Abstract
Epigenetic regulation of cellular function provides a mechanism for rapid organismal adaptation to changes in health, lifestyle and environment. Associations of cytosine-guanine di-nucleotide (CpG) methylation with clinical endpoints that overlap with metabolic phenotypes suggest a regulatory role for these CpG sites in the body’s response to disease or environmental stress. We previously identified 20 CpG sites in an epigenome-wide association study (EWAS) with metabolomics that were also associated in recent EWASs with diabetes-, obesity-, and smoking-related endpoints. To elucidate the molecular pathways that connect these potentially regulatory CpG sites to the associated disease or lifestyle factors, we conducted a multi-omics association study including 2474 mass-spectrometry-based metabolites in plasma, urine and saliva, 225 NMR-based lipid and metabolite measures in blood, 1124 blood-circulating proteins using aptamer technology, 113 plasma protein N-glycans and 60 IgG-glyans, using 359 samples from the multi-ethnic Qatar Metabolomics Study on Diabetes (QMDiab). We report 138 multi-omics associations at these CpG sites, including diabetes biomarkers at the diabetes-associated TXNIP locus, and smoking-specific metabolites and proteins at multiple smoking-associated loci, including AHRR. Mendelian randomization suggests a causal effect of metabolite levels on methylation of obesity-associated CpG sites, i.e. of glycerophospholipid PC(O-36: 5), glycine and a very low-density lipoprotein (VLDL-A) on the methylation of the obesity-associated CpG loci DHCR24, MYO5C and CPT1A, respectively. Taken together, our study suggests that multi-omics-associated CpG methylation can provide functional read-outs for the underlying regulatory response mechanisms to disease or environmental insults.
Introduction
Complex disorders including cancer, cardiovascular disease and diabetes, as well as exposure to environmental insults can lead to adjustments of the expression of corresponding enzymes, transporters and metabolic regulators (1). The organismal response to these challenges can be reflected by changes in DNA methylation (2). Individual differences in health, lifestyle and environmental exposure therefore leave their imprint on the individual’s epigenome (2). This has been documented in numerous recent epigenome-wide association studies (EWASs) with type 2 diabetes (T2D) (3–5), smoking (6–15), obesity (16–22), blood pressure (23) and protein markers of liver function (24). Methylation of CpG sites that associate with disease or lifestyle factors often also associates with changes in intermediate molecular phenotypes, in particular blood circulating metabolites, as we have previously shown (25).
Methylation of CpG cg05575921 at the aryl hydrocarbon receptor repressor (AHRR) gene locus was associated with tobacco smoking in numerous studies (6–9,11–14,26). Smoking during pregnancy also affected methylation at the same CpG site in newborns (15). We reported an association of this CpG site with blood circulating 4-vinylphenol, supporting the function of AHRR as a mediator of dioxin toxicity (25). Similarly, the robustly replicated associations of diabetes and obesity with differential CpG methylation near the genes that encode TXNIP (cg19693031), ABCG1 (cg06500161) and CPT1A (cg00574958) (3–5,17) likely reflect a gene regulatory response to diabetes and obesity induced metabolic dysregulations. TXNIP, for instance, plays an important role in glucose regulation by directly suppressing glucose uptake through binding to the glucose transporter GLUT1 (27). This idea is supported by our previously reported association of these three CpG sites with a diabetes-specific metabolic phenotype (metabotype), including changes in the well-established T2D biomarkers alpha-hydroxybutyrate (AHB), 3-methyl-2-oxovalerate, glycine and several diabetes-associated lipids (5,25). A recent obesity Mendelian randomization (MR) study by Wahl et al. (22) showed that adiposity was causal for changes in methylation of multiple CpG sites near obesity-related genes. Interestingly, several of the CpG sites identified in that study were also within a set of 20 CpG sites that we previously identified in an EWAS with blood metabolites (25) (Table 1).
Table 1.
Summary of CpG–intermediate trait–complex trait associations for the CpG sites from the Petersen et al. (25) study
| Intermediate traits in QMDiab |
Complex traits |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus name CpG Chr: Pos | Replication of Petersen studya | Metabolite (blood) | Metabolite (urine) | Metabolite (saliva) | Lipid | Protein | Glycan | T2D | BMI | Blood pressure | Liver functions | Smoking | Reference | |
| UGT2B15—cg09189601 | ||||||||||||||
| chr4: 69514031 | ✓✓ | ✓ | ||||||||||||
| TXNIP—cg19693031 | ||||||||||||||
| chr1: 145441552 | ✓ | ✓ | ✓ | ✓ | ✓b | c✓ | ✓* | ✓ | (3,23) | |||||
| DHCR24—cg17901584 | ||||||||||||||
| chr1: 55353706 | ✓✓ | ✓ | ✓ | (22) | ||||||||||
| MYO5C—cg06192883 | ||||||||||||||
| chr15: 52554171 | ✓✓ | ✓ | ✓ | (22) | ||||||||||
| ABCG1—cg06500161 | ||||||||||||||
| chr21: 43656587 | ✓✓ | ✓ | ✓ | ✓ | ✓ | (3,22) | ||||||||
| SLC25A22—cg09441501 | ||||||||||||||
| chr11: 798350 | ||||||||||||||
| CPT1A—cg00574958 | ||||||||||||||
| chr11: 68607622 | ✓✓ | ✓ | ✓ | ✓ | ✓ | (3,22,23) | ||||||||
| SLC7A11—cg06690548 | ||||||||||||||
| chr4: 139162808 | ✓ | ✓ | ✓ | ✓ | ✓ | (22–24) | ||||||||
| PHGDH—cg14476101 | ||||||||||||||
| chr1: 120255992 | ✓ | ✓ | (22,23) | |||||||||||
| LOC100132354—cg18120259 | ||||||||||||||
| chr6: 43894639 | ✓ | ✓ | ✓ | (22,23) | ||||||||||
| SLC1A5—cg22304262 | ||||||||||||||
| chr19: 47287778 | ✓ | ✓ | ✓ | (23,24) | ||||||||||
| cg13526915 | ✓✓ | |||||||||||||
| chr14: 24164078 | ||||||||||||||
| AHRR—cg05575921 | ||||||||||||||
| chr5: 373378 | ✓✓ | ✓ | ✓ | ✓ | b✓ | ✓* | (8) | |||||||
| ALPPL2—cg21566642 | ||||||||||||||
| chr2: 233284661 | ✓✓ | ✓ | ✓ | ✓ | ✓* | (8) | ||||||||
| F2RL3—cg03636183 | ||||||||||||||
| chr19: 17000585 | ✓ | ✓ | ✓ | b✓ | ✓* | (8) | ||||||||
| cg06126421 | ✓✓ | ✓ | ✓ | b✓ | ✓* | (8) | ||||||||
| chr6: 30720080 | ||||||||||||||
| RARA—cg19572487 | ||||||||||||||
| chr17: 38476024 | ✓✓ | ✓ | b✓ | ✓* | (8) | |||||||||
| GFI1—cg09935388 | ✓ | ✓* | (8) | |||||||||||
| chr1: 92947588 | ✓ | ✓ | ||||||||||||
| TPM1—cg10403394 | ||||||||||||||
| chr15: 63349192 | ||||||||||||||
| cg23079012 | ✓ | ✓ | ✓ | ✓* | (8) | |||||||||
| chr2: 8343710 | ||||||||||||||
Entries marked with asterisks indicate that these associations are genome-wide significant in QMDiab as well.
For the Replication of the Petersen study, 2 ticks indicate Bonferroni significance, and 1 tick indicates nominal significance.
These associations include a replication in KORA.
These associations include a replication in TwinsUK.
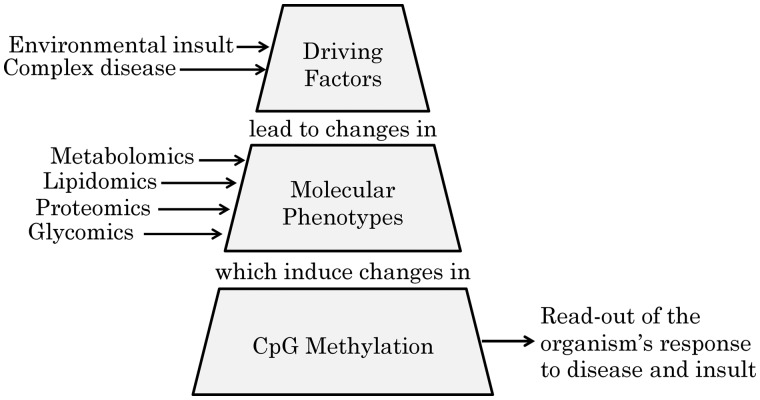
These observations clearly indicated a role of DNA methylation in the regulation of the cellular response to disease and environmental stress. We therefore hypothesized that the molecular pathways that constitute these organismal responses can be revealed by assessing the relationships between changes in intermediate molecular phenotypes and changes in gene regulation, in particular by studying their association with the DNA methylome (Fig. 1). As our study cohort is relatively small, but otherwise exceptionally deeply phenotyped at a multi-omics level, we focused on 20 CpG sites that we previously identified in our EWAS with metabolomics (25). Indeed, a review of recently published EWAS revealed that actually most of these 20 CpG sites were also associated with complex disease phenotypes including obesity, diabetes, blood pressure, and liver function, and or smoking (Table 1). As our previous EWAS with metabolomics did not contain a formal replication, we start by replicating the association of these CpG sites in a similar panel of blood metabolites in the QMDiab study, a diabetes cohort including Arab and South Asian ethnicities. We then focus our investigation on associations of these 20 CpG sites with a diverse set of almost 4000 deep molecular phenotypes, including blood, urinary and salivary metabolomics, lipidomics, proteomics and glycomics. We further replicate the newly discovered protein and glycan associations in independent studies. Finally, we use MR to evaluate the causal direction of selected CpG-metabolite associations.
Figure 1.
Hypothesis tested in this study. Exposure to physiological challenges, such as an increased BMI, smoking or dysregulated glycemic control leads physiological changes that translate into changes in intermediate molecular phenotypes, such as metabolite levels that are detectable in different body fluids, blood circulating lipids, proteins and protein glycosylation. These then further induce changes in DNA methylation at specific regulatory sites of genes that are required to counter this insult. Note that this view does not exclude that changes in the expression of certain genes may not also result in further changes in molecular phenotypes. Hence, despite the fact that we found here three cases of causality from metabolite to CpG, cases with reverse directionality are also likely to exist.
