Abstract
Vitamin B12 deficiency is common in older individuals. Circulating vitamin B12 concentration can be used to diagnose deficiency, but this test has substantial false positive and false negative rates. We conducted genome-wide association studies (GWAS) in which we resolved total serum vitamin B12 into the fractions bound to transcobalamin and haptocorrin: two carrier proteins with very different biological properties. We replicated reported associations between total circulating vitamin B12 concentrations and a common null variant in FUT2. This allele determines the secretor phenotype in which blood group antigens are found in non-blood body fluids. Vitamin B12 bound to haptocorrin (holoHC) remained highly associated with FUT2 rs601338 (p.Trp154Ter). Transcobalamin bound vitamin B12 (holoTC) was not influenced by this variant. HoloTC is the bioactive the form of the vitamin and is taken up by all tissues. In contrast, holoHC is only taken up by the liver. Using holoHC from individuals with known FUT2 genotypes, we demonstrated that FUT2 rs601338 genotype influences the glycosylation of haptocorrin. We then developed an experimental model demonstrating that holoHC is transported into cultured hepatic cells (HepG2) via the asialoglycoprotein receptor (ASGR). Our data challenge current published hypotheses on the influence of genetic variation on this clinically important measure and are consistent with a model in which FUT2 rs601338 influences holoHC by altering haptocorrin glycosylation, whereas B12 bound to non-glycosylated transcobalamin (i.e. holoTC) is not affected. Our findings explain some of the observed disparity between use of total B12 or holoTC as first-line clinical tests of vitamin B12 status.
Introduction
Severe deficiencies of vitamin B12 can lead to anemia and neurological symptoms, while more subtle and often asymptomatic deficiencies have been implicated in neural tube defects (1–3), other birth defects (4), cardiovascular diseases (5), and cancer (6,7). Moreover, vitamin B12 deficiency is common among the older, largely due to malabsorption (8). If left untreated, vitamin B12 deficiency may lead to irreversible neurologic damage and death.
Given the importance of vitamin B12, several groups have carried out genome-wide association studies (GWAS) using serum or plasma vitamin B12 as a quantitative trait (9–13). With the exception of a recent study on an older, ischemic stroke population of mixed race/ethnicity (14), these studies have found an association between circulating vitamin B12 concentration and a common single nucleotide polymorphism (SNP) in fucosyltransferase 2 (FUT2), a gene encoding a 1, 2-fucosyltransferase. This enzyme is involved in the synthesis of ABO(H) blood group antigens on the surface of blood and epithelial cells and is required for the presence of the soluble forms of these antigens in different secretions (15). The absence of functional FUT2 enzyme leads to so-called ‘non-secretor’ status in which homozygotes for null FUT2 alleles lack the A, B, and H antigens in saliva and other body fluids. The existence of ‘secretor’ and ‘non-secretor’ phenotypes in human populations was known long before its molecular basis was discovered (OMIM +182100). Significant numbers of individuals homozygous for null alleles of the FUT2 gene are present in all human populations examined and this locus shows evidence of positive selection for several different null alleles (16,17).
At least five vitamin B12 genome-wide association studies report that SNPs in FUT2 show the strongest statistical association with circulating vitamin B12 (9–13). Two of these studies also report significantly higher circulating vitamin B12 concentrations in homozygotes for the common, non-secretor allele of FUT2 in Caucasians [FUT2 rs601338 (9)] and Asians [FUT2 rs1047781 (10)]. However, exactly how a loss of FUT2 function influences vitamin B12 concentration is unknown. Helicobacter pylori (H. pylori) infection has been associated with vitamin B12 deficiency (18,19). Some authors have proposed that FUT2 genotype can influence the extent to which H. pylori attaches to gastric mucosa and influences vitamin B12 absorption (20,21). In contrast, a subsequent study found that secretor status as determined by FUT2 variation correlates with plasma vitamin B12 concentrations but is independent of H. pylori serotype (22). It is important to note that all previous studies measured total circulating vitamin B12. This measure does not resolve the proportion of the vitamin bound to its two distinct carrier proteins, transcobalamin and haptocorrin. These proteins carry significantly different quantities of vitamin B12 in blood and different biological properties; transcobalamin II delivers vitamins B12 to all tissues while vitamin B12 carried by haptocorrin is ultimately returned to the lumen of the gut (23,24).
Haptocorrin (HC) refers collectively to transcobalamin I (TCI) and transcobalamin III (TCIII), differentially glycosylated isoforms encoded by a single gene (TCN1), while holo-haptocorrin (holoHC) refers to the vitamin B12-bound HC. Although haptocorrin carries approximately 80% of vitamin B12 in blood, these holoHC complexes are not taken up by the cells of the peripheral tissues and persist in the circulation with a half-life measured in days (25). It is postulated that only the liver cells can endocytose holoHC (26). Despite intensive studies, the biological role of haptocorrin-bound vitamin B12 is not clear. Humans with very low concentrations of HC have been described and in many cases no clinical phenotypes are attributed to their ‘deficiency’ of HC (27). Interestingly, haptocorrin genes have been lost in several evolutionary lineages. It is present in most mammals, but not mouse and rat, and is found in reptiles, but not in birds or amphibians (28).
The bioavailable pool of vitamin B12 is carried by transcobalamin II (TC), encoded by the TCN2 gene. This pool represents ∼20% of circulating vitamin B12 and exhibits rapid turnover, exiting the circulation into the tissues (plasma half-life ∼10 min) (23,24). TC bound vitamin B12 enters cells via a specific receptor and is the main source for cellular vitamin B12-dependent reactions. Because the ratio of ‘inert’ vitamin B12 on holoHC to ‘bioavailable’ vitamin B12 on holoTC can be 4: 1 or higher, cellular vitamin B12 deficiency may be present even when the total circulating vitamin B12, as currently measured in clinical settings, is within the normal range. This potential masking of reduced concentrations of circulating bioavailable vitamin B12 bound to transcobalamin II may contribute to a delayed diagnosis of true deficiency.
In addition to confirming previously published GWAS of circulating vitamin B12, we also carried out a separate GWAS on vitamin B12 bound to each of its two circulating carrier proteins, i.e., holo-transcobalamin (holoTC) and holo-haptocorrin (holoHC) and we evaluated the influence of the FUT2 rs601338 secretor variant on vitamin B12 bound to each of these carrier proteins in a young, healthy Irish population as well as in an older Irish population. We also tested whether FUT2 rs601338 (p.Trp154Ter) genotype influences glycosylation of haptocorrin by examining the ratios of TCI and TCIII derived from individuals carrying different FUT2 alleles. Lastly, we developed a model system to ask whether FUT2-mediated glycosylation of HC influences holoHC concentration. These studies provide an avenue for understanding the independent observations that the FUT2 rs601338 secretor variant influences circulating vitamin B12.
Results
Common SNPs in the FUT2 gene are associated with serum vitamin B12 concentration in humans
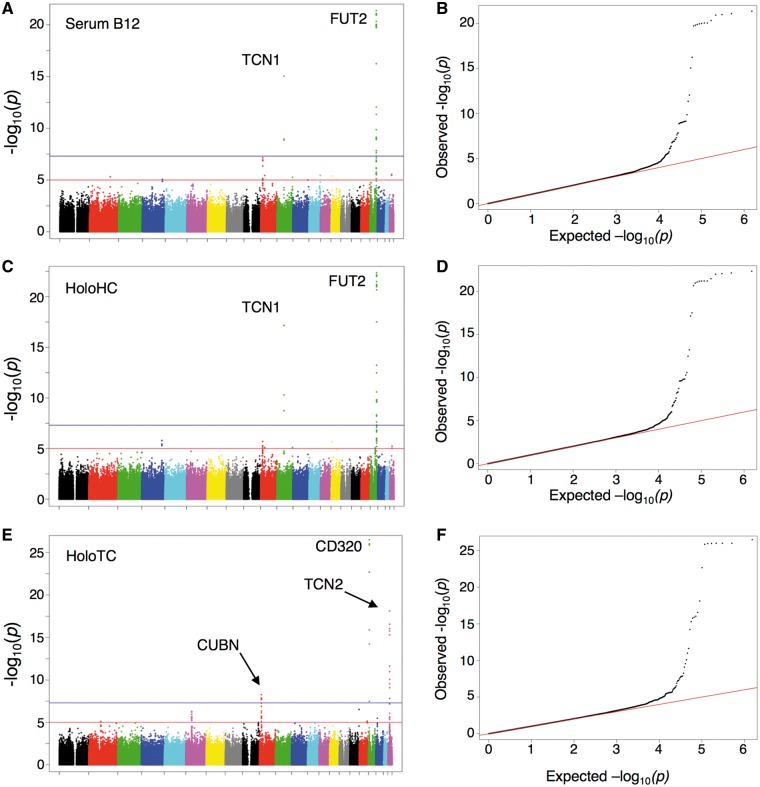
We performed a GWAS in 2232 young, healthy Irish adults by testing for associations between 757 533 genotyped SNPs and log-transformed serum vitamin B12 concentration using standard linear regression. The strongest association (P = 4.31×10−22) was between a synonymous SNP (rs681343, MAF = 0.4) in the FUT2 gene on chromosome 19 (Fig. 1, Table 1). This SNP is in nearly perfect linkage disequilibrium (LD; D’=1, r2=0.995) with an upstream nonsense variant (p.Trp154Ter, rs601338), a well-known secretor variant that renders the FUT2 enzyme inactive (15). Our results are consistent with the rs601338 AA genotype, encoding nonfunctional FUT2 enzyme (p.154Ter) and therefore non-secretor status, being associated with significantly higher serum vitamin B12 concentrations (Fig. 2A, P = 1.8×10−25). Mean serum vitamin B12 concentrations were 16–18% higher in non-secretors (rs601338 AA) compared with secretors (rs601338 GG or GA, Table 2). Previously published GWAS (9,11,12) have reported associations between vitamin B12 concentration and several SNPs [rs602662, rs492602, and rs601338 (p.Trp154Ter)] in FUT2. Our most significant association signal (rs681343) and these SNPs are in strong linkage disequilibrium (r2>0.9) with rs601338 (p.Trp154Ter) in this sample of the Irish population. Therefore, our GWAS result is consistent with these previous studies, and it appears that the FUT2 p.Trp154Ter secretor variant is the functional variant that most strongly affects circulating vitamin B12 concentration.
