Abstract
Objective
To implement a platform for co-localization of in vivo quantitative multi-parametric magnetic resonance imaging (mpMRI) features with ex vivo surgical specimens of patients with renal masses using patient-specific 3D-printed tumor molds, which may aid in targeted tissue procurement and radiomics/radiogenomic analyses.
Methods
Volumetric segmentation of 6 renal masses was performed with 3D Slicer (http://www.slicer.org) to create a three-dimensional (3D) tumor model. A slicing guide template was created with specialized software, which included notches corresponding to the anatomic locations of the MRI images. The tumor model was subtracted from the slicing guide to create a depression in the slicing guide corresponding to the exact size and shape of the tumor. A customized, tumor-specific, slicing guide was then printed using a 3D printer. After partial nephrectomy, the surgical specimen was bivalved through the preselected MRI plane. A thick slab of the tumor was obtained, fixed and processed as a whole-mount slide and correlated to mpMRI findings.
Results
All patients successfully underwent partial nephrectomy and adequate fitting of the tumor specimens within the 3D mold was achieved in all tumors. Distinct in vivo MRI features corresponded to unique pathologic characteristics in the same tumor. The average cost of printing each mold was US$ 160.7 ± 111.1 (range: US$ 20.9 – 350.7).
Conclusion
MRI-based pre-operative 3D printing of tumor-specific molds allow for accurate sectioning of the tumor after surgical resection and co-localization of in vivo imaging features with tissue-based analysis in radiomics/radiogenomic studies.
Keywords: 3D printing, multiparametric MRI, tumor mold, radiomics, renal masses
Introduction
Radiomics and radiogenomics, relatively new evolving fields, are starting to show their potential in the characterization of various cancers in vivo.1, 2 Radiomics involves the extraction of quantitative features of medical images using computational algorithms while radiogenomics provides the relationship between imaging features and their gene expression patterns.2 Renal cell carcinoma (RCC) is a highly heterogeneous tumor.3 While approximately 80% of the clear cell carcinomas are associated to a mutation or inactivation of the von Hippel-Lindau (VHL) tumor suppression gene, additional mutations differentiate the biological behavior of these tumors.3 Initial reports have illustrated the potential of radiogenomics in differentiating among different subtypes of clear cell renal cell carcinoma.4, 5
The last few decades have been noteworthy for the exceptional increase in the detection of renal masses due to the widespread use of cross-sectional imaging.6 Imaging techniques such as computed tomography (CT), ultrasound (particularly with intravenous contrast), and magnetic resonance imaging (MRI) can accurately characterize a renal lesion as cystic or solid. Furthermore, both CT and MRI may offer a means to differentiate among different histologic subtypes and are highly accurate in the diagnosis of fat-containing angiomiolipoma.7, 8 Multiparametric MRI (mpMRI) is particularly well-suited for evaluation of renal masses because the ability to combine different image contrast mechanisms such as T1- and T2-weighted images with multiphasic contrast-enhanced imaging. 9, 10 More recently, the potential of quantitative multi-parametric MRI (mpMRI) techniques including diffusion weighted imaging (DWI), arterial spin labelling (ASL), and high-temporal resolution dynamic contrast enhanced (DCE) MRI to improve the characterization and subtyping in RCC and to provide information about tumor micro-environment and aggressiveness non-invasively has been reported.9–15
The use of quantitative MRI techniques provides a unique opportunity to correlate objective imaging data with histologic and molecular alterations in the same tumor. However, these correlations are challenged by the complex anatomy, variability of location and size of renal tumor, and physiologic respiratory motion during MRI acquisitions. Three-dimensional (3D) printing has gained much attention in the medical sciences particularly in making models for planning of surgical procedures, development of patient-specific implants, and creating of 3D printed models to facilitate the education of patients and trainees.16–23 Previous studies have also focused on patient-specific MRI-based prostate molds for accurate correlation between the MRI features and histopathologic findings.24, 25 To the best of our knowledge, an approach to generate MRI-based patient-specific 3D kidney tumor molds has not been reported. The use of such molds to allow accurate co-localization of imaging features in vivo with histologic and molecular characteristics in the same tumor provides a platform for MRI-based tissue procurement for radiomics and radiogenomic studies. The purpose of our study was to generate an MRI-based patient-specific 3D mold for renal tumors and to assess our preliminary experience correlating MRI with histopathologic findings.
