Abstract
FRMPD4 (FERM and PDZ Domain Containing 4) is a neural scaffolding protein that interacts with PSD-95 to positively regulate dendritic spine morphogenesis, and with mGluR1/5 and Homer to regulate mGluR1/5 signaling. We report the genetic and functional characterization of 4 FRMPD4 deleterious mutations that cause a new X-linked intellectual disability (ID) syndrome. These mutations were found to be associated with ID in ten affected male patients from four unrelated families, following an apparent X-linked mode of inheritance. Mutations include deletion of an entire coding exon, a nonsense mutation, a frame-shift mutation resulting in premature termination of translation, and a missense mutation involving a highly conserved amino acid residue neighboring FRMPD4-FERM domain. Clinical features of these patients consisted of moderate to severe ID, language delay and seizures alongside with behavioral and/or psychiatric disturbances. In-depth functional studies showed that a frame-shift mutation, FRMPD4p.Cys618ValfsX8, results in a disruption of FRMPD4 binding with PSD-95 and HOMER1, and a failure to increase spine density in transfected hippocampal neurons. Behavioral studies of frmpd4-KO mice identified hippocampus-dependent spatial learning and memory deficits in Morris Water Maze test. These findings point to an important role of FRMPD4 in normal cognitive development and function in humans and mice, and support the hypothesis that FRMPD4 mutations cause ID by disrupting dendritic spine morphogenesis in glutamatergic neurons.
Introduction
Intellectual disability (ID) affects 1–3% of the general population and is genetically heterogeneous (1,2) X-linked ID (XLID) conditions, resulting from mutations in genes on the X-chromosome, affect approximately 1.7 per 1, 000 males (3–5). More than 120 such conditions have been described thus far, although only a part of these has been confirmed by appropriate replication and functional studies (6). Understanding genetic causes and mechanisms of ID is likely to provide valuable insights into cognitive development and function in humans and suggest drug targets for a rational development of effective therapies (7,8). Identification of novel ID genes is complicated by extreme clinical and genetic heterogeneity (7). With the characterization of those relatively common disease-causing genes to ID, it is anticipated that the majority of the remaining ID genes are rare and/or even private in a single family. The pace of new discoveries in the field has been accelerated through whole exome sequencing (WES) and whole genome sequencing (WGS) (8,9).
FRMPD4 is a neural scaffolding protein with abundant expression in the brain cortex and hippocampus (10,11). FRMPD4 consists of the N-terminal WW, PDZ, and FERM domains and the C-terminal PDZ-binding domains that interact with actin filaments, β-PIX, and PSD-95 (10,11). Studies have shown that the interaction of the C-terminal PDZ domain with PSD-95 is crucial for the formation of the excitatory synapses and dendritic spines (10). Over-expression of FRMPD4 in hippocampal neurons was shown to increase dendritic spine density while decreased expression using gene knockdown methods was shown to reduce spine density of excitatory neurons (10). Additional studies found that FRMPD4 creates a micro-domain that assembles mGluR1/5, Homer1, and proline-directed kinases (PDK) at the postsynaptic density of the excitatory synapses (11). Direct FRMPD4 binding to Homer is necessary for the formation of a FRMPD4-Homer1-PDKs-mGluR1/5 complex (11). PDK-mediated site-specific phosphorylation of mGluR1/5 enhances Homer1 interaction with mGluR1/5 and down-regulates mGluR5 signaling. Consistent with this hypothesis, frmpd4-KO mice exhibit enhanced mGluR5 signaling in the hippocampus (11).
In this study, we describe systematic genetic and functional characterization of deleterious FRMPD4 mutations in 4 unrelated XLID families, and behavioral analysis of a line of frmpd4-KO mice on learning and memory function. Our studies indicate that these disease-causing FRMPD4 mutations result in a severe defect in dendritic spinogenesis in excitatory neurons and frmpd4-KO mice show deficits in hippocampus-dependent spatial learning and memory. Our results implicate a mechanism in which deleterious FRMPD4 mutations cause ID through a defect of dendritic spinogenesis in glutamatergic neurons resulting in a disruption of the excitatory synaptic transmission in the brain.
Results
Clinical and genetic characterization of a FRMPD4 mutation in a large XLID family
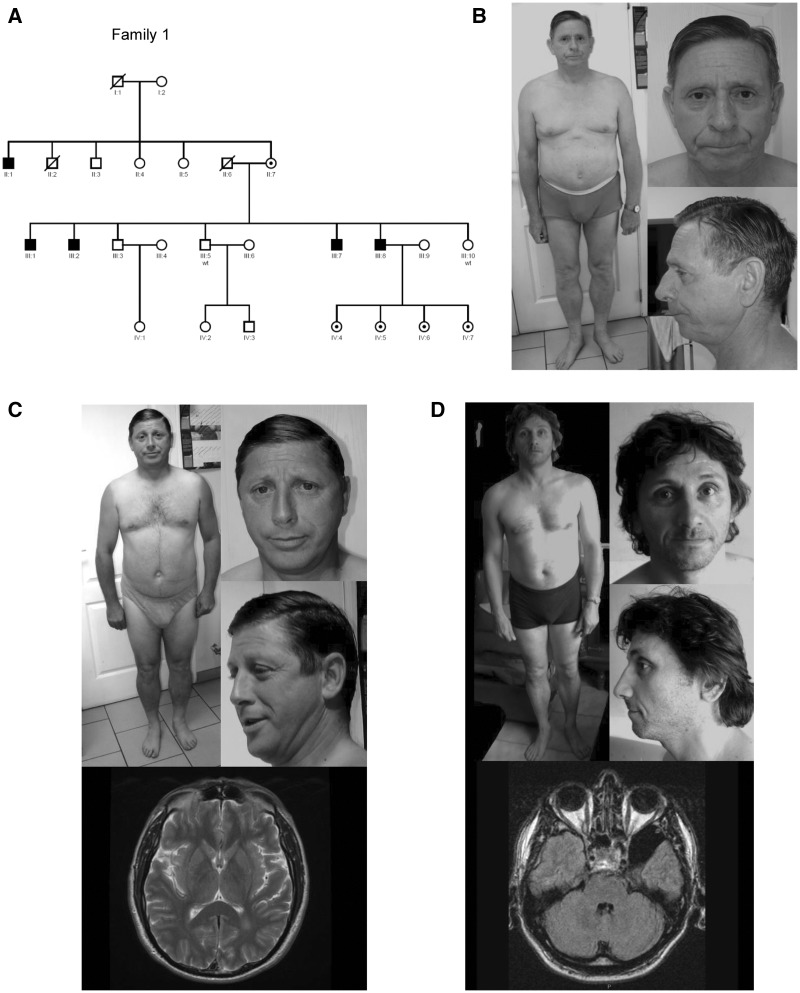
A recent exome-sequencing study of 405 XLID families identified a single base deletion of a cytosine at ChrX: 12734425 [hg19] in FRMPD4 in five male individuals of a large Caucasian family (Fig. 1) and a FRMPD4p.Cys553Arg missense mutation in a singleton (12). Resulting FRMPD4p.Cys618ValfsX8 mutation is predicted to cause a frame-shift and premature termination of translation 8 amino acids downstream (Fig. 2A; 3A). In this family, FRMPD4p.Cys618ValfsX8 was found to segregate with mild to severe ID in four affected male siblings and one affected maternal uncle while their mother is unaffected (Fig. 1A). These five patients (P1, P2, P3, P4 and P5) presented with developmental delay, mainly affecting their speech and language skills (Table 1). All patients were slow learners and required special education. Four of them were oriented towards supervised employment. They remain dependent on their family members for daily living. Two siblings had tremor (P1, P2). Psychiatric disturbances consisting of aggressiveness were noted in P3 and P5. Generalized epilepsy of adult-onset was noted in one patient (P5). General growth parameters including occipitofrontal circumference (OFC) were normal. No distinct dysmorphic craniofacial profile was consistently appreciated in all five patients (Fig. 1B, C, and D). Brain MRI studies were completed in two patients indicating nonspecific anomalies: a white matter hypersignal in the left occipital horn in P2 and a left temporal arachnoid cyst (axial T1) in P5 (Fig. 1C and D;Supplementary Material, Table S1).