Figure 1.
Genome-wide association analysis for serum vitamin B12 concentration. Manhattan plots and Q-Q plots for genome-wide association with log-transformed serum metabolite concentration based on 757, 533 genotyped SNPs. X-axis of Manhattan plots: SNP position, with color-differentiated chromosomes ordered. Y-axis of Manhattan plots: -log10(p) for each SNP. Horizontal red line at –log10(p)=5.0: Suggestive significance (P = 1 × 10−05). Horizontal blue line at –log10(p)=7.3: Genome-wide significance (P = 5 × 10−08). (A) Manhattan plot and (B) Q-Q plot for log-transformed serum B12. The two genome-wide significant signals are in FUT2 and TCN1. (C) Manhattan plot and (D) Q-Q plot for log-transformed serum holoHC. The two genome-wide significant signals are in FUT2 and TCN1. (E) Manhattan plot and (F) Q-Q plot for log-transformed serum holoTC. The three genome-wide significant signals are in CD320, TCN2 and CUBN.
Table 1.
Genome-wide association signalsa in a young, healthy Irish population for serum B12 and its carrier proteins
| Vitamin B12 GWAS | ||||||||
| GENEb | SNP | CHR | BP | BETA | SE | R2 | T | P |
| FUT2 | rs681343 | 19 | 53898274 | −0.0526 | 0.0054 | 0.0411 | −9.767 | 4.31E-22 |
| FUT2 | rs601338 | 19 | 53898486 | −0.0523 | 0.0054 | 0.0412 | −9.700 | 8.23E-22 |
| TCN1 | rs34324219 | 11 | 59379954 | −0.0732 | 0.0090 | 0.0286 | −8.099 | 9.03E-16 |
| CUBN | rs12243895 | 10 | 17198979 | 0.0343 | 0.0064 | 0.0131 | 5.404 | 7.24E-08 |
| HoloHC GWAS | ||||||||
| GENE | SNP | CHR | BP | BETA | SE | R2 | T | P |
| FUT2 | rs681343 | 19 | 53898274 | −0.0609 | 0.0061 | 0.0432 | −10.01 | 4.39E-23 |
| FUT2 | rs601338 | 19 | 53898486 | −0.0607 | 0.0061 | 0.0435 | −9.959 | 7.06E-23 |
| TCN1 | rs34324219 | 11 | 59379954 | −0.0890 | 0.0102 | 0.0329 | −8.690 | 6.91E-18 |
| CPE | rs3775315 | 4 | 166618915 | −0.0577 | 0.0120 | 0.0104 | −4.812 | 1.60E-06 |
| CUBN | rs12243895 | 10 | 17198979 | 0.0342 | 0.0072 | 0.0102 | 4.768 | 1.98E-06 |
| HoloTC GWAS | ||||||||
| GENE | SNP | CHR | BP | BETA | SE | R2 | T | P |
| CD320 | rs2232783 | 19 | 8274794 | 0.1646 | 0.0150 | 0.0514 | 10.950 | 3.16E-27 |
| TCN2 | rs5749135 | 22 | 29341906 | 0.0554 | 0.0062 | 0.0348 | 8.945 | 7.67E-19 |
| CUBN | rs12261966 | 10 | 17183006 | −0.0378 | 0.0065 | 0.0154 | −5.857 | 5.43E-09 |
| MUT* | rs3930746 | 6 | 49437212 | −0.0318 | 0.0063 | 0.0115 | −5.037 | 5.12E-07 |
Based on analyses using log-transformed values for each metabolite and directly genotyped SNPs.
Column headers: GENE, gene; SNP, single nucleotide polymorphism identifier; CHR, chromosome: BP, base position; BETA, regression coefficient; SE, standard error of the coefficient; R2, regression r-squared (multiple correlation coefficient); T, t-statistic for regression of phenotype on allele count; P, significance value for coefficient. Gene abbreviations: FUT2, fucosyltransferase 2; TCN1, transcobalamin 1; CPE, carboxypeptidase E; CUBN, cubilin; CD320, CD320 molecule (transcobalamin receptor); TCN2, transcobalamin 2; MUT, methylmalonyl-CoA mutase.
MUT is the nearest gene (68 kb) to this intergenic SNP.
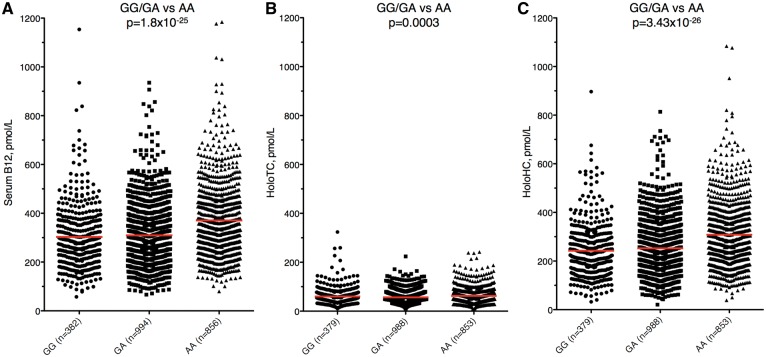
Figure 2.
Serum vitamin B12, holoTC and holoHC plotted by FUT2 rs601338 (p.Trp154Ter) genotype. (A) Total serum vitamin B12 concentration. (B) Serum holo-transcobalamin (holoTC2) concentration. (C) Serum holo-haptocorrin (holoHC) concentration. Association results are shown; linear regression using a recessive model was used to compare secretors (GG/GA) vs. nonsecretors (AA) for each metabolite.
Table 2.
Influence of FUT2 secretor variant genotype on circulating metabolite concentrations
| Current GWAS: Mean serum concentrations (± standard deviation) in young Irish adults (TSS). | |||||||
| FUT2 rs601338 (p.Trp154Ter) | GG | GA | AAb | A freq. | n | AAb vs GG | AAb vs GA |
| B12 (pmol/l) | 303 ± 139 | 310 ± 131 | 371 ± 154 | 0.60 | 2196 | 18.4% | 16.6% |
| HoloTC (pmol/l) | 59.5 ± 36.5 | 56.3 ± 26.8 | 62.2 ± 32.7 | 0.60 | 2184 | 4.3% | 9.4% |
| HoloHC (pmol/l) | 240.4 ± 118.4 | 250.8 ± 118.2 | 307.7 ± 140.0 | 0.60 | 2184 | 21.9% | 18.5% |
| Current replication: Mean serum concentration (± standard deviation) in older Irish adults (TUDA). | |||||||
| FUT2 rs601338 (p.Trp154Ter) | GG | GA | AAb | A freq. | n | AAb vs GG | AAb vs GA |
| B12 (pmol/l) | 266 ± 188 | 270 ± 13 | 324 ± 279 | 0.59 | 4868 | 21.9% | 20.0% |
| HoloTC (pmol/l) | 69.8 ± 71.0 | 71.0 ± 61.6 | 70.6 ± 53.4 | 0.59 | 4888 | 1.2% | −0.5% |
| HoloHC (pmol/l) | 189.5 ± 99.4 | 198.8 ± 98.0 | 246.1 ± 115.8 | 0.59 | 4817 | 29.9% | 23.8% |
| Hazra et al., 2008: Mean (95% CI) plasma vitamin B12 concentration in women of European ancestry. | |||||||
| FUT2 rs601338a (p.Trp154Ter) | GG | GA | AAb | A freq. | n | AAb vs GG | AAb vs GA |
| B12 (pg/ml) | 418.32 (403.36-433.84) | 418.7 (407.76-429-92) | 496.6 (477.89-516.04) | 0.49 | 1658 | 15.8% | 15.7% |
| Lin et al. (2012): Mean serum vitamin B12 concentration in Chinese adults. | |||||||
| FUT2 rs1047781 (p.Ile140Phe) | AA | AT | TTb | T freq. | n | TTb vs AA | TTb vs AT |
| B12 (pg/ml) | 652 | 692 | 776 | 0.46 | 1999 | 16.0% | 10.8% |
| B12 (pg/ml) | 619 | 683 | 826 | 0.47 | 1496 | 25.1% | 17.3% |
| Collin et al. (2011): Median (5th–95th percentile) plasma concentration in adult men from the United Kingdom. | |||||||
| FUT2 rs492602 (p.Ala68=) | AA | GA | GGb | G freq. | n | GGb vs GA | GGb vs AA |
| B12 (pmol/l) | 286 (159–473) | 300 (163–514) | 329 (194–575) | 0.50 | 1507 | 9.7% | 15.0% |
| HoloTC (pmol/l) | 56 (22–119) | 57 (23–125) | 57 (22–130) | 0.50 | 1507 | 0 | 1.8% |
| HoloHC (pM) | 230 (119–382) | 240 (123–417) | 286 (150–513) | 0.50 | 1507 | 19.2% | 24.3% |
imputed genotypes.
non-secretor genotype.
