Abstract
Although Zika virus (ZIKV) infection is often asymptomatic, in some cases, it can lead to birth defects in newborns or serious neurologic complications in adults. However, little is known about the interplay between immune and neural cells that could contribute to the ZIKV pathology. To understand the mechanisms at play during infection and the antiviral immune response, we focused on neural precursor cells (NPCs)-microglia interactions. Our data indicate that human microglia infected with the current circulating Brazilian ZIKV induces a similar pro-inflammatory response found in ZIKV-infected human tissues. Importantly, using our model, we show that microglia interact with ZIKV-infected NPCs and further spread the virus. Finally, we show that Sofosbuvir, an FDA-approved drug for Hepatitis C, blocked viral infection in NPCs and therefore the transmission of the virus from microglia to NPCs. Thus, our model provides a new tool for studying neuro-immune interactions and a platform to test new therapeutic drugs.
Introduction
Zika virus (ZIKV) is an arbovirus belonging to the genus Flavivirus first described in 1947 in Uganda during blood analyses of sentinel Rhesus monkeys (1). Until the 21st century, African and Asian lineages of ZIKV did not cause substantial human infections. However, in 2007, vectored by Aedes aegypti mosquitoes, the first noteworthy epidemic of ZIKV occurred on the island of Yap in Micronesia (2). The recent dramatic increase in newborns with microcephaly and other congenital malformations in Brazil has been associated with an outbreak of ZIKV (3,4). The outbreak has further been linked to the autoimmune neurological disorder Guillain-Barré syndrome (GBS) in older children and adults (5). The circulating Asian-lineage strain of ZIKV in Brazil (ZIKVBR) has been detected in the placenta and amniotic fluid of women with microcephalic fetuses (6–8) and in the blood of microcephalic newborns. Curiously, although the African MR-766 strain of ZIKV (ZIKVMR766, strain MR-766) was first identified in the 1950s, no birth defects in newborns or neurological complications in adults have been associated with this strain. Thus, recent publications focused on the mechanisms of transmission and spreading of ZIKV in the Americas (9–12).
Earlier this year, Liu and colleagues reported that ZIKV evolved to acquire a spontaneous mutation in its NS1 (A188V mutation) domain leading to increased antigenaemia (12). This enhancement of NS1 antigenaemia promoted ZIKV infectivity and prevalence in mosquitoes that could ease the transmission and possibly explain the recent ZIKV outbreaks. Following this publication, another group suggested that the A188V mutation on NS1 domain arose in Southeast Asia at the early 2000s and circulated in that region several years before spreading to South Pacific Islands and the Americas (13). Moreover, there is now converging evidence that the African strains lead to higher infection rate and viral production as well as stronger cell death in cellular and immunocompetent adult mouse models leading to the increased mortality while the Asian strains did not (14–16). This feature of the Asian strain might be responsible for causing chronic infections observed in congenital microcephaly cases and explain why no other diseases were observed until recently. Hence, there is an urgent need to develop model systems to examine the complex relationship between ZIKVBR infection and brain malformations.
In order to model ZIKVBR infectivity, we previously showed that ZIKVBR crosses the placenta to infect the fetus in vivo, causing microcephaly and birth defects (17). In addition, we showed that ZIKVBR targeted human neural precursor cells (NPCs), growing as neurospheres or organoids, induced cell death and led to a reduction of proliferative zones and a disruption of cortical layers as previously observed in vivo (17). Despite advances regarding the causal relationship between the ZIKVBR and birth defects, little is known about the pathogenic interactions of this virus with different cell types and its vertical transmission to fetal brain.
Since the onset of the current Brazilian outbreak in 2015, most ZIKV studies have focused on neurons and glial cells (17,18). However, further examination of ZIKV interaction with fetal circulating monocytes and the interplay between microglia and neural cells are important steps for understanding ZIKV pathophysiology. A recent publication, using the contemporary ZIKV strain PRVABC59 (PR2015), showed that the virus can infect and replicate in Hofbauer cells (HCs), a type of primary human placental macrophage (19). Hence, one possibility for intrauterine transmission allowing ZIKV to gain access to the fetal compartment is by direct infection of cells comprising the placental barrier (19). Yolk sac macrophages invade the brain parenchyma through the blood vessels during embryogenesis to become resident macrophages of the central nervous system (CNS), microglia (20). Considering the timing during embryogenesis of the entry of myeloid precursors, we hypothesized that microglia could also actively participate during ZIKV infection, perhaps acting as a Trojan horse by transporting the ZIKV during CNS invasion (21). Thus, in this study, using a ZIKV strain isolated from a clinical case in northeast of Brazil (17), ZIKVBR, we aimed to mimic those early interactions between NPCs and macrophage/microglia.
Here, we used human-induced pluripotent stem cells (hiPSCs) to model immune interactions in the developing central nervous system of the fetal brain during ZIKV infection by generating human macrophages/microglia and NPCs from the same healthy donor. Then, we established a co-culture system to model the cellular interplay that naturally occurs in the developing human brain. Our data reveal that a similar immune response was elicited in developing human microglia upon infection with the ZIKVBR as was found in the humoral immune response present in ZIKV-infected human tissues (22–25). Upon co-culture, microglia interacted with ZIKVBR-infected NPCs. Our data further show that ZIKV-infected macrophages/microglia transmit the virus to naïve NPCs, increasing the apoptosis of NPCs. These results imply that during embryogenesis, myeloid precursors could be responsible for ZIKV transmission to the CNS during their normal process of brain invasion. Finally, we tested Sofosbuvir (SOF), an FDA-approved drug against Hepatitis C infection, previously tested against ZIKV (26–28), and showed that SOF was able to decrease the cell death of ZIKV-infected NPCs. Our findings reveal that human microglia are profoundly influenced by their microenvironment and can sense ZIKV-infected cells in their immediate surroundings, providing insights on the mechanism of infection. Thus, our co-culture system could be used for studying neuro-immune interactions in vitro and testing new therapeutic candidates against ZIKV.
Results
Characterization of hiPSC-derived macrophage/ microglia (MΦ)
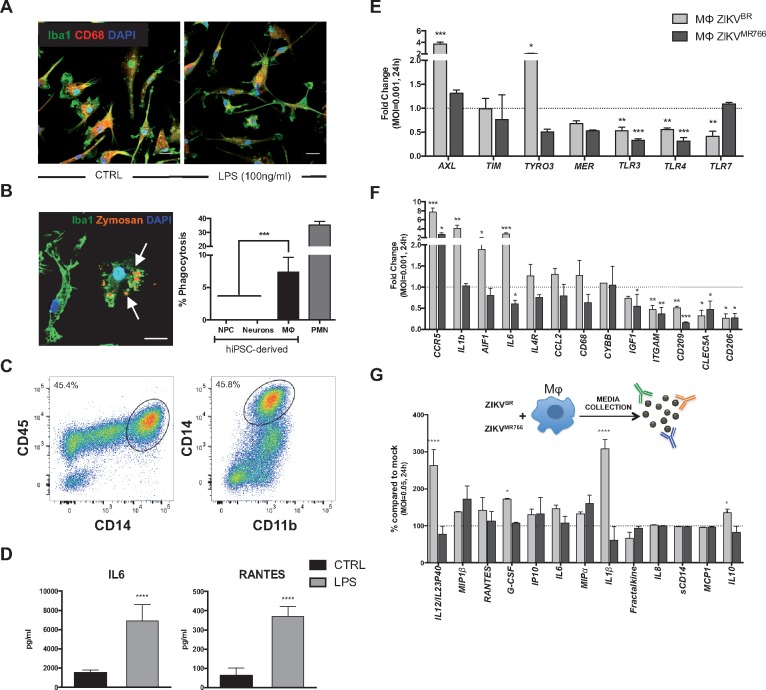
To understand the immune component of neurologic abnormalities observed in ZIKV-infected newborns, we first generated MΦ from hiPSCs using an edited version by Douvaras and colleagues of the previously published protocol by Yanagimachi et al. (29,30). This serum-free, feeder-free method gave rise to CD14+ monocytes, precursors of MΦ, in suspension starting at 15 days in vitro. CD14+ cells were then sorted using magnetic-activated cell sorting (MACS) system, treated with M-CSF and IL-34 for a week as previously published (29), and used in the subsequent experiments (Supplementary Material, Fig. S1A). These monocytes expressed classical MΦ markers such as Iba1 and CD68 (Fig. 1A). The analysis of the cells in suspension, prior to CD14 sorting, revealed that they expressed monocyte, macrophage and myeloid cell markers such as CD45/CD14, CD14/HLA-DRDPDQ, CD14/CD11b and CD14/CD68, (Fig. 1C, Supplementary Material, Fig. S1B). One week after sorting, CD14+ cells established a homogeneous macrophage population (Supplementary Material, Fig. S1C) and had similar cellular morphology and expression of cell markers such as CD68 and Iba1 to blood monocyte-derived macrophages (Supplementary Material, Fig. S1C and D). We next assessed whether the MΦ were functional by measuring their capacity for phagocytosis (Fig. 1B). In a classical assay (31–33), hiPSC-derived MΦ engulfed the yeast particle zymosan (Fig. 1B). The MΦ were also able to phagocytose living cells of the leading neonatal bacterial pathogen group B Streptococcus (GBS) whereas hiPSC-derived NPCs or 4-week-old neurons did not internalize the bacterium (Fig. 1B); primary human neutrophils were used as positive control of phagocytosis. In addition, we measured cytokine (IL-6, IL-1β, IL12/IL23p40), chemokine (MIP1α, MIP1β, RANTES, Fractalkine, IL8 and IP10) and the growth factor G-CSF release by hiPSC-derived MΦ after activating them with the classical pro-inflammatory TLR-4 ligand lipopolysaccharide (LPS) using multiplex cytometric bead based-assay (Fig. 1D, Supplementary Material, Fig. S1E). All cytokines/chemokines tested were readily detected in the MΦ-conditioned media. LPS treatment for 24 h significantly increased levels of IL-6, IL-1β, IL12/IL23p40, Fractalkine, G-CSF, IP10, RANTES, MIP1α and MIPβ (Fig. 1D, Supplementary Material, Fig. S1E).