Figure 1.
Pedigree, photographs, and brain MRIs of Affected Males with ID and FRMPD4p.Cys618ValfsX8 in an XLID Family. (A) Pedigree shows 5 affected males from a multigenerational family. Filled square: affected males; circle with a center dot, carrier females; unfilled square: unaffected males; unfilled circle: unaffected females. Symbols with a line crossed: deceased. (B) Note absence of distinct dysmorphic features except for bifid tip of the nose in P1 (II: 1). (C) Note absence of distinct dysmorphic features and a white matter hyper-signal in the left occipital horn on brain MRI (axial T2) in P2 (III: 1). (D) Note absence of distinct dysmorphic features and a left temporal arachnoid cyst (axial T1) in P5 (III: 8).
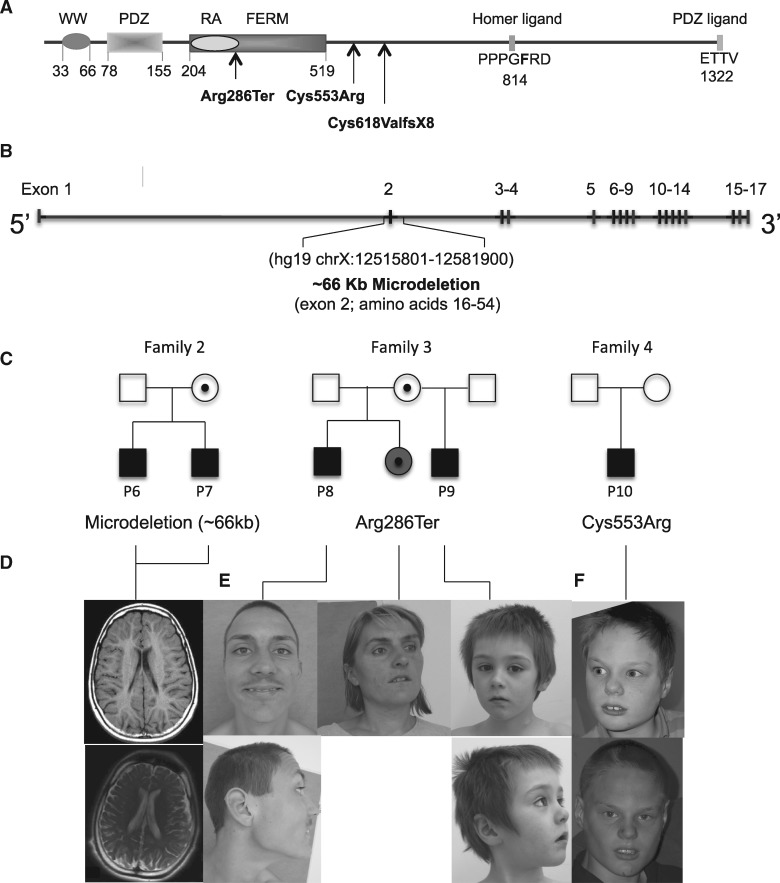
Figure 2.
Additional FRMPD4 Mutations in Affected Individuals with XLID. (A) Established domain structure of FRMPD4 and nature and locations of FRMPD4 mutations found in patients with ID. WW (domain with two conserved Trp residues): PDZ (PSD-95/Dlg/ZO-1): RA; FERM (four-point-one, ezrin, radixin, and moesin). (B) Genomic structure of FRMPD4 and genomic location of family 3 microdeletion. (C) Pedigrees and segregation analysis of FRMPD4 mutations identified in families 2, 3 and 4. Filled square, affected males; open square, unaffected males; circle with a center dot, unaffected carrier female; filled circle with a center dot, mildly disabled carrier female. (D) Brain MRI scans of P6 (at 12 years) and P7 (at 17 years) from Family 2. Note diffuse atrophy of white matter principally in the periventricular region, delayed myelination with periventricular white matter atrophy of P6 (top panel, axial T1), and compare to cortical atrophy and enlarged ventricles of P7 (bottom panel, axial T2). (E) Photographs of family 3 probands and their mildly disabled sister. (F) Proband of family 4 at different ages. Note frontal upsweep, trigonocephaly and broad nasal bridge.
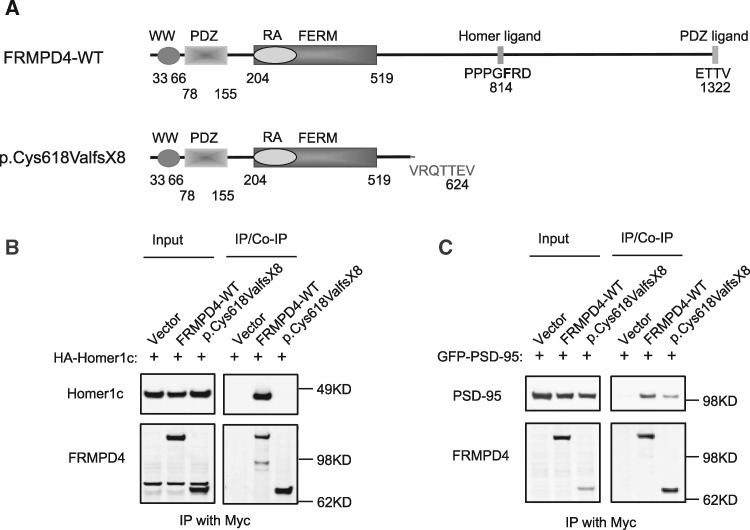
Figure 3.
