Abstract
Mucolipidosis type IV (MLIV) is a lysosomal storage disease characterized by neurologic and ophthalmologic abnormalities. There is currently no effective treatment. MLIV is caused by mutations in MCOLN1, a lysosomal cation channel from the transient receptor potential (TRP) family. In this study, we used genome editing to knockout the two mcoln1 genes present in Danio rerio (zebrafish). Our model successfully reproduced the retinal and neuromuscular defects observed in MLIV patients, indicating that this model is suitable for studying the disease pathogenesis. Importantly, our model revealed novel insights into the origins and progression of the MLIV pathology, including the contribution of autophagosome accumulation to muscle dystrophy and the role of mcoln1 in embryonic development, hair cell viability and cellular maintenance. The generation of a MLIV model in zebrafish is particularly relevant given the suitability of this organism for large-scale in vivo drug screening, thus providing unprecedented opportunities for therapeutic discovery.
Introduction
Mucolipidosis type IV (MLIV) is an autosomal recessive lysosomal storage disease (LSD) characterized by severe psychomotor developmental delay, progressive loss of vision, and gastric achlorydria (1–3). MLIV is caused by mutations in the MCOLN1 gene, which encodes a lysosomal cation channel from the transient receptor potential (TRP) family (4,5). MCOLN1, also named TRPML1, mediates efflux of calcium, sodium and certain heavy metals from lysosomes (6–9), and regulates fusion of lysosomes with other intraorganellar compartments, including autophagosomes (10,11), phagosomes (12) and plasma membrane (13,14).
The first signs of the disease are usually identified in infancy and include psychomotor delays, corneal clouding, and strabismus. The visual abnormalities are usually progressive, leading to retinal degeneration, pallor of the optic nerve, corneal dysplasia and, ultimately, blindness (3,5,15,16). Neurological symptoms include mental retardation, hypotonia, spasticity, and a severe delay in major motor milestones, with most patients not progressing beyond the 15–18 month levels in their language and motor functions (3). Achlorhydria is another common manifestation in MLIV patients leading to increased levels of gastrin in blood and iron deficiency anemia (17). Compared to other LSDs, the clinical progression of MLIV is relatively slow and patients can live for over four or five decades. It is still unclear whether life expectancy is decreased in MLIV patients.
MLIV is considered an infancy or early-childhood onset disorder; however, in some cases patients present symptoms at birth, suggesting a prenatal initiation of the disease (18). In addition, brain magnetic resonance imaging studies revealed dysmyelination and hypoplastic corpus callosum, a structure formed between the 13–20th week of gestation; the thinness of the corpus callosum has been shown to correlate with the severity of the neurological impairment (19). Therefore, a critical question that requires further characterization is whether MCOLN1 plays a role during prenatal development.
Cellular manifestations of the disease include the accumulation of storage or “compound” bodies filled with granular and lamellar electron-dense content (3,20). These storage bodies correspond to abnormal lysosomes and autolysosomes with reduced degradative capability, leading to the luminal accumulation of undegraded gangliosides, acidic mucopolysaccharides, and phospholipids (10,15). It is well established that MCOLN1 is involved in the regulation of lysosomal pH and homeostasis (21). MCOLN1 is also critical for the efficient fusion of autophagosomes with lysosomes and subsequent degradation of the autophagosome content. Fibroblasts from MLIV patients show a significant accumulation of autophagosomes and autolysosomes and a reduction of autophagy flux (10). Defective autophagy then results in an accumulation of protein aggregates and damaged mitochondria (22).
Recent evidence suggests that MCOLN1 channel activity is stimulated by reactive oxygen species (ROS). MCOLN1-mediated release of calcium from lysosomes is not only critical for facilitating autophagosome/lysosome fusion but also for triggering activation of the transcription factor TFEB, a master regulator of lysosomal biogenesis and autophagy (23). Therefore, MCOLN1 plays an essential role in the cellular adaptation to mitochondrial damage and its role may be particularly important in cell types that endure high levels of oxidative stress.
Generation of animal models is essential for better understanding the molecular bases of MLIV as well as for testing of potential drugs. MCOLN1 deficiency has been induced in C. elegans and D. melanogaster (24,25). However, invertebrates have only one MCOLN gene, whereas three different MCOLN genes (MCOLN1-3) are present in vertebrates. The characterization of a murine model of MLIV has provided important clues to understanding the biochemical and molecular bases of the disease (26). Nevertheless, studies in mice are costly and laborious, and the intrauterine gestation complicates the analysis of possible early developmental alterations. Here we report the generation of a novel MLIV model in Danio rerio (zebrafish). Similar to what has been observed in humans and mice, mutations in mcoln1 resulted in visual and muscular abnormalities that became more severe over time, indicating that our model successfully reproduces the MLIV pathology. Remarkably, we found that the MLIV animals exhibit additional pathological changes very early during embryonic development, including increased apoptosis in the eye and the brain, as well as severe alteration in the number and morphology of hair cells leading to perturbations in the structure of the neuromasts. Therefore, our work describes, for the first time, a role for mcoln1 during embryonic development and suggests that sensory cells, such as photoreceptors and hair cells, may be particularly vulnerable to the absence of mcoln1.
Results
Targeted mutation of mcoln1 genes in zebrafish
Two orthologs of MCOLN1 have been identified in zebrafish, Mcoln1a and Mcoln1b (27). The predicted protein sequence of Mcoln1a and Mcoln1b shares 62% and 57% identity with human MCOLN1, respectively. Multiple sequence analysis of the three proteins revealed that Mcoln1b lacks one of the two di-leucine motifs implicated in the delivery of MCOLN1 to late endosomal/lysosomal compartments (Fig. 1A) (28); however, given that the di-leucine motifs are partially redundant, it is expected that both Mcoln1a and Mcoln1b localize to late endosomes/lysosomes (27,28).
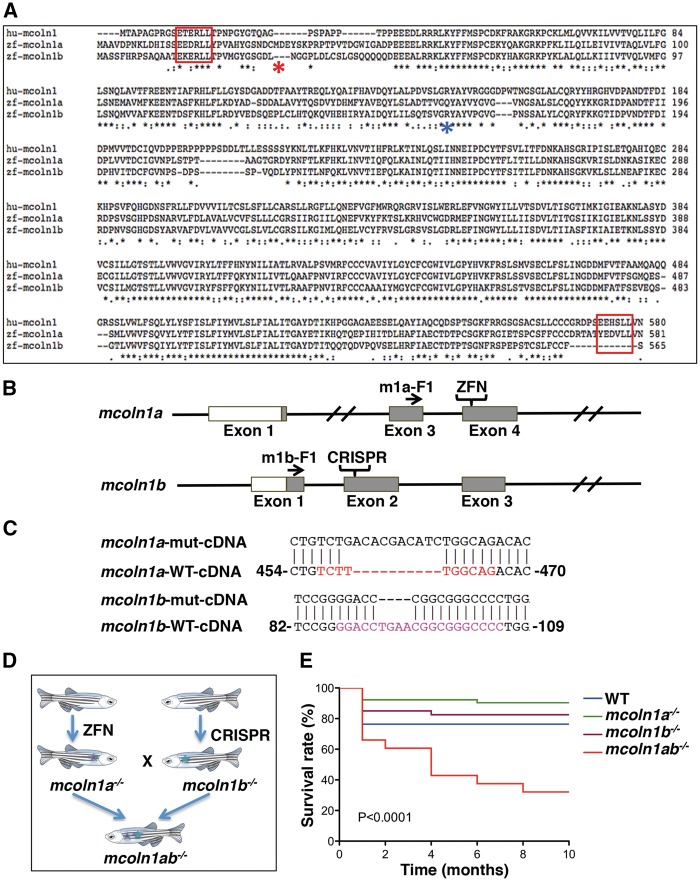
Figure 1.
Generation of mcoln1 mutants in zebrafish. (A) Alignment of human (hu) MCOLN1, zebrafish (zf) Mcoln1a and zf Mcoln1b protein sequences using online software T-COFFEE. The underline black asterisks mark the consensus sequences among these proteins. The two red rectangular brackets indicate the di-leucine lysosomal targeting motifs. The blue and red asterisks mark the mismatch start for Mcoln1a and Mcoln1b mutants, respectively. (B) Strategy and method used to create mcoln1a and mcoln1b mutants. ZFN target site for mcoln1a is on exon 4, while CRISPR/Cas9 target site for mcoln1b is on exon 2. F1 indicated the primers used to sequence the corresponding cDNAs and confirm the mutations (C) mcoln1a−/− and mcoln1b−/− cDNA sequences showing the introduction of a frameshift that results in a premature stop codon for both proteins. (D) Strategy used to create mcoln1ab−/− animals. (E) Kaplan-Meier survival curve of a cohort study (50 animals per genotype) showing the survival rate of each genotype at 0, 2, 4, 6 and 8 months. Differences between groups were calculated using the log-rank test.
To generate a zebrafish model for mucolipidosis IV, we first introduced mutations into the genomic locus of mcoln1a by using custom-engineered Zinc Finger Nucleases (ZFNs) targeting exon 4 (Fig. 1B). mRNAs coding for mcoln1a ZFNs were injected into one-cell-stage wild-type (WT) embryos. After screening the F0 founders and F1 progeny, we identified two mutant lines carrying a 10 bp insertion (#8) and a 10 bp deletion (#44), respectively. Both mutations resulted in a frameshift and a premature stop codon that were confirmed by sequencing their cDNA products (Fig. 1C).
The ZFN strategy for mcoln1b showed a very low efficiency as measured by the yeast MEL-1 reporter assay (29). Therefore, we used Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated system (CRISPR-Cas9) technology to create a mcoln1b mutant line. Firstly, we designed the guide RNA (gRNA) oligos using ZiFiT targeting exon 2 (30,31) (Fig. 1B). Next we made synthetic RNA for both gRNA and Cas9, and co-injected them into one-cell-stage embryos. After screening both the F0 founders and F1 progeny, we found that 8 out of 10 founders carried mutations, and 2 of them carried mutations that were predicted to cause a frameshift and premature stop codon. One of the mutant lines carried a 4 bp deletion (#9), whereas the other mutant line carried a 13 bp deletion (#10). We then extracted total RNA and made cDNA from the F1 progeny of these two lines to confirm the mutations in the transcripts (Fig. 1C).
To create a double mutant line for both mcoln1a and mcoln1b, we crossed mcoln1a−/− (#8) to mcoln1b−/− (#9). After two generations, we generated a double mutant line mcoln1ab−/−, which carries two mutant alleles for each mcoln1a or mcoln1b for the zygotic genes but not the maternal genes. We then intercrossed two mcoln1ab−/− mutant fish to get double mutant for both zygotic and maternal genes (Fig. 1D). All embryos and adult zebrafish used in this study were offspring from the intercrossed fish. mcoln1ab−/− fish were fertile and the morphology of the larvae and juvenile appeared normal; however, we identified a gradual loss of the mcoln1ab−/− fish at adult stage based on a cohort study for all four genotypes, 50 fish for each genotype (Fig. 1E). By 8 months of age, only 30% of the original 50 mcoln1ab−/− fish survived, whereas the numbers of mcoln1a−/− and mcoln1b−/− fish were not significantly different from the wild-type animals (Fig. 1E). This was the first indication that mcoln1a and mcoln1b may have redundant functions and work together to play important roles during adult homeostasis.
