Abstract
Oligopeptides are important markers of protein metabolism, as they are cleaved from larger polypeptides and proteins. Genetic association studies may help elucidate their origin and function. In 1,552 European Americans and 1,872 African Americans of the Atherosclerosis Risk in Communities study, we performed whole-genome and whole-exome sequencing and measured serum levels of 25 peptides. Common variants (minor allele frequency > 5%) were analysed individually. We grouped low-frequency variants (minor allele frequency ≤ 5%) by a genome-wide sliding window using region-based aggregate tests. Furthermore, low-frequency regulatory variants were grouped by gene, as were functional coding variants. All analyses were performed separately in each ancestry group and then meta-analysed. We identified 22 common variant associations with peptide levels (P-value < 4.2 × 10−10), including 16 novel gene-peptide pairs. Notably, variants in kinin-kallikrein genes KNG1, F12, KLKB1, and ACE were associated with several different peptides. Variants in KLKB1 and ACE were associated with a fragment of complement component 3f. Both common variants and low-frequency coding variants in CPN1 were associated with a fibrinogen cleavage peptide. Four sliding windows were significantly associated with peptide levels (P-value < 4.2 × 10−10). Our results highlight the importance of the kinin-kallikrein system in the regulation of serum peptide levels, strengthen the evidence for a broad link between the kinin-kallikrein and complement systems, and suggest a role of CPN1 in the conversion of fibrinogen to fibrin.
Introduction
Oligopeptides are short peptides consisting of two to twenty amino acids. Circulating peptides are generally formed when they are cleaved from larger polypeptides, and as a consequence they are important markers of protein metabolism. Although numerous peptides have documented biological roles (1), many others remain poorly characterized. Genetic association studies may help elucidate their origin and function.
Several studies have examined the association of serum peptides with common genetic variants characterized using genotyping arrays and subsequent imputation (2–4). These studies have resulted in important insights into the genetic factors regulating the production and degradation of peptides as well as the metabolic systems underlying disease (5). Further resolution in examining the genetic determinants of peptides may be gained through sequencing. Although several studies involving genotyped or sequenced exomes have been performed (6,7), much of the causal genetic variants associated with quantitative traits such as peptides are thought to reside in noncoding regions, meaning that exome sequencing studies may miss important effects. This limitation can be overcome by using whole-genome sequencing, although the analysis of whole-genome sequencing data in association studies remains at an early stage compared to that of genotyping array data (8–14).
In this study, we thus aimed to identify new associations between common, low-frequency, and rare genetic variants and serum levels of 25 peptides using whole-genome and whole-exome sequencing data from 3424 participants of the Atherosclerosis Risk in Communities (ARIC) study.
Materials and Methods
Study population
We used the ARIC study, a prospective epidemiological study designed to investigate the etiology and predictors of cardiovascular disease. At the baseline exam (1987–1989), 15,792 middle-aged men and women were recruited from four communities across the United States: Forsyth County, North Carolina; Jackson, Mississippi; northwest suburbs of Minneapolis, Minnesota; and Washington County, Maryland. The participants were mostly of European (EA) or African American (AA) ancestry. The design of the ARIC study has been described in further detail elsewhere (15). The ARIC study has been approved by the Institutional Review Board at all participating institutions. All participants included in this study provided written informed consent. Participants were excluded if they did not give consent for use of DNA information.
Peptide profiling
Non-targeted metabolomic profiling was performed on fasting serum samples using a protocol based on both gas and liquid chromatography mass spectrometry (Metabolon Inc., Durham, USA)) (16,17). The samples had been stored at -80 °C between the time of collection (1987–1989) and the analysis. Metabolites were measured in two phases: a total of 1,880 AA samples were analysed in 2010 and 2,170 EA and AA samples were analysed in 2014. Among metabolites that were measured both in 2010 and 2014, the subset of 49 metabolites classified as peptides were included in the present study. Peptides were excluded if over 25% of the values were below the detection limit were excluded, as well as if there was a low correlation between 2010 and 2014 measurements (Pearson correlation coefficient < 0.3). After this quality control, 25 were peptides and were included primary analyses in the present study. In secondary exploratory analyses, we go beyond these 25 peptides and also use the 24 peptides did not pass quality control. The reason that each of these peptides was excluded is listed in Supplementary Material, Table S1. Levels below the detectable limit were imputed with the lowest detected value for that peptide in all samples. Peptide levels outside the 1st–99th percentile were winsorized. Finally, all peptide levels were natural-log transformed in order for their distribution to approximate the normal distribution.
Whole-genome sequencing
Methods for the whole-genome sequencing of ARIC study participants have been described in detail in Morrison et al. (8). In brief, Hiseq 2000 instruments (Illumina Inc., San Diego, CA) were used to successfully sequence the genomes at a 7.4-fold average depth. The sequencing was performed by the Baylor College of Medicine Human Genome Sequencing Center. As described previously (13,18), 72.8 million genetic variants were called using goSNAP, which employed three calling algorithms (GATK, SNPTools, and GotCloud), each in joint calling mode, and a consensus approach was used to generate a high-quality variant call set (18). Variants with site-level inbreeding coefficient < 0.9 were excluded, as were variants not in Hardy-Weinberg equilibrium in ancestry-specific groups (P-value < 1 × 10−14). Principal-component analysis was used to identify 40 individuals as outliers within their ancestry group, and these individuals were excluded. 1,458 EA and 1,679 AA participants with both whole-genome sequencing data and serum peptide levels were included in this study. Whole-genome sequencing variants were annotated to functional domains using the Whole Genome Sequencing Annotation (WGSA) pipeline based on the reference genome GRCh37 (19). The 3’ and 5’ UTRs of genes were determined using ANNOVAR (20) annotations based on the RefSeq gene model (21). Promotors were defined based on the overlap between 1) the permissive set of CAGE peaks reported by the FANTOM5 project and 2) the 5 kb upstream region determined by the ANNOVAR annotation based on the RefSeq gene model (22). Enhancers and the target genes of the enhancers were defined based on the permissive set of enhancers and enhancer-promoter pairs reported by the FANTOM5 project. We assigned undesignated enhancers to the nearest gene. Association between significant variants and expression levels of nearby genes were obtained from the GTEx portal (GTEx Analysis Release V6p, dbGaP Accession phs000424.v6.p1). We used the single nucleotide polymorphism annotator (SNiPA) tool to identify associations of our lead variants (along with correlated variants with r2 > 0.8) with serum metabolite levels (mQTLs) and protein levels (pQTLs) (23,24).