Binding of FRMPD4p.Cys618ValfsX8 with known interacting proteins. (A) Domain structure of human FRMPD4 and its mutant protein. (B) Various FRMPD4 and Homer1c constructs were transfected into HEK293T cells. Anti-myc antibody immunoprecipitated FRMPD4-WT and FRMPD4p.Cys618ValfsX8. Homer1C was co-immunoprecipitated by FRMPD4-WT, but not FRMPD4p.Cys618ValfsX8. (C) Various FRMPD4 and PSD-95 constructs were transfected into HEK293T cells. Anti-myc antibody immunoprecipitated FRMPD4-WT and FRMPD4p.Cys618ValfsX8. PSD-95 was co-immunoprecipitated by FRMPD4-WT while this binding was significantly reduced between FRMPD4p.Cys618ValfsX8 and PSD-95.
Table 1.
Shared clinical features of patients with FRMPD4 mutations
| Family 1 |
Family 2 |
Family 3 |
Family 4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients | P1 (II-1) |
P2 (III-1) | P3 (III-2) | P4 (III-7) | P5 (III-8) | P6 |
P7 | P8 |
P9 | P10 |
| Mutation | p.Cys618ValfsX8 | Deletion of exon 2 | p.Arg286Ter | p.Cys553Arg | ||||||
| Mutation type | Frame-shift | Frame-shift | Frame-shift | Frame-shift | Frame-shift | Exon Deletion | Exon Deletion | Nonsense | Nonsense | Missense |
| Gender | Male | Male | Male | Male | Male | Male | Male | Male | Male | Male |
| Age at Examination (year) | 66 | 48 | NA | NA | 42 | 17 | 17 | 18 | 4 | 11 |
| Height | −0.1SD | −1.5SD | NA | NA | −0.9SD | NA | NA | +0.5SD | NA | +0.5SD |
| OFC | +0.2SD | −2SD | NA | NA | +0.7SD | −1.75SD | −0.07SD | −0.3SD | +0.1SD | −0.3SD |
| Delayed Gross Motor Development | No | No | No | NA | Yes | Yes | Yes | Yes | Yes | Yes |
| Delayed Speech and Language | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Intellectual Disability | Moderate | Moderate to severe | Moderate to severe | Mild to moderate | Mild to moderate | Severe to profound | Severe | Moderate | NA | Severe |
| Behavioural Disturbances | No | No | Aggressiveness | NA | Hyperactivity, Aggressiveness | Autism | Autism | Hyperactivity, Aggressiveness | Hyperactivity, Autism | Autism |
| Neurological Examination | Tremor | Tremor; brisk deep tendon reflexes | NA | NA | NA | Hypotonia; spasticity, brisk deep tendon reflexes; ataxia | Hypotonia; spasticity, brisk deep tendon reflexes; ataxia | NA | NA | NA |
| Epilepsy | No | No | No | No | Yes | Yes | Yes | No | No | No |
| Strabismus | No | No | NA | NA | No | Yes | Yes | No | NA | Yes |
FRMPD4 mutations were found in five affected males from three additional XLID families
To further establish genetic evidence that deleterious FRMPD4 mutations cause ID in humans, we identified 3 additional mutations in five affected males belonging to three unrelated families through an international collaboration (Fig. 2).
Family 2
Two affected boys (P6 and P7) with severe ID were found to have a ∼66 Kb microdeletion (chrX: 12515801–12581900 [hg19]) that removes the entire exon 2 of FRMPD4. Their unaffected mother is a heterozygote deletion carrier. This mutation is predicted to cause a FRMPD4 in-frame 40 amino acids deletion [14–54] that includes the entire WW domain at the N-terminus (Fig. 2B and C).
Family 3
X-chromosome exome sequencing was performed on probands from a cohort of XLID families (n = 50). In one family, two maternal male half-brothers with severe ID were found to harbor a nonsense mutation, FRMPD4c.856C>T resulting in FRMPD4p.Arg286*. This mutation is predicted to disrupt FRMPD4 FERM domain with subsequent truncation of the protein or reduced transcript levels caused by nonsense-mediated RNA decay (Fig. 2A and C). The unaffected mother carries the mutation as well as the mildly disabled sister of the probands (Fig. 2C).
Family 4
X-chromosome exome sequencing identified a de novo missense mutation, c.1657T > C, p.Cys553Arg in a single male (P10) with severe ID (12) (Fig. 2A and C). This single base substitution involves a highly conserved amino acid residue at 3’ of the FRMPD4-FERM domain and is predicted to be deleterious to the protein function (12).
Thus, our studies identified three additional ID-associated FRMPD4 mutations which are also located in the N-terminal half of the protein like in the princeps family. All inherited mutations segregate with ID phenotype and are predicted deleterious.
Clinical phenotype of the patients
All affected males carrying hemizygous FRMPD4 mutations presented ID ranging from mild to severe (Table 1) and normal growth parameters including height (5/5) and OFC (6/6). Language disorders were consistent findings in our patients: all of them had delayed or absent speech. The mean age at independent walking was slightly delayed (18.5 months; min: 12 months; max: 30 months). Behavioral or psychiatric disturbances were described in seven patients, consisting of hyperactivity and/or aggressive behavior (4/9) or an autism spectrum disorder (4/9). Epilepsy of the grand-mal type or early-onset focal seizures were observed in 3 of 10 patients, while neurological examination was altered in some: tremor (2/6), spasticity with increased deep tendon reflexes (3/6) and ataxia (2/6). Strabismus was also reported (3/7). Nonspecific brain anomalies were observed by imaging studies in 4 of 5 patients (Supplementary Material, Table S1). Mild and variable dysmorphic features were noted (6/8). However, no distinct craniofacial gestalt was consistently observed in all patients (Figs 1B, C, and D, 2E and F, Supplementary Material, Table S1).
FRMPD4p.Cys618ValfsX8 shows reduced interaction with PSD-95 and Homer1
The FRMPD4p.Cys618ValfsX8 mutation is caused by a single base deletion of a cytosine at position 12734425 of X-Chromosome. This deletion is predicted to cause a frame-shift resulting in a truncated protein of 624 amino acids instead of 1322 (Fig. 3A), which retains the N-terminal domains including WW, PDZ, and FERM but lacks the C-terminal Homer binding site and the PDZ ligand. Previous studies have shown that FRMPD4 binding to Homer via the Homer-binding motif (PPXXF) and PSD-95 via the C-terminal PDZ ligand are crucial to its regulatory function on mGluR1/5 signaling (11), and production and maintenance of spine density of excitatory neurons (10). To assess potential deleterious effects of this mutation, we investigated FRMPD4p.Cys618ValfsX8 interactions with Homer1 and PSD-95 by co-immunoprecipitation.
