Abstract
Vitamin B12 is an important cofactor in one-carbon metabolism whose dysregulation is associated with various clinical conditions. Indians have a high prevalence of B12 deficiency but little is known about the genetic determinants of circulating B12 concentrations in Indians. We performed a genome-wide association study in 1001 healthy participants in the Pune Maternal Nutrition Study (PMNS), replication studies in 3418 individuals from other Indian cohorts and by meta-analysis identified new variants, rs3760775 (P = 1.2 × 10−23) and rs78060698 (P = 8.3 × 10−17) in FUT6 to be associated with circulating B12 concentrations. Although in-silico analysis replicated both variants in Europeans, differences in the effect allele frequency, effect size and the linkage disequilibrium structure of credible set variants with the reported variants suggest population-specific characteristics in this region. We replicated previously reported variants rs602662, rs601338 in FUT2, rs3760776, rs708686 in FUT6, rs34324219 in TCN1 (all P < 5 × 10−8), rs1131603 in TCN2 (P = 3.4 × 10−5), rs12780845 in CUBN (P = 3.0 × 10−3) and rs2270655 in MMAA (P = 2.0 × 10−3). Circulating B12 concentrations in the PMNS and Parthenon study showed a significant decline with increasing age (P < 0.001), however, the genetic contribution to B12 concentrations remained constant. Luciferase reporter and electrophoretic-mobility shift assay for the FUT6 variant rs78060698 using HepG2 cell line demonstrated strong allele-specific promoter and enhancer activity and differential binding of HNF4α, a key regulator of expression of various fucosyltransferases. Hence, the rs78060698 variant, through regulation of fucosylation may control intestinal host-microbial interaction which could influence B12 concentrations. Our results suggest that in addition to established genetic variants, population-specific variants are important in determining plasma B12 concentrations.
Introduction
Vitamin B12 (B12) is a water soluble vitamin essential for two important pathways, methylmalonyl-CoA synthesis and one-carbon metabolism (OCM), the latter being crucial in regulating key biological processes including DNA and protein synthesis, epigenetic regulation, and oxidative pathways (1–3). It acts as a cofactor for the ubiquitous reaction in OCM catalysed by methionine synthase which converts homocysteine to methionine using methyl group from 5-methyltetrahydrofolate and thus plays an important role in regulating homocysteine concentrations (2,4). Hyperhomocysteinemia is a risk factor for neural tube defects, fetal growth restriction, and cardiovascular diseases (1,2). Vitamin B12 cannot be synthesized in humans and is available only through food and the intestinal microbiota (1,5,6). Low dietary intake, defective absorption, changes in microbiota and genetic factors predispose to B12 deficiency (1,7). In the western world, pernicious anaemia is the commonest cause of B12 deficiency and leads to a severe clinical condition (8). On the other hand, a large number of apparently healthy Indians, both vegetarians and non-vegetarians show B12 deficiency due to low intake of animal origin foods (9–11). We have shown an association between low maternal B12 status and increased risk of neural tube defects, fetal growth restriction, neurocognitive developmental deficits, and increased insulin resistance in the offspring (12–14).
Several studies have demonstrated strong heritability of plasma B12 concentrations suggesting a significant genetic contribution (15). Consistent with these observations, genome-wide association studies (GWAS) in Europeans and Chinese have identified several loci associated with plasma B12 concentrations (16–20). Indians have a unique dietary, socio-cultural and genetic diversity that may influence B12 concentrations but the genetic contribution to plasma B12 concentrations has hardly been investigated. We conducted a GWAS of plasma B12 concentrations in 1001 healthy individuals of Indo-European origin from Western India and replicated top hits as well as previously reported loci in 3418 individuals of different ages and of both Indo-European and Dravidian ethnicity. The overall aim was to identify new signals, carry out fine mapping by defining a credible set of variants to prioritize possible causal variants and understand the molecular mechanism through which the variants influence B12 concentrations.
Results
Clinical and demographic details of the stage I study samples, parents of children in Pune Maternal Nutrition Study (PMNS) (21), are shown in Table 1. The average age of these parents was 36 years (range 23 to 56 years), 46.8% were men. Almost half were B12 deficient (47.4%; <148 pmol/l), only 2.2% were folate deficient (<7 nmol/l) and ∼57% were hyperhomocysteinemic (>15 mmol/l) (Table 1). In the replication cohorts, the adult group had a similar picture to the stage I individuals but the children had lower levels of B12 deficiency. In the pregnant women, a large percentage of Parthenon Study (PS) mothers (41.6%) were B12 deficient but only 15.6% of women in Mumbai Maternal Nutrition Project (MMNP) cohort showed B12 deficiency (Table 1) (22,23).
Table 1.
Demographic and clinical characteristics of stage I and stage II samples
| Characteristics | Stage I | Stage II |
||||
|---|---|---|---|---|---|---|
| GWAS | Adultsa | PMNS children | PS Children | PS Mothers | MMNP Mothers | |
| N (male/female) | 1001 (468/533) | 724 (346/378) | 690 (352/338) | 534 (263/271) | 481 | 989 |
| Age in years | 36.0 (5.2) | 37.8 (11.2) | 11.2 (1.3) | 5.00 (0.1) | 28.9 (4.2) | 25.8 (4.0) |
| Gestational age (weeks)b | NA | NA | NA | NA | 30 | 10.7 (2.2) |
| BMI in Kg/m2 | 21.0 (3.6) | 23.3 (4.7) | 14.7 (2.0) | 13.6 (1.1) | 23.6 (4.6) | 21.0 (3.9) |
| Plasma B12 in pmol/l | 175.3 (160.5) | 191.8 (145.4) | 207.1 (87.2) | 361.6 (175.6) | 185.3 (100.0) | 266.2 (184.6) |
| B12 deficiency (%) (<148 pmol/l) | 47.4 | 38.0 | 22.7 | 4.5 | 41.6 | 15.6 |
| Serum folate in nmol/l | 18.7 (10.9) | 18.6 (15.1) | 23.0 (10.7) | 20.4 (9.6) | 35.3 (19.6) | 40.7 (27.7) |
| Folate deficiency (%) (<7 nmol/l) | 2.2 | 7.4 | 0.7 | 0.4 | 4.6 | 1.5 |
| Plasma Homocysteine in umol/l | 21.9 (16.2) | 23.2 (17.4) | 13.0 (6.8) | 6.58 (1.7) | 6.4 (2.4) | NA |
| Hyperhomocysteinemia (%) (>15 umol/l) | 56.9 | 56.6 | 25.5 | 0.2 | 0.4 | NA |
Data are presented as mean (SD); GWAS, Genome Wide Association Study; individuals in stage 1 GWAS were parents of the PMNS cohort.
aAdults group comprised samples from the remaining parents of PMNS, parents of Pune Children Study (PCS), PCS children at 21 years follow-up, and individuals from Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) cohorts.
bGestational age in weeks represents the time point at which blood samples were collected from pregnant mothers in PS and MMNP cohorts. PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; NA, not applicable.
