Abstract
Chronic obstructive pulmonary disease (COPD) is among the major health burdens in adults. While cigarette smoking is the leading risk factor, a growing number of genetic variations have been discovered to influence disease susceptibility. Epigenetic modifications may mediate the response of the genome to smoking and regulate gene expression. Chromosome 19q13.2 region is associated with both smoking and COPD, yet its functional role is unclear. Our study aimed to determine whether rs7937 (RAB4B, EGLN2), a top genetic variant in 19q13.2 region identified in genome-wide association studies of COPD, is associated with differential DNA methylation in blood (N = 1490) and gene expression in blood (N = 721) and lungs (N = 1087). We combined genetic and epigenetic data from the Rotterdam Study (RS) to perform the epigenome-wide association analysis of rs7937. Further, we used genetic and transcriptomic data from blood (RS) and from lung tissue (Lung expression quantitative trait loci mapping study), to perform the transcriptome-wide association study of rs7937. Rs7937 was significantly (FDR < 0.05) and consistently associated with differential DNA methylation in blood at 4 CpG sites in cis, independent of smoking. One methylation site (cg11298343-EGLN2) was also associated with COPD (P = 0.001). Additionally, rs7937 was associated with gene expression levels in blood in cis (EGLN2), 42% mediated through cg11298343, and in lung tissue, in cis and trans (NUMBL, EGLN2, DNMT3A, LOC101929709 and PAK2). Our results suggest that changes of DNA methylation and gene expression may be intermediate steps between genetic variants and COPD, but further causal studies in lung tissue should confirm this hypothesis.
Introduction
Chronic obstructive pulmonary disease (COPD) is a common, systemic, lung disease, mainly characterized by airway obstruction and inflammation (1). COPD often develops as a response to chronic exposure to cigarette smoke, fumes and gases (2,3). There is significant inter-individual variability in the response to these environmental exposures (4,5) that has been attributed to genetic factors (6,7). Genome-wide association studies (GWAS) have identified genetic variants associated with COPD susceptibility on chromosomes 4q31, 4q22, 15q25 and 19q13 (8–11). However, the mechanism explaining how these variants are involved in the pathogenesis of COPD remains elusive (12).
As is the case for many complex diseases, many single nucleotide polymorphisms (SNPs) associated with COPD and lung function by GWAS are located in non-protein coding intergenic and intronic regulatory regions (13,14). It has been hypothesized that these SNPs may modulate regulatory mechanisms, such as RNA expression, splicing, transcription factor binding and epigenetic modifications (e.g. DNA methylation). Changes in RNA expression as well as in DNA methylation regulating expression have recently been associated with COPD, suggesting that genetic and epigenetic factors are working in concert in the pathogenesis of COPD (15,16). Emerging evidence suggests that differential methylation sites (CpGs) are potentially important for COPD susceptibility (15–17), but their location was not linked to the GWAS loci. However, important associations in COPD genomic regions may have been missed as arrays with limited coverage (27K) were used in the studies conducted to date.
The 19q13.2 region is associated with COPD and cigarette smoking (18,19), lung function (20) and emphysema patterns (21). Genes in this region include RAB4B (member RAS oncogene family), EGLN2 (Egl-nine homolog 2), MIA (melanoma inhibitory activity) and CYP2A6 (cytochrome P450 family 2 subfamily A member 6). The top variant in the region, rs7937:C > T, has been identified by Cho et al. (9). This SNP (RAB4B, EGLN2) was associated with COPD (OR = 1.37, P = 2.9 × 10−9), but not with smoking. Nevertheless, in a study of 10 healthy non-smokers and 7 healthy smokers, EGLN2 was found to be expressed at a higher level in airway epithelium of smokers compared with non-smokers (22). In this small, underpowered study of airway epithelial DNA, there was no significant evidence for differential DNA methylation of EGLN2 between smokers and non-smokers.
In this study, we set out to determine whether rs7937 is involved in regulatory mechanisms like DNA methylation and gene expression and whether these mechanisms are also associated with COPD. We further evaluated the role of smoking in these regulatory mechanisms. For that purpose, we performed an epigenome-wide association study (EWAS) of rs7937 in blood using an array with high coverage (450K) and a transcriptome-wide association study of rs7937 in blood and lung tissues.
Results
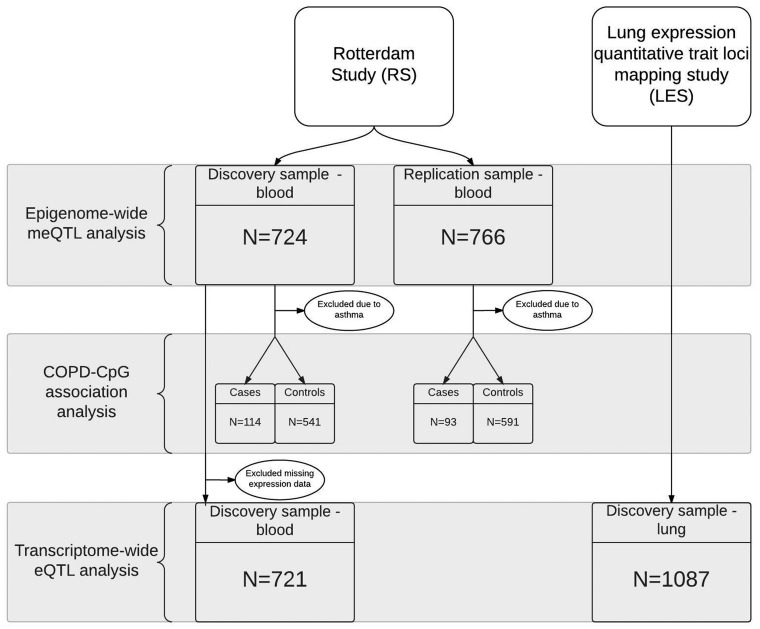
Our discovery cohort comprised 724 participants with genotype and DNA methylation data, while the replication cohort comprised 766 participants from the Rotterdam Study (RS) (23). The summary statistics of the discovery and replication cohorts are shown in Table 1. As expected, the prevalence of males, smokers and the average pack-years of smoking were higher in cases, compared with controls. Compared with the replication cohort, participants in the discovery cohort were on average 8 years younger and included significantly more COPD cases and current smokers, although the pack-years of smoking were comparable. The overview of the analysis pipeline and sample sizes used is presented in Figure 1.
