Abstract
Background:
The intratumoural heterogeneity, often driven by epithelial-to-mesenchymal transition (EMT), significantly contributes to chemoresistance and disease progression in adenocarcinomas.
Methods:
We introduced a high-throughput screening platform to identify surface antigens that associate with epithelial–mesenchymal plasticity in well-defined pairs of epithelial cell lines and their mesenchymal counterparts. Using multicolour flow cytometry, we then analysed the expression of 10 most robustly changed antigens and identified a 10-molecule surface signature, in pan-cytokeratin-positive/EpCAM-positive and -negative fractions of dissociated breast tumours.
Results:
We found that surface CD9, CD29, CD49c, and integrin β5 are lost in breast cancer cells that underwent EMT in vivo. The tetraspanin family member CD9 was concordantly downregulated both in vitro and in vivo and associated with epithelial phenotype and favourable prognosis.
Conclusions:
We propose that overall landscape of 10-molecule surface signature expression reflects the epithelial–mesenchymal plasticity in breast cancer.
Keywords: breast cancer, epithelial-mesenchymal plasticity, intratumoural heterogeneity, CD9
The intratumoural heterogeneity, determined by the presence of multiple cancer cell phenotypes within a single tumour, represents a major obstacle in effective cancer treatment (Marusyk et al, 2012). Intratumoural heterogeneity is pursued by distinct mechanisms of cancer plasticity and clonal evolution and selection. These events are reversible, dynamic and context-dependent. The ability of a fraction of cancer cells to reprogram their transcriptional state and escape or even evolve under the pressure of therapy is striking and still not well understood (Malladi et al, 2016). Phenotype of such rare cells seems to be very specific, but remains poorly defined. The newly developed advantages, such as acquisition of invasive phenotype, entrance into proliferative quiescence and dormancy, activation of stem-like state and, after some time, re-awakening and formation of overt metastases, are often driven or accompanied by the epithelial–mesenchymal transition (EMT), an important mechanism that contributes to cancer plasticity (Tsai and Yang, 2013).
Here, we identified 10 surface antigens that reflect epithelial–mesenchymal plasticity in vitro using an antibody-based high-throughput profiling and verified their expression in dissociated breast cancer patient samples. All depicted antigens were heterogeneously expressed in clinical specimens, and four of these, CD9, CD29, CD49c, and integrin β5 (ITGB5), were significantly downregulated in a subpopulation of breast cancer cells that underwent EMT in vivo. CD9, a marker decreased both in vitro and in vivo, is a tetraspanin with pleiotropic functions in development and disease, commonly overexpressed in breast cancer (Mimori et al, 2005). We further associated its expression with epithelial phenotype, epithelial–mesenchymal plasticity and favourable prognosis in large cohorts of breast cancer patients.
Uncovering the molecules that accompany, mirror and regulate cancer plasticity and mechanisms responsible for transcriptional reprogramming of cancer cells might help in the future to identify potential biomarkers and therapies that specifically target tumour- and metastasis-initiating cells and prevent cancer relapse and macrometastatic outgrowth.
Materials and methods
Cell culture
BPH-1 and CAFTD03 (a kind gift from Prof. Simon Hayward, Vanderbilt University Nashville, TN, USA), HMLE (a kind gift from Prof. Robert Weinberg, Whitehead Institute, Cambridge, MA, USA), MCF10A-LXSN and MCF10A-V12 cells (a kind gift from Dr Ben Ho Park, The Sidney Kimmel Comprehensive Cancer Center at John Hopkins, Baltimore, MD, USA) were cultured as described previously (Slabakova et al, 2015). HMLE-EMT cells were prepared in house using sequential trypsin fractionation from parental HMLE cell line as a pool, as previously described in Scheel et al (2011) and cultured as parental HMLE cells. Mesenchymal phenotype of HMLE-EMT cells was confirmed with western blot and flow cytometric analysis (Supplementary Figure S1A–C). All cell lines were routinely tested for mycoplasma contamination. The AmpFLSTR Identifiler PCR Amplification Kit (Applied Biosystems, Thermo Fisher Scientific, Prague, Czech Republic) was used to verify the origin of cell lines.