Figure 1.
Characterization of functional human induced pluripotent stem cell (hiPSC)- derived macrophages/microglia (MΦ) and their Innate immune system receptors and inflammatory response upon Zika virus (ZIKV) infection. (A) Representative images of hiPSC-derived MΦ stained with anti-Iba1 (green), anti-CD68 (red) and fluorescent nuclear DAPI stain (blue): control MΦ untreated CTRL, (left) and treated with lipopolysaccharides (LPS, 100 ng/ml, right). Scale bar: 20 μm. (B) pHrodo red zymosan (orange) particles engulfed by hiPSC-derived MΦ, stained with anti-Iba1 (green). Scale bar: 50 μm. Phagocytosis percentage determined by group B streptococcus (GBS) internalization. Note that hiPSC-derived neural precursors (NPC) or neurons do not engulf any GBS. Human primary polymorphonuclear neutrophils (PMN) were used as an experimental positive control. One-way ANOVA with Tukey multiple comparison tests were performed, bars represent means of the percentage of phagocytosis ± SD ***P < 0.001. (C) Fluorescent activated cell-sorting (FACS) analysis of monocytic lineage cells derived sequentially from pluripotent stem cells. (D) Cytometric Bead Array (CBA) performed on conditioned media from control or 1 μg/ml LPS-treated MΦ for 24–30 h. Student’s t-tests were performed to compare the two groups. Bars represent means of the amount of cytokines/chemokines released in the media (pg/ml) ± SD, ****P < 0.0001 compared with control (CTRL) untreated cells. (E) The expression of potential Zika virus entry receptors including AXL, TIM, TYRO3, MER and several TLR receptors (TLR3, TLR4, TLR7) was measured by qRT-PCR in MΦ infected with either Brazilian (ZIKVBR) or the highly in vitro passaged MR766 ZIKV strain (ZIKVMR766) (MOI=0.001). RNA was analysed 24 h p.i. One-way ANOVA with Tukey multiple comparison tests were performed. The bars represent means of the mRNA fold change compared with the mock-infected MΦ, shown by a dashed line on the y-axis at 1 ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. (F) Inflammatory response was measured by qRT-PCR in hiPSC- MΦ infected with either the ZIKVBR or ZIKVMR766 (MOI=0.001). RNA was extracted 24 h after infection. One-way ANOVA with Tukey multiple comparison tests were performed, bars represent means of the mRNA fold change compared with the mock-infected MΦ, shown by a dashed line on the y-axis at 1 ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, MΦ ZIKVBR is represented in light grey and ZIKVMR766 in dark grey. (G) Inflammatory cytokines/chemokines were measured in the media conditioned by MΦ for 24–30 h p.i (MOI=0.05) with ZIKVBR or ZIKV MR766. One-way ANOVA with Sidak multiple comparisons tests were performed. Bars represent average of fold change (in %) compared with the mock-infected MΦ (as shown by the dashed line at 100%) ± SD, *P < 0.05, ****P < 0.0001, MΦ ZIKVBR is represented in light grey and ZIKVMR766 in dark grey.
In sum, hiPSC-derived MΦ displayed monocyte and myeloid markers (Supplementary Material, Fig. S1D), phagocytosed zymosan particles and whole bacteria and responded to a pro-inflammatory stimulus as measured by cytokine/chemokine release similar to primary blood-derived monocytes/macrophages (34).
Immune response of macrophages/microglia (MΦ) upon infection with the ZIKV
Next, we studied the immune response of MΦ upon infection with ZIKVBR as well as with ZIKVMR766, a lab-adapted, highly in vitro passaged African MR766 strain, presumably different from the circulating African strain at 12 and 24 h post-infection (p.i) (Supplementary Material, Fig S2A and B). We selected a panel of potential virus entry receptor candidates based on recent publications including AXL, TIM, TYRO3, MER and several TLR receptors previously linked to ZIKV and arboviruses such as DENV (32,33,39); TLR3, TLR4, TLR7, to measure the expression levels by qRT-PCR (17,18,35,36). At 12h p.i., we did not observe any significant changes between the strains when compared with mock-infected samples (Supplementary Material, Fig. S2A). At 24 h p.i., both strains of the virus triggered a TLR7 up-regulation and only ZIKVMR766 -infected MΦ expressed higher levels of the MER receptor when compared with mock-infected conditions (Supplementary Material, Fig. S2B). Next, we used a lower MOI aiming to mimic the ZIKV infection in vivo and to unveil the most sensitive changes without inducing a dramatic inflammatory response that could mask small and strain-specific changes (37–41). Thus, these ranges of MOIs and 24h p.i. time point were used in the subsequent experiments (Fig. 1E and F).
TYRO3 and AXL were upregulated upon the ZIKVBR infection, with AXL gene expression increased by three-fold compared with the mock-infected MΦ (Fig. 1E). In contrast, expression of MER and TIM did not differ between ZIKVBR-infected and mock-infected MΦ (Fig. 1E). As for the TLR receptors expression, we focused on three relevant for our study: TLR3, a sensor of RNA viruses, which was recently linked to ZIKV infection (35); TLR4, which plays a central role in recognition of LPS from Gram-negative bacteria, which was previously shown to be upregulated in ZIKV-infected human neurospheres (36); and TLR7, a sensor of ssRNA involved in the detection of other arboviruses such as DENV (42). Surprisingly, all were downregulated when compared with mock-infected controls (Fig. 1E). Interestingly, the corresponding changes seen in AXL, TYRO3, and TLR7 gene expression upon infection with the ZIKVBR were absent in parallel infections with the ZIKVMR766, which did not differ from mock-infected MΦ in expression of all three receptor genes (Fig. 1E).
The geographic and symptomatology overlap between ZIKV and other Flaviviruses such as Dengue (DENV) initially complicated diagnosis (42). The vector by which it is propagated (Aedes aegypti) is shared by both viruses and clinically presented serological cross-reactivity. In addition, DENV is known to target primary blood cells such as monocytes and macrophages and induces an inflammatory response (42). Thus, we next measured the expression of genes encoding three receptors described for DENV infection: CLEC5A, CD209/DC-SIGN, and CD206/MRC1 (Fig. 1F). All three viral receptors, CLEC5A, CD209/DC-SIGN, and CD206/MRC1, were downregulated in ZIKVBR or ZIKVMR766-infected MΦ compared with mock-infected MΦ, suggesting that ZIKV does not implicate these DENV-related receptors in iPSCs-derived MΦ (Fig. 1F).
Next, we studied the expression of the MΦ markers such as AIF1/Iba1, CD68, and ITGAM/CD11b by qRT-PCR upon infection with the ZIKV strains. AIF1/Iba1 expression was increased in the MΦ infected with ZIKVBR compared with the mock-infected or ZIKVMR766-infected cells, whereas similar changes in CD68 expression did not achieve statistic significance (Fig. 1F). Notably, pro-inflammatory genes encoding IL6, IL1B, and CCR5 were markedly induced upon infection by the ZIKVBR, but not ZIKVMR766. In contrast, the MΦ marker ITGAM/CD11b and the anti-inflammatory factor IGF1 were decreased by infections with either ZIKV strain, whereas expression of CYBB/NOX2 remained unchanged upon infection with the viruses (Fig. 1F). These results indicate that the inflammatory response of MΦ infected with two different ZIKV strains at the same viral MOI elicited different inflammatory responses.