Table 1.
Characteristics of the discovery and replication cohorts and per COPD status
| Discovery cohort |
Replication cohort |
|||||
|---|---|---|---|---|---|---|
| COPD | Controls | All | COPD | Controls | All | |
| N (% of all) | 114 (15.7)a | 541 (74.7) | 724 | 93 (12.1)a | 591 (77.2) | 766 |
| Age (years)a | 61.9±8.6 | 59.3±7.9 | 59.9±8.2 | 68.2±5.7 | 67.6±5.9 | 67.7±5.9 |
| Males (%) | 68 (59.6) | 233 (43.1) | 331 (45.7) | 54 (58.1) | 249 (42.1) | 324 (42.3) |
| FEV1/FVC(% of all) | 0.63±0.07(71.9) | 0.78±0.04(66.0) | 0.76±0.08(67.0) | 0.63±0.07(95.7) | 0.79±0.05(91.0) | 0.76±0.08(91.8) |
| Current smokers, n (%)a | 43 (37.7) | 107 (19.8) | 168 (23.2) | 20 (21.5) | 52 (8.8) | 80 (10.4) |
| Ex-smokers, n (%) | 55 (48.2) | 239 (44.2) | 322 (44.5) | 51 (54.8) | 326 (55.2) | 427 (55.7) |
| Never smokers, n (%) | 16 (14.0) | 195 (36.0) | 234 (32.3) | 22 (23.7) | 213 (36.0) | 259 (33.8) |
| Pack-yearsb | 34.3±26.9 | 19.9±19.6 | 23.2±22.0 | 33.7±18.7 | 19.6±20.1 | 21.9±20.6 |
Data for quantitative measures presented as mean ± SD. COPD: Chronic Obstructive Pulmonary Disease cases; All: all participants included in EWAS. For traits that were not available for all participants (COPD status and FEV1/FVC), the valid percentage is denoted in brackets (% of all).
Significantly different between the discovery and replication cohort.
Pack-years data were available for all participants (mean and SD calculated in current and ex-smokers only).
Figure 1.
Analysis pipeline and datasets overview.
Methylation quantitative trait locus analysis
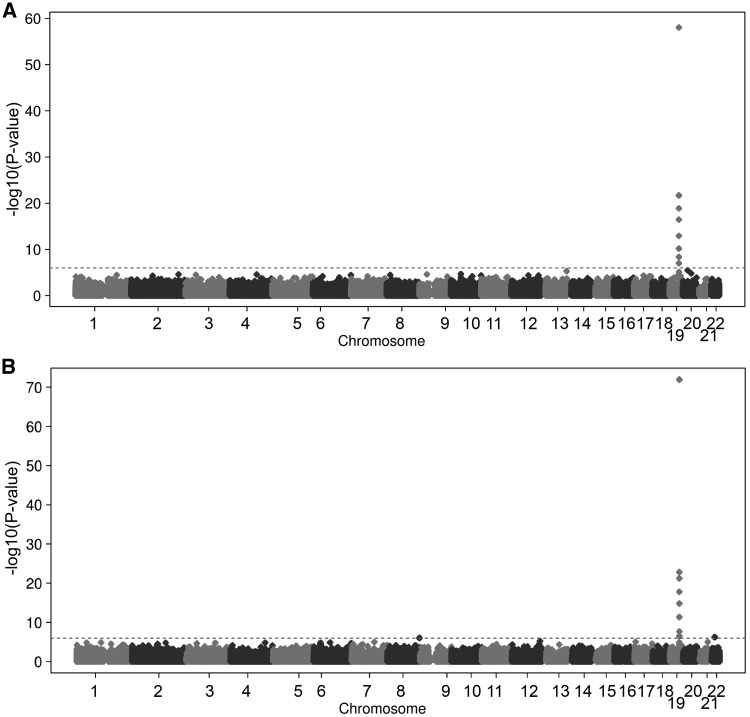
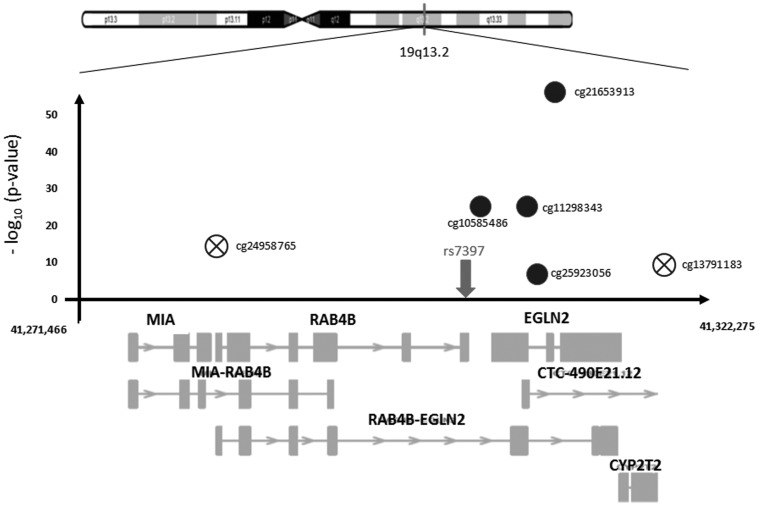
In the genome-wide blood methylation quantitative trait locus (meQTL) analysis of rs7937 in the discovery cohort, rs7937 was significantly [False Discovery Rate (FDR) <0.05] associated with differential DNA methylation at 6 CpG sites in the genes ITPKC and EGLN2, located within the same 19q13.2 region (Model 1, Table 2, Fig. 2A). Five of the six methylation sites were available in the replication dataset and four were significantly replicated with the same direction as found in the discovery cohort (Table 2, Figs 2B and 3). Adding smoking as a confounder (Model 2, Table 2) and testing interaction with smoking (Model 3, data not shown) did not change the results, suggesting that the association between rs7937 and DNA methylation at these sites is independent of smoking. In an additional Model 4, we show that adding COPD to the model did not change the effect of rs7937 on DNA methylation (Supplementary Material, Table S1).