Mcoln1 deficiency results in progressive accumulation of autophagosomes in skeletal muscle
One of the most prominent symptoms in MLIV patients is the presence of severe motor deficits that may, at least in part, be due to a muscle pathology (32). To determine whether our mutants constitute a good model for studying MLIV, we started by analysing skeletal muscle both in embryos and adult animals. Whole-mount immunostaining of wild-type, mcoln1a−/−, mcoln1b−/− and mcoln1ab−/− animals at 3 dpf did not reveal major changes in general muscle morphology. Staining using actinin antibody indicated that myofibers were regularly aligned within the somites in all the genotypes, and vinculin was localized to myotome boundaries resulting in a regular immunostaining pattern of chevrons (Fig. 2A). To visualize neuromuscular junctions (NMJ) we used α-bungarotoxin that binds to the α-subunit of the nicotinic acetylcholine receptor (AChR). No major differences were observed between wild-type and mutant embryos (Fig. 2A).
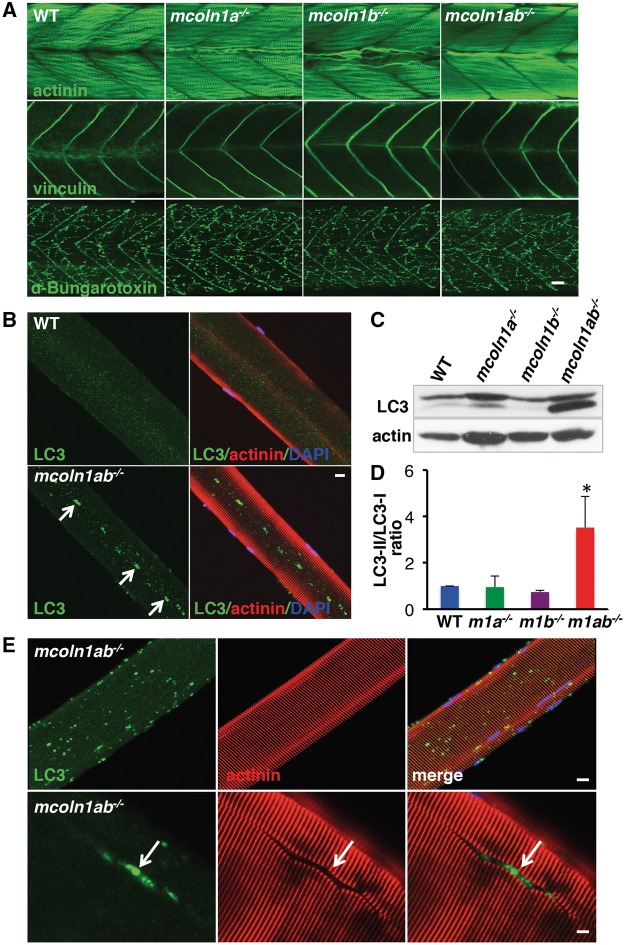
Figure 2.
Skeletal muscle pathology in mcoln1 mutant zebrafish at both embryonic and adult stages. (A) Whole mount immunostaining for actinin, vinculin, and α-Bungarotoxin in WT, mcoln1a−/−, mcoln1b−/−, and mcoln1ab−/− zebrafish embryos at 3 dpf. Scale bar, 20 μm. (B) Co-immunostaining for LC3, actinin and DAPI in single muscle fiber dissected from 8-month-old WT and mcoln1ab−/− zebrafish. White arrows mark the accumulation of LC3 in the middle of the single muscle fiber. Scale bar, 10 μm. (C) Western blot analysis showing LC3 expression levels in skeletal muscle lysates of 8-month-old WT, mcoln1a−/−, mcoln1b−/−, and mcoln1ab−/− zebrafish. Upper band shows the level of LC3-I and lower nad indicated the level of LC3-II. Actin is used as a loading control. (D) Quantification of LC3-II/LC3-I ratio shown in (C). Data are shown as mean ± SD and represent 4 independent experiments. The data were analyzed using paired t test (*P < 0.05). m1a−/− stands for mcoln1a−/−, m1b−/− stands for mcoln1b−/−, and m1ab−/− stands for mcoln1ab−/−. (E) Co-immunostaining for LC3, actinin and DAPI in single muscle fiber dissected from 12-month-old mcoln1ab−/− zebrafish. White arrow marks the accumulation of LC3 in a split muscle fiber. Scale bar for upper panel is 10 μm, for lower panel is 5 μm. All the images are representative of at least three independent experiments.
Mcoln1 knockout mice exhibit progressive age-dependent mobility decline (26,33). A measurable loss of strength is observed at 2–3 months, and by ages 7–9 months the animals become severely paralyzed. In contrast, no obvious reduction in movement was observed in mutant zebrafish. mcoln1ab−/− animals did not exhibit uncoordinated movements and showed escape reaction in response to a tactile stimulus. To gain further insights into the changes associated with loss of mcoln1 expression, we isolated single muscle fibers from eight-month-old wild-type and mcoln1ab−/− animals. Isolated muscle fibers preserve muscle morphology and allow for the detailed examination of fiber structure. Actinin staining did not reveal major sarcomere assembly abnormalities (Fig. 2B). However, we found an excessive accumulation of LC3-positive autophagosomes in muscle fibers of mcoln1ab−/− fish (Fig. 2B). This is consistent with the finding of autophagic defects in human and mice.
To further confirm these results, we isolated muscle from wild-type, mcoln1a−/−, mcoln1b−/− and mcoln1ab−/− animals, and assessed them by immunoblot for the levels of lipidated LC3 (LC3-II), which marks autophagosomes. As seen in Figure 2C and 2D, LC3-II levels were significantly upregulated in mcoln1ab−/− muscle, corroborating impaired autophagosome degradation. Moreover, the fact that autophagic defects were observed only in the mcoln1ab−/− animals suggests that Mcoln1a and Mcoln1b can compensate for each other and have, at least, partially redundant functions in muscle.
In older (one year of age) animals, the accumulation of autophagosomes in muscle fibers was much more dramatic (Fig. 2E, upper panels) and in many fibers, muscle architecture was disrupted by massive clustering of autophagosomes (Fig. 2E, lower panels). This suggests that progressive autophagy dysfunction contributes to muscle abnormalities in mcoln1ab−/− animals.
Similar to what has been described in other lysosomal storage diseases associated with muscle dystrophy, such as Pompe disease (34), the distribution of microtubules in mcoln1ab−/− muscle fibers was strikingly altered when compared to wild-type; microtubules appeared shorter and less organized in mcoln1ab−/− animals (Supplementary Material, Fig. S1A). Given the importance of intact microtubules for the formation and movement of lysosomes and autophagosomes, it is likely that cytoskeleton abnormalities contribute to the autophagic build-up seen in mcoln1ab−/− fish. Microtubules also play an essential role in the transport and positioning of mitochondria, mediating their anchoring at sites of high energy demand (35). Interestingly, immunostaining of single muscle fibers with two different mitochondrial markers, cytochrome C and AlF, revealed severely altered mitochondrial positioning and increased mitochondrial fragmentation in the mutant fish (Fig. 3A and Supplementary Material, Fig. S1B). Although not as pronounced as in older fish, mitochondrial alterations were already visible in 4-month-old animals, thus suggesting that mitochondrial dysfunction is an early event in the MLIV pathology (Supplementary Material, Fig. S1C).
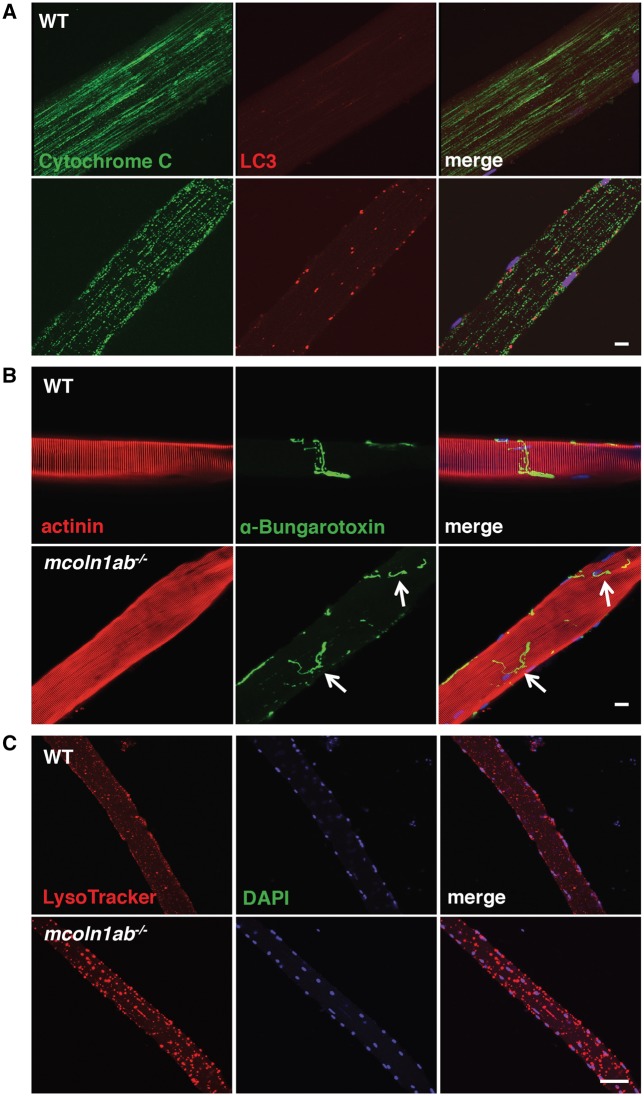
Figure 3.
Altered morphology and distribution of microtubule, mitochondria, and acethylcholine receptor in mcoln1 zebrafish adult mutant. (A) Co-immunostaining for Cytochrome C, LC3, and DAPI in single muscle fiber dissected from 12-month-old WT and mcoln1ab−/− zebrafish. (B) Co-staining for actinin, α-Bungarotoxin, and DAPI in single muscle fiber dissected from 12-month-old WT and mcoln1ab−/− zebrafish. Arrows mark the acetylcholine receptor distribution in mcoln1ab−/− zebrafish muscle fiber. Scale bar, 10 μm. (C) Co-staining for LysoTracker and DAPI in live single muscle fiber isolated from 12-month-old WT and mcoln1ab−/− zebrafish. All the images are representative of at least three independent experiments. Scale bar, 50 μm.
Significant pathological alterations at the neuromuscular junctions (NMJ) were detected in mcoln1ab−/− animals by immunolabeling single muscle fibers with α-bungarotoxin (Fig. 3B). In addition, lysosomal staining of cultured live muscle fibers from wild-type and mcoln1ab−/− adult fish with LysoTracker revealed accumulation of abnormally enlarged lysosomes in mcoln1ab−/− animals (Fig. 3C). In contrast, no noticeable skeletal defects in the axial skeleton were observed in mcoln1ab−/− fish (Supplementary Material, Fig. S2). Altogether, our data indicate that accumulation of autophagosomes and abnormal lysosomes, alterations in microtubule dynamics, mitochondrial fragmentation, and aberrant synaptic transmission at the NMJ are likely contributors to the motor dysfunction observed in the disease.
