Abstract
The rare recessive developmental disorder Trichothiodystrophy (TTD) is characterized by brittle hair and nails. Patients also present a variable set of poorly explained additional clinical features, including ichthyosis, impaired intelligence, developmental delay and anemia. About half of TTD patients are photosensitive due to inherited defects in the DNA repair and transcription factor II H (TFIIH). The pathophysiological contributions of unrepaired DNA lesions and impaired transcription have not been dissected yet. Here, we functionally characterize the consequence of a homozygous missense mutation in the general transcription factor II E, subunit 2 (GTF2E2/TFIIEβ) of two unrelated non-photosensitive TTD (NPS-TTD) families. We demonstrate that mutant TFIIEβ strongly reduces the total amount of the entire TFIIE complex, with a remarkable temperature-sensitive transcription defect, which strikingly correlates with the phenotypic aggravation of key clinical symptoms after episodes of high fever. We performed induced pluripotent stem (iPS) cell reprogramming of patient fibroblasts followed by in vitro erythroid differentiation to translate the intriguing molecular defect to phenotypic expression in relevant tissue, to disclose the molecular basis for some specific TTD features. We observed a clear hematopoietic defect during late-stage differentiation associated with hemoglobin subunit imbalance. These new findings of a DNA repair-independent transcription defect and tissue-specific malfunctioning provide novel mechanistic insight into the etiology of TTD.
Introduction
Trichothiodystrophy (TTD) is a rare recessive disorder, characterized by brittle hair and nails, due to a low content of sulfur-rich proteins in keratinocytes. Patients present a variable combination of additional symptoms including, ichthyosis, impaired intelligence, decreased fertility, microcephaly, developmental delay, anemia and progeroid features (1). Approximately, 50% of the TTD patients are sun(photo)-sensitive, caused by mutations in the xeroderma pigmentosum group B (XPB/ERCC3), group D (XPD/ERCC2) or trichothiodystrophy group A (TTDA/GTF2H5) genes (2,3), each encoding for subunits of the transcription factor IIH (TFIIH) (4). In addition to an essential role of TFIIH in transcription initiation, this complex is also pivotal for nucleotide excision repair (NER). These mutations impair NER, which is the only DNA repair system in mammals capable of removing sun-induced DNA damage and thus easily explain photo-sensitivity. The additional features are thought to be derived by subtle defects in the transcription function of TFIIH.
We previously proposed that reduced stability of mutant TFIIH in TTD patients may cause some of the TTD-specific clinical features, which are mainly apparent in terminally differentiated tissues (3). Within late-stage differentiated cells, the majority of genes become transcriptionally silenced, including genes encoding for basal transcription factors such as TFIIH. The amount of proteins encoded by transcriptionally “switched off” genes in these cells thus depends on the stability of their residual mRNAs and proteins produced at prior stages. We have shown that TTD-causing mutations in TFIIH genes affect the stability of the entire complex and that the steady-state level of each subunit becomes strongly reduced (2,5,6). In cultured fibroblast, this reduced amount appeared sufficient to provide normal transcription levels. However, at the final stages of cellular differentiation when de novo synthesis is switched off, the reduced TFIIH stability in TTD cells may result in too low amounts of TFIIH to support sufficient transcription of tissue-specific proteins. The observed reduced transcription of the SPRR2 gene in terminally differentiated keratinocytes from a TTD-mimicking mouse-model (7) supported this hypothesis. SPRR2 is a sulfur-rich matrix protein, only expressed at very late stages of differentiation, which crosslinks keratin filaments to provide mechanical strength to skin and hair cells. In terminally differentiated red blood cells, TTD-specific TFIIH mutations may cause imbalanced globin mRNA expression, explaining the relatively frequent anemic features of TTD (1,8). It is however not excluded that unrepaired (endogenously produced) DNA lesions in transcription units, as a consequence of the DNA repair defect associated with mutated TFIIH, may also contribute to reduced transcription in late-stage differentiated cells. Indeed, endogenous metabolic stress and environmental cues do generate a variety of DNA lesions, which may play a role in the pathogenesis of accelerating aging (9) and other degenerative diseases (10,11).
We hypothesize that a large of part of the clinical features associated with repair-proficient non-photosensitive TTD (NPS-TTD) are based on gene expression defects (12). In only a minority of NPS-TTD patients causative mutations were found in genes encoding for the M-phase-specific PLK1 interacting protein (MPLKIP/TTDN1) (13) or the β subunit of the transcription factor IIE (GTF2E2/TFIIEβ) (14) and in an isolated case in the RNF113A gene (15). Nevertheless, thus far no obvious experimental evidence for transcription malfunctioning in NPS-TTD patient cells has been provided. Here, we present the genetic and functional analysis of NPS-TTD cases with homozygous GTF2E2 mutations. We showed that fibroblasts derived from these patients exhibit a clear transcription defect, however only when cultured at elevated temperatures. We used induced pluripotent stem cell (iPS) reprogramming of patient fibroblasts followed by in vitro erythroid differentiation to show hemoglobin protein imbalance in late-stage differentiated erythroid cells, in line with patients’ anemic features.
Results
Genetic analysis of non-photosensitive TTD patients
Recently, two unrelated NPS-TTD patients were identified with different mutations in the gene encoding for the beta subunit of transcription initiation factor II E (TFIIEβ/GF2E2) (14), suggesting impaired transcription in these patients. To provide functional evidence for affected transcription in NPS-TTD, we first performed genetic analyses among a selected group of NPS-TTD patients to identify possible other mutations in transcription factors.
