Abstract
Li-Fraumeni Syndrome (LFS) results from heterozygous germline mutations of TP53, encoding a key transcriptional factor activated in response to DNA damage. We have recently shown, from a large LFS series, that dominant-negative missense mutations are the most clinically severe and, thanks to a new p53 functional assay in lymphocytes, that they alter the p53 transcriptional response to DNA damage more drastically than null mutations. In this study, we first confirmed this observation by performing the p53 functional assay in lymphocytes from 56 TP53 mutation carriers harbouring 35 distinct alterations. Then, to compare the impact of the different types of germline TP53 mutations on DNA binding, we performed chromatin immunoprecipitation-sequencing (ChIP-Seq) in lymphocytes exposed to doxorubicin. ChIP-Seq performed in wild-type TP53 control lymphocytes accurately mapped 1287 p53-binding sites. New p53-binding sites were validated using a functional assay in yeast. ChIP-Seq analysis of LFS lymphocytes carrying TP53 null mutations (p.P152Rfs*18 or complete deletion) or the low penetrant ‘Brazilian’ p.R337H mutation revealed a moderate decrease of p53-binding sites (949, 580 and 620, respectively) and of ChIP-Seq peak depths. In contrast, analysis of LFS lymphocytes with TP53 dominant-negative missense mutations p.R273H or p.R248W revealed only 310 and 143 p53-binding sites, respectively, and the depths of the corresponding peaks were drastically reduced. Altogether, our results show that TP53 mutation carriers exhibit a constitutive defect of the transcriptional response to DNA damage and that the clinical severity of TP53 dominant-negative missense mutations is explained by a massive and global alteration of p53 DNA binding.
Introduction
Li-Fraumeni syndrome (LFS; OMIM#151623) represents a paradigm in genetic predisposition to cancer considering the central role of p53 in the response to DNA damage and because our perception of the syndrome, as defined by the presence of germline TP53 [MIM*191170, NM_000546.5] mutations, has considerably evolved since its original definition in 1969 by Frederick Li and Joseph Fraumeni (1–8). Indeed, a remarkable feature of the LFS is the heterogeneity of its clinical presentation, in terms of tumour type and age of tumour onset. In clinical practice, the presence of a germline TP53 mutation should be considered in 4 different situations: (i) a proband with a LFS spectrum tumour (premenopausal breast cancer, soft tissue sarcoma, brain tumour, adrenocortical carcinoma (ACC)) before age 46 and at least one first or second-degree relative with a LFS tumour (except breast cancer, if the proband has breast cancer) before the age of 56 or with multiple tumours; (ii) a proband with multiple malignancies (except recurring breast cancer), two of which belonging to the LFS spectrum, the first being developed before 46 years; (iii) patients with rare early-onset tumours such as ACC, choroid plexus carcinoma (CPC) or embryonal anaplastic rhabdomyosarcoma, irrespective of the family history; and (iv) breast cancer before age 31 (1).
If germline TP53 mutations are identified in familial aggregations of childhood and adult tumours, they can also be detected in patients and families who have developed only adult cancers and in patients without a familial history of cancer (1,9). With the exponential use, in cancer patients, of TP53 tests performed with gene panels, it is likely that the percentage of germline TP53 mutations detected in such patients or families will increase. This provides clinical evidence that germline TP53 mutations display a heterogeneous penetrance. The diversity of the clinical presentations associated with germline TP53 mutations suggests that it should be appropriate to include Li-Fraumeni syndrome within an expanded category designated ‘TP53-related inherited cancers’ characterized by a broader tumour spectrum and age of tumour onset.
All types of TP53 alterations can be observed at the germline and at the somatic level. Nevertheless the TP53 mutation spectrum is characterized by the predominance of missense mutations (above 65%), most of the mutations reported so far in LFS corresponding to dominant-negative mutations (1,10). LFS animal models and clinical data support the existence of genotype-phenotype correlations in LFS: (i) mouse models of LFS have shown the phenotypic severity of dominant-negative mutations, as compared with null mutations (11–14); (ii) from a large series of 415 TP53 mutation carriers, we have recently reported that the mean age of tumour onset was statistically lower in carriers harbouring dominant-negative missense mutations than in carriers with loss of function mutations and that dominant-negative missense mutations represent the predominant germline alterations in carriers who developed childhood cancers, except ACC. In contrast, null mutations such as nonsense mutations, frameshift mutations or genomic rearrangements are predominantly detected in pedigrees characterized by cancers occurring in adulthood (1).
Thanks to the development of a new p53 functional assay in lymphocytes, based on the measurement of p53 transcriptional response to DNA damage, we provided a first molecular explanation of the dominant-negative missense mutation severity (15). Indeed, analysis of the p53 transcriptional response after doxorubicin exposure of lymphocytes derived from three TP53 wild-type control subjects, 3 carriers of dominant-negative missense mutations and 3 carriers of null mutations, revealed that in lymphocytes with germline dominant-negative missense mutations, the number of induced genes and the level of p53 target gene induction were drastically reduced, as compared to controls and LFS lymphocytes with null mutations. We subsequently developed a simpler version of the assay, based on the transcriptional induction of a limited number of p53 target genes in lymphocytes exposed to DNA damaging agents. This assay, performed in wild-type TP53 lymphocytes, can be used to determine and quantify the genotoxic effect of chemical or physical agents, p53 being a universal sensor of genotoxic stress (16). It can alternatively be used to determine the functionality of p53 protein in LFS patients harbouring germline TP53 mutations.
In this study, to deeply characterize the biological impact of the different classes of germline TP53 mutations, we first analysed, using a modified version of this functional assay, the effect of 35 distinct germline alterations on p53 transcriptional response to DNA damage and then characterized the consequences of germline TP53 mutations on DNA binding, using chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-Seq).
Results
p53 transcriptional response to DNA damage in lymphocytes constitutes an endophenotype of germline TP53 mutation severity
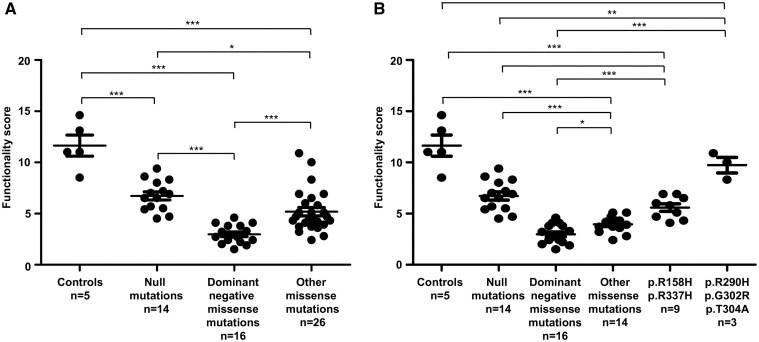
To confirm the results obtained in our previous study performed on a limited number of samples and showing that dominant-negative missense mutations had a more drastic impact on p53 transcriptional response compared to null mutations (15), we selected 35 EBV-immortalized lymphocytes from 5 control subjects with wild-type TP53 genotype, 14 LFS patients with TP53 null mutations and 16 LFS patients with missense mutations previously classified as dominant-negative mutations according to the IARC (International Agency for Research on Cancer) TP53 database (http://p53.iarc.fr/; date last accessed March 22, 2017) (17) (Table 1). We analysed the p53 transcriptional response to DNA damage in these EBV-immortalized lymphocytes, using our simplified p53 functional assay, based on the transcriptional induction of six p53 target genes after exposure of lymphocytes to doxorubicin, a genotoxic agent inducing DNA double-strand breaks, and on the subsequent comparison of p53 target gene expression levels between doxorubicin-treated and non-treated cells (Supplementary Material, Fig. S1). We checked that p53 was efficiently activated following exposure to doxorubicin, by monitoring p53 protein stabilization (Supplementary Material, Fig. S2). In this assay, the p53 functional activity is expressed as an arbitrary functionality score corresponding to the average of the six p53 target gene induction levels. As shown in Figure 1, in control wild-type TP53 lymphocytes, we obtained a mean p53 functionality score of 11.6 ± 1.04, indicating a strong induction of p53 transcriptional activity in response to DNA damage. In LFS lymphocytes with null mutations, we observed a 50% decrease of the mean score (6.7 ± 0.4) (Fig. 1A) illustrating the p53 haplodeficiency (Table 1). In LFS lymphocytes with dominant-negative missense mutations, we detected a drastic reduction of the p53 transcriptional activity (mean score =3.0 ± 0.2) (Fig. 1A). The mean scores were statistically different between control wild-type TP53 lymphocytes and those with null mutations (P-value < 0.0001, Student t-test) or with dominant-negative missense mutations (P-value< 0.0001, Student t-test) (Fig. 1A). As shown in Table 1, we obtained similar results for EBV-immortalized lymphocytes derived from different individuals harbouring the same germline TP53 mutation, indicating the reproducibility of the assay. These results therefore confirm our previous results showing that germline TP53 mutations detected in LFS patients result in a constitutive reduction of p53 transcriptional activity and that dominant-negative missense mutations have a more severe impact on this transcriptional activity than null mutations.