We next compared cytokine/chemokine release by the hiPSC-derived MΦ 24 h p.i. with the ZIKVBR or ZIKVMR766, (Fig. 1G, Supplementary Material, Fig. S4) using the same multiplex cytometric bead array that was used previously (Fig. 1D, Supplementary Material, Fig. S1E). Using several inocula of virus (MOI = 0.05, MOI = 0.01 and MOI = 0.001), we detected changes in release of several pro-inflammatory cytokines/chemokines (Fig. 1G, Supplementary Material, Fig. S4B and C). MΦ release of pro-inflammatory cytokines IL12/IL23p40, IL1β, IL10 and the growth factor G-CSF was significantly higher in response to ZIKVBR compared with ZIKVMR766 infection; whereas MIP1β, RANTES, IP-10, IL6, and MIP1α release from ZIKV-infected MΦ were comparable between the two strains (Fig. 1G, Supplementary Material, Fig. S4B and C), arguing for a ZIKVBR-specific response by the hiPSC-derived MΦ.
Differential expression of TAM/TIM and TLR receptors in human NPCs
Human NPCs were shown to be targeted by ZIKVBR (17). Similar to MΦ, we first infected NPCs at an MOI of 0.1 with either strains and studied the expression of different viral entry receptors at 12 and 24 h p.i. by qRT-PCR (Supplementary Material, Fig. S2C and D), however, we did not detect any differences between the two different strains at 12h p.i. (Supplementary Material, Fig. S2C). At 24h p.i., TYRO3 and TLR4 were both upregulated in NPCs upon infection with both ZIKVBR and ZIKVMR766 strains (Supplementary Material, Fig. S2D). Given that we did not see major differences in gene expression in both of the strains tested, we focused on the same MOI (0.001) determined from the MΦ expression analysis and the time points (24 and 96 h p.i.) (Fig. 1A and B, Supplementary Material, Fig. S2A and B) for the NPCs (Supplementary Material, Fig. S3A and B). Of four receptors analysed, only the MER was increased in the ZIKVBR 24 h p.i. compared with mock but returned to mock levels by 96 h p.i. (Supplementary Material, Fig. S3A). Expression of TYRO3 was downregulated in both mock- and ZIKVBR-infected NPCs at 96 h p.i. compared with 24 h p.i. (Supplementary Material, Fig. S3A). TLR4 was also decreased as in MΦ, while TLR7 expression was increased compared with the mock-infected controls at 24 h p.i. (Supplementary Material, Fig. S3B). The expression of TAM/TIM receptors and TLRs observed in NPCs at 96 h p.i. were accompanied by a decreased expression of cell markers such as NES/Nestin and PAX6 in both mock- and ZIKVBR-infected NPCs (Supplementary Material, Fig. S3C).
To summarize, we did not observe great changes in ZIKVBR-infected NPCs except for MER and TLR7, which were significantly upregulated compared with the mock-infected NPCs at 24-h post-infection.
The interplay between NPCs with macrophages/microglia (MΦ) during ZIKV infection
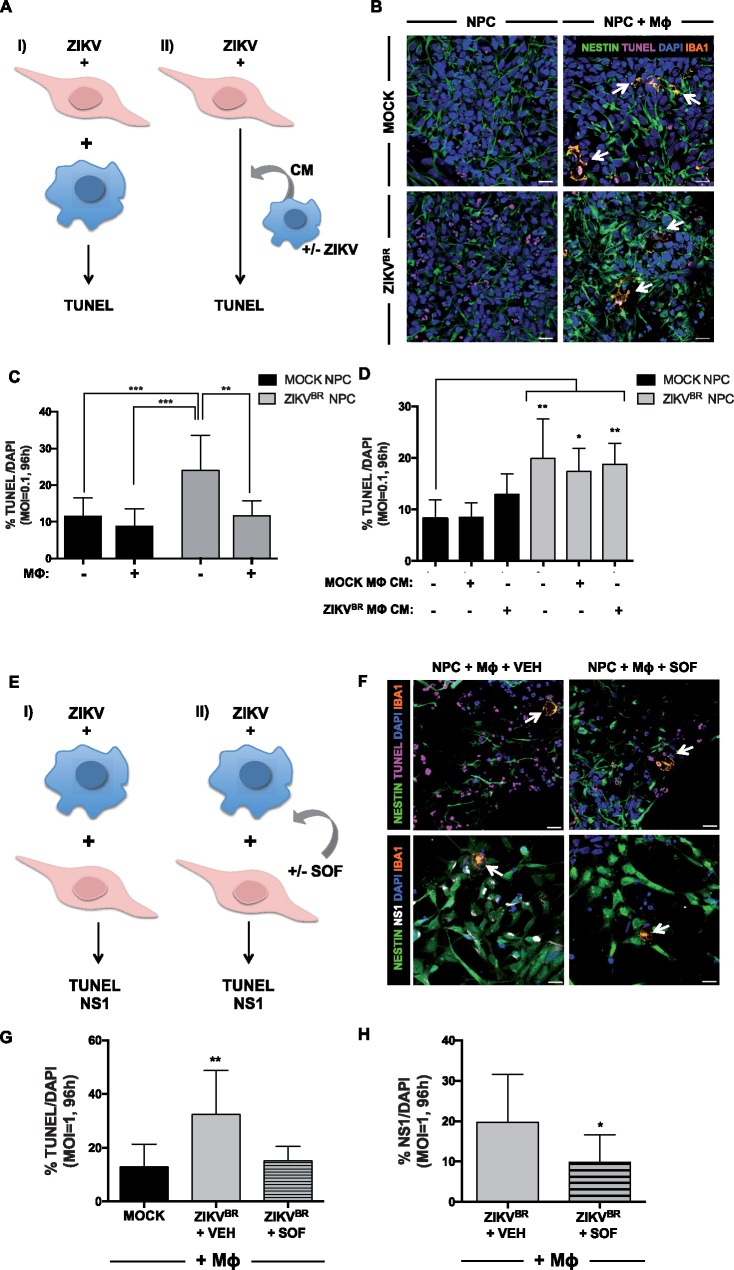
To investigate interactions of hiPSC-derived MΦ with ZIKVBR-infected NPCs we established a co-culture experimental platform. NPCs were infected (MOI = 0.1) for 2 h with the ZIKVBR or ZIKVMR766, media replaced, and MΦ overlaid 2 h after the media change. MΦ and NPCs were maintained in co-culture for 96 h, and the fraction of cells undergoing cell death examined by DNA fragmentation using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (Fig. 2A and B, Supplementary Material, Fig. S5). We initially seeded the same amount of MΦ and NPCs, however, the proliferation rate of the NPCs was higher and thus the percentage of MΦ in the co-culture with NPCs was around 2% at 96 h p.i. (Supplementary Material, Fig. S5C). In the absence of infection, addition of MΦ in co-culture atop NPCs did not influence the number of TUNEL+ cells (∼10%, see Fig. 2C). However, as previously shown (17), ZIKVBR infection at MOI = 0.1 doubled the percentage of TUNEL+ NPCs (Fig. 2B and C). No increase in TUNEL+ was detected in NPCs infected with ZIKVBR at MOI = 0.01 for 96 h (Supplementary Material, Fig. S5A). Infection with ZIKVMR766 had similar effect by increasing the amount of TUNEL+ NPCs (Supplementary Material, Fig. S5B). In addition, the amount of MΦ undergoing apoptosis (TUNEL+) remained under ∼5% in presence or absence of ZIKVBR indicating that the ZIKVBR was not cytotoxic to MΦ under our experimental conditions (Fig. 2B and C, Supplementary Material, Fig. S5D). Interestingly, adding MΦ to ZIKV-infected NPCs reduced the percentage of TUNEL+ cell levels similar to mock-infection (Fig. 2B and C, Supplementary Material, Fig. S5B). Thus, while adding naïve MΦ to uninfected NPCs did not increase cell death, infection of NPCs with the ZIKVBR increased cell death, and the presence of MΦ reduced cell death in ZIKVBR-infected NPCs (Fig. 2B and C). To elucidate the mechanism by which MΦ presence could decrease TUNEL+ ZIKVBR-infected NPCs, whether it is through phagocytosis or through protection by release of neurotrophic factors by MΦ, we used conditioned media (CM) from mock or ZIKVBR- infected MΦ and added on top of NPCs (Fig. 2D). Mock MΦ CM did not have any effect on the basal level of apoptosis in mock-infected NPCs whereas ZIKVBR MΦ CM addition tended to increase the level of TUNEL+ NPCs (Fig. 2D). Although the addition of mock or ZIKVBR MΦ CM on top of ZIKVBR-infected NPCs had a small tendency to decrease the amount of TUNEL+ cells, there was no statistically significant change compared with ZIKVBR-infected NPCs in the absence of any CM (Fig. 2D). Thus, MΦ most likely need to be in contact with NPCs to decrease the amount of TUNEL+ NPCs, most likely by phagocytosing the apoptotic cells.