Breast cancer tissue processing
Fresh breast cancer tissues, unused for diagnostic and therapeutic procedures and evaluated by licensed pathologist, were obtained from Masaryk Memorial Cancer Institute from patients undergoing surgical breast cancer removal. All human tissue samples were obtained based on approval of the Masaryk Memorial Cancer Institute Ethical committee (2017/1894/MOU) from donors that signed written informed consent. Clinical annotation for patient samples is listed in Supplementary Table 1. Tissue samples were minced to 1–2 mm pieces. Mechanical dissociation was performed before enzymatic digestion with GentleMACS Dissociator (Miltenyi, Bergish Gladbach, Germany; programs h_tumor_01–02, combined). Tissue pieces were digested in DMEM/F12 (Gibco, TFS) containing 2% bovine serum albumin (BSA; Serva, Heidelberg, Germany), 5 μg ml−1 recombinant human insulin (Sigma-Aldrich, Prague, Czech Republic), 0.5 μg ml−1 hydrocortisone (Sigma-Aldrich), 50 μg ml−1 gentamicin (Serva), 2 mg ml−1 collagenase type I (cat. no. LS 004194; Worthington, Lakewood, NJ, USA), 0.6 U ml−1 dispase II (cat. no. 04942078001; Roche, Prague, Czech Republic) and 10 μM Y-27632 dihydrochloride (SCBT, Dallas, TX, USA), for 14 h at 37 °C. Gentle agitation (40 rpm) was used to prevent viable cell loss and non-specific surface epitope cleavage. Samples were then treated with 15 μg ml−1 DNase I (Roche) for 5 min/37 °C, washed several times with sterile PBS, filtered through 70 μm strainer and cryopreserved in 90 FBS/10% DMSO. After fast recovery, cells were washed with DMEM/F12 containing 2% BSA, filtered through 70 μm strainer and processed to extracellular staining.
Antibody-based cell surface screening and flow cytometry
For high-throughput cell surface screening, suspensions of cell lines were barcoded with various dilutions of CellTrace Violet or/and CellTrace DDAO (Far Red) amine-reactive fluorescent dyes (Molecular Probes, TFS) and dispensed into LEGENDScreen Human Cell Screening PE Kit 96-well plates (cat. no. 700001; Biolegend, San Diego, CA, USA) and stained as recommended. Details regarding preparation of suspension, barcoding and staining are provided in Supplementary Materials and Methods. Compensation values for latter multicolour analyses were calculated automatically in FACSDiva Software (Becton Dickinson Biosciences/BD, Franklin Lakes, NJ, USA) or FlowJo (v10.0.7; TreeStar, Ashland, OR, USA) from single-conjugate stained UltraComp eBeads (eBioscience, San Diego, CA, USA) and/or cell lines (positive controls). Dilution, clonality, fluorochrome information, catalogue numbers of the antibodies used for flow cytometric analyses together with simplified protocol are provided in Supplementary Materials and Methods. In all flow cytometric experiments, dead cells were excluded from analysis based on their positivity to LIVE/DEAD Fixable Dead Cell Stains (various dyes, Invitrogen, TFS) or propidium iodide (final concentration 1 μg ml−1; Sigma-Aldrich). Cell aggregates and debris were excluded from analysis based on a dual-parameter dot plot in which the pulse ratio (signal height/y-axis vs signal area/x-axis) was displayed. Representative gating strategy is shown in Supplementary Figure S1D.
Western blot analysis
Western blot analysis of E-cadherin (cat. no. 610182, clone 36, BD), vimentin (cat. no. V6389, clone V9, Sigma-Aldrich) and α-tubulin (cat. no. T9026, clone DM1A, Sigma-Aldrich) was performed as described in Slabakova et al (2015).