Table 1.
Functionality scores in control subjects and TP53 mutation carriers
| Individual (gendera, ageb) | Family | Phenotypec (age d) | Germline TP53 mutatione | p53 mRNA expressionf | Scoreg |
|---|---|---|---|---|---|
| Controls | |||||
| Control 1 (M, 34) | F1 | No tumour | None | 79 | 11 |
| Control 2 (F, 37) | F2 | No tumour | None | 100 | 13.1 |
| Control 3 (F, 30) | F3 | No tumour | None | 100 | 14.6 |
| Control 4 (M, 29) | F4 | No tumour | None | 71 | 11 |
| Control 5 (M, 28) | F5 | No tumour | None | 87 | 8.5 |
| Missense mutations with dominant negative effect (DNE) h | |||||
| Patient 1 (F, 29) | F155 | Bilat. ACC (1 + 2), SC (28) | c.524G>A, p.R175H | 93 | 2.9 |
| Patient 2 (F, 2) | F65 | anRMS (1) | c.524G>A, p.R175H | 86 | 4.1 |
| Patient 3 (F, 14) | F24 | GBM (14), OS (16) | c.535C>T, p.H179Y | 78 | 3.8 |
| Patient 4 (M, 1) | F225 | CPC (0.3) | c.638G>A, p.R213Q | 98 | 4.1 |
| Patient 5 (F, 23) | F185 | HD (13), BC (21), BS (21) | c.659A>G, p.Y220C | 91 | 3.3 |
| Patient 6 (F, 23) | F161 | BC (22) | c.733G>A, p.G245S | 93 | 2.4 |
| Patient 7 (M, 52) | F241 | No tumour (52) | c.733G>A, p.G245S | 85 | 3.8 |
| Patient 8 (F, 26) | F254 | Bilat. BC (25) | c.736A>G, p.M246V | 90 | 2.7 |
| Patient 9 (F, 29) | F267 | OS (16), BC (28) | c.743G>A, p.R248Q | 76 | 2.2 |
| Patient 10 (F, 37) | F212 | BC (25) | c.742C>T, p.R248W | 70 | 1.5 |
| Patient 11 (F, 35) | F141 | STS (19), Bilat. BC (29 + 34) | c.742C>T, p.R248W | 73 | 3.2 |
| Patient 12 (M, 22) | F141 | Bilat. tes. GCT (17 + 19), BCC (19) | c.818G>A, p.R273H | 104 | 1.9 |
| Patient 13 (F, 3) | F209 | STS (2) | c.818G>A, p.R273H | 88 | 2.4 |
| Patient 14 (F, 9) | F145 | No tumour (10) | c.844C>T, p.R282W | 104 | 2 |
| Patient 15 (F, 35) | F79 | No tumour (35) | c.844C>T, p.R282W | 116 | 2.7 |
| Patient 16 (F, 9) | F79 | No tumour (9) | c.844C>T, p.R282W | 76 | 4.6 |
| Null mutations | |||||
| Patient 17 (F, 20) | F26 | No tumour (20) | c.(?_-202)_(*1207_?) del, p.0(comp del)i | 57 | 7.2 |
| Patient 18 (F, 66) | F26 | BC (46), STS (49) | c.(?_-202)_(*1207_?) del, p.0(comp del)i | 42 | 6.5 |
| Patient 19 (M, 16) | F35 | No tumour (16) | c.(?_-202)_(-29 + 1_-28-1)del, p.0 (prom-ex1 del)j | 53 | 5.4 |
| Patient 20 (M, 60) | F167 | STS (59) | c.(?_-202)_(-29 + 1_-28-1)del, p.0 (prom-ex1 del)j | 60 | 8 |
| Patient 21 (M, 28) | F56 | No tumour (28) | c.216dup, p.V73Rfs*76 | 51 | 4.5 |
| Patient 22 (F, 28) | F203 | BC (27) | c.216dup, p.V73Rfs*76 | 44 | 6.5 |
| Patient 23 (F, 31) | F208 | BC (28) | c.323_329dup, p.L111Ffs*40 | 59 | 6.9 |
| Patient 24 (F, 29) | F211 | Bilat. BC (26 + 29) | c.455del, p.P152Rfs*18 | 63 | 4.7 |
| Patient 25 (M, 12) | F223 | OS (11) | c.491_494del, p.K164Sfs*5 | 50 | 8.6 |
| Patient 26 (M, 40) | F223 | Tes. GCT (26), BCC (40) | c.491_494del, p.K164Sfs*5 | 68 | 8.3 |
| Patient 27 (F, 27) | F186 | Bilat. BC (25) | c.632_641del, p.T211Ifs*33 | 50 | 5.7 |
| Patient 28 (F, 29) | F146 | No tumour (29) | c.690del, p.T231Pfs*16 | 52 | 9.4 |
| Patient 29 (F, 12) | F162 | ACC (11) | c.820del, p.V274Ffs*71 | 53 | 7 |
| Patient 30 (F, 35) | F28 | No tumour (35) | c.673-2A>G, p.? | 42 | 5.5 |
| Missense mutations with no detectable DNE or whose dominant activity had not been assessed | |||||
| Patient 31 (M, 3) | F324 | NB (2) | c.31G>C, p.E11Q | 120 | 3 |
| Patient 32 (F, 58) | F150 | No tumour (58) | c.323G>A, p.G108D | 81 | 4.2 |
| Patient 33 (F, 2) | F204 | ACC (0.9) | c.374C>T, p.T125M | 92 | 5.1 |
| Patient 34 (F, 16) | F149 | ACC (0.5) | c.473G>A, p.R158Hk | 83 | 4.1 |
| Patient 35 (M, 3) | F188 | CPC (2) | c.473G>A, p.R158Hk | 93 | 4.3 |
| Patient 36 (F, 18) | F41 | No tumour (18) | c.473G>A, p.R158Hk | 89 | 4.7 |
| Patient 37 (M, 18) | F41 | No tumour (18) | c.473G>A, p.R158Hk | 72 | 5.1 |
| Patient 38 (F, 18) | F166 | No tumour (18) | c.473G>A, p.R158Hk | 83 | 6.9 |
| Patient 39 (F, 24) | F2 | OS (18), bilat. BC (27 + 29), LC (45) | c.523C>G, p.R175Gk | 101 | 3.2 |
| Patient 40 (F, 33) | F206 | STS (24), BC (26) | c.577C>G, p.H193D | 91 | 3.7 |
| Patient 41 (M, 57) | F181 | STS (49), LC (54), MM (55) | c.578A>C, p.H193P | 109 | 3.7 |
| Patient 42 (M, 30) | F144 | ARMS (3), STS (17) | c.646G>A, p.V216M | 94 | 4.3 |
| Patient 43 (M, 34) | F179 | No tumour (37) | c.646G>A, p.V216M | 82 | 4.3 |
| Patient 44 (F, 37) | F238 | BC (34) | c.685T>C, p.C229R | 82 | 4.3 |
| Patient 45 (F, 36) | F198 | No tumour (36) | c.761T>C, p.I254T | 72 | 2.4 |
| Patient 46 (F, 29) | F198 | No tumour (29) | c.761T>C, p.I254T | 72 | 2.8 |
| Patient 47 (M, 1) | F198 | ACC (0.9) | c.761T>C, p.I254T | 104 | 3.6 |
| Patient 48 (F, 32) | F198 | No tumour (32) | c.761T>C, p.I254T | 74 | 3.9 |
| Patient 49 (F, 24) | F148 | CPC (17), BS (24) | c.845G>C, p.R282P | 105 | 4.7 |
| Patient 50 (F, 67) | F160 | BC (47) | c.869G>A, p.R290Hk | 85 | 10.9 |
| Patient 51 (F, 44) | F142 | BC (41) | c.904G>A, p.G302R | 71 | 10 |
| Patient 52 (F, 33) | F77 | Ov. GCT+STS (28) | c.910A>G, p.T304A | 78 | 8.3 |
| Patient 53 (M, 2) | F195 | ACC (2) | c.1010G>A, p.R337H | 81 | 5.8 |
| Patient 54 (M, 38) | F259 | No tumour (38) | c.1010G>A, p.R337H | 85 | 6 |
| Patient 55 (F, 1) | F216 | ACC (0.6) | c.1010G>A, p.R337H | 81 | 6.5 |
| Patient 56 (M, 4) | F265 | ACC (3) | c.1010G>A, p.R337H | 101 | 6.9 |
M, male; F, female.