Table 2.
Association of rs7937-T with epigenome-wide DNA methylation in discovery (N = 724) and replication (N = 766) cohorts
| Discovery |
Replication |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Chr | Position | Gene | Model | β | SE | P | FDR | β | SE | P | FDR |
| cg21653913 | 19 | 41307778 | EGLN2 | 1 | −0.08925 | 0.00496 | 9.17 × 10−59 | 4.35 × 10−53 | −0.10030 | 0.00491 | 1.13 × 10−72 | 4.74 × 10−67 |
| 2 | −0.08956 | 0.00497 | 5.70 × 10−59 | 2.71 × 10−53 | −0.10059 | 0.00492 | 1.17 × 10−72 | 4.92 × 10−67 | ||||
| cg11298343 | 19 | 41306150 | EGLN2 | 1 | −0.02036 | 0.00201 | 1.94 × 10−22 | 3.53 × 10−17 | −0.01410 | 0.00141 | 5.42 × 10−22 | 6.36 × 10−17 |
| 2 | −0.02070 | 0.00199 | 1.37 × 10−23 | 3.25 × 10−18 | −0.01402 | 0.00140 | 5.30 × 10−22 | 7.42 × 10−17 | ||||
| cg10585486 | 19 | 41304133 | EGLN2 | 1 | −0.00901 | 0.00089 | 2.23 × 10−22 | 3.53 × 10−17 | −0.01349 | 0.00130 | 1.47 × 10−23 | 3.09 × 10−18 |
| 2 | −0.00902 | 0.00089 | 2.19 × 10−22 | 3.47 × 10−17 | −0.01347 | 0.00130 | 2.25 × 10−23 | 4.72 × 10−18 | ||||
| cg24958765 | 19 | 41283667 | RAB4B | 1 | 0.00235 | 0.00031 | 1.17 × 10−13 | 9.27 × 10−9 | −0.00427 | 0.00075 | 2.02 × 10−8 | 1.06 × 10−3 |
| 2 | 0.00237 | 0.00031 | 1.02 × 10−13 | 8.06 × 10−9 | −0.00433 | 0.00075 | 1.26 × 10−8 | 6.62 × 10−4 | ||||
| cg13791183 | 19 | 41316697 | CYP2A6- AK097370 | 1 | 0.01244 | 0.00209 | 4.23 × 10−9 | 2.51 × 10−4 | NA | |||
| 2 | 0.01255 | 0.00209 | 3.28 × 10−9 | 1.94 × 10−4 | ||||||||
| cg25923056 | 19 | 41306455 | EGLN2 | 1 | −0.00952 | 0.00177 | 1.00 × 10−7 | 5.30 × 10−3 | −0.01153 | 0.00141 | 1.50 × 10−15 | 1.05 × 10−10 |
| 2 | −0.00985 | 0.00175 | 2.54 × 10−8 | 1.34 × 10−3 | −0.01140 | 0.00141 | 2.73 × 10−15 | 1.91 × 10−10 | ||||
β: Regression coefficient estimates from linear regression model regressing DNA methylation levels on indicated SNPs. In Model 1 coefficients are corrected for sex, age, technical covariates and different white blood cellular proportions. In Model 2 coefficients are additionally adjusted for current smoking and pack-years smoked. SE: standard error of the effect, P: P-value of the significance, FDR: False discovery rate value. NA: Not available in the replication dataset.
Figure 2.
Association of the rs7937 with DNA methylation across the genome. In circles are represented all CpGs throughout the genome. X-axis shows chromosome locations; Y-axis shows negative logarithm of the P-value of the associations of the SNP with each CpG site. Dotted line represents the significance threshold (FDR < 0.05). (A) Discovery analysis; (B) Replication analysis.
Figure 3.
Region plot of chromosome 19q13.2 with significant SNP-CpG associations. The circles represent SNP-CpG associations; X-axis shows all genes in the region; Y-axis shows negative logarithm of the P-values of the associations of CpGs with the SNP. Crossed circles represent the non-replicated associations.
COPD and FEV1/FVC analyses
When testing for association of DNA methylation at the four replicated differentially methylated CpG sites with COPD, we observed a significant association with cg11298343 (EGLN2) in Model 1 [β (SE)=−7.080 (2.16), P = 0.001] (Table 3, Supplementary Material, Table S2), which remained nominally significant with diminished but still strong and concordant negative effect, after adjusting for smoking [Model 2; β (SE) = −4.924 (2.25), P = 0.029]. Further, we show that additionally adjusting for rs7937 slightly deteriorated the effect of cg11298343 on COPD (Model 4; Supplementary Material, Table S1).
Table 3.
