Abstract
The interaction of chromatin with the nuclear matrix via matrix attachment regions (MARs) on the DNA is considered to be of fundamental importance for higher-order chromatin organization and the regulation of gene expression. We have previously isolated a novel nuclear matrix-localized protein (MFP1) from tomato (Lycopersicon esculentum) that preferentially binds to MAR DNA. Tomato MFP1 has a predicted filament-protein-like structure and is associated with the nuclear envelope via an N-terminal targeting domain. Based on the antigenic relationship, we report here that MFP1 is conserved in a large number of dicot and monocot species. Several cDNAs were cloned from tobacco (Nicotiana tabacum) and shown to correspond to two tobacco MFP1 genes. Comparison of the primary and predicted secondary structures of MFP1 from tomato, tobacco, and Arabidopsis indicates a high degree of conservation of the N-terminal targeting domain, the overall putative coiled-coil structure of the protein, and the C-terminal DNA-binding domain. In addition, we show that tobacco MFP1 is regulated in an organ-specific and developmental fashion, and that this regulation occurs at the level of transcription or RNA stability.
The interaction of chromatin with the nuclear matrix via matrix attachment regions (MARs) on the DNA is considered to be of fundamental importance for higher-order chromatin organization and the regulation of gene expression. MARs have recently attracted much attention in the plant field because they appear capable of drastically increasing levels of transgene expression and reducing transformant-to-transformant variation of transgene expression (Allen et al., 1993; Allen et al., 1996; Mlynarova et al., 1996). These observations have led to models that attempt to explain the effects of MARs on gene expression by their interaction with the nuclear matrix (reviewed in Spiker and Thompson, 1996). The MAR-nuclear matrix interactions are believed to create independent chromatin loop domains, and the location of a gene with respect to this domain structure is thought to influence its expression level (Spiker and Thompson, 1996). The nuclear matrix has been biochemically defined as the insoluble component that remains after treatment of isolated nuclei with nucleases and extraction of proteins with different methods (Berezney and Coffey, 1974; Mirkovitch et al., 1984). Electron micrographs of the nuclear matrix show a dense network of fibers, similar in appearance to the cytoplasmic cytoskeleton (He et al., 1990). Chromatin loops are presumed to attach to these matrix fibers by protein-DNA interactions with the MARs.
Most investigations of structural components of the nucleus have focused on vertebrates and Drosophila, but even in these organisms, our knowledge about the molecular constituents of the nuclear matrix is sparse. In order to better understand, and, therefore, better predict the effects of MARs on gene expression, it will be necessary to isolate and characterize those proteins of the nuclear matrix that interact with MARs. A small number of MAR-binding proteins have been identified from animal nuclei, and have subsequently been shown to be components of the nuclear matrix (von Kries et al., 1991; Dickinson et al., 1992; Renz and Fackelmayer 1996; Göhring et al., 1997). No homologs to these proteins have been identified yet in plants. Previously, we reported the cloning of the first plant MAR-binding protein, MFP1, from tomato (Meier et al., 1996). MFP1 has the structural features of a filament-like protein, has similarity to nuclear and cytoplasmic filament proteins, and preferentially binds to MAR sequences from both animals and plants. It is thus a first candidate for a protein in plants that acts as a molecular anchor between chromatin and the filaments of the nuclear matrix.
We have extended our studies to investigate MFP1-like proteins in other plant species, and show that a single, immunologically related protein of comparable size is present in a variety of higher plant species, including important crop plants. As tobacco has evolved as the model plant for studies of both the nuclear matrix and the effects of MARs on transgene expression (Hall et al., 1991; Allen et al., 1993; Allen et al., 1996), we have isolated the cDNAs corresponding to two tobacco MFP1 genes and characterized the MFP1 gene family in tobacco. Interestingly, we found that the expression of tobacco MFP1 is not constitutive, but is regulated in an organ-specific and developmental fashion.
MATERIALS AND METHODS
cDNA Library Screening
An oligo(dT)-primed lambda-ZAP library made from tobacco (Nicotiana tabacum var SR1) leaf tissue was purchased from Stratagene (La Jolla, CA). Approximately 600,000 recombinants were screened according to the manufacturer's instructions. 32P-labeled probes were prepared using a DNA-labeling system (Random Primer, BRL, Gaithersburg, MD). Washes were performed at high stringency (0.1× SSC [Sambrook et al., 1989] and 0.1% [w/v] SDS at 65°C). Positive plaques were detected by autoradiography and carried through two subsequent rounds of purification. In vivo excision of positive phage was performed according to the manufacturer's protocol (Stratagene).
PCR
PCR reactions were carried out in a thermocycler (model 9600, Perkin Elmer, Foster City, CA). A 2-min 96°C denaturation cycle was followed by 30 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 90 s, and ended with an 8-min 72°C final extension cycle.
Cloning Techniques
Standard cloning techniques were performed according to the method of Sambrook et al. (1989). The PCR#1 PCR product was cloned into pBluescript KS (Stratagene) utilizing a BamHI and a XbaI site in the respective PCR primers.
Sequencing
DNA sequencing was carried out using an ABI model 377 sequencer (Perkin-Elmer/Applied Biosystems, Foster City, CA). Sequencing reactions utilized fluorescent sequencing techniques with d-rhodamine and Big Dye terminator chemistry (Perkin-Elmer/Applied Biosystems) and were performed according to the manufacturer's protocol.