Whole-exome sequencing
Whole-exome sequencing was performed on 8,544 individuals from the ARIC study, including 5,718 EA and 2,836 AA participants. DNA sequencing was performed using Hiseq 2000 instruments (Illumina Inc., San Diego, CA) after exome capture with VCRome 2.1 (NimbleGen, Inc., Madison, WI). Sequence alignment and variant calling were carried out via the Mercury pipeline (25). Variant-level quality control steps excluded variants outside the exon capture regions, multi-allelic sites, missing rate >20%, and mean depth of coverage >500-fold. Variants not meeting Hardy-Weinberg equilibrium expectations in ancestry-specific groups (P-value < 5x10−6) were also excluded. A sample was excluded for missingness >20%, or if compared to the other samples in the same ancestry group it fell outside of 6 standard deviations (SD) from the mean depth, mean singleton count, mean heterozygote to homozygote ratio, or mean Ti/Tv ratio. After quality control, the mean sequencing depth of the included variants was 87x.
We included 1,330 EA and 1,850 AA participants with both whole-exome sequencing and serum peptide levels. Variants were annotated using ANNOVAR (20) and dbNSFP v2.0 according to the reference genome GRCh37 and RefSeq (26).
Statistical analysis
Both single variant and burden test analysis were performed using the R package ‘seqMeta’ (27). EA and AA participants were analysed separately and then meta-analysed. Associations were classified as consistent across the two ethnicities when the P-value was below 0.05 and the effect direction was the same in both ethnicities. All analyses were adjusted for age, sex, estimated glomerular filtration rate (28), and the first three ancestry-informative principal components. The analyses in AA participants were additionally adjusted for phase, a variable indicating whether measurements took place in 2010 or 2014. All EA participants were profiled in 2014, so analyses in EA participants were not adjusted for phase. Whole-genome sequencing data was used for the single variant analysis, and only genetic variants with a minor allele frequency (MAF) greater than 5% in the meta-analysis of EA and AA participants were included. Out of 8,434,536 common variants, 7,567,083 were present in both EA and AA participants were included in the meta-analysis of the two ancestry groups.
Three approaches were used to define the regions used to aggregate genetic variants. For all approaches, only regions with a cumulative MAF > 1% were included. First, a sliding window approach involved windows of physical distance of 4 kb in length, beginning at position 0 bp for each chromosome and sliding 2 kb onwards with each step (8), leading to 1,336,946 sliding windows across the genome. Secondly, we defined 18,575 regulatory regions using information on the position of enhancers, promotors, and 3’ and 5’ UTRs around each gene. Thirdly, 11,551 protein-coding regions were investigated. The analysis of sliding windows and regulatory regions was based on whole-genome sequencing data, while the analysis of protein-coding regions was based on whole-exome sequencing data. These data were thus not aggregated, but were used in separate analyses. Only nonsynonymous, stop-gain, stop-loss, splicing, and frameshift variants were included in aggregate tests of protein-coding regions based on whole-exome sequencing data. On each of the sliding windows and regulatory regions we conducted a unidirectional aggregate test of association, the T5 (29), and a bidirectional aggregate test of association, the sequence kernel association test (SKAT) (30). The T5 burden was defined as the total number of minor alleles among variants in the window with a MAF less than or equal to 5%. As previously estimated using 1000 genomes data (31,32), we considered the single common variants to correspond to 2 million independent variants after accounting for linkage disequilibrium. The single variant analysis thus consisted of 50 million independent tests (2,000,000 × 25), while the region-based aggregate tests consisted of 68,353,600 independent tests (1,367,072 × 2 × 25). We therefore used a unified Bonferroni-corrected P-value threshold of 4.2 × 10−10.
In order to study the clinical relevance of the 25 peptides, we used linear regression models to examine the association of the peptides with five quantitative cardio-metabolic phenotypes: high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, triglycerides (TG), systolic blood pressure (SBP), and diastolic blood pressure (DBP). These phenotypes were modeled as the dependent variable. Analyses were stratified by ancestry group and adjusted for age, gender, measurement phase, estimated glomerular filtration rate, body mass index, current smoking, prevalent diabetes, antihypertensive medication, and cholesterol-lowering medication. Ancestry-specific results were then meta-analysed using fixed effects inverse-variance weighted meta-analysis as implemented in the ‘meta’ R package. We used a Bonferroni-correct P-value threshold of 4.0 × 10−4.
Although 24 of the 49 peptides did not pass quality control for inclusion in our primary analyses, we performed an exploratory analysis to examine the association between levels of these peptides and the lead variants at loci identified in our primary single variant analysis of common variants. Statistical methods described for the primary analyses were also used in this exploratory analysis. We used the same Bonferroni-corrected P-value as in the primary analysis.
Additional analyses that were undertaken specifically to further explore the identified associations are described in the Supplementary Methods.
Results
Baseline characteristics
Baseline characteristics of the 1552 EA and 1872 AA participants from the ARIC study having peptide levels and either whole-genome or whole-exome sequence data are shown in Supplementary Material, Table S2. The mean age was 53.6 years old, 60.0% percent of the participants were female, and the mean body-mass index (BMI) was 28.7. We used a Bonferroni-corrected significance threshold of P-value < 4.2 × 10−10 for all discovery analyses.