Identification of New Variants Associated with Plasma B12 Concentrations
The genome-wide SNP data were generated on 1122 subjects of Indo-European origin from Western India using Affymetrix SNP 6.0. Sixty and 48 individuals were excluded based on cryptic relatedness plus duplicated samples, or due to unavailable phenotype data, respectively (Supplementary Material, Fig. S1). Principal component analysis (PCA) of stage I samples showed no evidence of population stratification (Supplementary Material, Fig. S2). Thirteen and seven individuals respectively, with standard deviation (SD) ±4 of the first two components of PCA and extreme B12 values were removed, leaving 1001 individuals with complete genotype and phenotype data. The median genomic inflation factor was 1.028 and quantile-quantile (Q-Q) plot did not show any systemic bias in the association results (Supplementary Material, Fig. S2).
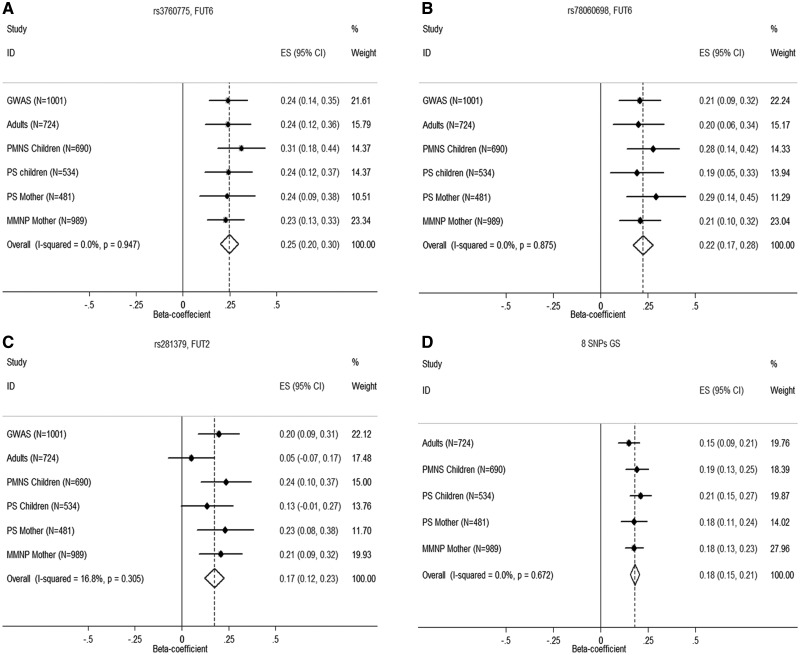
We did not find any SNP reaching GWAS significance (P = 5×10−8) in the stage I analysis but several SNPs were associated with plasma B12 concentrations at P < 10−4 (Supplementary Material, Fig. S3). Out of 41 SNPs taken for replication analysis in independent samples (Methods, SNP Selection and Replication Analysis), two new variants, rs3760775 and rs78060698, lying in a previously reported gene FUT6 were replicated and reached GWAS significance on meta-analysis (P = 1.2×10−23 and P = 8.3×10−17, respectively) (Table 2; Fig. 1A and B). Irrespective of age, gender, pregnancy status and ethnicity; the association of these two SNPs was replicated in all the groups (p-values range from 8.0×10−3 to 3.0×10−6; Table 2). We also observed statistically significant associations of previously reported FUT6 variants, rs3760776 (P = 4.5×10−14) and rs708686 (P = 5.7×10−15) (Table 2). Conditional analysis of rs3760776 and rs708686 suggested independent associations of the new variants rs3760775 (Pcond=2.6×10−7 & Pcond=2.4×10−6) and rs78060698 (Pcond=2.0×10−3 and Pcond=3.8×10−5) with B12 concentrations (Table 3). However, conditioning of rs3760775 abolished the association of rs3760776 (Pcond=0.086) and rs78060698 (Pcond=0.377) with B12 concentrations, indicating that rs3760775 may be the primary signal responsible for the observed association with other two SNPs. The variant rs780686 retained its association with B12 concentrations irrespective of other SNPs (Table 3). Additional adjustment for the first ten principal components as well as mixed linear model based association analysis to account for ancestry and population structure did not alter the results (Supplementary Material, Table S1)
Table 2.
Association analysis of stage I and II samples from independent cohorts and final meta-analysis
| Stage I |
Stage II |
Meta-analysis (N = 4419) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS (N = 1001) |
Adultsa (N = 724) |
PMNS Children (N = 690) |
PS Children (N = 534) |
PS Mothers (N = 481) |
MMNP Mothersb (N = 989) |
|||||||||||||||
| Status | CHR | SNP | BP | Gene | EA | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | I2 |
| New SNPs | 19 | rs3760775 | 5841356 | FUT6/3 | A | 0.24 | 6.0 × 10−6 | 0.24 | 9.9 × 10−5 | 0.31 | 2.9 × 10−6 | 0.24 | 2.1 × 10−4 | 0.24 | 2.0 × 10−3 | 0.23 | 1.0 × 10−5 | 0.25 | 1.2 × 10−23 | 0 |
| 19 | rs78060698 | 5832773 | FUT6 | A | 0.21 | 2.9 × 10−4 | 0.20 | 3.7 × 10−3 | 0.27 | 1.2 × 10−4 | 0.19 | 8.2 × 10−3 | 0.29 | 3.0 × 10−4 | 0.21 | 1.9 × 10−4 | 0.22 | 8.3 × 10−17 | 0 | |
| 19 | rs281379 | 49214274 | FUT2 | A | 0.20 | 4.6 × 10−4 | 0.05 | 0.42 | 0.24 | 4.5 × 10−4 | 0.13 | 0.06 | 0.23 | 2.8 × 10−3 | 0.21 | 4.7 × 10−4 | 0.17 | 3.3 × 10−11 | 16.8 | |
| Reported SNPs | 4 | rs2270655 | 146576418 | MMAA | C | −0.07 | 0.27 | 0.00 | 0.96 | −0.09 | 0.21 | −0.20 | 0.02 | −0.14 | 0.13 | −0.09 | 0.11 | −0.09 | 2.0 × 10−3 | 0 |
| 10 | rs12780845 | 17223244 | CUBN | G | 0.09 | 0.11 | 0.09 | 0.15 | 0.08 | 0.26 | 0.03 | 0.69 | 0.13 | 0.14 | 0.07 | 0.20 | 0.08 | 3.0 × 10−3 | 0 | |
| 11 | rs34324219 | 59623378 | TCN1 | A | NA | NA | −0.30 | 0.02 | −0.14 | 0.30 | −0.65 | 9.5 × 10−7 | −0.26 | 0.08 | −0.29 | 0.03 | −0.34 | 4.0 × 10−8 | 50.73 | |
| 11 | rs34528912 | 59631535 | TCN1 | T | NA | NA | −0.79 | 0.01 | 0.38 | 0.24 | −0.47 | 0.03 | −0.16 | 0.50 | −0.81 | 3.2 × 10−3 | −0.39 | 1.0 × 10−3 | 62.62 | |
| 11 | rs526934 | 59633493 | TCN1 | G | NA | NA | −0.07 | 0.27 | −0.10 | 0.12 | −0.16 | 0.02 | −0.16 | 0.05 | −0.03 | 0.56 | −0.09 | 1.0 × 10−3 | 0 | |
| 19 | rs3760776 | 5839746 | FUT6 | T | 0.10 | 0.06 | 0.23 | 4.4 × 10−4 | 0.30 | 3.3 × 10−6 | 0.18 | 6.5 × 10−3 | 0.33 | 3.0 × 10−5 | 0.11 | 0.03 | 0.19 | 4.5 × 10−14 | 55.19 | |