Association of DNA methylation at significant CpG sites in 19q13.2, with COPD and FEV1/FVC ratio – meta-analysis results
| Trait | CpG | N | Model | β | SE | P |
|---|---|---|---|---|---|---|
| COPD | cg21653913 | 1339 | 1 | −0.084 | 0.711 | 0.906 |
| 2 | 0.175 | 0.738 | 0.812 | |||
| cg11298343 | 1339 | 1 | −7.080 | 2.163 | 0.001 | |
| 2 | −4.924 | 2.254 | 0.029 | |||
| cg10585486 | 1339 | 1 | −2.215 | 3.657 | 0.545 | |
| 2 | −3.110 | 3.811 | 0.415 | |||
| cg25923056 | 1339 | 1 | −0.368 | 2.153 | 0.864 | |
| 2 | 1.551 | 2.255 | 0.492 | |||
| FEV1/FVC | cg21653913 | 1188 | 1 | −0.008 | 0.020 | 0.676 |
| 2 | −0.017 | 0.020 | 0.382 | |||
| cg11298343 | 1188 | 1 | 0.138 | 0.065 | 0.035 | |
| 2 | 0.047 | 0.064 | 0.464 | |||
| cg10585486 | 1188 | 1 | 0.096 | 0.093 | 0.304 | |
| 2 | 0.094 | 0.090 | 0.299 | |||
| cg25923056 | 1188 | 1 | −0.021 | 0.064 | 0.743 | |
| 2 | −0.098 | 0.062 | 0.114 |
N: number of participants in the meta-analysis; Model 1 is adjusted for age, sex, technical covariates and different white blood cellular proportions, Model 2 is additionally adjusted for smoking; β: Regression coefficient estimates from logistic/linear regression models; SE: standard error of the effect; P: P-value of the significance. In bold: nominally significant associations.
In the association with the quantitative determinant of COPD (Table 3, Supplementary Material, Table S2), the ratio of forced expiratory volume in 1 s (FEV1) over the forced vital capacity (FVC), we observed nominal significance for the same site [Model 1; β (SE)=0.138 (0.07), P = 0.04], which deteriorated with adjusting for smoking [Model 2; β (SE)=0.047 (0.06), P = 0.46].
Blood and lung expression quantitative trait loci analysis
In the genome-wide blood expression quantitative trait loci (eQTL) analysis in RS, rs7937 was significantly associated with differential expression of the ILMN_2354391 probe in the EGLN2 gene [β (SE)=0.064 (0.01), P = 9.3 × 10−9], (Table 4). The risk allele (T) was associated with increased expression of EGLN2 (Supplementary Material, Fig. S1). The association signal dropped albeit remained significant [β (SE)=0.058 (0.01), P = 1.9 × 10−6] after adjusting for the top CpG site cg11298343 in the same gene (Table 4). This suggests that differential DNA methylation at cg11298343 is partly responsible for the differential expression of ILMN_2354391 in EGLN2 in blood. In further investigation, we performed the formal mediation analysis where we show (Table 5) that 42% of the association between rs7937 and EGLN2 expression is indeed mediated through cg11298343 (P = 0.04).
Table 4.
Association of rs7937 with transcriptome-wide gene expression in blood (N = 721)
| SNP | A1 | A2 | Probe | Chr | Position (GRCh37/hg19) | Gene | Model | β | SE | P | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7937 | T | C | NM_080732.1 | 19 | 46006086-46006135 | EGLN2 | 1 | 0.0635 | 0.0109 | 9.29 × 10−9 | 0.000197 |
| 2 | 0.0577 | 0.0120 | 1.88 × 10−6 | 0.039942 |
A1: effect allele (tested allele), A2: alternative allele, Chr: chromosome of the probe, Model 1 is adjusted for age, sex, current smoking, technical covariates and different white blood cellular proportions, Model 2 is additionally adjusted for cg11298343; β: Regression coefficient estimates from linear regression models regressing gene expression on indicated SNP, SE: standard error, P: P-value of the significance, FDR: False discovery rate value.
Table 5.
Mediation of the rs7937-EGLN2 expression association through cg11298343 (N = 721)
| Estimate | 95%CI Lower | 95%CI Upper | P-value | |
|---|---|---|---|---|
| ACME | 0.02 | 0.005 | 0.032 | 0.01 |
| ADE | 0.02 | −0.014 | 0.068 | 0.22 |
| Total Effect | 0.04 | 0.005 | 0.080 | 0.03 |
| Proportion Mediated | 0.42 | 0.058 | 1.995 | 0.04 |
ACME: average causal mediation effect by DNA methylation at cg11298343; ADE: average direct effect of rs7937 on EGLN2 expression; Total: total effect rs7937 on EGLN2 expression; Proportion Mediated: proportion of the association between rs7937 and EGLN2 expression, explained by methylation at cg11298343; Regression models adjusted for sex, age, current smoking, pack-years, technical variance and estimated blood cell composition; In bold: significant results with P-value < 0.05.
Moreover, genome-wide eQTL analysis of rs7937 in lung tissue of 1087 participants from the Lung expression quantitative loci mapping study (LES), showed significant associations (P < 1.36 × 10−6) with 5 probes in the same region [in cis; AK097370(EGLN2), NUMBL] and other chromosomes [in trans; LOC101929709, DNMT3A and PAK2] (Table 6). In all cases, except one (LOC101929709 at chromosome 8), the T allele of rs7937 was consistently associated with decreased expression of the genes in lung tissue (Table 6, Supplementary Material, Fig. S2).
Table 6.
Association of rs7937 with transcriptome-wide gene expression in lung tissue (N = 1087)
| SNP | A1 | A2 | Probe | Chr | Position (GRCh37/hg19) | Annotation | β | SE | P | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7937 | T | C | NM_004756 | 19 | 40665906-40690658 | NUMBL | −0.099 | 0.013 | 4.39 × 10−15 | 1.17 × 10−11 |
| BC037804 | 19 | 40808443-40810818 | AK097370 (EGLN2) | −0.077 | 0.012 | 3.77 × 10−10 | 1.01 × 10−6 | |||
| BX330016 | 8 | 89720919-89724906 | LOC101929709 | 0.049 | 0.011 | 6.15 × 10−6 | 0.016 | |||
| AK025230 | 2 | 25233434-25246179 | DNMT3A | −0.028 | 0.006 | 8.00 × 10−6 | 0.021 | |||
| BQ445924 | 3 | 196829093-196830253 | PAK2 | −0.028 | 0.006 | 1.36 × 10−5 | 0.036 |
A1: effect allele (tested allele), A2: alternative allele, β: Regression coefficient estimates from linear regression models regressing gene expression on indicated SNP, SE: standard error, P: P-value of the significance, FDR: False discovery rate value.