We also replicated two previously reported secondary associations between vitamin B12 and SNPs in TCN1 and the intrinsic factor-cobalamin receptor, cubilin (CUBN) (10–14) (Fig. 1, Table 1), in this young, healthy Irish population. After the signal in FUT2, TCN1 rs34324219 was the SNP most significantly associated with log-transformed serum vitamin B12 in the current study (P = 9.03×10−16), as previously observed (12,14,29). This SNP is in the same LD block [1000 Genomes Project European superpopulation (1000G EUR, 30)] as the strongest signal observed in other studies [D'=1.0, r2=0.33 (10,11,13)]. We found a third genome-wide association between serum vitamin B12 and CUBN rs12243895 (P = 7.42×10−8), confirming a previously reported association (10). This CUBN SNP shares varying degrees of LD with the top CUBN signals reported for genome-wide circulating vitamin B12 association [rs1801222: D’=1.0, r2=0.24; 1000 G EUR (12–14); rs11254363: D’=0.77, r2=0.48; 1000 G EUR (11)].
FUT2 rs601338 (p.Trp154Ter) correlates with serum holo-haptocorrin concentrations
To determine whether FUT2 rs601338 (p.Trp154Ter) influences total vitamin B12 concentration via its circulating carrier proteins, we tested whether the FUT2 rs601338 (p.Trp154Ter) genotype influences vitamin B12 bound to haptocorrin or transcobalamin (holoHC or holoTC, respectively; Fig. 2). All analyses subsequent to the initial vitamin B12 GWAS employ a recessive model since FUT2 rs601338 (p.Trp154Ter) is a known null allele, and only homozygosity of the null allele produces the non-secretor phenotype. Linear regression using a recessive model revealed the significant association of the non-secretor genotype with concentrations of either holoTC (P = 0.0003) or holoHC (P = 3.4×10−26). Although statistically significant, the modest magnitude of the effect on holoTC concentration may not be biologically relevant, prompting further investigation of the influence of FUT2 rs601338 (p.Trp154Ter) on holoHC.
Mean serum holoHC concentrations were higher in non-secretors (rs601338 AA) compared with secretors of either genotype (18.5% higher compared with rs601338 GG; 21.9% higher compared with rs601338 GA; Table 2). FUT2 rs601338 (p.Trp154Ter) also exhibits one of the strongest genome-wide significant associations (P = 7.06×10−23) in a genome-wide association analysis of log-transformed holoHC in this population (Table 1, Fig. 1C), providing further evidence of this functional variant‘s influence on total serum B12 via holoHC. In contrast, the top genome-wide significant association signals for log-transformed holoTC were in CD320, the receptor responsible for cellular uptake of circulating holoTC, and TCN2 itself (Table 1, Fig. 1E). We conclude that FUT2 rs601338 (p.Trp154Ter) exerts its influence on total vitamin B12 primarily through holoHC.
Replication of the association of FUT2 rs601338 (p.Trp154Ter) with vitamin B12 and holoHC in older Irish adults
Replication was performed by testing whether the non-secretor genotype influences vitamin B12, holoTC or holoHC in a cohort of 4941 older Irish adults successfully genotyped for FUT2 rs601338 (p.Trp154Ter, Table 2). Again, a test of linear regression assuming a recessive genetic model was performed. The non-secretor genotype was significantly associated with log-transformed serum vitamin B12 concentration (P = 1.26×10−46) and holoHC (P = 6.34×10−37) but not holoTC (P = 0.46), similar to the strong effect of the non-secretor genotype on vitamin B12 and holoHC observed in young, healthy Irish adults. In this sample of older adults, mean serum holoHC concentrations were higher in non-secretors (rs601338 AA) compared with secretors of either genotype (29.8% higher compared with rs601338 GG; 23.8% higher compared with rs601338 GA; Table 2). In contrast, non-secretors showed very little difference from secretors for mean serum holoTC concentration. We conclude that the FUT2 rs601338 AA non-secretor genotype influences vitamin B12 concentration via changes in the level of circulating holoHC, and not holoTC.
Replication of other genome-wide associations for vitamin B12, holoHC and holoTC in older Irish adults
Independent from the FUT2 rs601338 signal, five genome-wide significant associations (P < 5×10−08, Table 1) were observed in young Irish adults, and subsequently tested for replication in the study of older Irish adults (Table 3, Supplementary Material, Table S5). These SNP-metabolite associations were significantly associated and genotype effects on mean metabolites concentration were similar in the primary and replication cohorts (Table 3). TCN1 rs34324219 is a coding SNP (p.Asp301Tyr), and like FUT2 rs601338 its association with both serum vitamin B12 and holoHC was strongly associated in the replication cohort. Similarly, the associations between CD320 rs2232783, TCN2 rs5749135 and CUBN rs12261966 and holoTC were replicated in this older Irish cohort (Table 3). The signal observed with CD320 rs2232783 is the top genome-wide association with holoTC (Fig. 1D). This SNP shares strong linkage disequilibrium with a 3 bp indel (CD320 rs150384171 p.Glu88del) that has been associated with NTD risk (31) and elevated MMA concentrations in newborns (32). We have previously reported that CD320 rs150384171 has a strong influence on serum vitamin B12 and holoTC (33), and have confirmed this association in both Irish cohorts (Table 3). We note that CD320 rs2232783 and CD320 rs150384171 share strong LD in the Irish population (D’= 0.91; r2 = 0.44), and the larger effect size observed for the latter (beta = 0.26) supports its being the functional variant driving the association. However, due to its low minor allele frequency (MAF ∼0.01), the effect of CD320 rs150384171 on holoTC in the population would be limited to <5% of individuals.
Table 3.
Replication in an older Irish cohort of genome-wide significant SNP-metabolite associations initially observed in the TSS
| TSS | TUDA | ||||||
|---|---|---|---|---|---|---|---|
|
TSS SB12, pmol/l |
TUDA SB12, pmol/l |
Replicationa |
|||||
| TCN1 rs34324219 | TT (23) | GT (395) | GG (1814) | TT (51) | GT (868) | GG (3966) | |
| Mean ±SD | 247.8 ± 78.7 | 289.3 ± 132.1 | 342.2 ± 146.1 | 212.7 ± 105.8 | 264.3 ± 135.5 | 295.8 ± 223.6 | P = 1.08 × 10−13 |
|
TSS holoHC, pmol/l |
TUDA holoHC, pmol/l |
Replicationa |
|||||
| TCN1 rs34324219 | TT (22) | GT (393) | GG (1805) | TT (51) | GT (862) | GG (3929) | |
| Mean ±SD | 182.4 ± 67.76 | 230.1 ± 114.4 | 282.2 ± 130.2 | 149.8 ± 91.06 | 191 ± 99.58 | 219.8 ± 108.9 | P = 7.83 × 10−20 |
|
TSS holoTC, pmol/l |
TUDA holoTC, pmol/l |
Replicationa |
|||||
| CD320 rs2232783 | TT (1) | TG (189) | GG (2027) | TT (10) | TG (340) | GG (4737) | |
| Mean ±SD | 172.4 ± 0 | 86.55 ± 49.38 | 56.83 ± 26.93 | 294.6 ± 289.5 | 102.4 ± 75.34 | 67.97 ± 56.47 | P = 4.42 × 10−39 |
| CD320 rs150384171 | del/del (0) | GAG/del (87) | GAG/GAG (2108) | del/del (2) | GAG/del (146) | GAG/GAG (4802) | |
| Mean ±SD | NA | 108.1 ± 52.7 | 57.3 ± 27.59 | 575.2 ± 429.2 | 129.8 ± 100.4 | 68.61 ± 56.5 | P = 9.40 × 10−35 |
| TCN2 rs5749135 | TT (361) | TC (1092) | CC (767) | TT (811) | TC (2330) | CC (1805) | |
| Mean ±SD | 69.27 ± 35.63 | 60.4 ± 30.06 | 53.45 ± 27.89 | 81.73 ± 73.21 | 72.46 ± 56.54 | 63.11 ± 57.25 | P = 6.00 × 10−24 |
| CUBN rs12261966 | TT (287) | TC (1049) | CC (860) | TT (612) | TC (2246) | CC (2050) | |
| Mean ±SD | 51.2 ± 27.43 | 59.46 ± 31.05 | 62.01 ± 31.17 | 64.41 ± 49.14 | 68.88 ± 55.37 | 73.87 ± 65.11 | P = 8.08 × 10−07 |
Replication was performed for each SNP by linear regression using an additive genetic model on log-transformed, unadjusted metabolite values measured in TUDA participants. See Supplementary Material, Table S5 for more details.