| 19 | rs708686 | 5840619 | FUT6 | T | NA | NA | 0.13 | 0.01 | 0.22 | 2.2 × 10−4 | 0.23 | 2.7 × 10−4 | 0.17 | 0.02 | 0.22 | 1.1 × 10−6 | 0.20 | 5.7 × 10−15 | 0 | |
| 19 | rs601338 | 49206674 | FUT2 | A | NA | NA | 0.05 | 0.46 | 0.25 | 3.8 × 10−5 | 0.18 | 4.3 × 10−3 | 0.29 | 2.2 × 10−3 | 0.16 | 1.4 × 10−3 | 0.17 | 6.7 × 10−10 | 47.01 | |
| 19 | rs602662 | 49206985 | FUT2 | A | NA | NA | 0.10 | 0.09 | 0.25 | 1.9 × 10−5 | 0.20 | 1.4 × 10−3 | 0.21 | 1.8 × 10−3 | 0.18 | 1.8 × 10−4 | 0.18 | 5.8 × 10−13 | 1.96 | |
| 19 | rs838133 | 49259529 | FUT2 | A | NA | NA | 0.05 | 0.48 | 0.27 | 2.0 × 10−4 | 0.06 | 0.45 | 0.31 | 5.7 × 10−4 | 0.10 | 0.08 | 0.14 | 8.6 × 10−6 | 60.95 | |
| 22 | rs1131603 | 31018975 | TCN2 | C | NA | NA | 0.43 | 0.04 | 0.05 | 0.81 | 0.44 | 0.05 | 0.77 | 0.01 | 0.63 | 3.4 × 10−3 | 0.41 | 3.4 × 10−5 | 29.65 | |
| 8 SNPs GSc | rs2270655, rs12780845, rs34324219, rs34528912, rs708686, rs3760775, rs602662, rs1131603 | 0.15 | 5.5 × 10−7 | 0.19 | 6.3 × 10−10 | 0.21 | 3.0 × 10−12 | 0.18 | 6.8 × 10−7 | 0.18 | 1.9 × 10−12 | 0.18 | 2.0 × 10−43 | 0 | ||||||
CHR, chromosome number; SNP, Single Nucleotide Polymorphism; BP, base position based on Build 37 hg19; EA, effect allele; PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; NA, not available; P, P-value; I2, heterogeneity. Beta is the effect size on an inverse normalized scale of residual standardized B12 concentrations adjusted for age and sex, fitted in linear regression with additive model of effect allele.
aAdults group comprised samples from the remaining parents of PMNS, parents of Pune Children Study (PCS), PCS children at 21 years follow-up, and individuals from Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) cohorts.
bFor MMNP additional adjustment for gestational age.
cGS represents the weighted genetic scores from eight SNPs selected based on their independent association in the conditional analysis (Table 3).
Figure 1.
Meta-analysis forest plots of association of plasma B12 concentrations with (A) rs3760775 in FUT6, (B) rs78060698 in FUT6, (C) rs281379 in FUT2 and (D) genetic score (GS) derived from eight independently associated SNPs (rs12780845 in CUBN, rs602662 in FUT2, rs708686 and rs3760775 in FUT6, rs2270655 in MMAA, rs34324219 and rs34528912 in TCN1 & rs1131603 in TCN2) identified from conditional analysis. GWAS, genome-wide association study; PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; ES, effect size.
Table 3.
Conditional analysis of various SNPs in FUT6, FUT2, and TCN1 associated with plasma B12 concentrations
| FUT6 SNPs | Unconditional |
Conditional |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs78060698 |
rs3760776 |
rs708686 |
rs3760775 |
|||||||
| Beta | P | Beta | Pcond | Beta | Pcond | Beta | Pcond | Beta | Pcond | |
| rs78060698 | 0.22 | 8.3 × 10−17 | NA | NA | 0.14 | 2.0 × 10−3 | 0.15 | 3.8 × 10−5 | 0.04 | 0.38 |
| rs3760776 | 0.19 | 4.5 × 10−14 | 0.12 | 5.0 × 10−3 | NA | NA | 0.11 | 3.0 × 10−3 | 0.07 | 0.09 |
| rs708686 | 0.2 | 5.7 × 10−15 | 0.13 | 4.8 × 10−6 | 0.14 | 1.9 × 10−5 | NA | NA | 0.09 | 9.0 × 10−3 |
| rs3760775 | 0.25 | 1.2 × 10−23 | 0.22 | 1.2 × 10−6 | 0.2 | 2.6 × 10−7 | 0.18 | 2.4 × 10−6 | NA | NA |
| FUT2 SNPs |
Unconditional |
Conditional |
||||||||
|
rs601338 |
rs602662 |
rs281379 |
rs838133 |
|||||||
| Beta | P | Beta | Pcond | Beta | Pcond | Beta | Pcond | Beta | Pcond | |
| rs601338 | 0.17 | 6.7 × 10−10 | NA | NA | −0.15 | 0.16 | 0.13 | 0.02 | 0.15 | 4.0 × 10−5 |
| rs602662 | 0.18 | 5.8 × 10−13 | 0.3 | 1.0 × 10−3 | NA | NA | 0.2 | 2.2 × 10−4 | 0.18 | 3.1 × 10−7 |
| rs281379 | 0.17 | 3.3 × 10−11 | 0.07 | 0.23 | −0.01 | 0.86 | NA | NA | 0.14 | 1.0 × 10-3 |
| rs838133 | 0.14 | 8.6 × 10−6 | 0.03 | 0.48 | 0 | 0.93 | 0.04 | 0.42 | NA | NA |
| TCN1 SNPs |
Unconditional |
Conditional |
||||||||
|
rs34324219 |
rs34528912 |
rs526934 |
||||||||
| Beta | P | Beta | Pcond | Beta | Pcond | Beta | Pcond | |||
| rs34324219 | 0.34 | 4.0 × 10−8 | NA | NA | 0.33 | 8.8 × 10−8 | 0.3 | 6.4 × 10−6 | ||
| rs34528912 | 0.39 | 1.0 × 10−3 | 0.3 | 0.01 | NA | NA | 0.44 | 3.0 × 10−4 | ||
| rs526934 | 0.09 | 1.0 × 10−3 | 0.06 | 0.07 | 0.1 | 1.0 × 10−3 | NA | NA | ||
Beta is the effect size accounted for allele with positive direction and on inverse normalized scale of residual standardized plasma B12 concentrations adjusted for age and sex along with the conditioning SNP on multivariate linear regression model. All the beta and P-values were from the meta-analysis of stage I and stage II analysis. NA, not applicable; P, P-value for unconditional analysis; Pcond, P-value for conditional analysis of respective SNPs; SNP, Single Nucleotide Polymorphism.