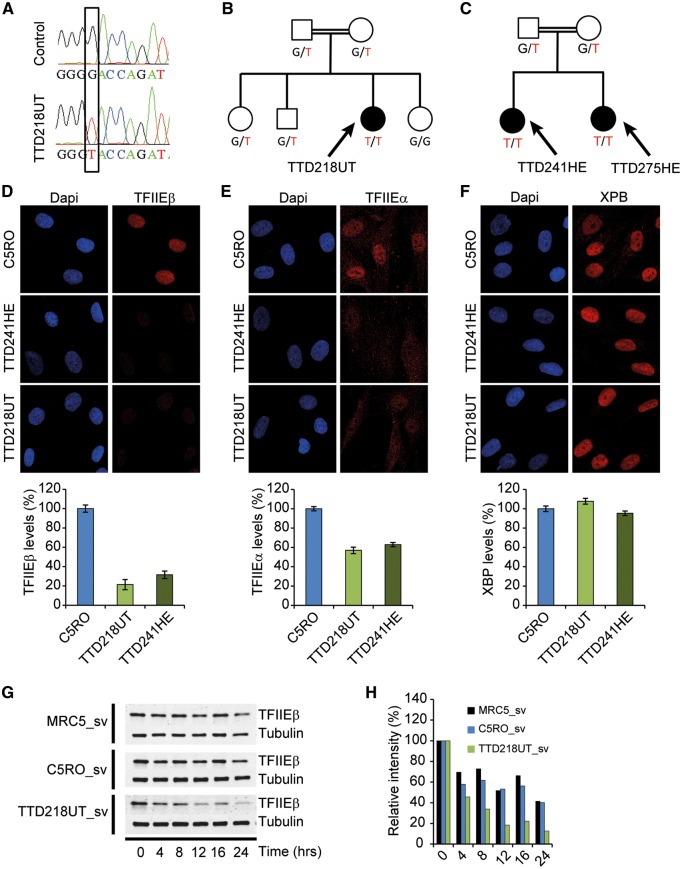
Patient TTD218UT is of Moroccan origin and was born to consanguineous parents. She was referred to the clinical geneticist at 1.5 years of age with brittle hair (low cysteine content and tiger-tail banding pattern), ichthyosis, microcephaly (−2.5 SD), psychomotor retardation, short stature (−2.5 SD) and microcytic anemia. She showed recurrent aggravation of hair-loss by tufted breakage at the scalp boundary immediately following episodes of infection-induced fever. Also upon long-term exposure to the heat of the sun, she appears to have increased hair-loss. At the age of 8 years, she shows no signs of progressive deterioration. She has a happy and friendly personality with an IQ of 54 (at 7 years of age). She has a limited physical endurance. Laboratory investigation revealed microcytic hypochromic anemia (Hb 6.5) without sequence anomalies nor deletions or duplications of the genes encoding for the alpha or beta globin chains (not shown). We performed whole genome sequencing on patient’s DNA, using the sequencing-by-ligation method from Complete Genomics (16,17). Surprisingly, we identified a homozygous variant in the GTF2E2/TFIIEβ gene, creating a missense mutation (c.C559T [p.Asp187Tyr]). This mutation is identical to one of the mutations in GTF2E2/TFIIEβ previously described by Kuschal et al. (14). Its homozygous presence was confirmed by Sanger sequencing of cDNA generated from patient’s cells and found to co-segregate with the phenotype within the family (Fig. 1A and B).
Figure 1.
TFIIE protein levels and stability is reduced in TFIIEβ-mutated TTD cells. (A) Sanger sequencing profiles of TFIIEβ cDNA. TTD218UT shows homozygosity for the c.G559T missense mutation. (B,C) TFIIEβ mutations segregating in the patients’ families. (D–F) Immuno-fluorescence analysis of TTD218UT and TTD251HE fibroblasts compared with wild-type control (C5RO) cells, stained for (D) TFIIEβ, (E) TFIIEα or (F) XPB (TFIIH) and DNA was stained with DAPI (blue). Quantification of the mean intensities (n = 50 nuclei), expressed as percentage of the mean intensity in normal cells, is shown beneath the representative images. Error bars indicate SEM. (G) Immuno-blot analysis to determine protein stability of TFIIEβ after cycloheximide (100 µM) treatment (for indicated times in hours) of Sv40-immoratlized patient cells (TTD218UT_sv) compared with two wild-type controls (MRC5_sv, C5RO-sv). (H) Quantification of TFIIEβ band intensities normalized to tubulin were expressed as percentages of t=0 values.
Additional Sanger sequencing of the TFIIE subunits among another group of selected NPS-TTD patients revealed the identification of the same homozygous mutation (c.G559T in GTF2E2/TFIIEβ) in two sibs, TTD241HE and TTD275HE. These patients are born to consanguineous parents of Moroccan origin, which are both heterozygous for this mutation (Fig. 1C) and are not related to patient TTD218UT. Patient TTD241HE was diagnosed at 18 months of age with brittle hair (tiger-tail banding pattern), dysmorphic features, developmental delay, moderate intellectual disability, microcephaly, difficulty swallowing and failure to thrive, cheerful character, mild Ichthyosis, growth retardation (length −4.7 SD, weight −2.6 SD), occipitofrontal circumference (OFC) (−3.1 SD), hypertonia, microcytic hypochromic anemia and recurrent infections. At 6 years of age, she still showed spasticity, ataxia and mild cognitive retardation without significant speech, but no signs of progressive deterioration. She has one affected younger sib (TTD275HE), also diagnosed with brittle hair, dysmorphic features, mild developmental delay, microcephaly, mild difficulty swallowing, no failure to thrive, cheerful character, mild ichthyosis, additional left thumb, no recurrent infections and growth retardation were noticed.
Functional analysis of DNA repair capacity
We consider this TFIIEβ mutation as likely causative for some of the NPS-TTD patients, in line with our hypothesis that one of the prime causes for this form of TTD is based on subtle transcriptional defects. Since TFIIE is closely associated with TFIIH’s transcription function (18,19), it is not excluded that mutant TFIIE may influence TFIIH’s repair function. To that aim, we thoroughly examined the cellular NER capacity in fibroblasts of patient TTD218UT and TTD241HE. Patients’ fibroblast were not hypersensitive to UV-light and exhibited proficient unscheduled DNA repair synthesis (UDS) and recovery of RNA synthesis after UV irradiation (RRS), indicating that both global-genome and transcription-coupled NER are not affected (Supplementary Material, Figs S1 and S2).