FUT2 rs601338 (p.Trp154Ter) correlates with glycosylation status of haptocorrin
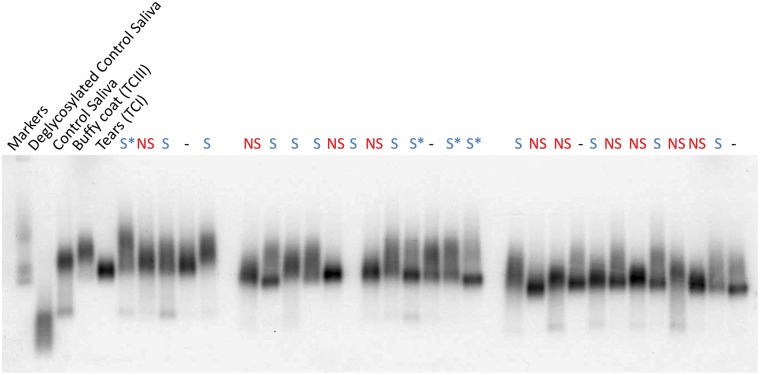
Based on the strong, replicated association of FUT2 rs601338 with holoHC, we hypothesized that the expression of functional FUT2 enzyme could influence total circulating vitamin B12 concentration by altering the glycosylation of haptocorrin. This hypothesis was based on the observation that haptocorrin consists of two alternatively glycosylated isoforms. The TCI isoform has more negatively charged sialic acid residues and migrates faster than TCIII by non-denaturing (native) gel electrophoresis (34). In contrast, TCIII contains more fucose residues than TCI and migrates more slowly by native gel electrophoresis. We took advantage of these differences in electrophoretic mobility to ask if FUT2 rs601338 (p.Trp154Ter) genotype influences the ratio of TCI to TCIII concentration as detected by direct autoradiography of radiolabeled (57Co) vitamin B12 bound to haptocorrin. In blood derived samples, holoTC obscures holoHC when analysing samples by this method (data not shown). Therefore, we screened haptocorrin derived from saliva rather than blood (n = 29; 25 of which were successfully genotyped) since saliva does not contain significant TC (34). Our results indicate that there is a correlation between the relative ratios of TCI/TCIII and secretor status as determined by FUT2 rs601338 (p.Trp154Ter) genotype (Fig. 3). In non-secretor individuals that do not express functional FUT2 enzyme (rs601338 AA genotype) the TCI isoform appears to be prevalent (Fig. 3). In contrast, in the secretor individuals that express functional FUT2 enzyme (rs601338 GG or GA genotypes) it appears that both TCI and TCIII isoforms are present (Fig. 3).
Figure 3.
Differential glycosylation of holoHC in secretors vs. non-secretors. Saliva samples from 29 individuals were incubated with radiolabeled (57Co) vitamin B12, run on native gel electrophoresis and visualized by autoradiography. Samples labeled ‘S’ in blue represent secretor genotypes (FUT2 rs601338 GG and GA; S* denotes homozygosity), samples labeled ‘NS’ in red represent non-secretor genotypes (FUT2 rs601338 AA), and the samples labeled ‘−’ in black failed genotyping. Controls lanes are labeled. This image consists of three aligned autoradiographs.
A functional assay of asialoglycoprotein receptor-mediated endocytosis of holoHC in HepG2 cells
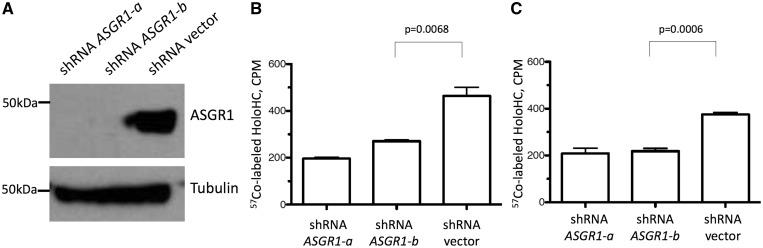
The association between FUT2 rs601338 (p.Trp154Ter) genotype and holoHC concentration could be driven by changes in the amount of vitamin B12 entering the circulation from the intestines, changes in resorption or catabolism by the kidneys (35), or by changes in the rates at which holoHC is removed from the bloodstream. Plasma clearance studies suggest that holoHC is removed from the blood by the liver via the asialoglycoprotein receptor (36). Because we found that FUT2 rs601338 (p.Trp154Ter) genotype appears to influence the glycosylation state of HC, we hypothesized that these differences in glycosylation could be associated with a change in the rate at which HC is bound and internalized by the asialoglycoprotein receptor. To test this, we developed a cell-based functional assay to measure the holoHC uptake via the asialoglycoprotein receptor. Our aim was to produce cells lacking the receptor to compare with cells expressing the native receptor. We created liver hepatocellular carcinoma HepG2 cells expressing two different short hairpin RNAs (shRNAs) targeting ASGR1, one of the subunits of the asialoglycoprotein receptor. Asialoglycoprotein receptor protein was undetectable by immunoblot in HepG2 cells expressing either shRNA targeting ASGR1 (lane 1 or lane 2, Fig. 4A). We assayed these asialoglycoprotein receptor knock-down cell lines for their ability to import the holoHC complex. HC is abundant in saliva and saliva does not contain transcobalamin (34). We mixed saliva samples with radiolabeled (57Co) vitamin B12 to obtain holoHC. Regardless of HC donor, HepG2 cells expressing either asialoglycoprotein receptor shRNA (column 1 or column 2) showed significantly reduced uptake compared with HepG2 cells expressing empty vector (Fig. 4B and C).
Figure 4.
ASGR is responsible for the cellular uptake of holoHC in HepG2 cells. (A) A Western blot analysis for ASGR1 in HepG2 cells expressing shRNAs against ASGR1 (shRNA ASGR1-a and shRNA ASGR1-b) or vector alone (shRNA vector). Tubulin was used as a loading control. (B, C) HoloHC uptake in HepG2 cells expressing shRNAs or vector alone. Saliva samples from different control individuals were used as a source of 57Co-labeled holoHC. One of two independent experiments is shown. For each experiment, triplicate wells of HepG2 cells were analyzed and the mean and standard error for each sample is shown. The Student t-test was used to compare holoHC uptake in HepG2 cells expressing shRNA ASGR1-b vs. shRNA vector.
FUT2 genotype does not influence holoHC uptake in a cell culture based assay
It was demonstrated in a rabbit model system that human TCIII/vitamin B12 complex is cleared from the plasma more rapidly than TCI/vitamin B12 complex (26). Clearance rates are dependent on the asialoglycoprotein receptor in the liver and the amount of sialic acid residues on the vitamin B12 transporters (26). The hepatic asialoglycoprotein receptor has a low affinity for highly sialated proteins, which would presumably result in decreased clearance of highly sialated HC in FUT2 null individuals. We hypothesized that in secretor individuals expressing enzymatically active FUT2, HC would be fully fucosylated. This posttranslational modification would prevent the attachment of terminal sialic acid residues (26,37), facilitating the receptor-mediated endocytosis of holoHC via the asialoglycoprotein receptor in the liver. In contrast, non-secretors lacking functional FUT2 enzyme cannot fucosylate HC, presumably resulting in more heavily sialated holoHC. Such a mechanism could explain the observation of higher amounts of holoHC in the blood of FUT2 null individuals.
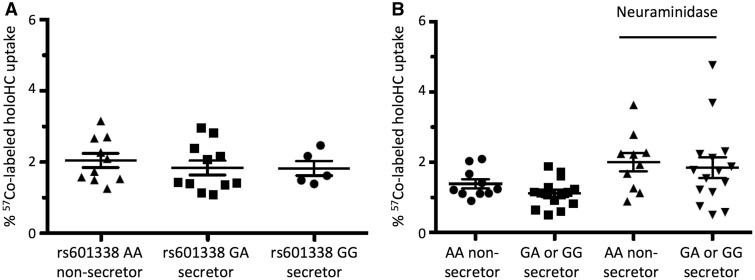
To investigate whether FUT2 genotype correlates with holoHC uptake we utilized the HepG2 based functional assay. Based on our hypothesis, we expected that the HC from non-secretors would show less ASGR-mediated uptake of the holoHC complex than HC from the secretors. We compared the HepG2 cellular uptake of holoHC complex using saliva from individuals that express functional and non-functional FUT2. As shown in Figure 5, we did not detect differences in the uptake of the holoHC complex by FUT2 rs601338 (p.Trp154Ter) genotype (Fig. 5A), or by secretor status as assigned by genotype (Fig. 5B). We also observed that neuraminidase treatment (Fig. 5B), which removes sialic acid residues from HC, did not reveal any differences in the uptake of HC derived from secretors vs non-secretors.
Figure 5.
Functional assay for cellular vitamin B12 uptake by HepG2 cells. (A) Cellular vitamin B12 uptake assay by FUT2 rs601338 W154X genotype. Saliva from 25 individuals of known secretor status was used as a source of 57Co-labeled holoHC. The group mean and standard error is included with each scatterplot. A Student t-test was performed to compare HepG2 uptake of holoHC from non-secretors (FUT2 rs601338 AA) and either secretor genotype group (FUT2 rs601338 GG, P = 0.5004; GA, P = 0.471). (B) Cellular vitamin B12 uptake assay of 57Co-labeled holoHC from non-secretors (FUT2 rs601338 AA) and secretors (FUT2 rs601338 GG, GA) before and after neuraminidase treatment. The group mean and standard error is included with each scatterplot. A Student t-test was performed to compare vitamin B12 uptake in secretors and non-secretors in the absence (P = 0.114) or presence of neuraminidase treatment (P = 0.712).