In-silico exploration of the published GWAS data from two European studies revealed that rs3760775 had a suggestive association with B12 concentrations in the Icelandic subjects (β = 0.07; P = 1.8×10−5; N = 23493) (Grarup, 2013), but not in the InCHIANTI (β = 0.16; P = 0.06; N = 1200) and in Baltimore Longitudinal Study of Aging (BLSA) (β=-0.21; P = 0.07; N = 641) (Tanaka, 2009) (17,20). The variant rs78060698 was also significantly associated with circulating B12 concentrations in the Icelandic subjects (β = 0.11; P = 1.1×10−7N = 23493); the data for rs78060698 was not available in other cohorts (Supplementary Material, Table S2). The effect allele frequency (EAF) of both variants in these studies was significantly lower than in the present study, further substantiated by the 1000 Genomes phase 3 data in different sub-populations (Supplementary Material, Table S3). The EAFs of all four SNPs in FUT6 were the highest in this study (EAF = 0.21-0.42) and this was especially noticeable for the new variants, rs3760775 and rs70860698 (0.27 and 0.21 vs 0.06 and 0.03 in CEU, respectively). The linkage disequilibrium (LD) pattern between the above SNPs was also quite variable in different populations as per the 1000 Genomes phase 3 data (Supplementary Material, Table S3). The r2 value for LD between rs3760775 and rs3760776 was 0.47 in the present study but 0.21 in CEU and 0.14 in CHB and the same between rs78060698 and rs3760776 was 0.54 in the present study and 0.39 and 0.66 in CEU and CHB populations respectively.
Fine Mapping of Genomic Regions, Credible Set Analysis and Functional Annotation
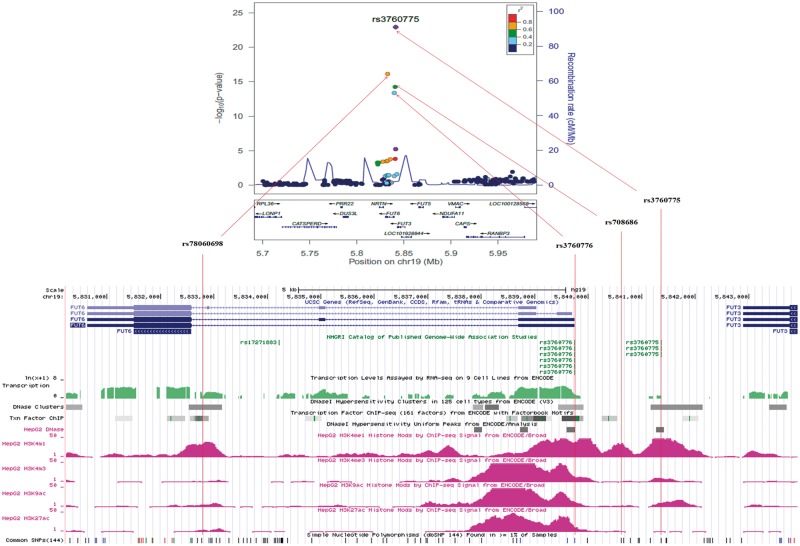
Using the Bayesian approach implemented in the SNPTEST, we constructed the 95% and 99% credible set of variants, adjusted for first ten principal components in the genomic region spanning 100 kb on either side of FUT6. This region encompassed FUT3, FUT5 and FUT6 cluster along with 9 other genes. Out of 170 variants in this region, 8 variants localized to the 95% credible set mapping to ∼20 kb region around the index SNP rs3760775, whereas 79 variants mapping in a 108 kb region around the index SNP formed the 99% credible set (Supplementary Material, Table S4). The index SNP rs3760775 was the strongest association signal accounting for 0.722 posterior probability. The remaining SNPs in the 95% and the 99% credible set explained 0.241 and 0.037 of the posterior probability respectively. The 8 SNPs in the 95% credible set were in LD with the index SNP rs3760775 (r2 range, 0.57-0.84) in South Asians in the 1000 Genomes phase 3 data (Supplementary Material, Fig. S4). The eight SNPs make two distinct LD blocks, one comprising the top two SNPs (rs3760775 and rs10409772) and the second with the remaining SNPs. The LD structure of these 8 credible set SNPs with three previously reported SNPs, rs3760776, rs798686 and rs778805 were different in South Asians, East Asians and Europeans (Supplementary Material, Fig. S5).
Except rs10409772 and rs12019136, all SNPs were enriched with functional regulatory marks (Supplementary Material, Fig. S6). The index SNP rs3760775 is positioned intergenic between 5’-upstream of FUT6 and 3’-downstream of FUT3 and marked with both promoter and enhancer histone marks. In the second LD block of 6 SNPs, rs12019136 and rs78060698 are located in the intronic region of FUT6, rs17855739 (E247K) and rs79744308 (A59T) are missense variants in FUT6 and Neurturin genes (NRTN) respectively and rs7250982 and rs8111600 are non-genic and located 5’-upstream of NRTN. The FUT6 intronic SNP, rs78060698 showed enrichment for enhancer histone marks and also altered the HNF4α motif. The missense FUT6 variant rs17855739 has been reported as a pathogenic variant causing FUT6 deficiency (24). The three variants in NRTN region were marked with enhancer histone marks in various tissues. None of the SNPs are represented as eQTLs in the GTEx portal database.
