Since malignancies develop by serial acquisition of somatic mutations over years or decades,1 cells transitioning from normalcy to malignancy need a sufficiently long life span to accumulate the required mutations. For cancers of the hematopoietic system, these cells can be self-renewing hematopoietic stem cells (HSCs) or cancer-initiating cells that pass genetic alterations down to their progeny during cell division and differentiation.
Chronic lymphocytic leukemia (CLL), the most common adult leukemia among Caucasians, results from expansion of clonal CD5+ B lymphocytes with a mature phenotype growing in bone marrow (BM), blood and lymph nodes.2 Consistent with stepwise disease development,3, 4 CLL is preceded by a pre-leukemic state, monoclonal B-lymphocytosis.5, 6 Although a series of genomic abnormalities has been defined in CLL, there is no single lesion or combination of genetic lesions common to every patient.7 Xenotransplantation studies suggest HSCs isolated from CLL patients are biased to developing mono-/oligo-clonal B cells with a CLL-like phenotype, thereby implicating HSCs in disease development.8 Supporting this implication is the report of mutations in immature hematopoietic cells (CD34+CD19−) in the blood of CLL patients.9 Here, we investigated this issue further by probing the presence of somatic mutations in defined immature cell populations of the hematopoietic lineage (Supplementary Figure 1). These populations were isolated by FACS from granulocyte-colony stimulating factor (G-CSF)-mobilized peripheral blood (MPB) of CLL patients in remission for whom autologous stem cell transplantation was anticipated but not carried out (Supplementary Table 1).
First, using the strategy indicated in Supplementary Figure 2, we purified CD5+CD19+ cells from six patients and then identified somatic mutations specific to each leukemic clone by performing whole-exome sequencing (WES) of CLL cells (mean read depth: 107.5) and paired buccal cells (mean read depth: 61.5). This revealed a total of 87 coding mutations, with an average of 14.5 (range 7–21) variants per sample (Supplementary Tables 2 and 3), in line with previous reports.10 In three additional cases for which matched normal DNA was not available (mean read depth: 91.8), mutation calls were limited to known CLL driver genes,7 accounting for a total of 95 substitutions and small indels (Supplementary Table 3). The sequences reported in this paper have been deposited in the NCBI's BioProject database under project PRJNA411889.
Next, we assessed the sensitivity for detecting small, subclonal mutations using amplicon-based deep sequencing, the approach we would subsequently use to identify the above mutations in immature hematopoietic cells from the patients. Detection sensitivity was determined using serial dilutions of a cell line harboring a known heterozygous TP53 mutation (Supplementary Methods). This indicated that a sequencing depth of 20 000 × to 100 000 × was sufficient to detect >1 mutated alleles out of 10 000 wild-type alleles, against background sequencing errors (Supplementary Figure 3). Increasing sequencing depth to 1 000 000 × did not improve sensitivity, as sequencing errors occurred at rates similar to those of true somatic mutations (Supplementary Figure 3).
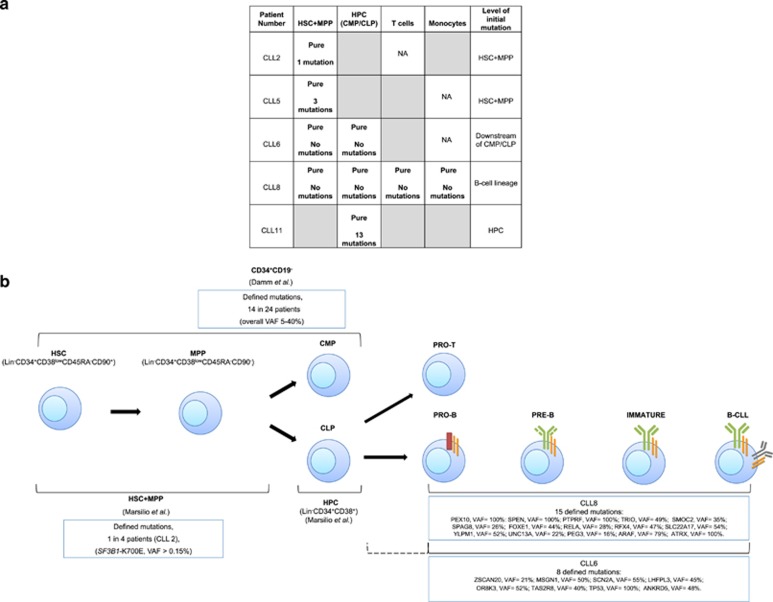
Next from the nine CLL patients’ G-CSF MPB, we isolated by FACS the following fractions: HSC plus MPP (Multi-Potent Progenitor cell) (HSC+MPP), defined as Lin−CD34+CD38lowCD45RA−CD90+/CD90−; downstream Hematopoietic Progenitor Cell populations (HPC) containing Common Lymphoid Progenitors and Common Myeloid Progenitors, marked as Lin−CD34+CD38+; and mature T cells (CD3+CD19−) and monocytes (CD14+CD19−) (Supplementary Figure 2). Post-sorting, the average purity of the HSC+MPP fractions was ⩾99%. Nevertheless, to evaluate contamination of the cell fractions with mature CLL B cells, we searched for tumor-specific immunoglobulin VH CDR3 sequences by employing a qPCR protocol that is used to detect minimal