Abstract
Small heat shock proteins (sHsps) are a diverse group of heat-induced proteins that are conserved in prokaryotes and eukaryotes and are especially abundant in plants. Recent in vitro data indicate that sHsps act as molecular chaperones to prevent thermal aggregation of proteins by binding non-native intermediates, which can then be refolded in an ATP-dependent fashion by other chaperones. We used heat-denatured firefly luciferase (Luc) bound to pea (Pisum sativum) Hsp18.1 as a model to define the minimum chaperone system required for refolding of a sHsp-bound substrate. Heat-denatured Luc bound to Hsp18.1 was effectively refolded either with Hsc/Hsp70 from diverse eukaryotes plus the DnaJ homologs Hdj1 and Ydj1 (maximum = 97% Luc reactivation with kob = 1.0 × 10−2/min), or with prokaryotic Escherichia coli DnaK plus DnaJ and GrpE (100% Luc reactivation, kob = 11.3 × 10−2/min). Furthermore, we show that Hsp18.1 is more effective in preventing Luc thermal aggregation than the Hsc70 or DnaK systems, and that Hsp18.1 enhances the yields of refolded Luc even when other chaperones are present during heat inactivation. These findings integrate the aggregation-preventive activity of sHsps with the protein-folding activity of the Hsp70 system and define an in vitro system for further investigation of the mechanism of sHsp action.
Plants and other organisms respond to high-temperature stress with the induction of heat shock proteins (Hsps) (Lindquist and Craig, 1988; Vierling, 1991), a number of which are now known to function as molecular chaperones (Georgopoulos and Welch, 1993; Hendrick and Hartl, 1993; Boston et al., 1996). Although molecular chaperones are structurally diverse, they share the property of binding other proteins that are in non-native structural states and thereby facilitate many basic cellular processes, including protein folding, targeting, and degradation. During high-temperature stress, molecular chaperones are believed to act in preventing irreversible protein denaturation that would otherwise be detrimental to the cell (Parsell and Lindquist, 1993). While the structure and mechanism of some chaperones, such as Hsp70 and GroE (or Hsp60), have been investigated extensively (Bukau and Horwich, 1998), much less is known about the action of other Hsps, in particular the small Hsps (sHsps), which are the most diverse and abundant Hsps synthesized by plants (Waters et al., 1996). Defining the function of sHsps is clearly critical to understanding the response of plants to high-temperature stress.
Plant sHsps are members of a diverse group of proteins that are conserved in both eukaryotes and prokaryotes (Gaestel et al., 1997; Vierling, 1997) and share a characteristic approximately 100-residue carboxyl-terminal domain with signature amino acid motifs (Plesofsky-Vig et al., 1992; deJong et al., 1993; Waters et al., 1996; Gaestel et al., 1997). While sHsps have monomeric molecular masses in the range of 15 to 40 kD, they typically form oligomers of nine to 32 subunits in the native state, with the oligomeric size specific to the individual sHsp. Plant sHsps can be divided into at least five gene families, two families of proteins found in the cytoplasm (class I and II), plus three organelle-localized (mitochondrion, chloroplast, and endoplasmic reticulum) families (Waters et al., 1996).
Recently, we and others have proposed a model for the molecular chaperone activity of sHsps (Waters et al., 1996; Ehrnsperger et al., 1997; Lee et al., 1997; Veinger et al., 1998). This model proposes that sHsps act to bind non-native proteins, preventing their aggregation and maintaining them in a state competent for ATP-dependent refolding by other chaperones. Multiple studies support the first part of this model. Experiments performed in vitro using sHsps from a variety of organisms have demonstrated that sHsps are particularly effective in preventing thermal aggregation of other proteins by an ATP-independent mechanism (Horwitz, 1992; Jakob et al., 1993; Jinn et al., 1995; Chang et al., 1996; Kim et al., 1998). We have characterized this activity for pea (Pisum sativum) Hsp18.1, a dodecameric, cytosolic, class-I sHsp, using a range of heat-sensitive model proteins as substrates (Lee et al., 1995, 1997). When malate dehydrogenase, citrate synthase, glyceraldehyde-3-P dehydrogenase, or firefly luciferase (Luc) are heat-inactivated in the presence of Hsp18.1, they bind the sHsp and form soluble high-Mr complexes. In contrast, in the absence of the sHsps, these heat-sensitive proteins form insoluble aggregates.
Support for the second part of the model, that sHsp-bound proteins are effective substrates for refolding by other chaperones plus ATP, is very limited. Ehrnsperger et al. (1997) reported that heat-inactivated citrate synthase bound to murine Hsp25 could be partially reactivated in the presence of bovine Hsp70 and ATP. However, over 2 h were required to achieve less than 15% reactivation, which represented only a 2.5-fold stimulation over spontaneous refolding. We reported that complete reticulocyte lysate can rapidly reactivate Luc, which had been heat-denatured and complexed with pea Hsp18.1, to over 40% of the initial activity in an ATP-dependent reaction (Lee et al., 1997). Wheat germ lysate was also effective in Luc refolding. However, the active components of neither lysate were defined.
In vivo interaction of Hsp70 and an sHsp in renaturation of heat-denatured Luc has been implicated by experiments performed by Forreiter et al. (1997), who showed that Luc expressed in Arabidopsis cells remained more active and recovered faster from heat denaturation in vivo when protoplasts were first transfected with plasmids directing the expression of Hsp70 and an sHsp. Experiments by Goloubinoff and colleagues (Veinger et al., 1998) extended the sHsp model to prokaryotes, with investigations of the chaperone function of the Escherichia coli sHsp IbpB. Their experiments indicated that up to 85% of malate dehydrogenase bound to IbpB could be reactivated by being passed from IbpB to the prokaryotic Hsp70 system (DnaK, DnaJ, and GrpE system (Liberek et al., 1991) and then to GroE, which perform ATP-dependent protein folding.