Replication of Reported Loci Associated with Plasma B12 Concentrations
In addition to replicating the association of the SNPs rs602662 (P = 5.8×10−13) and rs601338 in FUT2 (P = 6.7×10−10), we identified a new variant, rs281379 to be significantly associated with B12 concentrations (P = 3.3×10−11) (Table 2, Fig. 1C). However, rs281379 is in strong LD with the two previous SNPs (r2=0.75 in the present study, 0.92 in CEU and 1.0 in CHB). Conditional analysis of rs602662 and rs601338 confirmed that the association of rs281379 is explained by its strong LD with them (Table 3). Interestingly, both rs602662 and rs601338 deviated from Hardy Weinberg Equilibrium (HWE) in all the cohorts (P < 10−3). Cross-checking the genotype cluster (Supplementary Material, Fig. S7) and consistency of the genotypes using different genotyping platforms (including massarray-based genotyping using Sequenom and Sanger sequencing) in subsets of samples ruled out genotyping artefacts as possible reasons for deviation from HWE. We could not replicate the unique FUT2 SNP, rs1047781 identified in the Chinese population (P = 0.85) (Supplementary Material, Table S5). The fine mapping in 100kb upstream and downstream of FUT2 region showed close to 100 credible set variants in the 95% and 99% credible set with rs281379 as the index SNP .
We also replicated the association of previously reported transcobalamin 1 gene (TCN1) variants rs34324219 (P = 4.0×10−8), rs526934 and rs34528912 (P = 0.001 for both) with B12 concentrations (Table 2). Conditional analysis of the top SNP, rs34324219 on other two SNPs rs526934 and rs34528912 in this region confirmed that rs34528912 (P = 0.01) is an independent signal but not rs526934 (P = 0.07) (Table 3). While associations of other reported loci; cubulin (CUBN) rs12780845 (P = 0.003), methylmalonic aciduria (cobalamin deficiency) cblA type (MMAA) rs2270655 (P = 0.002) and transcobalamin 2 (TCN2) rs1131603 (P = 3.4×10−5) with B12 concentrations were replicated (Table 2), the same could not be confirmed for rs1047891 in carbamoyl-phosphate synthase 1 (CPS1), rs41281112 in citrate lyase beta like (CLYBL), rs1801222 in CUBN, rs117456053 in TCN1, rs2336573 in transcobalamin receptor (CD320) and rs5753231 in TCN2 (P > 0.01 for all) but the direction of the effect was similar to earlier reports (Supplementary Material, Table S5). Overall, we were able to replicate the association of 8 independent variants with plasma B12 concentrations. The association became more significant using the weighted genetic score (GS) calculated from the above 8 SNPs (P = 2.0×10−43) (Table 2, Fig. 1D).
Tracking Analysis of Plasma B12 Concentrations at Multiple Timepoints
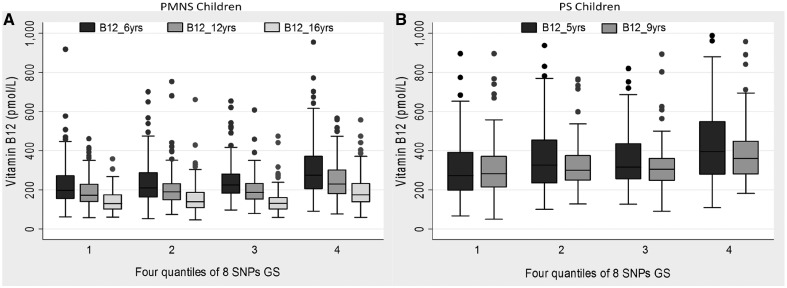
Comparison of plasma B12 concentrations in the PMNS children at 6yrs, 12yrs and 16yrs showed a statistically significant serial reduction in plasma B12 concentrations with progression of age (P < 0.001) (Table 4, Fig. 2). The significant association of the new FUT6 SNPs (rs3760775, rs78060698) and previously reported SNPs (rs3760776 in FUT6 and rs602662 in FUT2), as well as the GS calculated from 8 independently associated SNPs with B12 concentrations remained consistent at all three time points (P range from 5.2×10−7 to 3.2×10−8) (Table 4, Fig. 2). These observations were replicated in a similar analysis of the two time-point data (5 and 9.5 yrs) in PS children (P range from 1.0×10−3 to 3.0×10−7) (Table 4, Fig. 2). Individuals homozygous for the B12-raising alleles of the FUT6 and FUT2 variants had ∼1.4 times higher B12 concentrations than those homozygous for the other allele, at all ages in both cohorts. This was further substantiated using an 8 SNP-based GS which showed a similar magnitude of effect between the lowest and the highest quantiles of the GS. While this confirms the strong contribution of the identified SNPs on B12 concentrations, progressive fall in B12 concentration with age indicates environmental influence.
Table 4.