To confirm our observations, we performed transmission electron microscopy (TEM) of muscle samples isolated from 1-year-old mcoln1ab−/− fish (Fig. 4). The results clearly confirmed accumulation of autophagic debris in mutant animals, leading to disruption of the muscle fibers organization. Accumulation of storage material occurred both within the intermyofibrillar (Fig. 4A and B), as well as in the subsarcolemmal regions of muscle fibers (Fig. 4C and D). High magnification images revealed the presence of multi-lamellar structures (Fig. 4E), which is a very distinctive characteristic of MLIV cells, as well as abnormal mitochondria (Fig. 4F) and autophagosomes (Fig. 4G). Therefore, our results confirm that disruption of autophagy is a major contributor to the skeletal muscle pathology in mcoln1ab−/−.
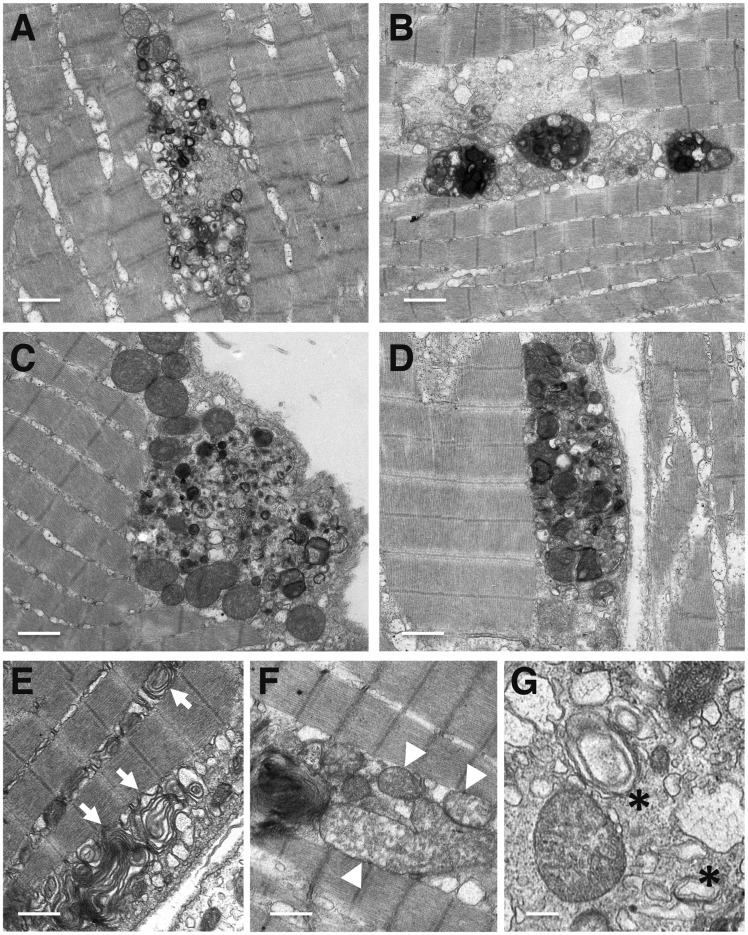
Figure 4.
Autophagy disruption in skeletal muscle of mcoln1ab−/− fish (A–G) Muscle samples of 12-month-old mcoln1ab−/− zebrafish were analyzed by transmission electron microscopy (TEM). Images show autophagic debris containing electro-dense inclusions, organelles and other cytoplasmic contents in mcoln1ab−/− skeletal muscle. Arrows in (E) indicate multi-lamellar structures; arrowheads in (F) point to abnormal mitochondria; and the asterisks in (G) denotes autophagosomes. Scale bar for (A–F), 1 μm. Scale bar for (G), 250 nm.
Retinal and corneal abnormalities in mcoln1 mutants
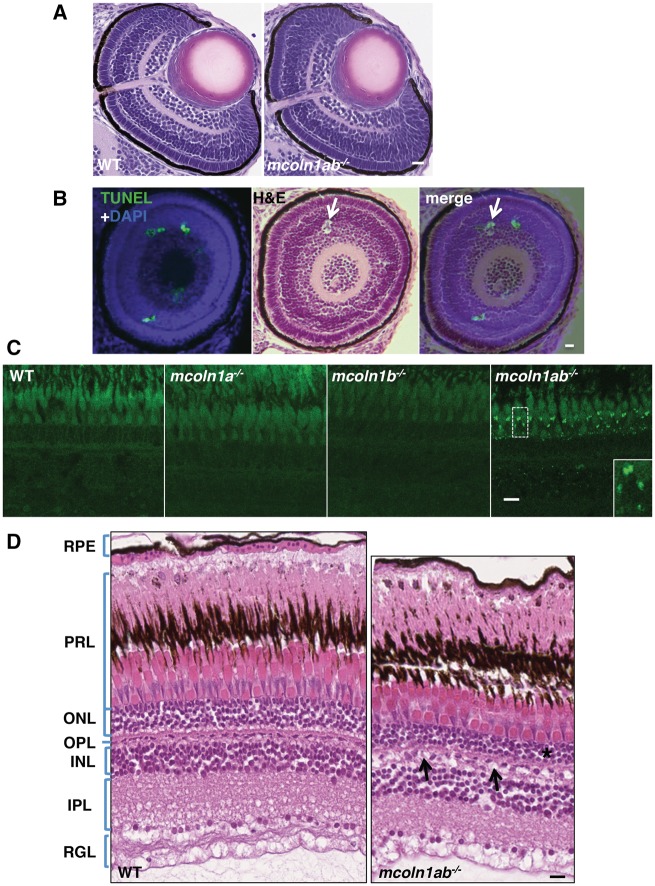
Next, we addressed eye development in mcoln1 mutants. Zebrafish is an ideal model system to study retinal development and regeneration of the damaged adult retina. The retinal structure starts developing at 32 hpf and embryos show visual responses by 3 dpf. Examination of hematoxylin and eosin (H&E) stained transverse (Fig. 5A) and parasagittal (Supplementary Material, Fig. S3) retinal sections did not reveal major differences in eye structure among wild-type, mcoln1a−/−, mcoln1b−/− and mcoln1ab−/− 3 dpf embryos. Immunostaining with markers for cone photoreceptors (zrp-1), rod photoreceptors (zrp-3), and amacrine cells (parvalbumin) confirmed the absence of apparent changes in morphology between wild-type and mcoln1 mutants (Supplementary Material, Fig. S4). The only noticeable alteration observed in the retina of mcoln1 mutants was the presence of small cell-devoid areas that resemble apoptotic bodies (Fig. 5B and Supplementary Material, Fig. S3). To confirm this observation, consecutive serial eye sections were stained with either H&E or TUNEL. As expected, we found TUNEL-positive cells in these areas, thus suggesting an increased rate of apoptosis in mcoln1ab−/− retinas (Fig. 5B). However, it is important to note that these apoptotic events were sporadic and did not seem to cause significant morphological retinal alterations.
Figure 5.
Eye pathology in mcoln1 mutant zebrafish at both embryonic and adult stages. (A) H&E staining on paraffin sections of 3 dpf WT and mcoln1ab−/− zebrafish embryos showing the eye structure. Scale bar, 20 μm. (B) TUNEL and DAPI co-staining on a 3dpf eye paraffin section of mcoln1ab−/− zebrafish is merged together with an H&E stained section that is 20 μm apart, the white arrow shows a TUNEL positive apoptotic body-like structure. Scale bar, 20 μm. (C) Auto-fluorescence of 8-month-old zebrafish retina on cryostat sections for WT, mcoln1a−/−, mcoln1b−/−, and mcoln1ab−/−. Insets show a twofold magnification of the indicated region. Scale bar, 5 μm. (D) H&E staining on paraffin sections of 8-month-old WT and mcoln1ab−/− zebrafish retina showing the structure of different retinal layers. The black asterisk indicates thinning of the ONL in mcoln1ab−/− zebrafish. The black arrows mark cell loss in the INL. RPE, Retinal Pigment Epithelium; PRL, Photoreceptor Layer; ONL, Outer Nuclear Layer; OPL, Outer Plexiform Layer; INL, Inner Nuclear Layer; IPL, Inner Plexiform Layer; RGL, Retinal Ganglion Layer. Scale bar, 10 μm. All the images are representative of at least three independent experiments.
A recent study has shown that the eye pathology in Mcoln1−/− mice resembles the ophthalmic features described in MLIV patients and includes a progressive accumulation of inclusion bodies in photoreceptors and ganglion cells that results in loss of photoreceptors and thinning of the outer nuclear layer (ONL) (36). Accordingly, we observed accumulation of auto-fluorescent structures in the retina of mcoln1ab−/− adult zebrafish (Fig. 5C). Auto-fluorescent structures are often seen in MLIV tissues and are considered to be aberrant lysosomes, which accumulate lipofuscins due to their reduced degradative capabilities. Accumulation of lipofuscins was more prominent in the photoreceptor layer, although storage inclusions were also observed in the inner nuclear layer (INL) and the inner plexiform layer (IPL). Similarly to what we observed in muscle, depletion of either Mcoln1a or Mcoln1b alone did not result in lipofuscin accumulation (Fig. 5C), suggesting some level of redundancy between the two mcoln1 genes in zebrafish retina. Also in agreement with what has been described in human and mice, we observed that the ONL was thinner in mcoln1ab−/− adult fish when compared with wild-type (Fig. 5D), which is indicative of increased retinal degeneration in mcoln1 mutants. Cell loss also seemed evident in the INL, further supporting the idea that Mcoln1 is important for retinal maintenance and function (Fig. 5D). Interestingly, we found increased Alcian Blue staining in the cornea of mcoln1a−/− and mucoln1ab−/− mutants, which is consistent with the accumulation of glycosaminoglycans in this tissue (Supplementary Material, Fig. S5). Therefore, our zebrafish model satisfactorily recapitulates the main retinal and corneal abnormalities described in MLIV patients.
Increased apoptosis in Mcoln1 deficient embryos
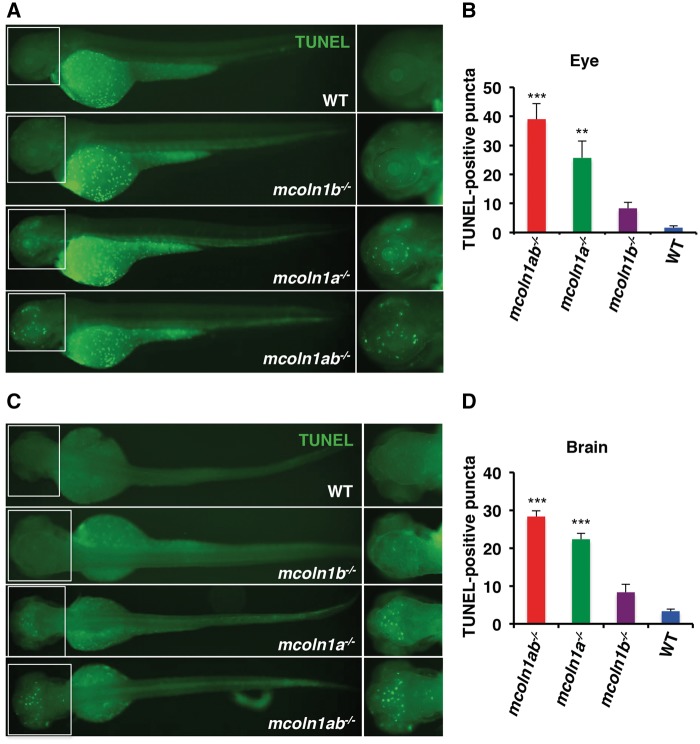
The presence of TUNEL-positive cells in the retina of 3 dpf mcoln1 deficient embryos prompted us to measure apoptosis in the whole embryo. To identify cells undergoing apoptosis, we performed whole-mount TUNEL staining of wild-type, mcon1a−/−, mcoln1b−/−, and mcoln1ab−/− 3 dpf embryos. The whole-mount analysis has the advantage of providing spatial information with regard to tissue specificity of apoptotic cells. Notably, TUNEL staining was detected in scattered cells within the brain and eye of mcoln1 mutants, while wild-type embryos showed significantly less apoptosis in these regions (Fig. 6A and C). Of note, the TUNEL-positive cells detected outside the brain and the eye are considered to be yolk debris. Quantification analysis revealed that apoptosis was more prominent in mcoln1ab−/− embryos (Fig. 6B and D). In addition, the number of TUNEL-positive cells was significantly higher in mcoln1a−/− compared to mcoln1b−/− embryos, suggesting a more prominent role of Mcoln1a in cell viability during embryonic development (Fig. 6B and D). These observations provide new and important insights into the pathogenesis of MLIV. First, the study uncovers a novel role of Mcoln1 in early embryonic development of the brain and retina; and second, the data suggest that some of the pathological consequences of Mcoln1 loss-of-function in these tissues may be independent of the progressive intralysosomal accumulation of undegraded material.