We sought to gain further support for the model of sHsp function and to define more precisely the chaperone requirements for efficient refolding of a heat-denatured protein bound to an eukaryotic sHsp. For this purpose, we used heat-denatured Luc complexed with pea Hsp18.1 as a substrate for in vitro reconstitution of protein folding. Although Luc is not a plant protein, because of the availability of a highly sensitive assay for Luc activity, Luc has been used extensively as a model substrate for protein folding in vitro (Schumacher et al., 1994; Levy et al., 1995; Buchberger et al., 1996), and has also been studied in vivo in prokaryotes (Schröder et al., 1993), animals (Nguyen et al., 1989; Pinto et al., 1991; Prip-Buus et al., 1996), and, as mentioned above, in plants (Forreiter et al., 1997). These data, as well as the fact that Luc is inactivated at moderate temperatures (39°C–42°C), corresponding to the temperature of Hsp induction in plants (Vierling, 1991), make Luc an excellent model protein for chaperone studies.
We report that >80% of heat-inactivated, Hsp18.1-bound Luc can be reactivated in a reaction that requires eukaryotic Hsc70/Hsp70, co-chaperones (Caplan et al., 1993; Silver and Way, 1993) and ATP. Reactivation is even more efficient with the prokaryotic Hsp70 (DnaK) system (100% yield), suggesting that the refolding mechanism does not require specific interactions between sHsps and Hsp70 systems. We also define functional differences between sHsps and Hsc70/Hsp70 systems in terms of preventing protein aggregation and facilitating refolding. These data extend the model of sHsp function and define an in vitro system for further mechanistic studies of sHsp action.
MATERIALS AND METHODS
Materials
ATP disodium salt, bovine serum albumin (BSA) (no. A-3803), protein A agarose (no. P-7786), apyrase, and reagent grade bovine or rabbit IgG were purchased from Sigma (St. Louis). Phosphoenolpyruvate and pyruvate kinase were obtained from Boehringer Mannheim (Basel). Recombinant firefly luciferase was purchased from Promega (Madison, WI) and rabbit reticulocyte lysate from Green Hectares (Oregon, WI).
Chaperone Proteins
Recombinant Hsp18.1 was expressed in Escherichia coli cells and purified as described previously (Lee et al., 1995; Lee and Vierling, 1998). The plasmid constructs for recombinant human Hsc70 and Hdj1, obtained from Dr. R. Morimoto (Northwestern University, Evanston, IL), were expressed in E. coli, and the proteins purified as described previously (Freeman et al., 1995). Human Hop, Hsp90, and yeast Ydj1 were provided by Dr. D. Toft (Mayo Clinic, Rochester, MN). Yeast Ssa1 was provided by Drs. J. Glover and S. Lindquist (Howard Hughes Medical Institute, University of Chicago). The plasmid constructs for E. coli DnaK and DnaJ (gift of Dr. C. Georgopoulos, University of Geneva), were expressed in E. coli, and the proteins purified as described previously (Cegielska and Georgopolous, 1989; Zylicz et al., 1989). E. coli GrpE was purchased from StressGen Biotechnologies (Victoria, British Columbia).
Wheat germ Hsp70 was isolated from wheat germ purchased at a grocery store and ground with a mortar, pestle, and sand in 25 mm Tris, pH 7.5. The clarified extract was precipitated between 20% to 50% (w/v) ammonium sulfate, resuspended, and dialyzed against 25 mm Tris, 50 mm NaCl, pH 7.5, then isolated by DEAE Sepharose fast-flow chromatography with a 50 to 200 mm NaCl gradient. Hsp70 was further purified by ATP-agarose (C-8 linkage, Fluka, Milwaukee, WI) chromatography as described previously (Freeman et al., 1995), then purified to greater than 95% homogeneity on a strong anion-exchange HPLC column (Rainin Instrument, Woburn, MA) with a 0 to 0.4 m NaCl gradient.
Protein concentrations were determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA) using BSA as the standard. Hsp18.1 concentrations in the text and figures refer to the 12-subunit oligomer. Concentrations of Luc and all other chaperone proteins refer to monomers.
Formation of sHsp/Luciferase Complexes
Hsp18.1/Luc complexes were formed basically as described previously (Lee et al., 1997; Lee and Vierling, 1998) by heating 1 μm Luc with 1 μm Hsp18.1 (50 μL) in 25 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES),5 mm MgCl2, 150 mm KCl, and 2 mm dithiothreitol, pH 7.5 (denaturation buffer) for 8 min at 42°C in siliconized 0.65-mL tubes. After heating, samples were immediately cooled on ice for 30 s. When complexes were formed in the presence of ATP, samples were supplemented with 2 mm ATP and the heating period was extended to 30 min. As determined by size exclusion chromatography, less than 5% of Luc remained in the free form in these reactions (Lee et al., 1997). Where indicated, Hsp18.1 was substituted with 2 μm DnaK, 0.4 μm DnaJ, and 0.8 μm GrpE.
Luc Refolding Experiments
Hsp18.1/Luc complexes or Luc heated in the presence of DnaK, DnaJ, and GrpE were prepared as described above. Refolding was initiated by adding complexes (25 nm Luc) to 50% (v/v) rabbit reticulocyte lysate or various combinations of chaperones at the indicated concentrations in denaturation buffer with 50 mm KCl (instead of 150 mm) plus 2 mm ATP (40-μL total) at 30°C in siliconized 0.65-mL tubes. When purified chaperones were substituted for reticulocyte lysate, the samples were supplemented with freshly prepared 0.5 mg/mL BSA to prevent Luc adsorption to tubes, and the Hsc70/Hsp70/Ssa1, Hop, or Hsp90 concentrations were 1.5, 0.3, or 1.5 μm, respectively, while the concentration of Hdj1 or Ydj1 was either 0.15 or 0.30 μm as indicated. The concentration of IgG was 1.25 mg/mL. When the DnaK system was used, the DnaK, DnaJ, and GrpE concentrations were 1.5, 0.3, and 0.6 μm, respectively, and the KCl concentration was 150 mm. Luc activities were determined by adding 2.5 μL of the refolding reaction to 50 μL of Luciferase Assay Mix (Promega) and monitoring light emission in a TD-20/20 luminometer (Turner, Sunnyvale, CA). Luc activities are expressed as percentages relative to that of an equivalent amount of native Luc measured prior to the heating step in the formation of Hsp18.1/Luc complexes.
