X.D., Lei H.X., Zhang Z., Zhao W.M. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]