Protein Expression, Purification, and Antibody Production
pRSETA-MFP1 (Meier et al., 1996) was digested with HindIII and religated to create pRSETA-HindIII, the expression vector for the H-183 fragment (Fig. 1). pRSETC-EcoRI and pRSETA-HincII, the expression vectors for the E-196 and H-207 fragments (Fig. 1), respectively, have been described previously (Meier et al., 1996). Expression of recombinant fusion proteins was induced by isopropyl β-d-thiogalactoside in Escherichia coli BL21 cells according to the instructions of the protein expression manual (Qiagen, Chatsworth, CA). The amount of fusion protein present in the different total E. coli protein extracts was determined by immunoblotting (Sambrook et al., 1989) with a monoclonal antibody directed against the T7 tag (Novagen, Madison, WI). For immunization, the fusion proteins were purified by nickel-affinity chromatography followed by SDS-PAGE. The bands corresponding to the fusion proteins were excised from the gel, and the gel slices were air-dried. Antibodies were produced in rabbits by Eurogentec (Seraing, Belgium) using the company's standard immunization protocols. The a288 antibody has been described previously (Meier et al., 1996).
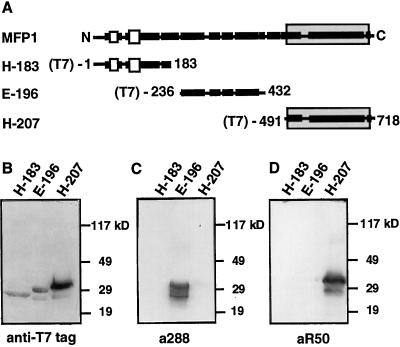
Figure 1.
Specificity of antibodies directed against LeMFP1 subdomains. A, Schematic representation of the subfragments of LeMFP1 that were expressed in E. coli. Black bars indicate predicted α-helical regions; white bars indicate hydrophobic domains. The shaded box marks the DNA-binding domain. Numbers indicate the positions of the first and last amino acid of each subfragment. (T7) indicates the N-terminal T7 tag. B to D, Approximately equal amounts of total protein extracts from E. coli BL21 strains expressing the T7-tagged and His-tagged protein subfragments indicated in A were separated on three replica SDS-PAGE gels and subjected to immunoblotting with a monoclonal antibody directed against the T7 tag (B), the a288 antiserum (E-196) (C), and the aR50 antiserum (H-207) (D). Molecular mass markers are indicated on the right.
Southern-Blot Analysis
Aliquots (20 μg) of DNA, digested with various restriction enzymes and separated on a 0.8% (w/v) agarose gel, were transferred to Immobilon N hydrophobic filters (Millipore, Bedford, MA). Hybridization techniques were essentially as described by Sambrook et al. (1989). The probe was prepared by purification of a 391-bp XhoI/SpeI fragment from the tobacco cDNA clone T3, as described by Sambrook et al. (1989). The probe was labeled with 32P using the DNA labeling system according to the manufacturer's instructions (BRL).
RNA Gel-Blot Analysis
Total RNA (20 or 30 μg) was separated on a formaldehyde gel, blotted onto an Immobilon N membrane (Millipore), and hybridized with a probe prepared by digesting the T1 cDNA clone with EcoRI and XhoI and purifying the 389-bp fragment, essentially as described by Sambrook et al. (1989). Signals were detectable after a 1-week exposure of the blot to x-ray film using an intensifying screen.
Immunoblot Analysis
A 1:3,000 dilution of a288 or aR50 antiserum and a 1:5,000 dilution of horseradish peroxidase-coupled anti-rabbit secondary antibody (Amersham, Buckinghamshire, UK) were used to perform immunoblot analyses as described in Sambrook et al. (1989). Enhanced chemiluminescence detection was performed using the ECL detection kit (Amersham) as described by the manufacturer.
Isolation of Total Protein, Genomic DNA, Total RNA, and Poly(A+) RNA
Total protein extracts were prepared from a variety of plant tissues. Tissue (100 mg) were ground to a fine powder in liquid nitrogen, resuspended in 0.5 mL of extraction buffer (62.5 mm Tris-Cl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, and 1.4 m β-mercaptoethanol), and incubated at 70°C for 10 min. The debris were removed by centrifugation at 15,000 rpm for 10 min at 4°C. The supernatants were transferred to new tubes, frozen in liquid nitrogen, and stored at −80°C. Genomic DNA was isolated from tobacco leaf tissue using a urea buffer extraction method, essentially as described by Chen and Dellaporta (1994). Total RNA was isolated from a variety of tobacco tissues using the TRIZOL reagent (BRL) according to the manufacturer's protocol.
Plant Material and Growth Conditions
All plants were grown in a growth chamber with a 12-h, 24°C light cycle followed by a 12-h, 20°C dark cycle.
Database Searches, Sequence Comparison, and Secondary Structure Prediction
The AtMFP1 genomic DNA sequence was accessed through the Arabidopsis database (http://genome-www.stanford.edu/Arabidopsis/). The deduced protein sequences of the MFP1 proteins were determined and compared using Lasergene software (DNASTAR, Madison, WI). The secondary structures of the proteins, hydrophobicity, α-helical, and coiled-coil regions were analyzed using Protean software (DNASTAR).
RESULTS
Domain-Specific Antibodies Against LeMFP1
Tomato MFP1 (LeMFP1) consists of an extended coiled-coil like α-helical domain and a shorter, N-terminal, non-α-helical region containing two hydrophobic domains (Fig. 1A). The MAR-binding domain has been localized to the C-terminal 226 AA of LeMFP1 (Meier et al., 1996) (shaded in Fig. 1A). To obtain antibodies specifically directed against different domains of the protein, fusion proteins containing an N-terminal 6-His tag and T7 tag fused to the protein subfragments E-196 and H-207 indicated in Figure 1A were expressed in E. coli, purified, and used to raise two rabbit antisera (a288 against E-196 and aR50 against H-207). To test the specificity of the antisera for their antigens, the three fusion proteins H-183, E-196, and H-207 (Fig. 1A) were expressed in E. coli. Figure 1B shows each fusion protein in the respective E. coli protein extracts, detected with an antibody directed against the N-terminal T7 tag. Figure 1, C and D, show replica blots probed with the a288 (Fig. 1C) and aR50 (Fig. 1D) antisera. Both antibodies were found to specifically recognize their antigen and show no cross-reactivity with the other MFP1 domains.