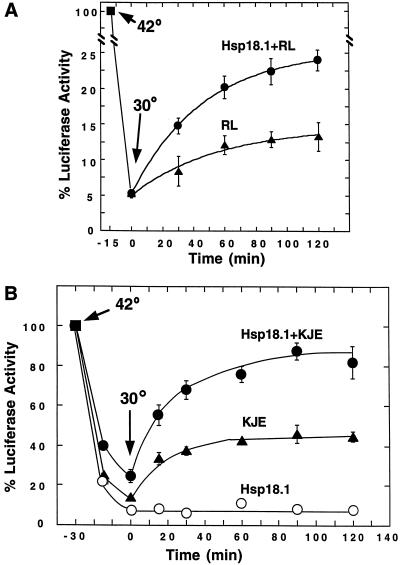
For the experiment presented in Figure 4A, 1 μm Luc was mixed with 50% (v/v) rabbit reticulocyte lysate with or without 0.5 μm Hsp18.1 in denaturation buffer with 50 mm KCl containing an ATP regeneration system consisting of 2 mm ATP, 10 mm phosphoenolpyruvate, and 0.2 unit of pyruvate kinase. The 50-μL samples were heated for 15 min at 42°C, then shifted to 30°C. Prior to measuring Luc activities, aliquots were first diluted 1:50 in 1 mg/mL BSA in 20 mm HEPES and 50 mm NaCl, pH 7.5. For the experiments presented in Figure 4B, reticulocyte lysate was replaced by 2 μm DnaK/0.4 μm DnaJ/0.8 μm GrpE or 1 μm Hsp18.1, or with the DnaK and sHsp systems combined, and the KCl concentration was 150 mm.
Figure 4.
Hsp18.1 enhances the ability of reticulocyte lysate and the DnaK system to refold Luc. A, 1 μm Luc in the presence of ATP was mixed with 50% (v/v) reticulocyte lysate (RL, ▴) or 0.5 μm Hsp18.1/50% (v/v) reticulocyte lysate (Hsp18.1 + RL, ●), heated for 15 min at 42°C, and then shifted to 30°C to allow refolding. B, Luc in the presence of ATP was mixed with Hsp18.1 (○), DnaK/DnaJ/GrpE (KJE, ▴), or Hsp18.1/DnaK/DnaJ/GrpE (Hsp18.1 + KJE, ●), denatured for 30 min, and then shifted to 30°C to allow refolding. Concentrations in B were 1 μm Luc, 1 μm Hsp18.1, 2 μm DnaK, 0.4 μm DnaJ, and 0.8 μm GrpE.
Data points and associated error bars represent the means and sd from at least three replicates. Apparent final Luc refolding yields and rate constants were determined by fitting data points to single exponential equations using least squares analysis. Associated errors reflect the error derived in the least squares analysis. Luc reactivation rates in all assays obeyed pseudo-first-order kinetics (R ≥ 0.988).
Luc Aggregation
Luc (1 μm) was heated with 0.7 μm Hsp18.1, 2 μm DnaK/0.4 μm DnaJ/0.8 μm GrpE, 2 μm Hsc70/0.2 μm Hdj1/0.2 μm Ydj1, or 0.80 mg/mL IgG in the presence of 2 mm ATP for 30 min at 42°C as described for the formation of complexes. After heating, samples (50 μL) were centrifuged for 15 min at 16,250g and the supernatant fractions removed. Both the supernatant and pellet fractions were treated with SDS sample buffer and analyzed by SDS-PAGE and Coomassie Blue staining.
Immunoprecipitation
Prior to immunoprecipitation, samples were ATP depleted with 0.5 unit of apyrase. Samples were precipitated for 30 min on ice with protein A agarose coupled to rabbit anti-Luc IgG (Cortex Biochem, San Leandro, CA) or an equivalent weight of non-specific rabbit IgG. Samples were washed three times with 25 mm HEPES, 200 mm NaCl, and 0.5% (v/v) Triton X-100, pH 7.5. Samples were treated with SDS sample buffer, separated by SDS-PAGE, and then subjected to western-blot analysis probed with anti-Luc, anti-Hsp18.1, or anti-Hsc70 (StressGen Biotechnologies) antibodies detected by chemiluminescence (Amersham-Pharmacia Biotech, Uppsala).
RESULTS
Eukaryotic or Prokaryotic Hsp70 and Co-Chaperones Support High Refolding Yields from Hsp18.1/Luc Complexes
To begin to define the basic mechanism by which sHsps could facilitate refolding of denatured proteins, we sought to reconstitute a refolding system using other known eukaryotic molecular chaperones in conjunction with pea Hsp18.1. Since our previous experiments had shown that Luc refolding was effectively accomplished with reticulocyte lysate (Lee et al., 1997), we first tested whether the Hsc/Hsp70 present in the lysate was essential for refolding activity. Immune-depletion experiments confirmed that Hsc/Hsp70 was required for Luc reactivation from Hsp18.1/Luc complexes in the lysate-supported reaction (data not shown).
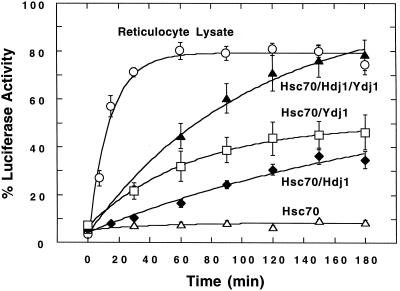
We then evaluated purified Hsc70 for its ability to reactivate Luc bound to Hsp18.1. When Hsp18.1/Luc complexes were added to excess human Hsc70 plus ATP, essentially no Luc reactivation was observed (Fig. 1). In contrast, when 1.5 μm Hsc70 was supplemented with 0.3 μm of the Hsc/Hsp70 DnaJ co-chaperones human Hdj1 or yeast Ydj1, which are required for maximum Hsp70 ATPase activity (Caplan et al., 1993; Silver and Way, 1993), significant Luc activity could be recovered. Curve fitting indicated final yields of 64% or 50%, with rate constants of 0.4 × 10−2 or 1.5 × 10−2 min−1 with added Hdj1 of Ydj1, respectively (Table I). Surprisingly, substantial enhancement of the refolding yield was observed when the two DnaJ homologs were combined with Hsc70 (Fig. 1). The combination of Hdj1 plus Ydj1 (both 0.15 μm) supported an extrapolated endpoint yield of 97% Luc activity (Table I). This enhancement in Luc refolding was not due to the increased ratio of Hsc70 to either Hdj1 or Ydj1, since the same refolding yield and rate was observed when either 0.15 or 0.3 μm Hdj1 or Ydj1 was added separately to 1.5 μm Hsc70 (data not shown).