To assess functional effects of Cys618ValfsX8 on FRMPD4 interaction with Homer 1, we co-transfected the expression vectors of HA-Homer1c with either myc-FRMPD4 or myc-FRMPD4p.Cys618ValfsX8 in HEK293T cells and performed co-immunoprecipitation studies using a α-myc antibody. As shown in Figure 3B, Homer1C can be efficiently co-immunoprecipitated by FRMPD4-WT, but not by FRMPD4p.Cys618ValfsX8. This is consistent with a lack of Homer binding site in this mutant. To assess functional effects of FRMPD4p.Cys618ValfsX8 on FRMPD4 interaction with PSD-95, we co-transfected expression constructs of GFP-PSD-95 with myc-FRMPD4 or myc-FRMPD4p.Cys618ValfsX8 in HEK293T cells and performed co-immunoprecipitation using a α-myc antibody. As shown in Figure 3C, PSD-95 can be co-immunoprecipitated efficiently by FRMPD4-WT, but with a significantly reduced efficiency by FRMPD4p.Cys618ValfsX8. This is consistent with lack of the C-terminal PDZ ligand in the FRMPD4 mutant. Furthermore, co-expression of FRMPD4p.Cys618ValfsX8 appears to reduce binding of FRMPD4 with Homer1 (Supplementary Material, Fig. S1A) and with PSD95 (Supplementary Material, Fig. S1B) in transfected HEK293T cells.
FRMPD4p.Cys618ValfsX8 fails to increase dendritic spine density in neurons
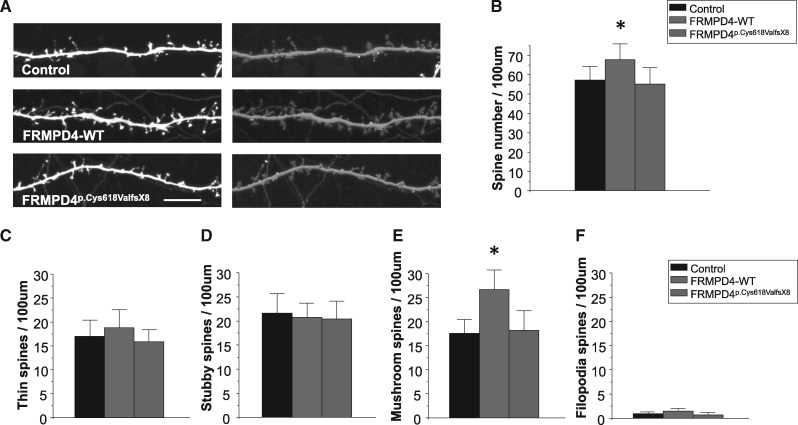
FRMPD4 overexpression in transfected neurons was shown to increase the density of dendritic spines (10). This action of FRMPD4 requires its WW, PDZ and FERM domains, and the C-terminal PDZ ligand (10). Correspondingly, decreased FRMPD4 expression in neurons by either knockdown or dominant-negative mutations have been shown to reduce dendritic spine density and excitatory synaptic transmission (10). To determine the effect of FRMPD4p.Cys618ValfsX8 on spine morphology and density, we transfected rat hippocampal neurons at DIV14 with myc-FRMPD4-WT or myc-FRMPD4p.Cys618ValfsX8 plus EGFP or EGFP alone and visualized dendritic spines by immunofluorescence staining of EGFP (Fig. 4A). We first compared spine density between the FRMPD4-WT and FRMPD4Cys618ValfsX8 transfected neurons. As shown in Figure 4B, we found that overexpression of FRMPD4-WT construct increased spine density while FRMPD4p.Cys618ValfsX8 failed to do so (Mean ± SEM, n = 19 neurons for EGFP only, 18 for WT, and 20 for Cys618ValfsX8, *P < 0.05, t-test). We next investigated changes in spine morphology between FRMPD4 WT and FRMPD4p.Cys618ValfsX8 transfected neurons. We classified dendritic spines into four categories based on their morphology and maturity: mushroom spines, thin spines, stubby spines, and filopodia. Spine shape is an indicator of the strength of synaptic input (13) with mushroom spines thought to represent long-lasting synaptic inputs associated with learning and memory. As shown in Figure 4C–F, we found that overexpression of FRMPD4-WT resulted in an increase in mushroom spines but not in thin spines, stubby spines, or filopodia. FRMPD4p.Cys618ValfsX8 did not increase mushroom or other spine categories. These results support the idea that FRMPD4p.Cys618ValfsX8 disrupts FRMPD4 function of regulating both spine density and morphology through a loss of function mechanism.
Figure 4.
FRMPD4p.Cys618ValfsX8 losses the ability to increase spine density. (A) Cultured hippocampal neurons were transfected with myc–FRMPD4 (WT or FRMPD4p.Cys618ValfsX8) plus EGFP or EGFP alone (control) (DIV 14) and visualized by immunofluorescence staining for EGFP. Images were analyzed by Neurolucida 360 to show different spine shapes. Scale bar, 10 μm. (B) FRMPD4-WT overexpression increased spine density but FRMPD4p.Cys618ValfsX8 did not. Mean ± 95% confidence interval (n = 19 neurons for EGFP, 18 for WT, and 20 for mutant, *P < 0.05, t-test). (C–F) Spines were classified into four categories. FRMPD4-WT overexpression increased mushroom spines but not thin spines, stubby spines and filopodia spines. FRMPD4p.Cys618ValfsX8 did not increase mushroom or other spine categories. Mean ± 95% confidence interval (n = 19 neurons for EGFP, 18 for WT, and 20 for mutant, *P < 0.05, t-test).