MFP1 Is Conserved among Higher Plants
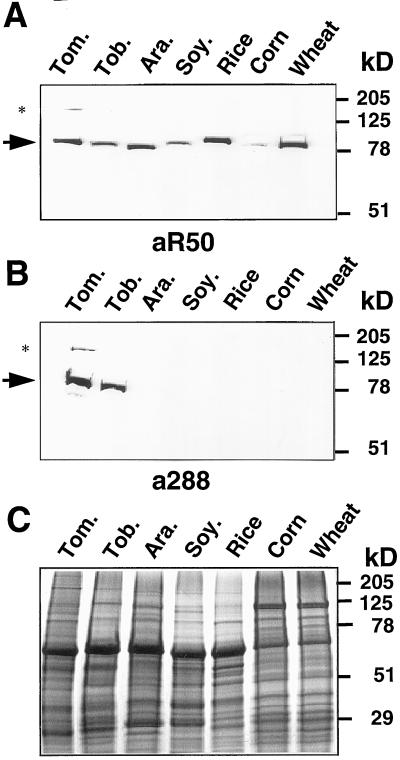
a288 and aR50 were then used to detect proteins with antigenic similarity to MFP1 in other plant species. Total protein extracts were prepared from leaf tissues of tomato (Lycopersicon esculentum L.), tobacco (Nicotiana tabacum L.), Arabidopsis, soybean (Glycine max L.), rice (Oryza sativa L.), corn (Zea mays L.), and wheat (Triticum aestivum L.). Approximately equal amounts of total protein, as determined by Coomassie Brilliant Blue staining of a replica gel (Fig. 2C), were probed in immunoblot experiments with aR50 and a288. Figure 2A shows that aR50 detects a single protein of slightly variable size in all species tested. A second band of higher molecular mass was only occasionally observed in tomato or tobacco extracts (asterisk in Fig. 2A), and might represent an aggregate of MFP1. In contrast, a288 detected a protein of about 80 kD only in tomato and tobacco extracts (Fig. 2B), suggesting that the DNA-binding domain of MFP1 that is recognized by aR50 is more highly conserved than the part of the coiled-coil domain recognized by a288 (Fig. 1).
Figure 2.
Identification of MFP1-like proteins in different plant species. Approximately equal amounts of total protein extracts from leaf tissue of tomato (Tom.), tobacco (Tob.), Arabidopsis (Ara.), soybean (Soy.), rice, corn, and wheat were subjected to immunoblot analysis using the aR50 (A) and the a288 (B) antisera. The arrow indicates the position of the MFP1-like proteins of approximately equal size. The asterisk shows the position of a second, minor band of higher molecular mass that was occasionally observed in tomato and tobacco extracts. C, Coomassie Brilliant Blue staining of a replica gel. Molecular mass markers are indicated on the right.
These data indicate that a protein of similar size, containing a domain related to the LeMFP1 DNA-binding domain, is conserved among higher plant species, and that among the plants investigated, the highest degree of similarity to LeMFP1 is expected from the protein in tobacco.
Isolation of NtMFP1 cDNAs
Tobacco has become the model organism for plant nuclear matrix biochemistry and for functional studies of MARs (Hall et al., 1991; Allen et al., 1993, 1996; Mlynarova et al., 1996). To conduct our future studies within a homologous system, we cloned and characterized the cDNAs encoding tobacco MFP1. A tobacco lambda-ZAP cDNA library was screened by DNA-hybridization with a 1.6-kb partial cDNA clone representing the 3′ two-thirds of the LeMFP1 cDNA (p7-2, Meier et al., 1996). Two positive plaque-forming units (pfus) were detected among approximately 600,000 pfus. After in vivo excision, sequence analysis of the two excised cDNAs (T6 and T1) showed that they represented 1103 and 912 bp of sequence with similarity to the 3′ part of the tomato MFP1 sequence, respectively (Fig. 3A).
Figure 3.
Structural analysis of the cloned NtMFP1 cDNAs. A, Schematic structure of the partial cDNAs isolated from a tobacco lambda ZAP cDNA library. T3, T1, and PCR#1 are shown as white boxes, represent overlapping fragments of the same gene (NtMFP1-1). T2 and T6 are shown as black boxes, represent overlapping fragments of a second gene (NtMFP1-2). The fragments derived from the NtMFP1 cDNA that were used as probes for Southern and RNA blots are indicated. B, Nucleotide sequence and deduced amino acid sequence of NtMFP1-1. Boxed regions indicate the two hydrophobic domains. Horizontal arrows indicate the oligonucleotide sequences used to amplify the PCR#1 fragment. Vertical arrows indicate the positions of intron sequences. The GenBank accession numbers for NtMFP1-1 and NtMFP1-2 are AF131231 and AF131232, respectively.