Age (in years) at blood sampling for EBV-immortalization of lymphocytes.
ACC, adrenocortical carcinoma; anRMS, anaplastic rhabdomyosarcoma; ARMS, alveolar rhabdomyosarcoma; BC, breast cancer; BCC, basal cell carcinoma; Bilat, bilateral; BS, breast sarcoma; CPC, choroïd plexus carcinoma; GCT, germ cell tumour; GBM, glioblastoma multiforme; HD, Hodgkin’s disease; LC, lung cancer; MM, malignant melanoma; NB, neuroblastoma; OS, osteosarcoma; Ov, ovary; SC, skin cancer; STS, soft tissue sarcoma; Tes, testis.
Age (in year) corresponds either to the age at tumour diagnosis or to the age of the unaffected individual.
cDNA numbering with the first nucleotide corresponding to the A of the ATG translation initiation codon in the reference sequence (GenBank RefSeq-file accession number NM_000546.5), according to the Human Genome Variation Society guidelines (www.hgvs.org/mutnomen; date last accessed March 22, 2017). The initiation codon is numbered as codon 1 (accession number NP_000537.3).
Indicated values correspond to p53 mRNA level; data are expressed as percentage of the value observed in control 2.
Indicated values correspond to the mean of the average fold-changes of target genes expression obtained from two independent p53 functional assays.
According to the IARC TP53 database.
Complete deletion.
Promoter and exon 1 deletion.
With no detectable DNE according to the IARC TP53 database.
Figure 1.
Comparative analysis of p53 transcriptional activity in wild-type and mutant TP53 EBV-immortalized lymphocytes exposed to doxorubicin. (A) Dot plot depicting the comparison of p53 functionality scores obtained using p53 functional assay, in lymphocytes from 5 wild-type TP53 controls and from 56 TP53 mutation carriers. For each type of mutations ‘n’ indicates the number of carriers analysed. (B) Subdivision of the 26 patients harbouring 15 different missense mutations into 3 groups: (i) mutations classified according to the IARC database, as supertrans or functional (p.R290H, p.G302R, p.T304A) (ii) p.R158H and p.R337H mutations, and (iii) other missense mutations (p.E11Q, p.G108D, p.T125M, p.R175G, p.H193D, p.H193P, p.V216M, p.C229R, p.I254T, p.R282P). Scores correspond to the average of two independent experiments; the error bars represent mean ± standard error of the mean (SEM). Statistical significance of the comparisons was evaluated using the unpaired t-test and represented by an asterisk (*P < 0.05; **P < 0.01; ***P < 0.001).
We then extended this functional analysis to 26 patients suspected to present with LFS and harbouring 15 additional different missense mutations. According to the IARC TP53 database (17), 3 had no detectable dominant-negative effect, and for 12 mutations the dominant-negative activity had not been assessed (Table 1). For these 15 mutations we observed, as shown in Figure 1A and Table 1, a dispersion of the p53 functionality scores, indicating that these mutations have a heterogeneous effect on p53 transcriptional activity, which led us to subdivide these mutations into 3 groups (Fig. 1B). The first group corresponded to 3 mutations (p.R290H, p.G302R, p.T304A) previously classified as ‘supertrans or functional’ in a yeast-based functional assay (18). The scores for these mutations (mean score = 9.7 ± 0.8) were very close to the control lymphocyte scores (Fig. 1B, Table 1), confirming the absence of the deleterious effect of these mutations on the p53 transcriptional activity. When we looked at the TP53 missense mutations p.R158H and p.R337H associated with adrenocortical carcinoma or adult tumours (1,19), we obtained scores close to the ones of the null mutations (mean score = 5.6 ± 0.4) (Fig. 1B, Table 1). Finally, when we analysed the other missense mutations (p.E11Q, p.G108D, p.T125M, p.R175G, p.H193D, p.H193P, p.V216M, p.C229R, p.I254T, p.R282P), we observed a drastic alteration of the p53 transcriptional activity (mean score = 3.8 ± 0.2), similar to that observed with well characterized dominant-negative missense mutations suggesting that most of these mutations are also bona fide dominant-negative mutations (Fig. 1B, Table 1). These results indicate that the impact of the mutations on p53 transcriptional response to DNA damage in LFS lymphocytes can be considered as an endophenotype of the clinical severity of germline TP53 mutations.
Genome-wide mapping of p53-binding sites in control lymphocytes exposed to DNA damage
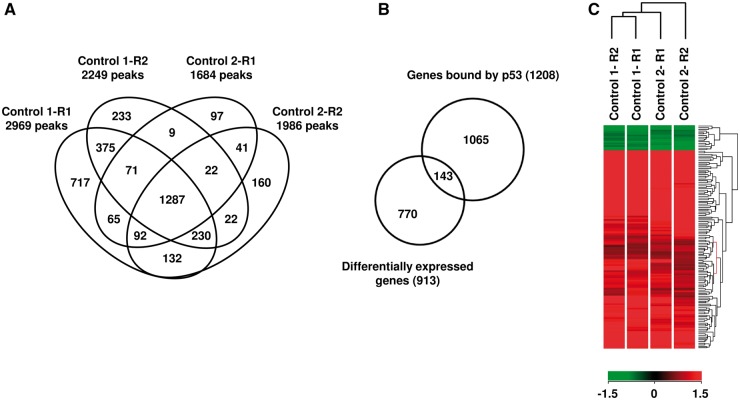
In order to evaluate the impact of germline TP53 mutations on p53 DNA binding, we did a genome-wide mapping of p53-binding sites in response to DNA damage, by performing ChIP-Seq in control EBV-immortalized lymphocytes exposed to doxorubicin. We used EBV-immortalized lymphocytes derived from two TP53 wild-type individuals in order to limit a potential bias due to inter-individual variability and we performed each experiment in duplicate. In control subject 1, we identified 2969 and 2249 peaks (FDR = 0.005) in replicates R1 and R2, respectively, with 1963 peaks in common between the two replicates. In control subject 2, 1684 and 1986 peaks were identified in replicates R1 and R2, with 1442 common peaks (Figs 2A and 3A). As expected, few peaks were detected when the same experiment was performed without doxorubicin treatment (Supplementary Material, Fig. S3). In order to identify the most relevant p53-binding sites, we selected peaks detected in common between replicates performed on lymphocytes from the two controls, which allowed us to identify 1287 high-confidence peaks (FDR = 0.005) (Fig. 2A, Supplementary Material, Table S1), among which 65% (842/1287) and 26% (337/1287) were located within 50 kb and 10 kb from transcription start sites (TSS), respectively. Moreover, 90% (1158/1287) of these high-confidence ChIP-Seq peaks harbour the previously characterized p53-binding motif (20) (Supplementary Material, Table S1).