Materials and Methods
The institutional review board approved this Health Insurance Portability and Accountability Act-compliant prospective study. Written informed consent was obtained from all patients prior to MR imaging. Inclusion criteria: patients >18 years of age, willing to sign informed consent, known renal mass > 2.5 cm in size scheduled for partial or radical nephrectomy, and confirmation of RCC diagnosis after surgery. Exclusion criteria: contraindication for MRI, prior local or systemic therapeutic intervention including chemotherapy for other known neoplasm, confirmation of non-ccRCC diagnosis after nephrectomy, and estimated glomerular filtration rate (eGFR) < 30 ml/min/1.73 m2.
MRI Acquisition
All MR examinations were performed on a 3T whole-body MRI system (Ingenia, Philips Healthcare, Best, The Netherlands) using a dStream anterior and posterior torso coils. The mpMRI protocol has been previously described. 14 Briefly, axial and coronal T2-weighted anatomic images were acquired followed by quantitative MRI techniques including a coronal 2-dimensional (2D) ASL sequence, DWI, and DCE MRI before and after the administration of a bolus 0.1 mmol/kg body weight of gadobutrol (Gadavist; Bayer Healthcare Pharmaceuticals, Wayne, NJ) at a rate of 2 mL/s followed by a 20 mL saline flush at 2 mL/s.13, 14 The basic imaging parameters for the MRI acquisitions are shown in Table 1 (Supplementary Table 1). A table location in the coronal plane located approximately in the center of the mass was selected for radiomics analysis and recorded.
Tumor Segmentation and 3D Printing of Patient-specific Mold
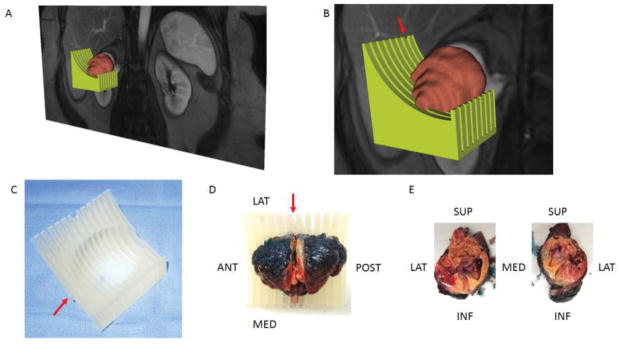
A set of coronal 3D post-contrast images from the DCE acquisition demonstrating a good contrast between the tumor and the renal parenchyma were selected and transferred into 3D Slicer (http://www.slicer.org), a free open source software for medical image computing.26 The editor tools in 3D Slicer were utilized to manually segment the entire tumor in the Digital Imaging and Communications in Medicine (DICOM) images (i.e. in every slice including portions of the tumor) into a ‘label map’. A 3D model of the tumor was generated by the model maker module in 3D Slicer and saved in the standard tessellation language (STL) file format. A slicing guide template was created in the computer aided design (CAD) software (SolidWorks; Dassault Systemes, Velizy-Villacoublay, France), which has notches corresponding to the anatomic locations of the MRI images to enable accurate sectioning of the tumor after surgical resection. In addition, the pre-selected center slice on the 3D acquisition is marked in the template with a deeper notch (Figs. 1A, 1B). This template was then exported as an STL file, and both the 3D tumor and slicing guide STL models were imported into Netfabb software (Netfabb GmbH, Parsberg, Germany). In Netfabb, a Boolean operation was performed, where the anatomic tumor model was subtracted from the slicing guide model, leaving a depression in the slicing guide corresponding to the exact size and shape of the tumor. The mold was marked with anterior, posterior, and/or lateral labels depending of the location of the primary tumor. Then, this customized, tumor-specific, slicing guide was printed as a 3D mold using a 3D printer (Projet 3512HD, 3D Systems, Rock Hall, SC) (Fig. 1C).