Figure 6.
Apoptosis in mcoln1 mutant embryos at 3 dpf. (A) Whole mount TUNEL staining in zebrafish embryos at 3 dpf for WT, mcoln1a−/−, mcoln1b−/−, and mcoln1ab−/− with the orientation to show the eye staining. (B) Quantification of TUNEL positive signals in the eyes of different genotypes. Data are shown as mean ± SD and represent 6 eyes counted. The data were analyzed using paired t test (***P < 0.001; **P < 0.01). (C) Whole mount TUNEL staining in zebrafish embryos at 3 dpf with different genotypes with the orientation to show the brain staining. (D) Quantification of TUNEL positive signals in the head of different genotypes. Data are shown as mean ± SD and represent 3 heads counted. The data were analyzed using paired t test (***P < 0.001).
Absence of mcoln1 affects hair cell viability and morphology
The accumulation of lipofuscin in mcoln1ab−/− photoreceptors, together with the reduced thinness of the ONL and the increased retinal apoptosis, suggest that sensory cells may be especially dependent on Mcoln1 function. To explore this possibility we turned to analysing sensory hair cells. Zebrafish possess sensory hair cells not only in the ear but also in the lateral line that extends along the head and the trunk (37). Lateral line hair cells are organized into distinct clusters named neuromasts that are regularly positioned along the axis of the animal (Fig. 7A). Each neuromast contains several mechanosensory hair cells that extend their stereocilia past the epidermis as a way to detect water movement. As such, neuromasts play an important role in mating, schooling and assisting fish to avoid predators (38). The neuromast hair cells in the lateral line are functionally and morphologically very similar to the hair cells in the inner ear of mammals and, in many cases, mutations that affect the function of the lateral line hair cells in fish cause deafness in humans (38).
Figure 7.
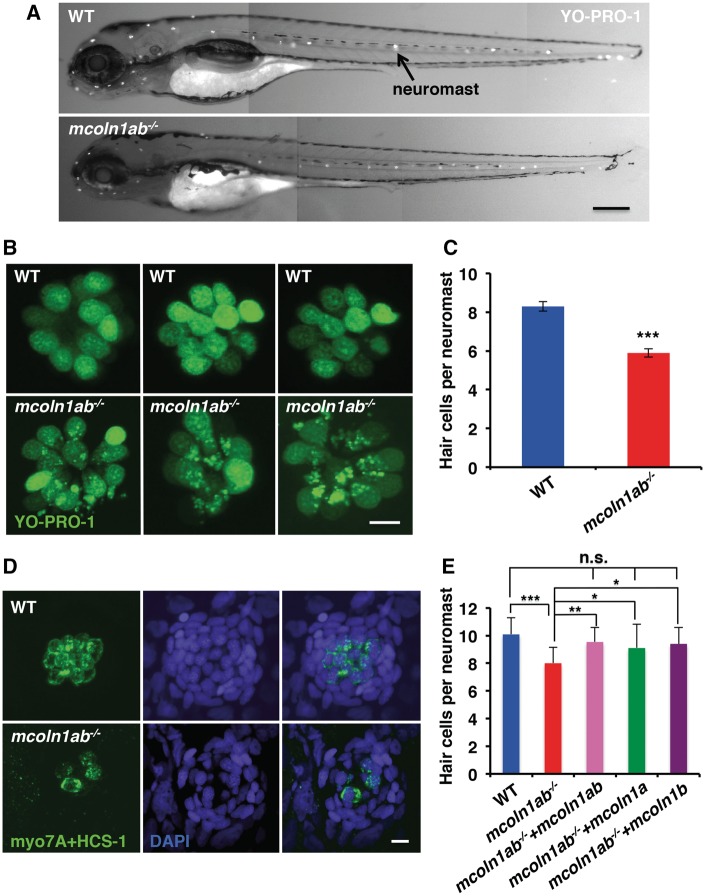
Hair cell viability and morphology in mcoln1 mutant larvae at 5 dpf. (A) Whole mount YO-PRO-1 staining in live zebrafish larvae at 5 dpf. The positive signal highlighted by the arrow indicates a typical neuromast. Scale bar, 250 μm. (B) A higher magnification of neuromast staining from (A) to show hair cell morphology. Scale bar, 5 μm. (C) Quantification of hair cell number per lateral line neuromast in YO-PRO-1 stained larvae for both WT and mcoln1ab−/− at 5 dpf. Data are shown as mean ± SD and represent 30 neuromast from 10 embryos counted per genotype in one representative experiment. Three independent experiments were performed. The data were analyzed using paired t test (***P < 0.001). (D) Co-immunostaining for myo7A/HCS-1 and DAPI for fixed WT and mcoln1ab−/− larvae at 5 dpf. Scale bar, 5 μm. All the images are representative of at least three independent experiments. (E) Reduced number of hair cells in mcoln1ab−/− head neuromast was rescued by mcoln1a and mcoln1b RNA co-injection, as well as by injection of either mcoln1a RNA or mcoln1b RNA 4 dpf. Data are shown as mean ± SD and represent 10 neuromasts from 10 different embryos per genotype. The result was analyzed using paired t test (***P < 0.001; **P < 0.01; *P < 0.05; n.s. not significant).
To test whether Mcoln1 is important for hair cell viability and function, we stained wild-type and mcoln1ab−/− 5 dpf larvae with YO-PRO-1, a vital dye that specifically accumulates in the nuclei of hair cells, allowing easy visualization of these hair cells in live animals. We found that the lateral line neuromasts of mcoln1ab−/− larvae showed a significant reduction in the total number of YO-PRO-1-positive hair cells, although overall morphology appeared normal (Fig. 7A and B). Less than 6 hair cells were found in mcoln1ab−/− animals (5.89 SEM 0.21, n = 30), whereas the average number of hair cells per neuromast was 8 in wild-type larvae (8.29 SEM 0.24, n = 30) (Fig. 7C). In addition to the nucleus, the YO-PRO-1 dye labeled what looks like cytosolic aggregates in mcoln1ab−/− cells (Fig. 7B). Given the ability of YO-PRO-1 to bind DNA, it is possible that these aggregates correspond to damaged mitochondria (see below). Hair cell numbers were not altered in mcoln1a−/− and mcoln1b−/− mutants, again implying a redundant function for the two proteins in different cell types (Supplementary Material Fig. S6A–C). Reduction of hair cells in mcoln1ab−/− larvae was confirmed by immunostaining with hair cell specific antibodies (Fig. 7D). Finally, a significant reduction in hair cell numbers was also observed in mcoln1ab−/− head neuromasts (Fig. 7E). Importantly, these defects were rescued by co-injection with mcoln1a and mcoln1b mRNAs, confirming the specificity of the observed phenotype (Fig. 7E). Moreover, rescue was also achieved by injection of either mcoln1a or mcoln1b mRNA, further suggesting that the two genes function redundantly (Fig. 7E).
In contrast to mammals, zebrafish hair cells have the ability to regenerate either spontaneously or following insult-driven hair cell loss (39,40). To address whether mcoln1ab−/− mutants have a hair cell regeneration defect, 5 dpf wild type and mcoln1ab−/− larvae were treated with 10 µM copper sulfate as a way to synchronize neuromast hair cell death (Supplementary Material, Fig. S7A). Hair cell regeneration was monitored by YO-PRO-1 staining at 7 dpf. The neuromasts of mcoln1ab−/− larvae were able to fully replace hair cells to the same level observed before the treatment (Supplementary Material, Fig. S7B and C), indicating that mcoln1 is not required for hair cell regeneration. Hair cells are also sensitive to antibiotics, such as neomycin. Treatment with low neomycin concentrations affected control and Mcoln1-depleted cells to the same extent, suggesting that the absence of Mcoln1 does not alter antibiotic susceptibility (Supplementary Material, Fig. S7D–F). Altogether, our results suggest that Mcoln1 does not affect hair cell regeneration or sensitivity to antibiotics but is important for hair cell homeostasis.
Massive accumulation of autophagosomes and autolysosomes in mcoln1ab−/− hair cells
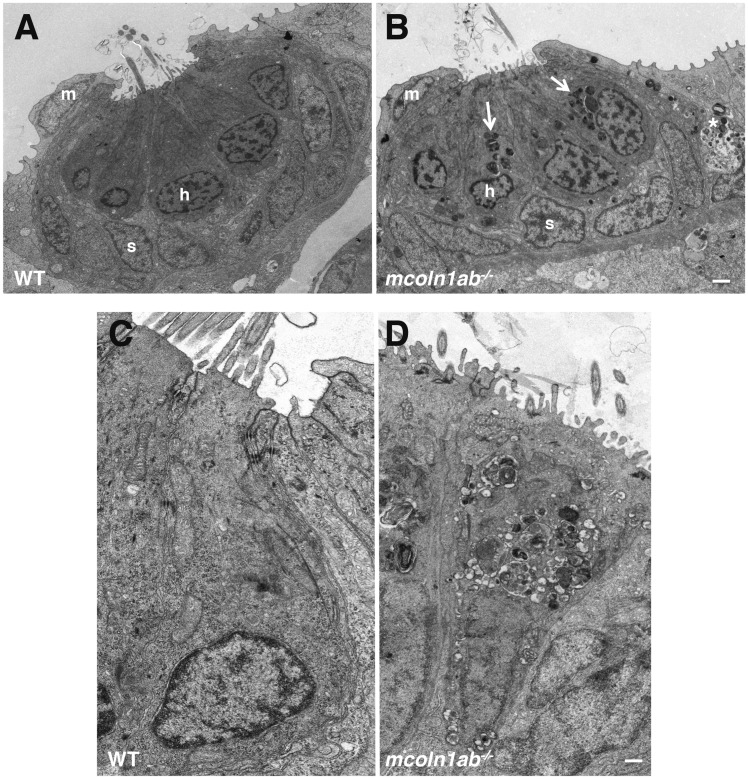
To further assess hair cell alterations in mcoln1ab−/− larvae, we performed transmission electron microscopy (TEM). Figure 8A shows TEM of a wild-type lateral line neuromast. The hair cells occupied the central region of the neuromast; they had an elongated pear-like shape with a large circular nucleus basally placed. The cytoplasm contained numerous elongated mitochondria with well-defined cristae (Fig. 8A and C). Note that the surface of hair cells showed kinocilia and stereocilia. Hair cells were surrounded by at least two other distinct cell populations, inner support cells underneath and adjacent to hair cells and mantle cells that form a ring encircling the entire neuromast (Fig. 8A and Supplementary Material, Fig. S8A). Support cells were elongated, with their nuclei occupying a significant portion of the cytoplasm, which appeared more electron‐lucid than that of hair cells. Support cells had processes surrounding the hair cells, thus preventing hair cell to hair cell contact (Fig. 8A).