Figure 2.
The impact of ZIKV on macrophage/microglia (MΦ) and NPCs co-culture. (A) Schematic of the experimental design. ZIKV-infected NPCs are co-cultured with either naïve MΦ (I), or mock or ZIKVBR-infected MΦ-conditioned media (MΦ CM) (II), followed by an analysis of cell death measured by the percentage of TUNEL positive cells. (B) Images of mock and ZIKVBR-infected NPCs stained with Nestin (green), TUNEL (pink) and fluorescent nuclear staining, DAPI (blue), and MΦ stained with Iba1 (orange) in control (upper left), ZIKVBR-infected NPCs (bottom left), in control NPCs co-cultured MΦ (upper right) and in ZIKVBR-infected NPCs co-cultured with MΦ (bottom right). White arrows point to MΦ (orange). Scale bar: 20 μm. (C) TUNEL images of NPCs infected with the ZIKVBR (MOI =0.1) were acquired, and the percentages of dying cells were calculated, averaged, and graphed accordingly. One-way ANOVA tests with Tukey multiple comparison were performed to compare different groups. The presented values are means of TUNEL+/DAPI+ percentage ± SD, **P < 0.01, ***P < 0.001. Note that infected NPCs have increased TUNEL+/DAPI+ percentage compared with the mock and the addition of MΦ decreases TUNEL+/DAPI+ percentage. (D) TUNEL images of NPCs infected with the ZIKVBR and treated with mock or ZIKVBR-infected MΦ conditioned media (MΦ CM) (MOI=0.1) were acquired, and the percentages of apoptotic cells were calculated, averaged, and graphed accordingly. One-way ANOVA with Tukey multiple comparisons tests were performed to compare different groups. The presented values are means of TUNEL+/DAPI+ percentage ± SD, *P < 0.05, **P < 0.01. (E) Schematic of the experimental design: ZIKV-infected MΦ are co-cultured with NPCs (I), treated with either vehicle (VEH) or Sofosbuvir (SOF) (II) for 96 h before quantification of TUNEL and NS1 positive cells. (F) Images of ZIKVBR-infected cells stained with Nestin (green), TUNEL (pink), Iba1 (orange) and DAPI (blue) (upper), and images of ZIKVBR-infected cells stained with Nestin (green), NS1 (white) and Iba1 (orange) (lower). MΦ were treated with either VEH (left), or 20μM SOF (right) after ZIKV-infection and co-cultured with NPCs. White arrows point to MΦ (orange). Scale bar: 20 μm. (G) TUNEL images of cells infected with the ZIKVBR (MOI =1) were acquired, and the percentages of dying cells were calculated, averaged, and graphed accordingly. One-way ANOVA tests with Tukey multiple comparison were performed to compare different groups. The presented values are means of TUNEL+/DAPI+ percentage ± SD, **P < 0.01. Note that MΦ treated with SOF and co-cultured infected NPCs have decreased TUNEL+/DAPI+ percentage compared with the vehicle (VEH). (H) NS1 images were acquired, and the percentages of NS1 positive cells were calculated, averaged, and graphed accordingly. One-way ANOVA with Tukey multiple comparisons tests were performed to compare different groups. The presented values are means of NS1+/DAPI+ percentage ± SD, *P < 0.05.
Finally, to assess whether ZIKV-infected MΦ could transmit the virus to the NPCs, we first infected MΦ with ZIKV (ZIKVBR or ZIKVMR766, Supplementary Material, Fig. S5), washed and added on top of NPCs the next day (Fig. 2E). We co-cultured ZIKV-infected MΦ with NPCs for 96 h and analysed the amount of TUNEL+ cells (Fig. 2E and F, Supplementary Material, Fig. S5E). Importantly, upon addition of ZIKV-infected MΦ on top of NPCs, the amount of TUNEL+ cells increased significantly (Fig. 2F and G, Supplementary Material, Fig. S5E). Keeping in mind that MΦ represent around 2% of the cells in total (Supplementary Material, Fig. S5C) and that ZIKV infection did not increase the MΦ in apoptosis (Supplementary Material, Fig. S5D), the increase in TUNEL can be attributed to the increase of cell death of NPCs (Fig. 2G, Supplementary Material, Fig. S5E). More importantly, we tested an FDA-approved drug, SOF, which was previously shown efficient against ZIKV infection in our model (26–28).
Next, in a following experiment, we added 20 μM SOF at the same time as the addition of ZIKV-infected MΦ on top of uninfected NPCs (Fig. 2E, G and H, Supplementary Material, Fig. S5E and F). Importantly, addition of SOF decreased significantly the amount of TUNEL+ cells in the case of ZIKVBR-infected MΦ addition (Fig. 2F and G) and showed a tendency to decrease upon infection with ZIKVMR766-infected MΦ (Supplementary Material, Fig. S5E). In addition, this decrease was accompanied by a decrease the amount of the flavivirus NS1+ domain specific staining, suggesting that SOF was able to limit the ZIKVBR replication (Fig. 2F–H). As for the MR766, SOF had only a tendency to decrease the amount of NS1+ cells (Supplementary Material, Fig. S5F). Thus, SOF was able to block the increased cell death of NPCs due to the infection by ZIKV-infected MΦ.
Altogether, our model was able to mimic neuro-immune interactions, which is likely to occur during human neurodevelopment and was proven to be useful for testing new therapeutic drugs to block ZIKV-associated phenotypes.
Discussion
In this study, we modeled immune interactions between NPCs and macrophages/microglia (MΦ) that would occur in the developing brains of the ZIKVBR-infected fetuses/newborns, using an induced pluripotent stem cell experimental platform. A fundamental strength of induced pluripotent stem cells is the possibility of deriving different cell types from individuals in vitro. Generation of MΦ using this technology will circumvent the need for MHC-matched bone marrow-derived MΦ from a healthy donor. In addition, by generating NPCs and MΦ from the same donor, we established an autologous approach to the study of immune interactions between different cell types derived from the same genetic background. The novelty of our study relies on the establishment of an in vitro platform, able to mimic human neuro-immune interactions. In particular, these interactions in human cells were studied by co-culturing hiPSC-derived NPCs together with hiPSC-derived MΦ. To our knowledge this is the first study enabling the analysis of human neuro-immune interactions, unlike the studies using 3D organoids, where the mesoderm-derived immune cells such as MΦ of the central nervous system are lacking, which is a major limitation of this technology (17,35,43–45). Thus, we strongly believe that our platform could be further used in other studies implicating MΦ to model the interplay among the different cell types and their impact on neurodevelopmental diseases.
In this study, using low MOI infections to mimic the ZIKV infection in vivo, we analysed the cytokine and immune receptor responses to infections with the ZIKVBR or ZIKVMR766 strains. We then investigated cellular receptors implicated in ZIKV cell entry in developing human MΦ, beginning with C-type lectin receptors known to bind DENV on the surface of macrophages and dendritic cells and modulate inflammatory responses (42,46). Unlike in DENV infections, CLEC5A, CD209/DC-SIGN and CD206/MRC1 all had reduced expression in ZIKVBR and ZIKVMR766-infected MΦ compared with controls (42,46). TAM/TIM receptors and TLRs have been recently implicated in ZIKV infection (18,35). We found that MΦ and NPCs had differential expressions of the genes encoding these receptors upon infection with ZIKVBR: TYRO3 and AXL were upregulated in MΦ infected with ZIKVBR unlike NPC, which had only an upregulation of MER, suggesting that AXL does not have a central role in ZIKV infections in NPCs. Indeed, several publications focused on AXL receptors over the last year since it was suggested as a potential viral entry receptor (18), however, two independent studies showed that blocking AXL does not prevent ZIKV infection of NPCs (46,47). But a recent publication claims that selective expression of Musashi in NPCs could explain their increased vulnerability to ZIKV (48). In the future, experiments such as RNAseq on ZIKV-infected MΦ and NPCs could be more informative to comprehend mechanisms underlying ZIKV-infections. Thus, more research is needed to unveil the different mechanisms underlying ZIKV infection of immune and neural cells.