residual disease (MRD) in patients with leukemia,11 achieving a sensitivity of ⩽10−4 (7 patients), ⩽10−5 (1 patient), and 5 × 10−4 (1 patient) (Supplementary Table 4). Out of a total of 32 populations analyzed, 9 were ‘pure’, that is, below the level of detection of the patient-specific VH CDR3 sequences; these included 4/8 HSC+MPP, 3/9 HPC Population initially indicated as HPC, 1/8 CD3+CD19− and 1/6 CD14+CD19− (Figure 1a). In the remaining 23 fractions, contamination with mature CLL cells was found at various levels, ranging from low (between 5 × 10−5 and 5 × 10−4; n=13) to higher (>5 × 10−4; n=10) grade contamination (Supplementary Table 4).
Figure 1.
Fractions within the hematopoietic differentiation pathway containing somatic mutations in CLL patients. (a) Cell fractions free of mature CLL-cell contamination as assessed by patient-specific VH CDR3 qPCR. Nine fractions were scored as ‘pure’ based on the inability to detect patient-specific VH CDR3 signatures by qPCR. In the remaining 23 fractions (not shown here, see Supplementary Table 4), mature CLL cell VH CDR3 signatures were found at various levels indicating low-grade (5 × 10−5−5 × 10−4; n=13) to higher-grade (>5 × 10−4; n=10) contamination. Specific mutations are listed in those pure fractions in which they were found. The contaminated fractions are highlighted in gray. NA: sample not available. (b) Assignment of somatic genomic changes to the earliest cells in the hematopoietic lineage (HSC+MPP of CLL2 SF3B1-K700E) or to cells at or downstream of the CMP/CLP stages (CLL8 and CLL6). Assignments restricted to those fractions noted in A as ‘pure’. For CLL 2, initial mutation occurred in the HSC+MPP fraction. Whereas in CLL 8, all mutations occurred within the B-cell lineage (between pro-B and mature B-cell stages), since the patient’s companion T cells did not contain the same mutation. For CLL6, the precise relationship cannot be defined because the companion T-cell population was contaminated with mature CLL B cells. Hence, the mutation for CLL6 occurred downstream of the combined CMP/CLP stage, possibly just upstream of the T- and B-cell bifurcation or within the B-cell lineage; this equivocation is indicated by the dotted line. For CLL5 and CLL11, more than one mutation was detected in the indicated fraction (see panel a).
Having measured the levels of contaminating CLL cells and the sequencing sensitivity, we performed multiplex deep sequencing targeting 92 unique loci in patient-matched immature and mature hematopoietic populations; this interrogated 91% of the mutations (n=86/95) previously identified in the corresponding CLL dominant clone (Supplementary Table 5). The contaminated populations were also included as controls in the deep sequencing experiment. This approach yielded an average of 56 572 pairs of reads per sample per locus (range: 289–182 851). We then developed a Bayesian approach to assess the presence of true mutations at very low abundance. Each mutation was tested against 18 negative control samples from HSC+MPP and HPC, isolated from G-CSF MPB of nine healthy adult donors (40–57 years of age) who underwent stem cell mobilization as donors for allogeneic transplantation. Sequencing DNA from the same immature cell fractions of normal individuals at identical genomic regions and at the same depth provided a true assessment of the error rate at a given position. This indicated significant mutation-dependent variation in sensitivity. Specifically, when the sequencing error rate for a given nucleotide change was low (for example, as for transversions), we could discern mutations present at ⩾1 in 10 000 alleles (Supplementary Figure 3). In contrast, for variants associated with a high sequencing error rate (for transitions), the sensitivity was reduced to 5–10 in 10 000 alleles, independent of sequencing depth. As expected, targeted deep sequencing of ‘tumor contaminated’ samples identified all events detected by WES in the CLL clone (not shown), documenting the sensitivity and accuracy of the approach.