Figure 8.
Ultrastructure of hair cell in mcoln1ab−/− larvae at 5 dpf. Transmission Electron Microscopy (TEM) for one neuromast in both WT (A) and mcoln1ab−/− (B) larvae at 5 dpf. m stands for mantle cell, h stands for hair cell, and s stands for support cell. White arrows mark the inclusion bodies in mcoln1ab−/− hair cells, and the asterisk marks inclusion bodies surrounding the hair cells. Scale bar, 1 μm. (C) and (D) Higher magnification showing more details of one of the hair cells in both WT (C) and mcoln1ab−/− (D). Scale bar, 200 nm.
The neuromast structure appeared profoundly disorganized in mcoln1ab−/− animals. The position of the hair cells was less well defined and the cells seem to contact each other. The shape of the nucleus was irregular and the cytosol contained a very substantial accumulation of electron-dense vacuoles (Fig. 8B and D).
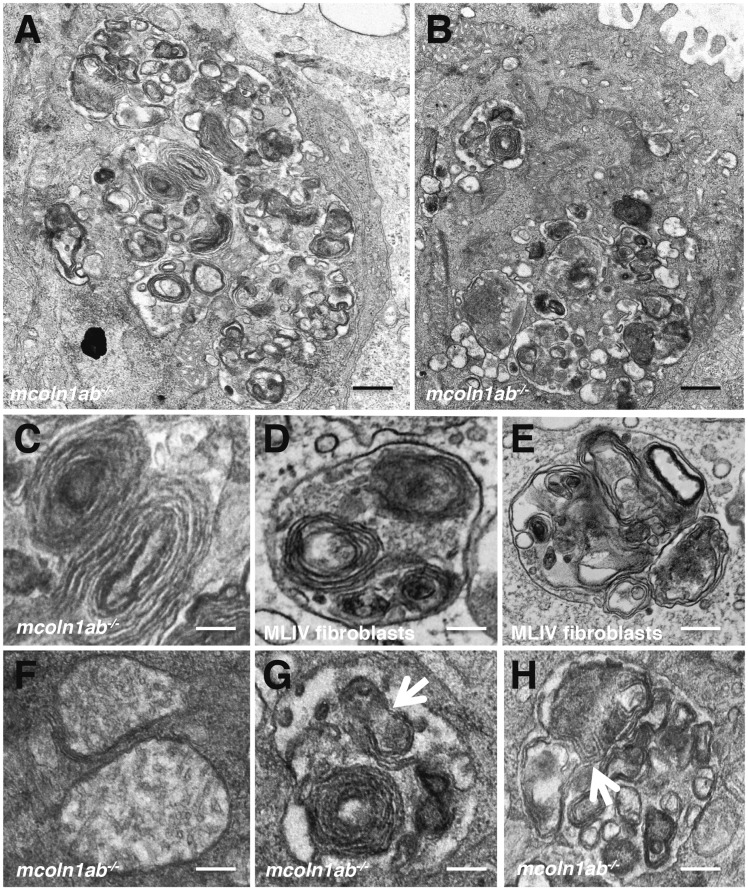
Higher magnification revealed that the cytosolic vacuoles likely correspond to autophagosomes and autolysosomes. Autophagic vacuoles contained electron-dense inclusions, organelles and other cytoplasmic contents (Fig. 9A and B). Notably, concentric multi-lamellar structures were often observed in mcoln1ab−/− hair cells (Fig. 9C). These remarkably resemble the characteristic lamellar structures previously described in MLIV cells and tissues (10,41) (Fig. 9D and E). Vesicles containing both, lamellar structures and partially degraded material, were also found and probably correspond to autolysosomes.
Figure 9.
Autophagic dysfunction in mcoln1ab−/− hair cells at 5 dpf. (A,B) Higher magnification of TEM showing autophagic vacuoles containing electro-dense inclusions, organelles and other cytoplasmic contents in mcoln1ab−/− hair cells. (C) Typical lamellar structure in mcoln1ab−/− hair cells. (D,E) Typical lamellar structures in MLIV fibroblasts. (F) Electron-transparent matrix and reduced cristae in mcoln1ab−/− hair cell mitochondria. (G,H) Autolysosomes containing partially degraded mitochondria inside in mcoln1ab−/− hair cells. Scale bar for (A) and (B), 200 nm. Scale bar for (C) to (H), 80 nm.
Accumulation of damaged mitochondria was common in mcoln1ab−/− hair cells. These organelles exhibited an electron-transparent matrix and reduced cristae (Fig. 9F). In many instances, partially degraded mitochondria were observed inside autophagosomes and autolysosomes (Fig. 9G and H). Accumulation of fragmented mitochondria was confirmed by DASPEI staining, a vital dye that preferentially labels hair cell mitochondria (Supplementary Material, Fig. S8B). While no co-localization between YO-PRO-1 and DASPEI was observed in WT cells, numerous cytosolic puncta contained both markers in mcoln1ab−/− cells (Supplementary Material, Fig. S8C). We suggest that these puncta indicate access of YO-PRO-1 to damaged mitochondria; however, we cannot rule out that the YO-PRO-1 dye gets non-specifically trapped in the inclusion bodies in mcoln1ab−/− cells. Mitochondria are a potent source of reactive oxygen species (ROS) and loss of mitochondrial integrity can lead to increased ROS production and its release into the cytoplasm (42). Accordingly, we found an increased production of mitochondrial ROS in mcoln1ab−/− cells (Supplementary Material, Fig. S8D).
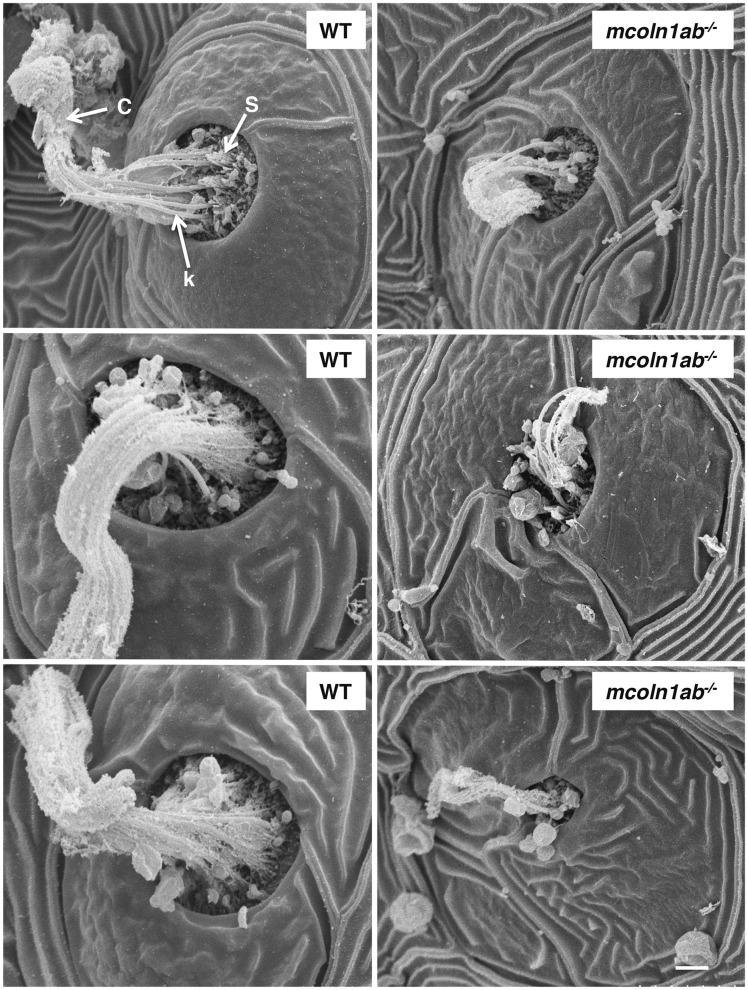
Finally, lateral line neuromasts were examined by scanning electron microscopy (SEM). At 5 dpf the center of WT neuromasts displayed hair bundles with short stereocilia and long kinocilia covered by a gelatinous-like material or cupula (Fig. 10). Notably, less kinocilia were detected in mcoln1ab−/− neuromasts and they were much shorter than those seen in the wildtype, further supporting the idea that Mcoln1 plays an important role in hair cell development and function (Fig. 10).
Figure 10.
Lateral line neuromast structure of WT and mcoln1ab−/− 5 dpf larvae by scanning electron microscopy (SEM). c, cupula, k, kinocilia; s, stereocilia. Scale bar, 10 μm.
Discussion
In this study, we report the generation of a new MLIV model in zebrafish. Our model replicates the main alterations previously reported in MLIV patients and Mcoln1 KO mice, including the presence of muscular and ophthalmologic abnormalities. The striking similarities in the structure and organization of muscle and eye between zebrafish and human, makes this model an excellent system to further uncover the molecular mechanisms of MLIV. Moreover, the ability to visualize these processes in zebrafish, an organism known for its facile large-scale in vivo drug screening, will likely provide unique opportunities for discovering new therapeutic approaches that restore muscle and retina function, with potential implications for clinical intervention.
Our model revealed novel insight into origins and progression of the MLIV pathology. MLIV patients present with motor disabilities. Whereas it has been suggested that these defects may be due to neurodegeneration and decreased synaptic transmission at the NMJ (25,43), a recent study reported that Mcoln1 KO mice develop primary, early-onset muscular dystrophy independent of neuronal degeneration (32). It was further suggested that defective muscle membrane repair might contribute to the muscle pathology observed in Mcoln1 KO mice (32). Here we used single muscle fiber analysis to address the pathological stages of progression of MLIV in skeletal muscle. Notably, we found a progressive accumulation of autophagosomes in muscle fibers, eventually leading to dystrophy in mcoln1 mutants. Therefore, similarly to what has been described in other lysosomal storage diseases, such as Pompe disease, the autophagic build-up resulting from defective lysosomal function greatly contributes to the muscle pathology in MLIV. It is important to note that the NMJ structure also appeared altered in mcoln1ab−/− muscle fibers, thus suggesting that a combination of reduced synaptic transmission and autophophagic build-up likely accounts for the motor impairment.
Whereas the progressive accumulation of autophagosomes and lipofuscins in muscle and eye are consistent with the idea of MLIV as a degenerative disease, we unexpectedly found that alterations in the mcoln1ab−/− mutants started very early during embryonic development. These alterations included increased apoptosis in eye and brain detected at 3 dpf as well as decreased hair cell viability detected a 5 dpf. Therefore, it is likely that both, defective prenatal development and progressive degeneration, are components of the MLIV pathology and contribute to the reduced lifespan observed in mcoln1ab−/− mutants.