Common variant analyses
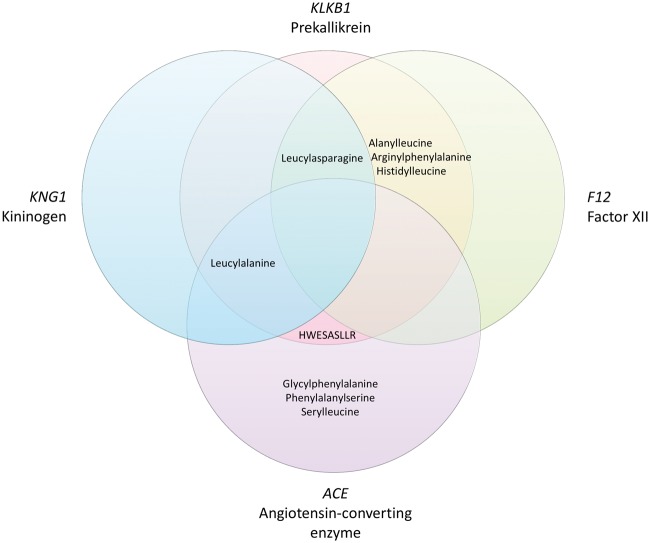
In the meta-analysis of EA and AA participants, 22 independent single variant associations between genetic variants and peptides were identified (Table 1), of which all but one were consistently associated (P-value < 0.05 in both ethnicities and same effect direction) across the two ethnicities (Supplementary Material, Fig. S1). Even for the inconsistent association (variants at SRSF12/PM20D2 and N-acetylcarnosine) the effect direction was consistent across the ethnicities, but the allele frequency in EA participants was very low (MAF = 7 × 10−4). Functional annotations of the 22 lead variants, including associations with expression of nearby genes obtained from the GTEx portal are shown in Supplementary Material, Table S3 (33). Of these associations, six have previously been reported (2,4). Out of the 25 peptides that were included in this study, eleven were affected by at least one genetic variant. A heatmap representing the correlations among these eleven peptides is shown in Supplementary Material, Figure S2. Most peptides were only moderately correlated with each other (Pearson’s r < 0.6), although a cluster of peptides including alanylleucine, arginylphenylalanine, histidylleucine, leucylalanine, and leucylasparagine showed stronger pairwise correlations (Pearson’s r = 0.6 - 0.8). Several loci in the kinin-kallikrein pathway were related to more than one peptide: the KLKB1 locus contributed to levels of six peptides, the ACE locus was associated with five peptides, the F12 locus influenced four peptides, and the KNG1 locus was associated with two peptides. There was considerable overlap among the peptides related to each these genes, as shown in Figure 1.
Table 1.
Single variant associations
| Meta-Analysis |
European Americans |
African Americans |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant rsID | Chr:Pos | Peptide | Closest Genes | Alleles | MAF | Beta | P-value | Beta | P-value | Beta | P-value |
| rs3733402 | 4:187158034 | Alanylleucine | KLKB1 | A/G | 0.37 | 0.19 | 3.6 × 10−27 | 0.22 | 5.7 × 10−20 | 0.15 | 1.8 × 10−9 |
| rs1801020 | 5:176836532 | Alanylleucine | F12 | G/A | 0.35 | 0.12 | 1.7 × 10−12 | 0.13 | 2.5 × 10−6 | 0.12 | 1.4 × 10−7 |
| rs3733402 | 4:187158034 | Arginylphenylalanine | KLKB1 | A/G | 0.37 | 0.16 | 7.0 × 10−20 | 0.16 | 1.1 × 10−10 | 0.16 | 1.1 × 10−10 |
| rs1801020 | 5:176836532 | Arginylphenylalanine | F12 | G/A | 0.35 | 0.15 | 9.0 × 10−19 | 0.16 | 4.2 × 10−8 | 0.15 | 3.8 × 10−12 |
| rs11592631 | 10:101878219 | DSGEGDFXAEGGGVR | CPN1/ERLIN1 | A/G | 0.22 | 0.28 | 7.4 × 10−12 | 0.24 | 4.0 × 10−6 | 0.34 | 2.1 × 10−7 |
| rs4335 | 17:61565025 | Glycylphenylalanine | ACE | A/G | 0.38 | −0.14 | 1.7 × 10−20 | −0.13 | 6.7 × 10−11 | −0.16 | 2.5 × 10−11 |
| rs3733402 | 4:187158034 | Histidylleucine | KLKB1 | A/G | 0.37 | 0.25 | 1.6 × 10−44 | 0.29 | 7.3 × 10−31 | 0.21 | 1.8 × 10−16 |
| rs1801020 | 5:176836532 | Histidylleucine | F12 | G/A | 0.35 | 0.17 | 1.6 × 10−22 | 0.18 | 1.3 × 10−9 | 0.17 | 2.0 × 10−14 |
| rs4363 | 17:61574492 | HWESASLLR | ACE* | A/G | 0.48 | 0.31 | 1.5 × 10−23 | 0.32 | 7.5 × 10−10 | 0.31 | 3.1 × 10−15 |
| rs4253311 | 4:187174683 | HWESASLLR | KLKB1 | G/A | 0.44 | 0.22 | 9.8 × 10−13 | 0.19 | 2.7 × 10−4 | 0.24 | 5.9 × 10−10 |
| rs3733402 | 4:187158034 | Leucylalanine | KLKB1 | A/G | 0.37 | 0.15 | 2.4 × 10−26 | 0.14 | 2.9 × 10−14 | 0.15 | 1.1 × 10−13 |
| rs4363 | 17:61574492 | Leucylalanine | ACE* | A/G | 0.48 | −0.11 | 2.1 × 10−17 | −0.10 | 1.7 × 10−7 | −0.12 | 1.5 × 10−11 |
| rs5030081 | 3:186458910 | Leucylalanine | KNG1 | A/G | 0.44 | 0.09 | 4.2 × 10−12 | 0.10 | 5.7 × 10−8 | 0.08 | 1.4 × 10−5 |
| rs3733402 | 4:187158034 | Leucylasparagine | KLKB1 | A/G | 0.37 | 0.33 | 3.2 × 10−36 | 0.37 | 2.1 × 10−21 | 0.30 | 6.8 × 10−17 |
| rs1801020 | 5:176836532 | Leucylasparagine | F12 | G/A | 0.35 | 0.24 | 2.1 × 10−20 | 0.31 | 1.6 × 10−11 | 0.20 | 3.8 × 10−11 |
| rs5030082 | 3:186458949 | Leucylasparagine | KNG1 | G/A | 0.44 | 0.16 | 4.8 × 10−11 | 0.15 | 1.7 × 10−4 | 0.17 | 6.0 × 10−8 |
| rs9524869 | 13:95913675 | N-acetylcarnosine | ABCC4* | G/C | 0.43 | 0.08 | 1.4 × 10−13 | 0.10 | 1.5 × 10−11 | 0.06 | 2.2 × 10−4 |
| rs144330743 | 6:89852656 | N-acetylcarnosine | SRSF12/PM20D2* | A/T | 0.08 | −0.15 | 3.0 × 10−13 | −0.46 | 0.052 | −0.15 | 8.6 × 10−13 |
| rs6800284 | 3:30758956 | N-acetylcarnosine | TGFBR2/GADL1* | T/C | 0.37 | −0.07 | 1.7 × 10−12 | −0.08 | 6.5 × 10−10 | −0.07 | 5.6 × 10−4 |
| rs4363 | 17:61574492 | Phenylalanylserine | ACE* | A/G | 0.48 | −0.15 | 5.2 × 10−24 | −0.12 | 3.2 × 10−9 | −0.16 | 9.7 × 10−17 |
| rs4363 | 17:61574492 | Serylleucine | ACE | A/G | 0.48 | −0.21 | 8.7 × 10−50 | −0.22 | 3.4 × 10−25 | −0.20 | 2.3 × 10−26 |