Data reproducibility, bioinformatic and statistical analysis
In high-throughput antibody-based screen, all cell lines were barcoded with fluorescent dyes, pooled and analysed in parallel, one well per antibody. Initial screen was performed once. All further cell line-based experiments were performed independently at least three times. Analyses of multiparametric cytometric data were performed in FlowJo and Cytobank, as described in Supplementary Materials and Methods. Clinical data sets (accession numbers GSE2603, GSE6061, GSE12276, GSE2109, GSE20271, GSE8465, GSE5764, and GSE349) were retrieved via GEO (NCBI) and Oncomine (TFS). Kaplan–Meier plots were assessed via KM plotter and PROGGene (accession numbers GSE2603, GSE42568, GSE37946, GSE7309, GSE25055, and GSE4922_U133B), median was set as cutoff (Gyorffy and Schafer, 2009; Goswami and Nakshatri, 2013). Heat map generation and cluster analyses were performed with Morpheus (Broad Institute, Cambridge, MA, USA). Statistical analyses were performed in Prism (v6, GraphPad, La Jolla, CA, USA). P values were calculated with paired t test and ratio paired t test (two-tailed), if not stated otherwise.
Results
Distinct cell surface signature reflects epithelial–mesenchymal plasticity in vitro
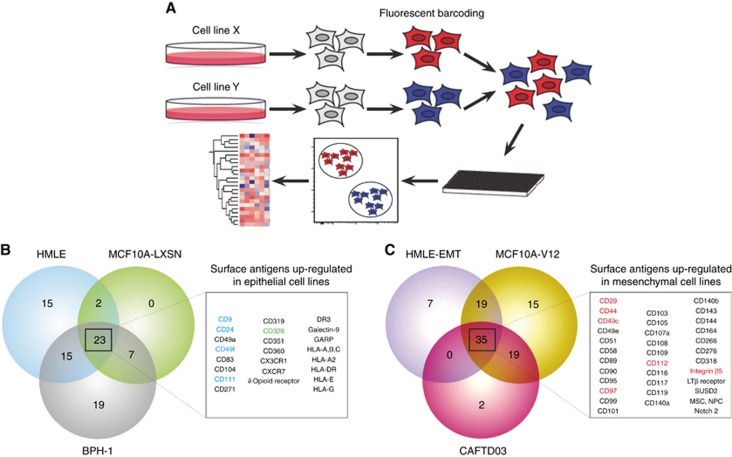
The lack of new surface antigens that associate with cancer plasticity and enable sorting of viable cancer cells with distinct cellular phenotypes significantly limits our understanding of tumour heterogeneity to several, well-described molecules (Medema, 2013). Commonly used proteomic high-throughput approaches, such as mass spectrometry, are valuable tools for discovery of novel and rare antigens. However, in such cases, only limited (if any) number of validated reagents is available for immediate use by the scientific community. To enable high-throughput analysis of ‘EMT-surfaceome’ with validated antibodies, we introduced flow cytometric, fluorescent barcode-based platform, which allowed simultaneous analysis of up to six cell lines in parallel (Figure 1A). Analysis of three well described model epithelial cell lines and their isogenic mesenchymal counterparts (Supplementary Figure S1C) revealed that from 332 markers analysed, 120 antigens were expressed on the surface of at least one cell line (Supplementary Figure S2A), 23 markers were upregulated on the surface of all epithelial cell lines (Figure 1B) and 35 markers were upregulated on the surface of all mesenchymal cell lines (Figure 1C). From these, we selected four markers that most robustly associated with epithelial phenotype (Supplementary Figure S2B) and six that changed with acquisition of mesenchymal phenotype (Supplementary Figure S2C) for subsequent validation. Further bioinformatic analysis of screen results suggested that these novel epithelial- and mesenchymal-like markers clustered together as well as clustered cell lines based on their EMT-surface phenotype (Supplementary Figure S2D), suggesting association of these molecules with cellular phenotype (Supplementary Figure S2E). Independent flow cytometric validation revealed that pan-epithelial marker EpCAM and 10 novel surface antigens, related to EMT-state of permanent cell lines, are differentially expressed in at least two model pairs of ‘in vitro plasticity’ (Supplementary Figure S3 and Supplementary Table S5). We, thus, proposed that these 10 markers, further denoted as ‘10-molecule surface signature’, are coupled with the epithelial and mesenchymal state in vitro (Supplementary Figure S2E). To assess the significance of the 10-molecule signature in dissociated clinical specimens, we established multicolour flow cytometric protocol enabling parallel analysis of their surface expression at a single-cell level. We tested this panel in HMLE cell line, known for the presence of at least two surface phenotypes. While epithelial-like, CD24hi/CD44lo subpopulation showed enrichment in epithelial markers EpCAM, CD9, CD49f, and CD111; mesenchymal-like, CD24lo/CD44hi cells retained mesenchymal signature (Supplementary Figure S4A; signature is shown in Supplementary Figure S2E). Multidimensional data analyses suggested the presence of several phenotypically distinct subpopulations (Supplementary Figure S4B). Such findings implicated that even simplified systems, such as permanent cell lines, preserve a significant degree of surface heterogeneity.