Frmpd4-KO mice show deficits in hippocampus-dependent spatial learning and memory
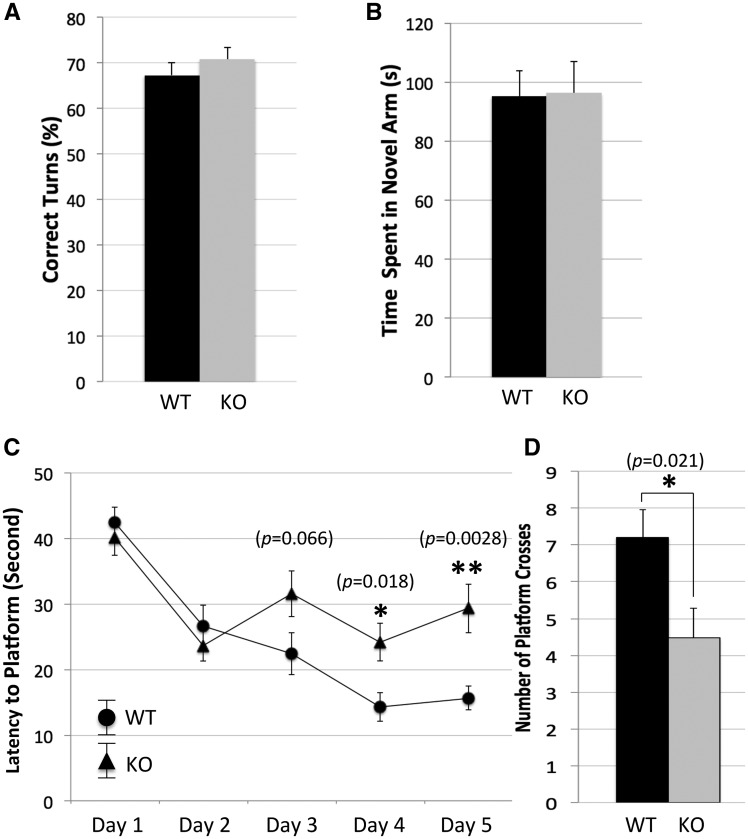
To implicate a role of FRMPD4 in normal cognitive function, we conducted a panel of rodent behavioral tests on a cohort of adult male frmpd4-KO mice and their WT male littermates with an emphasis on their learning and memory functions. Compared with WT, frmpd4-KO mice showed a moderate increase in the total ambulatory activities (total beam breaks in 30 min: Mean ± SEM: WT, 2, 254 ± 235; KO, 3, 025 ± 167; n = 12 for WT; n = 12 for KO; t-test, P < 0.05), normal total fine movements (total beam breaks in 30 min: Mean ± SEM: WT, 339 ± 47; KO, 352 ± 62; n = 12 for WT; n = 12 for KO; t-test, P > 0.05) and normal peripheral activities (total beam breaks in 30 min: Mean ± SEM: WT, 387 ± 14; KO, 387 ± 18; n = 12 for WT; n = 12 for KO; t-test, P > 0.05), in Open Field test. No significant difference was identified between WT and KO mice in Elevated Plus Maze (time in open arm in seconds: Mean ± SEM: WT, 62.5 ± 2.8; KO, 78.6 ± 3.7; n = 11 for WT; n = 12 for KO; t-test, P > 0.05) suggesting that FRMPD4-KOmice have normal anxiety levels. FRMPD4-KOmice showed no significant deficit in spatial working memory function in the Y-maze tests including spontaneous alternation (% of correct alternations, time in novel arm in seconds: Mean ± SEM: WT, 67.3 ± 2.8; KO, 70.8 ± 2.6; n = 15 for WT; n = 15 for KO; t-test, P > 0.05) (Fig. 5A) and blocked arm tests (time in novel arm in seconds: Mean ± SEM: WT, 95.2 ±8.5; KO, 96.2 ±10.9; n = 15 for WT; n = 15 for KO; t-test, P > 0.05) (Fig. 5B). Frmpd4-KO mice were found to have significant deficits in the Morris Water Maze indicating a hippocampus-dependent spatial reference memory deficit (Fig. 5C and D). As shown in Figure 5C, during the 5 days of training trials, the average time for mice to reach the platform decreased for both WT and KO groups. However, the time for the KO mice plateaus after day 2 whereas the time for the WT mice continues to decrease until day 4, when it also plateaus. The average time to reach the platform begins to diverge on day 3 (t-test, P = 0.066) and is significantly different on days 4 and 5 (P = 0.018 and P = 0.0028, respectively), with the KO mice taking significantly longer time to find the platform. For the final probe day, the number of times the mice crossed over the previous location of the platform was significantly lower in the KO mice (number of platform crosses: Mean ± SEM: WT, 7.2 ±0.75; KO, 4.5 ±0.82; n = 15 for WT; n = 15 for KO; t-test, P = 0.02) (Fig. 5D).
Figure 5.
Frmpd4-KO mice show normal spatial working memory but a defective spatial reference memory. The Y-maze tests for spatial working memory include Spontaneous Alternation and Blocked Arm tests. (A) Spontaneous Alternation: the test mouse was placed at the end of one arm and remained in the maze for 5 min. The total number of correct alternations divided by the number of total possible alternations was recorded and analyzed. (B) Blocked Arm test: one of the three arms was blocked, the test mouse was allowed to explore the 2 unblocked arms for 5 min followed by rest for 10 min. The test mouse was returned to the maze with all 3 arms open and allowed to explore for another 5 min. Data was analyzed for time spent in the arm blocked during the second trial (novel arm) and presented as mean ± SEM. t-test was used to compare data between frmpd4-KO mice (n = 15) and WT littermates (n = 15). *P < 0.05 was considered statistically significant. Note that no significant difference was identified between WT and frmpd4-KO in these tests suggesting a normal working memory function. Morris water maze is used to test hippocampus dependent spatial reference memory. (C) Training trials: mice were subjected to four, 1-min swim trials each day in the water maze to locate a hidden platform using multiple spatial signs. The time to reach the platform is an average of 4 trials for each mouse, n = 15 mice per group. Note that frmpd4-KO mice showed slow progression in reducing time needed to locate the platform compared with WT. (D) Probe trial: after the training trial, mice were subjected to one 3-min trial of free swimming in maze without the platform. Time spent probing the quadrant where the platform was located during training trial and total number of crosses to the platform region were automatically tracked and compared between frmpd4-KO and WT control mice. Note the reduced number of crosses to the platform site in the frmpd4-KO. MEAN ± SEM was presented. t-test was used to compare data at each test point between WT and frmpd4-KO mice. *p< 0.05 was considered statistically significant. Note frmpd4-KO mice (n = 15) showed significantly reduced number of crosses to the platform site compared with WT littermates (n = 15) suggesting a deficit in the spatial reference memory function.
Discussion
We report on the human phenotype associated with 4 FRMPD4 mutations identified in ten affected males belonging to four unrelated families, describe functional consequences of FRMPD4p.Cys618ValfsX8 mutation and provide evidence for alteration of hippocampus-dependent behavior in a KO mouse model. All pedigrees are consistent with X-linked transmission. FRMPD4 deficiency-related phenotype includes variable degree of ID and language disorders in patients presenting with normal growth and OFC parameters, nonspecific brain anomalies and mild dysmorphic features. Psychiatric disturbances, strabismus and seizures are present in some patients. All four mutations are rare and absent from public variant databases including dbSNP, the male-restricted portion of the 1000 Genomes, and the Exome Variant Server datasets (see web resources). Three FRMPD4 mutations including an exon deletion, a nonsense mutation, and a frame-shift mutation caused by a single base deletion, are predicted to result in FRMPD4 deleterious transcripts and/or protein products. By contrast, FRMPD4p.Cys553Arg involves a highly conserved amino acid residue near the FERM domain predicted to be deleterious by protein function alteration. Furthermore, behavioral analyses of FRMPD4-KOmice identified significant defects in the hippocampus-dependent spatial learning and memory in Morris Water maze test. Together, these results provide strong evidence that deleterious FRMPD4 mutations are causal for the phenotype observed in our families.
FRMPD4p.Cys618ValfsX8 is predicted to result in a truncated protein that potentially reduces FRMPD4 function due to a lack of HOMER-binding site and C-terminal PDZ domain or through a dominant negative mechanism. Our data suggest that presence of this truncated protein likely interferes with binding of FRMPD4 with Homer1 and PSD95, respectively, in transfected HEK293T cells. Alternatively, this mutation may result in a significant reduction of total FRMPD4 transcript and/or protein levels due to a nonsense mediated RNA decay mechanism. Since FRMPD4 does not express in lymphoblasts or fibroblast cell lines, testing neurons differentiated from iPSC cells from patients and/or studying brain tissues from mutant mice carrying FRMPD4p.Cys618ValfsX8 should help to test these hypotheses directly.