Figure 1.
A 3D object was generated by the model maker module in 3D Slicer and saved in the standard tessellation language (STL) file format. (A) The green-colored and brown-colored objects represent the STLs for the mold and tumor, respectively. (B) Zoomed-in image illustrating the notch in the mold (arrow) corresponding to the location of the preselected MRI image plane for radiomic analysis. (C) Printed mold using 3D printer with labeling on the anterior side (arrow). (D) Slicing of tumor positioned within the 3D mold at the level of the notch (arrow). (E) Tumor specimen after sectioning is positioned anatomically. Tissue samples can then be obtained from specific co-localized areas to the MRI findings.
Radio-pathologic correlation
After partial nephrectomy, each surgical specimen was positioned spatially to match its anatomic orientation in vivo using fiducial markers placed during the surgery as previously described 14 and placed on the 3D mold. The tumor was bivalved through the center slice notch to match the MRI plane (Fig. 1D). Targeted tissue samples were then obtained from selected areas in the tumor for radiogenomic analysis (Fig 1E). A 4–5 mm thick slab of the other half of the tumor was obtained, which is inked in different colors for future spatial orientation, placed in whole mount blocks, fixed in 10% buffered formaldehyde solution, and processed as part of our routine whole-mount technique using a 7-hour schedule with automatic microwave assistance (Pathos Delta; Milestone, Italy), and embedded in paraffin. Tissue blocks were sliced in 4 μm thick sections and stained for hematoxylin/eosin (H&E) and for immunohistochemistry (IHC). For tumors exceeding a 4.5 × 4.5 cm in size, the thick tumor slab is sectioned and placed in multiple blocks and the H&E scanned images were put together to reconstruct the entire slab.
Results
We generated patient-specific 3D molds in 6 patients after an mpMRI examination. All patients successfully underwent partial nephrectomy with complete resection of the renal tumor. Patient demographics, tumor characteristics, and surgical data are presented in Table 2 (Supplementary Table 2). For the first patient we created two 3D molds, one of the outer contour of tumor and one of the internal surface of the tumor where the tumor contacts the renal parenchyma (Fig. 2). It became obvious that using the latter would result in more precise fitting of the tumor within the 3D mold because the amount of retroperitoneal fat resected during a partial nephrectomy is more variable from patient to patient. In contrast, the inner parenchymal surface of the tumors is usually smooth after careful enucleation of the tumor during a nephron-sparing partial nephrectomy. Furthermore, small amounts of uninvolved renal parenchyma that are resected during partial nephrectomies tend to remain on the side of the mass and do not interfere with the positioning of the tumor specimen in the mold’s depression. Accordingly, for the following 5 patients we only printed the 3D molds with indentations representing the inner contour of the tumor. The tumor specimen fitted accurately for the 6 patients included in this preliminary study. Representative images of the correlation between MRI and gross specimen are presented in Fig. 3 and 4. Distinct pathologic features were noted in the different pre-selected areas of the tumor corresponding to unique tumor characteristics on MRI. The average cost of printing each mold was US$ 160.7 ± 111.1 (range: US$ 20.9 – 350.7). In one patient, the entire kidney and a large renal tumor were segmented and printed and the cost was $1000.
Figure 2.