In a second round, the tobacco cDNA library was screened with a 1.0-kb 5′ fragment of the LeMFP1 cDNA (p1-3, Meier et al., 1996). Two additional positive pfus were detected among approximately 600,000 pfus. Sequencing of the excised cDNAs (T2 and T3) showed that they represented partial cDNAs, overlapping with T1 and T6 (Fig. 3A). The sequence similarity between the two 3′ fragments T1 and T6 is 91%, suggesting the presence of two MFP1 genes in tobacco. Sequence analysis of the T2 and T6 cDNAs showed that they shared 445 bp of identical overlapping sequence, but that an additional 119 bp at the 3′ end of the T2 cDNA sequence showed no relatedness to T6. To determine whether T2 represents a chimeric cDNA, a GenBank search was carried out with this 119-bp fragment. This search revealed 70% identity between this sequence and an RNA helicase from Arabidopsis. It was therefore eliminated from subsequent analysis of the T2 cDNA. We concluded that the remaining part of the T2 cDNA and the T6 cDNA represent overlapping portions of the same gene.
T3 and T1 share 70 bp of identical overlapping sequence, but within this area, there is only a single bp difference between T6 and T1. Therefore, we could not confidently conclude that T3 and T1 are derived from the same gene. To show whether this is the case, PCR primers (indicated in Fig. 3B) were designed from the T3 and T1 sequences, which would allow the amplification of a 397-bp fragment from a tobacco lambda-ZAP cDNA library overlapping both cDNAs. The sequence of the fragment PCR#1 (Fig. 3A) is 100% identical with both T1 and T3 and has 18 mismatches to the respective region of T2 and T6, confirming that T1 and T3 are derived from the same gene and represent a second type of MFP1 cDNA.
In summary, two distinct NtMFP1 cDNAs were isolated and named NtMFP1-1 (T3 and T1) and NtMFP1-2 (T2 and T6). NtMFP1-1 contains the full MFP1 open reading frame of 722 amino acids, while NtMFP1-2 is a partial cDNA and contains an open reading frame of 398 amino acids. NtMFP1-1 and NtMFP1-2 have 77% and 79% similarity on DNA level to LeMFP1, respectively. The similarity between the two tobacco sequences is 92%.
Sequence Analysis of NtMFP1-1 and NtMFP1-2
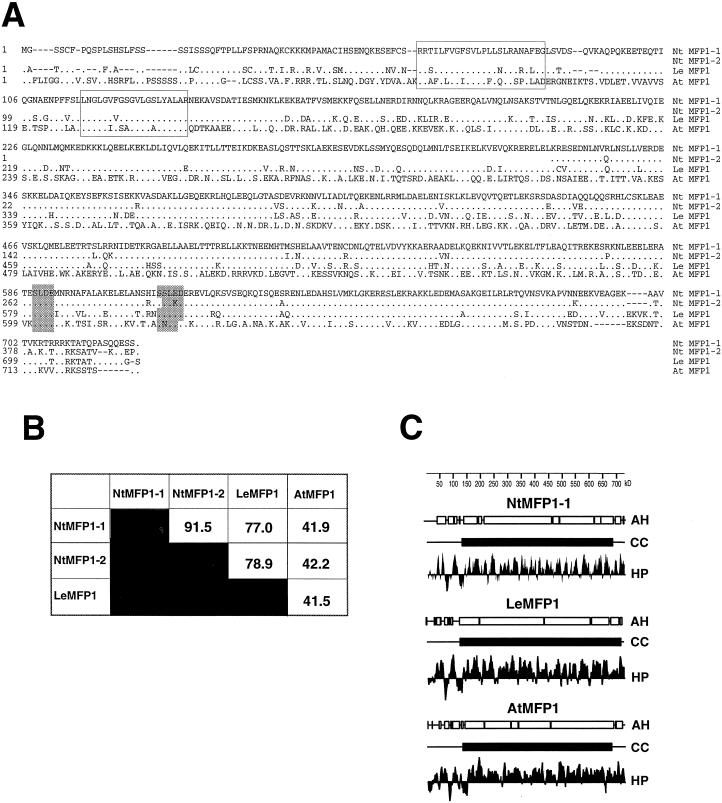
The DNA sequence and deduced amino acid sequence of NtMFP1-1 are shown in Figure 3B. NtMFP1-1 contains an open reading frame of 722 amino acids, preceded by 69 bp of 5′ non-coding sequence. Figure 4A shows the comparison of the amino acid sequences of LeMFP1, NtMFP1-1, and NtMFP1-2, and the Arabidopsis homolog of MFP1 (AtMFP1), that has been identified by the Arabidopsis genome project (GenBank accession no. AB012247). The percentage of amino acid identity between the different MFP1 proteins is indicated in Figure 4B. Based on amino acid sequence identity, NtMFP1-1 and NtMFP1-2 are most closely related. LeMFP1 is more closely related to the two tobacco MFP1s than to AtMFP1, reflecting the closer relationship of the two solanaceous species. The two N-terminal hydrophobic domains that have been shown to act as a targeting signal for LeMFP1 (Gindullis and Meier, 1999) are highly conserved between tomato, tobacco, and Arabidopsis. Two potential casein kinase II sites are conserved in all four sequences (Fig. 4A). In addition, a block of basic amino acids at the C terminus that could potentially act as a nuclear localization signal is present in all four polypeptides.
Figure 4.
Comparison of primary and predicted secondary structure of MFP1 proteins. A, Alignment of MFP1 protein sequences from tobacco (NtMFP1-1 and NtMFP1-2), tomato (LeMFP1), and Arabidopsis (AtMFP1). Dots represent amino acids identical to those of NtMFP1-1. Boxed regions indicate the positions of the two hydrophobic domains. Shaded regions at the C terminus show the conserved potential casein kinase II sites. Gaps are indicated by dashes. B, Percent identical amino acids in pairwise comparisons of the four MFP1 proteins. C, Hydrophilicity and secondary structure analysis of LeMFP1, NtMFP1-1, and AtMFP1. AH, α-Helical; CC, coiled-coil; HP, hydrophilicity plot.