FRMPD4 C-terminal PDZ ligand was shown to bind the PDZ domain of PSD-95 (10). This interaction is needed in the formation of excitatory synapses and regulation of dendritic spine density (10). Our studies confirmed that lack of the C-terminal PDZ ligand causes reduced mutant FRMPD4 binding to PSD-95. Consistent with this finding, we show that FRMPD4p.Cys618ValfsX8 lacks the ability to increase spine density in transfected neurons as compared with FRMPD4-WT. FRMPD4p.Cys618ValfsX8 appears to negatively regulate FRMPD4 function on generation and/or maintenance of dendritic spine density at neuron levels. Dynamic regulation of number and morphology of dendritic spines is essential for maintenance of short- and long-term plasticity at the excitatory synapses on pyramidal neurons in cerebral cortex, and deficits have important implications for normal cognition and memory function (14–16). Abnormalities of dendritic spine density and/or morphology are associated with several intellectual disability syndromes including Fragile-X syndrome (17,18), Down syndrome (19), Rett syndrome (20), CDKL5-, PAK3- and KIAA2022-related encephalopathies (21, 22) and several neuropsychiatric disorders such as autism spectrum disorders, schizophrenia, and Alzheimer’s disease (23) (for review, see Martinez-Cerdeno, 2017) (24). We speculate that FRMPD4p.Cys618ValfsX8 and other FRMPD4 loss-of-function mutations involving WW, FERM, and PDZ domains contribute to cognitive impairment in our patients through a similar mechanism.
FRMPD4 binding to Homer1 is required for the formation of a FRMPD4-Homer1-PDKs-mGluR1/5 complex, which is regulated by ERK phosphorylation at the Homer binding site (11). This site-specific phosphorylation enhances Homer and mGluR5 interaction, which down-regulates mGluR5 signaling. Consistent with this model, frmpd4-KO mice exhibit an exacerbated mGluR5 signaling in hippocampus (11) and deficits in hippocampus-dependent spatial learning in Morris Water Maze. Abnormally enhanced mGluR1/5 signaling has been implicated in the pathogenesis of human ID syndromes including Fragile-X syndrome (25,26) and SYNGAP1 haploinsufficiency (27). In our study, FRMPD4p.Cys618ValfsX8 is predicted to result in a truncated protein of 624 amino acids lacking the Homer1 binding site. Consistently, our studies confirm a lack of FRMPD4p.Cys618ValfsX8 binding with Homer1c in Co-IP assays. We speculate that defect in the proper assembly of the FRMPD4-Homer1-PDKs-mGluR1/5 complex, which leads to an enhanced mGluR1/5 signaling contributes to the cognitive impairment caused by FRMPD4 mutations in our patients.
In conclusion, our studies identified several FRMPD4 deleterious mutations in males with ID in the context of X-linked inheritance. Our data support the hypothesis that alteration of FRMPD4 function caused by these mutations contributes to cognitive impairment. Further supporting this hypothesis, frmpd4-KO mice exhibit a deficit in the hippocampus-dependent spatial learning and memory. Based on current understanding of FRMPD4 functions and data from functional studies hereby presented, we speculate that defects in dendritic spinogenesis and/or abnormally enhanced mGluR5 signaling are major determinants of ID in FRMPD4 deficiency-related XLID. Although two FRMPD4-related genes with similar domain structures, FRMPD3 (PRESO2; KIAA1817; Xq22.3) and FRMPD1 (PRESO3; 9p13.2) (28), are known to express abundantly in brain and spinal cord (11), our results suggest that they are not functionally redundant since FRMPD4 deficiency alone is sufficient to cause the phenotype in our patients.
Materials and Methods
Study patients
Patients with XLID and normal control males were recruited by investigators at the respective institutions in Belgium, Canada, France, Poland, and the United States. The human subject research protocols for these studies have been approved by the Institutional Review Boards (IRBs) at the respective institutions. An informed consent was obtained from each study patient and/or their parents or legal guardians. Patients enrolled in this study were evaluated by clinical geneticists and other specialists and underwent comprehensive laboratory studies for ID. All patients were found to have a normal karyotype, negative molecular testing for Fragile X syndrome, and a negative screen for common inborn errors of metabolism. A blood sample was collected from affected individuals and family members to establish EB-transformed lymphoblast cell lines and prepare genomic DNA for sequencing and mutation screening.
Exome-based sequencing and molecular cytogenetics screening
Whole exome sequencing and X-chromosome exome sequencing for mutation identification in the probands of XLID families were conducted as described (12,29). Sanger-based sequencing was used to confirm individual mutations and to perform segregation analysis in the respective probands’ families (12,29). Array Comparative Genomic Comparation (aCGH) was performed using Agilent’s CGX™ HD microarray (PerkinElmer) for identifying the microdeletion of FRMPD4-exon2 in Family 2, according to the manufacturer’s instructions.
Antibodies
The following antibodies were previously described or obtained commercially: mGluR5 from Upstate; pS-mGluR5 was described previously (30); myc (mouse monoclonal 9E10) from Millipore; FRMPD4 were generated and described before (11).
Cell culture and transfection
HEK293T cells were cultured in DMEM medium with 10% FBS. Transfections were performed with X-tremeGENE HP to manufacture’s specifications (SigmaAldrich). Cells were harvested 2 d after transfection.
Coimmunoprecipitation assay and immunobloting
HEK293T cells were used for the coimmunoprecipitation assay as previously reported. Briefly, 500 microliters of immunoprecipitation buffer (PBS, pH 7.4, with 5 mM EDTA, 5 mM EGTA, 1 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM NaF, and 1% Triton X-100) containing Complete EDTA-Free protease inhibitors (Roche) was added and samples were sonicated. After centrifugation, the supernatant (300 μl) was then mixed with 2 μg of myc antibody for 3 h at 4 °C. Then 50 μl of 1: 1 protein G–Sepharose slurry was added for an additional 1 h. The protein beads were washed three times with immunoprecipitation buffer containing 1% Triton X-100. The protein samples were eluted with SDS loading buffer and separated electrophoretically using NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred to an Immobilon-P PVDF membrane (Millipore). The membrane was blocked with blocking buffer for 1 h at room temperature, followed by incubation with primary antibody in PBS buffer overnight at 4 °C. After three washes with PBS buffer, membranes were incubated with 680-conjugated anti-rabbit, or anti-mouse antibody in PBS for another hour. After three washes with PBS buffer, the membrane was scanned with Licor imaging system according to the manufacturer’s protocol (LI-COR Biosciences).
Neuronal culture and transfection
Neuronal hippocampal cultures from embryonic day 18 pups were prepared as reported previously (31). 1 × 106 neurons were added to each 60mm plate (Corning) with 5 coverslips coated with poly-L-lysine. Growth medium consisted of MEM (Invitrogen) supplemented with 1% FBS (Hyclone), 2% B27, 1% glutamine (Invitrogen), 100 U/ml penicillin, and 100 U/ml streptomycin (Invitrogen). Neurons were fed twice per week with glia-conditioned growth medium. Neuronal transfections were performed with lipofectamine-2000 (Invitrogen) and analyzed 24 h later.