| rs2519093 | 9:136141870 | Serylleucine | ABO | T/C | 0.16 | −0.14 | 4.1 × 10−13 | −0.13 | 1.2 × 10−7 | −0.15 | 5.4 × 10−7 |
Locus-Peptide association found by previous studies.
For each significant locus only the most significant variant per peptide is shown. The Alleles column shows the coded allele/non-coded allele. MAF refers to the minor allele frequency in the meta-analysis of European and African American participants.
Figure 1.
Overlap between the peptides associated with KLKB1, F12, KNG1, and ACE.
The lead variants in the F12 and ACE genes influenced expression levels of F12 and ACE respectively (Supplementary Material, Table S3). The ACE-increasing alleles A of rs4335 and rs4363 were increased levels of glycylphenylalanine, leucylalanine, phenylalanylserine, and serylleucine, but lowered levels of HWESASLLR. The F12-increasing G allele of rs1801020 was associated with higher levels of alanylleucine, arginylphenylalanine, histidylleucine, and leucylasparagine. Lead variants at all of the identified loci were associated with serum levels of multiple metabolites by previous studies, and several were also associated with serum protein levels (Supplementary Material, Table S4). Notably, variants at the KNG1 locus were associated with serum levels of kininogen-1, coagulation Factor XI, and kallikrein levels. Variants at the KLKB1 locus were similarly associated with levels of kallikrein and coagulation factor XI. Variants at the ABO locus were associated with levels of 20 different proteins.
Low-frequency and rare variant analyses
Using SKAT tests we identified four significant associations between peptides and sliding windows (Table 2). All four of these associations overlap with loci from the single variant analysis. No sliding windows reached the significance threshold using T5 tests. No significant associations were detected by focusing on regulatory regions with either SKAT or T5 tests. Additionally, we applied gene-based SKAT tests to whole-exome sequencing data and identified an association between CPN1 and DSGEGDFXAEGGGVR levels, which overlapped with the findings from the single variant analysis (Table 2). No significant genes were found using T5 tests. Lachesis plots for these five loci are shown in Supplementary Materials, Figures S3–S7. Lachesis plots are regional association plots that provide a comprehensive view of the association of common variants, sliding windows, regulatory regions, and protein-coding regions in a given region (8,18). The contribution of single low-frequency and rare variants within these five regions to levels of the relevant peptides is shown in Supplementary Materials, Tables S5–S9. Most SKAT associations appeared to be driven by one or more low-frequency variants. In particular, the SKAT association involving CPN1 was driven by rs11592631, a single low-frequency variant that was more significant than the lead common variant. Because the minor allele frequency of rs11592631 was over 5% in EA participants, this variant was not included in the SKAT test when the analyses were restricted to EA participants. Even though the results of the SKAT test between CPN1 and DSGEGDFXAEGGGVR were therefore not consistent across the ancestry groups, the driving variant rs11592631 had a consistent direction and magnitude of effect (BetaEA = 0.80, P-valueEA = 3.9 × 10−11, BetaAA = 0.59, P-valueAA = 0.0019). Except for the association between chr4: 187114119 -187118118 and leucylasparagine, the remaining significant windows were nominally significant in both ancestry groups.
Table 2.
Significant SKAT associations with peptides in the meta-analysis of European and African American participants
| Meta-analysis |
European Americans |
African Americans |
||||||
|---|---|---|---|---|---|---|---|---|
| Region* | Peptide | Genes in region | cMAF | P-value | cMAF | P-value | cMAF | P-value |
| Whole-genome sequencing - Sliding windows | ||||||||
| chr4: 187114119 – 187118118 | Histidylleucine | CYP4V2 | 0.40 | 4.3 × 10−12 | 0.08 | 0.0033 | 0.27 | 1.4 × 10−9 |
| chr4: 187114119 – 187118118 | Leucylasparagine | CYP4V2 | 0.40 | 6.5 × 10−14 | 0.08 | 0.60 | 0.27 | 6.2 × 10−11 |
| chr6: 89814009 – 8981800 | N-acetylcarnosine | SRSF12 | 0.21 | 7.9 × 10−11 | 0.06 | 1.2 × 10−4 | 0.21 | 3.8 × 10−8 |
| chr17: 61152052 – 6115605 | Serylleucine | TANC2 | 0.31 | 2.1 × 10−11 | 0.21 | 1.2 × 10−7 | 0.18 | 0.0047 |
| Whole-exome sequencing – Genes | ||||||||
| CPN1 | DSGEGDFXAEGGGVR | CPN1 | 0.03 | 1.4 × 10−14 | 0.003 | 0.22 | 0.02 | 2.0 × 10−7 |
For each significant locus only the most significant region per peptide is shown. SKAT refers to sequence kernel association test. cMAF refers to the cumulative minor allele frequency of all variants within the region that were included in the analysis.