Figure 1.
Identification of novel antigens associated with in vitro plasticity of epithelial cells, identified with high-throughput surface profiling. (A) Schematic shows experimental workflow of high-throughput surface screen with fluorescently barcoded cell lines. Cell lines were harvested, barcoded with fluorescent amine-reactive dyes, pooled and stained in multiwell plates. After data acquisition, cell lines were deconvoluted based on their fluorescent barcode and processed to data analysis. (B) Antigens upregulated on the surface of epithelial cell lines. Antigens selected for further validation are highlighted in blue, pan-epithelial marker EpCAM is shown in green. (C) Antigens upregulated on the surface of mesenchymal cell lines. Antigens selected for further validation are highlighted in red.
Ten-molecule signature is heterogeneously expressed on surface of primary breast cancer cells
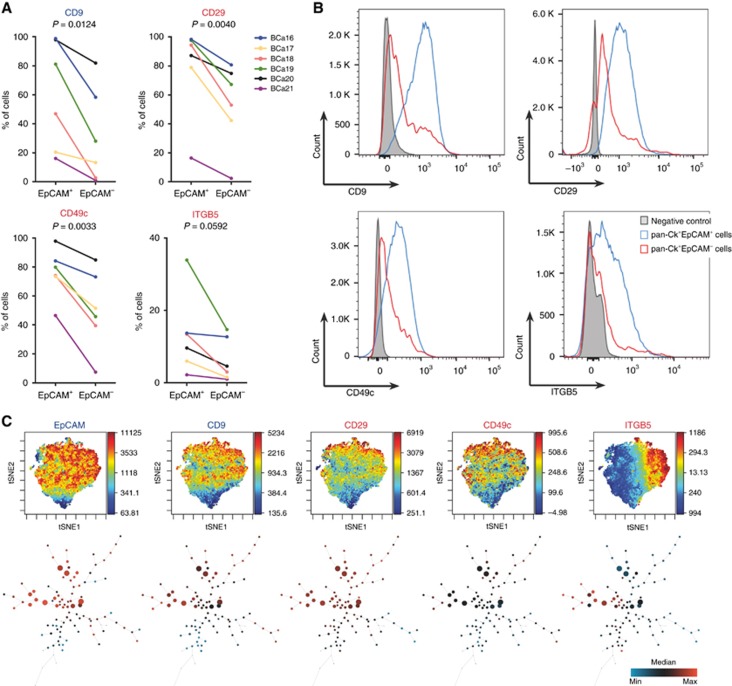
While EpCAM is one of the most rapidly downregulated genes during EMT, genes encoding several members of cytokeratin family remain steadily expressed even in cancer cells that have acquired mesenchymal phenotype (Schneck et al, 2015). We further extended the established multicolour flow cytometric panel for analysis of intracellular pan-cytokeratin (pan-Ck), as a surrogate marker for cancer cells in dissociated breast cancer samples. Assessment of membrane EpCAM immunophenotype of pan-Ck+ cells (Supplementary Figure S5A) within the dissociated specimen then allowed tracking of epithelial cancer cells (EpCAM+) and cancer cells that underwent EMT in vivo (EpCAM−; Supplementary Figure S5B). We analysed expression of EpCAM, 10-molecule signature and pan-Ck in six breast cancer samples that classified into PAM50 subtype Luminal A and triple-negative (Supplementary Table S1). All of the proposed antigens were detected the on surface of pan-Ck+EpCAM− and pan-Ck+EpCAM+ subpopulations. Moreover, pan-Ck+EpCAM+ subpopulations from Luminal A tumours (BCa16-19), identified by complex immunophenotype, clustered together (Supplementary Figure S5C). Subpopulations from triple-negative breast cancers (BCa20-21) clustered based on their origin, most likely due to already mesenchymal features of TNBC (Criscitiello et al, 2012). We, thus, propose that the landscape of 10-molecule signature expression powerfully reflects the plasticity in epithelial and mesenchymal-like cancer cells. However, each patient sample displayed a distinct 10-molecule signature profile, reflecting patient-to-patient variability (Supplementary Table S6). Despite such variability, four antigens – CD9, CD29, CD49c, and ITGB5 – were accordingly downregulated on surface of pan-Ck+EpCAM− cells in six out of six analysed