Immunocytochemistry, confocal imaging, and analysis
Neurons were washed in 4% sucrose in PBS twice and then fixed with 4% Paraformaldehyde (PFA) and 4% sucrose in PBS for 10 min. After washing with PBS twice, fixed neurons were permeabilized in 0.2% Triton X-100 for 10 min. They were subsequently blocked for 1 h at room temperature in 10% goat serum before incubating with anti-GFP-488 antibody for 1 h at RT. After washing, neurons were mounted on slides in Prolong gold (Invitrogen). Immunofluorescence was viewed with an LSM 710 confocal microscope using a 63x oil-immersion objective, 1024 x 1024 resolutions and scan speed of 7, 12-bit pixel depth and scan average of 8. The optimal pinhole diameter, gain and offset were determined for each fluorophore using the Palette function. Once the optimal pinhole, gain and offset were determined, these were kept constant throughout. Images were analyzed by Neurolucida 360 using the default settings with minor modifications.
Rodent behavioral testing
Animals
Frmpd4-KO mice were generated by the traditional gene-targeting method and bred onto C57BL6/J congenic strain background as described (11). A complete lack of FRMPD4 expression was verified by immunoblots of brain lysates of frmpd4-KO mice (11). Frmpd4-KO mice were viable and fertile, and show similar physical growth, and breeding behaviors compared with WT mice. Mice were genotyped using PCR of tail DNA following a published protocol (11). Adult male frmpd4-KO (n = 12–15) and male WT littermate controls (n = 12–15) between 2–4 months of age were studied using standard behavioral tests for the rodent. Mice were housed in temperature-controlled rooms with 12 h light/dark cycle (9: 00 and 21: 00) and had free access to water and standard mouse chow. Animal breeding and procedures were conducted in strict accordance with NIH Guide for Care and Use of Laboratory Animals. An animal research protocol for this project was approved by the Johns Hopkins University Animal Care and Use Committee.
Behavioral tests
Mouse behavioral tests were conducted at Animal Behavioral Core of the Johns Hopkins University School of Medicine following standard testing protocols (http://www.brainscienceinstitute.org/index.php/cores) as described (32–34). The test order and age of mice for the individual test (in parenthesis) are provided as follows: Open Field (2 months), elevated plus maze (2 months), Y-maze (2–3 months), Morris water maze (3–4 months). For individual test, WT and KO mice were always tested together to minimize potential variations. At least one week of rest was arranged between any two behavioral tests.
Open-field test
Each individual test mouse was placed in a photo-beam (n = 16 at an equal spacing of 2.5 cm) equipped clear plastic chamber [45 × 45 cm and was allowed to explore free from interference for 30 min. The peripheral area (425 sq cm)] was defined by the two side-photo beams, #1–2 and #15–16 while the central area (1600 sq cm) was defined by photo beams #3–14 at each direction. Their movements were tracked using an SDI Photobeam Activity System (San Diego Instruments). Their patterns of ambulatory movement, fine movement, and rearing behavior at central and peripheral areas were recorded and analyzed.
Elevated-plus maze
Elevated plus maze tests anxiety-related behavior in rodents. The maze, made of stainless steel, consists of two closed arms measuring 48 cm (L) × 10 cm (W) × 38 cm (H) and two open arms measuring 48 cm (L) × 10 cm (W) (San Diego Instruments). The four arms were connected by a middle 10 cm × 10 cm platform. The test mouse was placed on the middle platform and remained in the maze during the 5 min session. The total time spent and the number of entries into the closed and open arms were recorded and analyzed.
Y-maze
The Y-maze for spatial working memory in rodents consists of three identical arms [46 cm (L) × 6.25 cm (W) × 2.5 cm (H)] radiating at 120-degree angles from a central platform. The test was done in three trials. During the first trial, the test mouse was placed at the end of one arm, chosen at random prior to the test, and remained in the maze free from interference for 5 min. The total number of spontaneous alternations divided by the number of total possible alternations was recorded and analyzed. The second and third trials were run seven days after the first trial. During the second trial, one of the arms, chosen randomly for each mouse prior to the test, was blocked. The test mouse was allowed to explore the two unblocked arms for 5 min followed by a resting period of 10 min. During the third trial, the test mouse was returned to the maze with all three arms open and allowed to explore for another 5 min. The third trial was analyzed for time spent in the arm blocked during the second trial. The data were analyzed for the first 2 min and full 5 min of trial 3 and compared between the WT and KO cohorts.
Morris water maze
Morris water maze for spatial reference memory was tested in male Frmpd4-KO and male WT littermate control mice. A standard water maze (120 cm in diameter) containing deep, opaque water (25 °C) was set up with a rescuing platform (10 cm x 10 cm) ½ inch below the water surface and Four large Patterned signs as spatial cues were placed on the inner walls of the tub to distinguish each quadrant. An automatic tracking system was used to document latent and swimming time, swimming paths, time spent in each quadrant, and the number of crosses over hidden platform regions. On day 1, mice were subjected to four, 1-min swimming trials in the maze to locate the platform marked with a cue (flag). In each of the four trials, the platform was placed in a different quadrant of the maze. On each of the 5 consecutive training days, the platform cue was removed. Each of the 30 mice went through four trials starting in each of the four quadrants and was allowed to swim until it found the hidden platform placed at a fixed quadrant of the maze, or for 60 s. After finding the platform the mouse was left on the platform for 10 s or, if the mouse did not find the platform in 60 s, was placed on the platform for 10 s. After each trial, the mouse was dried and placed under a heat lamp for approximately 30 min until its next trial. The order of the mice was randomized each day but kept consistent within the four trials in one day, and the order of starting quadrants was different each day but consistent within one day. On the final probe day, the platform was removed and each mouse went through one trial starting in one of the two quadrants farthest from the platform, randomized so that 8 KO and 8 WT started in one quadrant and 7 KO and 7 WT started in the other. Each mouse was allowed to swim for 90 s and both the time spent in the platform quadrant and the number of times the mouse crossed over where the platform was previously situated were automatically recorded and compared between the cohorts of WT and KO mice.
Statistical analysis
Statistical analyses of behavioral testing data were performed using a two-tailed t-test for comparison of the means of two independent samples and two-way ANOVA followed by t-test post hoc for multiple comparisons. Behavioral data were presented as mean ± SEM; P < 0.05 was considered statistically significant.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. De Saedeleer (Mons, Belgium) for kindly providing clinical data on his patients. The cooperation of all patients and their families is gratefully acknowledged.
Conflict of Interest statement. None declared.
Funding
National Institute of Health (GM110265) to P.W. and T.W.; (MH100024) to P.W.; (NS073854) to C.E.S. and T.W.; the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health to D.H.; NARSAD Young Investigator Grant (#21203) to J.H. National Science Centre in Poland (2012/07/B/NZ4/01764). Department of Disabilities and Special Needs of South Carolina and Greenwood Genetic Center Foundation.