Results
Deep molecular phenotyping of 3996 multi-omics parameters in an Arab–Asian cohort
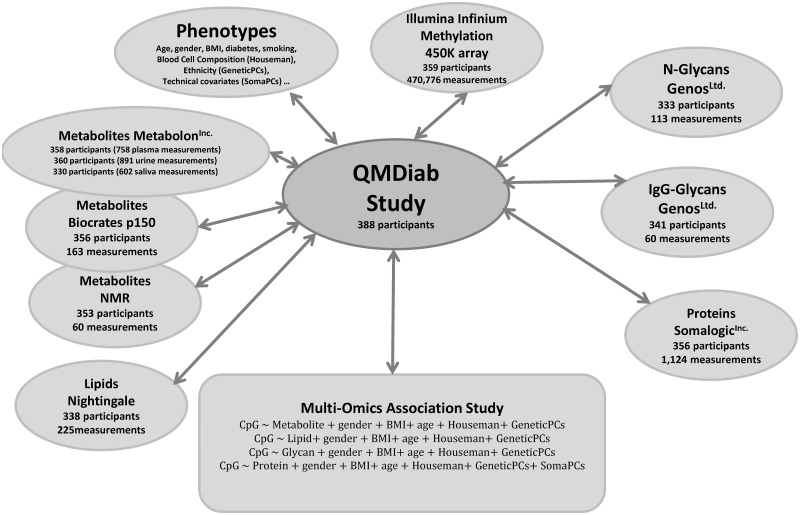
We determined 2251 metabolic traits (758 from plasma, 891 from urine and 602 from saliva) using a non-targeted metabolomics platform (Metabolon Inc., Durham, NC), 163 metabolites using a targeted metabolomics kit (Biocrates Life Sciences AG, Innsbruck, Austria), 60 urinary metabolite concentrations (Chenomx Inc., Edmonton, CA), 225 mostly lipid-related blood traits (Nightingale Ltd, Helsinki, Finland) based on 1H NMR measurements, 1124 blood circulating proteins using an aptamer-based technology (Somalogic Inc., Boulder, USA), 113 blood N-glycans using UPLC, 60 IgG-glycopeptides by liquid chromatography mass spectrometry lycol-profiling (Genos Ltd., Zagreb, Croatia) and methylation at 470 776 CpG sites using the Illumina Infinium HumanMethylation450 BeadChip platform (28) (see Materials and Methods), in up to 359 individuals from the QMDiab study (Table 2) (29). The methylation data overlapped with at least one type of proteomics, lipidomics, glycomics or metabolomics data in this study (Fig. 2). This diabetes case-control study comprises 50.7% individuals with diabetes and 17.3% individuals who are smokers. Taken together, we obtained a maximum of 3996 molecular phenotypes in saliva, blood and urine samples of up to 359 individuals (Supplementary Material, Table S1).
Table 2.
General characteristics of the QMDiab study participantsa
| Age (years) | 46.8±12.8 (mean ± s.d.) |
| Sex | 177 (49.3%) female |
| 182 (50.7%) male | |
| Body mass index (kg/m2) | 29.6±6.0 (mean ± s.d.) |
| Ethnicityb | 189 (52.6%) Arab |
| 106 (29.5%) South Asian | |
| 34 (9.5%) Filipino | |
| 13 (3.6%) other/mixed | |
| 17 (4.7%) missing | |
| T2D status | 182 (50.7%) having diabetes |
| 176 (49.0%) no diabetes | |
| 1 (0.03%) missing | |
| Smoking statusc | 62 (17.3%) smokers |
| 280 (78.0%) non-smokers | |
| 17 (4.7%) missing |
The QMDiab study has been described previously and comprises 388 study participants from Arab and Asian ethnicities (29). The statistics here are reported for the 359 samples with methylation data overlapping with at least one type of proteomics, lipidomics, glycomics, or metabolomics measurement.
Arab ethnicity includes participants from Bahrain, Egypt, Iraq, Jordan, Kuwait, Lebanon, Morocco, Oman, Palestine, Qatar, Saudi Arabia, Somalia, Sudan, Syria, Tunisia, United Arab Emirates and Yemen. South Asian ethnicity includes participants from Bangladesh, India, Nepal, Pakistan, and Sri Lanka.
Smoking status was determined based on the detection of cotinine in blood at the time of blood collection.
Figure 2.
Multi-omics dataset and study design. A total of 388 individuals participated in the initial QMDiab study. A total of 359 samples had DNA methylation data and at least one other deep-molecular trait.
Replication of a previous metabolomics EWAS
The first EWAS with metabolic traits assessed 649 blood metabolic traits from 1805 samples from the KORA study with methylation measurements for 457 004 CpG sites (25). The associations from this study have not been replicated. We therefore started by replicating the metabolomics–methylation associations reported in that study. We could replicate 10 out of the 20 lead methylation associations with metabolites at Bonferroni significance (P < 0.05/20) and found nominal significance (P < 0.05) for a further seven of them (Supplementary Material, Table S2).
Discovery of novel phenome-wide associations with CpG methylation
We then tested the 20 CpG sites for association with all available 2474 metabolites, 225 lipids, 1124 proteins and 173 glycan traits, requiring a Bonferroni level of significance that accounted for the respective number of tested molecular phenotypes and 20 CpG loci (see Materials and Methods). Loci were annotated following (25), using the most likely regulated gene, CpG identifier and phenotype (diabetes, smoking, obesity, steroids, other). We identified 138 associations between methylation and other phenotypes including numerous hits at the TXNIP-diabetes and at the AHRR-smoking loci (Table 3). Of the 138 associations, 12 involved proteins, 19 involved lipids, 91 involved metabolites and 16 involved glycans. We found multiple associations at the DHCR24 and ABCG1 obesity loci with various LDL lipid subclasses, consistent with previous studies (30,31). We also linked the kidney function marker myo-inositol (32), measured here in urine, to changes in methylation of the obesity locus ABCG1. Further highlights include the association of a new, yet unidentified metabolite (X-19141) with cg09189601 methylation at the UGT2B15 locus, of specific IgG glycopeptides with cg06192883 methylation at the MYO5C obesity locus, and of the blood circulating protein levels of Tumor necrosis factor ligand superfamily member 4 (TNFSF4) with cg00574958 at the diabetes and obesity locus CPT1A. Many of the CpG–blood metabolite associations previously reported in the supplement of the Petersen et al. study were also replicated here using different metabolomics technologies. We further observed for the first time associations of the same metabolites in urine and saliva, sometimes stronger than the associations in plasma. The complete set of significant associations is in Supplementary Material, Table S3.
Table 3.
Multi-omics associations with CpG methylation in QMDiab
| Locus | Group | Trait | Trend | P-Value |
|---|---|---|---|---|
|
Metabolites | X-19141 [plasma] | ↓ | 6.21 ×10−23 |
|
Metabolites | 1, 5-anhydroglucitol (1, 5-AG) [plasma]a | ↑ | 7.56 × 10−21 |
| Glucose [NMR] | ↓ | 1.17 × 10−14 | ||
| 2-hydroxybutyrate (AHB) [urine] | ↓ | 2.52 × 10−14 | ||
| 3-hydroxybutyrate (BHBA) [urine] | ↓ | 5.87 × 10−13 | ||
| glucose [plasma]a | ↓ | 3.15 × 10−12 | ||
| … (list truncated) | ||||
| Lipids | L-VLDL-CE_% | ↑ | 1.01 × 10−8 | |
| XL-VLDL-CE_% | ↑ | 2.05 × 10−8 | ||
| M-VLDL-CE_% | ↑ | 4.42 × 10−6 | ||
| XL-VLDL-C_% | ↑ | 5.66 × 10−6 | ||
| … (list truncated) | ||||
| Proteins | Transmembrane glycoprotein NMB (GPNMB) | ↓ | 1.30 × 10−8 | |
| Aminoacylase-1 (ACY1) | ↓ | 2.59 × 10−7 | ||
| Sex hormone-binding globulin (SHBG) | ↑ | 4.65 × 10−7 | ||
| Melanoma-derived growth regulatory protein (MIA) | ↑ | 6.88 × 10−7 | ||
| Glycans | PGP23 | ↓ | 2.75 × 10−8 | |
| PGP31 | ↓ | 9.31 × 10−8 | ||
| PGP29 | ↓ | 7.61 × 10−6 | ||
| PGP28 | ↓ | 8.42 × 10−6 | ||
|
Lipids | M-VLDL-C_% | ↑ | 6.62 × 10−9 |
| M-VLDL-TG_% | ↓ | 7.14 × 10−9 | ||
| M-VLDL-CE_% | ↑ | 1.87 × 10−8 | ||
| S-VLDL-TG_% | ↓ | 1.36 × 10−6 | ||
| … (list truncated) | ||||
|
Glycans | PGP58 | ↑ | 8.31 × 10−9 |
| PGP70 | ↑ | 6.91 × 10−8 | ||
| PGP1 | ↑ | 3.67 × 10−7 | ||
| PGP17 | ↓ | 8.34 × 10−7 | ||
| PGP99 | ↑ | 1.23 × 10−6 | ||
| … (list truncated) | ||||
| IgG1_G0F | ↑ | 9.46 × 10−7 | ||
| IgG4_G2FN | ↓ | 1.79 × 10−5 | ||
|
Metabolites | Myo-inositol [urine] | ↑ | 7.21 × 10−7 |
| Lipids | L-VLDL-CE_% | ↓ | 1.03 × 10−8 | |
| M-VLDL-CE_% | ↓ | 2.19 × 10−7 | ||
| M-VLDL-C_% | ↓ | 2.26 × 10−7 | ||
| XXL-VLDL-CE_% | ↓ | 8.73 × 10−7 | ||
| … (list truncated) | ||||
|
Proteins | Tumor necrosis factor ligand superfamily member 4 (TNFSF4) | ↑ | 1.61 × 10−6 |
|
Metabolites | Serine [plasma] | ↑ | 3.05 × 10−7 |
|
Metabolites | o-cresol sulfate [urine] | ↓ | 2.66 × 10−27 |
| 3-ethylphenylsulfate* [urine] | ↓ | 1.08 × 10−17 | ||
| X-17185 [urine]b X-17185 [plasma] | ↓ ↓ | 2.42 × 10−16, 1.52 × 10−7 | ||
| X-12161 [urine] | ↓ | 5.17 × 10−13 | ||
| X-17398 [urine] | ↓ | 1.36 × 10−12 | ||