Focusing on the contamination-free cell fractions (five patients; Figure 1a), several observations emerged. First, for CLL6 and CLL8, none of the WES-identified mutations could be found in the HSC+MPP and HPC fractions, and for CLL8 also not in the CD3+ and CD14+ fractions (Figure 1). These findings suggest that, for both patients, the genetic point variants found in the mature CLL cells occurred downstream of Common Lymphoid and Common Myeloid Progenitors (HPC Population initially indicated as HPC), and, for CLL8, after the bifurcation between T and B lymphocytes, that is, cells committed to the B-cell lineage (Figure 1b).
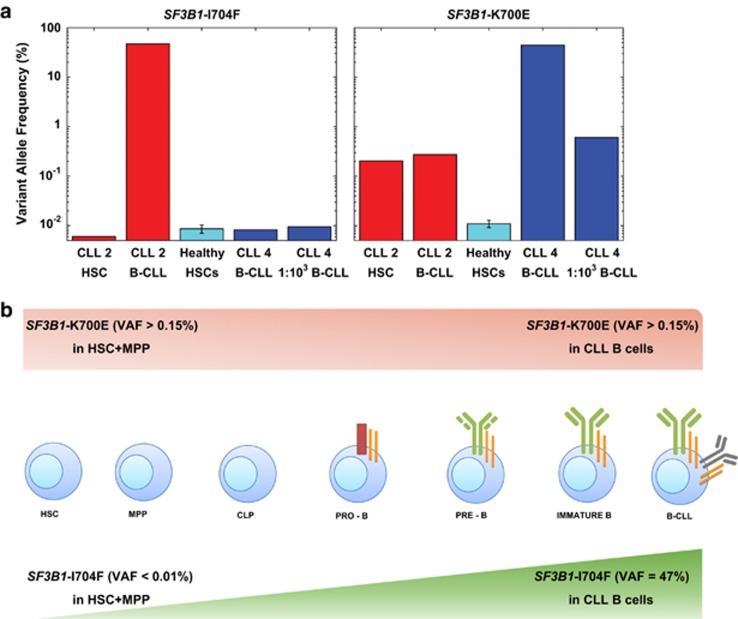
Second, although we did not detect the SF3B1-I704F mutation (sensitivity >0.01%) that had been observed by WES in the leukemic B cells of CLL2 (variant allele frequency, VAF =47%) (Figure 2), we did find an SF3B1-K700E mutation (VAF >0.15%) in the HSC+MPP fraction, which had not been identified by WES (Supplementary Table 3). Therefore, we repeated the deep sequencing experiment at 10-fold increased depth, testing undiluted mature CLL2 cells as well as undiluted and 1:103 diluted mature CLL4 cells as positive controls (since they harbored the SF3B1-K700E mutation, VAF=37%). This approach confirmed the SF3B1-K700E mutation (VAF >0.15%) and the absence of the SF3B1-I704F mutation (Figure 2; Supplementary Table 6) in the HSC+MPP fraction of CLL2. Interestingly, the SF3B1-K700E mutation was also detected in mature CLL2 cells (VAF >0.15%). Examining individual reads for these mutations indicated that SF3B1-K700E existed in clones mutually exclusive from those harboring SF3B1-I704F (chi-squared test P<0.0001; Supplementary Methods). This points out the high mutability and significance of this gene in CLL development, and is consistent with growth/survival advantages of subclones that acquire more mutations to become dominant populations. The presence of mutually exclusive clones with low-frequency SF3B1 mutations in newly diagnosed CLL has been reported.12
Figure 2.