An unexpected finding was the dramatic accumulation of inclusion bodies in hair cells. Similar to what we found in muscle and it has been reported in other lysosomal diseases, the accumulation of autophagosomes and autolysosomes is thought to be a gradual process that gets more severe as the animal gets older. However, in hair cells, we observed a severe autophagic build-up at 5 dpf. This suggests that hair cells might be particularly vulnerable to the absence of Mcoln1. Mechanosensory hair cells are densely packed with mitochondria to accommodate their high metabolic load. Mitochondria are also critical for the uptake of cytosolic calcium, thus preventing toxicity during the detection and transmission of acoustic information (44). Mitochondrial metabolism leads to the production of ROS and excessive production of ROS results in oxidative stress. Interestingly, hair cells seem to be especially susceptible to oxidative stress due to their lower content of antioxidants (45) and oxidative stress is prominent in the pathology of acquired hearing loss (46). Moreover, noise-induce elevation of calcium flow into mitochondria is sufficient to increase mitochondrial membrane permeability, increase ROS production and release into the cytoplasm, with subsequent hair cell damage and death (44). Recent evidence suggested that MCOLN1 is activated by oxidative stress (23). MCOLN1-mediated calcium release from lysosomes contributes to the activation of the transcription factor TFEB, which helps restoring cellular homeostasis by increasing lysosomal function and promoting autophagy-mediated elimination of damaged mitochondria (47). Therefore, we suggest that in cells lacking Mcoln1, defective mitophagy and TFEB activation in response to oxidative stress prevent restoration of cellular homeostasis, resulting in cell death. The accumulation of damaged mitochondria, ROS, and autophagosomes/autolysosomes observed in mcoln1ab−/− mutants agree with this model. Moreover, this could explain why neurons and other sensory cells, such as retinal cells, which must deal with elevation in cytosolic calcium and potential oxidative stress, show increased apoptosis in mcoln1−/− mutants.
It is important to note that MCOLN1 is a cation channel permeable not only to Ca2+ but also to Fe2+ and Zn2+ (6,48). In fact, lysosomal buildup of Fe2+ and Zn2+ has been reported in fibroblast isolated from MLIV patients and in the brain of Mcoln1−/− mice, and this buildup was linked to increased oxidative stress (49–51). Therefore, accumulation of heavy metals inside lysosomes in mcoln1ab−/− hair cells may also contribute to the observed increased ROS production, further leading to lysosomal dysfunction and autophagic clearance failure. Accordingly, it has been reported that heavy metals cause profound alterations on mechanosensory cells of the zebrafish lateral line (52). Altogether, our study indicates that Mcoln1 is required for the clearance of damaged mitochondria and removal of excess ROS and support a critical role of this protein mitigating oxidative stress in sensory cells.
Recent evidence has suggested that the auditory system may be compromised in several lysosomal storage diseases (53,54). This is usually an underreported manifestation due to the difficulty to perform audiological assessments in a pediatric neurologically compromised population. Our data showing that Mcoln1 plays an important role in hair cell viability suggest that future studies should monitor whether MLIV patients are at risk for developing progressive hearing loss.
In summary, our MLIV zebrafish model provides unique advantages to improve our understanding of the disease pathogenesis and ultimately develop new, effective therapies for the treatment of this devastating disorder.
Materials and Methods
Fish maintenance
Both adult and larval wild-type zebrafish (Danio rerio) were kept under a 14 h/10 h light/dark cycle at 28 °C. Larvae were staged according to days post fertilization and raised in E3 medium. All experiments were approved by the NHLBI Animal Care and Use Committee (protocol # H-0252), and all experiments were performed in accordance with relevant guidelines and regulations.
Zinc finger nucleases synthesis
CompoZrTM Custom Zinc Finger Nucleases (ZFNs) for mcoln1a were designed and synthesized by Sigma-Aldrich. The sequence for ZFN binding site/cut site is as following: GTCTTCCAGTATCTGTCTttggcaGACACGACAGTAGGC. mRNA for ZFN was generated by in vitro transcription using T7 RNA polymerase (Promega).
CRISPR/Cas9 RNA synthesis
pDR274 and pT3TS-nls-zCas9-nls constructs were kindly provided by Dr. Shao Jun Du from University of Maryland and Dr. Shawn Burgess from NHGRI (55,56). To make nls-zCas9-nls RNA, template DNA was linearized by XbaI digestion and purified using a QIAprep column (Qiagen). Capped nls-zCas9-nls RNA was synthesized using mMESSAGE mMACHINE T3 kit (Life Technologies) and purified using Centri-Sep columns (Princeton Separations). Oligo sequences for mcoln1b guide RNA were designed using ZiFiT (30,31). The oligo sequence for mcoln1b guide RNA: oligo1: TAGGCTCCGGGGACCTGAACGG; oligo 2: AAACCCGTTCAGGTCCCCGGAG. To make gRNA, pDR274 was linearized by BsaI digestion; the annealed oligos were inserted into the linearized vector. The vector with insertion was then digested by DraI. gRNA was generated by in vitro transcription using T7 RNA polymerase (Promega) or MAXIscript T7 kit (Life Technologies). After in vitro transcription, gRNA was purified using Centri-Sep columns. The size and quality of resulting gRNA was checked by electrophoresis through a 2% (wt/vol) formaldehyde agarose gel.
Microinjection
ZFN mRNA for mcoln1a was injected into one-cell-stage zebrafish embryos. A mixture of gRNA for mcoln1b and nls-zCas9-nls RNA was injected directly into one-cell–stage embryos. Embryos were derived from the wild-type AB cross.
Mutant Line Screening
Ten injected embryos at 24-day post-fertilization (dpf) from either ZFN or CRISPR/Cas9 injection were pooled. Genomic DNA was extracted using Extract-N-Amp Tissue PCR kit (Sigma-Aldrich). PCR product was sequenced to check whether mutations were created. To confirm germ line transmission, the F0 founders were crossed to WT and 10 embryos from the cross were pooled for DNA extraction: PCR product for the specific target sequence was cloned to pGEM-T vector (Promega) to identify mutations by single colony sequencing.
Primers used for mcoln1a genotyping:
M1-Ex3-F: CAGATGACGCACTCGCTGTTTACACACAGAG;
M1-Ex4-R: GATGAAGGTGTCGTTAGCCGGGTCGATCC.
Primers used for mcoln1b genotyping:
m1.2-t1-geno-F1: TTAGAGAAGGAGCGTCTGCTG;
m1.2-t1-geno-R1: GGCTTTCCCCATCTGTTTTT.
Total RNA extraction and cDNA synthesis
Ten zebrafish embryos at 24 hpf were euthanized and homogenized. Total RNA was extracted using TRIzol reagent (Thermo fisher scientific, NY). cDNA synthesis was carried out with the First-Strand cDNA synthesis kit (Thermo fisher scientific, NY) using 1–5 μg of total RNA as a template.
Primers used for mcoln1a cDNA cloning and sequencing:
m1.1-F1: GATGACGCACTCGCTGTTTA;
m1.1-R1: GCCAGATCAAACAACACACG
Primers used for mcoln1b cDNA cloning and sequencing:
m1.2-F1: AGATGGTGGTGGCGTTTAAG
m1.2-R1: CCAGACACATTGGGGTCTCT
TUNEL staining
TUNEL staining was conducted on both whole mount zebrafish embryos and cryopreserved tissue sections at 3 dpf using TUNEL Apoptosis Detection Kit (GenScript Cat. No. L00301) according to the manufacturer’s protocol. For the whole mount staining at 3 dpf, the embryos were treated with 0.1% (1 mg/ml) collagenase (Sigma C-9891) for 75 min at room temperature before the staining.
Whole mount immunostaining in 3 dpf embryos
Zebrafish embryos at 3 dpf were fixed with 4% paraformaldehyde (PFA) at room temperature for 1 h and washed in PBS with 0.1% TWEEN 20 (PBST). Fixed embryos were treated with 0.1% (1 mg/ml) collagenase for 75 min, washed three times with PBST, and treated with cold acetone at −20 °C for 10 min. After washing in PBST, the embryos were incubated with blocking buffer: BDP (0.1%BSA + 1%DMSO in 1xPBS)+5% goat serum for 1 h. The blocking buffer was then replaced with primary antibody in BDP at 4 °C overnight. After washing in PBST, the embryos were incubated in BDP with secondary antibody at room temperature in the dark for 1 h. After washing in PBST, the heads of the stained embryos were removed, and the trunks were mounted with antifade with DAPI (Thermo Fisher Scientific) on the slides (Daigger Scientific).
Cryostat sectioning and immunostaining
Zebrafish embryos or adult zebrafish eyes were fixed in 4% paraformaldehyde for 1 h at room temperature, washed with PBST for half an hour, soaked in 30% sucrose, and embedded in O.C.T. (Tissue-Tek) on dry ice. Cryostat sections were cut at 10 µm in the cryostat using Leica CM3050 under −20 °C, and sections were put on Superfrost plus slides (Daigger Scientific).
For immunostaining, the sections on slides were air dried for 30 min at room temperature, and washed with PBST for 15 min. The slides were then incubated in blocking buffer (see above) for 40 min at room temperature. The immunostaining, washing and mounting procedures were as described for the whole mount immunostaining.
Paraffin sectioning and H&E staining
Adult zebrafish or zebrafish embryos were dissected and fixed in 10% neutral buffered formalin. After dehydration with serial increased concentration of ethanol, the tissue was embedded in hot paraffin. Paraffin blocks were sectioned at 5 μm and put on glass slides. For H&E staining, paraffin sections were deparaffinized by Xylene, and rehydrated by ethanol. The slides were then stained with Hematoxylin and Eosin Y and washed with tap water. After dehydration with ethanol, the slides were incubated in Xylene. Slides were mounted using Permount (Electron Microscopy Sciences).
Alcian blue staining
Slides with paraffin sections were deparaffinized in Xylene and rehydrated in distilled water. Then they were incubated in Alcian Blue Solution (Alcian blue in 3% acetic acid, pH 2.5) for 30 min. After washing with running tap water and dehydration with ethanol, the slides were incubated in Xylene to dehydrate before mounting.
Isolation of single muscle fibers and immunostaining
Adult zebrafish single muscle fibers were isolated as described (57) with some modifications. Briefly, adult zebrafish were euthanized and fixed in 2% paraformaldehyde in phosphate buffer for 1 h at room temperature. Skin was removed during the fixation. Muscle bundles were dissected after fixation, and fibers were then gently pulled apart from muscle bundles with fine tweezers in a puddle of 0.04% saponin in PBS. For immunostaining, 30–40 isolated single muscle fibers were placed in one well of a 24-well plate. Blocking, antibody incubation, washes, and mounting were as described above for the whole mount immunostaining.
Whole mount hair cell immunostaining in 5 dpf larvae
Zebrafish larvae at 5 dpf were fixed with 4% paraformaldehyde (PFA) at room temperature for 1 h and washed with PBST. Fixed larvae were then treated with 10μg/ml proteinase K for 20 min, re-fixed with 4% PFA for 20 min, and washed three times with PBST. The samples were treated with cold acetone at −20 °C for 10 min. After washing in PBST, the larvae were incubated with blocking buffer (see above) for 1 h. The antibody incubation and washes were similar to those for the whole mount immunostaining. Finally, the stained larvae were embedded in low melting agarose gel and mounted with antifade with DAPI (Thermo Fisher Scientific) on the top of the agarose gel, then covered with coverslips.