Discussion
Our study shows that the rs7937 in 19q13.2 is associated with differential blood DNA methylation of 4 CpG sites located in the EGLN2 in the discovery and replication cohort. The COPD risk allele (T) is associated with lower DNA methylation at these sites. These relationships are independent of smoking and of COPD. We further show that DNA methylation in blood at cg11298343 (EGLN2) is associated with COPD, and remains nominally significant after adjusting for smoking. Finally, rs7937 is associated with differential expression in blood of EGLN2, 42% explained by EGLN2 DNA methylation at site cg11298343, and in lung tissue of NUMBL, AK097370 (EGLN2), LOC101929709, DNMT3A and PAK2.
EGLN2 is coding prolyl hydroxylase domain-containing protein 1 (PHD1) which regulates posttranscriptional modifications of hypoxia induced factor (HIF), a transcriptional complex involved in oxygen homeostasis. At normal oxygen levels, the alpha subunit of HIF is targeted for degradation by PHD1, which is an essential component of the pathway through which cells sense oxygen (24), is also known to be involved in activation of inflammatory and immune genes, including those implicated in COPD (25). Furthermore, read-through transcription exists between this gene and the upstream RAB4B, and together they were shown to be involved in invasive lung cancer (26).
Our study of DNA methylation in blood replicates the findings of a study on meQTLs in blood across the human life course: during pregnancy, at birth, childhood, adolescence and middle age (27) which reported three (cg10585486, cg11298343, cg25923056) out of our four replicated CpGs. We additionally report a novel finding in association with rs7937, our top hit, cg21653913. Interestingly, they report differential DNA methylation at cg11298343 to be associated with rs7937 at all five time points. In the present study, we now show that rs7937 is also associated with cg11298343 in our elderly sample, the age category at highest risk for developing COPD. This finding goes in line with our hypothesis that the life-long change in the DNA methylation is involved in the pathogenesis and onset of COPD in older age, rather than the other way around. However, further longitudinal studies are needed, testing this hypothesis in lung tissue.
In line with our findings, rs7937 has previously been associated with differential expression of EGLN2 in blood (28). Having both DNA methylation and transcription data available, we could further test whether the relation of rs7937 and EGLN2 expression could be explained by DNA methylation levels of EGLN2 at cg11298343 site. We show for the first time that in blood there is indeed a mediation of 42% through the DNA methylation at cg11298343, confirming our hypothesis. We report two novel findings in this region in the lung tissue. We found that rs7937 is involved in expression of AK097370 in lung tissue, a DNA clone in the proximity of EGLN2, as well as with NUMBL. NUMBL is known as a negative regulator of NF-kappa-B signaling pathway in neurons (29) and was also found to be expressed in the lungs in the GTEx database (30). The association between rs7937 and gene expression in the same dataset has been tested earlier by Lamontagne et al. (31) but no significant results were reported. In the present study, using a more powerful meta-analysis approach, rs7937 was associated with two loci in the region (NUMBL and AK097370, close to EGLN2) and three more loci in other chromosomes (LOC101929709, DNMT3A and PAK2). Taken together, our findings raise the hypothesis that the genetic effect of rs7937 on COPD might be mediated by DNA methylation at cg11298343 and subsequent alteration of expression of EGLN2 and other genes in this region, such as NUMBL. We show this in blood but further formal mediation analyses in lung tissue are needed to confirm this hypothesis, requiring the assembly of a large dataset of lung tissue characterized for genetic, epigenetic and transcriptomic data.
Furthermore, we have found significant associations in lungs of rs7937 in trans, i.e. with the expression of genes on other chromosomes. These effects include differential expression of PAK2 (chromosome 3), DNMT3A (chromosome 2) and long non-coding RNA on chromosome 8. The protein encoded by PAK2 gene is activated by proteolytic cleavage during caspase-mediated apoptosis, and may play a role in regulating the apoptotic events in the dying cell (32). DNMT3A is the gene encoding the DNA methyltransferase which plays a key role in de novo methylation. This may imply that rs7937 is involved in the pathogenesis of COPD through differential DNA methylation and regulation of expression throughout the genome, again asking for further research of DNA methylation.
The strength of our analysis is the use of large and unique samples of patients whose genetic, epigenetic and transcriptomic characteristics were assessed in detail. However, a limitation of our study is the use of blood tissue for the assessment of DNA methylation and gene expression. Nevertheless, our findings regarding the role of the genetic variants in blood corroborate with the changes in the transcriptome in lung tissue. It has been shown that blood can be used to evaluate methylation changes related to COPD and smoking, as the disease induces systemic changes associated with elevated markers of systemic inflammation in blood (25). A second limitation of our study is that we cannot distinguish the expression in lung parenchymal tissue, which comprises multiple cell types. It may be speculated that expression of only distinct cells is affected by rs7939. If this is the case, the most likely effect is that the power of our study is reduced, but has not biased our findings in the sense of generating false positives. However, although eQTLs are frequently cell- and tissue-specific (33), many eQTLs are also shared across tissues (34). Finally, the number of patients with COPD and spirometry measures were limited, which may have compromised the power of the study. Nevertheless, we observed significant findings that are relevant for COPD.