Figure 1.
Refolding of Hsp18.1-bound Luc by reticulocyte lysate and eukaryotic chaperones. Hsp18.1/Luc was added to Hsc70 (▵), Hsc70/Hdj1 (♦), Hsc70/Ydj1 (□), Hsc70/Hdj1/Ydj1 (▴), or 50% (v/v) reticulocyte lysate (○), and allowed to refold at 30°C. Concentrations were: 25 nm Hsp18.1/Luc, 1.5 μm Hsc70, 0.30 μm Hdj1, 0.30 μm Ydj1, or 0.15 μm each Hdj1 and Ydj1 when used in combination.
Table I.
Apparent Luc refolding yields and rate constants in the presence of eukaryotic folding components
| Folding Components | % Yield | kob (× 10−2 min−1) |
|---|---|---|
| Reticulocyte lysate | 79 ± 2 | 7.1 ± 0.9 |
| IgG | 7 ± 1 | ND |
| Hsc70 | 9 ± 1 | ND |
| Hsc70, Hdj1 | 64 ± 8 | 0.4 ± 0.2 |
| Hsc70, Ydj1 | 50 ± 1 | 1.5 ± 0.1 |
| Hsc70, Hdj1, Ydj1 | 97 ± 10 | 1.0 ± 0.2 |
| wHsp70, Hdj1, Ydj1 | 92 ± 10 | 0.8 ± 0.1 |
| Ssa1, Hdj1, Ydj1 | 74 ± 3 | 1.4 ± 0.2 |
| Hsc70, Hdj1, Ydj1, Hop | 86 ± 3 | 1.7 ± 0.2 |
| Hsc70, Hdj1, Ydj1, Hop, Hsp90 | 93 ± 3 | 1.7 ± 0.2 |
Luc (1 μm) was heated with 1 μm Hsp18.1 in the absence of ATP for 8 min at 42°C. Heated samples were diluted to 25 nm Luc into refolding reactions containing the indicated folding components plus ATP, then monitored for Luc refolding as shown in Figure 1. Data points were fitted to single exponential equations from which endpoint yields and observed rate constants (kob) were calculated. ND, Not determined. Concentrations of folding components were 50% (v/v) reticulocyte lysate, 1.5 μm Hsc70, wheat germ Hsp70 (wHsp70) or yeast Ssa1, 0.3 μm Hdj1 or Ydj1, or 0.15 μm each Hdj1 and Ydj1 when combined, 0.3 μm Hop, and 1.5 μm Hsp90.
Essentially no difference in refolding was observed when human Hsc70 was substituted with an equivalent amount of wheat germ Hsp70, while the yeast Hsp70 homolog Ssa1 supported a lower final yield, but with a somewhat higher rate constant (Table I). Thus, a highly functional eukaryotic refolding system can be assembled with a combination of plant, human, and yeast chaperone homologs without apparent species specificity. This result is consistent with the ability of heterologous DnaJ proteins to interact with and stimulate ATP hydrolysis by diverse Hsp70 proteins (Kroczynska et al., 1996; Takayama et al., 1999).
We considered that the addition of other eukaryotic chaperones or co-chaperones could be required for optimal refolding in conjunction with Hsc70/Hdj1/Ydj1. In some cellular processes, such as activation of steroid hormone receptors, Hsp70 is known to act in a large chaperone complex involving Hsp90 and a variety of co-chaperones including Hop (Hsp70–Hsp90 organizing protein) (Frydman and Höhfeld, 1997; Pratt and Toft, 1997). Hsp90 and Hop are found in plants (Hernandez Torres et al., 1995; Boston et al., 1996), and the same complex of chaperones exists in plants, as evidenced by the substitution of wheat germ lysate for reticulocyte lysate in steroid receptor activation reactions (Stancato et al., 1996). However, addition of human Hsp90 had no effect on the yield or rate of refolding, and supplementation of the system with Hop plus Hsp90 only moderately increased the apparent rate constant of refolding (Table I).
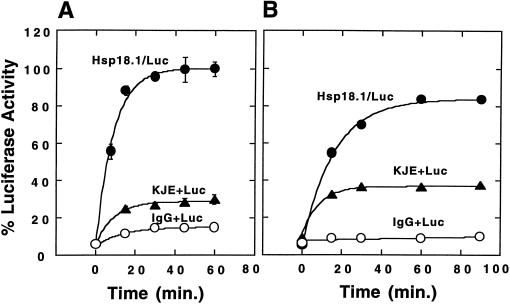
We next determined if refolding from this higher plant sHsp could be accomplished with the prokaryotic Hsp70 system comprised of DnaK, DnaJ, and GrpE. Interestingly, Hsp18.1/Luc complexes supported faster and higher refolding with the DnaK system compared with reticulocyte lysate; 100% of the original Luc activity was recovered with an observed rate constant of kob = 11.3 × 10−2/min (Fig. 2A).
Figure 2.
Refolding of Hsp18.1-bound Luc by the DnaK system. Luc (1 μm) was heated in the presence of 1 μm Hsp18.1 (Hsp18.1/Luc, ●), 2 μm DnaK/0.4 μm DnaJ/0.8 μm GrpE (KJE + Luc, ▴) or 0.21 mg/mL bovine IgG (IgG + Luc, ○) at 42°C for either 8 min in the absence of ATP (A) or for 30 min in the presence of ATP (B). Samples containing 25 nm Luc were then added to 1.5 μm DnaK/0.3 μm DnaJ/0.6 μm GrpE and ATP and allowed to refold at 30°C.