TFIIE protein levels and stability
The general transcription factor TFIIE consists of two subunits; GTF2E1/TFIIEα and GTF2E2/TFIIEβ (20–22) and is a crucial component of the transcription preinitiation complex required for transcription initiation and promoter opening by facilitating loading and stable binding of TFIIH (18,23,24). Since TTD-causing mutations in TFIIH destabilize the entire complex (25) and because this frail TFIIH was suggested to cause TTD-specific transcription features (3), we wondered whether also this TFIIEβ mutation affects TFIIE stability. Immuno-fluorescence analysis showed that the steady-state levels of TFIIEβ and TFIIEα were strongly reduced to approximately 20 and 60%, respectively, of TFIIE levels in wild-type cells assayed in parallel. Whereas the levels of TFIIH (assessed with anti-XPB antibody) appeared unaffected, as expected (Fig. 1D–F, Supplementary Material, Fig. S3A–C).
Immuno-blot analysis of cell-free extracts confirmed the low steady-state levels of both TFIIE subunits (Supplementary Material, Fig. S3D–E). Since quantitative RT-PCR showed that the mRNA levels of both TFIIE subunits were unaffected (data not shown), it is likely that the reduced TFIIE levels are derived from protein instability. Western blot analysis on protein extracts isolated from cells incubated for different times in the presence of the translation inhibitor cycloheximide showed a strongly reduced stability of mutated TFIIEß, as compared with wild-type TFIIEß (Fig. 1G and H). The levels of both subunits can be fully restored by reintroducing wild-type TFIIEβ cDNA in the patient cells (Supplementary Material, Fig. S4). Together, these data show that the single amino-acid substitution p.Asp187Tyr in the beta subunit of TFIIE causes instability of the entire complex.
Transcription levels
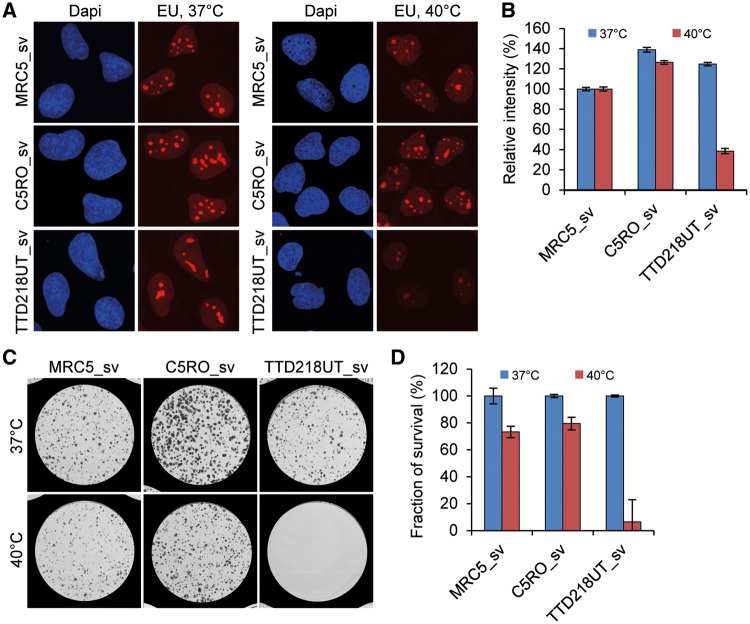
It is to be expected that with such a severe reduction of the cellular content of TFIIE, the overall transcription would be impaired. Moreover, previous studies (26) have shown that amino acid substitutions p.Ile171Ser and p.Ile189Ser, both in close proximity of patient’s TFIIEβ mutation (c.G559T [p.Asp187Tyr]), affect XPB helicase activity and severely reduce transcriptional capacity in cell-free in vitro assays. Surprisingly however, basal transcription levels, assayed by ethynyl uridine (EU) pulse labeling, were not affected in patient cells under standard culture conditions (Fig. 2A and B, Supplementary Material, Fig. S5A and B). Despite the strong reduction in steady-state levels of TFIIE, the remaining amount is apparently sufficient to support normal levels of transcription. Therefore, we searched for experimental conditions in which TFIIE levels may become limiting to transcription. We became aware that upon recurrent infections patient TTD218UT experiences reversible fever-dependent worsening of her brittle hair phenotype, most likely due to a further decrease or inactivation of the already low amounts of mutant TFIIE. Recurrent infections have also been reported for patient TTD241HE, however, fever-dependent worsening of any clinical feature could not be confirmed. Previously, we have found that a specific TTD-causative mutation in XPD (c.C2050T [p.Arg658Cys]) causes thermo-lability of the entire TFIIH complex, which was also accompanied by fever-dependent worsening of the TTD-specific hair phenotype in these patients (6). Incubation of cells derived from these XPD-TTD patients at elevated temperature showed a strong temperature-dependent reduction of transcription and severely reduced viability of the cells. Upon culturing of TFIIEβ-mutated TTD218UT and TTD241HE cells for 3 days at 40 °C, we observed a strikingly reduced transcription as compared with normal cells exposed to the same temperature (Fig. 2A and B, Supplementary Material, Fig. S5A and B).
Figure 2.
Transcription and cell survival is reduced at 40 °C in TFIIEβ-mutated TTD cells. (A) Transcription levels after incubation for 72 h at 37 °C or 40 °C, measured by pulse-labeling with ethynyl-uridine (EU) and subsequent fluorescent staining of incorporated EU, of Sv40-immortalized patient cells (TTD218UT_sv), compared with wild-type controls (MRC5_sv, C5RO-sv). DNA was stained with DAPI (blue). (B) Quantification of the mean intensities (n = 50), expressed as percentages of intensity in MRC5_sv. Error bars indicate SEM. (C,D) Coomassie staining and quantification of colonies (D) of TTD218UT_sv, MRC5_sv and C5RO-sv cells formed after 12 days culturing at 37 °C (blue bars) or 40 °C (red bars) (n = 2).
To determine the effect of prolonged exposure to elevated temperatures, we performed a colony survival assay on patient-derived cells that were incubated for 12 days at 40 °C. This extended culturing at higher temperature induced a dramatic reduction in colony-forming ability of the TTD218UT cells compared with control cells (Fig. 2C and D). The chronic exposure to higher temperatures likely depletes TFIIE levels to such an extent that they become too low to allow sufficient transcription to support cellular survival.