In conclusion, our findings suggest that genetic variations underlying EGLN2 methylation contribute to the risk of developing COPD. This finding adds insight into how genetic variants are involved in the pathogenesis of COPD, through differential DNA methylation and regulation of expression, irrespective of smoking. Future integrative studies involving genetics, epigenetics and transcriptomics in lung tissue are crucial to elucidate the molecular mechanisms behind COPD genetic susceptibility and to translate the findings to clinical care and prevention. This may lead to an increased specificity and sensitivity of diagnostic and prognostic tools. In addition, novel DNA methylation loci may be used as a target for future drug design in COPD. While smoking cessation is shown to be a useful prevention tool for disease risk and mortality reduction, DNA methylation loci independent of smoking may be used as a target for a more personalized and focused treatment approach.
Materials and Methods
Study population
For our analyses in blood we used two independent subsets of participants from the RS (23). The full discovery set for meQTL analysis was comprised of 724 participants with full genomic and epigenetic data, while replication set included 766 participants. The replication subset is part of the Biobanking and Biomolecular Resources Research Infrastructure for The Netherlands (BBMRI-NL), BIOS (Biobank-based Integrative Omics Studies) project (35). RS has been approved by the Medical Ethics Committee of the Erasmus MC and by the Ministry of Health, Welfare and Sport of the Netherlands, implementing the Population Studies Act: Rotterdam Study. All participants provided written informed consent to participate in the study and to obtain information from their treating physicians. The detailed information on our samples can be found in Supplementary Material.
Spirometry measures and COPD diagnosis
From the initial full datasets for meQTL analysis (ndiscovery=724, nreplication=766), after excluding participants with asthma, we used data from 655 participants in the discovery and 684 participants in the replication cohort for the association analyses with COPD. COPD diagnosis was defined as pre-bronchodilator FEV1/FVC < 0.7. More detailed information can be found in Supplementary Material.
COPD SNPs selection
Using the GWAS catalog (36) on 15 January 2017, we performed a search with the term ‘19q13.2’, additionally applying filters for the P-value ≤ 5 × 10−8 and for the trait to include term ‘Chronic obstructive pulmonary disease’. One GWAS study passed these filtering criteria, and reported the top SNP, rs7937-T (NC_000019.10: g.40796801C > T), to be associated with COPD (OR = 1.37) (9). We used this SNP to perform all analyses in our study.
Genotyping in RS
Genotyping was performed using 610K and 660K Illumina arrays for which whole blood genomic DNA was used. Detailed information can be found in Supplementary Material. Imputation was done using 1000 Genomes (1KG) phase I v3 reference panel, with measured genotypes that had minor allele frequencies (MAF)>1%, performed with MacH software and Minimac implementation. We extracted dosages of rs7937 [risk allele T, allele frequency = 0.54], from RS imputed data using DatABEL library of R-package (37).
DNA methylation array in RS
We used Illumina Infinium Human Methylation 450K array to quantify DNA methylation levels across the genome from whole blood in RS. The detailed QC and normalization criteria can be found in the Supplementary Material. Ultimately, after the QC and normalization steps our discovery set included 724 Caucasian participants and 463 456 probes, while the replication set included 766 Caucasian participants and 419 936 probes.
RNA array in blood in RS
In the discovery sample, we used the same blood samples at baseline to isolate RNA, which we hybridized to Illumina Whole-Genome Expression Beadchips Human HT-12 v4 array. Raw probe intensities were quantile-normalized and 2-log transformed and controlled for quality as described elsewhere (28). After all normalization and QC steps the sample consisted of 21 238 probes in 721 participants with available full data on SNP and RNA arrays and all covariates.
RNA array in lung tissue
Gene expression was quantified using lung tissue samples obtained from patients that underwent lung resection surgery at three facilities participating in the LES: University of Groningen (GRN), Laval University (Laval) and University of British Columbia (UBC) (38). Illumina Human1M-Duo BeadChip arrays were used for genotyping, and a custom Affymetrix microarray (GPL10379) for gene expression profiling. The final dataset for the eQTL analysis consisted of 1087 subjects. More detailed information can be found in Supplementary Material.
Statistical analyses
meQTL analysis
We performed EWAS in the discovery cohort using linear regression analysis with rs7937-T as independent variable and DNA methylation sites as dependent variable. We fitted two models; first adjusted for age, sex, technical covariates to correct for batch effects (array number and position on array) and the estimated white blood cell counts (39) (including monocytes, T-lymphocytes: CD4 and CD8, B-lymphocytes, natural killer cells, neutrophils and eosinophils) (Model 1); and second, for significant sites additionally adjusted for current smoking and pack-years smoked (Model 2). We used the FDR < 0.05 as an epigenome-wide significance threshold (40). Significant sites from Model 1 were then tested for association in the replication cohort using the same models as in the discovery. Since the 19q13.2 region was also implicated in smoking behavior, we also tested significant CpG sites in a third model including ‘rs7937 × smoking’ interaction term to assess possible interaction between rs7937 and smoking (for both current smoking and pack-years smoked), in the discovery and replication cohorts. In addition, for the significant sites we used another model additionally adjusted for COPD status which we compared with the Model 1 in attempt to further elucidate the direction of the effect between DNA methylation and COPD.
COPD and FEV1/FVC analysis
To test if the significantly associated methylation sites are also associated with the lung phenotypes, we performed logistic and linear regression analyses with COPD and FEV1/FVC ratio, respectively as dependent variables and DNA methylation as independent variable. In the first model we adjusted for age, sex, technical covariates and estimated white blood cell counts and additionally for current smoking and pack-years smoked in the second model, in both the discovery and the replication cohort. Results from the two cohorts were meta-analysed using fixed effect models with ‘rmeta’ package in R (41). Bonferroni correction was applied to adjust for multiple testing. In addition, for the significant sites we used another model additionally adjusted for rs7937 which we compared with the first model in attempt to further elucidate the direction of the effect between DNA methylation and COPD.