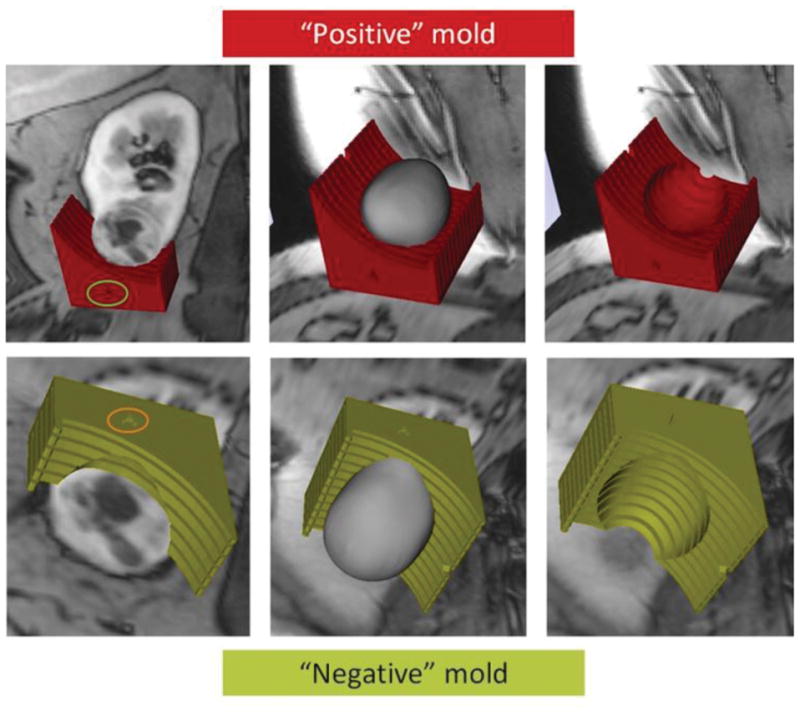
Building model for 3D printing. Representative images using a positive mold (red, top) versus a negative mold (green, bottom) of the tumor. The anterior side of the 3D mold is indicated by the green and orange circles, respectively. Fitting of the tumor on a positive mold may be challenged by various amounts of perirenal fat around the tumor specimen. In contrast, the inner aspect of the tumor (i.e. interface with the renal parenchyma) tends to be more predictable.
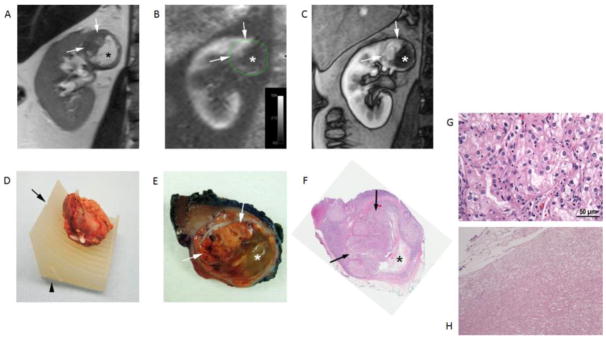
Figure 3.
Clear cell RCC (ISUP grade 3) in a 49-years-old male patient. Coronal T2-weighted single-shot turbo spin echo image (A), arterial spin labelled (ASL) perfusion map (B), and T1-weighted spoiled gradient echo image acquired during the corticomedulary phase (C) of a dynamic contrast enhanced examination show a heterogeneous mass in the upper pole of right kidney. Note the different morphologic features in the tumor with a solid hypervascular component superiorly (arrows) and a non-vascularized area inferiorly (*). (D) Tumor specimen positioned within the 3D mold indentation after resection and spatial orientation using fiducials (i.e. staples) placed during surgery. The desired location for cutting the specimen corresponding to the location of MRI images (A–C) is indicated by the notch (arrow) and the anterior aspect of the mass is also noted in the mold (arrowhead). (E) Section of gross specimen demonstrating golden yellow solid (arrows) and gelatinous friable (*) components in the mass corresponding to the MRI features in vivo. (F) Whole-mount histopathologic H&E slide confirms the presence of a large solid component superiorly (arrows) and necrosis inferiorly (*). Note that the necrotic space is partially collapsed after sectioning. (G) Histopathology of solid area of the tumor shows morphologic features characteristic of clear cell RCC. The tumor cells are arranged in nested architecture with vascular septae. The cells have eosinophilic to clear cytoplasm, round nucleus and prominent grade 3 nucleoli. (H) The gelatinous areas consist of eosinophilic amorphous cellular debris with cystic degeneration. No viable tumor cells or fibrosis is identified.