Since all five regions overlapped with loci identified in the analysis of common variants, we adjusted the analyses of these regions for the lead common variant. Four of the SKAT associations were attenuated after adjustment for the lead common variant (Table 3), but they remained significantly associated (P-value < 0.013). The SKAT test between CPN1 and DSGEGDFXAEGGGVR became more significant upon adjustment for the lead common variant.
Table 3.
Conditional analysis of significant SKAT associations adjusting for the lead common variants
| Region | Common variant | Peptide | cMAF | Unadjusted P-value | Adjusted P-value |
|---|---|---|---|---|---|
| Whole-genome sequencing - Sliding windows | |||||
| chr4: 187114119 - 187118118 | rs3733402 | Histidylleucine | 0.40 | 4.3 × 10−12 | 1.4 × 10−4 |
| chr4: 187114119 - 187118118 | rs3733402 | Leucylasparagine | 0.40 | 6.5 × 10−14 | 5.9 × 10−6 |
| chr6: 89814009 - 8981800 | rs144330743 | N-acetylcarnosine | 0.21 | 7.9 × 10−11 | 1.3 × 10−5 |
| chr17: 61152052 - 6115605 | rs4363 | Serylleucine | 0.31 | 2.1 × 10−11 | 1.0 × 10−6 |
| CPN1 | rs11592631 | DSGEGDFXAEGGGVR | 0.03 | 2.0 × 10−12 | 6.2 × 10−16 |
SKAT refers to sequence kernel association test. cMAF refers to the cumulative minor allele frequency of all variants within the region that were included in the analysis.
Association of peptides with cardio-metabolic phenotypes
Out of the 25 peptides, 22 were associated with one or more of the cardio-metabolic phenotypes (Supplementary Material, Table S10). Of the eleven peptides for which we identified genetic loci, nine were associated with one or more of the cardio-metabolic phenotypes. The two most significant associations among these nine peptides were N-acetylcarnosine with HDL cholesterol (Beta = −0.17, P-value = 3.1 × 10−21), and alanylleucine with TG (Beta = 0.13, P-value 9.6 × 10−23) (34).
Association of identified loci with levels of peptides that did not pass quality control
As an exploratory analysis, we examined common variant associations between lead variants of loci identified in the primary analysis and levels of the 24 peptides that did not pass quality control, and hence were not included in the primary analysis (Supplementary Material, Table S11). There were 26 peptide-locus associations below the 4.2 × 10−10 significance level. These associations included 15 of the 24 peptides, and five loci: ten associations mapped to the KLKB1 locus, eight to F12, five to ACE, two to KNG1, and one to ABO.
Bradykinin
Many of the identified genes were from the kinin-kallikrein pathway, which leads to the production of bradykinin. As described in the Supplementary Methods, bradykinin levels were available in a subset of 1,432 participants. As an exploratory analysis, we evaluated the association between the seven lead variants in kinin-kallikrein genes and bradykinin levels. As shown in Supplementary Material, Table S12, lead variants in ACE and KLKB1 were significantly associated with bradykinin levels (P-value < 0.01). We therefore examined whether bradykinin mediated the association between these variants and peptide levels. While the associations of lead variants in ACE with glycylphenylalanine, leucylalanine, and phenylalanylserine were not mediated by bradykinin, the associations with HWESASLLR and serylleucine were partially mediated (Supplementary Material, Table S13). For KLKB1, only the association with HWESASLLR was partially mediated by bradykinin.
Fibrinopeptide A
DSGEGDFXAEGGGVR, which was associated with low-frequency variants in CPN1, is a derivative of fibrinopeptide A, which in turn is cleaved from fibrinogen during conversion to fibrin by thrombin. To investigate whether variants in CPN1 broadly affect the formation of fibrinopeptide A and derivatives, or whether it specifically affects the conversion of fibrinopeptide A to DSGEGDFXAEGGGVR, we examined the association of the common and low-frequency lead variants in CPN1 with DSGEGDFXAEGGGVR, ADSGEGDFXAEGGGVR, and their ratio (Supplementary Methods). The minor allele of rs61751507, the low-frequency lead variant in CPN1, was associated with both increased DSGEGDFXAEGGGVR and ADSGEGDFXAEGGGVR (Supplementary Material, Table S14). However, it was also associated with a higher DSGEGDFXAEGGGVR to ADSGEGDFXAEGGGVR ratio. The common variant was not associated with any of the phenotypes in this subset.
Discussion
Using single variant analysis, we identified 22 peptide-locus pairs of which 16 were novel. Five of these associations were accompanied by low-frequency variant signals. Genes involved in the kinin-kallikrein system, including KLKB1, F12, KNG1, ACE, and CPN1 were strongly represented among the associated loci. Several associations reveal new insights into biological processes, including the association of DSGEGDFXAEGGGV with CPN1, as well as the association of HWESASLLR with KLKB1.