Erythroid differentiation of human iPS cells
Proliferating cultured fibroblasts are likely not representative to mimic the in vivo conditions that determine the TTD-specific symptoms, particularly since these are mainly apparent in terminally differentiated tissues, such as hair-follicles (brittle hair), epidermal keratinocytes (ichthyosis), neurons (impaired neurologic functions) and erythrocytes (anemia). Patients harboring a TFIIEβ mutation present microcytic anemia [this study and (14)], characterized by a reduced mean cell volume (MCV), which might be the consequence of reduced iron binding or a defect in late-stage erythroid differentiation driven by transcriptional problems.
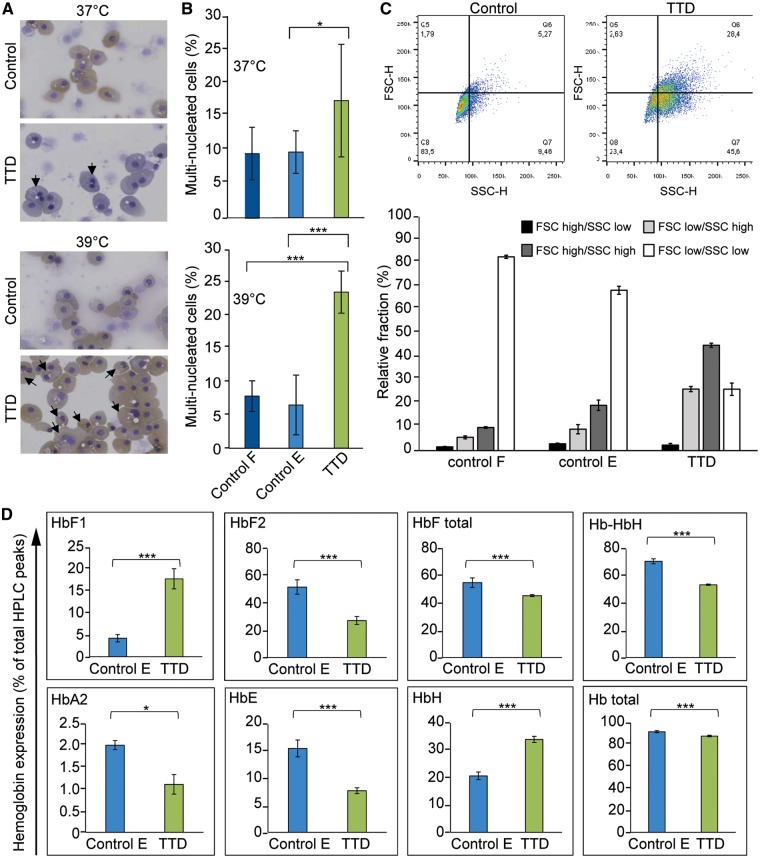
To recapitulate the TTD-phenotype in this patient, induced pluripotent stem (iPS) cells may offer a powerful platform to investigate genotype–phenotype relationship in relevant tissue. To this aim, we established an iPS clone reprogrammed from TTD218UT primary fibroblasts, and two control iPS cell lines obtained from healthy individuals, one reprogrammed from primary fibroblasts and one reprogrammed from a blood sample. Isolated iPS clones were selected on the basis of normal karyotypes, and thoroughly screened for expression of pluripotency markers (e.g. OCT4, Nanog) and absence of expression of differentiation markers (e.g. GFAP, AFP) (27), all of which were normal (data not shown). To investigate whether the TFIIEβ mutation could account for hematological malfunctions, we used a two-phased liquid culture system, including an expansion and a differentiation phase (28). Wild type and patient iPS cells were similar in their ability to differentiate into a homogenous population of CD71+/CD235+ erythroblasts (Supplementary Material, Fig. S7). Also, the cumulative and relative number of erythroid cells generated during the expansion phase, prior to erythroid differentiation, did not differ (data not shown), suggesting that proliferation is not affected. Comparing erythroblast differentiation at 37 °C and a fever-mimicking temperature of 39 °C indicated that differentiation to CD71+/CD235+ erythroblasts was still similar among all iPS-derived erythroblasts (Supplementary Material, Fig. S7). However, the cell size and number of multinuclear erythroblasts were increased in terminally differentiated TTD-erythroblasts, which were even more affected at 39 °C (Fig. 3A–C, Supplementary Material, Fig. S6). Flow cytometric analysis revealed an increase of cells with high forward and/or side scatter of TTD-derived erythroid cells at 37 °C compared with control cells (Fig. 3C), suggesting defective cytokinesis and cell size control, two important aspects heavily controlled during erythropoiesis (29). Several TTD patients have microcytic hypochromic anemia, which may suggest a disturbed hemoglobin synthesis. Hemoglobin consists of two subunits from the alpha locus and two subunits from the beta locus. The beta locus produces β and δ subunits for adult HbA and HbA2, respectively, γ1 and γ2 for fetal HbF1 and HbF2, and ε for embryonal HbE. Subunit imbalance can present as microcytic hypochromic anemias and thalassemia-like phenotypes (30), as observed among TTD patients without additional mutations in globin subunits (8). Hemoglobins cannot be compared between adult erythrocytes and iPS-derived mature erythroblasts because iPS-derived cells execute an embryonic/fetal program. However iPS-derived cells can be compared among each other. HPLC analysis (31) of iPS-derived erythroid cells showed reduced levels of HbF, HbA2 and HbE in TTD cells, with increased hemoglobin H (HbH/barts; tetramers of beta locus globins only) (Fig. 3D, Supplementary Material, Fig. S8). This may indicate an underproduction of alpha globin chains in TTD cells causing increased tetramerization of γ globin or β globin chains and reduced HbF, HbA2 and HbE. Control cells primarily express HBF2, and low levels of HBF1, contrary to TTD cells that express significantly higher levels of HBF1 with a concomitant reduction of HBF2. These results suggest a disturbed hemoglobin regulation during erythropoiesis in TTD iPS-derived erythroid cells. Reduced stability and functionality of TFIIE in TTD may lead to reduced expression of specific high abundant mRNAs e.g. the globin genes during erythroid differentiation. We suggest that in TTD patient-derived iPS cells the beta locus generates several subtypes of globin polypeptides, which cannot be matched by the reduced expression of the α-chain from the alpha locus, resulting in HbH formation, anemia and microcytic hypochromic erythrocytes.