Hsp70 Systems Do Not Substitute for sHsp Activity in Maintaining Folding Competence and Substrate Solubility
Like sHsps, the DnaK system has been previously demonstrated to prevent thermal aggregation of Luc (Schröder et al., 1993). Because of this apparent overlap in activity, we wanted to determine what functional attributes distinguish sHsps from the DnaK and Hsp70 systems. To investigate this, first the DnaK system (DnaK/DnaJ/GrpE at 2.0:0.4:0.8 μm) was used instead of 1 μm Hsp18.1 during Luc inactivation (42°C for 8 min, minus ATP). After dilution into excess DnaK system, Luc heated with DnaK/DnaJ/GrpE in the absence of ATP refolded to less than one-third of the activity relative to Luc heated in the presence of sHsps (100% versus 29%) (Fig. 2A).
As the chaperone activities of DnaK are ATP dependent (Wawrzynów et al., 1995), the experiment was repeated using heating reactions that were supplemented with ATP (Fig. 2B). ATP, a Luc substrate, was found to stabilize Luc during heating and, therefore, 30 min at 42°C was required to inactivate Luc to a similar extent (approximately 5% of the original activity) as in the absence of ATP. When the ATP-supplemented samples were refolded with excess DnaK/DnaJ/GrpE, Luc heated in the presence of sHsp gave rise to at least twice the Luc activity relative to that heated in the presence of DnaK/DnaJ/GrpE (84% versus 37%) (Fig. 2B).
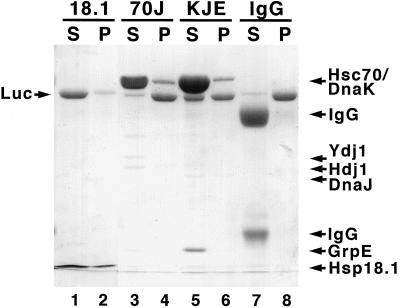
Since protection of substrates from aggregation is another assay for the chaperone activity of the sHsps (Lee et al., 1997), we analyzed the relative effectiveness of Hsp18.1 and Hsp70 systems in this type of assay. The extent of aggregation protection associated with equal weights of Hsp18.1, Hsc70/Hdj1/Ydj1, or DnaK/DnaJ/GrpE was compared. Luc was first heated with ATP in the presence of Hsp18.1 or either of the Hsp70 systems (same conditions used in Fig. 2B), then the samples were centrifuged to pellet insoluble Luc and analyzed by SDS-PAGE. Nearly all of the Luc heated in the presence of the Hsp18.1 remained soluble (Fig. 3). In contrast, virtually all of the Luc heated in the presence of Hsc70/Hdj1/Ydj1 partitioned into the pellet fraction, while Luc heated in the presence of DnaK/DnaJ/GrpE partitioned approximately equally between the supernatant and pellet fractions (Fig. 3). Therefore, the yields of refolding reflect the protection of the Luc from aggregation by the various chaperone systems.
Figure 3.
Hsp18.1 maintains higher Luc solubility than Hsp70 systems. Luc (1 μm) in the presence of ATP and 0.7 μm Hsp18.1, 2 μm Hsc70/0.15 μm Hdj1/0.15 μm Ydj1 (70J), 2 μm DnaK/0.4 μm DnaJ/0.8 μm GrpE (KJE), or 0.21 mg/mL bovine IgG was maintained at 42°C for 30 min. Samples were centrifuged for 15 min at 16,250g, separated into supernatant (S) or pellet (P) fractions, and then analyzed by SDS-PAGE and Coomassie Blue staining. The Hsp18.1 bands appear narrow because they are compressed at the buffer front.
Hsp18.1 Also Enhances Refolding When Combined with the Reticulocyte Lysate or DnaK Systems during Heating
We next asked if sHsps could also enhance Luc refolding in conditions more similar to those found in cells, i.e. when both the Hsp70 system and sHsp are present during heating and when refolding is carried out without dilution and the addition of excess chaperones. Despite the extensive endogenous chaperone network in reticulocyte lysate, 0.5 μm Hsp18.1 also enhanced refolding of Luc that was heat denatured directly in 50% (v/v) reticulocyte lysate (Fig. 4A). Taking into account the 5% residual Luc activity following 15 min of heat denaturation, the addition of Hsp18.1 doubled the amount of Luc reactivation over that obtained with the lysate alone.
We also addressed this question in the prokaryotic system. Luc (1 μm) was heated for 30 min at 42°C in the presence of 1 μm Hsp18.1 plus DnaK/DnaJ/GrpE (2.0:0.4:0.8 μm) and ATP, then cooled to 30°C to initiate Luc refolding. Under these conditions the inclusion of Hsp18.1 significantly increased the reaction yields; recovery of Luc activity was more than 2-fold higher in samples containing Hsp18.1 compared with samples containing only the DnaK system (Fig. 4B).
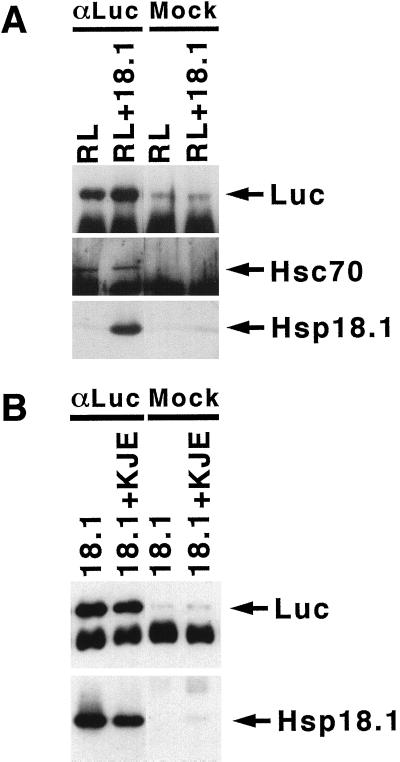
Immunoprecipitation with anti-Luc IgG, performed immediately after the heating step, was used to assess the relative amount of Luc bound to Hsp18.1 when either the reticulocyte lysate or purified DnaK components and ATP were also present during heating (Fig. 5). In reticulocyte lysate, endogenous Hsc70 and Hsp18.1 were associated with Luc immediately following heating (Fig. 5A), suggesting that both chaperones were involved in stabilizing heat-denatured Luc. Comparison of western-blot signals from immunoprecipitates to that of purified protein standards indicated that approximately equal weights of Luc, Hsp18.1, and Hsc70 were immunoprecipitated (not shown). Hsp18.1 was also found associated with Luc in the presence of the DnaK system (Fig. 5B) and, compared with in its absence, the presence of the DnaK system did not significantly reduce the amount of Hsp18.1 found associated with Luc. The amount of Luc associated with DnaK could not be determined because DnaK bound non-specifically to IgG-coupled protein A resin (not shown).