Several associations between kinin-kallikrein genes and the 25 peptides included in our analysis have been previously described beyond those that we identified, including KLKB1 and F12 for glycylphenylalanine (14). Previous studies have also identified associations between kinin-kallikrein genes and peptides that were not included in our analysis. Suhre et al. identified KLKB1 for bradykinin, des-arg (9) and ACE for aspartylphenylalanine (3). Shin et al. identified the ACE locus for aspartylphenylalanine to phenylalanylleucine ratio and alpha-glutamyltyrosine, as well as the KNG1 and F12 loci for bradykinin, des-arg (2,9). Yu et al. linked the ACE locus to threonylphenylalanine (4). Finally, Long et al. identified the ACE locus for alpha-glutamylglycine, glycylglycine, KLKB1 for isoleucylvaline, leucylphenylalanine to isoleucylphenylalanine ratio, and prolylproline, and F12 for isoleucylvaline, leucylphenylalanine to isoleucylphenylalanine ratio, and prolylproline (14). These results reinforce our finding that variants in kinin-kallikrein genes have widespread effects on serum peptide levels.
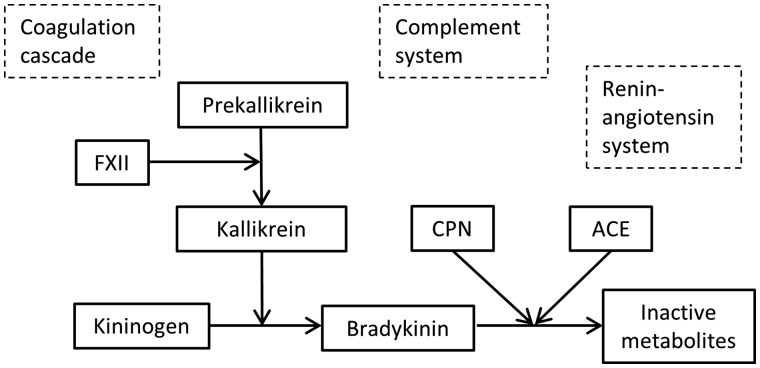
The kinin-kallikrein pathway is an enzymatic cascade related to coagulation and inflammation (Fig. 2). Kinin-kallikrein genes accounted for 16 of the 22 reported associations. In fact, when we expanded our analyses beyond the 25 peptides that passed quality control to the 24 peptides that did not pass quality control, we identified an additional 25 associations involving kinin-kallikrein genes. Prekallikrein, encoded by KLKB1, is converted into active kallikrein by factor VII, encoded by F12. The high molecular weight (HMW) form of kininogen, encoded by KNG1, interacts with factor VII and kallikrein to initiate the contact activated (also called intrinsic) coagulation pathway. Kallikrein also enhances the conversion of plasminogen into plasmin, which is required for the degradation of fibrin clots. Additionally, kallikrein breaks down HMW kininogen to form bradykinin, a peptide that dilates the blood vessels, decreasing blood pressure. The proteins encoded by ACE and CPN1, angiotensin I converting enzyme and carboxypeptidase N, both inactivate bradykinin (35).
Figure 2.
Interactions among proteins within the kinin-kallikrein system (shown in green) and their interactions with other pathways. Kininogen, kallikrein, and coagulation factor XII participate in the coagulation cascade by initiating the contact activation (intrinsic) pathway. CPN may play a role in coagulation through its interaction with fibrinogen. CPN interacts with the complement system by inactivating anaphylatoxins, which are derived from complement components. Our study suggests that kallikrein and ACE also interact with complement components. Finally, bradykinin and ACE play important roles in the regulation of blood pressure through their roles in the renin-angiotensin system and vasodilation.
Previous genetic association studies have identified variants for bradykinin in KLKB1, F12, and KNG1 (2). While the identification of these genes for bradykinin was expected based on known biology, our study identified novel associations between these genes and levels of a range of other peptides, including dipeptides alanylleucine, arginylphenylalanine, histidylleucine, leucylalanine, and leucylasparagine, as well as oligopeptide HWESASLLR. The biology underlying these associations is less clear, although two patterns emerge from our results. First, all peptides associated with KNG1 and F12 were also associated with KLKB1. While some peptides affected by ACE were also related to these genes, others were not. This suggests that KNG1, F12, and KLKB1 may affect peptide levels through a shared mechanism. Secondly, lead variants in these genes tended to affect peptides in the same direction: all associations with variants in KLKB1, F12, and KNG1 were positive, while most associations with variants in ACE were negative. The only exception was the relationship between HWESALLR and the lead variant in ACE.
HWESASLLR is derived from complement component 3 (C3). C3 is initially cleaved into C3a and C3b, and the C3b fragment is then further cleaved to produce both iC3b and C3f (36). HWESALLR makes up the final eight amino acids on the C-terminal of the C3f fragment. Our results thus suggest a link between the kinin-kallikrein system and the complement system. Such a link has been suggested by previous research: complement component 1 is known to inhibit the activation of Factor XII and kallikrein (37), and carboxypeptidase N, the same protein that inactivates bradykinin is also known to inactivate anaphylatoxins derived from complement components, including C3a (38). However, the association of HWESALLR with ACE and KLKB1 suggests that the link is much broader than previously described, while the mediation by bradykinin levels specifically implicates bradykinin.