Age-wise tracking association analysis of new and reported SNPs with plasma B12 levels in PMNS and PS children
| Cohort | Follow-up age | Median B12 (pmol/l) | rs78060698 (A) |
rs3760775 (A) |
rs3760776 (T) |
rs602662 (A) |
8 SNPs GS |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | Beta | P | GG | GA | AA | Beta | P | CC | CT | TT | Beta | P | GG | GA | AA | Beta | P | 1st quantile | 4th quantile | Beta | P | |||
| PMNS Children | 6yrs | 224 | 217 | 233 | 323 | 35.3 | 4.1 × 10−5 | 211 | 235 | 295 | 35.0 | 1.2 × 10−5 | 216 | 231 | 301 | 35.5 | 4.6 × 10−6 | 218 | 217 | 299 | 26.4 | 1.2 × 10−4 | 197 | 274 | 24.7 | 3.2 × 10−8 |
| 12yrs | 188 | 188 | 191 | 278 | 24.9 | 7.8 × 10−5 | 183 | 195 | 258 | 28.7 | 9.5 × 10−7 | 183 | 191 | 233 | 24.7 | 1.4 × 10−5 | 183 | 187 | 238 | 21.0 | 4.1 × 10−5 | 172 | 230 | 17.8 | 5.6 × 10−8 | |
| 16yrs | 138 | 134 | 146 | 213 | 29.3 | 1.5 × 10−6 | 134 | 141 | 180 | 21.9 | 9.2 × 10−5 | 131 | 146 | 170 | 19.8 | 3.4 × 10−4 | 136 | 139 | 167 | 22.3 | 7.8 × 10−6 | 128 | 174 | 15.7 | 5.2 × 10−7 | |
| PS Children | 5yrs | 322 | 311 | 320 | 443 | 41.0 | 1.0 × 10−3 | 310 | 317 | 442 | 48.8 | 1.4 × 10−5 | 310 | 318 | 433 | 39.3 | 1.0 × 10−3 | 308 | 324 | 418 | 38.7 | 3.4 × 10−4 | 272 | 396 | 35.1 | 3.0 × 10−7 |
| 9yrs | 314 | 306 | 305 | 430 | 39.3 | 3.0 × 10−3 | 314 | 296 | 424 | 33.8 | 6.0 × 10−3 | 306 | 295 | 419 | 31.6 | 0.01 | 294 | 313 | 359 | 39.5 | 1.0 × 10−3 | 283 | 362 | 25.5 | 1.0 × 10−3 | |
Tracking analysis of the new variants, rs78060698 and rs3760775 and previously reported SNPs rs3760776 in FUT6, rs602662 in FUT2 and genetic score calculated from eight independently associated SNPs, identified from conditional analysis. Alleles in parentheses represent the effect allele. Beta is the additive effect of the effect allele on plasma B12 levels adjusted for age and sex. PMNS; Pune Maternal Nutrition Study; PS, Parthenon Study; GS, weighted genetic score; SNP, Single Nucleotide Polymorphism. The values represented in the genotype columns of SNPs and 1st and 4th quantiles of 8 SNPs GS are median values of plasma B12 concentrations, P, P value.
Figure 2.
Trend of plasma B12 concentrations with increasing age in four quantiles derived from eight SNPs genetic score (GS) in (A) Pune Maternal Nutrition Study (PMNS) children and (B) Parthenon Study (PS) children. The horizontal midlines in each group indicate the median point.
Bioinformatics and Functional Study of New and Reported SNPs in FUT6
The region encompassing both the new variants, rs3760775 and rs78060698 showed DNase I hypersensitive sites as well as active histone modification marks, but only that around rs78060698 was enriched with strong transcription factor binding sites (Fig. 3 and Supplementary Material, Fig. S6). In-silico transcription factor binding analysis using JASPAR database demonstrated differential binding affinity of HNF4α and HNF4γ in the alternate allelic background of rs78060698. The binding score of HNF4α (15.554) for the B12 raising ‘A’ allele of rs78060698 was 1.18 fold higher against that of the ‘G’ allele (score = 13.20). We noted complete abolition of HNF4γ binding to the ‘G’ allele while the binding score for the ‘A’ allele was 13.973. But no allelic differences on transcription binding prediction was noted for rs3760775. Of the two previously reported SNPs, rs3760776 was positioned at the promoter region of FUT6, with strong enrichment of DNase I hypersensitive and transcription factor binding sites, but rs708686 was intergenic between 5’-FUT6 and 3’-FUT3 and not marked with DNase I hypersensitive or transcription factor binding sites (Fig. 3). No allele-specific differential binding of transcription factors were noted for either rs3760776 or rs780686 variants.
Figure 3.
Regional plot of FUT6 and functional regulatory marks. Genomic region of FUT6 after meta-analysis showing two new SNPs rs78060698 and rs3760775, and previously reported variants rs3760776 and rs708686 with functional regulatory mark of DNase I hypersensitive sites, transcription factor binding sites and active histone modification marks in UCSC genome browser. The r2 values in the regional plot were derived using hg19/1000 Genome Nov 2014 SAN Genome build and LD population in LocusZoom.
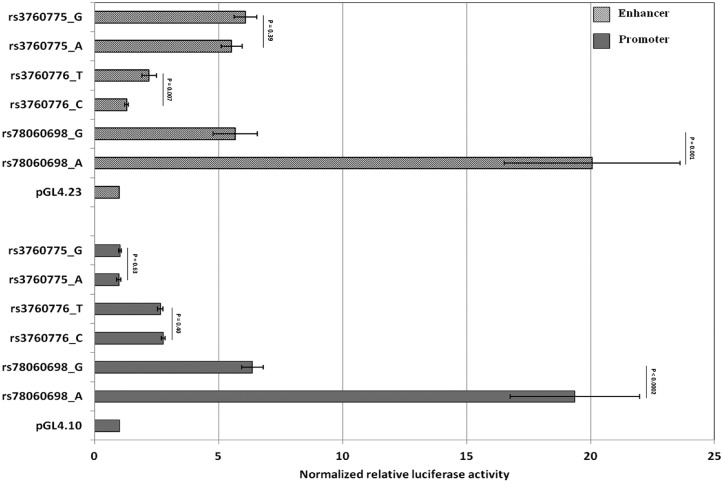
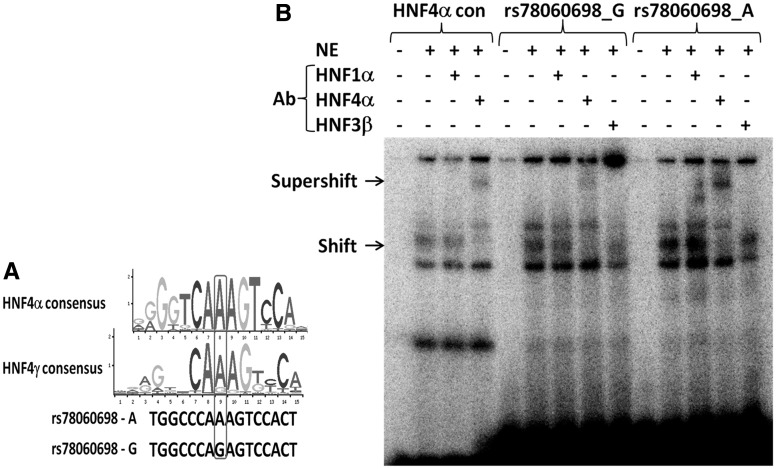
Observations from the bioinformatics prediction were further validated through pGL4 luciferase reporter assay in HepG2 cell line. The region encompassing both new variants, rs3760775 and rs78060698 showed enhancer activity compared to the basic enhancer vector pGL4.23, but allele-specific differences were noticed only for rs78060698 (Fig. 4). Compared to the basal promoter construct pGL4.23, the ‘A’ and ‘G’ alleles at rs78060698 showed 20.1 times and 5.7 times enhancer activity respectively, indicating a 3.5 fold higher enhancer activity for the B12 raising ‘A’ allele (P = 1.1×10−3) (Fig. 4). The rs78060698 region also showed strong promoter activity with the B12 raising ‘A’ allele having ∼3.0 times higher promoter activity than the ‘G’ allele (P = 1.6×10−4). The ‘A’ allele had 19.4 times and ‘G’ allele had 6.4 times higher promoter activity compared to the basal pGL4.10 vector (Fig. 4). Further investigation by EMSA revealed relatively higher protein binding (shift) to the ‘A’ allele than the ‘G’ allele and shift pattern was comparable to the HNF4α consensus binding (Fig. 5A). Using HNF4α antibody super-shift assay, we identified a strong super-shift band for the ‘A’ allele compared to the ‘G’ allele, confirming that the allelic differential activity of enhancer and promoter was due to differential binding of HNF4α at alternate alleles of the FUT6 variant, rs78060698 (Fig. 5B).