Figure 5.
Hsp18.1 binds heat-denatured Luc in the presence of other chaperones. A, Samples similar to those in Figure 4A containing Luc and reticulocyte lysate (RL) supplemented with or without Hsp18.1 were heated for 15 min at 42°C in the presence of ATP. B, Samples similar to those in Figure 4B containing Luc and Hsp18.1 with or without the DnaK system (KJE), were heated for 30 min at 42°C in the presence of ATP. After heating, Luc was immunoprecipitated with anti-Luc IgG (αLuc) or non-specific rabbit IgG (mock). Immunoprecipitates were analyzed by western blot for the presence of Luc, Hsp18.1, or Hsc70. Bands below Luc and Hsp70 are IgG heavy chains.
DISCUSSION
Current models derived from in vitro studies propose that sHsps prevent irreversible substrate aggregation by binding heat-denatured substrates, and then present substrate to other cellular components for ATP-dependent refolding (Waters et al., 1996; Ehrnsperger et al., 1997; Lee et al., 1997; Veinger et al., 1998). Our data extend this model for sHsp chaperone activity in several important ways. First, we determined that the chaperones required for high levels of refolding of Hsp18.1-bound Luc were Hsp/Hsc70 plus DnaJ homologs. The addition of Hsp90 and Hop gave minimal or no further enhancement of refolding. Despite the eukaryotic origin of Hsp18.1, the highest Luc refolding rates were observed when Hsp18.1-bound Luc was reactivated in the presence of the prokaryotic DnaK system. These findings imply that the mechanism of sHsp action in conjunction with Hsp70 systems is universal among eukaryotes and prokaryotes, and suggest that sHsps may not physically interact with the Hsp70 systems. Also, the present study clarified a distinction between the activities of sHsps and Hsp70 systems in the protection of substrate during heat denaturation. sHsps clearly enhanced recovery of substrate activity even in the presence of other chaperones, as would be typical of conditions within the cell. Moreover, the sHsps have a higher apparent substrate capacity than Hsp70 systems and, unlike the Hsp70 systems, substrate protection is ATP independent.
In defining the minimal components for refolding sHsp-bound Luc, we found a strict requirement for a DnaJ homolog in conjunction with eukaryotic Hsp70. Ehrnsperger et al. (1997) observed small amounts of citrate synthase reactivation when murine Hsp25/citrate synthase complexes were incubated with bovine Hsp70 in the absence of a DnaJ homolog. This difference in DnaJ homolog requirement may reflect differences in the substrates used and/or the sHsps themselves, or it may account for the low refolding yields reported by these researchers. Several other studies have reported the absolute requirement of DnaJ homologs to stimulate eukaryotic Hsp70-mediated refolding of chemically and heat-denatured proteins (Freeman and Morimoto, 1996; Schumacher et al., 1996; Johnson et al., 1998). We also found that the presence of two DnaJ homologs, Hdj1 and Ydj1, significantly increased the rate and yield of Luc refolding compared with that obtained with either DnaJ protein alone. Additive activities of different DnaJ proteins on Hsp70 function have not previously been reported. This effect may arise from structural differences between the two DnaJ proteins. Ydj1, Hdj2, and DnaJ all possess a Cys-rich region that in DnaJ forms a zinc finger motif capable of interacting with non-native substrates (Szabo et al., 1994). In contrast, Hdj1 lacks this domain. It is possible that Hdj1 and Ydj1, in conjunction with Hsc/Hsp70, stabilize different Luc intermediates or regulate the Hsc/Hsp70 ATPase differently so that a wider range of non-native Luc intermediates can be reactivated.
Our reconstituted Hsc70/Hdj1/Ydj1 system matched the final Luc yield of complete reticulocyte lysate, but operated at a 7-fold lower rate. We anticipated that the addition of other chaperones or co-chaperones would increase the refolding rate in this purified system; Johnson et al. (1998) found that refolding of heat-denatured Luc by Hsc70 and Ydj1 was stimulated by Hop and Hsp90. However, in our system, the addition of Hop or Hsp90 had little effect. Also, we found that the Hsp90-specific inhibitor geldanamycin (Whitesell et al., 1994) did not affect refolding from Hsp18.1/Luc complexes in reticulocyte lysate (G. J. Lee and E. Vierling, unpublished observations), further supporting a lack of requirement for Hsp90 in our system. It is possible that the identity or nature of the substrate may determine whether Hop and Hsp90 are required for folding. For example, in our studies, Luc was added to the refolding components as Hsp18.1/Luc complexes, while in the studies of Johnson et al. (1998), Luc was added in a heat-denatured, aggregation-prone form. For Luc in the latter form, Hop and Hsp90 may be instrumental in stabilizing the initial folding intermediates as well as in assisting Hsc70 in subsequent refolding steps. In total, our results suggest that other unidentified components are required for optimal rates of refolding of sHsp-bound Luc with eukaryotic Hsp70. This is not surprising in view of the continuing identification of new proteins that modulate eukaryotic Hsp70 activity (Takayama et al., 1999). Alternatively, the reaction conditions for the Hsp70/Hdj1/Ydj1 Luc refolding are not yet optimized or a fully homologous eukaryotic system could be required for maximal rates.