Besides its role within the kinin-kallikrein and complement systems, carboxypeptidase N also appears to bind to both fibrin and fibrinogen (39). This function may be particularly relevant to the identified association with DSGEGDFXAEGGGV, which is a fibrinogen cleavage peptide derived from fibrinopeptide A (ADSGEGDFLAEGGGV) (40). When thrombin converts fibrinogen into fibrin, fibrinopeptide A is cleaved off as a by-product (41). Fibrinopeptide A is therefore used as a marker of thrombin activity (42). The association between variants in CPN1 and increased DSGEGDFXAEGGGV levels suggests that carboxypeptidase N may have a role in regulating the cleavage of fibrinogen by thrombin. Although no further evidence for such a role exists, there is evidence for involvement of carboxypeptidase N in fibrinolysis, the degradation of fibrin clots. After activation through proteolytic cleavage, carboxypeptidase N gains the ability to inhibit fibrinolysis (43). It is doubtful that this mechanism explains the effect of CPN1 on DSGEGDFXAEGGGV, however, because fibrinopeptide A is cleaved from fibrinogen prior to the formation of fibrin clots. Although carboxypeptide N may cleave fibrinopeptides (44), it is equally unlikely that carboxypeptidase N cleaves the N-terminal alanine from fibrinopeptide A to form DSGEGDFXAEGGGV, since carboxypeptidase N specifically cleaves C-terminal arginine and lysine. Our additional analyses do not support an effect on conversion since the lead variant was related to increased levels of both DSGEGDFXAEGGGV and fibrinopeptide A, and these associations were more significant than the association with the ratio of DSGEGDFXAEGGGVR to fibrinopeptide A. Instead, it may be that upon binding to fibrinogen, carboxypeptidase N influences the rate at which fibrinogen is cleaved by thrombin. CPN1 specifically codes for the small subunit of carboxypeptidase N, which contains the enzymatic domain, and of which two are present in each protein. The lead variant, rs61751507, that appears to be driving the association of CPN1 with DSGEGDFXAEGGGV, is a nonsynonymous variant known as Gly178Asp. The minor allele of this variant has been implicated in carboxypeptidase N deficiency, most likely in combination with further rare variants (45).
ACE cleaves COOH-terminal dipeptides from larger peptides, and is therefore not a surprising locus for serum dipeptide levels. Most famously, it cleaves histidylleucine from angiotensin I to form angiotensin II (46). In our study, the lead variant in the ACE gene was only suggestively associated with histidylleucine (P-value = 3.9 × 10−5). Several other associations between variants in ACE and dipeptide levels have previously been described (2,4), and in this study we uncovered associations with two more dipeptides: glycylphenylalanine and serylleucine. The comparative prominence of these other associations reinforces the idea that ACE cleaves broad range of peptides, and suggests that the cleavage of other, so far unknown, peptides may belong to its core functions (47).
The loci did not overlap with those that we or others have previously identified for amino acids, suggesting that these peptides are primarily produced through protein catabolism rather than amino acid metabolism (2,13,14). In addition to the loci that we identified for peptides, several more have been described by previous studies. Suhre et al. identified the FUT2 and ABO loci for ADpSGEGDFXAEGGGVR to ADSGEGDFXAEGGGVR ratio as well as the ALPL and ENPEP loci for ADpSGEGDFXAEGGGVR to DSGEGDFXAEGGGVR ratio (3). Long et al. found additional associations between ENPEP and leucylalanine and between HIF1AN and phenylalanylserine (14).
In conclusion, our results provide novel insight into the origin and function of serum peptides, revealing genes in the kinin-kallikrein system to be the predominant genetic determinants of serum peptide levels. In addition to highlighting the important role of the kinin-kallikrein system in the regulation of serum peptide levels, this study provides additional evidence for a broad link between the kinin-kallikrein and complement systems, as well as an interaction between carboxypeptidase N and fibrinogen.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of the ARIC study for their important contributions.
Conflict of Interest statement. None declared.
Funding
The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Funding support for ‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium’ was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Metabolomics measurements were sponsored by the National Human Genome Research Institute (3U01HG004402-02S1). Sequencing was carried out at the Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 and R01HL086694).
References
- 1. Babizhayev M.A., Deyev A.I., Yermakova V.N., Semiletov Y.A., Davydova N.G., Kurysheva N.I., Zhukotskii A.V., Goldman I.M. (2001) N-Acetylcarnosine, a natural histidine-containing dipeptide, as a potent ophthalmic drug in treatment of human cataracts. Peptides, 22, 979–994. [DOI] [PubMed] [Google Scholar]
- 2. Shin S.Y., Fauman E.B., Petersen A.K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T.P.. et al. (2014) An atlas of genetic influences on human blood metabolites. Nat. Genet., 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B., Altmaier E. CardioGram Deloukas P., Erdmann J.. et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature, 477, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu B., Zheng Y., Alexander D., Morrison A.C., Coresh J., Boerwinkle E. (2014) Genetic determinants influencing human serum metabolome among African Americans. PLoS Genet., 10, e1004212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suhre K., Raffler J., Kastenmuller G. (2016) Biochemical insights from population studies with genetics and metabolomics. Arch Biochem. Biophys., 589, 168–176. [DOI] [PubMed] [Google Scholar]
- 6. Yu B., Li A.H., Metcalf G.A., Muzny D.M., Morrison A.C., White S., Mosley T.H., Gibbs R.A., Boerwinkle E. (2016) Loss-of-function variants influence the human serum metabolome. Sci. Adv., 2, e1600800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rhee E.P., Yang Q., Yu B., Liu X., Cheng S., Deik A., Pierce K.A., Bullock K., Ho J.E., Levy D.. et al. (2016) An exome array study of the plasma metabolome. Nat. Commun., 7, 12360.