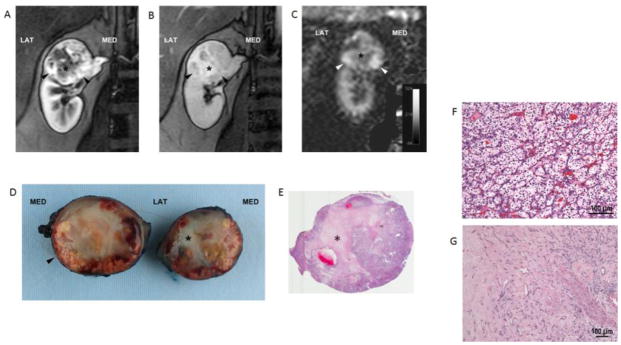
Fig. 4.
Clear cell RCC (ISUP grade 2) in a 50-year-old female patient. Coronal T1-weighted spoiled gradient echo images acquired during the corticomedulary (A) and delayed nephrographic (B) phases of a dynamic contrast enhanced examination show a heterogeneous mass at the upper pole of right kidney exhibiting a hypervascular component peripherally (arrowheads). A central stellate area with intense delayed enhancement (*) is present. Coronal perfusion map (C) generated with a 2D ASL acquisition shows high perfusion in the periphery of the mass inferiorly (arrowheads) with areas of decreased tumor perfusion centrally (*). Bivalved gross specimen confirms the presence of a rim of viable tumor tissue (arrowhead) around a central scar (*) (D). E) Histopathology in viable tumor showed prototypic clear cell renal cell carcinoma, ISUP nucleolar grade 2, with nests of clear cells surrounded by intricate branching vascular network (F). Sections from the central scar show hyalinization and fibrosis with branching capillary network. No viable tumor cells or necrosis is identified. (G).
Discussion
Several methods have been proposed to accurately register histopathologic slides to imaging findings.25, 27 Reports about the use of 3D molds for MRI-histopathology correlations have been primarily limited to the prostate. 25, 27 Costa et al. reported an improvement in the anatomical registration of the prostate contour of in vivo mpMRI datasets and ex vivo prostate whole-mounts using a patient-specific MRI-derived mold compared to conventional whole mount sectioning.28 Importantly, the improvement in the registration was also maintained for the prostate internal architecture including the delineation of the peripheral zone, transitional zone, as well as the tumor contour.28 Our preliminary evaluation using a similar approach for renal tumors indicates optimal registration of imaging and pathology slides.
Our renal tumor mold was created in a sequential manner, first with segmentation of the tumor on the acquired MRI datasets, then by creating a virtual model from the segmented anatomy, and finally by post processing and physical printing the mold. We found that creating a mold with an indentation that reflects the inner surface of the tumor (i.e. the interface between the tumor and the kidney) better accommodates the tumor specimen and thus facilitates the registration.