Western blot analysis
Adult zebrafish muscle tissue was homogenized in RIPA buffer (Thermo Fisher Scientific) with protease inhibitor cocktail (cOmplete Mini, EDTA-free, Roche) on ice for 30 min (vortexed every 5 min). 10 μl of buffer was used for 1 mg of tissue. Samples were analysed by SDS-PAGE (4–20% gradient gels, Thermo Fisher Scientific) under reducing conditions and transferred to nitrocellulose membrane (Thermo Fisher Scientific). Immunoblotting was done using LC3 antibody (Cell Signaling Technology, Cat. #4108) and Horseradish peroxidase-conjugated anti-rabbit antibody. HRP-chemiluminescence was developed using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). The protein band intensity was quantified using Image J.
Antibodies used for western blot and immunostaining
The following primary antibodies were used: LC3A/B antibody at 1:250 dilution for Western Blot, 1:500 for immunostaining (Cell Signaling Technology, Cat. #4108); MYO7A 138-1 (dilution 1:500) was deposited to the DSHB by Orten, D.J. (DSHB Hybridoma Product MYO7A 138-1); HCS-1 (dilution 1:50) was deposited to the DSHB by Corwin, J. (DSHB Hybridoma Product HCS-1); Alpha-actinin antibody at 1:1000 dilution (Clone EA-53, Sigma-Aldrich); zpr-1 and zpr-3 antibodies at 1:1000 dilution (Zebrafish International Resource Center, ZIRC); Parvalbumin antibody at 1:1000 dilution (Clone PARV-19, Sigma-Aldrich); alpha-tubulin antibody at 1:100 dilution, was deposited to the DSHB by Walsh, C. (DSHB Hybridoma Product AA4.3); Cytochrome C antibody at 1:500 dilution (Clone 6H2.B4, BD Biosciences); AIF antibody at 1:500 dilution (Cell Signaling Technology, Cat.#4642). The following secondary antibodies were used: anti-rabbit IgG, HRP-linked at 1:7000 dilution (Cell Signaling Technology, Cat.#7074); goat anti-Rabbit IgG (H + L), Alexa Fluor 488 at 1:1000 dilution (Thermo Fisher Scientific); goat anti-Mouse IgG (H + L), Alexa Fluor 568 at 1:1000 dilution (Thermo Fisher Scientific).
Scanning electron microscopy
5 dpf zebrafish larvae were fixed in 2.5% glutaraldehyde in 0.1 M Cacodylate buffer (pH 7.4) overnight at 4° C. The samples were washed three times in buffer, post fixed with 1% OsO4 for 2 h and washed three times in buffer. The tissue was stained en bloc with 1% uranyl acetate for 1 h and serially dehydrated in ethanol. The samples were critical point dried (Samdri-795, Tousimus, Rockville, MD), placed on carbon adhesive tape coated with 10nm gold in an EMS 575X sputter coater (Electron Microscopy Sciences, Hatfield, PA). The images were obtained on a Hitachi S3400-N1 SEM (Hitachi High Technologies, Pleasanton, CA).
Transmission electron microscopy
Zebrafish larvae at 5 dpf or muscle samples of 12-month-old animals were fixed for 48 h at 40 °C in 2% glutaraldehyde and 1% paraformaldehyde in 0.1M cacodylate buffer (pH 7.4) and washed with cacodylate buffer three times. The larvae were then fixed with 1% OsO4 for 2 h, washed again with 0.1 M cacodylate buffer three times, washed with water and placed in 1% uranyl acetate for 1 h. The pellets were subsequently serially dehydrated in ethanol and propylene oxide and embedded in EMBed 812 resin (Electron Microscopy Sciences, Hatfield, PA, USA). Thin sections, approx. 80 nm, were obtained by utilizing the Leica ultracut-UCT ultramicrotome (Leica, Deerfield, IL, USA), placed onto 300 mesh copper grids and stained with saturated uranyl acetate in 50% methanol and then with lead citrate. The grids were viewed in the JEM-1200EXII electron microscope (JEOL Ltd, Tokyo, Japan) at 80kV and images were recorded on the XR611M, mid mounted, 10.5Mpixel, CCD (charge-coupled device) camera (Advanced Microscopy Techniques Corp, Danvers, MA, USA).
Hair cell live staining and imaging
Zebrafish larvae at 5dpf were incubated with 1 μM YO-PRO-1 (DNA dye, Molecular Probes; excitation 491 nm, emission 509 nm) in E3 medium at 28 °C for 1 h, washed twice in E3 medium, anesthetized with MS222 (3-aminobenzoic acid ethyl ester, methanesulfonate salt; Sigma) diluted in embryo media, and the the number of hair cells per neuromast was counted. To check hair cell morphology, the stained larvae were anesthetized and embedded in low melting agarose gel for imaging (58). For YO-PRO-1 and DASPEI double labeling, the larvae were first stained with 1 μM YO-PRO-1 for 1 h prior to DASPEI staining (0.005% final concentration, 2-(4-(dimethylamino) styryl)-N-ethylpyridinium iodide; Molecular Probes, Eugene, OR) for 15 min. Following DASPEI labeling, larvae were washed twice with E3 medium and anesthetized with MS222 prior to embedding in low melting agarose gel for imaging.
Hair cell regeneration
5 dpf zebrafish larvae were treated with 10 μM of copper sulfate (Sigma) for 2 h at room temperature or with 10 μM or 20 μM neomycin for 30 min at room temperature. Larvae were allowed to recover for 48 h except when otherwise indicated and then used for hair cell staining and counting (59).
Micro-CT imaging
Micro-CT of zebrafish anatomy was performed with a SkyScan 1172 Micro Xray CT scanner (Micro Photonics, Inc. Allentown PA, USA, SkyScan, Kontich, Belgium) with the x-ray source (focal spot size, 4 micrometers, energy range 20–100 kV) biased at 60 kV/167 microamps. The images were acquired with a pixel size of 14.05 micrometers, with the camera to source distance of 283 mm and an object to source distance of 172 mm. Four hundred ninety-nine projections in each of 3 segments were acquired with an angular resolution of 0.4 degrees through 180 degrees rotation. Four frames were averaged for each projection radiograph with an exposure time of 295 ms per frame. The scan duration was approximately 25 min per segment, 1.2 h per samples. Tomographic images were reconstructed using vendor-supplied software based on the Feldkamp cone beam algorithm.
Microscopes and imaging
The images for H&E or Alcian blue staining were taken on either HAMAMATSU NANOZOOMER (Hamamatsu Photonics) or Leica DMB4000 Upright light Microscopy (Leica Biosystems). The images for whole mount TUNEL staining were taken on Zeiss SteREO Discovery V12 (Zeiss). Confocal fluorescence images were taken on Zeiss 510 or Zeiss 780 Confocal Microscope (Zeiss).
Single live muscle fiber isolation and staining
Single live myofibers from 12-month-old zebrafish were isolated as previously described (60). Briefly, muscles were digested in 0.2% (w/v) collagenase type 1 (Sigma-Aldrich) in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 1mg/ml of BSA for 1 h at 37 °C in an atmosphere of 5% CO2. Individual fibers were released under a dissecting microscope by gentle pipetting; translucent myofibers (∼20 fibers) were plated in Matrigel-coated µ-slide 8-well chambers (Ibidi GmbH, Martinsried, Germany). The fibers were incubated in growth medium [10% FBS, 0.1% Chicken Embryo Extract (CEE) in DMEM overnight at 28 °C to let the fibers attach to the surface. The fibers were then used for labelling LysoTracker1 Red DND-99 (Life Technologies); 100 nM; 30 min, followed by three washes with PBS; Hoechst 33342 (Thermo Fisher Scientific) was added to the second wash.
RNA rescue
Zebrafish mcoln1a and mcoln1b were amplified by Phusion® High-Fidelity DNA Polymerase (NEW ENGLAND BioLabs) from 1 dpf or 3 dpf WT cDNA libraries, respectively. Primers used: for mcoln1a: m1a-CDS-F1: atggccgcagtggaccccaataa; m1a-CDS-R1: gttgacaaggaggacgtcctc; for mcoln1b: m1b-CDS-F1: atggcgagctcttttcacaga; m1b-CDS-R1: aagcagcagaagagcgagcag. PCR products for both mcoln1a or mcoln1b were cloned to pGEM-T vector. Sequencing result showed the T7 to SP6 orientation for the constructs. Not I was used to linearize the vector and mMESSAGE mMACHINE® T7 ULTRA Transcription Kit (Thermo Fisher Scientific) was used to make ARCA-capped RNA. 200 pg of RNA for either mcoln1a or mcoln1b, or co-injection of 100 pg RNA for each was used for micro injection.
Statistical Analysis
The relative quantification of LC3II/I level, TUNEL positive signals and hair cell numbers from different genotypes were compared by student t test. All tests were two-tailed and P-values of P < 0.05 were considered significant. Kaplan-Meier survival curve was used to show the survival rate of each genotype group at 0, 2, 4, 6 and 8 months. Differences between groups were calculated using the log-rank test, P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We are grateful to Dr. Zu-xi Yu from NHLBI pathology core and Dr. Xufeng Wu from NHLBI microscopy core for processing the samples and microscopy help. We thank Raman Sood from the National Human Genome Research Institute (NHGRI) zebrafish core and Charles River contractors for zebrafish line maintenance and daily care. We thank Kade Pettie from NHGRI for assistance with hair cell phenotype analysis. We also thank Dr. Danielle Donahue and Dr. Brenda Klaunberg from MIF for their help with the microCT data analysis. Finally, we thank Dr. Milton English from National Eye Institute (NEI) for his helpful comments.
Conflict of Interest statement. None declared.
Funding
Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI).
Supplementary Material
Supplementary Material is available at HMG online.