Figure 3.
Affected Hematopoiesis in iPS-derived erythroid cells of TTD patients. In vitro erythroblast differentiation performed on iPS cells derived from TTD218UT (TTD) fibroblasts, control fibroblasts (control F) and peripheral blood mononuclear cell derived erythroblasts (control E).(A) Cytospins and Giemsa-Grünwald staining of in vitro differentiated erythroblasts. Arrows indicate multinucleated cells. (B) Quantification of multinucleated cells, expressed as a percentage of total cells [>500 cells, counted by two independent researchers (N = 2)]. (C) Dot plots of flow-cytometric analysis of differentiated erythroid control and TTD cells. Lower bar graph depicts the quantification of the gated dot-plots [Q8=FSClow/SSClow (normal size and intracellular structures); Q5=FSChigh/SSClow (bigger cells); Q6=FSChigh/SSChigh (bigger cells, more intracellular structures); Q7=FSClow/SSChigh (normal size, more intracellular structure); N = 5 for each. Error bars represent standard deviation] (D) HPLC analysis of different hemoglobin variants. Bar graphs depict the percentage of hemoglobin variants: “HbF total” = sum of HbF1 and HbF2, Hb-HbH = sum of all non-aberrant hemoglobin variants, HbA2, HbE, HbH represent the peaks identified as HbH, probably representing HbBarts (beta tetramers or gamma tetramers respectively) and “Hb Total” = sum of all the peaks. Experiments represent an N = 3 for TTD and N = 6 for control iPS-derived erythroid cells. Error bars represent standard deviation and student t-test was performed to calculate significance, * <0.05 and ***< 0.01.
Discussion
We have identified a homozygous missense mutation (c.G559T [p.Asp187Tyr]) in the beta subunit of TFIIE (GTF2E2/TFIIEβ) (Fig. 1A–C) in two non-related families of Moroccan origin with non-photosensitive TTD (NPS-TTD). This same mutation was earlier identified as causative for NPS-TTD in another non-related Moroccan NPS-TTD patient (14). In addition, in the same study by Kuschal et al. another homozygous missense mutation in TFIIEβ (c.448 G > C [p.Ala150Pro]) was identified in one NPS-TTD patient from Asian origin. Together, these TFIIEβ mutations in four non-related families firmly establish a causative genetic relationship between mutated TFIIEβ and NPS-TTD.
We further noticed that this mutation in TFIIEβ [c.G559T (p.Asp178Tyr)] causes a severe reduction in the cellular amount of this protein (Fig. 1D). Strikingly, also the alpha subunit of TFIIE (TFIIEα) appeared significantly reduced by the mutation in the beta subunit (Fig. 1E), eventually leading to a strong reduction of the entire two-subunit TFIIE complex in cultured cells of these patients. Since this amino acid substitution is in close proximity of the predicted interaction-site of TFIIEβ with the winged-helix domain of TFIIEα (32), it is anticipated to compromise complex formation, which may explain the observed fragility of the entire complex. In addition, it is likely that a mutation causing instability of one subunit renders the entire complex unstable, as previously found for other protein complexes, including TTD-causing mutations of TFIIH (5,25).
Strikingly, however, this decline in the amount of an essential transcription initiation factor did not result in a measurable effect on the overall transcription when cells were grown under standard optimal culturing conditions (Fig. 2A). Only under specific conditions, i.e. incubation at elevated temperatures (Fig. 2B) or terminal differentiation (Fig. 3), we were able to observe phenotypic consequences of this hypomorphic GTF2E2/TIIEβ allele. This fever-dependent worsening of clinical symptoms is apparently not solely dependent on the pathogenic mutation, since this peculiar phenotype was not clinically confirmed in patients TTD241HE and TTD275HE. This apparent discrepancy could be explained by an incomplete or reduced penetrance of the disease-causing allele. Also, the patient’s body temperature might not always reach the critical height and duration to observe worsening of TTD-specific features, which is off course much better controlled within a cellular in vitro assay. We previously reported on a similar thermo-sensitive TTD-causing mutation in the XPD subunit of TFIIH (6) that is also associated with fever-dependent aggravation of TTD-specific features. Hyperthermia generally causes partial denaturation of cellular proteins and may affect protein complex formation. We thus envisage that fever-driven hyperthermia will further reduce the already compromised, mutation-derived, protein stability of TFIIE complexes in TTD patient cells. The number of available TFIIE complexes will thus decline to such an extent that eventually transcription will be affected. The consistent presence of presumably hypomorphic variants in these TTD patients is expected, since general transcription factors, such as TFIIE, are highly conserved and essential proteins in mammals, so fully inactivating mutations (nonsense, frame-shift) are likely not tolerated. Similarly, all reported cases with TFIIH mutations, including the TTD-causative ones, carry at least one hypomorphic allele (33). This is in line with previous reports that complete ablation of the TFIIH subunits XPB and XPD in mice (Xpb−/− or Xpd−/−) resulted in very early embryonic lethality already at the two cell stage (34,35).
Until recently, it appeared rather difficult to extrapolate TTD-causing mutations to disease-specific phenotypic expression through available cellular and biochemical assays. However, with the here applied iPS reprogramming of patient-derived fibroblasts to stem cell-like cells and the subsequent in vitro differentiation into erythroid progenitors we were now able to show such a correlation. The observed disturbance of the delicate balance of hemoglobin-forming polypeptide production in late-stage erythroid progenitors underscores our hypothesis that at least part of the TTD-specific features are a consequence of affected transcription. It is further important to note that transcriptional defects associated with TTD mutations are mainly instigated by a stability problem of either TFIIE (this paper) or TFIIH (5,25), rather than a direct failure in transcription initiation. Hence, we speculate that fragility-causing mutations in other basal factors required for gene-expression, i.e. transcription-maturation, splicing [shown recently for mutations within RNF113A (36)] or even protein translation may result in TTD-like diseases. It is thus important to further investigate possible gene-expression functions of other identified genes (e.g. MPLKIP/TTDN1) and not-yet-identified NPS-TTD causing genes.