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morrison A.C., Voorman A., Johnson A.D., Liu X., Yu J., Li A., Muzny D., Yu F., Rice K., Zhu C.. et al. (2013) Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat. Genet., 45, 899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li A.H., Morrison A.C., Kovar C., Cupples L.A., Brody J.A., Polfus L.M., Yu B., Metcalf G., Muzny D., Veeraraghavan N.. et al. (2015) Analysis of loss-of-function variants and 20 risk factor phenotypes in 8,554 individuals identifies loci influencing chronic disease. Nat. Genet., 47, 640–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wood A.R., Tuke M.A., Nalls M., Hernandez D., Gibbs J.R., Lin H., Xu C.S., Li Q., Shen J., Jun G.. et al. (2015) Whole-genome sequencing to understand the genetic architecture of common gene expression and biomarker phenotypes. Hum. Mol. Genet., 24, 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danjou F., Zoledziewska M., Sidore C., Steri M., Busonero F., Maschio A., Mulas A., Perseu L., Barella S., Porcu E.. et al. (2015) Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat. Genet., 47, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. UK10K Consortium, Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M.. et al. (2015) The UK10K project identifies rare variants in health and disease. Nature, 526, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu B., de Vries P.S., Metcalf G.A., Wang Z., Feofanova E.V., Liu X., Muzny D.M., Wagenknecht L.E., Gibbs R.A., Morrison A.C.. et al. (2016) Whole genome sequence analysis of serum amino acid levels. Genome Biol., 17, 237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Long T., Hicks M., Yu H.C., Biggs W.H., Kirkness E.F., Menni C., Zierer J., Small K.S., Mangino M., Messier H.. et al. (2017) Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet., 49, 568–578. [DOI] [PubMed] [Google Scholar]
- 15.(1989) The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol., 129, 687–702. [PubMed] [Google Scholar]
- 16. Ohta T., Masutomi N., Tsutsui N., Sakairi T., Mitchell M., Milburn M.V., Ryals J.A., Beebe K.D., Guo L. (2009) Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol. Pathol., 37, 521–535. [DOI] [PubMed] [Google Scholar]
- 17. Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem., 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- 18. Morrison A.C., Huang Z., Yu B., Metcalf G., Liu X., Ballantyne C., Coresh J., Yu F., Muzny D., Feofanova E.. et al. (2017) Practical approaches for whole-genome sequence analysis of heart- and blood-related traits. Am. J. Hum. Genet., 100, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X., Wu C., Li C., Boerwinkle E. (2016) dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat., 37, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang K., Li M., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res., 38, e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.. et al. (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res., 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fantom Consortium and the RIKEN PMI and CLST (DGT), Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M.. et al. (2014) A promoter-level mammalian expression atlas. Nature, 507, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmuller G. (2015) SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics, 31, 1334–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K.. et al. (2017) Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun., 8, 14357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C.. et al. (2014) Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics, 15, 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Jian X., Boerwinkle E. (2013) dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat., 34, E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voorman A., Brody J., Chen H., Lumley T., Davis B. (2016), in press.
- 28. Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F. 3rd, Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T.. et al. (2009) A new equation to estimate glomerular filtration rate. Ann. Intern. Med., 150, 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B., Leal S.M. (2008) Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet., 83, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. (2011) Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet., 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J., Ellinghaus D., Franke A., Howie B., Li Y. (2012) 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet., 20, 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Vries P.S., Chasman D.I., Sabater-Lleal M., Chen M.H., Huffman J.E., Steri M., Tang W., Teumer A., Marioni R.E., Grossmann V.. et al. (2016) A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum. Mol. Genet., 25, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. GTEx Consortium. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. (1990) An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest., 86, 1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuoppala A., Lindstedt K.A., Saarinen J., Kovanen P.T., Kokkonen J.O. (2000) Inactivation of bradykinin by angiotensin-converting enzyme and by carboxypeptidase N in human plasma. Am. J. Physiol. Heart Circ. Physiol., 278, H1069–H1074. [DOI] [PubMed] [Google Scholar]
- 36. Ganu V.S., Muller-Eberhard H.J., Hugli T.E. (1989) Factor C3f is a spasmogenic fragment released from C3b by factors I and H: the heptadeca-peptide C3f was synthesized and characterized. Mol. Immunol., 26, 939–948. [DOI] [PubMed] [Google Scholar]
- 37. Kaplan A.P., Ghebrehiwet B. (2010) The plasma bradykinin-forming pathways and its interrelationships with complement. Mol. Immunol., 47, 2161–2169. [DOI] [PubMed] [Google Scholar]
- 38. Bokisch V.A., Muller-Eberhard H.J. (1970) Anaphylatoxin inactivator of human plasma: its isolation and characterization as a carboxypeptidase. J. Clin. Invest., 49, 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Talens S., Lebbink J.H., Malfliet J.J., Demmers J.A., Uitte de Willige S., Leebeek F.W., Rijken D.C. (2012) Binding of carboxypeptidase N to fibrinogen and fibrin. Biochem. Biophys. Res. Commun., 427, 421–425. [DOI] [PubMed] [Google Scholar]
- 40. Masuda Y., Sugiyama T. (2001) Human fibrinopeptide A mediates allergic reaction in mice in the acute phase. Peptides, 22, 1511–1513. [DOI] [PubMed] [Google Scholar]
- 41. Scheraga H.A. (2004) The thrombin-fibrinogen interaction. Biophys. Chem., 112, 117–130. [DOI] [PubMed] [Google Scholar]
- 42. Nossel H.L., Ti M., Kaplan K.L., Spanondis K., Soland T., Butler V.P. Jr. (1976) The generation of fibrinopeptide A in clinical blood samples: evidence for thrombin activity. J. Clin. Invest., 58, 1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker J.B., Binette T.M., Mackova M., Lambkin G.R., Mitchell L., Bajzar L. (2008) Proteolytic cleavage of carboxypeptidase N markedly increases its antifibrinolytic activity. J. Thromb. Haemost., 6, 848–855. [DOI] [PubMed] [Google Scholar]
- 44. Matthews K.W., Wetsel R.A. (2001) Characterization of mouse carboxypeptidase N small active subunit gene structure. J. Immunol., 166, 6196–6202. [DOI] [PubMed] [Google Scholar]
- 45. Cao H., Hegele R.A. (2003) DNA polymorphism and mutations in CPN1, including the genomic basis of carboxypeptidase N deficiency. J. Hum. Genet., 48, 20–22. [DOI] [PubMed] [Google Scholar]
- 46. Coates D. (2003) The angiotensin converting enzyme (ACE). Int. J. Biochem. Cell Biol., 35, 769–773. [DOI] [PubMed] [Google Scholar]
- 47. Altmaier E., Menni C., Heier M., Meisinger C., Thorand B., Quell J., Kobl M., Romisch-Margl W., Valdes A.M., Mangino M.. et al. (2016) The Pharmacogenetic Footprint of ACE Inhibition: A Population-Based Metabolomics Study. PLoS One, 11, e0153163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.