SF3B1-K700E mutation occurs in CLL2 HSC+MPP fraction, whereas SF3B1-I704F mutation is only detected further down in the hematopoietic differentiation pathway. (a) Detection of SF3B1-K700E in CLL2 HSC+MPP fraction. Although SF3B1-I704F was present in mature CLL2 B cells, deep sequencing analysis did not detect this mutation in the HSC+MPP population of CLL2, or in the control undiluted and 1:103 diluted mature CLL4 B-cells (VAF <0.01%). However, SF3B1-K700E was identified in both CLL2 HSC+MPP and in the companion mature B cells (VAF >0.15%), as well as in undiluted and 1:103 diluted mature CLL4 B-cells at expected VAFs. (b) Graphical representation for the pattern of SF3B1 mutations identified in CLL2 by targeted sequencing at >630 000 depth. SF3B1-I704F was identified with a VAF of 47% by WES in the leukemic clone but was not found in the HSC+MPP fraction (VAF <0.01%). SF3B1-K700E mutation was present in the HSC +MPP fraction (VAF >0.15%), and the same abundance was maintained in the mature CLL B-cell clone (VAF >0.15%).
Finally, in the HSC+MPP of CLL5 and HPC of CLL11a, deep sequencing detected 50% and 82% of the mutations identified by WES in the mature CLL cells, respectively (Figure 1a, Supplementary Table 7). These findings raise the concern of tumor contamination below the sensitivity of the VH CDR3 analysis but discernible by our sequencing approach. Indeed, deep sequencing of CLL11a HPC Population initially indicated as HPC detected 61% of the mutations identified by WES in a sample of non-mobilized PB obtained 4 years later (‘CLL11b’). Specifically, 88% of mutations shared by CLL11a and CLL11b, and 40% of mutations specific to CLL11b were found in the HPCs.
In summary, our findings confirm that CLL mutations can occur in CD34+ cells9 and precisely define HSC+MPP and HPC as the fractions containing these mutations. In addition, our results indicate that not all CLL clones develop mutations at these maturation stages. Although we cannot discount that mutations are acquired in non-coding genomic regions, of the four tumor samples with pure HPC+MPP fractions (Figure 1a), only one had a CLL exome mutation (25%). For two cases, none of the CLL mutations were found in these cell populations, and for a third all variants had to develop after commitment to the B-cell lineage (Figures 1a and b). Moreover, despite starting with more CD34-enriched samples (G-CSF MPB) than others9 and achieving >99% pure precursor populations by FACS, several fractions contained mature CLL-cell ‘contaminants’ that were detected by high sensitivity, ultra-deep sequencing and MRD methods. Indeed, only ~28% of the populations studied (9 of 32) were pure by the MRD approach, highlighting the need to rigorously control for tumor cell contamination when searching for infrequent variant alleles in normal cell fractions. Lastly, our results emphasize the value of utilizing a calibrated sequencing and sample-specific statistical approach for precise detection of mutations in minor cell populations during the course of CLL evolution.
Acknowledgments
This work was supported in part by philanthropic contributions from The Karches Foundation, Marks Foundation, Nash Family Foundation, Jerome Levy Foundation, Leon Levy Foundation, Frank and Mildred Feinberg Foundation, the Mona and Edward Albert Foundation, and the Jean Walton Fund for Leukemia, Lymphoma, and Myeloma Research.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Dr Steven Devereux: Member of Advisory Board of Janssen, Gilead and Merck Sharp and Dohme and recipient of honoraria and travel expenses from Janssen, Gilead and GlaxoSmithKline. Dr Nicholas Chiorazzi: Consultant fee from Janssen Pharmaceuticals, and honorarium from Nodality, Inc. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170. [DOI] [PubMed] [Google Scholar]
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med 2005; 352: 804–815. [DOI] [PubMed] [Google Scholar]
- Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008; 359: 575–583. [DOI] [PubMed] [Google Scholar]
- Wang J, Khiabanian H, Rossi D, Fabbri G, Gattei V, Forconi F et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. eLife 2014; 3: e02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med 2009; 360: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezzato M, Giaretta I, Madeo D, Rodeghiero F. Identical IGHV-D-J gene rearrangement may precede the clinical onset of chronic lymphocytic leukemia by several years. Am J Hematol 2010; 85: 868–871. [DOI] [PubMed] [Google Scholar]
- Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016; 16: 145–162. [DOI] [PubMed] [Google Scholar]
- Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 2011; 20: 246–259. [DOI] [PubMed] [Google Scholar]
- Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov 2014; 4: 1088–1101. [DOI] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015; 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden VHJ, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER et al. Analysis of minimal residual disease by Ig//TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007; 21: 604–611. [DOI] [PubMed] [Google Scholar]
- Rasi S, Khiabanian H, Ciardullo C, Terzi-di-Bergamo L, Monti S, Spina V et al. Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica 2016; 101: e135–e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.