Figure 2.
Identification by ChIP-Seq analysis of p53-binding sites in control EBV-immortalized lymphocytes exposed to doxorubicin. (A) Venn diagram illustrating the overlaps of peaks detected in common between the 2 replicates (R1 and R2) performed in 2 controls (control 1 and control 2). The numbers indicate the number of binding sites recognized by wild-type p53. (B) Overlap between genes containing p53-binding sites and genes differentially expressed upon doxorubicin treatment (note that the 1287 high-confidence peaks correspond to 1208 different genes). (C) Heat map, obtained in lymphocytes from two wild-type TP53 controls, showing differential expression upon doxorubicin treatment of 143 genes containing a p53-binding site. For each control, replicates are indicated (R1 and R2). Red color indicates up-regulated genes and green color down regulated genes.
In order to identify, among these genes containing p53-binding sites, those who are differentially expressed upon p53 activation, we completed the transcriptomic analysis previously performed on lymphocytes derived from the control 1 and exposed to doxorubicin (15), by a new transcriptomic analysis performed on lymphocytes derived from control 2. Global gene expression analysis allowed us to identify in exposed control lymphocytes a core of 913 differentially expressed genes, including 532 up and 381 down-regulated genes (P-value < 0.005) (Supplementary Material, Table S2). Comparison of ChIP-Seq data with gene expression profiles showed that 12% (143/1208) of the genes containing a p53-binding site were differentially expressed (127 up-regulated and 16 down-regulated genes) upon doxorubicin treatment (Fig. 2B and C). These 143 genes include 40 previously known p53 target genes (21,22), such as CDKN1A [MIM*116899, NM_000389] and MDM2 and 103 new putative p53 target genes (Supplementary Material, Table S2). As expected, functional clustering of gene annotations, using the DAVID algorithm, revealed an enrichment of genes involved in biological pathways known to be regulated by p53 in response to DNA damage (Supplementary Material, Table S2).
Functional validation of new putative p53 target genes using a p53-binding assay in yeast
To confirm the functional relevance of the p53-binding sites identified by ChIP-Seq within new putative p53 target genes, we constructed 2 yeast strains, one expressing human wild-type p53 (YPH500-TP53) and the other containing the empty vector (YPH500-Empty). p53-binding sites, detected by ChIP-Seq, were PCR-amplified and cloned by homologous recombination into the gapped plasmid pRS313-cyc-ADE2 in the minimal promoter upstream of the ADE2 open reading frame and transformed into the YPH500-TP53 and YPH500-Empty yeast strains. In this assay, the ADE2 gene expression is strictly dependent on p53-binding to the cloned fragment. Colonies of YPH500 strains cultured on media containing a limited amount of adenine are spontaneously red, whereas colonies expressing ADE2 turn white (Supplementary Material, Fig. S4).
From the ChIP-Seq data, we selected 4 binding sites in genes not known as p53 target genes and absent from previous ChIP-Seq studies (GCNT3, LINC01480, PLXNB1 and SRSF8) (23) and tested them in the yeast binding assay. As a positive control, we used the p53-binding site within MDM2 and, as a negative control, a sequence from the USH1G gene [MIM*607696, NM_173477] without any signal detected in the p53 ChIP-Seq experiments. All 4 putative p53-binding sites generated white colonies when transformed into YPH500-TP53 strain and red colonies when transformed into the YPH500-Empty yeast strain, confirming that they indeed correspond to functional p53-binding sites (Supplementary Material, Fig. S5). We then performed quantitative RT-PCR analysis to confirm the results obtained in the transcriptomic analyses for these genes. In accordance with the transcriptomic data, all the genes examined were differentially expressed in control lymphocytes exposed to doxorubicin except for SRSF8 (Supplementary Material, Fig. S5, Supplementary Material, Table S2). In lymphocytes harbouring the TP53 p.R248W mutation, the up-regulation of these genes was significantly reduced, demonstrating the specificity of the regulation by p53 (Supplementary Materials, Fig. S5, Table S2).
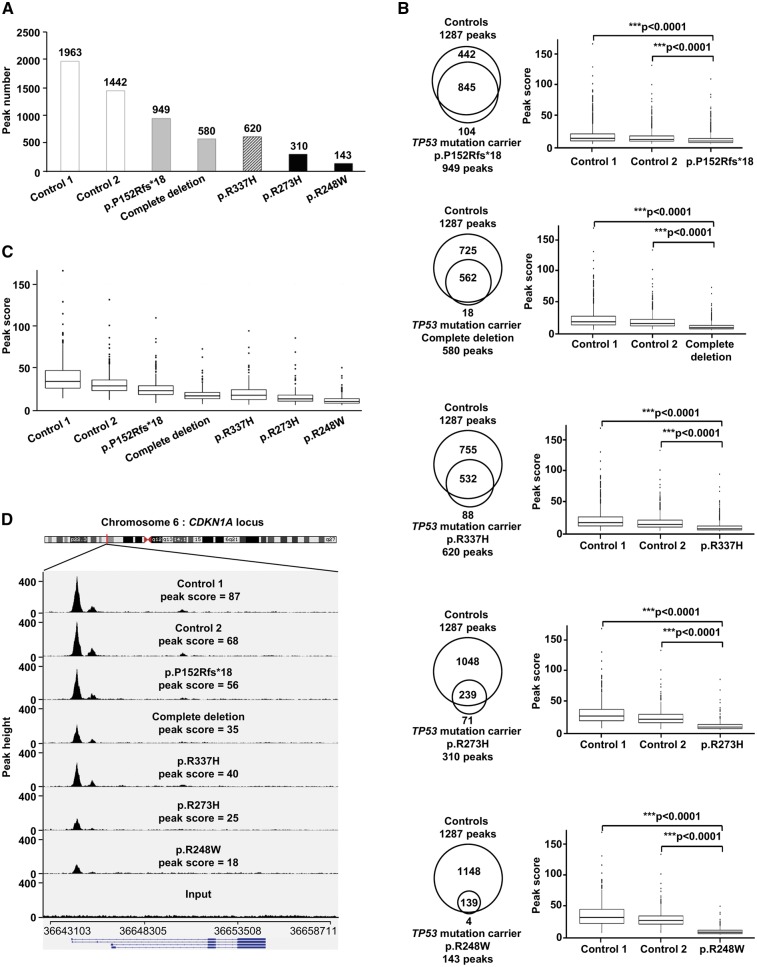
Drastic effect of TP53 dominant-negative missense mutations on p53 DNA binding
In order to study the functional consequences of germline TP53 mutations on p53 DNA binding in presence of DNA damage, we performed a ChIP-Seq analysis of EBV-immortalized lymphocytes from LFS patients harbouring heterozygous TP53 mutations, after exposure to doxorubicin. For this study, we selected (i) two null mutations, i.e. a frameshift mutation (p.P152Rfs*18) and a complete deletion, both resulting into p53 haplodeficiency, (ii) two canonical missense mutations, classified as dominant-negative (p.R248W, p.R273H), according to the IARC TP53 database and (iii) the p.R337H mutation, a low penetrance TP53 mutation associated with adrenocortical carcinomas or adulthood tumours (Table 1). As shown in Figure 3A, the total number of ChIP-Seq peaks detected was reduced approximately by half in LFS lymphocytes harbouring the p.P152Rfs*18 mutation or the complete TP53 deletion compared to the two controls. In LFS lymphocytes harbouring dominant-negative missense mutations, a massive reduction was observed (Fig. 3A). Interestingly, the number of peaks obtained for the p.R337H missense mutation was closer to those obtained for null mutations than those obtained for dominant-negative mutations (Fig. 3A). We then compared the peak scores of the p53-binding sites, detected in wild-type and mutant lymphocytes. We identified 845, 562, 532, 239 and 139 peaks in common between control lymphocytes and LFS lymphocytes harbouring p.P152Rfs*18, complete deletion, p.R337H, p.R273H and p.R248W mutations, respectively (Fig. 3B). In each case, the peak score in mutant lymphocytes was significantly reduced (P < 0.0001 unpaired t-test), as compared with control lymphocytes (Fig. 3B). As shown for the 123 peaks detected in common in wild-type and mutant lymphocytes, (Fig. 3C) and as illustrated by the visualization of peaks at the CDKN1A locus (Fig. 3D), a well-known p53 target gene, we observed a massive reduction of the peak scores in LFS lymphocytes harbouring dominant-negative missense mutations, as compared to control lymphocytes or LFS lymphocytes harbouring null mutations. Again, the results obtained for the p.R337H mutation were similar to those obtained with null mutations (Fig. 3C and D).