Regarding TLR receptors, we show that all three TLRs, TLR3, TLR4 and TLR7 were downregulated in ZIKVBR-infected MΦ. On closer examination of the inflammatory response, we showed that ZIKVBR-infected MΦ expressed higher levels of AIF1, IL6, IL1β and CCR5 and released higher levels of IL12/IL23p40, IL1β, IL10 and G-CSF upon infection with ZIKVBR but not with ZIKVMR766 suggesting that the Brazilian strain might induce a distinct inflammatory response at lower MOIs. In line with this, a recent publication showed that the Asian-lineage ZIKV infection of pregnant women‘s blood led CD14+ monocytes to a M2 phenotype, with a notable increase in IL-10 production (49), suggesting that our ZIKVBR-infected MΦ can, at least partially, recapitulate the strain-specific features of ZIKV infection. Noteworthy, as stated previously, the African strain MR766 of the ZIKV has been passaged multiple times in vitro, hence the differences seen in inflammatory response could be attributed to the artificial nature of MR-766 strain (50). Thus, this platform could be used to distinguish subtle differences between the several viruses. Interestingly, Manangeeswaran and colleagues found increased levels of IL1β in the CNS of IFNAR KO mice upon infection with an Asian strain of the ZIKV (24). Another study investigating the cytokine kinetics upon ZIKV infection of travelers from Thailand, Tahiti, Malaysia and Brazil, found also elevated levels of IL1β and IL10 in the blood (23). A Brazilian patient who developed with encephalomyelitis upon ZIKV infection, showed elevated levels of G-CSF in the cerebrospinal fluid (25). Finally, a recent study that evaluated the humoral response during ZIKV infection in the amniotic fluid of ZIKV-positive pregnant women with neonatal microcephaly reported elevated IP-10, IL6, IL8, MCP-1 and G-CSF (22). Altogether, we show that the Brazilian ZIKV-induced similar inflammatory response by hiPSC-derived MΦ, thus resembling the immune activation observed in patients, including pregnant women infected with ZIKV. Therefore, hiPSC-derived MΦ could be used as potential targets for drug development.
Finally, in order to mimic the immune interactions occurring between macrophages of the central nervous system, microglia, and NPCs in the fetal brain during ZIKV infection, we derived hiPSC-derived MΦ and NPCs from the same donor. We then established a co-culture system to study the multi-cellular interactions that will occur during infection, such as between naïve MΦ and ZIKVBR-infected NPCs. In our co-culture system, we sought to mimic the microglia percentage in vivo which in mouse cortex is around 5% (51–54). In our experiments, we obtained 2% MΦ after 96 h of co-culture both in ZIKVBR or mock-infection conditions, similar to the percentage of microglia found in vivo (51–54). When we added naïve MΦ to infected NPCs, the percentage of dying NPCs decreased significantly. Moreover, by using conditioned media from mock or ZIKVBR-infected MΦ, we explained our results mechanistically by identifying MΦ phagocytosis of apoptotic NPCs, demonstrating that hiPSC-derived MΦ are functional in their ability to recognize virus-infected cells. Furthermore, we showed that conditioned media from ZIKVBR-infected MΦ alone is not sufficient to cause apoptosis of NPCs in our experimental conditions and that physical contact is necessary. The MΦ inflammatory response to ZIKV infection together with the phagocytosis of ZIKV-infected NPCs by MΦ, could contribute to microcephaly in newborns.
Next, we hypothesized that ZIKV-infected MΦ could transmit the virus to the NPCs since the timing of microglial CNS invasion during neurodevelopment corresponds to the timing of ZIKV antigen detection in placental tissues (20,55). Indeed, our data show that ZIKV-infected MΦ transmit ZIKV to NPCs, increasing apoptosis. Importantly, an FDA-approved drug, SOF, was able to block the cell death of NPCs in this context.
Finally, one of the classic pathological findings in GBS, which has been linked to the ZIKV outbreak (5), is inflammatory infiltrates, mainly T cells and macrophages (56). Thus, our in vitro model using hiPSC-derived MΦ could be used to further investigate the inflammatory machinery involved in GBS. Our study sheds light on the immune implications of ZIKV infection and might give insights on the risk of developing ZIKV-associated GBS. Thus, we propose that MΦ could be a therapeutic target to limit the virus spreading amongst the most vulnerable and relevant cell types involved in microcephaly, such as NPCs, and that our co-culture model could be used for drug discovery against ZIKV infection.
Experimental procedures
Cell culture
hiPSC-derived macrophages/microglia (MΦ) differentiation protocol was adapted from (29) and (30). Briefly, iPSCs cell lines were generated as previously described, by reprogramming fibroblast from two healthy donors (57). The iPSC colonies were plated on Matrigel-coated (BD Biosciences) plates and maintained in mTESR media (Stem Cell Technologies). The protocol of myeloid cell lineage consisted of 4 sequential steps. In the first step, primitive streak cells were induced by BMP4 addition, which in step 2, were differentiated into hemangioblast-like hematopoietic precursors [VEGF (80 ng/ml, Peprotech), SCF (100 ng/ml, Gemini) and basic fibroblast growth factor (bFGF), (25 ng/ml, Life Technologies)]. Then, in the third step, the hematopoietic precursors were pushed towards myeloid differentiation [FLT-3 ligand (50 ng/ml, HumanZyme), IL-3 (50 ng/ml, Gemini), SCF (50 ng/ml, Gemini), Thrombopoietin, TPO (5 ng/ml), M-CSF (50 ng/ml)] and finally into the monocytic lineage in step 4 [FLT3-ligand (50 ng/ml), M-CSF (50ng/ml), GM-CSF (25 ng/ml)]. Cells produced in suspension in step 4 were recovered, sorted by using anti-CD14 magnetic microbeads (MACS, Miltenyi), treated with 50ng/ml M-CSF and 50ng/ml IL-34 for a week before being used in different experiments.
Monocyte-derived macrophages were isolated from the peripheral blood of a healthy donor as previously described (34). PBMCs were isolated from the blood by density gradient centrifugation. CD14+ monocytes were then purified by MACS using CD14 microbeads.
For the generation of NPCs, cell were differentiated and maintained as previously described (57–59). Two iPSCs lines obtained from healthy patients maintained in mTSER media were switched to N2 [DMEM/F12 media supplemented with 1xN2 NeuroPlex Serum-Free Supplement (Gemini) with the dual SMAD inhibitors 1 μM of dorsomorphin (Tocris) and 10uM of SB431542 (Stemgent) daily, for 48 h. After two days, colonies were scraped off and cultured under agitation (95rpm) as embryoid bodies for seven days using N2 media with dorsomorphin and SB431542. Media was changed every other day. Embryoid bodies were then plated on Matrigel-coated dishes, and maintained in DMEM/F12 supplemented with 0.5x of N2 supplement, 0.5x Gem21 NeuroPlex Serum-Free Supplement (Gemini), 20 ng/ml basic fibroblast growth factor (bFGF, LifeTechnologies) and 1% penicillin/streptomycin (P/S). After 7 days in culture, rosettes that arose from the plated EBs were manually selected, gently dissociated with StemPro Accutase (LifeTechnologies) and plated onto 10 μg/ml poly-L-ornithine (Sigma)/5 μg/ml Laminin-coated (LifeTechnologies) plates. Neuronal precursor cells (NPCs) were maintained in DMEM/F12 with N2, Gem21, bFGF and P/S. The medium was changed every other day. NPCs were split as soon as confluent, using StemPro Accutase for 5min at 37 °C, centrifuged and replated with NGF with a 1: 3 ratio in poly-L-ornithine/Laminin-coated plates.
Virus culture and amplification
Both ZIKVBR and ZIKVMR766 were obtained and amplified as in (17): lyophilized ZIKVBR, which was isolated from a clinical case in Brazil, was provided by the Evandro Chagas Institute in Belém, Pará and was reconstituted in 0.5 ml of sterile DEPC water. The African-lineage MR-766 (ZIKVMR766), a reference strain isolated in Uganda in 1947, used here as a control, was provided by the Institute Pasteur in Dakar, Senegal. C6/36 Aedes albopictus mosquito cells were used to culture both viruses. The culture medium used for C6/36 cells was Leibovitz‘s L-15 medium supplemented with 10% fetal bovine serum (FBS) (Gemini), 1% non-essential amino acids (Gibco), 1% sodium pyruvate (Gibco), 1% penicillin/streptomycine (Gibco) and 0.05% amphotericin B (Gibco). The cells were kept at 27 °C. When cells reached around 70% of confluency, 50μl of viral sample was added. The cells were subcultured three times before the supernatant was harvested, titrated and used for experimental inoculation.