| … (list truncated) | ||||
| Proteins | Polymeric immunoglobulin receptor (PIGR) | ↓ | 2.03 × 10−11 | |
|
Metabolites | o-Cresol sulfate [urine] | ↓ | 7.43 × 10−16 |
| 3-ethylphenylsulfate* [urine] | ↓ | 4.45 × 10−9 | ||
| X-17185 [plasma]b X-17185 [urine] | ↓ ↓ | 6.32 × 10−8, 1.15 × 10−6 | ||
| X-17398 [urine] | ↓ | 6.35 × 10−8 | ||
| 2-ethylphenylsulfate [urine] | ↓ | 4.54 × 10−7 | ||
|
Metabolites | o-Cresol sulfate [urine] | ↓ | 4.93 × 10−13 |
| 3-ethylphenylsulfate* [urine] | ↓ | 1.09 × 10−9 | ||
| X-17398 [urine] | ↓ | 1.41 × 10−8 | ||
| X-17185 [urine] | ↓ | 9.18 × 10−7 | ||
| Proteins | Polymeric immunoglobulin receptor (PIGR) | ↓ | 9.02 × 10−7 | |
|
Metabolites | X-17185 [urine]b X-17185 [plasma] | ↓ ↓ | 2.99 × 10−10, 6.23 × 10−7 |
| o-Cresol sulfate [urine] | ↓ | 2.23 × 10−9 | ||
| X-17398 [urine] | ↓ | 2.49 × 10−7 | ||
| 3-ethylphenylsulfate* [urine] | ↓ | 5.37 × 10−7 | ||
| X-17320 [urine] | ↓ | 5.47 × 10−7 | ||
| 3-methyl catechol sulfate 1 [urine] | ↓ | 5.53 × 10−7 | ||
| Proteins | Polymeric immunoglobulin receptor (PIGR) | ↓ | 3.36 × 10−7 | |
|
Metabolites | o-Cresol sulfate [urine] | ↓ | 3.29 × 10−7 |
| Proteins | Gelsolin (GSN) | ↑ | 1.89 × 10−6 | |
|
Metabolites | o-Cresol sulfate [urine] | ↓ | 2.90 × 10−7 |
|
Metabolites | o-Cresol sulfate [urine] | ↓ | 4.51 × 10−8 |
| Proteins | X-linked interleukin-1 receptor accessory protein-like 2 (IL1RAPL2) | ↓ | 4.35 × 10−9 | |
| Vascular endothelial growth factor A (VEGFA) | ↓ | 1.42 × 10−6 | ||
| NudC domain-containing protein 3 (NUDCD3) | ↓ | 1.55 × 10−6 |
Association data for 14 of the 20 CpG loci reported by Petersen et al. (25). P-values are for the reported phenotypes in linear regression models with the respective covariates (Fig. 2). Associations were required to reach a Bonferroni level of significance of pmetabolite < 1.01 × 10−6, plipid < 1.11 × 10−5, pprotein < 2.22 × 10−6, and pglycan < 2.21 × 10−5 for metabolites, lipids, proteins, and glycan traits, respectively. Genomic coordinates are based on human genome build 37. A positive association with methylation levels is indicated by (↑), while a negative is indicated by (↓). Full summary statistics are in Supplementary Material, Table S3.
This metabolite was already reported in Petersen et al.
This metabolite was measured on different platforms or in different fluids in QMDiab (indicated in square brackets).
Note: We only present the five most significant associations for each category in this table. For a more comprehensive list, see Supplementary Material, Table S3.
Replication of novel CpG-protein and CpG-glycan associations in independent studies
Next, we attempted replication of the novel protein–methylation associations in the KORA study (N = 997) and of the novel glycan–methylation associations in the TwinsUK study (N = 165). Six of the twelve protein–methylation associations replicated in KORA at a Bonferroni level of significance (P < 0.0041 = 0.05/12). All replicated associations showed the same direction of effect (Table 4). Of the 16 glycan–methylation associations, two were not measured in the TwinsUK study and could not be tested, four displayed nominal significance (P < 0.05) and one replicated at Bonferroni significance (P < 0.0035 = 0.05/14). All glycan–methylation associations showed concordant directions between the two studies (Table 5).
Table 4.
Replication of novel proteome-methylation associations in the KORA study
| QMDiab |
KORA |
||||
|---|---|---|---|---|---|
| Locus | Protein | P-value | Beta | P-value | Beta |
| TXNIP | Transmembrane glycoprotein NMB | 1.30 × 10−8 | −0.006 | 0.841 | −0.002 |
| cg19693031 | Aminoacylase-1 | 2.59 × 10−7 | −0.002 | 2.58 × 10−7 | −0.028 |
| Sex hormone-binding globulin | 4.65 × 10−7 | 0.002 | 0.002 | 0.019 | |
| Melanoma-derived growth regulatory protein | 6.88 × 10−7 | 0.006 | 0.106 | 0.021 | |
| CPT1A cg00574958 | Tumor necrosis factor ligand superfamily member 4 | 1.61 × 10−6 | 0.002 | 0.204 | 0.003 |
| AHRR cg05575921 | Polymeric immunoglobulin receptor | 2.03 × 10−11 | −0.004 | 3.30 × 10−27 | −0.153 |
| F2RL3 cg03636183 | Polymeric immunoglobulin receptor | 9.02 × 10−7 | −0.002 | 5.82 × 10−19 | −0.075 |
| cg06126421 | Polymeric immunoglobulin receptor | 3.36 × 10−7 | −0.003 | 8.29 × 10−11 | −0.065 |
| RARA cg19572487 | Gelsolin | 1.89 × 10−6 | 0.004 | 0.001 | 0.059 |
| cg23079012 | X-linked interleukin-1 receptor accessory protein-like 2 | 4.35 × 10−9 | −0.001 | 0.756 | −0.001 |
| Vascular endothelial growth factor A | 1.42 × 10−6 | −0.002 | 0.600 | −0.004 | |
| NudC domain-containing protein 3 | 1.55 × 10−6 | −0.001 | 0.338 | 0.003 | |
Six out of twelve protein-methylation associations were replicated in KORA (N = 997) at Bonferroni significance p < 0.0041 (0.05/12). All but one association showed concordant directions in the two studies.
Table 5.
Replication of novel N-glycan-methylation associations in the TwinsUK study
| QMDiab |
TwinsUK |
||||
|---|---|---|---|---|---|
| Locus | Glycan | P-value | Beta | P-value | Beta |
| TXNIP | PGP23 | 2.75 × 10−8 | −0.023 | 0.115 | −0.007 |
| cg19693031 | PGP31 | 9.31 × 10−8 | −0.025 | 0.035 | −0.009 |
| PGP29 | 7.61 × 10−6 | −0.019 | 0.135 | −0.007 | |
| PGP28 | 8.42 × 10−6 | −0.020 | 0.002 | −0.014 | |
| MYO5C | PGP58 | 8.31 × 10−9 | 0.013 | 0.064 | 0.009 |
| cg06192883 | PGP70 | 6.91 × 10−8 | 0.013 | 0.060 | 0.010 |
| PGP1 | 3.67 × 10−7 | 0.011 | 0.016 | 0.011 | |
| PGP17 | 8.34 × 10−7 | −0.011 | 0.175 | −0.006 | |
| PGP99 | 1.23 × 10−6 | 0.010 | 0.012 | 0.012 | |
| PGP77 | 4.00 × 10−6 | 0.009 | 0.073 | 0.008 | |
| PGP81 | 4.00 × 10−6 | −0.009 | 0.073 | −0.008 | |
| PGP64 | 6.88 × 10−6 | −0.009 | 0.266 | −0.005 | |
| PGP73 | 1.61 × 10−5 | 0.009 | 0.132 | 0.006 | |
| PGP72 | 1.72 × 10−5 | −0.010 | 0.030 | −0.013 | |
Four of the glycan-methylation associations displayed nominal significance P < 0.05 in the TwinsUK study (N = 165) and one was replicated at Bonferroni significance P < 0.0035 (0.05/14). All associations had the same direction of effect as in QMDiab. Glycan annotations are provided in Supplementary Material, Table S1.
Multi-omics associations of the TXNIP-diabetes locus
Methylation of CpG cg19693031 at the TXNIP locus showed 54 associations with metabolites, proteins and glycan traits (complete list in Supplementary Material, Table S3). TXNIP methylation was also associated with T2D as an endpoint in our study (P = 6.80 × 10−12), replicating previous reports (5). The most significant metabolic trait association with TXNIP methylation was with 1,5-anhydroglucitol (1,5-AG) in plasma (P = 7.56 × 10−21), which also showed a moderate association signal in saliva (P = 3.17 × 10−3). Cg19693031 further strongly associated with AHB (P = 2.52 × 10−14 in urine, P = 2.46 × 10−9 in plasma) and with glucose in urine (P = 1.17 × 10−14). Cg19693031 also associated with blood levels of several proteins, including transmembrane glycoprotein NMB (GPNMB) (P = 1.30 × 10−8), aminoacylase-1 (ACY1) (P = 2.59 × 10−7), sex hormone binding globulin (SHBG) (P = 4.65 × 10−7) and melanoma-derived growth regulatory (MIA) protein (P = 6.88 × 10−7), and with different complex N-glycan traits (PGP26 and PGP34). These glycans were recently reported to be associated with T2D (33). Sumer-Bayraktar et al. (34) reported that SHBG in serum is N-glycosylated by a glycan that corresponds to PGP18 in QMDiab. In QMDiab, PGP18 glycans associated with SHBG protein levels (P = 7.19 × 10−4), and methylation of cg19693031 was nominally associated with PGP18 (P = 0.011).