tumour samples (Figure 2A and B). While the downregulation of CD9 reflects the in vitro findings (Supplementary Figure S3); CD29, CD49c, and ITGB5 were upregulated in mesenchymal cells in vitro. Similar trends were observed for CD24 and CD111, which were downregulated in several pan-Ck+EpCAM− samples (Supplementary Figure S5D, E; data are summarised in Supplementary Table S6). Further analysis of pan-Ck+CD9+ breast cancer cells revealed that this fraction is enriched not only in EpCAM+ cells, but also in CD29+, CD49c+, and ITGB5+ cells and in cells positive for CD24 and CD111, markers of epithelial-like cells (Supplementary Figure S5F). Similarly to EpCAM, CD9+ subpopulations from Luminal A tumours (BCa16-19) mostly clustered together based on their surface signature (Supplementary Figure S5G), with some exceptions that first clustered based on their origin. Advanced analyses of multidimensional data revealed striking heterogeneity in distribution of all analysed antigens on surface of cancer cells. This surface heterogeneity also allowed for virtual subdivision of cancer cells into multiple (at least 100), phenotypically distinct subpopulations (Figure 2C and Supplementary Figure S6A and B). Tumour cell compartment, thus, represents unimaginably heterogeneous, plastic and versatile ecosystem.
Figure 2.
Validation of in vitro findings in dissociated primary breast tumour samples. (A) Plots show paired analysis of antigens CD9, CD29, CD49c, and ITGB5 that are significantly downregulated in cancer cells that underwent in vivo EMT (n=6; Supplementary Table S1). (B) Representative overlays of histograms from pan-Ck+EpCAM− and pan-Ck+EpCAM+ cells for downregulated antigens CD9, CD29, CD49c, and ITGB5. Histograms are derived from multicolour flow cytometric analysis of dissociated patient sample BCa19 (Luminal A subtype). (C) viSNE plots (upper part) show distribution of selected markers in population of cancer cells within the BCa19 sample (Luminal A subtype). SPADE trees (lower part) show cell abundance and contribution of each marker to calculated, phenotypically distinct subpopulations (see also Supplementary Figure S6; Colour legend: The viSNE plots are visualised in pseudocolour and each dot represents a single cell – the lowest expression of selected marker is in dark blue and the highest expression in dark red, the corresponding scale is next to each plot and reflects the level of marker expression. The SPADE plots are visualised as nodes, the colour ranges from dark blue for the lowest expression in such node to the highest expression in dark red, the scale is in the right corner of figure panel. The size of the node represents the number of cells that falls within the particular subpopulation).