Virus titration
The virus titration was performed as previously described (17). Briefly, porcine kidney epithelial (PS) cells were used for virus titration in L15 medium with 5% FBS. Serial dilution ranging from 10−1 to 10−11 for each virus stock were added onto 24-well plates, followed by PS cells seeding into each well of a 24-well plate for 3 h at 37 °C. Next, each well was overlaid with complete carboxymethyl cellulose medium (0.6% in L15 supplemented with 3% FBS). Following 5 days of incubation in 37 °C, the plaque assay was performed using 0.05% crystal violet solution. The viral dilution was estimated to determine the amount of infected cells (plaque forming units or PFU.ml−1).
In vitro infection
NPCs were infected with commercially available ZIKVMR766 (MR-766, obtained from ATCC) and ZIKVBR, which was isolated from a febrile case in the state of Paraiba (Brazil-ZKV2015) as previously described (17). Briefly, NPCs were seeded in 24-well, infected at different MOIs for an hour at 37 °C, then were washed and their media was changed. For NPC and MΦ, both of the viruses were used at MOIs of 0.001 for RNA, 0.001, 0.01 and 0.05 for CBA experiments, 0.1 for the first co-culture experiment and MOI of 1 for the second co-culture experiment.
Cytometric bead array (CBA)
The bead-based immunoassay was performed following manufacturer‘s instructions (BD Biosciences). Briefly, iPSCs-derived MΦ or monocyte-derived macrophages were seeded at a density of 5x104 cells/well. Media was conditioned with or without 1μg/ml lipopolysaccharides (LPS, Sigma Aldrich) for 24–30h. 50μl of conditioned media per sample were analysed by CBA. A customized array of cytokines and chemokines were measured in the conditioned media, including IL1β, IL-6, IL-8, IL-10, IL12/23p40, G-CSF, IP10, sCD14, RANTES, Fractalkine, MIP1α, MIP1β and MCP1, using a BD fluorescence activated cell sorting (FACS) Canto II instrument followed by an analysis on FCAP Array software (BD Biosciences).
Phagocytosis assay
For internalization experiments, hiPSCs-derived MΦ were seeded to 2 x104 cells per well. Human neutrophils (PMNs), used as positive controls, were isolated from healthy donors using PolyMorphPrep (Axis-Shield) in accordance with the University of California, San Diego, (UCSD) Human Research Protections Program, and seeded to 2 x104 cells per well.
Group B Streptococcus (GBS) strain COH1 was grown overnight at 37 °C in Todd-Hewitt broth (Difco), diluted in PBS to MOI = 10, and spun onto cell 167 g for 3 min. After 90 min, the media was supplemented with 100µg/ml gentamicin to kill extracellular bacteria. After 30 min, the cells were washed 1x in PBS and lysed with 0.05% Triton-X100 (Sigma) for dilution plating and CFU enumeration on Todd-Hewitt agar plates (Difco).
Co-culture assay
For the first co-culture experiment, 2 × 104 NPCs per well of a 96-well plate were plated 24 h prior to the ZIKV infection. After ZIKV infection for an hour at 37 °C, cells were washed and fresh media was added. Two hours later, 2 × 104 naïve hiPSCs-derived MΦ per well were plated on top of the NPCs. Cells were fixed 96 h post-infection for further analysis with 4% paraformaldehyde, for 20 min at room temperature. For the second co-culture experiment, 2 × 104 NPCs were plated. MΦ were infected at an MOI of 1 for an hour at 37 °C and the media was changed. The next day, MΦ were lifted off by adding accutase for 5 min, washed with PBS and centrifuged at 300g for 5 min. The cells were counted and 2 × 104 cells/well of MΦ were added onto NPCs with 20 μM of SOF (Acme Bioscience AB3793) or vehicle (DMSO). The dose of 20μM of SOF used on NPCs was optimized in our laboratory (Mesci et al. manuscript in revision). The cells were fixed after 96 h of incubation. For mock controls, the same volume of supernatant was added to each experiment.
Imaging analyses
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Next, samples were permeabilized in 1x-PBS containing 0.1% (v/v) Triton X-100 for 10 min. Cells were next incubated with blocking solution [1% fetal bovine serum, (Life Technologies) in 1xPBS]. After 1 h, the primary antibody was added (diluted in blocking solution) and samples were incubated overnight at 4 °C. Slides were then washed two times with 1x-PBS, and incubated with the secondary antibody for 30 min at 4 °C. Secondary antibodies (all conjugated to Alexa Fluor 488, 555 and 647) were purchased from Life Technologies and used at a 1: 1000 dilution. After the 30 min incubation, samples were washed twice (1x-PBS), incubated for 5 min with and fluorescent nuclear DAPI stain, and mounted with Prolong gold anti-fade reagent (Life Technologies). Samples were imaged using an Axio Observer Z1 Microscope with ApoTome (Zeiss). Captured images were analysed with Zen software from Zeiss. Antibodies and dilutions used: Monoclonal Mouse Anti-Human CD68, (1: 500, Dako); Polyclonal Rabbit Anti-Iba-1, (1: 500, Wako). For TUNEL analysis, NPCs were plated, infected after 24 h, and fixed 96 h p.i with 4% paraformaldehyde for 20 min. Samples were permeabilized with 0.25% Triton X-100 for 15 min, and stained for fragmented DNA using TUNEL (Click-iT TUNEL 647 assay kit from Life Technologies). The cells were then blocked 1% bovine serum albumin for 60 min. Cells were incubated in primary antibodies: Monoclonal Mouse Nestin, Polyclonal anti-Chicken or Polyclonal Rabbit Nestin (1: 500, Abcam); Polyclonal Rabbit Anti-Iba-1, (1: 500, Wako) and monoclonal mouse NS1 (1: 250, Millipore) overnight at 4 °C and stained with secondary antibodies conjugated to Alexa Fluor and DAPI the following day prior to mounting. Images were blindly collected using an Axio Observer Z1 Microscope with ApoTome (Zeiss) and blindly analysed with ImageJ software.
Fluorescence-activated cell sorting analyses
Using previously mentioned protocol to generate monocytes/macrophages (29), we have collected the cells in the supernatant at the step 4 and labeled them with the appropriate antibodies (CD45 BV786, CD14 PE, CD11b APC, CD68 PE-Cy7, HLA-DRDPDQ FITC all from BD Biosciences). Briefly, cells were harvested and counted, then washed twice with the stain buffer, containing PBS and 1% heat inactivated FBS, at 300g for 5 min. The antibodies were added in the cell mixture and incubated in RT in the dark for 20–30 min. The cells were then washed twice with the stain buffer. The cells were resuspended in 500 μl stain buffer and the viability dye, 7-AAD was added. The data were acquired in BD LSRFORTESSA instrument and the plots were generated using the software FlowJo.
RNA extraction and expression analyses by qPCR
Total RNA was extracted from cells using RNeasy Micro and Mini kit for iPSC-derived MΦ and NPCs respectively (Qiagen), following manufacturer’s instructions. Next, 900 ng of total RNA was treated RNase-free DNaseI (Qiagen), and was reverse transcribed using QuantiTect Reverse Transcription Kit (Qiagen). Approximately, 15 ng of cDNAs was used per reaction and PCRs were carried out in a final volume of 10 μl. Triplicate samples were analysed in a CFX384 Touch Real-Time PCR Detection System (Bio-rad) using iQ™ SYBR® Green Supermix (Bio-rad). GADPH was used as internal normalization control. For the complete list of primers, please refer to Table 1. The run method was as follows: 3min at 95 °C, 40 cycles of 10s at 95 °C followed by 30s at 58 °C, and a melting curve was performed to confirm the identity of the amplified product.
Table 1.