Multi-omics associations of the smoking loci
We found 17 Bonferroni-significant multi-omics associations at the AHRR smoking locus (Table 3 and Supplementary Material, Table S3). AHRR methylation was also associated with smoking in the QMDiab study (P = 1.89 × 10−25). AHRR (cg05575921) and several of the other smoking associated CpG sites (ALPPL2—cg21566642, F2RL2—cg03636183, cg06126421, RARA—cg19572487 and GFI1—cg09935388) associated with o-cresol sulfate in urine (P = 2.66 × 10−27–3.29 × 10−7). The strongest CpG–protein association for smoking loci was for the polymeric immunoglobulin receptor (PIGR) and CpG sites cg05575921 (AHRR), cg03636183 (F2RL2) and cg06126421 (P = 2.03 × 10−11–3.36 × 10−7). Methylation of cg01965508 at the PIGR locus showed a nominally significant negative association with smoking (P = 0.016). CpG cg01965508 lies in a promotor region less than 1500 bp upstream of the transcription start site of PIGR. PIGR is known to be a heavily N-glycosylated protein (35). The plasma N-glycome is known to associate with smoking (36) and we also found several nominal associations of the PIGR protein levels with numerous N-glycans (PGP4, PGP5, PGP10, PGP13, PGP16, PGP20, PGP23, PGP26, PGP31, PGP32, PGP34 and PGP35; p < 0.05). Finally, we found a CpG–protein association at the cg19572487 (RARA) smoking locus with the actin-regulatory protein Gelsolin (P = 1.89 × 10−6).
Mendelian randomization
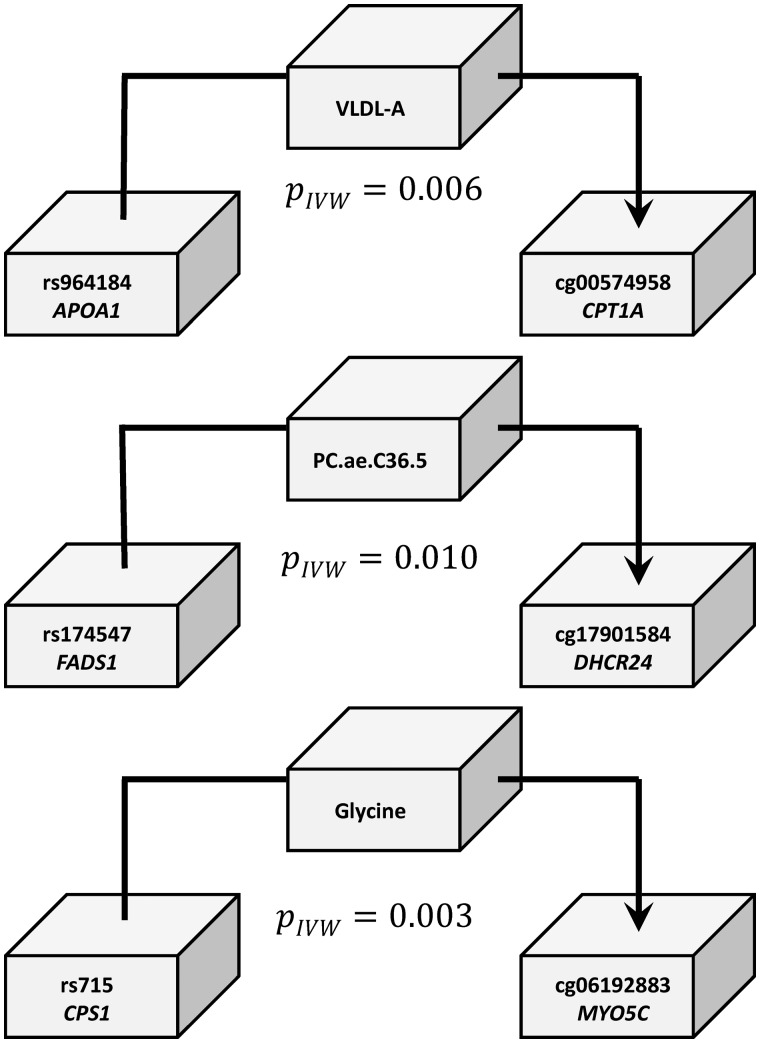
To determine whether changes in metabolite levels are causal for changes in CpG methylation, we conducted an MR study (see Materials and Methods). As the QMDiab study was too small to obtain meaningful results, we used the KORA study instead. To limit the multiple testing burden, we limited our MR analysis to the top CpG–metabolite associations previously reported by Petersen et al. (25). We further required that the SNP-metabolite (mQTL) and the SNP-methylation (meQTL) associations be reported previously in mGWAS and meGWAS (see Materials and Methods). We identified three suitable SNP-CpG-metabolite trios and verified that the genetic instruments were valid in the causal direction from the metabolite to the CpG methylation: SNP rs174547 at FADS1 was an mQTL for the metabolite PC(O-36: 5) (a glycerophospholipid) and a meQTL for cg17901584 at DHCR24, SNP rs715 at the CPS1 locus associated with glycine and cg06192883 methylation at the MYO5C locus, and SNP rs964184 at the APO cluster gene locus associated with VLDL-A (very low-density lipoprotein A) and cg00574958 methylation of the CPT1A locus (Fig. 3; Table 6). Since complete summary statistics were not available for all associations from these GWASs, we could not use a two-sample MR approach and used the KORA data instead. In all three cases, we observed a significant (P < 0.05/3) causal effect of metabolite levels on CpG methylation (Table 6). We found no valid instrument that would have allowed testing of the reverse causal direction, from methylation to metabolite.
Figure 3.
Evidence supporting the hypothesis that genetically induced changes in metabolite levels are causal to the associated changes in methylation levels. The instrumental variables here were identified using the BIOS server (75) and SNiPA (76). The three-way associations were evaluated using the KORA dataset (N∼1800). The P-values (PIVW) shown are associated with the estimate (Wald test). In all three cases presented here (see Table 6 for details), the associations between SNP and CpG methylation can be fully explained via the metabolite. This suggests that the metabolic trait is causal to the association between metabolite and CpG.
Table 6.
Causality analysis using Mendelian randomization
| Triangle associations |
MR (IVW method) |
|||
|---|---|---|---|---|
| Metabolite∼SNP (instrument) | CpG∼SNP | CpG∼Metabolite (observed) | CpG∼Metabolite (predicted) | |
| CPT1A | ||||
| (cg00574958) | P = 3.48 × 10−9 | P = 0.00589 | P = 5.89 × 10−14 | PMR = 0.006 |
| APO-cluster | β = 0.254 | β= −0.124 | β = −0.186 | β= −0.489 |
| (rs964184) | SE = 0.0427 | SE = 0.0450 | SE = 0.0246 | SE = 0.177 |
| VLDL-A | CI95 = [0.170, 0.338] | CI95 = [−0.212, −0.0358] PIV = 0.080 | CI95 = [−0.234, −0.138] | CI95 = [−0.837,-0.141] |
| DHCR24 | ||||
| (cg17901584) | P = 1.63 × 1 0−23 | P = 0.0103 | P = 3.43 × 10−18 | PMR = 0.010 |
| FADS1 | β = −0.344 | β = −0.0886 | β = 0.202 | β = 0.258 |
| (rs174547) | SE = 0.0339 | SE = 0.0345 | SE = 0.023 | SE = 0.100 |
| PC.ae.C36.5 | CI95 = [−0.411, −0.277] | CI95 = [−0.156, −0.0209] PIV = 0.563 | CI95 = [0.157, 0.248] | CI95 = [0.061, 0.454] |
| MYO5C | ||||
| (cg06192883) | P = 4.45 × 10−42 | P = 0.00318 | P = 7.69 × 10−15 | PMR = 0.003 |
| CPS1 | β = 0.455 | β = −0.107 | β= −0.199 | β = −0.235 |
| (rs715) | SE = 0.0325 | SE = 0.0361 | SE = 0.0254 | SE = 0.079 |
| glycine | CI95 = [0.391, 0.519] | CI95 = [−0.178, 0.00359] PIV = 0.633 | CI95 = [−0.249,-0.149] | CI95 = [−0.390,-0.079] |
KORA data (N∼1800) was used for MR analysis using the inverse-variance weighted method. All three MR analyses suggest that changes in metabolites are causal for the observed changes in CpG methylation with Bonferroni significance PMR < 0.017 (0.05/3).
Abbreviations: β = effect size (units: s.d./s.d. or s.d./minor allele copy), SE = standard error, P = P-value, CI95 = 95% confidence intervals, PIV = the P-value for the association of the CpG to the metabolite conditioned on the SNP; this association must not be significant for a valid instrument.
Discussion
To the best of our knowledge, this is the first study to analyze such a large number of multi-omics phenotypes with CpG methylation in a single study, providing a deeper insight into the molecules that may be involved of the underlying mechanisms of the organismal response to disease and environmental insult. Our study emphasizes the power of linking the methylome to the phenome (smoking, diabetes or obesity) by deep molecular phenotyping of multiple body fluids in a multi-omics approach. We replicated and uncovered novel associations of a wide range of metabolite, protein and glycan traits with smoking-, diabetes- and obesity-associated CpG loci in a novel multi-ethnic cohort. Many of the multi-omics associations showed strong biological evidence to be linked to pathways involved in both diabetes and smoking.
For instance, 1,5-AG associated with CpG methylation at the TXNIP locus, is an established marker of glycemic control in patients with diabetes (37) and is utilized in the FDA-approved GlycoMarkTM test (GlycoMark Inc., New York, NY). AHB, also associated with methylation at the TXNIP locus, is a key biomarker of pre-diabetes and is utilized in the QuantoseTM test (Metabolon, Morrisville, NC) for pre-diabetes monitoring. Most of the other metabolites associated with cg19693031 at the TXNIP locus were also directly associated with multiple diabetes phenotypes in our previous analysis of this dataset (38). The presence of glucose in urine (glucosuria) is also a common characteristic of diabetes.
Similarly, o-cresol sulfate, a metabolite associated with methylation of multiple smoking associated CpG sites, is a known biomarker for smoking and also associated with colorectal cancer (39). The protein associations at the AHRR locus included blood circulating levels of the PIGR protein. PIGR facilitates the secretion of soluble polymeric isoforms of immunoglobulins A and M. PIGR transcription was previously reported up-regulated in smokers (40). Another CpG–protein association of interest at a smoking locus is the actin-regulatory protein Gelsolin with cg19572487 (RARA). Gelsolin expression is down-regulated in heavy smokers (41). Gelsolin controls the length of actin polymers and mediates multiple cellular functions including cell motility, morphogenesis and actin cytoskeletal remodeling. It also regulates signal transduction through the integrin and small GTPase (Rac-Rac)-mediated pathways (42). Gelsolin was also differentially expressed in patients with heart failure (42) and in several types of cancers (43).