CD9 associates with epithelial phenotype and favourable prognosis in breast cancer
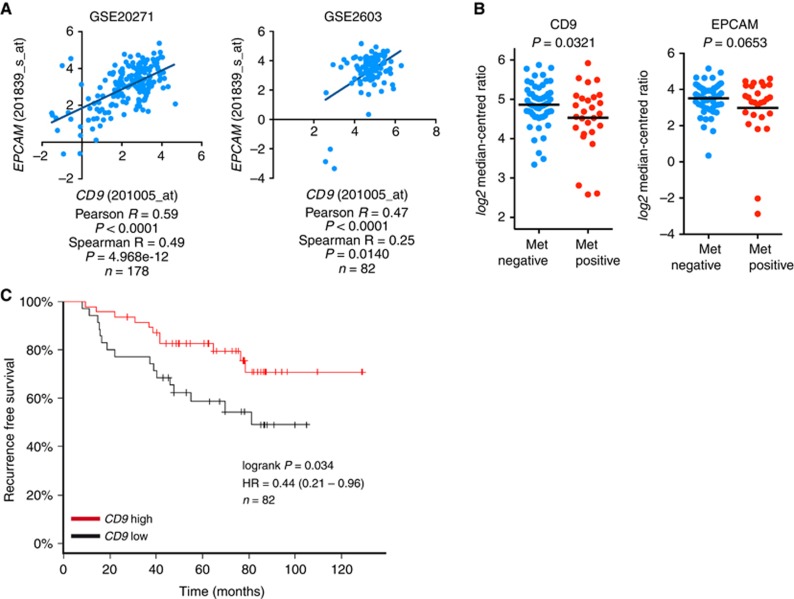
As CD9 was the only antigen downregulated in EMT-ed cells both in vitro and in vivo, we further focused on its expression in breast cancer data sets and on its role in epithelial–mesenchymal plasticity. We found that CD9 mRNA expression positively correlated with EPCAM mRNA in multiple data sets (Figure 3A and Supplementary Figure S7A) and that both CD9 and EPCAM expression was downregulated in primary tumours that formed metastasis in distant organs (Figure 3B and Supplementary Figure S7B). Moreover, high CD9 expression showed as a favourable, in terms of recurrence-free survival (Figure 3C and Supplementary Figure S7C). Expression of EpCAM was not prognostic in any of these data sets (not shown).
Figure 3.
CD9 expression associates with epithelial phenotype, cancer progression and recurrence-free survival in a cohort of breast cancer patients. (A) Correlation analysis shows relationship between CD9 and EPCAM mRNA expression in clinical samples. (B) A plot shows expression of CD9 and EpCAM mRNA in non-metastatic primary tumours (met negative n=55) and primary tumours that disseminated to distant sites (met positive n=27; GSE2603). (C) The graph shows recurrence-free survival analysis in cohort of breast cancer patients according to high or low CD9 expression levels (GSE2603).
Discussion
Since the very first observation of EMT phenomenon in cancer cells, it has been doubted for years whether EMT really occurs in patient samples and whether it has some functional consequences in vivo. Thanks to extensive studies, EMT was observed both in murine models of cancer and clinical settings (Ye and Weinberg, 2015). Similarly, besides epithelial EpCAM+ cancer cells, we detected an EpCAM− fraction of cancer cells in all analysed samples.
Nowadays, EMT is considered as one of the master regulators and drivers of solid cancer plasticity, and thus, significantly contributes to intratumoural heterogeneity, but its role in recurrence and dissemination of cancer cells remains questionable (Tsai and Yang, 2013). The lack of reliable, surface pan-carcinoma markers that are not lost during the EMT of cancer cells is still a major obstacle in analysis of viable EMT-ed cells in patient samples. Novel technologies, such as reliable and functionally validated reagents for detection of epithelial mRNAs (such as cytokeratins) in viable cells and in dissociated suspensions from patient samples, are greatly expected (Halo et al, 2014). This approach would enable sorting of in vivo EMT-ed cells, based on their positivity for cytokeratin family transcripts and negative EpCAM immunophenotype, and their subsequent transcriptomic analysis and in vivo studies, such as tumourigenicity.