Figure 3.
Impact of germline TP53 mutations on DNA-binding in lymphocytes exposed to doxorubicin, revealed by ChIP-Seq analysis. (A) Histograms presenting the number of peaks detected by ChIP-Seq performed on lymphocytes derived from two wild-type TP53 controls (control 1 and control 2) (open bars), two TP53 mutation carriers harbouring null mutations (p.P152Rfs*18 and complete deletion) (grey bars), one carrier harbouring the p.R337H mutation (hatched bars), and two carriers harbouring dominant-negative missense mutations (p.R273H and p.R248W) (black bars); (B) Venn diagrams showing, for each TP53 mutation, the overlaps of ChIP-Seq peaks detected in common between control and mutant lymphocytes. Box plot analysis comparing, for each mutation, the average ChIP-Seq peak scores of the peaks detected in common between control lymphocytes and mutant lymphocytes. The ends of the boxes correspond to 25 and 75% quartiles, the median is marked by a horizontal line inside the box, the lower whiskers on the box plots represent the smallest peak score ≥ 25% quartile – 1.5 (IQR) (inter-quartile range), and the upper whisker represent the largest peak score ≤ 75% quartile + 1,5 (IQR).Peak scores beyond the end of the whiskers are the outliers and plotted as individual points. Unpaired t-tests were performed to compute significance. (C) Box plot analysis of the average peak scores of the 123 peaks detected in common between all control and TP53 mutant lymphocytes. (D) IGV visualization of p53 ChIP-Seq results at the CDKN1A locus in control lymphocytes, lymphocytes harbouring null mutations, the p.R337H mutation or dominant negative missense mutations. The average ChIP-Seq peak scores, detected at the CDKN1A locus, in both replicates of ChIP-Seq performed on lymphocytes from the two controls (control 1 and control 2) and each the TP53 mutation carriers (p.P152Rfs*18, complete deletion, p.R337H, p.R273H and p.R248W) are indicated.
No evidence for gain of function in lymphocytes with missense mutations
In addition to loss of function and dominant-negative effect, some mutant p53 can exert additional oncogenic activity, by gain-of-function (GOF) mechanism. Because some studies have shown that some dominant-negative missense mutations, such as the p.R248W mutation, may result in GOF by modulating expression of target genes involved in oncogenesis (24,25), we explored thanks to the ChIP-Seq data, the putative GOF activity of the p.R248W mutant in lymphocytes not exposed to doxorubicin. As expected in this basal condition, only 17 peaks were detected (Supplementary Materials, Fig. S3A, Table S1). Among these peaks, none was specific to the mutant. These results indicate that in non-malignant cells, such as lymphocytes, there is no evidence that missense mutations induce GOF.
Discussion
In this study, we explored the impact of germline TP53 mutations on the transcriptional activity of the protein in response to DNA damage. Since 1992, it has been established that the tumour suppressor activity of p53 relies mostly on its ability, when DNA damage occur, to upregulate the transcription of numerous target genes involved in many pathways including cell cycle arrest, DNA repair, apoptosis and metabolism. The use of the simple p53 functional assay that we developed in lymphocytes, based on the semi-quantitative measurement of the induction of six p53 target genes, confirms that in TP53 mutation carriers there is a constitutive defect in the transcriptional response to DNA damage, detectable in most of the cases. Indeed, the results of the p53 functional assay obtained for 30 carriers with TP53 null or dominant-negative missense mutations clearly showed, except for 4 carriers harbouring null mutations, that the transcriptional activity was altered as compared to controls (Table 1). It should be highlighted that the main advantage of this assay developed in lymphocytes, although it requires immortalization of lymphocytes by EBV, is that it allows one to assess the biological impact of heterozygous mutations in the genetic context of the patients, in contrast to in vitro and ex vivo assays, such as those developed in yeast (18,26,27).We have compared the p53 transcriptional response to DNA damage between peripheral blood mononuclear cells and EBV-immortalized lymphocytes, and found that the transcriptional response to doxorubicin was similar (data not shown). Moreover, the absence of p53 stabilization in unexposed EBV-immortalized lymphocytes from 5 controls (Supplementary Material, Fig. S2) confirms that EBV immortalization does not introduce genetic changes activating p53 pathway.
In TP53 mutation carriers, this constitutive alteration of the transcriptional response to DNA damage, detectable in non-tumour cells, constitutes an endophenotype before the occurrence of cancer and confirms that germline TP53 mutations represent a genetic permissive context facilitating the malignant transformation of cells in which DNA damage has occurred. This suggests that the core LFS tumour spectrum is probably explained by the differential sensitivities of tissues to oncogenic stresses and that the tumour spectrum associated with germline TP53 mutations is much broader than initially considered. This also represents an additional argument supporting that genotoxic treatments, such as radiotherapy, contribute to secondary tumours occurring in LFS patients and therefore should be avoided, if possible. Thus, LFS represents a rare example of autosomal dominant predisposition to cancers, in which a biological endophenotype can be detected in mutation carriers. As shown in Table 1 and Figure 1, the results obtained with the functional assays confirm the difference in the biological severity of dominant-negative missense mutations versus the other types of missense mutations and the null mutations. This assay should help the classification of TP53 mutations and the interpretation of variants of unknown significance. We anticipate that this challenge will increase with the detection of such germline TP53 variants in patients with atypical clinical presentations, performed in the context of next-generation sequencing (NGS) cancer gene panels. A normal score obtained with this p53 functional assay will not exclude that the variant is deleterious, since the assay explores only the p53-mediated transcriptional response to damage in lymphocytes and, furthermore, we observed for some null mutations a score close to that obtained in controls (Table 1). In contrast, an abnormal score should constitute a strong argument supporting the deleterious effect of the variant.
Several studies have previously analysed p53 DNA-binding in cells exposed to genotoxic or non-genotoxic agents, using ChIP-Seq. Most of these studies have been performed in established cancer cell lines, such as HCT116 or U2OS derived from human colon carcinoma and osteosarcoma, respectively (28–31). Our study provides new insights concerning p53 binding to DNA in non-cancer cells, when DNA damage occurs. First, ChIP-Seq performed in replicates on control lymphocytes allowed us to detect 1287 high-confidence p53-binding sites in the presence of DNA damage, and 90% of these high-confidence peaks harboured a p53-binding motif (20). We showed, by gene expression analyses and yeast assays, that a fraction of these p53-binding sites corresponds to real p53 target genes. The comparison of p53 gene occupancy and gene expression profiles in control lymphocytes exposed to doxorubicin reveals that 12% of the genes bound by p53 were differentially expressed upon doxorubicin treatment (Fig. 2B and C). The 770 differentially expressed genes not bound by p53 could represent genes regulated by a distal p53-binding site or genes indirectly regulated by p53 through other transcription factors. On the other hand, we showed that 1065 genes specifically bound by p53 were not differentially expressed in response to doxorubicin. This lack of induction could either reflect a bias in the annotation of the peaks which were arbitrary associated with the nearest transcription start site since the bound elements could represent distal enhancers for another gene; it could also be explained by the lack of a cell-specific or context-specific cofactor needed for p53 transcriptional activity. Indeed, several p53 ChIP-Seq studies have shown that in different cells treated with different genotoxic (doxorubicin, 5-Fluorouracil) or non-genotoxic agents (RITA, Nutlin-3a), the p53-transcriptional response is different despite a mostly similar binding profile (30–32). This observation suggests that changes in gene expression induced by p53 require other factors, such as cofactors recruited in a cell type-specific and stress response-specific manner, indicating that p53 binding is necessary, but not sufficient for transactivation. Alternatively, p53 chromatin occupancy in response to DNA lesions without transactivation may reflect other p53 roles in chromatin integrity (33).