Discussion
We carried out a detailed study of how genetic variation influences the concentration of vitamin B12 in blood. In contrast to previous work, we measured vitamin B12 while preserving the original biological context of its association with its circulatory carrier proteins. We provide evidence that the FUT2 secretor status variant correlates with serum holoHC concentration and influences HC glycosylation.
A 10–25% increase in circulating total vitamin B12 concentration in non-secretors as determined by FUT2 rs601338 genotype has been shown in multiple studies with two different population-specific null FUT2 alleles (Table 2). This confirms that it is the loss of FUT2 function driving these quantitative differences. Prior hypotheses regarding the mechanism of action have focused on influencing absorption of dietary vitamin B12, either through intrinsic factor or H. pylori infection. It has been demonstrated that SNPs in FUT2 can influence the attachment of H. pylori to the gastric mucosa (20,21,38). This led others to hypothesize that the difference in vitamin B12 concentrations between secretors and non-secretors is via the increased susceptibility secretors have for H. pylori infection (9,13). In this model, the secretors would have lower plasma vitamin B12 concentration due to attenuated processing of vitamin B12 in gastric juice and subsequent less efficient extraction/absorption of the vitamin in the terminal segment of the ileum. However, a recent study demonstrated that there is no correlation between secretor status and H. pylori infection, indicating that FUT2 genotype influences vitamin B12 concentration via a different mechanism (22). In a subsequent study, the same group explored the possibility that FUT2 genotype could affect the glycosylation status of another vitamin B12 transporter, gastric intrinsic factor (GIF) (39). However, this study was restricted to evaluating a small number of case and control individuals from three unrelated families. Although a potential FUT2 rs601338 genotype effect was observed on gastric intrinsic factor secretion and glycosylation, the GIF phenotypes of the FUT2 rs601338 GA heterozygotes more closely aligned with those of the non-secretor genotype (AA) than those with the secretor genotype (GG). This does not reflect the recessive mode of FUT2 rs601338 influence observed on serum vitamin B12, in which mean vitamin B12 concentrations are elevated solely in non-secretors (Table 2). It remains unclear to what extent FUT2 genotype influences GIF secretion and thereby alters vitamin B12 concentration in the general population. In both models described above, FUT2 genotype would be predicted to alter the pool of vitamin B12 absorbed from the gut. As vitamin B12 transported from the gut binds to transcobalamin in plasma, these models are not consistent with our data, which show that FUT2 genotype influences the concentration of haptocorrin-bound vitamin B12 to a far greater extent than transcobalamin-bound vitamin B12. This is consistent with FUT2 exerting influence via its fucosylation function, as haptocorrin is a glycosylated protein and transcobalamin is not. It also suggests that FUT2 activity impacts the intra-organismal recycling of vitamin B12, not the absorption and assimilation of the vitamin from the diet.
If the mechanism by which fucosyltransferase (FUT2) affects vitamin B12 concentration can be elucidated, it could shed light on why the standard clinical test for vitamin B12 status suffers such significant false positive and false negative rates. Our data provide an important part of the answer: we demonstrated that FUT2 exerts its effect on total vitamin B12 concentration in the blood via one of the vitamin B12 carriers, HC. Non-secretor status as determined by FUT2 rs601338 AA genotype is associated with mean total vitamin B12 concentrations that are 10–20% higher with no expected impact on cellular sufficiency. This was also observed in a candidate variant study assessing prostate cancer risk of vitamin B12-related SNPs, where FUT2 rs492602 was seen to significantly influence holoHC but not holoTC in a control UK population (40). Our results replicate this finding, and establish that the FUT2 secretor status variant as determined by FUT2 rs601338 genotype is by far the strongest genome-wide association signal influencing holoHC concentration. To investigate the mechanism behind this effect, we hypothesized that as a fucosyltransferase, FUT2 might influence holoHC concentration by altering the post-translation modifications of the heavily glycosylated HC. Indeed, our results are consistent with genotypic secretor status influencing the glycosylation status of HC (Fig. 3). How this translates into differences in circulating holoHC (and therefore total circulating vitamin B12) remains to be determined. HoloHC uptake by ASGR did not differ based on FUT2 rs601338 genotypic secretor status in our in vitro system. A possible source of noise is misclassification of secretors since we relied solely on the variant identified in our vitamin B12 GWAS to assign secretor or non-secretor status. However, FUT2 rs601338 (p.Trp154Ter) is the only significant secretor variant in populations of European descent (16,17,41).
Several important questions remain. Our holoHC uptake assay does not precisely mimic the HC uptake in blood. We used saliva as our source of HC. The glycosylation of HC in saliva can differ from HC in blood (34). To investigate a genetic association observed in serum samples, the ideal source of HC would be blood, but the interference of circulating TC and availability of TC-free saliva samples led us to begin with this source of HC. Also, we only tested a small number of available samples (n = 25) but ideally this in vitro assay would be carried out on a large scale to confirm that the secretor status as determined by FUT2 rs601338 (p.Trp154Ter) might slightly influence ASGR uptake rates of holoHC. In addition, whether there are other receptors besides ASGR that can bind the holoHC complex remains obscure, as does the biological role of the holoHC complex, which carries the majority (∼80%) of vitamin B12 in the circulation. Our model system may not be sensitive enough to detect small differences in hepatic uptake. As holoHC has a very long half-life in plasma, small differences in uptake rates could translate into larger differences in steady state. To fully characterize steady state vitamin B12 in the circulation, it would be of great interest to capture all the players that contribute (e.g., TCI, TCIII, apoHC, holoHC, total HC, apoTC, holoTC, total TC, total vitamin B12) in a significant number of individuals genotyped for FUT2 rs601338. We and others do not find that FUT2 rs601338 influences levels of apoHC, apoTC, or total TC [(39), Supplementary Material, Table S4]. Analysis of all involved factors in a single study would allow better modeling of the system dynamics and perhaps reveal how individual factors contribute to the most commonly measured analyte - total circulating vitamin B12. Such a model may still be incomplete. It is also possible that the renal clearance rate for vitamin B12 is influenced by the differential glycosylation of haptocorrin associated with FUT2 alleles, since the heavy glycosylation of HC is likely an important factor in renal handling (35). Alternately, differential glycosylation of HC could also affect its secretion and ultimate contribution to total circulating vitamin B12. Lastly, it remains unknown whether FUT2 directly fucosylates haptocorrin. Further confirmation of serum holoHC fucosylation and any correlation with FUT2 rs601338 genotype would be best addressed with functional studies and lectin binding assays.
Understanding how FUT2 genotype influences vitamin B12 concentration could directly impact clinical testing. The interpretation of vitamin B12 tests can be difficult for those presenting with low/normal borderline concentration of the vitamin. It is well recognized that measuring circulating vitamin B12 by the current clinical test is inadequate and has both low sensitivity and specificity (42). Only ∼20% of circulating vitamin B12 (holoTC) represents the ‘active’ bioavailable form meaning that the most commonly ordered clinical test for vitamin B12 mainly measures the holoHC, which can mask an existing vitamin B12 deficiency. This has led to the promotion of tests that measure holoTC directly (43). Recommendations to use additional markers of vitamin B12-dependent enzyme activity such as methylmalonic acid (MMA) and total homocysteine when evaluating or confirming vitamin B12 deficiency are also problematic (44). For example, homocysteine is also elevated in folate deficiency and renal dysfunction (44). At this time measuring MMA in clinical samples is expensive and not widely available (45). To complicate this picture, we recently described a genetic variant that exerts a strong influence on MMA concentration in a vitamin B12 independent fashion (46). Furthermore, there is no common agreement on the reference ranges of MMA concentration that should be used in the clinic (47,48). Based on our findings, it would be interesting to examine if secretor status, as determined by FUT2 genotype or antigen testing of saliva, could increase the sensitivity and specificity of clinical evaluation of vitamin B12 deficiency. Understanding how FUT2 contributes to the normal vitamin B12 biology could lead to the creation of predictive algorithms that enhance the current methods used to evaluate individuals for vitamin B12 deficiency.
Materials and Methods
Study population
The Trinity Student Study (TSS) population has been previously described (46). Briefly, 2524 participants were enrolled over one academic year (2003–2004) from the student population attending the University of Dublin, Trinity College (TCD). Analyses were restricted to participants in good health and of Irish ethnicity, as defined as having all four grandparents born in Ireland. Study inclusion required completion of a health and lifestyle questionnaire, and donation of a non-fasting blood sample. There were 2508 participants who met the inclusion criteria and their blood samples were used to extract DNA for genotyping and measure metabolites. Participants gave written informed consent upon enrollment, and all samples were anonymized. Ethical approval of the study was granted by the Dublin Federated Hospitals Research Ethics Committee and the Office of Human Subjects Research at the National Institutes of Health.
Metabolite measurements
Blood samples were collected into EDTA and clotting tubes. Samples were processed within 3 h and stored at − 80 °C until analysed. Serum vitamin B12 from clotting tubes was measured by microbiological assay using colistin-resistant L. delbreukii as described (49). The inter-assay coefficient of variability (CV) was 10.6%. Serum holoTC was measured with an immunofluorescence method as quantitated on an Abbott AxSym (50,51) with an inter-assay CV of 9.4%. Total TC was measured by subsequent saturation of these samples and requantitation. ApoTC was calculated as the difference between total TC and holoTC in individuals with both measures. HoloHC was calculated as the difference between serum vitamin B12 and serum holoTC in individuals with both measures.