Materials and Methods
Massive parallel sequencing
Massive parallel sequencing (software version 2.5.0.37) was done as described previously (16). Briefly, the human genome sequencing procedures include DNA library construction, DNA Nano-Ball (DNB) generation, DNB array self-assembling, cPAL-based sequencing and imaging. Image data analyses including base calling, DNB mapping, and sequence assembly. Reads were mapped to the National Center for Biotechnology Information (NCBI) reference genome, build 37. Variants were annotated using NCBI build 37 and dbSNP build 137. Data were provided as lists of sequence variants (SNPs and short indels) relative to the reference genome. Analysis of the massive parallel sequencing data was performed using Complete Genomics analysis tools (cga tools version 1.8.0 build 1; http://www.completegenomics.com/sequence-data/cgatools/) and TIBCO/Spotfire version 7.0.1 (http://spotfire.tibco.com/).
Ethics statement
Prior to our experimental onset, we obtained written informed consent from the patient’s family and all clinical investigations have been conducted according to the Declaration of Helsinki, developed by The World Medical Association (WMA).
Cell culture
Primary fibroblasts: TTD218UT (TTD), TTD241HE (TTD), C4RO (wild-type) and C5RO (wild-type), were cultured in Ham’s F10 medium (BE02-014 F, Lonza) supplemented with 10% fetal bovine serum (S1810, Biowest) and 1% penicillin-streptomycin (P0781, Sigma-Aldrich) at 37 °C, 20% O2 and 5% CO2.
SV40-immortalized human fibroblasts: TTD218UT-sv (TTD), TTD218UT-sv stably expressing TFIIEβWT-GFP, MRC5_sv (wild-type) and C5RO_sv (wild-type), were cultured in a 1: 1 mixture of DMEM (BE12-604 F/U1) and Ham’s F10 medium (BE02-014 F, Lonza) supplemented with 10% fetal bovine serum (S1810, Biowest) and 1% penicillin-streptomycin (P0781, Sigma-Aldrich) at 37 °C, 20% O2 and 5% CO2.
Human iPS cells: were cultured feeder free in E8 medium (Thermo Fisher Scientific), on matrigel (BD-biosciences)-coated plates. Medium was changed every other day and cells were passaged every 4–5 days, using ReLeSR (Stem cell technologies) as described by manufacturers with a split-ratio of 1: 20. iPS were differentiated as whole colonies derived from single cells (28). In short, iPS were made into single cells using TrypLselect (Lifetech) and 100–150 cells were plated onto 6 cm dishes. Colonies were allowed to expand to 400 μm size upon which media was changed to differentiation media containing 20 ng/ml BMP4, 40 ng/ml VEGF and 50 ng/ml bFGF in Stemline II (sigma-aldrich), media was refreshed after 3 days. At day 6, this medium was replaced by serum- free humanized IMDM-based medium (Cellquin) (37) supplemented with 1 μg/ml interleukin 3, 10 μg/ml interleukin 6, 100 ng/ml SCF, 25 ng/ml Nplate (TPO agonist), 40 ng/ml VEGF, 20 ng/ml BMP4 and 1 U/ml Erythropoietin. Cells were refreshed every 2 days and erythroid progenitors harvested between day 9 and 15 were cultured as described before in (28).
Antibodies
Primary antibodies used: α-TFIIEβ (ab187143; Abcam), α-TFIIEα (H00002960-B01, Abnova), α-TFIIEα (#1G6; gift from J.M. Egly), α-XPB (sc-293, Santa Cruz Biotechnology), α-GFP (ab290, Abcam) and α-Tubulin (T5168, Sigma-Aldrich).
Secondary antibodies used: CF770 anti-rabbit (SAB4600215, Sigma-Aldrich), CF680 anti-mouse (SAB4600199, Sigma-Aldrich), IRDye 800CW Donkey anti-mouse (926-32212, LI-COR), Alexa Fluor 555 goat anti-mouse (A21424, Invitrogen), Alexa Fluor 555 goat anti-rabbit (A21429, Invitrogen) and Alexa Fluor 488 goat anti-rabbit (A11034, Invitrogen), α-CD71 (Miltenyi Biotech), α-CD235 (BD Biosciences).
Western blot analysis
Whole cell extracts were prepared by direct lysis of isolated cell pellets in SDS–PAGE protein sample buffer reagent, separated on 8% SDS-PAGE gel, blotted onto Immobilon-FL membrane (IPFL00010, Merck Millipore Ltd.), stained with specific primary and secondary antibodies and analyzed using an Odyssey imager (LI-COR).
Protein stability
Cells were plated on 6 cm dishes and incubated overnight under normal culture conditions. Cells were treated with 100 µM Cycloheximide (CHX) and harvested at different time-points (0, 4, 8, 12, 16 and 24 h) after treatment. Whole-cell extracts were obtained by direct lysis in sample buffer. The signal intensities were measured and normalized to Tubulin. Signal intensities were plotted as the percentage of TFIIEβ levels after CHX treatment compared with the signal intensities of the non-treated samples (set at 100%).
Colony-forming ability/survival
Cells were plated on 10 cm dishes (1500 cells/dish, 40 °C survival), in triplicate. After 24 h, cells were continuously incubated at 37 °C or 40 °C for approximately 2 weeks. Colonies were fixed and stained with 0.1% Brilliant Blue R (Sigma) and counted (Gelcount, Oxford Optronix Ltd.). The survival was plotted as the percentage of colonies obtained after treatment compared with the mean number of colonies from the mock-treated cells (set at 100%).
Transcription capacity measured by EU incorporation
Cells were grown onto 24 mm cover slip and cultured for 1 day prior to the experiments. The cells were washed once with PBS and incubated for 2 h in culture medium containing 100 μM 5-ethynyl-uridine (EU). After EU incorporation, cells were fixed in 4% formaldehyde/PBS, washed twice with 3% BSA/PBS, permeabilized for 20 min in 0.5% Triton/PBS and washed once with PBS. Cells were incubated for 30 min with fluorescent dye coupling buffer containing 10 mM CuSO4 and Alexa Fluor 594 azide (Click-iT, Thermo Fisher Scientific). After washing with PBS, cells were mounted in vectashield, containing 1.5 μg/ml DAPI. The mean fluorescence is determined with a confocal microscope (Zeiss LSM 700) from at least 50 cells. Images were processed using ImageJ and the average fluorescence intensity in the nucleus of MRC5_sv wild-type cells was set at 100%.