References
- 1. Amir N., Zlotogora J., Bach G. (1987) Mucolipidosis type IV: clinical spectrum and natural history. Pediatrics, 79, 953–959. [PubMed] [Google Scholar]
- 2. Wakabayashi K., Gustafson A.M., Sidransky E., Goldin E. (2011) Mucolipidosis type IV: an update. Mol. Genet. Metab., 104, 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bach G. (2001) Mucolipidosis type IV. Mol. Genet. Metab., 73, 197–203. [DOI] [PubMed] [Google Scholar]
- 4. Sun M., Goldin E., Stahl S., Falardeau J.L., Kennedy J.C., Acierno J.S. Jr., Bove C., Kaneski C.R., Nagle J., Bromley M.C.. et al. (2000) Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet., 9, 2471–2478. [DOI] [PubMed] [Google Scholar]
- 5. Altarescu G., Sun M., Moore D.F., Smith J.A., Wiggs E.A., Solomon B.I., Patronas N.J., Frei K.P., Gupta S., Kaneski C.R.. et al. (2002) The neurogenetics of mucolipidosis type IV. Neurology, 59, 306–313. [DOI] [PubMed] [Google Scholar]
- 6. Dong X.P., Cheng X., Mills E., Delling M., Wang F., Kurz T., Xu H. (2008) The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature, 455, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Puertollano R., Kiselyov K. (2009) TRPMLs: in sickness and in health. Am. J. Physiol. Renal Physiol., 296, F1245–F1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M.. et al. (2010) PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun., 1, 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eichelsdoerfer J.L., Evans J.A., Slaugenhaupt S.A., Cuajungco M.P. (2010) Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J. Biol. Chem., 285, 34304–34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vergarajauregui S., Connelly P.S., Daniels M.P., Puertollano R. (2008) Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet., 17, 2723–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curcio-Morelli C., Charles F.A., Micsenyi M.C., Cao Y., Venugopal B., Browning M.F., Dobrenis K., Cotman S.L., Walkley S.U., Slaugenhaupt S.A. (2010) Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol. Dis., 40, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samie M., Wang X., Zhang X., Goschka A., Li X., Cheng X., Gregg E., Azar M., Zhuo Y., Garrity A.G.. et al. (2013) A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell, 26, 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LaPlante J.M., Sun M., Falardeau J., Dai D., Brown E.M., Slaugenhaupt S.A., Vassilev P.M. (2006) Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab., 89, 339–348. [DOI] [PubMed] [Google Scholar]
- 14. Medina D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M.. et al. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell, 21, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith J.A., Chan C.C., Goldin E., Schiffmann R. (2002) Noninvasive diagnosis and ophthalmic features of mucolipidosis type IV. Ophthalmology, 109, 588–594. [DOI] [PubMed] [Google Scholar]
- 16. Bonavita S., Virta A., Jeffries N., Goldin E., Tedeschi G., Schiffmann R. (2003) Diffuse neuroaxonal involvement in mucolipidosis IV as assessed by proton magnetic resonance spectroscopic imaging. J. Child Neurol., 18, 443–449. [DOI] [PubMed] [Google Scholar]
- 17. Schiffmann R., Dwyer N.K., Lubensky I.A., Tsokos M., Sutliff V.E., Latimer J.S., Frei K.P., Brady R.O., Barton N.W., Blanchette-Mackie E.J.. et al. (1998) Constitutive achlorhydria in mucolipidosis type IV. Proc. Natl Acad. Sci. U S A, 95, 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chitayat D., Meunier C.M., Hodgkinson K.A., Silver K., Flanders M., Anderson I.J., Little J.M., Whiteman D.A., Carpenter S. (1991) Mucolipidosis type IV: clinical manifestations and natural history. Am. J. Med. Genet., 41, 313–318. [DOI] [PubMed] [Google Scholar]
- 19. Frei K.P., Patronas N.J., Crutchfield K.E., Altarescu G., Schiffmann R. (1998) Mucolipidosis type IV: characteristic MRI findings. Neurology, 51, 565–569. [DOI] [PubMed] [Google Scholar]
- 20. Berman E.R., Livni N., Shapira E., Merin S., Levij I.S. (1974) Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J. Pediatr., 84, 519–526. [DOI] [PubMed] [Google Scholar]
- 21. Soyombo A.A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. (2006) TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem., 281, 7294–7301. [DOI] [PubMed] [Google Scholar]
- 22. Jennings J.J. Jr., Zhu J.H., Rbaibi Y., Luo X., Chu C.T., Kiselyov K. (2006) Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem., 281, 39041–39050. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X., Cheng X., Yu L., Yang J., Calvo R., Patnaik S., Hu X., Gao Q., Yang M., Lawas M.. et al. (2016) MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun., 7, 12109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fares H., Greenwald I. (2001) Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet., 28, 64–68. [DOI] [PubMed] [Google Scholar]
- 25. Venkatachalam K., Long A.A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. (2008) Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell, 135, 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venugopal B., Browning M.F., Curcio-Morelli C., Varro A., Michaud N., Nanthakumar N., Walkley S.U., Pickel J., Slaugenhaupt S.A. (2007) Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. Am. J. Hum. Genet., 81, 1070–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benini A., Bozzato A., Mantovanelli S., Calvarini L., Giacopuzzi E., Bresciani R., Moleri S., Zizioli D., Beltrame M., Borsani G. (2013) Characterization and expression analysis of mcoln1.1 and mcoln1.2, the putative zebrafish co-orthologs of the gene responsible for human mucolipidosis type IV. Int. J. Dev. Biol., 57, 85–93. [DOI] [PubMed] [Google Scholar]
- 28. Vergarajauregui S., Puertollano R. (2006) Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic, 7, 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doyon Y., McCammon J.M., Miller J.C., Faraji F., Ngo C., Katibah G.E., Amora R., Hocking T.D., Zhang L., Rebar E.J.. et al. (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol., 26, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sander J.D., Zaback P., Joung J.K., Voytas D.F., Dobbs D. (2007) Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res., 35, W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., Dobbs D. (2010) ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res., 38, W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng X., Zhang X., Gao Q., Ali Samie M., Azar M., Tsang W.L., Dong L., Sahoo N., Li X., Zhuo Y.. et al. (2014) The intracellular Ca(2)(+) channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nat. Med., 20, 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walker M.T., Montell C. (2016) Suppression of the motor deficit in a mucolipidosis type IV mouse model by bone marrow transplantation. Hum. Mol. Genet., 25, 2752–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shea L., Raben N. (2009) Autophagy in skeletal muscle: implications for Pompe disease. Int. J. Clin. Pharmacol. Ther., 47 Suppl 1, S42–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miragoli M., Sanchez-Alonso J.L., Bhargava A., Wright P.T., Sikkel M., Schobesberger S., Diakonov I., Novak P., Castaldi A., Cattaneo P.. et al. (2016) Microtubule-Dependent Mitochondria Alignment Regulates Calcium Release in Response to Nanomechanical Stimulus in Heart Myocytes. Cell Rep., 14, 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grishchuk Y., Stember K.G., Matsunaga A., Olivares A.M., Cruz N.M., King V.E., Humphrey D.M., Wang S.L., Muzikansky A., Betensky R.A.. et al. (2016) Retinal Dystrophy and Optic Nerve Pathology in the Mouse Model of Mucolipidosis IV. Am. J. Pathol., 186, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wada H., Kawakami K. (2015) Size control during organogenesis: Development of the lateral line organs in zebrafish. Dev. Growth Differ., 57, 169–178. [DOI] [PubMed] [Google Scholar]
- 38. Whitfield T.T. (2002) Zebrafish as a model for hearing and deafness. J. Neurobiol., 53, 157–171. [DOI] [PubMed] [Google Scholar]
- 39. Behra M., Bradsher J., Sougrat R., Gallardo V., Allende M.L., Burgess S.M. (2009) Phoenix is required for mechanosensory hair cell regeneration in the zebrafish lateral line. PLoS Genet., 5, e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piotrowski T., Baker C.V. (2014) The development of lateral line placodes: taking a broader view. Dev. Biol., 389, 68–81. [DOI] [PubMed] [Google Scholar]
- 41. Tellez-Nagel I., Rapin I., Iwamoto T., Johnson A.B., Norton W.T., Nitowsky H. (1976) Mucolipidosis IV. Clinical, ultrastructural, histochemical, and chemical studies of a case, including a brain biopsy. Arch. Neurol., 33, 828–835. [DOI] [PubMed] [Google Scholar]
- 42. Batandier C., Leverve X., Fontaine E. (2004) Opening of the mitochondrial permeability transition pore induces reactive oxygen species production at the level of the respiratory chain complex I. J. Biol. Chem., 279, 17197–17204. [DOI] [PubMed] [Google Scholar]
- 43. Grishchuk Y., Sri S., Rudinskiy N., Ma W., Stember K.G., Cottle M.W., Sapp E., Difiglia M., Muzikansky A., Betensky R.A.. et al. (2014) Behavioral deficits, early gliosis, dysmyelination and synaptic dysfunction in a mouse model of mucolipidosis IV. Acta Neuropathol. Commun., 2, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Esterberg R., Hailey D.W., Rubel E.W., Raible D.W. (2014) ER-mitochondrial calcium flow underlies vulnerability of mechanosensory hair cells to damage. J. Neurosci., 34, 9703–9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sha S.H., Taylor R., Forge A., Schacht J. (2001) Differential vulnerability of basal and apical hair cells is based on intrinsic susceptibility to free radicals. Hear. Res., 155, 1–8. [DOI] [PubMed] [Google Scholar]
- 46. Bottger E.C., Schacht J. (2013) The mitochondrion: a perpetrator of acquired hearing loss. Hear. Res., 303, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raben N., Puertollano R. (2016) TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Ann. Rev. Cell Dev. Biol., 32, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cuajungco M.P., Basilio L.C., Silva J., Hart T., Tringali J., Chen C.C., Biel M., Grimm C. (2014) Cellular zinc levels are modulated by TRPML1-TMEM163 interaction. Traffic, 15, 1247–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kukic I., Lee J.K., Coblentz J., Kelleher S.L., Kiselyov K. (2013) Zinc-dependent lysosomal enlargement in TRPML1-deficient cells involves MTF-1 transcription factor and ZnT4 (Slc30a4) transporter. Biochem. J., 451, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coblentz J., St Croix C., Kiselyov K. (2014) Loss of TRPML1 promotes production of reactive oxygen species: is oxidative damage a factor in mucolipidosis type IV? Biochem. J., 457, 361–368. [DOI] [PubMed] [Google Scholar]
- 51. Grishchuk Y., Pena K.A., Coblentz J., King V.E., Humphrey D.M., Wang S.L., Kiselyov K.I., Slaugenhaupt S.A. (2015) Impaired myelination and reduced brain ferric iron in the mouse model of mucolipidosis IV. Dis. Model Mech., 8, 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Froehlicher M., Liedtke A., Groh K.J., Neuhauss S.C., Segner H., Eggen R.I. (2009) Zebrafish (Danio rerio) neuromast: promising biological endpoint linking developmental and toxicological studies. Aquat. Toxicol., 95, 307–319. [DOI] [PubMed] [Google Scholar]
- 53. Wold S.M., Derkay C.S., Darrow D.H., Proud V. (2010) Role of the pediatric otolaryngologist in diagnosis and management of children with mucopolysaccharidoses. Int. J. Pediatr. Otorhinolaryngol., 74, 27–31. [DOI] [PubMed] [Google Scholar]
- 54. King K.A., Gordon-Salant S., Yanjanin N., Zalewski C., Houser A., Porter F.D., Brewer C.C. (2014) Auditory phenotype of Niemann-Pick disease, type C1. Ear. Hear., 35, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol., 31, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jao L.E., Wente S.R., Chen W. (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl Acad. Sci. U S A, 110, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raben N., Shea L., Hill V., Plotz P. (2009) Monitoring autophagy in lysosomal storage disorders. Methods Enzymol., 453, 417–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Owens K.N., Cunningham D.E., MacDonald G., Rubel E.W., Raible D.W., Pujol R. (2007) Ultrastructural analysis of aminoglycoside-induced hair cell death in the zebrafish lateral line reveals an early mitochondrial response. J. Comp. Neurol., 502, 522–543. [DOI] [PubMed] [Google Scholar]
- 59. Pei W., Huang S.C., Xu L., Pettie K., Ceci M.L., Sanchez M., Allende M.L., Burgess S.M. (2016) Loss of Mgat5a-mediated N-glycosylation stimulates regeneration in zebrafish. Cell Regen (Lond), 5, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anderson J.E., Wozniak A.C., Mizunoya W. (2012) Single muscle-fiber isolation and culture for cellular, molecular, pharmacological, and evolutionary studies. Methods Mol. Biol., 798, 85–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.