Blood eQTL analysis
In the discovery cohort, we tested whether rs7937 is associated with differential expression in the whole blood. We used linear regression analysis with rs7937 as independent variable and genome-wide normalized gene expression as dependent variable. For this analysis, we used model adjusted for age, sex, current smoking, technical batch effects (plate ID and RNA quality) and white blood cell counts (lymphocytes, monocytes and granulocytes). For significant (FDR < 0.05) probes, the second model was additionally adjusted for significant DNA methylation levels.
Mediation analysis
We have performed formal mediation analysis using the bootstrapping method in the ‘mediation’ package in R (42), to assess the potential mediator role of significant DNA methylation in the SNP-expression association. One thousand bootstraps were run to estimate the confidence intervals (43). We used models adjusted for age, sex, current smoking, pack-years, expression technical batch effects (plate ID and RNA quality), methylation technical batch effects (position on array and array number) and estimated blood cell composition.
Lung eQTL analysis
To test if the SNP rs7937 is associated (FDR < 0.05) with differential expression in lung tissue in LES, we performed a genome-wide linear regression analysis with the SNP as the independent variable and 2-log transformed gene expression levels as dependent variable. This analysis was performed for each of the three participating cohorts (GRN, Laval and UBC) separately, adjusted for lung disease status, age, sex, smoking status and cohort-specific principal components (PCs). The inverse-variance weighted fixed effect meta-analysis of the results obtained from the three cohorts was performed with ‘rmeta’ package in R software. The detailed overview of the fitted models can be found in the Supplementary Material.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The generation and management of the Illumina 450K methylation array data and Illumina Whole-Genome Expression Beadchips Human HT-12 v4 array data for the Rotterdam Study were executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, The Netherlands. We thank Michael Verbiest, Mila Jhamai, Sarah Higgins, Marijn Verkerk and Lisette Stolk for their help in creating the methylation database. We also thank Mila Jhamai, Sarah Higgins, Marjolein Peters, Marijn Verkerk and Jeroen van Rooij for their help in creating the RNA array expression database. The authors are grateful to the study participants, the participating general practitioners and pharmacists and the staff from the Rotterdam Study as well as the staff from the Respiratory Health Network Tissue Bank of the FRQS for their valuable assistance with the Lung eQTL dataset at Laval University. Finally, we would like to acknowledge members of the BIOS Consortium (https://www.bbmri.nl/? p=259; date last accessed May 26, 2017).
Conflict of Interest statement. None declared.
Funding
This study is sponsored by Lung Foundation (Longfonds), the Netherlands under grant number 4.1.13.007 (D.A.v.d.P., K.d.J. and N.A. were supported by the same grant). I.N. was supported by the ERAWEB scholarship. L.L. was a Postdoctoral Fellow of the Research Foundation-Flanders (FWO). E.C.-M. was funded by a TALENTIA fellowship program from Andalusian Region Government, Spain. M.O. is a fellow of the Parker B. Francis Foundation. Y.B. holds a Canada Research Chair in Genomics of Heart and Lung Diseases. G.B. coordinates the Concerted Research Action BOF14/GOA/027, funded by Ghent University. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The generation and management of the EWAS and RNA-expression data of Rotterdam Study were executed and funded by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC and by the Netherlands Organization for Scientific Research (NWO; project number 184021007). The Lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l'Université Laval, the Fondation de l'Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP - 123369), and the Cancer Research Society and Read for the Cure. The sponsors of this study played no role in the design of the study, data collection, analysis, interpretation or in the writing and submission of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Erasmus Medical Center, Rotterdam and Longfonds consortium.
References
- 1. Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S.Y.. et al. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet, 380, 2095.. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Jong K., Vonk J.M., Timens W., Bossé Y., Sin D.D., Hao K., Kromhout H., Vermeulen R., Postma D.S., Boezen H.M. (2015) Genome-wide interaction study of gene-by-occupational exposure and effects on FEV1 levels. J. Aller. Clin. Immunol., 136, 1664–1672. e1614. [DOI] [PubMed] [Google Scholar]
- 3. Gibson G.J., Loddenkemper R., Lundbäck B., Sibille Y. (2013) European Respiratory Society (in press). [DOI] [PubMed]
- 4. Burrows B., Knudson R.J., Cline M.G., Lebowitz M.D. (1977) Quantitative Relationships between Cigarette Smoking and Ventilatory Function 1, 2. Am. Rev. Respir. Dis., 115, 195–205. [DOI] [PubMed] [Google Scholar]
- 5. Higgins M.W., Keller J.B., Landis J.R., Beaty T.H., Burrows B., Demets D., Diem J.E., Higgins I.T., Lakatos E., Lebowitz M.D.. et al. (1984) Risk of chronic obstructive pulmonary disease. Collaborative assessment of the validity of the Tecumseh index of risk. Am. Rev. Respir. Dis., 130, 380–385. [DOI] [PubMed] [Google Scholar]
- 6. McCloskey S.C., Patel B.D., Hinchliffe S.J., Reid E.D., Wareham N.J., Lomas D.A. (2001) Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am. J. Respir. Crit. Care Med., 164, 1419–1424. [DOI] [PubMed] [Google Scholar]
- 7. Silverman E.K., Chapman H.A., Drazen J.M., Weiss S.T., Rosner B., Campbell E.J., O'Donnell W.J., Reilly J.J., Ginns L., Mentzer S.. et al. (1998) Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med., 157, 1770–1778. [DOI] [PubMed] [Google Scholar]
- 8. Boezen H.M. (2009) Genome-wide association studies: what do they teach us about asthma and chronic obstructive pulmonary disease?. Proc. Am. Thorac. Soc., 6, 701–703. [DOI] [PubMed] [Google Scholar]