In agreement with recent studies (34,35), our data show that, in non-damaged cells, p53 is already bound to a small proportion of its genomic targets, and 100% of these p53-binding sites remain when DNA damage occurs (Supplementary Material, Fig. S3B), suggesting that this constitutive fixation of p53 to target genes allows a rapid response to genotoxic stress. As shown by the results of the ChIP-Seq performed on control lymphocytes exposed to doxorubicin, induction of DNA damage results not only in an increase of peak scores (a measure of p53-binding strength), at sites detectable in basal conditions, but also into a massive increase of the total number of p53-binding sites (Supplementary Material, Table 1, Supplementary Material, Fig. S3). Our results contrast with those obtained in cancer cell lines exposed to doxorubicin (30). Indeed, in that study, 90% of the p53-binding sites detected in untreated cells did not contain the p53-binding motif, and in treated cells many of these sites were not bound by p53. The differences between both studies might be explained by the numerous genetic and epigenetic modifications occurring in cancer cell lines (36), which deeply modify the landscape of p53-binding sites (28,29). Indeed, most cancer cell lines harbour either a TP53 mutated gene or an alteration in the p53 signalling pathway (37,38).
In lymphocytes from LFS patients harbouring null mutations, the p53 functionality scores obtained with the p53 functional assay, the number and scores of ChIP-Seq peaks were reduced by half in average, as compared to control lymphocytes (Figs 1 and 3). These results are those expected for haplodeficiency due to a 50% reduction of the mRNA expression level (Table 1). This observation suggests that the DNA binding of wild-type p53 protein, encoded by the wild-type allele allows maintaining a partial p53 transcriptional response to DNA damage. This probably explains why TP53 null mutations are associated with later tumour-onset and why in tumours with such mutations, LOH affecting the wild-type allele is more commonly observed than in tumours with other types of TP53 mutations (1,39).
The drastic effect of dominant-negative missense mutations on p53 transcriptional activity and DNA binding in response to DNA damage (Figs 1A, 3A–D) is probably explained by the ability of the p53 dominant-negative mutants to disrupt the wild-type p53 function by forming inactive heterotetramers. Indeed, it has been suggested that p53 dimers are formed co-translationally, resulting in either mutated or wild-type homo-dimers, whereas tetramers are formed post-translationally by dimerization of the existing dimmers (40). According to this model, the mutant p53 protein would inactivate 75% of p53 tetramers (41). This is consistent with our data showing a reduction close to 75% of the functionality score and the number of detected peaks in LFS lymphocytes with dominant-negative missense mutations in comparison to those observed in control lymphocytes. Thus, we provide a biological explanation of the clinical severity of the dominant-negative missense mutations which drastically alter the p53 transcriptional activity in response to DNA damage by abrogating binding to DNA.
The observation of germline TP53 null mutations strongly supports that the primum movens of LFS is the loss of p53 function. Therefore, the predominance of missense mutations and, in particular of those with dominant-negative activities, constitutes an apparent paradox. The drastic effect of these mutations on DNA binding and transcriptional response to DNA, that we report here, probably may explain only in part their predominance and clinical severity. The second part of the explanation probably corresponds to the gain of multiple neomorphic properties, TP53 missense mutations being able to transform the tumour suppressor protein into an oncogenic protein, providing a selective advantage during cancer progression. As highlighted by numerous studies, GOF includes alteration of chromatin structures, genomic instability, inhibition of DNA repair, interaction with transcription factors, aberrant upregulation of genes and subversion of molecular partners of wild-type p53 (42,43). A recent study, also based on ChIP-Seq but performed in cancer cell lines, has reported that missense mutations with GOF cause aberrant DNA binding and upregulation of genes involved in chromatin conformation (25). However, our analyses performed in lymphocytes with missense mutations did not provide any evidence for aberrant transcription or DNA binding. As previously shown by transcriptomic analyses performed in non-cancer tissues from p53−/−and p53 mutant/mutant mice, which did not reveal any significant changes (44), our study confirms that GOF is not observed in non-malignant cells and suggests that GOF occurs when cells are transformed.
The clinical management of LFS represents a real challenge for physicians, considering the wide tumour spectrum associated with germline TP53 mutations, and the diversity of the clinical severity and age of tumour onset observed among families. The ‘Toronto protocol’ including annual total body MRI and brain MRI, from the first year of life, has recently been elaborated to ensure early tumour detection in germline TP53 mutation carriers (45). In the context of the increased detection of germline TP53 mutations in atypical LFS presentations, our study strengthens the idea that it should be appropriate to adapt the medical management of TP53 mutation carriers to the severity of the mutation.
Materials and Methods
p53 functional assay
This assay was derived from those previously described (15,16). EBV-Immortalized lymphocytes from wild-type TP53 control individuals and from germline TP53 mutation carriers (Table 1) were cultured in RPMI 1640 medium (GIBCO, Invitrogen), supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) at 37 °C with 5% CO2. The TP53 genotype was systematically assessed by Sanger sequencing. For the p53 functional assay, cells were seeded in duplicates in 12-well plates (Corning) at a density of 106 cells/well and treated or not with 0.3 µM of doxorubicin (Sigma-Aldrich) for 8 h. After treatment, cells were harvested by centrifugation and total RNA extraction was performed with Nucleospin RNAII kit (Macherey Nagel) according to the manufacturer’s instructions and quantified using a ND-1000 UV-Vis Spectrophotometer (NanoDrop technologies).
Reverse transcription (RT) was performed on 100 ng of total RNA using the Verso cDNA synthesis kit (Thermo scientific). A RT-Quantitative Multiplex PCR of Short Fluorescent fragments (RT-QMPSF) was performed to measure the transcriptional induction of 6 p53 target genes selected from transcriptomic analysis (CEP170B [NM_015005], PODXL [MIM*602632, NM_001018111], RRAD [MIM*179503, NM_004165], GLS2 [MIM*606365, NM_013267], CABYR [MIM*612135, NM_012189], TP53I3 [MIM*605171, NM_004881]) using two control amplicons (SF3A1 [MIM*605595, NM_005877] and TBP [MIM*600075, NM_003194]) to normalize the data. Fluorescent amplicons were separated by size on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA), and resulting fluorescent profiles were analysed using the GeneScan 3.7 software (Applied Biosystems). RT-QMPSF profiles from exposed and unexposed cells were superimposed after adjustment of control amplicons to the same height, the fold-change was determined for each p53 target gene and an arbitrary p53 functionality score corresponding to the average of fold-changes was defined (Supplementary Material, Fig. S1). For each TP53 mutation, a functionality score was determined from two independent assays and compared to the scores obtained in TP53 wild-type cells, using two-sided Student's t-test.
Comparative gene expression profiling
Comparative gene expression profiling of EBV-immortalized lymphocytes treated or not with doxorubicin, for 8 h was performed using Whole Human Genome Oligo 4 × 44K Microarray (G4112F, Agilent), according to the Agilent Two-Color Gene Expression workflow, as previously described (15,16). Briefly, starting from 100 ng of total RNA,cRNA was synthesized and labelled using the low input Quick Amp Labeling Kit (Agilent), with Cy3 (for untreated cells) and Cy5 (for treated cells). Following co-hybridization, microarrays were scanned (DNA microarray scanner G2565CA, Agilent Technologies) with a resolution of 5 µm. For each experiment, comparative gene expression profiling was performed in 2 replicates. GeneSpring GX 14.5 software (Agilent Technologies) was used to select the differentially expressed genes between treated and untreated cells in both replicates (Student t-test, P-value < 0.005).