Associations between SHBG, MIA and different complex N-glycan traits (PGP26 and PGP34), in addition to the same glycans being recently reported to be associated with T2D (44) support the potential involvement of glycans in defining the posttranslational modifications that alter or enrich the function of the involved proteins. Likewise, the smoking associated PIGR protein being a heavily N-glycosylated protein (35) and smoking being associated with the plasma N-glycome (36) are consistent with this.
CpG methylation involvement with other phenotypes such as protein markers of liver function and blood pressure has also been documented. Liver enzyme levels, for example, gamma-glutamyl transferase (GGT), may alter epigenetic mechanisms involved in genes that regulate liver function and enzyme levels leading to differential methylation at the PHGDH locus (24). Also, DNA methylation in inflammatory genes with known vascular function, or previously related to cardiovascular disease may be driven by mechanisms involved in blood pressure regulation (23). Association studies alone cannot conclude on causality and may not provide a final answer here. However, they are an important hypothesis-generating tool that can direct further investigation by dedicated experimentation and support from existing literature, as exemplified here in the cases of the TXNIP diabetes and the AHRR–smoking associations.
In an attempt to conclude on causality in a few sufficiently powered examples, we used MR to determine directionality between some obesity-associated metabolites and methylation sites. The direction of association we found between the methylation of the obesity-associated locus CPT1A with VLDL-A is consistent with the Dekkers et al. study (30) and goes from metabolite to methylation. Similarly, the direction of the associations between the methylation of DHCR24 with PC(O-36: 5), and of MYO5C with glycine also go from metabolite to methylation, both of which are obesity-associated loci and metabolites, respectively. Still, these results should be interpreted with caution since the validity of MR analyses is based on assumptions and has several limitations as outlined in a recent review (45).
We are aware of some limitations to this study. Correction for cell proportions, ethnicity and cell abuse have all been taken into consideration (see Materials and Methods). Medication was not accounted for in the statistical analysis and may potentially confound some of the associations. In addition, as the participants of QMDiab were not fasting prior to sample collection as in KORA and TwinsUK, decreased replicability power may be implied. However, as we have shown in previous work using the same data (38), this increased variability is random and does not tend to bias the associations. Thus non-replication in QMDiab does not suggest that the association in Petersen et al. was a false positive. The replication of many previously reported CpG-metabolite, CpG-diabetes and CpG-smoking associations supports the robustness and hence biological relevance of these signals. Finally, although we replicated the majority of our novel protein–methylation associations in KORA, only some of the novel glycan-methylation associations were replicated in TwinsUK due to the smaller sample size.
Conclusion
With over 2700 studies published to date, genome-wide association studies (GWASs) with clinical endpoints and intermediate risk factors have reached maturity (46). The field of EWAS, however, is just emerging and only recently started to generate relevant biomedical results. In contrast to GWAS, where the causal direction of the association is always from the genetic variant (SNP) to the phenotype, causality cannot be inferred directly from an EWAS, and the determination of causality is vulnerable to potential confounding and reverse causation (47).
Taken together, our study supports the view that changes in health, lifestyle and environment can lead to differential regulation of a plethora of molecular phenotypes. A holistic multi-omics view of the organism’s response to environmental and disease-induced stress then emerges. Using MR approaches, causal networks connecting environmental insults and lifestyle factors to disease end points through multi-omics read-outs can now be delineated. This information can generate new insights into the affected pathways suggesting that multi-omics-associated CpG methylation is a consequence of the underlying disease pathway or an environmental insult.
Materials and Methods
Study population
QMDiab is a cross-sectional case-control study that was conducted in 2012 at the Dermatology Department in Hamad Medical Corporation (HMC Doha, Qatar). This study has been described previously and comprises 388 study participants from Arab and Asian ethnicities (29) (Table 1 contains the statistics for the subset of 359 samples that were selected for this study). The initial study was approved by the Institutional Review Boards of HMC and Weill Cornell Medicine—Qatar (WCM-Q) under research protocol number 11131/11). All study participants provided written informed consent. In addition to the 374 study participants reported in (29), we included 14 additional samples from individuals who were not sent to metabolomics analysis, bringing the total participant number in QMDiab to 388. Data used in this study was then limited to individuals who agreed to have their data and samples used for research beyond the initial scope of QMDiab, and for whom there was still sufficient material available for further analysis. For smoke exposure, the cotinine measurement (a major metabolite of nicotine from tobacco smoke observed in blood, urine or saliva) was used as a more objective indicator than self-reported smoking (48). Cotinine-derived smoking status highly overlaps with self-reported smoking status (Spearman correlation coefficient of 0.92) (49).
Sample collection
Non-fasting saliva, urine and plasma samples were collected and processed using standardized protocols. Saliva was obtained using the Salivette system following the manufacturer’s recommendations. Identical protocols, instruments and study personnel were used to randomly collect cases and controls as they appeared at the clinic. After collection, the samples were stored in ice for transportation to WCM-Q. Within 6 h of sample collection, all samples were centrifuged at 2500g for 10 min, aliquoted, and stored at −80°C until analysis.
DNA extraction and quantification
Blood samples were thawed at 37°C in a water bath for 5–10 min. Samples were then left to cool down to room temperature and 400 ul of blood was transferred to a 2 ml cryotubes. A total of 400 ul of phosphate buffered saline (PBS) was added to the blood and mixed by pipetting back and forth. The mixture was transferred to a 2 ml Sarstedt 72.694 tube and loaded to the QIA Symphony for DNA extraction. The QUBIT kit was then used for DNA quantification.
DNA methylation
Genome-wide DNA methylation profiling was performed using the Illumina Infinium HumanMethylation450 (450K) BeadChip array (28) for interrogating over 485 000 methylation sites per sample. DNA methylation was determined for 359 samples which all passed initial quality assessment of assay performance using the Genome Studio software integrated controls dashboard. A total of 500 ng genomic DNA from each sample was bisulfite-converted using the EZ DNA Methylation Kit (Zymo Research, catalog No. D5002) according to the manufacturer’s procedure, with the recommended incubation conditions when using the Infinium Methylation Assay. DNA methylation was assessed following the Infinium HD Methylation protocol. This consisted of a whole-genome amplification step using 4 ul of each bisulfite-converted sample, followed by enzymatic fragmentation and application of the samples to BeadChips. The arrays were fluorescently stained and scanned with the Illumina iScan system. Genome Studio (version 2011.1) with methylation module (version 1.9.0) was used to process the raw image data generated by the BeadArray Reader. Initial quality assessment of the assay performance was conducted using the Genome Studio software integrated controls dashboard. All 359 samples were processed with Genome Studio (background subtraction and control normalization). Beta-values, raw signals and detection P-values were extracted also using Genome Studio.
Initial quality checks were performed on the methylation data to confirm data integrity. Sex checks were first performed by verifying the distribution of CpG methylation on the X-chromosome and matching against the sex specification in the manifest. Females had a characteristic peak in the distribution around a b-value of 0.5 while males had two peaks at b-values 0 and 1, which is attributed to the X-chromosome in-activation property. Next, the overall beta distribution and intensity distributions were visually inspected for any abnormalities in all subjects. Two individuals had a slightly left-skewed intensity plot and their beta distributions showed a slight shift in the fully methylated peak toward 0.6–0.8 as opposed to the common case where the peak is around 0.8–0.9 but were not eliminated from the study. Measurements from non-CpG probes and the 65 probes targeting SNPs (as identified in the Illumina manifest) were excluded, leaving methylation readouts from 482 421 probes. Further filtering included methylation sites whose detection P-values were greater than 0.01 in more than 5% of the samples (121 probes) and non-autosomal probes (11 135 on the X-chromosome and 416 on the Y-chromosome). This left 470 776 methylation sites for data analysis. Normalization was carried out on data from these probes using the Lumi: BMIQ pipeline, which includes color bias adjustment, QN (quantile normalization), and BMIQ (beta mixture quantile dilation). Normalization matched the centers and peaks of the methylation profiles no longer necessitating the elimination of any samples from the study. The corrected b-values ranged from 9.663 × 10−4 to 0.9997. White blood cell fractions (granulocytes, monocytes, B-cells, NK-cells, CD8+-T-cells and CD4+-T-cells) were estimated from the methylation data using the method described by Houseman et al. (50). Computation of the white blood cell Houseman coefficients included batch adjustment by modeling the batch number as a random effect. Thus technical variation was accounted for through the white blood cell percentages.
Non-targeted metabolomics
The semi-quantitative non-targeted UPLC-MS/MS and GC-MS platform from Metabolon Inc. was used, yielding measurements of 2251 metabolic traits (758 from plasma, 891 from urine and 602 from saliva). The platform has been described in detail previously (51,52). Briefly, non-targeted metabolic profiling at Metabolon was achieved in 330 saliva, 358 in blood plasma and 360 urine samples using ultrahigh-performance liquid-phase chromatography and gas chromatography separation, coupled with tandem MS using established procedures (51). Osmolality in saliva and urine were measured and used for normalization. The median process variability in saliva was 15.3%, in plasma was 15.8% and in urine was 9.8%, which was determined by repeated measurements of pooled samples.
Targeted metabolomics
A total of 26 quantified and 137 semi-quantified metabolites (due to lack of standards) were measured in 356 plasma samples using a commercially available FIA-MS metabolomics kit (AbsoluteIDQTM kit p150, Biocrates Life Sciences AG, Innsbruck, Austria). The kit was run on the metabolomics platform of the Helmholtz Center Munich. Assay procedures and the full biochemical names have been described in detail in our previous work (53).
NMR urine metabolomics
1H-NMR spectra were acquired for 353 urine samples on a Bruker DRX-400 NMR spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) operating at 400.13 MHz 1H frequency. Samples were measured at 300 K. The Fourier-transformed and baseline-corrected NMR spectra were manually annotated by spectral pattern matching using the Chenomx Worksuite 7.0 by Chenomx, Inc. (Edmonton, Canada) to deduce absolute urinary metabolite concentrations for 60 compounds as described previously.