Although sHsps and Hsp70 systems share the common ability to prevent thermal aggregation of proteins, Hsp18.1 appears more effective than Hsp70 systems alone. Equimolar Hsp18.1 oligomer to Luc was sufficient to almost completely prevent Luc aggregation during heat denaturation. Previous studies utilizing Hsp70 systems alone have achieved Luc protection only with a large molar excess of Hsc70/Hdj1 (47- and 32-fold, respectively) (Minami et al., 1996) or DnaK/DnaJ (5- and 2-fold, respectively) (Schröder et al., 1993). These observations are consistent with our previous findings that the binding capacity of Hsp18.1 is exceptionally large, and in the case of malate dehydrogenase, up to one malate dehydrogenase subunit can be bound per Hsp18.1 subunit (Lee et al., 1997), which corresponds to twice the mass of substrate as sHsp. Other studies have also observed high substrate binding capacities for sHsps and the related α-crystallins from a variety of different organisms (Horwitz, 1992; Chang et al., 1996; Ehrnsperger et al., 1997; Leroux et al., 1997). This high substrate capacity contrasts sharply with substrate binding to Hsp70, which binds only a single substrate polypeptide (Palleros et al., 1991), or to GroEL, which can bind a maximum of two polypeptides per oligomer (Hartl, 1996).
The positive effect of Hsp18.1 on Luc reactivation was also observed in experiments that placed Luc, Hsp18.1, and the reticulocyte lysate or DnaK systems together during heat denaturation as well as refolding. These experiments eliminated the ordered addition of specific, pre-formed complexes to excess refolding system. In these experiments, we found that DnaK alone could stabilize and refold Luc, as has also been reported by Schröder et al. (1993). However, Hsp18.1 significantly enhanced the DnaK-mediated Luc refolding, with yields greater than 80%. A similar effect was also observed when reticulocyte lysate was supplemented with Hsp18.1. Although we have not directly determined the relative affinity of heat-denatured Luc for sHsps versus Hsp70s in these mixtures, co-precipitation of both Hsp18.1 and Hsc70 with Luc directly after heating demonstrates that the substrate associates with similar amounts of either protein.
A more comprehensive model of sHsp activity can now be developed based on data presented here. We propose that during high-temperature stress, sHsps act to bind substrate proteins in an ATP-independent manner, even in the presence of other chaperones, and that specific substrates may preferentially bind to sHsps. The high capacity of sHsps for substrate compared with the Hsc70 (or the DnaK) system, and the fact that sHsps can accumulate to over 1% of total protein in plant (Waters et al., 1996) and other cells (Arrigo and Landry, 1994), indicates that sHsps are likely an important reservoir for folding-competent, denatured proteins, as was previously suggested (Waters et al., 1996; Ehrnsperger et al., 1997; Lee et al., 1997).
In eukaryotes, substrate is then passed to Hsc70 and co-chaperones for ATP-dependent folding, and can undergo multiple rounds of interaction with the Hsc70 system in order to achieve the final native state. It is unclear whether substrate transfer to the folding system is an active process. However, the ability of diverse combinations of prokaryotic and eukaryotic components to effect folding of Hsp18.1-bound Luc, albeit with differing efficiencies, suggests that no highly specific, direct interactions of sHsps themselves with the folding system components are required. Altogether, optimal reactivation of heat-denatured proteins can be achieved by combining the functional specialization of sHsps and the Hsc70 system. The data of Forreiter et al. (1997) showing that transient expression of Hsp70 and an sHsp in Arabidopsis protoplasts can enhance Luc protection from heat can now be explained in terms of this model. Our reconstituted system for sHsp-facilitated substrate refolding will allow further studies of the mechanisms involved in this process.
A considerable amount of data support a role for sHsps in the acquisition of thermotolerance to high temperature (Vierling, 1991, 1997; Arrigo and Landry, 1994), although in some organisms or under some environmental conditions their function may be redundant with other cellular components (Petko and Lindquist, 1985; Servant and Mazodier, 1995; Wotton et al., 1996). Gain-of-function experiments in mammalian and Drosophila cells have shown that constitutive expression of sHsps confers some thermotolerance (Arrigo and Landry, 1994), and expression of plant sHsps in E. coli has also been reported to enhance high-temperature tolerance (Yeh et al., 1997; Soto et al., 1999). Direct genetic evidence comes from the cyanobacterium Synechocystis sp. PCC6803, which has reduced thermotolerance when its single sHsp is deleted (Lee et al., 1998), and from Neurospora crassa, which has reduced thermotolerance under conditions of carbohydrate limitation when the Hsp30 gene is disrupted (Plesofsky-Vig and Brambl, 1995). The role of sHsps in thermotolerance could easily be explained by our model, which states that sHsps prevent irreversible heat-inactivation of other proteins. Based on in vitro evidence demonstrating that sHsps are capable of interacting with a broad range of model substrates (Horwitz, 1992; Jakob et al., 1993; Lee et al., 1995; Chang et al., 1996; Lee et al., 1997; Veinger et al., 1998), the sHsp/Hsc70 system could provide a general pathway for the reactivation of many different heat-labile proteins.
Hsp18.1 represents only a single class of sHsps found in plants, the cytosolic-localized, class I proteins (Waters et al., 1996). Thus, although there is good evidence for chaperone activity for Hsp18.1, as well as many cytosolic sHsps from other organisms, there is a lack of data exploring the chaperone activity of the other cytosolic and organelle-localized classes of plant sHsps. A pea cytosolic class II protein has also been shown to prevent substrate aggregation, although less effectively than Hsp18.1 (Lee et al., 1995). The extent to which our model can be extrapolated to these other sHsps remains to be investigated. Identifying critical in vivo sHsp substrates and defining the functional distinctions between different sHsp classes are important next steps in resolving the role of sHsps in plant thermotolerance.
ACKNOWLEDGMENTS
We are indebted to Drs. Costa Georgopoulos, John Glover, Susan Lindquist, Richard Morimoto, and David Toft for providing plasmid constructs and proteins. We thank Drs. Kim Giese, Luke Whitesell, and Bruce Patterson for critical reading of versions of this manuscript.
Footnotes
This work was supported by the National Institutes of Health (grant no. RO1 GM42762 to E.V. and postdoctoral fellowship no. 5F32–GM16748 to G.J.L.) and by the American Cancer Society Faculty Research Award (no. FRA–420 to E.V.).