- 9. Cho M.H., Castaldi P.J., Wan E.S., Siedlinski M., Hersh C.P., Demeo D.L., Himes B.E., Sylvia J.S., Klanderman B.J., Ziniti J.P.. et al. (2012) A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet., 21, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho M.H., McDonald M.-L.N., Zhou X., Mattheisen M., Castaldi P.J., Hersh C.P., DeMeo D.L., Sylvia J.S., Ziniti J., Laird N.M.. et al. (2014) Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med., 2, 214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A.. et al. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet., 5, e1000421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang Q. (2015) Genetic study of complex diseases in the post-GWAS era. J. Genet. Genomics, 42, 87–98. [DOI] [PubMed] [Google Scholar]
- 13. Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. U S A, 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J.. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu W., Baccarelli A., Carey V.J., Boutaoui N., Bacherman H., Klanderman B., Rennard S., Agusti A., Anderson W., Lomas D.A.. et al. (2012) Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am. J. Respir. Crit. Care Med., 185, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vucic E.A., Chari R., Thu K.L., Wilson I.M., Cotton A.M., Kennett J.Y., Zhang M., Lonergan K.M., Steiling K., Brown C.J.. et al. (2014) DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am. J. Respir. Cell Mol. Biol., 50, 912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiu W., Wan E., Morrow J., Cho M.H., Crapo J.D., Silverman E.K., DeMeo D.L. (2015) The impact of genetic variation and cigarette smoke on DNA methylation in current and former smokers from the COPDGene study. Epigenetics, 10, 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S.. et al. (2010) Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet., 42, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tobacco and Genetics, C. (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet., 42, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Artigas M.S., Wain L.V., Miller S., Kheirallah A.K., Huffman J.E., Ntalla I., Shrine N., Trochet H., McArdle W.L., Alves A.C. (2015) Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat. Commun., 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castaldi P.J., Cho M.H., San José Estépar R., McDonald M.-L.N., Laird N., Beaty T.H., Washko G., Crapo J.D., Silverman E.K. (2014) Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am. J. Respir. Crit. Care Med., 190, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryan D.M., Vincent T.L., Salit J., Walters M.S., Agosto-Perez F., Shaykhiev R., Strulovici-Barel Y., Downey R.J., Buro-Auriemma L.J., Staudt M.R.. et al. (2014) Smoking dysregulates the human airway basal cell transcriptome at COPD risk locus 19q13.2. PLoS One, 9, e88051.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hofman A., Brusselle G.G., Darwish Murad S., van Duijn C.M., Franco O.H., Goedegebure A., Ikram M.A., Klaver C.C., Nijsten T.E., Peeters R.P.. et al. (2015) The Rotterdam Study: 2016 objectives and design update. Eur. J. Epidemiol., 30, 661–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruick R.K., McKnight S.L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science, 294, 1337–1340. 10.1126/science.1066373 [DOI] [PubMed] [Google Scholar]
- 25. Cummins E.P., Berra E., Comerford K.M., Ginouves A., Fitzgerald K.T., Seeballuck F., Godson C., Nielsen J.E., Moynagh P., Pouyssegur J.. et al. (2006) Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad. Sci. U S A, 103, 18154–18159. 10.1073/pnas.0602235103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu Y.C., Yuan S., Chen H.Y., Yu S.L., Liu C.H., Hsu P.Y., Wu G., Lin C.H., Chang G.C., Li K.C.. et al. (2009) A four-gene signature from NCI-60 cell line for survival prediction in non-small cell lung cancer. Clin. Cancer Res., 15, 7309–7315. [DOI] [PubMed] [Google Scholar]
- 27. Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K.. et al. (2016) Systematic identification of genetic influences on methylation across the human life course. Genome Biol., 17, 61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westra H.-J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E.. et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Q., Zhou L., Shi H., Huo K. (2008) NUMBL interacts with TAB2 and inhibits TNFalpha and IL-1beta-induced NF-kappaB activation. Cell Signal, 20, 1044–1051. 10.1016/j.cellsig.2008.01.015 [DOI] [PubMed] [Google Scholar]
- 30. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N.. et al. (2013) The Genotype-Tissue Expression (GTEx) project. 45, Nat. Genet., 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamontagne M., Couture C., Postma D.S., Timens W., Sin D.D., Paré P.D., Hogg J.C., Nickle D., Laviolette M., Bossé Y., Miao X.-P. (2013) Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One, 8, e70220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marlin J.W., Eaton A., Montano G.T., Chang Y.W., Jakobi R. (2009) Elevated p21-activated kinase 2 activity results in anchorage-independent growth and resistance to anticancer drug-induced cell death. Neoplasia, 11, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu J., Wolfs M.G.M., Deelen P., Westra H.-J., Fehrmann R.S.N., te Meerman G.J., Buurman W.A., Rensen S.S.M., Groen H.J.M., Weersma R.K.. et al. (2012) Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet., 8, e1002431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A.. et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet., 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biobanking and BioMolecular Resources research Infrastructure , BIOS project (2016), http://www.bbmri.nl/?p=259.
- 36. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J.. et al. (2017) The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research, 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aulchenko Y., Struchalin M., Ripke S., Johnson T. (2010) GenABEL: genome-wide SNP association analysis. R Package Version, 1.6-4 (in press). [Google Scholar]
- 38. Hao K., Bossé Y., Nickle D.C., Paré P.D., Postma D.S., Laviolette M., Sandford A., Hackett T.L., Daley D., Hogg J.C.. et al. (2012) Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet, 8, e1003029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hochberg Y., Benjamini Y. (1990) More powerful procedures for multiple significance testing. Stat Med, 9, 811–818. 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- 41. Lumley T., Lumley M.T. (2006) The rmeta Package (in press).
- 42. Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. (2014) Mediation: R Package for Causal Mediation Analysis (in press).
- 43. Mayer A., Thoemmes F., Rose N., Steyer R., West S.G. (2014) Theory and analysis of total, direct, and indirect causal effects. Multivariate Behav. Res., 49, 425–442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.