The similarity between AtMFP1 and LeMFP1 is highest in the two hydrophobic domains and within a stretch of approximately 100 amino acids close to the C terminus. The conservation of this latter sequence could explain the ability of aR50, but not a288, to detect AtMFP1, and supports the assumption that the DNA-binding domain of MFP1 is more conserved than the non-DNA-binding part of the coiled-coil domain.
Figure 4C shows the comparison of the predicted secondary structure of LeMFP1, NtMFP1-1, and AtMFP1. Like LeMFP1 (Meier et al., 1996), NtMFP1-1 and AtMFP1 are predicted to contain an extended α-helical, coiled-coil like domain, and the shorter N-terminal, non-α-helical region that contains the two hydrophobic domains. These predicted structural features are extremely well conserved between tomato, tobacco, and Arabidopsis MFP1, despite a relatively low degree of similarity on the amino acid level in some areas. The distance between the first and second hydrophobic domains is very similar in all three proteins (29 amino acids for tomato, 31 amino acids for tobacco, and 33 amino acids for Arabidopsis MFP1), suggesting a functional relevance of the spacing between the two hydrophobic domains. The length of the N-terminal domain preceding the first hydrophobic domain varies between 56 amino acids for tomato, 61 amino acids for tobacco, and 72 amino acids for Arabidopsis MFP1. The common feature of this domain in all three proteins is a relatively high content of Ser and Thr residues (27%–28%).
Two MFP1 Genes Are Present in the Amphidiploid Tobacco Genome
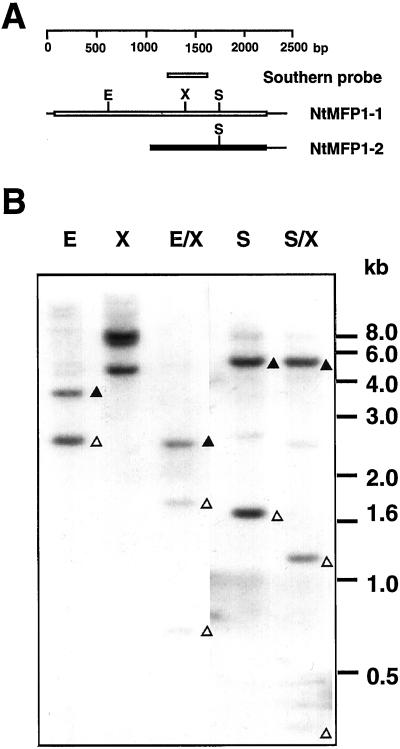
The divergence between the two tobacco MFP1 cDNAs suggests that they are derived from two different genes. It has been previously shown that a single gene codes for MFP1 in tomato (Meier et al., 1996), and we have found AtMFP1 to be a single gene in Arabidopsis (data not shown). To analyze the gene copy number of MFP1 in tobacco, tobacco genomic DNA was digested with the enzyme combinations indicated in Figure 5B and probed with a 391-bp XhoI/SpeI fragment derived from NtMFP1-1 (Figs. 3A and 5A). The Southern-blot experiment was carried out under stringency conditions that allow for hybridization with both genes (see “Materials and Methods”).
Figure 5.
Genomic organization of tobacco MFP1. A, Schematic representation of the two isolated tobacco cDNAs. EcoRI (E), XbaI (X), and SspI (S) sites are indicated. The NtMFP1-1-derived Southern-blot probe that spans the XbaI site of NtMFP1-1 is shown. B, Tobacco genomic DNA was digested with the indicated restriction enzymes, separated by agarose gel electrophoresis, and hybridized in a genomic Southern blot with the 391-bp XhoI/SpeI fragment from the tobacco cDNA clone T1 (A, and Fig. 3A). E, EcoRI; X, XbaI; E/X, EcoRI/XbaI; S, SspI; S/X, SspI/XbaI. The position of DNA size markers is indicated on the right. ▵, Fragments most likely representing NtMFP1-1; ▴, fragments most likely representing NtMFP1-2.
In the region overlapping the probe, NtMFP1-1 contains a single XbaI site, whereas NtMFP1-2 contains no XbaI site. NtMFP1-1 contains an EcoRI site at position 657, and both cDNAs contain an SspI site at identical positions (Fig. 5A). Two fragments (approximately 3.5 and 2.5 kb) were detected in the lane loaded with EcoRI-digested DNA, and three (approximately 8.0, 7.5, and 4.5 kb) were seen in the lane containing XbaI-digested DNA. This is consistent with the presence of two genes, one representing NtMFP1-1 and being cleaved by XbaI and one representing NtMFP1-2 and not being cleaved. In the lane containing the EcoRI/XbaI double digest, the 2.5-kb EcoRI fragment appears to be cleaved by XbaI, leading to two smaller fragments of approximately 1.8 and 0.7 kb. This suggests that this fragment contains a portion of the NtMFP1-1 gene. The 3.5-kb EcoRI fragment, which most likely represents the NtMFP1-2 gene, also appears to be cleaved by XbaI. Only one smaller fragment is detected, suggesting the presence of an XbaI site outside of the region hybridizing with the probe. In addition, SspI and SspI/XbaI digests were analyzed. Two prominent bands were detected in the SspI digest, the smaller of which is cleaved in the SspI/XbaI double digest, indicating that this band corresponds to NtMFP1-1. The observed patterns are all consistent with the presence of two genes in the tobacco genome, represented by the two isolated cDNAs.