To the best of our knowledge, this is the first report of 3D-printing based patient-specific molds to correlate MRI features with histopathology characteristics in the same kidney tumors. Furthermore, the mold also allows for collection of targeted tissue samples in areas of the tumors that exhibit specific characteristics in vivo such as different blood flow, water diffusivity, or fat content. The combination of quantitative MRI techniques with the 3D patient-specific tumor mold offers a new opportunity for radiomic and radiogenomic analysis based on objective quantitative imaging- and tissue-based data. Further refinements in the processing of MRI and histopathology data with texture analysis and machine learning algorithms would likely benefit from the improved registration achieved with patient-specific 3D-printed molds.29, 30
Our proposed methodology is not without challenges. First, the printing cost for our 3D molds varied from $20 to $400 depending upon the size of the tumor; however, lower costs may be feasible using different materials and 3D printers. Second, it takes approximately 12–14 hours to print a patient-specific mold, which may limit the ability of creating a mold in cases with limited available time between the imaging examination and the surgery. Third, the tumor segmentation on the MRI images and converting these into the proper format for 3D printing is somewhat time-consuming. Fourth, the effect of motion during the image acquisition and specimen deformation and/or size change (e.g. shrinkage due to lack of blood flow) were not evaluated. However, we found optimal fitting of the tumor in the mold indentation on all tumors. Finally, the correlation in tumors with cystic components may be more challenging because of leakage of the fluid content and subsequent collapse of the tumor after sectioning.
In conclusion, we present a workflow for generating MRI-based patient-specific 3D molds of renal tumors that allow proper co-localization of imaging features in vivo with histopathologic characteristics in the same tumor and facilitates correlations with tissue-based analyses for radiomics and radiogenomic studies.
Supplementary Material
Acknowledgments
Funding:
NIH # P50CA196516 (J.A.C., P.K., I.P.); R01 5RO1CA154475 (D.D., Y.Z., I.P.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 2.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinagare AB, Vikram R, Jaffe C, et al. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group. Abdom Imaging. 2015;40:1684–1692. doi: 10.1007/s00261-015-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamshidi N, Jonasch E, Zapala M, et al. The Radiogenomic Risk Score: Construction of a Prognostic Quantitative, Noninvasive Image-based Molecular Assay for Renal Cell Carcinoma. Radiology. 2015;277:114–123. doi: 10.1148/radiol.2015150800. [DOI] [PubMed] [Google Scholar]
- 6.Sun M, Thuret R, Abdollah F, et al. Age-adjusted incidence, mortality, and survival rates of stage-specific renal cell carcinoma in North America: a trend analysis. Eur Urol. 2011;59:135–141. doi: 10.1016/j.eururo.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Bosniak MA, Megibow AJ, Hulnick DH, Horii S, Raghavendra BN. CT diagnosis of renal angiomyolipoma: the importance of detecting small amounts of fat. AJR Am J Roentgenol. 1988;151:497–501. doi: 10.2214/ajr.151.3.497. [DOI] [PubMed] [Google Scholar]
- 8.Hindman N, Ngo L, Genega EM, et al. Angiomyolipoma with minimal fat: can it be differentiated from clear cell renal cell carcinoma by using standard MR techniques? Radiology. 2012;265:468–477. doi: 10.1148/radiol.12112087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Leon AD, Costa D, Pedrosa I. Role of Multiparametric MR Imaging in Malignancies of the Urogenital Tract. Magn Reson Imaging Clin N Am. 2016;24:187–204. doi: 10.1016/j.mric.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canvasser NE, Kay FU, Xi Y, et al. Diagnostic Accuracy of Multiparametric Magnetic Resonance Imaging to Identify Clear Cell Renal Cell Carcinoma in cT1a Renal Masses. J Urol. 2017 doi: 10.1016/j.juro.2017.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandarana H, Rosenkrantz AB, Mussi TC, et al. Histogram analysis of whole-lesion enhancement in differentiating clear cell from papillary subtype of renal cell cancer. Radiology. 2012;265:790–798. doi: 10.1148/radiol.12111281. [DOI] [PubMed] [Google Scholar]