We used trio of unrelated, well-described, normal or pre-neoplastic epithelial cell lines and their (partially) EMT-ed, isogenic counterparts for identification of novel surface antigens, that associate with plasticity of epithelial cells. Though their mesenchymal phenotype was driven and preserved by distinct mechanisms (stem-like state in HMLE-EMT cells, oncogenic K-Ras in MCF10A-V12 cells and microenvironment in CAFTD03 cells), we found several antigens that were accordingly up- and downregulated in all models. Beside commonly used markers of mammary (cancer) stem cells, CD24 and CD44 (Al-Hajj et al, 2003), we identified several surface proteins that have never been linked to EMT and cancer plasticity before (CD9, CD97, CD111, and CD112). Their function and signalling in cancer cells remain mostly unknown. Nonetheless, we used validated reagents for our discoveries, which can be immediately used by the scientific community to answer these crucial questions.
Interestingly, CD9 was identified as the only antigen that acted similarly in vitro and in vivo. CD9 is a member of the tetraspanin family and is commonly present on the surface of extracellular vesicles (Andreu and Yanez-Mo, 2014). This marker is often overexpressed in breast cancer and was shown to play important role in invasiveness and development of metastasis (Rappa et al, 2015). However, similarly to our findings, high expression of CD9 also associated with better prognosis (Mimori et al, 2005). Though CD9 was downregulated in EMT-ed cancer cells, we also identified a panCk+EpCAM−CD9+ subpopulation of cancer cells. And, importantly, function of these cells may be completely unrelated to their EpCAM-CD9− or EpCAM+CD9+ counterparts. As advanced mechanistic studies are not available, their role in tumour microenvironment remains unresolved. Moreover, CD9 was shown to interact with other downregulated molecules, CD29 and CD49c (Berditchevski, 2001). Such relation suggests that these molecules may be co-regulated by the EMT machinery or even interact in breast cancer cells in vivo. Besides CD9, CD29, and CD49c; ITGB5 was also significantly downregulated on surface of EMT-ed cancer cells. Despite the association of CD29, CD49c and ITGB5 with mesenchymal-like phenotype in vitro, integration of cacophony of microenvironmental signals in patient-derived cancer cells may lead to complex and more physiological disturbances on cellular surface in comparison to cultured cell lines. The different levels of complexity in in vitro monolayer culture and in in vivo settings with a distinct tissue architecture and cellular polarity, thus, may have contributed to the differences between in vitro/in vivo approaches (Camphausen et al, 2005). Moreover, besides the complex effects of microenvironment on cellular phenotype, it was shown that the ex vivo expansion of both non-malignant and cancer cells leads to propagation of clonogenic cells with significantly different surface phenotype, in comparison to the tissue of origin (Anderson et al, 2002; Zhang and Lodish, 2005). The early events during the ex vivo expansion of cell lines used in our cell surface screen, followed by the events accompanying their transformation, may have contributed to stabilisation of the ‘in vitro’ phenotype that represents only a fraction of cells from the tissue of their origin. These reasons may be at least in part responsible for such discrepancy, and thus, validation of our results in clinical setting was necessary.
Complex, multiparametric analysis of the surface immunophenotype together with simultaneous detection of intracellular pan-Ck revealed that several markers from the 10-molecule surface signature showed high patient-to-patient variability. However, the presence of these subpopulations within the tumour mass of every analysed sample connects them with a special function and phenotype. We believe that future studies will reveal mechanisms responsible for heterogeneity in their expression and their role in tumour microenvironment.
Acknowledgments
We would like to thank Iva Lišková, Martina Urbánková and Kateřina Svobodová for technical assistance; to the team of surgeons, namely Drs Lucie Mouková, Vuk Fait and Richard Feranc (Department of Oncological Surgery, Masaryk Memorial Cancer Institute, Brno, Czech Republic), for collection of patient samples; to Prof. Radek Vrtěl for cell line authentication; to Monika Hortová (née Smějová) for derivation of HMLE-EMT cell line; to Jakub Lajmon (Lajmon Network, Czech Republic) for help with data processing and digital art; and to Prof. Simon Hayward (Vanderbilt University, Nashville, TN, USA), Prof. Robert Weinberg (Whitehead Institute, Cambridge, MA, USA) and Dr Ben Ho Park (The Sidney Kimmel Comprehensive Cancer Center at John Hopkins, Baltimore, MD, USA) for providing model cell lines. This work was supported by Ministry of Health of the Czech Republic, grant no. 15-33999A, all rights reserved (to KS and MS); project no. LQ1605 from the National Program of Sustainability II (MEYS CR).