List of primers
| AXL-F | GTTTGGAGCTGTGATGGAAGGC |
| AXL-R | CGCTTCACTCAGGAAATCCTCC |
| CCR5-F | CCTGCTGCTTTGCCTACATTGC |
| CCR5-R | ACACACTTGGCGGTTCTTTCGG |
| CD11b-F | GGAACGCCATTGTCTGCTTTCG |
| CD11b-R | ATGCTGAGGTCATCCTGGCAGA |
| CD206-F | AGCCAACACCAGCTCCTCAAGA |
| CD206-R | CAAAACGCTCGCGCATTGTCCA |
| CD209-F | GCAGTCTTCCAGAAGTAACCGC |
| CD209-R | GCTCTCCTCTGTTCCAATACTGC |
| CD68-F | CGAGCATCATTCTTTCACCAGCT |
| CD68-R | ATGAGAGGCAGCAAGATGGACC |
| CLEC5A-F | TTGTCAACACGCCAGAGAAACTG |
| CLEC5A-R | CAACGCCACCTTTTCTCTTCACG |
| IBA1-F | CCCTCCAAACTGGAAGGCTTCA |
| IBA1-R | CTTTAGCTCTAGGTGAGTCTTGG |
| IGF1-F | CTCTTCAGTTCGTGTGTGGAGAC |
| IGF1-R | CAGCCTCCTTAGATCACAGCTC |
| IL1B-F | CCACAGACCTTCCAGGAGAATG |
| IL1B-R | GTGCAGTTCAGTGATCGTACAGG |
| IL6-F | AGACAGCCACTCACCTCTTCAG |
| IL6-R | TTCTGCCAGTGCCTCTTTGCTG |
| MCP1-F | AGAATCACCAGCAGCAAGTGTCC |
| MCP1-R | TCCTGAACCCACTTCTGCTTGG |
| MERTK-F | CAGGAAGATGGGACCTCTCTGA |
| MERTK-R | GGCTGAAGTCTTTCATGCACGC |
| NOX2-F | CTCTGAACTTGGAGACAGGCAAA |
| NOX2-R | CACAGCGTGATGACAACTCCAG |
| TIM-F | CTTCACCTCAGCCAGCAGAAAC |
| TIM-R | GCCATCTGAAGACTCTGTCACG |
| TLR3-F | GCGCTAAAAAGTGAAGAACTGGAT |
| TLR3-R | GCTGGACATTGTTCAGAAAGAGG |
| TLR4-F | CCCTGAGGCATTTAGGCAGCTA |
| TLR4-R | AGGTAGAGAGGTGGCTTAGGCT |
| TLR7-F | CTTTGGACCTCAGCCACAACCA |
| TLR7-R | CGCAACTGGAAGGCATCTTGTAG |
| TYRO3-F | GCAAGCCTTTGACAGTGTCATGG |
| TYRO3-R | GTTCATCGCTGATGCCCAAGCT |
| NES-F | CCATAGAGGGCAAAGTGGTAA |
| NES-R | TTCTTCCCATATTTCCTGCTGC |
| PAX-F | TGTCCAACGGATGTGTGAGTA |
| PAX-R | CAGTCTCGTAATACCTGCCCA |
| GAPDH-F | TGCACCACCAACTGCTTAGC |
| GAPDH-R | GGCATGGACTGTGGTCATGAG |
Statistical analysis
One-way ANOVA or two-way ANOVA (if two or more variables) tests followed by a Tukey or Sidak multiple comparison tests were used to compare groups and Student’s t-test to compare means of two groups.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We would like to thank Dr. Edison Durigon and his group for the ZIKVBR aliquots, the UCSD stem cell core and Dr. Christopher Alfonso from BD Biosciences. We would like to thank Spencer M. Moore for critical reading of the manuscript.
Conflict of Interest statement. None declared.
Funding
This work was supported by grants from the California Institute for Regenerative Medicine (DISC2–09649), the National Institutes of Health through the U19MH107367, Zika Network FAPESP projects 2011/18703–2 and 2014/17766–9, the NGO “the tooth fairy project”, a fellowship from the A.P. Giannini Foundation to C.N.L., and an NARSAD Independent Investigator Grant to A.R.M. Dr. Mesci has an International Rett syndrome foundation (IRSF) mentored training fellowship. Dr. Macia has a NARSAD Young Investigator grant.
References
- 1. Dick G.W.A., kitchen S.F., haddow A.J. (1952) Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg., 46, 509–520. [DOI] [PubMed] [Google Scholar]
- 2. Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J., Stanfield S.M., Duffy M.R. (2008) Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis., 14, 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campos G.S., Bandeira A.C., Sardi S.I. (2015) Zika virus outbreak, Bahia, Brazil. Emerg. Infect. Dis., 21, 1885–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V.. et al. (2016) Zika virus associated with microcephaly. N. Engl. J. Med., 374, 951–958. [DOI] [PubMed] [Google Scholar]
- 5. Beckham J.D., Pastula D.M., Massey A., Tyler K.L. (2016) Zika virus as an emerging global pathogen: neurological complications of Zika virus. JAMA Neurol., 10.1001/jamaneurol.2016.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calvet G., Aguiar R.S., Melo A.S.O., Sampaio S.A., de Filippis I., Fabri A., Araujo E.S.M., de Sequeira P.C., de Mendonça M.C.L., de Oliveira L.. et al. (2016) Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet. Infect. Dis., 16, 653–660. [DOI] [PubMed] [Google Scholar]
- 7. Martines R.B., Bhatnagar J., Keating M.K., Silva-Flannery L., Muehlenbachs A., Gary J., Goldsmith C., Hale G., Ritter J., Rollin D.. et al. (2016) Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses—Brazil, 2015. MMWR. Morb. Mortal. Wkly. Rep., 65, 1–2. [DOI] [PubMed] [Google Scholar]
- 8. Sarno M., Sacramento G.A., Khouri R., do Rosário M.S., Costa F., Archanjo G., Santos L.A., Nery N., Vasilakis N., Ko A.I.. et al. (2016) Zika virus infection and stillbirths: a case of hydrops fetalis, hydranencephaly and fetal demise. PLoS Negl. Trop. Dis., 10, e0004517.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metsky H.C., Matranga C.B., Wohl S., Schaffner S.F., Freije C.A., Winnicki S.M., West K., Qu J., Baniecki M.L., Gladden-Young A.. et al. (2017) Zika virus evolution and spread in the Americas. Nature, 546, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faria N.R., Quick J., Claro I.M., Thézé J., de Jesus J.G., Giovanetti M., Kraemer M.U.G., Hill S.C., Black A., da Costa A.C.. et al. (2017) Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature, 546, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grubaugh N.D., Ladner J.T., Kraemer M.U.G., Dudas G., Tan A.L., Gangavarapu K., Wiley M.R., White S., Thézé J., Magnani D.M.. et al. (2017) Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature, 546, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Liu J., Du S., Shan C., Nie K., Zhang R., Li X.-F., Zhang R., Wang T., Qin C.-F.. et al. (2017) Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature, 545, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delatorre E., Mir D., Bello G. (2017) Tracing the origin of the NS1 A188V substitution responsible for recent enhancement of Zika virus Asian genotype infectivity. Mem. Inst. Oswaldo Cruz., 10.1590/0074-02760170299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis B.S., Duggal N.K., Chang G.-J.J., Ritter J.M., McDonald E.M., Romo H., Brault A.C., Guirakhoo F. (2017) Differential neurovirulence of African and Asian genotype Zika virus isolates in outbred immunocompetent mice. Am. J. Trop. Med. Hyg., 10.4269/ajtmh.17-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anfasa F., Siegers J.Y., van der Kroeg M., Mumtaz N., Stalin Raj V., de Vrij F.M.S., Widagdo W., Gabriel G., Salinas S., Simonin Y.. et al. (2017) Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere, 2, e00292-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simonin Y., Loustalot F., Desmetz C., Foulongne V., Constant O., Fournier-Wirth C., Leon F., Molès J.-P., Goubaud A., Lemaitre J.-M.. et al. (2016) Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine, 12, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L.M., Guimarães K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S.. et al. (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature, 534, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowakowski T.J., Pollen A.A., Di Lullo E., Sandoval-Espinosa C., Bershteyn M., Kriegstein A.R. (2016) Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell, 18, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quicke K.M., Bowen J.R., Johnson E.L., McDonald C.E., Ma H., O’Neal J.T., Rajakumar A., Wrammert J., Rimawi B.H., Pulendran B.. et al. (2016) Zika virus infects human placental macrophages. Cell Host Microbe, 20, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R.. et al. (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science, 330, 841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lum F.-M., Low D.K.S., Fan Y., Tan J.J.L., Lee B., Chan J.K.Y., Rénia L., Ginhoux F., Ng L.F.P. (2017) Zika virus infects human fetal brain microglia and induces inflammation. Clin. Infect. Dis., 64, 914–920. [DOI] [PubMed] [Google Scholar]
- 22. Ornelas A.M.M., Pezzuto P., Silveira P.P., Melo F.O., Ferreira T.A., Oliveira-Szejnfeld P.S., Leal J.I., Amorim M.M.R., Hamilton S., Rawlinson W.D.. et al. (2017) Immune activation in amniotic fluid from Zika virus-associated microcephaly. Ann. Neurol., 81, 152–156. [DOI] [PubMed] [Google Scholar]