Figure 4.
Functional analysis of SNPs, rs78060698, rs3760775 and rs3760776 in the FUT6 region by luciferase assay for promoter and enhancer activity. The Y-axis shows different constructs carrying the specific allele of each variant, and the X-axis represents normalized relative luciferase activity to the basal promoter pGL4.10 and enhancer pGL4.23 constructs. Except rs3760775 promoter constructs, each construct carrying the specific allele showed significant promoter or enhancer activity compared to respective basal constructs (P < 0.01).
Figure 5.
Consensus binding motif of HNF4α and HNF4γ and electrophoretic-mobility shift assay of rs78060698 alternate alleles. (A) Change in binding motif of HNF4α and HNF4γ due to A and G alleles at rs78060698. (B) Gel shift and super-shift of HNF4α consensus sequence, oligonucleiotide probes of A and G alleles of rs78060698. Arrows indicate the shift and supershift bands. NE, nuclear extract from HepG2 cells; HNF4α con; HNF4α consensus sequence; Ab, Antibody.
The region bearing the other new variant, rs3760775 demonstrated enhancer activity (∼ 6 times) compared to basic pGL4.23, but no significant allele-specific differences were noted (P = 0.39) (Fig. 4). Although, a significant promoter activity compared to the basic pGL4.10 (∼2.7 times) was noted for the previously reported SNP rs3760776, allele specific differences were absent (P = 0.40) (Fig. 4). However, the region showed weak enhancer activity with significant allelic differences; the vitamin B12 raising ‘T’ allele had 2.2 times and ‘C’ allele had 1.3 times higher enhancer activity compared to basal pGL4.23 thus, the B12-raising ‘T’ allele had 1.7 times the enhancer activity of the other allele (P = 0.007) (Fig. 4).
Discussion
In the first two-stage genome-wide association study of circulating vitamin B12 concentrations in the Indian population, we have made two significant observations. First, we identified GWAS-significant new variants, rs78060698 and rs3760775 in FUT6 gene and rs281379 in FUT2 gene and secondly, demonstrated a functional mechanism through which one of the new variant, rs78060698 in FUT6 may influence plasma B12 concentrations. These variants had similar effect size across ethnicity, age and gender, indicating that they are likely to play a causal role in determining B12 concentrations. This is in addition to replicating associations of many of the variants in 13 established plasma B12-associated loci.
Genome-wide association studies in Europeans and Chinese have made significant contributions towards understanding genetic determinants of B12 concentrations (16–20). However, the new FUT6 variants, rs3760775 and rs78060698 identified as independent signals through conditional analysis in this study were not reported by either of them, although we replicated the association of previously reported FUT6 variants, rs3760776 in Chinese and rs708686 in Europeans. While the new variants were replicated in-silico with a suggestive association in Icelandic subjects (Grarup, 2013), the association was not replicated in the Europeans from InCHIANTI and BLSA cohorts (Tanaka 2009) (17,20). We can only speculate the possible reasons. Both variants have significantly lower frequency of the effect allele and effect size compared to the present study, hence, they were not identified as primary GWAS signals in small sample size in the InCHIANTI and BLSA cohorts but reached significance in a study on a large number of Icelandic subjects. The allele frequencies of these four SNPs were highest in Indians, possibly providing adequate power to detect the association at GWAS significance. In addition, the LD structure of the credible set variants with the previously reported Chinese and European SNPs are different in South Asian, East Asian and European populations with the strongest overall correlation in South Asians. We speculate that the population specificity of these variants might be due to differences in effect allele frequency and the LD structure in the three populations (25–27). Replication of these SNPs in three different populations suggests that there might be shared common causal variants, either coding or regulatory variants in this region with unique lead variants and variable LD structure (28). In addition to these two SNPs, credible set and fine mapping analysis of the region identified 6 other variants, of which rs17855739, a missense FUT6 variant that shows strongest association in Europeans is known to cause FUT6 deficiency (24).
We also replicated associations of previously reported loci, especially those involved in B12 absorption and transport such as TCN1, TCN2, CUBN and MMAA, with similar direction of effect (16–20). Some of the reported variants could not be replicated, possibly due to the modest effect size or low power of the study samples and variability in LD structure. It is noteworthy that effect of genetic factors on B12 concentration was consistent despite the differences in age, gender and the presence or absence of physiological state like pregnancy and persisted despite a progressive fall in concentration with increasing age in two independent cohorts which have different ethnic and socioeconomic characteristics (21,22). These observations support a strong contribution of genetic factors in the determination of circulating B12 levels. Overall, the study provides evidence that there are common genetic determinants of vitamin B12 concentrations in different ethnic groups, although population-specific variants also exist and should be investigated for their causal role.
We investigated the possible molecular mechanism by which rs78060698 and rs3760775, and the earlier reported SNP rs3760776 might influence B12 concentrations. True to the bioinformatic prediction, all three FUT6 SNPs showed promoter and/or enhancer activity but significant allele-specific differences were noted only for rs78060698. This variant, in alternate allelic states, also differentially influences the binding affinity of HNF4α and HNF4γ. HNF4α is a key regulator of FUT6 expression and its knockdown represses the expression of FUT6, and nearby genes FUT3 and FUT5 in HepG2 cell line (29,30). This cluster of FUT genes encodes the α1,3/4 fucosyltransferase family of proteins which add fucose to the GlcNAc moiety in the glycan chain (31). Fucose moiety in the intestinal tract has an important role in maintaining host-microbial interaction, microbial abundance and diversity (32–34). Thus, the rs78060698 variant might play a crucial role in maintaining the glycan structure and their metabolism thereby mediating intestinal host-microbial interaction leading to alteration of plasma B12 concentrations (35,36). The other possibility, of an influence of fucosyltransferases on B12 concentrations, may be mediated through the glycosylation of B12 binding proteins and their receptors including TCN1, GIF, CUBN and CD320 etc. (37–39). However, the mechanism through which the fucosyltransferases act upon these B12 binding proteins and their receptors is unclear and needs to be investigated further. Our observations add to the growing evidence of the existence of multiple functional regulatory variants in an association signal (40–42).