DNA extraction and genotyping
Qiagen QIAamp DNA Blood Mini Kits were used to extract genomic DNA from all samples. Eighteen samples were excluded based on low quantity, and the remaining 2490 samples were genotyped at the Center for Inherited Disease Research (CIDR, Baltimore, MD) using Illumina 1 M Human Omni1-Quad_v1–0_B chips. High quality genotypes were returned for 2438 samples, 14 blind duplicates and HapMap controls. Concordance rates were 99.997% for the blind duplicates and 99.71% for the HapMap controls.
Genotyping quality control and data preparation for GWAS
Further sample exclusion was based on the following criteria. Ten participants were excluded based on sex discrepancies between self-report and genotypes or abnormal sex chromosomes. Sixteen subjects missing anthropometric data were excluded. Family relatedness was another basis for exclusion. Sibling relationships were self-reported, accounting for participation of two siblings from 74 families and three siblings from three families. These relationships were confirmed using pair-wise tests for relatedness and population stratification. PLINK (52) was used to analyse a test set of 25 718 SNPs that are common (MAF > 0.1), high quality (genotype call rates > 0.99) and essentially independent [all SNP pairs have very low linkage disequilibrium (r2 < 0.04)]. In addition to the known sibships, these analyses revealed other individuals that appeared to be related at the level of first cousins. A total of 175 individuals were excluded for reported or detected relatedness while maintaining inclusion of one individual from each participating family. Lastly, the same test set of SNPs were used to evaluate potential population stratification. Principal components analysis performed using EIGENSOFT 3.0 (53) did not detect any population stratification, although two individuals were further excluded as outliers (6 standard deviations from the mean) leaving 2235 samples. Within the group of 2235 samples an additional sample was removed for a low call rate compared with the remaining samples and two additional samples were removed for being phenotype outliers. After completion of sample exclusions, 2232 study samples were retained for analysis in the GWAS.
Quality control assessment was also performed on the set of 1 008 829 SNPs. In the first stage, SNPs with genotyping call rates < 95% were dropped. Remaining samples with < 97% call rates or cryptic relatedness (see above) were dropped (57 samples total). In the second stage, SNPs were dropped based on the following criteria: a) call rate < 98%; b) Mendelian errors using HapMap trios; c) discordant markers using HapMap controls, and; d) discordant markers detected in any study duplicate pair. SNPs deviating from Hardy-Weinberg equilibrium (HWE, P < 1×10−4) were flagged for future reference and retained for analysis (n = 3512).
LD was measured in the entire dataset, and LD plots were reviewed using Haploview (54). Multiple methods to examine batch effects were employed, and no significant effects were detected. These methods included descriptive statistics per batch (average MAF, average genotyping call rate across all SNPs for each plate) and association test results comparing each batch to the others (average P-value, number of significant results at a particular threshold). After excluding monomorphic and low MAF (MAF < 0.01) SNPs, the final dataset for association testing consisted of 757 533 genotyped SNPs for 2232 samples. TSS genotype and phenotype data have been deposited in dbGaP ([dbGaP: phs000789.v1.p1] Collaborative Study of Genes, Nutrients and Metabolites [CSGNM]).
SNP imputation
A set of 757 533 autosomal SNPs was used to impute non-genotyped SNPs in MaCH 1.0 (55), using the HapMap II reference panel (56). Imputed SNPs were only retained for analysis if the squared correlations between imputed and actual genotypes for a set of masked genotypes (R2) was greater than 0.5. This resulted in 2 350 763 imputed genotypes per person. Two hundred individuals were used to estimate model parameters with 100 iterations for the Monte Carlo procedure using all available haplotypes for each update and then all SNPs in the HapMap II reference panel were imputed.
GWAS statistical analysis
GWAS was performed for three quantitative traits: serum vitamin B12, holoTC, and holoHC. Serum vitamin B12 and holoTC were directly measured, and holoHC was calculated as their difference. In the final dataset, serum vitamin B12 measurements were available for 2230 of the 2232 samples, holoTC measurements were available for 2219 of the 2232 samples, and holoHC measures were calculated for the 2219 samples that had both serum vitamin B12 and holoTC measurements. Standard descriptive statistics were performed on each of the three metabolite datasets both with and without transformation. The distributions of these measurements deviated significantly from a normal distribution and so log transformation was used to normalize each metabolite‘s dataset. Each GWAS was performed using genotyped autosomal SNPs to test for association using the log-transformed metabolite datasets while adjusting for age and sex (see Results and Supplementary Material, Tables S1–S3). For comparison, each GWAS was also performed using the untransformed metabolite datasets (Supplementary Material, Fig. S1). Lastly, each GWAS was performed using the set of genotyped plus imputed autosomal SNPs to test for association using both log transformed and the untransformed metabolite datasets (Supplementary Material, Figs S2 and S3).
In the simple linear regression model, each marker was tested for association using an additive genetic model (i.e. number of minor alleles, 0, 1, and 2) using PLINK v2. The genomic control (λ) values for the log-transformed and untransformed datasets for each of the three metabolites do not show evidence of inflation of test statistics (Supplementary Material, Table S5). Each SNP’s contributed variation to a metabolic trait was calculated by regression R-squared (R2). R scripts written within our institution were used to generate Manhattan plots and Q-Q plots. Additionally, Q-Q plots were generated without the markers on chromosomes 11 (TCN1 signal) and 19 (FUT2 signal) so that the distribution of GWAS p-values obtained for log-transformed measures of serum vitamin B12 and holoHC could be inspected in the absence of these highly significant associations, and similarly Q-Q plots for log-transformed measures of holoTC were generated without the markers on chromosome 10 (CUBN signal), chromosome 19 (CD320 signal), and chromosome 22 (TCN2 signal; see Supplementary Material, Fig. S4).
Replication association analyses
The Trinity, Ulster, Department of Agriculture (TUDA) Study has been previously described (46,57). Briefly, 5186 participants were enrolled in Dublin and in Northern Ireland from hospital outpatient clinics or the community between September 2008 and December 2012. Participants were eligible if aged over 60 years, with Irish parents, and without a prior diagnosis of dementia. Information on participant health, lifestyle and medical history was obtained. Participants provided written informed consent upon enrollment. Ethical approval of the study was granted based on the relevant jurisdiction: The Research Ethics Committee of St. James‘s Hospital and The Adelaide and Meath Hospital, Dublin, and the office for Research Ethics Committees, Northern Ireland (ORECNI; reference 08/NI/RO3113) with corresponding approvals from The Northern and Western Health and Social Care Trusts, Northern Ireland.
The TUDA cohort was genotyped for FUT2 rs601338 (p.Trp154Ter) by LGC Genomics using KASP genotyping chemistry with a success rate of 96% (4941/5142). Duplicate samples (2%) were also typed with a concordance of >99%. Serum vitamin B12 and serum holoTC were measured in the TSS cohort as described above, with holoHC calculated as their difference.
The non-secretor genotype (homozygous FUT2 rs601338 p.154Ter) was tested for association with log-transformed unadjusted serum vitamin B12, holoTC and holoHC values in the young adult and older adult Irish cohorts. These association tests were performed with PLINK v.2 by simple linear regression using a recessive genetic model.
Cell lines
HepG2 cells were grown in DMEM media supplemented with 10% FBS.
Autoradiographic analysis
Twenty-nine human saliva samples [kind gift of Drs. Gonzalez and Sanz (58)] were analysed for glycosylation states of HC. These anonymized saliva samples were incubated with 57Co-B12 tracer stock (MP Biomedicals) for 1 h at 37 °C, followed by addition of native polyacrylamide gel electrophoresis (PAGE) sample buffer (Life Technologies). Native PAGE was performed using Novex 4–20% Tris-glycine mini gels with Tris-glycine running buffer (Life Technologies). Autoradiographic films were exposed to the dried gels for approximately 6 days before development.
Western blot analyses
Native PAGE was performed as above to detect salivary HC. Denaturing PAGE was used to detect ASGR1 in HepG2 extracts. Briefly, the samples were mixed with NuPAGE LDS sample buffer and reducing agent (Life Technologies), boiled and run on a Nupage 4–12% Bis-Tris gradient gel using NuPAGE MOPS SDS Running buffer (Life Technologies). Gels were transferred onto a nitrocellulose membrane (0.45 μM pore size) using iBlot (Life Technologies). Membranes were incubated in blocking solution at room temperature, then incubated at room temperature for 1–2 h or overnight at 4 °C with a primary antibody against ASGR1 [anti-ASGR1 rabbit polyclonal antibody (11739–1-AP), Proteintech group, 1: 1000]. Immunoblotting was performed using a WesternBreeze Chemiluminescent Kit (anti-mouse or anti-rabbit, Invitrogen) per the manufacturer‘s instructions. To detect beta-tubulin, membranes were incubated in 5% milk in TBS buffer with 0.1% Tween (TBS-T). After washing, the membranes were incubated with primary antibody against beta-tubulin [anti-beta tubulin (ab21058), Abcam, 1: 1000] in a TBS-T/0.5% milk solution. After the final washes the membranes were incubated with ECL Prime Western Blotting Detection Reagent (GE Healthcare Amersham) per the manufacturer‘s instructions.