- 23. Tappe D., Pérez-Girón J.V., Zammarchi L., Rissland J., Ferreira D.F., Jaenisch T., Gómez-Medina S., Günther S., Bartoloni A., Muñoz-Fontela C.. et al. (2016) Cytokine kinetics of Zika virus-infected patients from acute to reconvalescent phase. Med. Microbiol. Immunol., 205, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manangeeswaran M., Ireland D.D.C., Verthelyi D. (2016) Zika (PRVABC59) infection is associated with T cell infiltration and neurodegeneration in CNS of immunocompetent neonatal C57Bl/6 mice. PLoS Pathog., 12, e1006004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galliez R.M., Spitz M., Rafful P.P., Cagy M., Escosteguy C., Germano C.S.B., Sasse E., Gonçalves A.L., Silveira P.P., Pezzuto P.. et al. (2016) Zika virus causing encephalomyelitis associated with immunoactivation. Open Forum Infect. Dis., 3, ofw203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sacramento C.Q., de Melo G.R., de Freitas C.S., Rocha N., Hoelz L.V.B., Miranda M., Fintelman-Rodrigues N., Marttorelli A., Ferreira A.C., Barbosa-Lima G.. et al. (2017) The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci. Rep., 7, 40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bullard-Feibelman K.M., Govero J., Zhu Z., Salazar V., Veselinovic M., Diamond M.S., Geiss B.J. (2017) The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res., 137, 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Onorati M., Li Z., Liu F., Sousa A.M.M., Nakagawa N., Li M., Dell’Anno M.T., Gulden F.O., Pochareddy S., Tebbenkamp A.T.N.. et al. (2016) Zika virus disrupts phospho-tbk1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep., 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yanagimachi M.D., Niwa A., Tanaka T., Honda-Ozaki F., Nishimoto S., Murata Y., Yasumi T., Ito J., Tomida S., Oshima K.. et al. (2013) Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS One, 8, e59243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Douvaras P., Sun B., Wang M., Kruglikov I., Lallos G., Zimmer M., Terrenoire C., Zhang B., Gandy S., Schadt E.. et al. (2017) Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports, 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grozdanov V., Bliederhaeuser C., Ruf W.P., Roth V., Fundel-Clemens K., Zondler L., Brenner D., Martin-Villalba A., Hengerer B., Kassubek J.. et al. (2014) Inflammatory dysregulation of blood monocytes in Parkinson‘s disease patients. Acta Neuropathol., 128, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Underhill D.M. (2003) Macrophage recognition of zymosan particles. J. Endotoxin Res., 9, 176–180. [DOI] [PubMed] [Google Scholar]
- 33. Speert D.P., Silverstein S.C. (1985) Phagocytosis of unopsonized zymosan by human monocyte-derived macrophages: maturation and inhibition by mannan. J. Leukoc. Biol., 38, 655–658. [DOI] [PubMed] [Google Scholar]
- 34. Ohradanova-Repic A., Machacek C., Fischer M.B., Stockinger H. (2016) Differentiation of human monocytes and derived subsets of macrophages and dendritic cells by the HLDA10 monoclonal antibody panel. Clin. Transl. Immunol., 5, e55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dang J., Tiwari S.K., Lichinchi G., Qin Y., Patil V.S., Eroshkin A.M., Rana T.M. (2016) Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell, 19, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcez P.P., Nascimento J.M., de Vasconcelos J.M., Madeiro da Costa R., Delvecchio R., Trindade P., Loiola E.C., Higa L.M., Cassoli J.S., Vitória G.. et al. (2017) Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep., 7, 40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenberger C.M., Podyminogin R.L., Askovich P.S., Navarro G., Kaiser S.M., Sanders C.J., McClaren J.L., Tam V.C., Dash P., Noonan J.G.. et al. (2014) Characterization of innate responses to influenza virus infection in a novel lung type I epithelial cell model. J. Gen. Virol., 95, 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biacchesi S., Skiadopoulos M.H., Yang L., Lamirande E.W., Tran K.C., Murphy B.R., Collins P.L., Buchholz U.J. (2004) Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J. Virol., 78, 12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan M.C., Battini L., Tuyama A.C., Macip S., Melendi G.A., Horga M.A., Luca Gusella G. (2007) Characterization of human metapneumovirus infection of myeloid dendritic cells. Virology, 357, 1–9. 10.1016/j.virol.2006.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bayless N.L., Greenberg R.S., Swigut T., Wysocka J., Blish C.A. (2016) Zika virus infection induces cranial neural crest cells to produce cytokines at levels detrimental for neurogenesis. Cell Host Microbe, 20, 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M.. et al. (2016) Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell, 18, 587–590. 10.1016/j.stem.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen S., Lin Y., Huang M., Wu M., Cheng S., Lei H., Lee C., Chiou T., Wong C., Hsieh S. (2008) CLEC5A is critical for dengue-virus-induced lethal disease. 453, 672–676. [DOI] [PubMed] [Google Scholar]
- 43. Garcez P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science, 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 44. Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C.. et al. (2016) Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell, 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wells M.F., Salick M.R., Wiskow O., Ho D.J., Worringer K.A., Ihry R.J., Kommineni S., Bilican B., Klim J.R., Hill E.J.. et al. (2016) Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from Zika virus infection. Cell Stem Cell, 19, 703–708. 10.1016/j.stem.2016.11.011 [DOI] [PubMed] [Google Scholar]
- 46. Miller J.L., deWet B.J.M., Martinez-Pomares L., Radcliffe C.M., Dwek R. a., Rudd P.M., Gordon S. (2008) The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog, 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meertens L., Labeau A., Dejarnac O., Cipriani S., Sinigaglia L., Bonnet-Madin L., Le Charpentier T., Hafirassou M.L., Zamborlini A., Cao-Lormeau V.-M.. et al. (2017) Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep., 18, 324–333. [DOI] [PubMed] [Google Scholar]
- 48. Chavali P.L., Stojic L., Meredith L.W., Joseph N., Nahorski M.S., Sanford T.J., Sweeney T.R., Krishna B.A., Hosmillo M., Firth A.E.. et al. (2017) Neurodevelopmental protein Musashi 1 interacts with the Zika genome and promotes viral replication. Science (80-.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Foo S.-S., Chen W., Chan Y., Bowman J.W., Chang L.-C., Choi Y., Yoo J.S., Ge J., Cheng G., Bonnin A.. et al. (2017) Asian Zika virus strains target CD14(+) blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat. Microbiol., 10.1038/s41564-017-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haddow A.D., Schuh A.J., Yasuda C.Y., Kasper M.R., Heang V., Huy R., Guzman H., Tesh R.B., Weaver S.C., Olson K.E. (2012) Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis., 6, e1477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mandrekar-Colucci S., Landreth G.E. (2010) Microglia and inflammation in Alzheimer‘s disease. CNS Neurol. Disord. Drug Targets, 9, 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rezaie P., Patel K., Male D.K. (1999) Microglia in the human fetal spinal cord–patterns of distribution, morphology and phenotype. Brain Res. Dev. Brain Res., 115, 71–81. [DOI] [PubMed] [Google Scholar]
- 53. Aloisi F. (2001) Immune function of microglia. Glia, 36, 165–179. 10.1002/glia.1106 [DOI] [PubMed] [Google Scholar]
- 54. Lawson L.J., Perry V.H., Dri P., Gordon S. (1990) Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience, 39, 151–170. 10.1016/0306-4522(90)90229-W [DOI] [PubMed] [Google Scholar]
- 55. Martines R.B., Bhatnagar J., de Oliveira Ramos A.M., Davi H.P.F., Iglezias S. DAndretta., Kanamura C.T., Keating M.K., Hale G., Silva-Flannery L., Muehlenbachs A.. et al. (2016) Pathology of congenital Zika syndrome in Brazil: a case series. Lancet (London, England), 388, 898–904. [DOI] [PubMed] [Google Scholar]
- 56. Yuki N., Hartung H.-P. (2012) Guillain-Barré syndrome. N. Engl. J. Med., 366, 2294–2304. [DOI] [PubMed] [Google Scholar]
- 57. Marchetto M.C.N., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. (2010) A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell, 143, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nageshappa S., Carromeu C., Trujillo C.A., Mesci P., Espuny-Camacho I., Pasciuto E., Vanderhaeghen P., Verfaillie C.M., Raitano S., Kumar A.. et al. (2015) Altered neuronal network and rescue in a human MECP2 duplication model. Mol. Psychiatry, 21, 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Griesi-Oliveira K., Acab A., Gupta A.R., Sunaga D.Y., Chailangkarn T., Nicol X., Nunez Y., Walker M.F., Murdoch J.D., Sanders S.J.. et al. (2014) Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Mol. Psychiatry, 20, 1350–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.