MFP1 Expression Is Developmentally Regulated in Tobacco
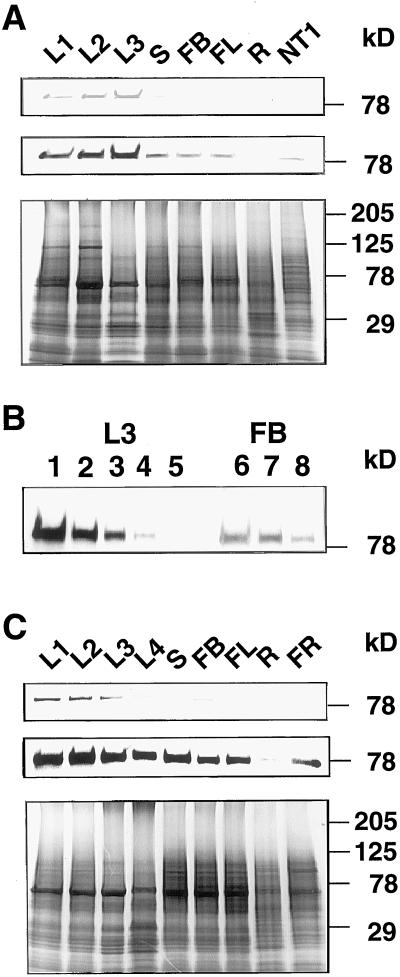
The expression levels of MFP1 were compared between organs and between different stages of leaf development in tobacco and tomato. Figure 6A shows the results of immunoblot experiments using the a288 antiserum and total protein extracts from tobacco leaves at three developmental stages (L1, 1.25 cm in length; L2, 12 cm in length; and L3, 35 cm in length), and from stems, flower buds, flowers, roots, and tobacco NT-1 suspension culture cells. Approximately equal amounts of protein, as determined by Coomassie Brilliant Blue staining of replica gels (Fig. 6A, bottom), were analyzed. MFP1 is expressed at a low level in suspension-cultured cells and in stems, flower buds, flowers, and roots, and at a significantly higher level in leaves. In leaves, MFP1 expression increases from an early developmental stage (L1) to later developmental stages (L2 and L3), and is highest in L3. Comparison of a dilution series of the L3 leaf extract and the flower bud extract indicated that the amount of MFP1 protein in the L3 extract is approximately 6- to 8-fold higher than in the flower bud extract (Fig. 6B). In a second experiment using a different set of plants, an even larger difference between the leaf extracts and the extracts from other organs was found (data not shown).
Figure 6.
Expression of MFP1 in tobacco and tomato. A, Approximately equal amounts of total protein extracts from various tobacco tissues were subjected to immunoblot analysis using the a288 antiserum. L1, Leaf 1.25 cm in length; L2, leaf 12 cm in length; L3, leaf 35 cm in length; S, stem; FB, flower bud; FL, flower; R, root; NT1, NT-1 suspension-cultured cells. The top and middle panels show a shorter and a longer exposure of the immunoblot, respectively. The bottom panel shows the Coomassie Brilliant Blue staining of a replica gel. B, Titration of L3 leaf extract and flower bud extract. Lane 1 was loaded with the same amount (1×) L3 protein extract that was used in A; lanes 2, 3, 4, and 5 were loaded with 0.5×, 0.25×, 0.125×, and 0.062× the amount loaded in lane 1, respectively. Lane 7 was loaded with the same amount (1×) of flower bud protein extract that was used in A. Lanes 6 and 8 were loaded with 2× and 0.5× the amount in lane 7, respectively. C, Approximately equal amounts of total protein extracts from various tomato tissues were subjected to immunoblot analysis using the a288 antiserum. L1, Leaf 1.5 cm in length; L2, leaf 9 cm in length; L3, leaf 13 cm in length; L4, leaf 21 cm in length; S, stem; FB, flower bud; FL, flower; R, root; FR, ripening fruit. The top and middle panels show a shorter and a longer exposure of the immunoblot, respectively. The bottom panel shows the Coomassie Brilliant Blue staining of a replica gel. Molecular mass markers are indicated on the right.
A similar, although somewhat less pronounced, expression pattern was observed in tomato (Fig. 6C); a lower level of expression was detected in stems, flower buds, flowers, roots, and young fruit compared with leaves. A slight decrease in the amount of MFP1 protein was observed in late leaf development, between stages L3 (13 cm) and L4 (21 cm).
To investigate if the organ-specific differences in NtMFP1 expression occur at the level of mRNA or protein synthesis, an RNA gel blot experiment with total RNA from different tobacco tissues was performed. Consistent with the length of the NtMFP1–1 cDNA and with the size of the MFP1 mRNA in tomato, a single 2.4-kb mRNA species was detected (Fig. 7). NtMFP1 mRNA was detected in all tissues investigated, with the lowest signal in roots and NT-1 cells visible only on a longer exposure of the RNA blot shown in Figure 7 (data not shown). NtMFP1 mRNA abundance in different tissues and developmental stages correlated with the abundance of the protein. A more pronounced difference in RNA abundance than in protein abundance was detected between leaves and the other tissues. In contrast to protein abundance, NtMFP1 mRNA abundance declined between leaf stages L2 and L3 (Fig. 7).
Figure 7.
RNA-blot analysis from various tobacco tissues. The top panel shows the hybridization signal obtained using a 389-bp EcoRI/XhoI fragment from clone T1 (shown in Fig. 3A) as a probe. The bottom panel shows the intensity of the rRNA bands after staining of a replica gel with ethidium bromide. The position of the 25S and 18S rRNA bands are indicated in both panels. L1, Leaf 1.25 cm in length; L2, leaf 12 cm in length; L3, leaf 35 cm in length; S, stem; FB, flower bud; FL, flower; R, root; NT1, NT-1 suspension-cultured cells.