- 12.Pedrosa I, Sun MR, Spencer M, et al. MR imaging of renal masses: correlation with findings at surgery and pathologic analysis. Radiographics. 2008;28:985–1003. doi: 10.1148/rg.284065018. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Q, Kapur P, Zhang Y, et al. Intratumor Heterogeneity of Perfusion and Diffusion in Clear-Cell Renal Cell Carcinoma: Correlation With Tumor Cellularity. Clin Genitourin Cancer. 2016;14:e585–e594. doi: 10.1016/j.clgc.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Kapur P, Yuan Q, et al. Tumor Vascularity in Renal Masses: Correlation of Arterial Spin-Labeled and Dynamic Contrast-Enhanced Magnetic Resonance Imaging Assessments. Clin Genitourin Cancer. 2016;14:e25–36. doi: 10.1016/j.clgc.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanzman RS, Robson PM, Sun MR, et al. Arterial spin-labeling MR imaging of renal masses: correlation with histopathologic findings. Radiology. 2012;265:799–808. doi: 10.1148/radiol.12112260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silberstein JL, Maddox MM, Dorsey P, Feibus A, Thomas R, Lee BR. Physical models of renal malignancies using standard cross-sectional imaging and 3-dimensional printers: a pilot study. Urology. 2014;84:268–272. doi: 10.1016/j.urology.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 17.Mehra P, Miner J, D’Innocenzo R, Nadershah M. Use of 3-d stereolithographic models in oral and maxillofacial surgery. J Maxillofac Oral Surg. 2011;10:6–13. doi: 10.1007/s12663-011-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sodian R, Schmauss D, Markert M, et al. Three-dimensional printing creates models for surgical planning of aortic valve replacement after previous coronary bypass grafting. Ann Thorac Surg. 2008;85:2105–2108. doi: 10.1016/j.athoracsur.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 19.McGurk M, Amis AA, Potamianos P, Goodger NM. Rapid prototyping techniques for anatomical modelling in medicine. Ann R Coll Surg Engl. 1997;79:169–174. [PMC free article] [PubMed] [Google Scholar]
- 20.Lim KH, Loo ZY, Goldie SJ, Adams JW, McMenamin PG. Use of 3D printed models in medical education: A randomized control trial comparing 3D prints versus cadaveric materials for learning external cardiac anatomy. Anat Sci Educ. 2016;9:213–221. doi: 10.1002/ase.1573. [DOI] [PubMed] [Google Scholar]
- 21.Naftulin JS, Kimchi EY, Cash SS. Streamlined, Inexpensive 3D Printing of the Brain and Skull. PLoS One. 2015;10:e0136198. doi: 10.1371/journal.pone.0136198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Urso PS, Barker TM, Earwaker WJ, et al. Stereolithographic biomodelling in cranio-maxillofacial surgery: a prospective trial. J Craniomaxillofac Surg. 1999;27:30–37. doi: 10.1016/s1010-5182(99)80007-9. [DOI] [PubMed] [Google Scholar]
- 23.Zerr J, Chatzinoff Y, Chopra R, Estrera K, Chhabra A. Three-dimensional printing for preoperative planning of total hip arthroplasty revision: case report. Skeletal Radiol. 2016;45:1431–1435. doi: 10.1007/s00256-016-2444-1. [DOI] [PubMed] [Google Scholar]
- 24.Trivedi H, Turkbey B, Rastinehad AR, et al. Use of patient-specific MRI-based prostate mold for validation of multiparametric MRI in localization of prostate cancer. Urology. 2012;79:233–239. doi: 10.1016/j.urology.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah V, Pohida T, Turkbey B, et al. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum. 2009;80:104301. doi: 10.1063/1.3242697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiessling F, Le-Huu M, Kunert T, et al. Improved correlation of histological data with DCE MRI parameter maps by 3D reconstruction, reslicing and parameterization of the histological images. Eur Radiol. 2005;15:1079–1086. doi: 10.1007/s00330-005-2701-5. [DOI] [PubMed] [Google Scholar]
- 28.Costa DN, Chatzinoff Y, Passoni NM, et al. Improved Magnetic Resonance Imaging-Pathology Correlation With Imaging-Derived, 3D-Printed, Patient-Specific Whole-Mount Molds of the Prostate. Invest Radiol. 2017 doi: 10.1097/RLI.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–223. doi: 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging. 2010;31:680–689. doi: 10.1002/jmri.22095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.