DNA isolation and FUT2 rs601338 genotyping of saliva samples
DNA from saliva was isolated as previously described (59) with minor modifications. In brief, 10 µl saliva sample and 50 µl Chelex100 reagent (Biorad) were incubated for 30 min at 56 °C, followed by 8 min of incubation at 100 °C. The preparation was spun down for 5 min at 10 000 g, and supernatant was used for PCR reactions as previously described (60) to sequence the whole coding exon (exon 2) of the FUT2 gene. The PCR products were subjected to Sanger sequencing and analysed with SeqMan Pro (DNASTAR, Inc.).
DNA constructs and generation of stable cell lines
pLKO.1 lentiviral vectors expressing shRNAs targeting ASGR1 (TRCN0000061480 and TRCN0000061482) were obtained from Thermo Scientific. Lentiviral particles were generated by transfecting a 293 T cell line with pLKO.1 construct (shRNA or empty vector), pVSV-G, and pCMV-HRdel8.2 using Lipofectamin reagent (Invitrogen). Virus-containing medium was filtered and stored at − 80 °C. Subsequently, HepG2 cells were infected in the presence of 8ug/µl polybrene (Sigma). Stable pools were selected with puromycin.
Vitamin B12 uptake assay
Human saliva was used as a source of HC. Saliva was incubated with 57Co-B12 tracer stock (MP Biomedicals) and PBS buffer for 1 h at room temperature. The unbound 57Co-B12 was filtered through Microcon column-30 (Millipore). The HC/57Co-B12 complex was added to the serum free media and incubated with hepatocellular carcinoma cell line HepG2 for 3 h at 37 °C. Following the removal of the medium and washing, cells were lysed in RIPA buffer [10 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and protease inhibitor cocktail (Sigma)]. The radioactivity of the RIPA lysed fraction was used to measure the cellular uptake of HC/57Co-B12 complex. The percentage of uptake was defined as the total number of counts found in cells compared with the total number counts placed in the incubation media.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors acknowledge the contributions made by the study participants in the Trinity Student Study and the replication cohort. We thank Dr. Yoonhee Kim for her contributions to quality control and preparation of the genotyping data for GWAS. We thank Dr. Per Ueland for his critical reading and constructive comments on the manuscript. We are grateful to Drs. Mireya Gonzalez-Begné and Ignacio Sanz of the University of Rochester for providing a subset of the saliva samples used in this work. The authors thank Axis-Shield Diagnostics Ltd. for providing the reagents used to measure holo-transcobalamin. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov; date last accessed October 3, 2017). The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about; date last accessed October 3, 2017.
Conflict of Interest statement. None declared.
Funding
Intramural Research Programs of the National Human Genome Research Institute, the Eunice Shriver National Institute of Child Health and Development, the Health Research Board (Dublin, Ireland), the Irish Department of Agriculture, Food & the Marine and Health Research Board: Food Institutional Research Measure (FIRM) initiative, and the Cross-Border Research and Development Programme: ‘Strengthening All- Island Research.’
References
- 1. Botto L.D., Moore C.A., Khoury M.J., Erickson J.D. (1999) Neural-tube defects. N. Engl. J. Med., 341, 1509–1519. [DOI] [PubMed] [Google Scholar]
- 2. Molloy A.M., Kirke P.N., Troendle J.F., Burke H., Sutton M., Brody L.C., Scott J.M., Mills J.L. (2009) Maternal vitamin B12 status and risk of neural tube defects in a population with high neural tube defect prevalence and no folic Acid fortification. Pediatrics, 123, 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ray J.G., Wyatt P.R., Thompson M.D., Vermeulen M.J., Meier C., Wong P.Y., Farrell S.A., Cole D.E. (2007) Vitamin B12 and the risk of neural tube defects in a folic-acid-fortified population. Epidemiology, 18, 362–366. [DOI] [PubMed] [Google Scholar]
- 4. Verkleij-Hagoort A.C., de Vries J.H., Ursem N.T., de Jonge R., Hop W.C., Steegers-Theunissen R.P. (2006) Dietary intake of B-vitamins in mothers born a child with a congenital heart defect. Eur. J. Nutr., 45, 478–486. [DOI] [PubMed] [Google Scholar]
- 5. Spence J.D., Stampfer M.J. (2011) Understanding the complexity of homocysteine lowering with vitamins: the potential role of subgroup analyses. JAMA, 306, 2610–2611. [DOI] [PubMed] [Google Scholar]
- 6. Dahlin A.M., Van Guelpen B., Hultdin J., Johansson I., Hallmans G., Palmqvist R. (2008) Plasma vitamin B12 concentrations and the risk of colorectal cancer: a nested case-referent study. Int. J. Cancer, 122, 2057–2061. [DOI] [PubMed] [Google Scholar]
- 7. Liu A.Y., Scherer D., Poole E., Potter J.D., Curtin K., Makar K., Slattery M.L., Caan B.J., Ulrich C.M. (2013) Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol. Nutr. Food Res., 57, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andres E., Loukili N.H., Noel E., Kaltenbach G., Abdelgheni M.B., Perrin A.E., Noblet-Dick M., Maloisel F., Schlienger J.L., Blickle J.F. (2004) Vitamin B12 (cobalamin) deficiency in elderly patients. CMAJ, 171, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. (2008) Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet., 40, 1160–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin X., Lu D., Gao Y., Tao S., Yang X., Feng J., Tan A., Zhang H., Hu Y., Qin X. (2012) Genome-wide association study identifies novel loci associated with serum level of vitamin B12 in Chinese men. Hum. Mol. Genet., 21, 2610–2617. [DOI] [PubMed] [Google Scholar]
- 11. Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M.G., Usala G., Lai S., Mulas A., Corsi A.M., Vestrini A.. et al. (2009) Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet., 84, 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grarup N., Sulem P., Sandholt C.H., Thorleifsson G., Ahluwalia T.S., Steinthorsdottir V., Bjarnason H., Gudbjartsson D.F., Magnusson O.T., Sparso T.. et al. (2013) Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. PLoS Genet., 9, e1003530.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hazra A., Kraft P., Lazarus R., Chen C., Chanock S.J., Jacques P., Selhub J., Hunter D.J. (2009) Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum. Mol. Genet., 18, 4677–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keene K.L., Chen W.M., Chen F., Williams S.R., Elkhatib S.D., Hsu F.C., Mychaleckyj J.C., Doheny K.F., Pugh E.W., Ling H.. et al. (2014) Genetic associations with plasma B12, B6, and folate levels in an ischemic stroke population from the vitamin intervention for stroke prevention (VISP) trial. Front. Public Health, 2, 112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelly R.J., Rouquier S., Giorgi D., Lennon G.G., Lowe J.B. (1995) Sequence and expression of a candidate for the human Secretor blood group alpha(1, 2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem., 270, 4640–4649. [DOI] [PubMed] [Google Scholar]
- 16. Ferrer-Admetlla A., Sikora M., Laayouni H., Esteve A., Roubinet F., Blancher A., Calafell F., Bertranpetit J., Casals F. (2009) A natural history of FUT2 polymorphism in humans. Mol. Biol. Evol., 26, 1993–2003. [DOI] [PubMed] [Google Scholar]
- 17. Koda Y., Tachida H., Pang H., Liu Y., Soejima M., Ghaderi A.A., Takenaka O., Kimura H. (2001) Contrasting patterns of polymorphisms at the ABO-secretor gene (FUT2) and plasma alpha(1, 3)fucosyltransferase gene (FUT6) in human populations. Genetics, 158, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaptan K., Beyan C., Ural A.U., Çetin T., Avcu F., Gülşen M., Finci R., Yalçín A. (2000) Helicobacter pylori–is it a novel causative agent in Vitamin B12 deficiency? Arch. Intern. Med., 160, 1349–1353. [DOI] [PubMed] [Google Scholar]
- 19. Carmel R., Perez-Perez G.I., Blaser M.J. (1994) Helicobacter pylori infection and food-cobalamin malabsorption. Dig. Dis. Sci., 39, 309–314. [DOI] [PubMed] [Google Scholar]
- 20. Ikehara Y., Nishihara S., Yasutomi H., Kitamura T., Matsuo K., Shimizu N., Inada K., Kodera Y., Yamamura Y., Narimatsu H. (2001) Polymorphisms of two fucosyltransferase genes (Lewis and Secretor genes) involving type I Lewis antigens are associated with the presence of anti-Helicobacter pylori IgG antibody. Cancer Epidemiol. Biomarkers Prev., 10, 971–977. [PubMed] [Google Scholar]
- 21. Magalhaes A., Rossez Y., Robbe-Masselot C., Maes E., Gomes J., Shevtsova A., Bugaytsova J., Boren T., Reis C.A. (2016) Muc5ac gastric mucin glycosylation is shaped by FUT2 activity and functionally impacts Helicobacter pylori binding. Sci. Rep., 6, 25575.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oussalah A., Besseau C., Chery C., Jeannesson E., Gueant-Rodriguez R.M., Anello G., Bosco P., Elia M., Romano A., Bronowicki J.P.. et al. (2012) Helicobacter pylori serologic status has no influence on the association between fucosyltransferase 2 polymorphism (FUT2 461 G->A) and vitamin B-12 in Europe and West Africa. Am. J. Clin. Nutr., 95, 514–521. [DOI] [PubMed] [Google Scholar]