Lipid-omics
Metabolite concentrations for 338 individuals were quantified from plasma samples using a high-throughput NMR metabolomics platform (Nightingale Ltd, Helsinki, Finland) (54,55). The experimental protocol, sample preparation, NMR spectroscopy and metabolite identification details are described previously in (54,56). A total of 225 metabolites were measured out of which 148 were directly measured and 77 were derived. The 148 metabolites include 14 lipoprotein subclasses (98 measurements), three sizes of lipoprotein particles, two apolipoproteins, eight fatty acids, eight glycerides and phospholipids, nine cholesterols, nine amino acids, one inflammatory marker and ten small molecules involved in glycolysis, citric acid cycle and urea cycle. The subclasses for the lipoproteins are categorized according to size following this classification: chylomicrons and extremely large VLDL particles (average particle diameter at least 75 nm); five VLDL subclasses—very large VLDL (average particle diameter of 64.0 nm), large VLDL (53.6 nm), medium VLDL (44.5 nm), small VLDL (36.8 nm) and very small VLDL (31.3 nm); intermediate-density lipoprotein (IDL; 28.6 nm); three LDL subclasses—large LDL (25.5 nm), medium LDL (23.0 nm) and small LDL (18.7 nm); and four HDL subclasses—very large HDL (14.3 nm), large HDL (12.1 nm), medium HDL (10.9 nm) and small HDL (8.7 nm). Measurements were log10 transformed and z-scored.
Proteomics
The SOMAscan platform was used to quantify a total of 1124 protein measurements in 356 plasma samples. Details of the SOMAscan platform have been described elsewhere (57–63). In brief, undepleted EDTA-plasma was diluted into three dilution bins (0.05%, 1% and 40%) and incubated with bin-specific collections of bead-coupled SOMAmers in a 96-well plate format. Subsequent to washing steps, bead-bound proteins were biotinylated and complexes comprising biotinylated target proteins and fluorescence-labeled SOMAmers were photo-cleaved off the bead support and pooled. Following recapture on streptavidin beads and further washing steps, SOMAmers were eluted and quantified as a proxy to protein concentration by hybridization to custom arrays of SOMAmer-complementary oligonucleotides. Based on standard samples included on each plate, the resulting raw intensities were processed using a data analysis work flow including hybridization normalization, median signal normalization and signal calibration to control for inter-plate differences. The 356 samples from QMDiab were analyzed at the WCM-Q proteomics core (64).
Glycomics
Unthawed aliquots of 356 samples were sent to Genos Ltd. (Zagreb, Croatia) for analysis of total plasma N-glycosylation using UPLC and IgG Fc N-glycosylation using liquid chromatography mass spectrometry glyco-profiling. Quantification of 113 N-glycan traits in 333 samples and 60 IgG-glycopeptides in 341 samples was achieved on this platform as follows.
Total plasma N-glycan release and labeling
Glycans were released from total plasma proteins and labeled as previously described (65). In brief, 10 µl of plasma was denatured by adding 20 µl 2% (w/v) SDS (Invitrogen, USA) and the N-glycans were released by adding 1.2 U of PNGase F (Promega, USA). The released N-glycans were labeled with 2-aminobenzamide (Sigma-Aldrich, USA). Hydrophilic interaction liquid chromatography solid-phase extraction was used to remove free labels and the reducing agent from the samples. In the stationary phase, 0.2 µm 96-well GHP filter-plates (Pall Corporation, USA) were used. After a short incubation and washing five times with cold 90% ACN, the samples were loaded into the wells. After 15 min of shaking at room temperature, glycans were eluted with 2 × 90 µl of ultrapure water and then the combined eluates were stored at −20°C until usage.
Total plasma N-glycome UPLC analysis
Total plasma N-glycans were analyzed by hydrophilic interaction ultra-performance liquid chromatography (HILIC-UPLC) as described previously (65). In brief, excitation and emission wavelengths of 250 and 428 nm, respectively, were used to separate fluorescently labeled N-glycans on an Acquity UPLC instrument (Waters, USA). The labeled N-glycans were separated on a Waters BEH Glycan chromatography column, 150 × 2.1 mm i.d., 1.7 µm BEH particles, with 100 mM ammonium formate having pH 4.4 as solvent A and acetonitrile (ACN) (Fluka, USA) as solvent B. The separation method works by using a linear gradient of 30–47% solvent A at a flow rate of 0.56 ml/min during a 23-min analytical run.
IgG isolation from plasma
IgG was isolated using protein G monolithic plates (BIA Separations, Slovenia) as described previously (66). In brief, approximately 70–100 µl of plasma was diluted 8× with 1× PBS having pH 7.4, applied to the protein G plate and instantly washed with 1× PBS having pH 7.4 to remove unbound proteins. IgG was then eluted with 1 ml of 0.1 M formic acid (Merck, Germany) and neutralized with 1 M ammonium bicarbonate (Merck, Germany).
IgG enzymatic cleavage and purification
A total of 25 µg of IgG was digested overnight at 37°C with 200 ng trypsin (Worthington, USA). Then Chromabond C18 ec beads (Macherey-Nagel, Germany) were used to purify IgG tryptic glycopeptides by reverse phase solid-phase extraction. C18 beads were activated with 80% ACN that contains 0.1% trifluoroacetic acid (TFA; Sigma-Aldich, USA) and conditioned with 0.1% TFA. Tryptic digests were diluted 10× with 0.1% TFA, loaded onto C18 beads, and washed with 0.1% TFA. Glycopeptides were eluted with 20% ACN containing 0.1% TFA. Eluates were dried by vacuum centrifugation and dissolved in 20 µl of ultrapure water.
Subclass-specific Fc IgG N-glycome liquid chromatography mass spectrometry (LC-MS) analysis
Tryptic digests were analyzed on a nanoACQUITY UPLC system (Waters, USA) coupled to micrOTOF-Q mass spectrometer (Bruker Daltonics, Germany). A total of 9 µl of glycopeptides was loaded into an Acclaim PepMap100 C8 (5 mm × 300 µm i.d.) trap column and washed for 1 min with 0.1% TFA (solvent A) at a flow rate of 40 µl/min. Separation was achieved on a Halo C18 nano-LC column (150 mm × 75 µm i.d., 2.7 µm HALO fused core particles; Advanced Materials Technology, USA) using a 3.5 min gradient at a flow rate of 1 µl/min from 18% to 25% solvent B (80% ACN). Column temperature was 30°C. Mass spectra were recorded from m/z 200 to 1900 with 2 averages at a frequency of 0.5 Hz. Quadrupole ion energy and collision energy of the MS were set to 4 eV. NanoACQUITY UPLC system and the Bruker microOTOF-Q were operated under HyStar software version 3.2. The same software was used for data extraction.
Glycan data was first normalized (total area normalization) and then batch corrected using Combat. Batch correction was performed on the log-transformed normalized data. After batch correction, the data was inverse transformed so all values were between 0 and 100. Finally the data was z-scored. Glycan structural features are given in terms of number of galactoses (G0, G1 and G2), fucose (F), bisecting N-acetylglucosamine (N) and N-acetylneuraminic acid (S).
Statistical analysis
Linear models were computed using the R function lm (67) with DNA methylation (B-values) as the dependent variable and the z-scored metabolite, lipid, protein or glycan levels as independent variables. After excluding metabolites with fewer than 50 valid detections (many of which were xenobiotics related to medication) for the 359 samples for which methylation data was available, 2474 metabolites were used for the analyses. Sex, BMI, age and Houseman-based white blood cell coefficients were used as covariates. DNA methylation can be cell-type dependent. As we only obtained DNA from blood cells, we may have missed organ-specific association signals. Furthermore, methylation profiles have been shown to vary with blood cell type (50). To account for cell type variability, we used the Houseman method (50) to determine white blood cell distribution using our 450K DNA methylation data. Also, the first three principle components of the genotyping data (GeneticPCs) were added as covariates, as they represent ethnicity more accurately than self-reported information. Mixed ethnicity in the QMDiab study may lead to population-specific stratification and result in inflated P-values. We have previously shown that the self-reported ethnicity of our study is well captured by the first three principal components (PCs) of the genotype variants (64). Details of genotyping data for QMDiab and the computed PCs that were used to account for ethnicity have been described previously (64).
For associations including proteins, the first three principle components of the proteomics data were also included (somaPC1, somaPC2 and soma PC3) to account for a moderate level of observed cell lysis. Although visual inspection of the blood plasma samples did not show any signs of hemolysis, principal component analysis of the protein data still suggested a moderate degree of cell lysis. This approach was shown to yield highly reproducible associations between the QMDiab study and KORA (64).
The multiple-testing Bonferroni corrected level of significance for metabolites was Pmetabolite =1.01 × 10−6 (0.05/2474/20), accounting for the number of metabolic traits (N = 2474) and the number of tested DNA methylation sites (N = 20). Similarly, for lipids (N = 225) the required Bonferroni level of significance was Plipids = 1.11 × 10−5 (0.05/225/20), for proteins (N = 1124) it was Pprotein = 2.22 × 10−6 (0.05/1124/20) and for glycan traits (N = 113) it was Pglycan = 2.21 × 10−5 (0.05/113/20).
Replication of CpG-glycan associations in the TwinsUK study
The TwinsUK study was established in 1992 to recruit monozygotic and dizygotic twins without selecting for particular diseases or traits (68). It has been used in many epidemiological studies and is representative of the general U.K. population for a wide range of diseases and traits (69). DNA methylation was measured for 808 individuals of European ancestry randomly selected from the TwinsUK cohort. The Infinium HumanMethylation450 BeadChip (Illumina Inc, San Diego, CA, USA) was used to measure DNA methylation. Details of experimental approaches have been previously described (70) and normalization was carried out using the ‘minfi’ R package (71). Blood cell-type coefficients were estimated from the methylation data using the method described by Houseman et al. (50). Total plasma glycans were prepared as described previously (72). Glycans were normalized, and all measurements were adjusted for age, sex and technical confounders. Total plasma proteins glycans were available for 2752 individuals of European ancestry, of whom 165 had also DNA methylation data. The TwinsUK dataset included 152 females and 13 males, whose median age was 58.30 (mean = 56.71, SD = 12.34). All individuals were of European ancestry. Association studies were conducted for individual CpG sites and glycans using a linear mixed model as implemented in the lme4 R packages (73), in order to keep into account the non-independence of twin data, and adjusting for BMI, age, sex, Houseman-based white blood cell coefficients and technical confounders. Outliers (measurements more than three standard deviations from the mean) were excluded from the analysis.