Immuno fluorescence
Cells were grown on glass cover slips (24 mm) for 1 day prior to the experiments and washed with PBS, fixed with 2% paraformaldehyde for 15 min, washed with PBS, washed 2 times 10 min with 0.1% Triton X-100/PBS and incubated for 15 min with PBS+ (PBS containing 0.15% glycine and 1% BSA). Cells were incubated overnight at 4 °C with primary antibodies in a moist chamber. The next day, cover slips were washed 2 times 10 min with PBS/Triton X-100 and washed once with PBS+. Cells were incubated for 1 h with secondary antibodies at room temperature in moist chamber and again washed three times in PBS/Triton X-100. Samples were embedded in Vectashield mounting medium (Vector Laboratories, containing 1.5 μg/ml DAPI). The mean fluorescence is determined with a confocal microscope (Zeiss LSM 700) from at least 50 cells. Images were processed using ImageJ.
Generation of human iPS cells
Primary fibroblasts from patient TTD218UT and control fibroblasts were reprogrammed through lentiviral transduction of human genes OCT4, SOX2, c-MYC and KLF4, using engineered color-coded lentiviral vectors (27).
Flow cytometry
Cells were washed once in PBS, resuspended in PBS with 0.5% BSA and stained with appropriate antibodies for 1 h as indicated in the figure legends. Cells were washed in PBS and flow cytometry experiments were performed with BD FACS CantoII or BD LSRII + HTS (BD Bioscsiences, Franklin Lakes, NJ). The data were analyzed with FACSDiva Software (BD Biosciences, Franklin Lakes, NJ) and FlowJo Software (Tree Star, Ashland, OR).
Cytospin preparation
Cells (5 × 105) were cytospun onto glass slides, fixed in methanol, and stained with benzidine to visualize haemoglobin and DIFCO B/C. Images were taken with a Zeiss Axioscope A1 macroscope (50x lens) and processed using Adobe Photoshop 9.0 (Adobe Systems Inc.; CA, USA).
HPLC to detect hemoglobin variants
Separation of the various Hb fractions was performed by high-performance cation-exchange liquid chromatography (CE-HPLC) on Waters Alliance 2690 equipment (Waters, Milford, MA, USA) according to (31). In short, the protocol consisted of a 30-min elution over a combined 20–200 mM NaCl and pH 7.0–6.6 gradient in 20 mM BisTris/HCl, 2 mM KCN. The column, a PolyCAT A 100/4.6-mm, 3 μm, 1500 Å column, was purchased from PolyLC (Columbia, MD, USA).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank the Optical Imaging Centre (OIC) of the Erasmus MC for microscopy support.
Conflict of Interest statement. None declared.
Funding
Dutch Science Organization (NWO), ALW division (grant numbers 864.13.004 and 854.11.002), ZonMW division (grant number 912.12.132) and by the European Research Council (ERC; grant numbers 233424 and 340988). Funding to pay the Open Access publication charges for this article was provided by the European Research Council, grant ERC-ID 340988.
References
- 1. Faghri S., Tamura D., Kraemer K.H., Digiovanna J.J. (2008) Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J. Med. Genet., 45, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stefanini M., Botta E., Lanzafame M., Orioli D. (2010) Trichothiodystrophy: from basic mechanisms to clinical implications. DNA Repair (Amst), 9, 2–10. [DOI] [PubMed] [Google Scholar]
- 3. Theil A.F., Hoeijmakers J.H., Vermeulen W. (2014) TTDA: big impact of a small protein. Exp. Cell Res., 329, 61–68. [DOI] [PubMed] [Google Scholar]
- 4. Compe E., Egly J.M. (2012) TFIIH: when transcription met DNA repair. Nat Rev Mol. Cell Biol., 13, 343–354. [DOI] [PubMed] [Google Scholar]
- 5. Vermeulen W., Bergmann E., Auriol J., Rademakers S., Frit P., Appeldoorn E., Hoeijmakers J.H., Egly J.M. (2000) Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet., 26, 307–313. [DOI] [PubMed] [Google Scholar]
- 6. Vermeulen W., Rademakers S., Jaspers N.G., Appeldoorn E., Raams A., Klein B., Kleijer W.J., Hansen L.K., Hoeijmakers J.H. (2001) A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat. Genet., 27, 299–303. [DOI] [PubMed] [Google Scholar]
- 7. de Boer J., de Wit J., van Steeg H., Berg R.J., Morreau H., Visser P., Lehmann A.R., Duran M., Hoeijmakers J.H., Weeda G. (1998) A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell, 1, 981–990. [DOI] [PubMed] [Google Scholar]
- 8. Viprakasit V., Gibbons R.J., Broughton B.C., Tolmie J.L., Brown D., Lunt P., Winter R.M., Marinoni S., Stefanini M., Brueton L.. et al. (2001) Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum. Mol. Genet., 10, 2797–2802. [DOI] [PubMed] [Google Scholar]
- 9. Vermeij W.P., Hoeijmakers J.H., Pothof J. (2016) Genome Integrity in Aging: Human Syndromes, Mouse Models, and Therapeutic Options. Annu. Rev. Pharmacol. Toxicol., 56, 427–445. [DOI] [PubMed] [Google Scholar]
- 10. Hasty P., Campisi J., Hoeijmakers J., van Steeg H., Vijg J. (2003) Aging and genome maintenance: lessons from the mouse?. Science, 299, 1355–1359. [DOI] [PubMed] [Google Scholar]
- 11. Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J. (2003) Oxidative DNA damage: mechanisms, mutation, and disease. faseb J., 17, 1195–1214. [DOI] [PubMed] [Google Scholar]
- 12. Vermeulen W., van Vuuren A.J., Chipoulet M., Schaeffer L., Appeldoorn E., Weeda G., Jaspers N.G., Priestley A., Arlett C.F., Lehmann A.R.. et al. (1994) Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb. Symp. Quant. Biol., 59, 317–329. [DOI] [PubMed] [Google Scholar]