LITERATURE CITED
- Arrigo A-P, Landry J. Expression and function of the low-molecular weight heat shock proteins. In: Morimoto R, Tissieres A, Georgopolous C, editors. The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 335–373. [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Schröder H, Hesterkamp T, Schönfeld HJ, Bukau B. Substrate shuttling between the DnaK and GroEL systems indicates a chaperone network promoting protein folding. J Mol Biol. 1996;261:328–333. doi: 10.1006/jmbi.1996.0465. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with HSP70 stress proteins. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielska A, Georgopolous C. Functional domains of the Escherichia coli dnaK heat shock protein as revealed by mutational analysis. J Biol Chem. 1989;264:21122–21130. [PubMed] [Google Scholar]
- Chang Z, Primm TP, Jakana J, Lee IH, Serysheva I, Chui W, Gilbert HF, Quiocho FA. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- deJong WW, Leunissen JA, Vooter CE. Evolution of the α-crystallin/small heat-shock protein family. Mol Biol Evol. 1993;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger M, Gräber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L. Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell. 1997;9:2171–2181. doi: 10.1105/tpc.9.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, Hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Höhfeld J. Chaperones get in touch: the hip-hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- Gaestel M, Vierling E, Buchner J. The small heat shock protein (sHsp) family: an overview. In: Gething M-J, editor. Guidebook to Molecular Chaperones and Protein-Folding Catalysts. New York: Oxford University Press; 1997. pp. 269–272. [Google Scholar]
- Georgopoulos C, Welch WJ. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hernandez Torres J, Chatellard P, Stutz E. Isolation and characterization of gmsti, a stress-inducible gene from soybean (Glycine max) coding for a protein belonging to the TPR (tetratricopeptide repeats) family. Plant Mol Biol. 1995;27:1221–1226. doi: 10.1007/BF00020896. [DOI] [PubMed] [Google Scholar]
- Horwitz J. α-Crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- Jinn T-L, Chen Y-M, Lin C-Y. Characterization and physiological function of class I low molecular weight heat shock protein complexes in soybean. Plant Phys. 1995;108:693–701. doi: 10.1104/pp.108.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates hsp70/hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- Kim R, Kim KK, Yokota H, Kim SH. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA. 1998;95:9129–9133. doi: 10.1073/pnas.95.16.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B, Zhou RG, Wood C, Miernyk JA. AtJ1, a mitochondrial homologue of the Escherichia coli DnaJ protein. Plant Mol Biol. 1996;31:619–629. doi: 10.1007/BF00042234. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Vierling E. Expression, purification, and molecular chaperone activity of plant recombinant small heat shock proteins. Methods Enzymol. 1998;290:350–365. doi: 10.1016/s0076-6879(98)90031-3. [DOI] [PubMed] [Google Scholar]
- Lee S, Prochaska DJ, Fang F, Barnum SR. A 16.6 kilodalton protein in the cyanobacterium, Synechocystis sp. PCC 6803, plays a role in the heat shock response. Curr Microbiol. 1998;37:403–407. doi: 10.1007/s002849900400. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Melki R, Gordon B, Batelier G, Candido EPM. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- Levy EJ, McCarty J, Bukau B, Chirico WJ. Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Minami Y, Höhfeld J, Ohtsuka K, Hartl FU. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Morange M, Bensaude O. Protein denaturation during heat shock and related stress. J Biol Chem. 1989;264:10487–10492. [PubMed] [Google Scholar]
- Palleros DR, Welch WJ, Fink AL. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci USA. 1991;88:5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Petko L, Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1985;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- Pinto M, Morange M, Bensaude O. Denaturation of proteins during heat shock: in vivo recovery of solubility and activity of reporter enzymes. J Biol Chem. 1991;266:13941–13946. [PubMed] [Google Scholar]
- Plesofsky-Vig N, Brambl R. Disruption of the gene for hsp30, an α-crystallin-related heat shock protein of Neurospora crassa, causes defects in thermotolerance. Proc Natl Acad Sci USA. 1995;92:5032–5036. doi: 10.1073/pnas.92.11.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesofsky-Vig N, Vig J, Brambl R. Phylogeny of the α-crystallin-related heat-shock proteins. J Mol Evol. 1992;35:537–545. doi: 10.1007/BF00160214. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Prip-Buus C, Westermann B, Schmitt M, Langer T, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homologue, Mdj1p, in the prevention of heat-induced protein aggregation. FEBS Lett. 1996;380:142–146. doi: 10.1016/0014-5793(96)00049-x. [DOI] [PubMed] [Google Scholar]
- Schröder H, Langer T, Hartl F-U, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, Toft DO. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry. 1996;35:14889–14898. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]
- Schumacher RJ, Hurst R, Sullivan WP, McMahon NJ, Toft DO, Matts RL. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- Servant P, Mazodier P. Characterization of Streptomyces albus 18-kilodalton heat shock-responsive protein. J Bacteriol. 1995;177:2998–3003. doi: 10.1128/jb.177.11.2998-3003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA, Way JC. Eukaryotic dnaJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- Soto A, Allona I, Collada C, Guevara M, Casado R, Rodriguez-Cerezo E, Aragoncillo C, Gomez L. Heterologous expression of a plant small heat-shock protein enhances Escherichia coli viability under heat and cold stress. Plant Physiol. 1999;120:521–528. doi: 10.1104/pp.120.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancato LF, Hutchison KA, Krishna P, Pratt WB. Animal and plant cell lysates share a conserved chaperone system that assembles the glucocorticoid receptor into a functional heterocomplex with hsp90. Biochemistry. 1996;35:554–561. doi: 10.1021/bi9511649. [DOI] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system: DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Xie ZH, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Vierling E. The small heat shock proteins in plants are members of an ancient family of heat induced proteins. Acta Physiol Plant. 1997;19:539–547. [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- Wawrzynów A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M. ATP hydrolysis is required for the DnaJ-dependent activation of DnaK chaperone for binding to both native and denatured protein substrates. J Biol Chem. 1995;270:19307–19311. doi: 10.1074/jbc.270.33.19307. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Freeman K, Shore D. Multimerization of Hsp42p, a novel heat shock protein of Saccharomyces cerevisiae, is dependent on a conserved carboxyl-terminal sequence. J Biol Chem. 1996;271:2717–2723. doi: 10.1074/jbc.271.5.2717. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Chang PFL, Yeh KW, Lin WC, Chen YM, Lin CY. Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci USA. 1997;94:10967–10972. doi: 10.1073/pnas.94.20.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz M, Ang D, Liberek K, Georgopolous C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]