Replication of CpG-protein associations and MR in the KORA study
The KORA F4 study is a population-based cohort of 3080 subjects living in southern Germany, who were recruited between 2006 and 2008. The DNA methylation dataset from KORA, which was determined using the Infinium HumanMethylation450 BeadChip platform, was described in detail previously (25) and comprises 1805 samples. The 1805 samples consisted of 880 males and 925 females whose median age was 61 (mean = 60.92, SD = 8.87). For replication of the novel methylation–proteomics associations, we used protein traits that were measured using the SOMAscan platform. Of the 1805 methylation samples, only 997 had the matching proteomics measurements available. The proteomics dataset has been described in detail previously (64). For the MR analysis, we used all 1805 methylation samples and their matching genotyping data for the selected instruments, and their matching metabolomics data for the selected metabolites. The KORA genotyping data was described previously in detail (64), and the metabolomics dataset was also described previously (25).
Mendelian randomization
We used the inverse-variance weighted method (74) as implemented in the R function ‘mr_ivw: MendelianRandomization’ to conduct MR on the original 20 CpG-metabolite associations reported in the Petersen et al. study (25), using inverse-normal scaled metabolite and CpG methylation data from the KORA study (N∼1800). To reduce the multiple-testing burden and avoid testing weak associations we only selected SNPs as instruments that showed an association with both, CpG methylation and metabolite levels. We used the BIOS QTL browser (http://genenetwork.nl/biosqtlbrowser; date last accessed May 10, 2017) (75) to retrieve all methylation-QTLs for the 20 CpGs investigated here. We then used the SNiPA server (http://snipa.org) (76) to identify all overlapping metabolite-QTLs on matching CpG-metabolite pairs. When multiple correlated SNPs were available (R2 > 0.8) we selected the one with strongest association.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank the staff from HMC dermatology department and WCM-Q clinical research core for their contribution to QMDiab. We thank Brian Sellers from SomaLogic for support with measuring the QMDiab samples at WCM-Q. Most of all we wish to thank the study participants for their contribution to this research study. We also wish to express our appreciation to all study participants of the TwinsUK cohorts. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union (EU) and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility, and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King′s College London.
Conflict of Interest statement. M.P. and G.L. are working for or have stakes in Genos Ltd. The other authors declare that they have no conflict of interest.
Funding
This work was supported by the Biomedical Research Program funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation. The statements made herein are solely the responsibility of the authors. Dennis Mook-Kanamori was supported by Dutch Science Organization (ZonMW-VENI Grant 916.14.023). Gordon Lauc was supported by the European Union H2020 grants SYSCID (contract #733100 ) and IMForFuture (contract #721815) and by the European Structural and Investment Funds IRI (grant #KK.01.2.1.01.0003) and Croatian National Centre of Research Excellence in Personalized Healthcare (grant #KK.01.1.1.01.0010). Funding to pay the Open Access publication charges for this article was provided by Weill Cornell Medicine – Qatar.
Ethics Approval and Consent to Participate
The QMDiab study was approved by the Institutional Review Boards of HMC and WCM-Q under research protocol number 11131/11). All study participants provided written informed consent.
Authors’ Contributions
Conceived and designed the study: S.B.Z., K.S.
Performed experiments: S.K., N.S., A.H., R.E., H.S., E.K.A., Y.M., W.R.M., J.A., G.K., N.F., M.P., G.L., J.M., J.G.
Performed statistical analysis: S.B.Z.
Analyzed data: S.B.Z., K.S.
Contributed reagents/materials/analysis tools: D.O.M., A.V., P.C.T., T.S., J.B., M.F., .A.W., M.W., A.P., C.G.
Wrote the paper: S.B.Z., K.S.
All authors discussed the results and reviewed the final manuscript.
References
- 1. Banovich N.E., Lan X., McVicker G., van de Geijn B., Degner J.F., Blischak J.D., Roux J., Pritchard J.K., Gilad Y.. et al. (2014) Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genetics, 10, e1004663.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J.. et al. (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America, 102, 10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chambers J.C., Loh M., Lehne B., Drong A., Kriebel J., Motta V., Wahl S., Elliott H.R., Rota F., Scott W.R.. et al. (2015) Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. The Lancet Diabetes & Endocrinology, 3, 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kulkarni H., Kos M.Z., Neary J., Dyer T.D., Kent J.W., Goring H.H.H., Cole S.A., Comuzzie A.G., Almasy L., Mahaney M.C.. et al. (2015) Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Human Molecular Genetics, 24, 5330–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Al Muftah W.A., Al-Shafai M., Zaghlool S.B., Visconti A., Tsai P.C., Kumar P., Spector T., Bell J., Falchi M., Suhre K. (2016) Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clinical Epigenetics, 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. (2013) Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Human Molecular Genetics, 22, 843–851. [DOI] [PubMed] [Google Scholar]
- 7. Zaghlool S.B., Al-Shafai M., Al Muftah W.A., Kumar P., Falchi M., Suhre K. (2015) Association of DNA methylation with age, gender, and smoking in an Arab population. Clinical Epigenetics, 7, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeilinger S., Kuhnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A.. et al. (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PloS One, 8, e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guida F., Sandanger T.M., Castagne R., Campanella G., Polidoro S., Palli D., Krogh V., Tumino R., Sacerdote C., Panico S.. et al. (2015) Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Human Molecular Genetics, 24, 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breitling L.P., Yang R.X., Korn B., Burwinkel B., Brenner H. (2011) Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. American Journal of Human Genetics, 88, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun Y.V., Smith A.K., Conneely K.N., Chang Q.Z., Li W.Y., Lazarus A., Smith J.A., Almli L.M., Binder E.B., Klengel T.. et al. (2013) Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Human Genetics, 132, 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elliott H.R., Tillin T., McArdle W.L., Ho K.R., Duggirala A., Frayling T.M., Smith G.D., Hughes A.D., Chaturvedi N., Relton C.L. (2014) Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clinical Epigenetics, 6, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harlid S., Xu Z.L., Panduri V., Sandler D.P., Taylor J.A. (2014) CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the sister study. Environmental Health Perspectives, 122, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dogan M.V., Shields B., Cutrona C., Gao L., Gibbons F.X., Simons R., Monick M., Brody G.H., Tan K., Beach S.R.H.. et al. (2014) The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics, 15, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joubert B.R., Haberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun O., Cupul-Uicab L.A.. et al. (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environmental Health Perspectives, 120, 1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Almen M.S., Nilsson E.K., Jacobsson J.A., Kalnina I., Klovins J., Fredriksson R., Schioth H.B. (2014) Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene, 548, 61–67. [DOI] [PubMed] [Google Scholar]
- 17. Demerath E.W., Guan W.H., Grove M.L., Aslibekyan S., Mendelson M., Zhou Y.H., Hedman A.K., Sandling J.K., Li L.A., Irvin M.R.. et al. (2015) Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Human Molecular Genetics, 24, 4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aissi D., Wahl S., Meduri E., Morange P.E., Gagnon F., Grallert H.. et al. (2014) DNA methylation and body-mass index: a genome-wide analysis. The Lancet, 383, 1990–1998. [DOI] [PubMed] [Google Scholar]
- 19. Feinberg A.P., Irizarry R.A., Fradin D., Aryee M.J., Murakami P., Aspelund T., Eiriksdottir G., Harris T.B., Launer L., Gudnason V.. et al. (2010) Personalized epigenomic signatures that are stable over time and covary with body mass index. Science Translational Medicine, 2, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mendelson M.M., Marioni R.E., Joehanes R., Liu C., Hedman A.K., Aslibekyan S., Demerath E.W., Guan W., Zhi D., Yao C.. et al. (2017) Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Medicine, 14, e1002215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., Zhu H., Snieder H., Su S., Munn D., Harshfield G., Maria B.L., Dong Y., Treiber F., Gutin B.. et al. (2010) Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Medicine, 8, 87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wahl S., Drong A., Lehne B., Loh M., Scott W.R., Kunze S., Tsai P.C., Ried J.S., Zhang W., Yang Y.. et al. (2017) Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature, 541, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richard M.A., Huan T., Ligthart S., Dehghan A., Marioni R.E., Brody J.A., Sotoodehnia N., Jhun M.A., Kardia S.L., Smith J.A.. et al. (2016) Abstract 32: Novel genetic loci for blood pressure regulation identified by the analysis of DNA methylation. Circulation, 133, A32–A32. [Google Scholar]
- 24. Nano J., Ghanbari M., Wang W., de Vries P.S., Dhana K., Muka T., Uitterlinden A.G., van Meurs J.B.J., Hofman A., consortium B.. et al. (2017) Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis. Gastroenterology 154. [DOI] [PubMed] [Google Scholar]
- 25. Petersen A.K., Zeilinger S., Kastenmuller G., Romisch-Margl W., Brugger M., Peters A., Meisinger C., Strauch K., Hengstenberg C., Pagel P.. et al. (2014) Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Human Molecular Genetics, 23, 534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. (2012) Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 159B, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu N., Zheng B., Shaywitz A., Dagon Y., Tower C., Bellinger G., Shen C.H., Wen J., Asara J., McGraw T.E.. et al. (2013) AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Molecular Cell, 49, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L.. et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics, 98, 288–295. [DOI] [PubMed] [Google Scholar]