- 23. England J.M., Down M.C., Wise I.J., Linnell J.C. (1976) The transport of endogenous vitamin B12 in normal human serum. Clin. Sci. Mol. Med., 51, 47–52. [DOI] [PubMed] [Google Scholar]
- 24. Hall C.A. (1977) The carriers of native vitamin B12 in normal human serum. Clin. Sci. Mol. Med., 53, 453–457. [DOI] [PubMed] [Google Scholar]
- 25. Hall C.A. (1975) Transcobalamins I and II as natural transport proteins of vitamin B12. J. Clin. Invest., 56, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burger R.L., Schneider R.J., Mehlman C.S., Allen R.H. (1975) Human plasma R-type vitamin B12-binding proteins. II. The role of transcobalamin I, transcobalamin III, and the normal granulocyte vitamin B12-binding protein in the plasma transport of vitamin B12. J. Biol. Chem., 250, 7707–7713. [PubMed] [Google Scholar]
- 27. Carmel R. (2003) Mild transcobalamin I (haptocorrin) deficiency and low serum cobalamin concentrations. Clin. Chem., 49, 1367–1374. [DOI] [PubMed] [Google Scholar]
- 28. Greibe E., Fedosov S., Nexo E., Gasset M. (2012) The cobalamin-binding protein in zebrafish is an intermediate between the three cobalamin-binding proteins in human. PLoS One, 7, e35660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nongmaithem S.S., Joglekar C.V., Krishnaveni G.V., Sahariah S.A., Ahmad M., Ramachandran S., Gandhi M., Chopra H., Pandit A., Potdar R.D. (2017) GWAS identifies population specific new regulatory variants in FUT6 associated with plasma B12 concentrations in Indians. Hum Mol Genet, 26, 2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The 1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pangilinan F., Mitchell A., VanderMeer J., Molloy A.M., Troendle J., Conley M., Kirke P.N., Sutton M., Sequeira J.M., Quadros E.V.. et al. (2010) Transcobalamin II receptor polymorphisms are associated with increased risk for neural tube defects. J. Med. Genet., 47, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quadros E.V., Lai S.C., Nakayama Y., Sequeira J.M., Hannibal L., Wang S., Jacobsen D.W., Fedosov S., Wright E., Gallagher R.C.. et al. (2010) Positive newborn screen for methylmalonic aciduria identifies the first mutation in TCblR/CD320, the gene for cellular uptake of transcobalamin-bound vitamin B(12). Hum. Mutat., 31, 924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stone N., Pangilinan F., Molloy A.M., Shane B., Scott J.M., Ueland P.M., Mills J.L., Kirke P.N., Sethupathy P., Brody L.C., Oresic M. (2011) Bioinformatic and genetic association analysis of microRNA target sites in one-carbon metabolism genes. PLoS One, 6, e21851.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang S.Y., Coleman P.S., Dupont B. (1982) The biochemical and genetic basis for the microheterogeneity of human R-type vitamin B12 binding proteins. Blood, 59, 747–755. [PubMed] [Google Scholar]
- 35. Birn H. (2006) The kidney in vitamin B12 and folate homeostasis: characterization of receptors for tubular uptake of vitamins and carrier proteins. Am. J. Physiol. Renal. Physiol., 291, F22–F36. [DOI] [PubMed] [Google Scholar]
- 36. Ashwell G., Morell A. (1974) The dual role of sialic acid in the hepatic recognition and catabolism of serum glycoproteins. Biochem. Soc. Symp., 40, 117–124. [PubMed] [Google Scholar]
- 37. Burger R.L., Mehlman C.S., Allen R.H. (1975) Human plasma R-type vitamin B12-binding proteins. I. Isolation and characterization of transcobalamin I. TRANSCOBALAMIN III. and the normal granulocyte vitamin B12-binding protein. J. Biol. Chem., 250, 7700–7706. [PubMed] [Google Scholar]
- 38. Carmel R., Sinow R.M., Karnaze D.S. (1987) Atypical cobalamin deficiency. Subtle biochemical evidence of deficiency is commonly demonstrable in patients without megaloblastic anemia and is often associated with protein-bound cobalamin malabsorption. J. Lab. Clin. Med., 109, 454–463. [PubMed] [Google Scholar]
- 39. Chery C., Hehn A., Mrabet N., Oussalah A., Jeannesson E., Besseau C., Alberto J.-M., Gross I., Josse T., Gérard P.. et al. (2013) Gastric intrinsic factor deficiency with combined GIF heterozygous mutations and FUT2 secretor variant. Biochimie, 95, 995–1001. [DOI] [PubMed] [Google Scholar]
- 40. Collin S.M., Metcalfe C., Palmer T.M., Refsum H., Lewis S.J., Smith G.D., Cox A., Davis M., Marsden G., Johnston C.. et al. (2011) The causal roles of vitamin B(12) and transcobalamin in prostate cancer: can Mendelian randomization analysis provide definitive answers? Int. J. Mol. Epidemiol. Genet., 2, 316–327. [PMC free article] [PubMed] [Google Scholar]
- 41. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60, 706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yetley E.A., Pfeiffer C.M., Phinney K.W., Bailey R.L., Blackmore S., Bock J.L., Brody L.C., Carmel R., Curtin L.R., Durazo-Arvizu R.A.. et al. (2011) Biomarkers of vitamin B-12 status in NHANES: a roundtable summary. Am. J. Clin. Nutr., 94, 313S–321S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herrmann W., Obeid R., Schorr H., Geisel J. (2005) The usefulness of holotranscobalamin in predicting vitamin B12 status in different clinical settings. Curr. Drug Metab., 6, 47–53. [DOI] [PubMed] [Google Scholar]
- 44. Bjorke Monsen A.L., Ueland P.M. (2003) Homocysteine and methylmalonic acid in diagnosis and risk assessment from infancy to adolescence. Am. J. Clin. Nutr., 78, 7–21. [DOI] [PubMed] [Google Scholar]
- 45. Klee G.G. (2000) Cobalamin and folate evaluation: measurement of methylmalonic acid and homocysteine vs vitamin B(12) and folate. Clin. Chem., 46, 1277–1283. [PubMed] [Google Scholar]
- 46. Molloy A.M., Pangilinan F., Mills J. L., Shane B., O’Neill M.B., McGaughey D. M., Velkova A., Abaan H. O., Ueland P. M., McNulty H.. et al. (2016) A Common Polymorphism in HIBCH Influences Methylmalonic Acid Concentrations in Blood Independently of Cobalamin. Am. J. Hum. Genet., 98, 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vogiatzoglou A., Oulhaj A., Smith A.D., Nurk E., Drevon C.A., Ueland P.M., Vollset S.E., Tell G.S., Refsum H. (2009) Determinants of plasma methylmalonic acid in a large population: implications for assessment of vitamin B12 status. Clin. Chem., 55, 2198–2206. [DOI] [PubMed] [Google Scholar]
- 48. Lloyd-Wright Z., Hvas A.M., Moller J., Sanders T.A., Nexo E. (2003) Holotranscobalamin as an indicator of dietary vitamin B12 deficiency. Clin. Chem., 49, 2076–2078. [DOI] [PubMed] [Google Scholar]
- 49. Kelleher B.P., Walshe K.G., Scott J.M., O'Broin S.D. (1987) Microbiological assay for vitamin B12 with use of a colistin-sulfate-resistant organism. Clin. Chem., 33, 52–54. [PubMed] [Google Scholar]
- 50. Brady J., Wilson L., McGregor L., Valente E., Orning L. (2008) Active B12: a rapid, automated assay for holotranscobalamin on the Abbott AxSYM analyzer. Clin. Chem., 54, 567–573. [DOI] [PubMed] [Google Scholar]
- 51. Lonati S., Novembrino C., Ippolito S., Accinni R., Galli C., Troonen H., Campolo J., Della Noce C., Lunghi G., Catena F.B. (2004) Analytical performance and method comparison study of the total homocysteine fluorescence polarization immunoassay (FPIA) on the AxSYM analyzer. Clin. Chem. Lab. Med., 42, 228–234. [DOI] [PubMed] [Google Scholar]
- 52. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patterson N., Price A.L., Reich D. (2006) Population structure and eigenanalysis. PLoS Genet., 2, e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 55. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. International HapMap, C. (2003) The International HapMap Project. Nature, 426 789–796. [DOI] [PubMed] [Google Scholar]
- 57. Laird E., McNulty H., Ward M., Hoey L., McSorley E., Wallace J.M., Carson E., Molloy A.M., Healy M., Casey M.C.. et al. (2014) Vitamin D deficiency is associated with inflammation in older Irish adults. J. Clin. Endocrinol. Metab., 99, 1807–1815. [DOI] [PubMed] [Google Scholar]
- 58. Ching K.H., Burbelo P.D., Gonzalez-Begne M., Roberts M.E., Coca A., Sanz I., Iadarola M.J. (2011) Salivary anti-Ro60 and anti-Ro52 antibody profiles to diagnose Sjogren's Syndrome. J. Dent. Res., 90, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matto J., Saarela M., Alaluusua S., Oja V., Jousimies-Somer H., Asikainen S. (1998) Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J. Clin. Microbiol., 36, 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yip S.P., Lai S.K., Wong M.L. (2007) Systematic sequence analysis of the human fucosyltransferase 2 (FUT2) gene identifies novel sequence variations and alleles. Transfusion, 47, 1369–1380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.