Quantitative real-time PCR
Cells were treated or not with doxorubicin (0.3 µM) for 8h. For quantitative real-time PCR (qRT-PCR) analysis, total RNA was extracted from cells using a Nucleospin RNAII kit (Macherey Nagel), and cDNA was obtained using the Verso cDNA synthesis kit (Thermo scientific). qPCR was performed using the SsoFast EvaGreen supermix (Bio-Rad) detection method on a CFX96 real-time PCR detection system (Bio-Rad). Three biological replicates were performed for each experiment. The comparative Ct method was used for quantification of MDM2 [MIM*164785, NM_002392], GCNT3 [MIM*606836, NM_004751], LINC01480 [NR_110724], PLXNB1 [MIM*601053, NM_002673] and SRSF8 [MIM*603269, NM_032102] gene expression compared to that of the GAPDH [MIM*138400, NM_002046] gene, used as control. The fold-changes between treated and untreated conditions were then calculated.
Western blot analysis
For total protein extraction, cells were pelleted and homogenized in 100 µl of RIPA buffer (25mM Tris–HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) from Pierce Biotechnology supplemented with cocktails of protease inhibitors (Sigma-Aldrich) and phosphatase inhibitors (Pierce Biotechnology). Samples were centrifuged at 11,300 g for 20 min at 4 °C to remove cellular debris and the supernatant was collected. Thirty µg of protein were resolved by 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes (Hybond-C Extra; Amersham Biosciences). The following primary antibodies were used: mouse monoclonal anti-p53 DO-1 antibody (1/2000; Santa Cruz Biotechnology, Inc.), anti-actin JLA20 antibody (1/10000; Sigma Aldrich). Membranes were incubated with peroxidase-labelled anti-mouse or anti-rabbit antibodies (1/10000) from Jackson Immunoresearch Laboratories, and signals were detected with chemiluminescence reagents using G:Box (Syngene) and GeneSnap software.
p53-binding assay in yeast
Putative p53-binding sites detected by ChIP-Seq were PCR-amplified and sequences required for homologous recombination in yeast were added to PCR products using a second PCR (primer sequences available upon request). The ADE2-YPH500-TP53 yeast strain, containing the pLS76 plasmid and expressing human p53, and the ADE2- YPH500-Empty yeast strain, containing the pSS16 vector, were transformed, using the lithium acetate procedure, with PCR products and a linearized pRS313-cyc-ADE2 vector, containing the ADE2 [NM_001183547.3] open reading frame downstream of a minimal promoter (cyc) (26,46). Transformed yeast cells were plated on a synthetic minimal medium minus leucine and histidine, to select for cells containing the pLS76 or pSS16 and pRS313 vectors, respectively, plus a limited amount of adenine (10 µg/ml) and incubated for 2 days at 30 °C (Supplementary Material, Fig. S4).
Chromatin Immunoprecipitation
EBV-immortalized lymphocytes from wild-type TP53 subjects or from germline TP53 mutation carriers were seeded in duplicates in 15cm dishes, at a density of 20 x 106 cells/dish and treated with 0.3 µM of doxorubicin for 8h (Sigma-Aldrich). Cells were fixed by the addition of 1% formaldehyde for 10 min (Sigma-Aldrich) and then fixation was stopped by the addition of 0.125 M glycine for 5 min (Millipore). Cells were washed 3 times in PBS and then suspended in SDS lysis buffer supplemented with a protease inhibitor cocktail (EZ-ChIP kit,Millipore). Chromatin from 20 x 106 cells was fragmented by sonication using a S220 ultrasonicator (Covaris) yielding genomic DNA fragments with an average size of 250 bp. Chromatin immunoprecipitation was performed with the EZ-ChIP kit (Millipore), according to the manufacturer’s instructions, using 4 µg of mouse monoclonal anti-p53 DO-1 antibody (Santa Cruz Biotechnology, Inc.). DNA was purified on Millipore columns (EZ-ChIP kit, Millipore) and then concentrated on columns from DNA clean and concentrator-10 kit (Zymo Research).
High-throughput Sequencing
Libraries were prepared from 4 ng of immunoprecipitated DNA or input DNA, using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® and the NEBNext® Multiplex Oligos for Illumina® (NEB). Briefly, after end-repair, A-tailing and adaptor ligation, libraries were PCR-amplified using 10 cycles and size-selected. Library concentrations and sizes were checked on a TapeStation 2200 system (Agilent). Libraries were then subjected to a 2x75bp paired-end sequencing on an Illumina NextSeq500 sequencer to yield an average of 50 million paired-end reads per sample.
Bioinformatic Analyses
Paired-end 75 bp reads were aligned against the Human Reference Genome (assembly hg19, UCSC), using the BWA MEM algorithm (v0.7.10) (47) and sorted with Picard SortSam (v1.141) (http://broadinstitute.github.io/picard; date last accessed March 22, 2017). Detection and annotation of the peaks were performed using the HOMER (v4.8) algorithm (48). Peak false discovery rate (FDR) was set to 0.005 (i.e 5 false positive peaks every 1000 called peaks), analysis was performed with default Poisson P-value and fold change parameters, and retained peaks were filtered against the input and against their local background. Peak sequences were checked for significant p53-binding motif enrichment, using HOMER (P-value < 0.05). Each peak was assigned to the gene with the nearest transcription start site (TSS) during the annotation process (HOMER). Intersections of the peaks detected in the different ChIP-Seq experiments were performed using the Odysseus software that we developed. Venn diagrams were plotted using the BioVenn web application (http://www.cmbi.ru.nl/cdd/biovenn/; date last accessed March 22, 2017). DAVID (v6.7) (49) was used for the functional annotation of genes with a detectable p53-binding site. The peaks were inspected using the IGV Genome Browser (v2.3) (50).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to Mario Tosi for critical review of the manuscript.
Conflict of Interest statement. None declared.
Funding
This work was supported by the French National Cancer Institute (INCa), the French Association for Cancer Research (Fondation ARC), the League against cancer (Ligue contre le Cancer) and by the European Regional Development Fund (European Union and Région Normandie). Funding to pay the Open Access publication charges for this article was provided by the French Institute for Health and Medical Research (Inserm).