- 29. Mook-Kanamori D.O., Selim M.M.E., Takiddin A.H., Al-Homsi H., Al-Mahmoud K.A.S., Al-Obaidli A., Zirie M.A., Rowe J., Yousri N.A., Karoly E.D.. et al. (2014) 1, 5-Anhydroglucitol in saliva is a noninvasive marker of short-term glycemic control. The Journal of Clinical Endocrinology and Metabolism, 99, E479–E483. [DOI] [PubMed] [Google Scholar]
- 30. Dekkers K.F., van Iterson M., Slieker R.C., Moed M.H., Bonder M.J., van Galen M., Mei H., Zhernakova D.V., van den Berg L.H., Deelen J.. et al. (2016) Blood lipids influence DNA methylation in circulating cells. Genome Biology, 17, 138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfeiffer L., Wahl S., Pilling L.C., Reischl E., Sandling J.K., Kunze S., Holdt L.M., Kretschmer A., Schramm K., Adamski J.. et al. (2015) DNA methylation of lipid-related genes affects blood lipid levels. Circulation. Cardiovascular Genetics, 8, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sekula P., Goek O.N., Quaye L., Barrios C., Levey A.S., Romisch-Margl W., Menni C., Yet I., Gieger C., Inker L.A.. et al. (2016) A metabolome-wide association study of kidney function and disease in the general population. Journal of the American Society of Nephrology, 27, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keser T., Gornik I., Vuckovic F., Selak N., Pavic T., Lukic E., Gudelj I., Gasparovic H., Biocina B., Tilin T.. et al. (2017) Increased plasma N-glycome complexity is associated with higher risk of type 2 diabetes. Diabetologia 33. [DOI] [PubMed] [Google Scholar]
- 34. Sumer-Bayraktar Z., Nguyen-Khuong T., Jayo R., Chen D.D.Y., Ali S., Packer N.H., Thaysen-Andersen M. (2012) Micro- and macroheterogeneity of N-glycosylation yields size and charge isoforms of human sex hormone binding globulin circulating in serum. Proteomics, 12, 3315–3327. [DOI] [PubMed] [Google Scholar]
- 35. Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D.T., Loo J.A. (2006) Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. Journal of Proteome Research, 5, 1493–1503. [DOI] [PubMed] [Google Scholar]
- 36. Knezevic A., Gornik O., Polasek O., Pucic M., Redzic I., Novokmet M., Rudd P.M., Wright A.F., Campbell H., Rudan I.. et al. (2010) Effects of aging, body mass index, plasma lipid profiles, and smoking on human plasma N-glycans. Glycobiology, 20, 959–969. [DOI] [PubMed] [Google Scholar]
- 37. Buse J.B., Freeman J.L., Edelman S.V., Jovanovic L., McGill J.B. (2003) Serum 1, 5-anhydroglucitol (GlycoMark): a short-term glycemic marker. Diabetes Technology & Therapeutics, 5, 355–363. [DOI] [PubMed] [Google Scholar]
- 38. Yousri N.A., Mook-Kanamori D.O., Selim M.M., Takiddin A.H., Al-Homsi H., Al-Mahmoud K.A., Karoly E.D., Krumsiek J., Do K.T., Neumaier U.. et al. (2015) A systems view of type 2 diabetes-associated metabolic perturbations in saliva, blood and urine at different timescales of glycaemic control. Diabetologia, 58, 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cross A.J., Boca S., Freedman N.D., Caporaso N.E., Huang W.Y., Sinha R., Sampson J.N., Moore S.C. (2014) Metabolites of tobacco smoking and colorectal cancer risk. Carcinogenesis, 35, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gohy S.T., Detry B.R., Lecocq M., Bouzin C., Weynand B.A., Amatngalim G.D., Sibille Y.M., Pilette C. (2014) Polymeric immunoglobulin receptor down-regulation in chronic obstructive pulmonary disease. Persistence in the cultured epithelium and role of transforming growth factor-beta. American Journal of Respiratory and Critical Care Medicine, 190, 509–521. [DOI] [PubMed] [Google Scholar]
- 41. Dosaka-Akita H., Hommura F., Fujita H., Kinoshita I., Nishi M., Morikawa T., Katoh H., Kawakami Y., Kuzumaki N. (1998) Frequent loss of gelsolin expression in non-small cell lung cancers of heavy smokers. Cancer Research, 58, 322–327. [PubMed] [Google Scholar]
- 42. Li G.H., Shi Y., Chen Y., Sun M., Sader S., Maekawa Y., Arab S., Dawood F., Chen M.Y., De Couto G.. et al. (2009) Gelsolin regulates cardiac remodeling after myocardial infarction through DNase I-mediated apoptosis. Circulation Research, 104, 896–U131. [DOI] [PubMed] [Google Scholar]
- 43. Winston J.S., Asch H.L., Zhang P.J., Edge S.B., Hyland A., Asch B.B. (2001) Downregulation of gelsolin correlates with the progression to breast carcinoma. Breast Cancer Research and Treatment, 65, 11–21. [DOI] [PubMed] [Google Scholar]
- 44. Keser T., Gornik I., Vuckovic F., Selak N., Pavic T., Lukic E., Gudelj I., Gasparovic H., Biocina B., Tilin T.. et al. (2017) Increased plasma N-glycome complexity is associated with higher risk of type 2 diabetes. Diabetologia, 60, 2352–2360. [DOI] [PubMed] [Google Scholar]
- 45. Davey Smith G., Hemani G. (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics, 23, R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L.. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research, 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Relton C.L., Smith G.D. (2012) Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. International Journal of Epidemiology, 41, 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Florath I., Saum K.U., Brenner H. (2016) Self-reported smoking, serum cotinine, and blood DNA methylation. Environmental Research, 146, 395–403. [DOI] [PubMed] [Google Scholar]
- 49. McDonald S.D., Perkins S.L., Walker M.C. (2005) Correlation between self-reported smoking status and serum cotinine during pregnancy. Addictive Behaviors, 30, 853–857. [DOI] [PubMed] [Google Scholar]
- 50. Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry, 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- 52. Dehaven C.D., Evans A.M., Dai H., Lawton K.A. (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. Journal of Cheminformatics, 2, 9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Romisch-Margl W., Prehn C., Bogumil R., Rohring C., Suhre K., Adamski J. (2012) Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics, 8, 133–142. [Google Scholar]
- 54. Soininen P., Kangas A.J., Wurtz P., Tukiainen T., Tynkkynen T., Laatikainen R., Jarvelin M.R., Kahonen M., Lehtimaki T., Viikari J.. et al. (2009) High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst, 134, 1781–1785. [DOI] [PubMed] [Google Scholar]
- 55. Soininen P., Kangas A.J., Wurtz P., Suna T., Ala-Korpela M. (2015) Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circulation: Cardiovascular Genetics, 8, 192–206. [DOI] [PubMed] [Google Scholar]
- 56. Inouye M., Silander K., Hamalainen E., Salomaa V., Harald K., Jousilahti P., Mannisto S., Eriksson J.G., Saarela J., Ripatti S.. et al. (2010) An immune response network associated with blood lipid levels. PLoS Genetics, 6, e1001113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N., Carter J., Dalby A.B., Eaton B.E., Fitzwater T.. et al. (2010) Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One, 5, e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hathout Y., Brody E., Clemens P.R., Cripe L., DeLisle R.K., Furlong P., Gordish-Dressman H., Hache L., Henricson E., Hoffman E.P.. et al. (2015) Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proceedings of the National Academy of Sciences of the United States of America, 112, 7153–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sattlecker M., Kiddle S.J., Newhouse S., Proitsi P., Nelson S., Williams S., Johnston C., Killick R., Simmons A., Westman E.. et al. (2014) Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimer's & Dementia, 10, 724–734. [DOI] [PubMed] [Google Scholar]
- 60. Kraemer S., Vaught J.D., Bock C., Gold L., Katilius E., Keeney T.R., Kim N., Saccomano N.A., Wilcox S.K., Zichi D.. et al. (2011) From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One, 6, e26332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kiddle S.J., Sattlecker M., Proitsi P., Simmons A., Westman E., Bazenet C., Nelson S.K., Williams S., Hodges A., Johnston C.. et al. (2014) Candidate blood proteome markers of Alzheimer's disease onset and progression: a systematic review and replication study. Journal of Alzheimer's Disease, 38, 515–531. [DOI] [PubMed] [Google Scholar]
- 62. Lourdusamy A., Newhouse S., Lunnon K., Proitsi P., Powell J., Hodges A., Nelson S.K., Stewart A., Williams S., Kloszewska I.. et al. (2012) Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Human Molecular Genetics, 21, 3719–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Menni C., Kiddle S.J., Mangino M., Vinuela A., Psatha M., Steves C., Sattlecker M., Buil A., Newhouse S., Nelson S.. et al. (2015) Circulating proteomic signatures of chronological age. The Journals of Gerontology: Series A, 70, 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K.. et al. (2017) Connecting genetic risk to disease end points through the human blood plasma proteome. Nature Communications, 8, 14357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Akmacic I.T., Ugrina I., Stambuk J., Gudelj I., Vuckovic F., Lauc G., Pucic B.M. (2015) High-throughput glycomics: optimization of sample preparation. Biochemistry. Biokhimiia, 80, 934–942. [DOI] [PubMed] [Google Scholar]
- 66. Pucic M., Knezevic A., Vidic J., Adamczyk B., Novokmet M., Polasek O., Gornik O., Supraha-Goreta S., Wormald M.R., Redzic I.. et al. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Molecular & Cellular Proteomics, 10. doi: 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. R Development Core Team. (2010). R Foundation for Statistical Computing.
- 68. Moayyeri A., Hammond C.J., Valdes A.M., Spector T.D. (2013) Cohort profile: TwinsUK and healthy ageing twin study. International Journal of Epidemiology, 42, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Andrew T., Hart D.J., Snieder H., de Lange M., Spector T.D., MacGregor A.J. (2001) Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Research: The Official Journal of the International Society for Twin Studies, 4, 464–477. [DOI] [PubMed] [Google Scholar]
- 70. Tsaprouni L.G., Yang T.P., Bell J., Dick K.J., Kanoni S., Nisbet J., Vinuela A., Grundberg E., Nelson C.P., Meduri E.. et al. (2014) Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics: Official Journal of the DNA Methylation Society, 9, 1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Keser T., Vuckovic F., Barrios C., Zierer J., Wahl A., Akinkuolie A.O., Stambuk J., Nakic N., Pavic T., Perisa J.. et al. (2017) Effects of statins on the immunoglobulin G glycome. BBA General Subjects, 1861, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bates D., Machler M., Bolker B.M., Walker S.C. (2015) Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- 74. Burgess S., Butterworth A., Thompson S.G. (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic Epidemiology, 37, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J.. et al. (2016) Disease variants alter transcription factor levels and methylation of their binding sites. Nature Genetics, 49, 131–138. [DOI] [PubMed] [Google Scholar]
- 76. Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmuller G. (2015) SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics, 31, 1334–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.