Our results indicate that NtMFP1 expression is highest in leaves, and is largely regulated at the level of transcription or mRNA stability. In addition, the higher abundance of MFP1 protein but reduced accumulation of MFP1 mRNA at leaf stage L3 suggests that the protein has a relatively slow turnover rate in tobacco leaves.
DISCUSSION
MFP1 Is Conserved among Higher Plants
MFP1 was originally identified as a MAR-binding filament-like protein from tomato (Meier et al., 1996). It has no sequence similarity to the animal MAR-binding proteins that have been identified so far (von Kries et al., 1991; Dickinson et al., 1992; Renz and Fackelmayer, 1996; Göhring et al., 1997), none of which has a putative filament-like structure. The animal nuclear matrix proteins with the greatest structural similarity to MFP1 are the nuclear lamins. Lamin A, B, and C are a group of intermediate filament proteins that form the nuclear lamina, a filamentous protein network that lines the inner membrane of the nuclear envelope (McKeon et al., 1986). Lamin B is attached to the inner nuclear membrane by a C-terminal farnesyl group and the interaction with integral membrane proteins (Schafer and Rine, 1992; Gerace and Foisner, 1994). Lamin A and C bind to lamin B. Recent studies have demonstrated that lamins A and B can specifically bind MAR DNA, suggesting a role of the lamina in anchoring chromatin loops to the nuclear envelope (Zhao et al., 1996).
Despite several efforts, no lamins have been identified from plants. Proteins with antigenic relationship to animal lamins have been identified in pea and onion nuclei (McNulty and Saunders, 1992; Minguez and Moreno Diaz de la Espina, 1993), but in contrast to animal lamins, they were detected throughout the nuclear matrix, and were not confined to a lamina at the nuclear envelope (Minguez and Moreno Diaz de la Espina, 1993). It is thus possible that the anti-lamin antibodies detect filament-like proteins in plants that share antigenic determinants with lamins, but might have additional functions in the plant nuclear matrix. Interestingly, no open reading frames for lamin-like proteins are present in the yeast genome (Mewes et al., 1998). We have shown that MFP1 is associated with the nuclear envelope in tobacco NT-1 cells and that the N-terminal domain is necessary for this localization (Gindullis and Meier, 1999). It is therefore conceivable that MFP1 represents a protein that is involved in attaching chromatin to the nuclear envelope in plants and that is different from the animal lamins. These findings imply that different classes of MAR-binding filament-like proteins might have evolved in the different kingdoms. It was therefore of interest to determine if MFP1-like proteins would be generally conserved among higher plants and, if so, how conserved the different functional domains would be.
Here we show that an MFP1-like protein is present in a variety of dicot and monocot species, including several important crop plants. Using two domain-specific antibodies, we have demonstrated that the DNA-binding domain of MFP1 is more highly conserved between species than the central part of the putative coiled-coil domain. An antibody against the central domain recognizes MFP1 only in tobacco, the species most closely related to tomato.
Cloning of the two tobacco MFP1 cDNAs and comparison of the sequence of NtMFP1-1 with tomato MFP1 and with the published Arabidopsis MFP1 sequence confirmed that the DNA-binding domain is more highly conserved between tomato, tobacco, and Arabidopsis than the central putative coiled-coil domain. This result is consistent with the hypothesis that a conserved sequence and structure is necessary for the specific recognition of MAR DNA. In contrast, coiled-coil domains might be less conserved, as an amphipathic α-helix can be created by the same distribution of hydrophobic and charged amino acids without direct sequence conservation (Boice et al., 1996). This is evident by the relative low degree of similarity between the animal lamins. Drosophila and mouse lamins are only 37% identical, and even lamins in species as closely related as human and mouse share only 49% of the residues (Weber et al., 1989). Nevertheless, all animal lamins have a conserved secondary structure with a globular head domain, a central coiled-coil rod domain and a globular tail domain (Stuurman et al., 1998). Similarly, LeMFP1, NtMFP1-1 and AtMFP1 have the same structural features, based on structural predictions from their amino acid sequences.
Conserved between all three proteins is an N-terminal domain that contains two stretches of hydrophobic amino acids of similar length and sequence and with very similar spacing, followed by a long putative coiled-coil domain of relatively low sequence similarity, and the C-terminal more highly conserved DNA-binding domain. The high degree of conservation of sequence and spacing of the two hydrophobic domains suggests that they are indeed functionally relevant. A likely function for this protein domain is to provide membrane attachment to anchor MFP1 to the nuclear envelope, analogous to the lamins, but utilizing direct membrane attachment instead of farnesylation and interaction with a membrane protein. The recent finding that the N-terminal domain of LeMFP1 is necessary for the specific targeting of the protein to the nuclear rim (Gindullis and Meier, 1999) supports this hypothesis.
Tobacco MFP1 Is Regulated in an Organ-Specific and Developmental Fashion
We have found that tobacco MFP1 expression is regulated in an organ-specific and developmental fashion. The highest expression of both protein and mRNA was found in leaves, where MFP1 expression increased between an early developmental stage (1.25 cm in length) and a medium developmental stage (12 cm in length). While MFP1 expression was lower in other organs, a small amount of either protein or mRNA could be detected in all organs investigated. Earlier results had shown that MFP1 is ubiquitously expressed in tomato, although a somewhat higher level of mRNA was detected in leaves than in other tissues examined (Meier et al., 1996). We have now corroborated these findings with respect to protein abundance.