Ethical approval and consent to participate
All human tissue samples were obtained at Masaryk Memorial Cancer Institute, Brno, Czech Republic based on approval of the Masaryk Memorial Cancer Institute Ethical committee (2017/1894/MOU) from donors that signed written informed consent.
Availability of data and materials
Preprocessed data are available in Supplementary Information. Raw data and materials used in this study, except the dissociated patient samples, are available upon reasonable request. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karel Souček.
Author contribution
JR and KS conceptualised and designed the study, analysed and interpreted the data and wrote and reviewed the manuscript. JR developed methodology. JN, PF and MS managed clinical samples. JR, RF, LB and ES acquired the data. All authors approved the final version of manuscript. KS supervised the study.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100(7): 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RC, Elder JB, Brown MD, Mandigo CE, Parsa AT, Kim PD, Senatus P, Anderson DE, Bruce JN (2002) Changes in the immunologic phenotype of human malignant glioma cells after passaging in vitro. Clin Immunol 102(1): 84–95. [DOI] [PubMed] [Google Scholar]
- Andreu Z, Yanez-Mo M (2014) Tetraspanins in extracellular vesicle formation and function. Front Immunol 5: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski F (2001) Complexes of tetraspanins with integrins- more than meets the eye. J Cell Sci 114(23): 4143–4151. [DOI] [PubMed] [Google Scholar]
- Camphausen K, Purow B, Sproull M, Scott T, Ozawa T, Deen DF, Tofilon PJ (2005) Influence of in vivo growth on human glioma cell line gene expression- Convergent profiles under orthotopic conditions. Proc Natl Acad Sci USA 102(23): 8287–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello C, Azim HA Jr., Schouten PC, Linn SC, Sotiriou C (2012) Understanding the biology of triple-negative breast cancer. Ann Oncol 23(Suppl 6): vi13–vi18. [DOI] [PubMed] [Google Scholar]
- Goswami C, Nakshatri H (2013) PROGgene - gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma 3(1): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Schafer R (2009) Meta-analysis of gene expression profiles related to relapse-free survival in 1,079 breast cancer patients. Breast Cancer Res Treat 118(3): 433–441. [DOI] [PubMed] [Google Scholar]
- Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, Malin D, Strekalova E, Cryns VL, Cheng C, Mirkin CA, Thaxton CS (2014) NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci USA 111(48): 17104–17109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, de Stanchina E, Massague J (2016) Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 165(1): 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12(5): 323–334. [DOI] [PubMed] [Google Scholar]
- Medema JP (2013) Cancer stem cells: the challenges ahead. Nat Cell Biol 15(4): 338–344. [DOI] [PubMed] [Google Scholar]
- Mimori K, Kataoka A, Yoshinaga K, Ohta M, Sagara Y, Yoshikawa Y, Ohno S, Barnard GF, Mori M (2005) Identification of molecular markers for metastasis-related genes in primary breast cancer cells. Clin Exp Metastasis 22(1): 59–67. [DOI] [PubMed] [Google Scholar]
- Rappa G, Green TM, Karbanova J, Corbeil D, Lorico A (2015) Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 6(10): 7970–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA (2011) Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145(6): 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneck H, Gierke B, Uppenkamp F, Behrens B, Niederacher D, Stoecklein NH, Templin MF, Pawlak M, Fehm T, Neubauer H Disseminated Cancer Cell Network D (2015) EpCAM-independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS ONE 10(12): e0144535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabakova E, Kharaishvili G, Smejova M, Pernicova Z, Suchankova T, Remsik J, Lerch S, Strakova N, Bouchal J, Kral M, Culig Z, Kozubik A, Soucek K (2015) Opposite regulation of MDM2 and MDMX expression in acquisition of mesenchymal phenotype in benign and cancer cells. Oncotarget 6(34): 36156–36171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Yang J (2013) Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 27(20): 2192–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Weinberg RA (2015) Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol 25(11): 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lodish H (2005) Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood 105(11): 4314–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.