Web Resources
UCSC Genome Browser, http://genome.ucsc.edu
1000 Genomes Project, http://www.1000genomes.org/
dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
ExAC Browser, http://exac.broadinstitute.org/
RefSeq, https://www.ncbi.nlm.nih.gov/refseq/
HomoloGene, http://www.ncbi.nlm.nih.gov/homologene/
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
References
- 1. Deciphering Developmental Disorders Study. (2015) Large-scale discovery of novel genetic causes of developmental disorders. Nature, 519, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maulik P., Mascarenhas M., Mathers C., Dua T., Saxena S. (2011) Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil., 32, 419–436. [DOI] [PubMed] [Google Scholar]
- 3. Stevenson R. (2000) Splitting and lumping in the nosology of XLMR. Am. J. Med. Genet., 97, 174–182. [DOI] [PubMed] [Google Scholar]
- 4. Ropers H., Hamel B. (2005) X-linked mental retardation. Nat. Rev. Genet., 6, 46–57. [DOI] [PubMed] [Google Scholar]
- 5. Chelly J., Mandel J. (2001) Monogenic causes of X-linked mental retardation. Nat. Rev. Genet., 2, 669–680. [DOI] [PubMed] [Google Scholar]
- 6. Piton A., Redin C., Mandel J. (2013) XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am. J. Hum. Genet., 93, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Bokhoven H. (2011) Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet., 45, 81–104. [DOI] [PubMed] [Google Scholar]
- 8. de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C.. et al. (2012) Diagnostic exome sequencing in persons with severe intellectual disability. New Engl. J. Med., 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
- 9. Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A.. et al. (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature, 511, 344–347. [DOI] [PubMed] [Google Scholar]
- 10. Lee H., Choi J., Shin H., Kim K., Yang J., Na M., Choi S., Kang G., Eom S., Kim H., Kim E. (2008) Preso, a novel PSD-95-interacting FERM and PDZ domain protein that regulates dendritic spine morphogenesis. J. Neurosci., 28, 14546–14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu J., Yang L., Kammermeier P., Moore C., Brakeman P., Tu J., Yu S., Petralia R., Li Z., Zhang P.. et al. (2012) Preso1 dynamically regulates group I metabotropic glutamate receptors. Nat. Neurosci., 15, 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu H., Haas S.A., Chelly J., Van Esch H., Raynaud M., de Brouwer A.P.M., Weinert S., Froyen G., Frints S.G.M., Laumonnier F.. et al. (2016) X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry, 21, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuste R., Bonhoeffer T. (2001) Morphological changes in dendritic spines associated with long-term synaptic plasticity. Ann. Rev. Neurosci., 24, 1071–1089. [DOI] [PubMed] [Google Scholar]
- 14. Okamoto K., Nagai T., Miyawaki A., Hayashi Y. (2004) Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci., 7, 1104–1112. [DOI] [PubMed] [Google Scholar]
- 15. Matsuzaki M., Honkura N., Ellis-Davies G., Kasai H. (2004) Structural basis of long-term potentiation in single dendritic spines. Nature, 429, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zito K., Scheuss V., Knott G., Hill T., Svoboda K. (2009) Rapid functional maturation of nascent dendritic spines. Neuron, 61, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(1994) The Dutch-Belgian Fragile X Consortium. (1994) Fmr1 knockout mice: a model to study fragile X mental retardation. Cell, 78, 23–33. [PubMed] [Google Scholar]
- 18. Nimchinsky E., Oberlander A., Svoboda K. (2001) Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci., 21, 5139–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haas M.A., Bell D., Slender A., Lana-Elola E., Watson-Scales S., Fisher E.M.C., Tybulewicz V.L.J., Guillemot F., Kihara A.H. (2013) Alterations to dendritic spine morphology, but not dendrite patterning, of cortical projection neurons in Tc1 and Ts1Rhr mouse models of down syndrome. PloS ONE, 8, e78561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Belichenko P., Wright E., Belichenko N., Masliah E., Li H., Mobley W., Francke U. (2009) Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J. Comp. Neurol., 514, 240–258. [DOI] [PubMed] [Google Scholar]
- 21. Ricciardi S., Ungaro F., Hambrock M., Rademacher N., Stefanelli G., Brambilla D., Sessa A., Magagnotti C., Bachi A., Giarda E.. et al. (2012) CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol., 14, 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Maldergem L., Hou Q., Kalscheuer V.M., Rio M., Doco-Fenzy M., Medeira A., de Brouwer A.P.M., Cabrol C., Haas S.A., Cacciagli P.. et al. (2013) Loss of function of KIAA2022 causes mild to severe intellectual disability with an autism spectrum disorder and impairs neurite outgrowth. Hum. Mol. Genet., 22, 3306–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Penzes P., Cahill M., Jones K., VanLeeuwen J., Woolfrey K. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci., 14, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martínez-Cerdeño V. (2017) Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol., 77, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huber K., Gallagher S., Warren S., Bear M. (2002) Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl Acad. Sci. USA, 99, 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bear M., Huber K., Warren S. (2004) The mGluR theory of fragile X mental retardation. Trends Neurosci., 27, 370–377. [DOI] [PubMed] [Google Scholar]
- 27. Barnes S., Wijetunge L., Jackson A., Katsanevaki D., Osterweil E., Komiyama N., Grant S., Bear M., Nagerl U., Kind P., Wyllie1 D. (2015) Convergence of hippocampal pathophysiology in syngap +/- and fmr1-/y mice. J. Neurosci., 35, 15073–15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. An N., Blumer J., Bernard M., Lanier S. (2008) The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G proteins. J. Biol. Chem., 283, 24718–24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niranjan T.S., Skinner C., May M., Turner T., Rose R., Stevenson R., Schwartz C.E., Wang T., Bardoni B. (2015) Affected kindred analysis of human X chromosome exomes to identify novel X-linked intellectual disability genes. PLoS One, 10, e0116454.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park J., Hu J., Milshteyn A., Zhang P., Moore C., Park S., Datko M., Domingo R., Reyes C., Wang X.. et al. (2013) A Prolyl-isomerase mediates dopamine-dependent plasticity and cocaine motor sensitization. Cell, 154, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu J., Park J., Park S., Xiao B., Dehoff M., Kim S., Hayashi T., Schwarz M., Huganir H., Seeburg P., Linden D., Worley P. (2010) Homeostatic Scaling requires Group I mGluR activation mediated by Homer1a. Neuron, 68, 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pletnikov M., Ayhan Y., Nikolskaia O., Xu Y., Ovanesov M., Huang H., Mori S., Moran T., Ross C. (2008) Indicible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol. Psychiatry, 13, 173–186. [DOI] [PubMed] [Google Scholar]
- 33. Ayhan Y., Abazyan B., Nomura J., Kim R., Ladenheim B., Krasnova I., Sawa A., Margolis R., Cadet J., Mori S.. et al. (2011) Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence of neurodevelopmental origin of major psychiatric disorders. Mol. Psychiatry, 16, 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adamczyk A., Mejias R., Takamiya K., Yocum J., Krasnova N., Calderon J., Cadet J., Huganir R., Pletnikov M., Wang T. (2012) GluA3-deficiency in mice is associated with increased social and aggressive behavior and elevated dopamine in striatum. Behav. Brain Res., 229, 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.