We cannot distinguish at this point whether the accumulation of NtMFP1 at later stages of leaf development is correlated with the photosynthetic activity of the tissue or if it is regulated by a developmental program. However, the fact that its abundance in light-grown seedlings (data not shown) and in stems is significantly lower than in leaves might indicate that its accumulation is not simply light induced or correlated with green tissue. We have previously shown that MFP1 is predominantly located in speckle-like structures at the nuclear rim of tobacco NT-1 suspension-cultured cells (Gindullis and Meier, 1999). It will now be of great interest to investigate if the protein has the same subcellular localization in tobacco leaves, or if the higher abundance of MFP1 correlates with a different localization pattern.
Several nuclear matrix proteins from animals, such as lamins A and B, nucleolin, ARBP, and SAF-B, are expressed as “housekeeping” genes, as might be expected for a protein that has a general function in chromatin organization. However, there is at least one exception, which has spiked interest in potential roles of MAR-binding proteins in tissue-specific gene expression. SATB1 is a nuclear matrix-localized MAR-binding protein that is predominantly expressed in thymocytes, but is also present at a low level in other cell types (Dickinson et al., 1992; Kohwi-Shigematsu et al., 1997). It can suppress promoter activity in a MAR-dependent fashion (Kohwi-Shigematsu et al., 1997) and it has been suggested that the function of SATB1 is to silence genes during certain stages of thymocyte development. How this effect on gene expression is connected with the proposed function of MAR-binding proteins in the organization of chromatin is not known, but it is possible that tissue-specific MAR-binding proteins could be involved in creating a specific chromatin environment for genes that are either highly expressed or silenced in a given tissue. The question of whether MFP1 is involved in a chromatin-based mechanism for tissue-specific gene regulation in plants will have to await further study.
ACKNOWLEDGMENTS
We thank Drs. Shawn Anderson and Timothy Caspar for critical reading of the manuscript.
Footnotes
This work was supported in part by the German Science Foundation (grant no. ME 1133/2–1 to I.M.).
LITERATURE CITED
- Allen GC, Hall GE, Jr, Childs LC, Weissinger AK, Spiker S, Thompson WF. Scaffold attachment regions increase reporter gene expression in stably transformed plant cells. Plant Cell. 1993;5:603–613. doi: 10.1105/tpc.5.6.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, Hall G, Jr, Michalowski S, Newman W, Spiker S, Weissinger AK, Thompson WF. High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell. 1996;8:899–913. doi: 10.1105/tpc.8.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1470. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Boice JA, Dieckmann GR, DeGrado WF, Fairman R. Thermodynamic analysis of a designed three-stranded coiled-coil. Biochemistry. 1996;35:14480–14485. doi: 10.1021/bi961831d. [DOI] [PubMed] [Google Scholar]
- Chen JC, Dellaporta S. Urea-based plant DNA miniprep. In: Walbot V, Freeling M, editors. The Maize Handbook. New York: Springer Verlag; 1994. pp. 526–527. [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Gerace L, Foisner R. Integral membrane proteins and dynamic organization of the nuclear envelope. Trends Cell Biol. 1994;4:127–131. doi: 10.1016/0962-8924(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Gindullis F, Meier I. Matrix attachment region-binding protein MFP1 is localized in discrete domains at the nuclear envelope. Plant Cell. 1999;11:1117–1128. doi: 10.1105/tpc.11.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhring F, Schwab BL, Nicotera P, Leist M, Fackelmayer FO. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target for apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall G, Jr, Allen GC, Loer DS, Thompson WF, Spiker S. Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc Natl Acad Sci USA. 1991;88:9320–9324. doi: 10.1073/pnas.88.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- McNulty AK, Saunders MJ. Purification and immunological detection of pea nuclear intermediate filaments: evidence for plant nuclear lamins. J Cell Sci. 1992;103:407–414. doi: 10.1242/jcs.103.2.407. [DOI] [PubMed] [Google Scholar]
- Meier I, Phelan T, Gruissem W, Spiker S, Schneider D. MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell. 1996;8:2105–2115. doi: 10.1105/tpc.8.11.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Hani J, Pfeiffer F, Frishman D. MIPS: a database for protein sequences and complete genomes. Nucleic Acids Res. 1998;26:33–37. doi: 10.1093/nar/26.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguez A, Moreno Diaz de la Espina S. Immunological characterization of lamins in the nuclear matrix of onion cells. J Cell Sci. 1993;106:431–439. doi: 10.1242/jcs.106.1.431. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Mirault M-E, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffolds. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mlynarova L, Keizer LCP, Stiekema WJ, Nap J-P. Approaching the lower limits of transgene variability. Plant Cell. 1996;8:1589–1599. doi: 10.1105/tpc.8.9.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24:843–849. doi: 10.1093/nar/24.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EM, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schafer WR, Rine J. Protein prenylation: genes, enzymes, targets and functions. Annu Rev Genet. 1992;30:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- Spiker S, Thompson WF. Nuclear matrix attachment regions and transgene expression in plants. Plant Physiol. 1996;110:15–21. doi: 10.1104/pp.110.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- von Kries JP, Buhrmeister H, Strätling WH. A matrix/scaffold attachment region binding protein: identification, purification and mode of binding. Cell. 1991;64:123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Ulrich W. Cytoplasmic intermediate filament proteins of invertebrates are closer to nuclear lamins than are vertebrate intermediate filament proteins: sequence characterization of two muscle proteins of a nematode. EMBO J. 1989;8:3221–3227. doi: 10.1002/j.1460-2075.1989.tb08481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Harel A, Stuurman N, Guedalia D, Gruenbaum Y. Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett. 1996;12:161–164. doi: 10.1016/0014-5793(96)00034-8. [DOI] [PubMed] [Google Scholar]