References
- 1. Bougeard G., Renaux-Petel M., Flaman J.-M., Charbonnier C., Fermey P., Belotti M., Gauthier-Villars M., Stoppa-Lyonnet D., Consolino E., Brugières L., et al. (2015) Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J. Clin. Oncol., 33, 2345–2352. [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez K.D., Noltner K.A., Buzin C.H., Gu D., Wen-Fong C.Y., Nguyen V.Q., Han J.H., Lowstuter K., Longmate J., Sommer S.S., et al. (2009) Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J. Clin. Oncol., 27, 1250–1256. [DOI] [PubMed] [Google Scholar]
- 3. Kamihara J., Rana H.Q., Garber J.E. (2014) Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Hum. Mutat., 35, 654–662. [DOI] [PubMed] [Google Scholar]
- 4. Li F.P., Fraumeni J.F., Mulvihill J.J., Blattner W.A., Dreyfus M.G., Tucker M.A., Miller R.W. (1988) A cancer family syndrome in twenty-four kindreds. Cancer Res., 48, 5358–5362. [PubMed] [Google Scholar]
- 5. Li F.P., Fraumeni J.F. (1969) Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann. Intern. Med., 71, 747–752. [DOI] [PubMed] [Google Scholar]
- 6. Malkin D., Li F.P., Strong L.C., Fraumeni J.F., Nelson C.E., Kim D.H., Kassel J., Gryka M.A., Bischoff F.Z., Tainsky M.A. (1990) Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science, 250, 1233–1238. [DOI] [PubMed] [Google Scholar]
- 7. Srivastava S., Zou Z.Q., Pirollo K., Blattner W., Chang E.H. (1990) Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature, 348, 747–749. [DOI] [PubMed] [Google Scholar]
- 8. Tinat J., Bougeard G., Baert-Desurmont S., Vasseur S., Martin C., Bouvignies E., Caron O., Bressac-de Paillerets B., Berthet P., Dugast C., et al. (2009) 2009 version of the Chompret criteria for Li Fraumeni syndrome. J. Clin. Oncol., 27, 108–109. [DOI] [PubMed] [Google Scholar]
- 9. Gonzalez K.D., Buzin C.H., Noltner K.A., Gu D., Li W., Malkin D., Sommer S.S. (2009) High frequency of de novo mutations in Li-Fraumeni syndrome. J. Med. Genet., 46, 689–693. [DOI] [PubMed] [Google Scholar]
- 10. Bougeard G., Sesboüé R., Baert-Desurmont S., Vasseur S., Martin C., Tinat J., Brugières L., Chompret A., de Paillerets B.B., Stoppa-Lyonnet D., et al. (2008) Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J. Med. Genet., 45, 535–538. [DOI] [PubMed] [Google Scholar]
- 11. Hanel W., Marchenko N., Xu S., Yu S.X., Weng W., Moll U. (2013) Two hot spot mutant p53 mouse models display differential gain of function in tumourigenesis. Cell Death Differ., 20, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lang G.A., Iwakuma T., Suh Y.-A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. (2004) Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell, 119, 861–872. [DOI] [PubMed] [Google Scholar]
- 13. Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. (2004) Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell, 119, 847–860. [DOI] [PubMed] [Google Scholar]
- 14. Song H., Hollstein M., Xu Y. (2007) p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol., 9, 573–580. [DOI] [PubMed] [Google Scholar]
- 15. Zerdoumi Y., Aury-Landas J., Bonaïti-Pellié C., Derambure C., Sesboüé R., Renaux-Petel M., Frebourg T., Bougeard G., Flaman J.-M. (2013) Drastic effect of germline TP53 missense mutations in Li-Fraumeni patients. Hum. Mutat., 34, 453–461. [DOI] [PubMed] [Google Scholar]
- 16. Zerdoumi Y., Kasper E., Soubigou F., Adriouch S., Bougeard G., Frebourg T., Flaman J.-M. (2015) A new genotoxicity assay based on p53 target gene induction. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 789–790, 28–35. [DOI] [PubMed] [Google Scholar]
- 17. Petitjean A., Achatz M.I.W., Borresen-Dale A.L., Hainaut P., Olivier M. (2007) TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene, 26, 2157–2165. [DOI] [PubMed] [Google Scholar]
- 18. Kato S., Han S.-Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. (2003) Understanding the function-structure and function-mutation relationships of p53 tumour suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad., 100, 8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Achatz M.I.W., Olivier M., Le Calvez F., Martel-Planche G., Lopes A., Rossi B.M., Ashton-Prolla P., Giugliani R., Palmero E.I., Vargas F.R., et al. (2007) The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett., 245, 96–102. [DOI] [PubMed] [Google Scholar]
- 20. Koeppel M., van Heeringen S.J., Kramer D., Smeenk L., Janssen-Megens E., Hartmann M., Stunnenberg H.G., Lohrum M. (2011) Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Res., 39, 6069–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol., 9, 402–412. [DOI] [PubMed] [Google Scholar]
- 22. Zeron-Medina J., Wang X., Repapi E., Campbell M.R., Su D., Castro-Giner F., Davies B., Peterse E.F.P., Sacilotto N., Walker G.J., et al. (2013) A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell, 155, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tebaldi T., Zaccara S., Alessandrini F., Bisio A., Ciribilli Y., Inga A. (2015) Whole-genome cartography of p53 response elements ranked on transactivation potential. BMC Genomics, 16, 464.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Do P.M., Varanasi L., Fan S., Li C., Kubacka I., Newman V., Chauhan K., Daniels S.R., Boccetta M., Garrett M.R., et al. (2012) Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev., 26, 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu J., Sammons M.A., Donahue G., Dou Z., Vedadi M., Getlik M., Barsyte-Lovejoy D., Al-awar R., Katona B.W., Shilatifard A., et al. (2015) Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature, 525, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flaman J.M., Frebourg T., Moreau V., Charbonnier F., Martin C., Chappuis P., Sappino A.P., Limacher I.M., Bron L., Benhattar J. (1995) A simple p53 functional assay for screening cell lines, blood, and tumours. Proc. Natl. Acad. Sci., 92, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frebourg T., Barbier N., Kassel J., Ng Y.S., Romero P., Friend S.H. (1992) A functional screen for germ line p53 mutations based on transcriptional activation. Cancer Res, 52, 6976–6978. [PubMed] [Google Scholar]
- 28. Botcheva K., McCorkle S.R., McCombie W.R., Dunn J.J., Anderson C.W. (2011) Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle Georget. Tex., 10, 4237–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Botcheva K., McCorkle S.R. (2014) Cell context dependent p53 genome-wide binding patterns and enrichment at repeats. PloS One, 9, e113492.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menendez D., Nguyen T.-A., Freudenberg J.M., Mathew V.J., Anderson C.W., Jothi R., Resnick M.A. (2013) Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res., 41, 7286–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nikulenkov F., Spinnler C., Li H., Tonelli C., Shi Y., Turunen M., Kivioja T., Ignatiev I., Kel A., Taipale J., et al. (2012) Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ., 19, 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smeenk L., van Heeringen S.J., Koeppel M., Gilbert B., Janssen-Megens E., Stunnenberg H.G., Lohrum M. (2011) Role of p53 serine 46 in p53 target gene regulation. PloS One, 6, e17574.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lieberman P.M. (2016) Retrotransposon-derived p53 binding sites enhance telomere maintenance and genome protection. BioEssays, 38, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kenzelmann Broz D., Spano Mello S., Bieging K.T., Jiang D., Dusek R.L., Brady C.A., Sidow A., Attardi L.D. (2013) Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev., 27, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tonelli C., Morelli M.J., Bianchi S., Rotta L., Capra T., Sabò A., Campaner S., Amati B. (2015) Genome-wide analysis of p53 transcriptional programs in B cells upon exposure to genotoxic stress in vivo. Oncotarget., 6, 24611–24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferraro A. (2016) Altered primary chromatin structures and their implications in cancer development. Cell. Oncol. Dordr., 39, 195–210. [DOI] [PubMed] [Google Scholar]
- 37. Leroy B., Girard L., Hollestelle A., Minna J.D., Gazdar A.F., Soussi T. (2014) Analysis of TP53 mutation status in human cancer cell lines: a reassessment. Hum. Mutat., 35, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herrero A.B., Rojas E.A., Misiewicz-Krzeminska I., Krzeminski P., Gutiérrez N.C. (2016) Molecular mechanisms of p53 deregulation in cancer: an overview in multiple myeloma. Int. J. Mol. Sci., 17, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Varley J.M., Evans D.G., Birch J.M. (1997) Li-Fraumeni syndrome–a molecular and clinical review. Br. J. Cancer, 76, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nicholls C.D., McLure K.G., Shields M.A., Lee P.W.K. (2002) Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J. Biol. Chem., 277, 12937–12945. [DOI] [PubMed] [Google Scholar]
- 41. Aramayo R., Sherman M.B., Brownless K., Lurz R., Okorokov A.L., Orlova E.V. (2011) Quaternary structure of the specific p53-DNA complex reveals the mechanism of p53 mutant dominance. Nucleic Acids Res., 39, 8960–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haupt S., Raghu D., Haupt Y. (2016) Mutant p53 drives cancer by subverting multiple tumour suppression pathways. Front. Oncol., 6, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mantovani F., Walerych D., Sal G.D. (2016) Targeting mutant p53 in cancer: a long road to precision therapy. Febs J., 10.1111/febs.13948. [DOI] [PubMed] [Google Scholar]
- 44. Sabapathy K. (2015) The contrived mutant p53 oncogene - beyond loss of functions. Front. Oncol., 5, 276.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Villani A., Shore A., Wasserman J.D., Stephens D., Kim R.H., Druker H., Gallinger B., Naumer A., Kohlmann W., Novokmet A., et al. (2016) Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol., 17, 1295–1305. [DOI] [PubMed] [Google Scholar]
- 46. Sikorski R.S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl., 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang D.W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 50. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.