- 13. Heller E.R., Khan S.G., Kuschal C., Tamura D., DiGiovanna J.J., Kraemer K.H. (2015) Mutations in the TTDN1 gene are associated with a distinct trichothiodystrophy phenotype. J. Invest. Dermatol., 135, 734–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuschal C., Botta E., Orioli D., Digiovanna J.J., Seneca S., Keymolen K., Tamura D., Heller E., Khan S.G., Caligiuri G.. et al. (2016) GTF2E2 mutations destabilize the general transcription factor complex TFIIE in individuals with DNA repair-proficient trichothiodystrophy. Am. J. Hum. Genet., 98, 627–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corbett M.A., Dudding-Byth T., Crock P.A., Botta E., Christie L.M., Nardo T., Caligiuri G., Hobson L., Boyle J., Mansour A.. et al. (2015) A novel X-linked trichothiodystrophy associated with a nonsense mutation in RNF113A. J. Med. Genet., 52, 269–274. [DOI] [PubMed] [Google Scholar]
- 16. Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G.. et al. (2010) Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science, 327, 78–81. [DOI] [PubMed] [Google Scholar]
- 17. Swagemakers S.M., Jaspers N.G., Raams A., Heijsman D., Vermeulen W., Troelstra C., Kremer A., Lincoln S.E., Tearle R., Hoeijmakers J.H.. et al. (2014) Pollitt syndrome patients carry mutation in TTDN1. Meta. Gene, 2, 616–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sainsbury S., Bernecky C., Cramer P. (2015) Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol., 16, 129–143. [DOI] [PubMed] [Google Scholar]
- 19. Nogales E., Louder R.K., He Y. (2017) Structural insights into the eukaryotic transcription initiation machinery. Annu. Rev. Biophys., 46, 59–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Itoh Y., Unzai S., Sato M., Nagadoi A., Okuda M., Nishimura Y., Akashi S. (2005) Investigation of molecular size of transcription factor TFIIE in solution. Proteins, 61, 633–641. [DOI] [PubMed] [Google Scholar]
- 21. Hayashi K., Watanabe T., Tanaka A., Furumoto T., Sato-Tsuchiya C., Kimura M., Yokoi M., Ishihama A., Hanaoka F., Ohkuma Y. (2005) Studies of Schizosaccharomyces pombe TFIIE indicate conformational and functional changes in RNA polymerase II at transcription initiation. Genes Cells, 10, 207–224. [DOI] [PubMed] [Google Scholar]
- 22. Jawhari A., Uhring M., De Carlo S., Crucifix C., Tocchini-Valentini G., Moras D., Schultz P., Poterszman A. (2006) Structure and oligomeric state of human transcription factor TFIIE. EMBO Rep., 7, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H.T., Warfield L., Hahn S. (2007) The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol., 14, 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grunberg S., Warfield L., Hahn S. (2012) Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol., 19, 788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Botta E., Nardo T., Lehmann A.R., Egly J.M., Pedrini A.M., Stefanini M. (2002) Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet., 11, 2919–2928. [DOI] [PubMed] [Google Scholar]
- 26. Lin Y.C., Gralla J.D. (2005) Stimulation of the XPB ATP-dependent helicase by the beta subunit of TFIIE. Nucleic Acids Res., 33, 3072–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Warlich E., Kuehle J., Cantz T., Brugman M.H., Maetzig T., Galla M., Filipczyk A.A., Halle S., Klump H., Scholer H.R.. et al. (2011) Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther., 19, 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van den Akker E., Satchwell T.J., Pellegrin S., Daniels G., Toye A.M. (2010) The majority of the in vitro erythroid expansion potential resides in CD34(-) cells, outweighing the contribution of CD34(+) cells and significantly increasing the erythroblast yield from peripheral blood samples. Haematologica, 95, 1594–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dolznig H., Bartunek P., Nasmyth K., Mullner E.W., Beug H. (1995) Terminal differentiation of normal chicken erythroid progenitors: shortening of G1 correlates with loss of D-cyclin/cdk4 expression and altered cell size control. Cell Growth Differ., 6, 1341–1352. [PubMed] [Google Scholar]
- 30. Cao A., Galanello R. (2010) Beta-thalassemia. Genet. Med., 12, 61–76. [DOI] [PubMed] [Google Scholar]
- 31. van Zwieten R., Veldthuis M., Delzenne B., Berghuis J., Groen J., Ait Ichou F., Clifford E., Harteveld C.L., Stroobants A.K. (2014) Hemoglobin analyses in the Netherlands reveal more than 80 different variants including six novel ones. Hemoglobin. 38, 1–7. [DOI] [PubMed] [Google Scholar]
- 32. Miwa K., Kojima R., Obita T., Ohkuma Y., Tamura Y., Mizuguchi M. (2016) Crystal Structure of Human General Transcription Factor TFIIE at Atomic Resolution. J. Mol. Biol., 428, 4258–4266. [DOI] [PubMed] [Google Scholar]
- 33. Hashimoto S., Egly J.M. (2009) Trichothiodystrophy view from the molecular basis of DNA repair/transcription factor TFIIH. Hum. Mol. Genet., 18, R224–R230. [DOI] [PubMed] [Google Scholar]
- 34. de Boer J., Donker I., de Wit J., Hoeijmakers J.H., Weeda G. (1998) Disruption of the mouse xeroderma pigmentosum group D DNA repair/basal transcription gene results in preimplantation lethality. Cancer Res., 58, 89–94. [PubMed] [Google Scholar]
- 35. Andressoo J.O., Weeda G., de Wit J., Mitchell J.R., Beems R.B., van Steeg H., van der Horst G.T., Hoeijmakers J.H. (2009) An Xpb mouse model for combined xeroderma pigmentosum and cockayne syndrome reveals progeroid features upon further attenuation of DNA repair. Mol. Cell Biol., 29, 1276–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu N.Y., Chung C.S., Cheng S.C. (2017) Role of Cwc24 in the First Catalytic Step of Splicing and Fidelity of 5' Splice Site Selection. Mol. Cell Biol., 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Migliaccio G., Sanchez M., Masiello F., Tirelli V., Varricchio L., Whitsett C., Migliaccio A.R. (2010) Humanized culture medium for clinical expansion of human erythroblasts. Cell Transplant, 19, 453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.