The three new variants in FUT6 and FUT2 together explained 3.85% and the eight independently associated SNPs explained 8.36% of the variability in plasma B12 concentrations, indicating a large undetermined proportion of the heritability of total vitamin B12 concentrations. Although we identified three new variants and replicated many previously reported variants, the study had some limitations. Our discovery stage sample size is relatively small and hence we may have failed to discover novel genetic regions in a population with a high prevalence of vitamin B12 deficiency. Secondly, a large proportion of low vitamin B12 concentrations in Indians might be contributed by various factors including undernutrition and strict vegetarian habits that might mask the effect of genetic factors on vitamin B12 concentrations.
In summary, this study reports new variants in a previously reported genomic region of FUT6 and stresses the importance of population-specific genetic variants in the background of common genetic determinants of vitamin B12 concentrations. We also demonstrate their possible functional mechanism through differential binding with HNF4α and thus regulation of expressions of FUT6 and other fucosyltransferases. Further functional analysis of other reported variants and their interaction with environmental factors will help elucidate the complex interplay between fucosyltransferases, glycan metabolism, host-microbe interaction and determination of B12 concentrations.
Materials and Methods
Study Participants
We followed a two-stage study design to conduct a GWAS of plasma B12 concentrations. Stage I comprised 1122 Indo-European healthy parents of children in the PMNS cohort from Western India (Supplementary Material, Fig. S1) (21). The stage II replication study included subjects of different age groups of Indo-European (n = 2403) and Dravidian (n = 1015) ethnicities. The adult group comprised Pune adults (n = 724), which included the remaining parents of children in PMNS, parents from Pune Children Study (PCS), PCS children at 21 years follow-up and individuals from the Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) study (43,44). Samples from pregnant mothers from the MMNP (n = 989) and PS (n = 481) were also included in the replication analysis (22,23). The children’s group comprised children from the PMNS (n = 690) and PS (n = 534). Ethnicity was delineated using geographical location, the mother tongue and place of origin of their parents. Subjects in the PMNS, PCS, CRISIS and MMNP belonged to Indo-European ethnicity while those from PS were Dravidian in origin. Detailed phenotypic information was available at different time points through investigator-administered structured questionnaire. Vitamin B12 concentrations and other biochemical parameters like folate and homocysteine were measured using standard techniques as described earlier (9,14,45). All participants gave informed consent and the Institutional Ethics Committee of all participating institutes approved the study following guidelines for human research by Indian Council of Medical Research, Government of India.
Stage I GWAS Analysis
Genome-wide data on stage I samples were generated using Affymetrix SNP 6.0 Chips (Affymetrix, CA, USA) and 93% (n = 8,07,908) of autosomal single nucleotide polymorphisms (SNPs) were successfully genotyped. Samples and SNPs with a call rate <95% and SNPs with minor allele frequency (MAF) <5% and HWE P < 0.001 were excluded and a total of 6,23,209 SNPs were used for further analysis. Genome-wide imputation was carried out by IMPUTE2 (v2.2; https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) using all the individuals across 26 populations in the 1000 Genomes phase 3 (November 2015 release) as the reference panel (46). For imputation quality control, we applied impute2 info score ≥ 0.3, genotype probability threshold of 90%, call rate of 95% and 99% for SNPs with MAF > 5% and MAF < 5%, respectively. Finally, 8.9 million QC passed SNPs having MAF > 0.001were used for association analysis. Association analysis of genotyped and imputed SNPs was performed using PLINK (v1.07) and SNPTEST (v2.5.2), respectively (47,48). We applied Frequentist association test with score method to account for probable uncertainty in the imputed genotypes. To account for ancestry and population structure, we further adjusted the association analysis for first ten principal components and finally, compared the results with mixed linear model association analysis using mlma-loco in GCTA software (49).
SNP Selection and Replication Analysis
We used various criteria for choosing SNPs from stage I data for replication analysis. SNPs with P < 10−4 were considered, and in a region harboring multiple SNPs in strong LD (r2>0.8), the most significant ones were selected. We further performed gene based association analysis using Versatile Gene-based Association Studies (VEGAS) (50) and genes with P < 0.01 and/or their proxy SNPs with P < 10−4 were included for replication genotyping. A more stringent P < 10−5 was used for selecting SNPs from the gene-desert regions. A total of 41 SNPs including 24 SNPs from stage I, and 17 previously reported variants, were selected for replication (Supplementary Material, Fig. S1). Genotyping was performed on Fluidigm SNP 96.96 genotyping system and both samples and SNPs with <90% call rate, and SNPs with HWE P < 0.001 were removed from the association analysis. We carried out independent association analysis on replication genotype data for each cohort and final results were combined by meta-analysis using fixed effects models.
Statistical Analyses
PCA on LD-pruned SNPs (r2<0.2) was performed by SVS Golden Helix and extreme outliers (with ±4 standard deviation (SD) of the first two principal components were excluded from the association analysis (Supplementary Material, Fig. S1). Plasma B12 concentrations were adjusted for age and sex, and the residual standardized B12 was inverse normal transformed using STATA. Associations between inverse normal transformed B12 concentrations with SNPs were performed by linear regression using an additive model of minor allele by PLINK (v.1.07) (47). Meta-analysis of summary statistics from stage I and replication data was performed using an inverse variance-weighted approach implemented in STATA using command ‘metan’. Conditional analysis was performed in the regions where multiple SNPs showed an association by adjusting the top SNP or the previously established variant in the regression model. The results after the conditional analysis from each of the replication cohorts were combined by meta-analysis. The variants showing meta-analysis P ≤ 0.01 were considered independent signals. A genetic score (GS), weighted for the respective effect size (beta coefficient from the meta-analysis stage) of each SNP, was calculated using 8 independently associated SNPs identified from the conditional analysis. The SNPs used were rs12780845 in CUBN, rs602662 in FUT2, rs708686 and rs3760775 in FUT6, rs2270655 in MMAA, rs34324219 and rs34528912 in TCN1 & rs1131603 in TCN2 and the genotypes were coded according to the additive effect of the B12-raising alleles. The weighted GS was calculated as follows:
The GS was further grouped into quartiles and the B12 concentrations were compared between these quartiles in PMNS and PS children at different ages. HaploView (v.4.2) (51) was used to perform LD analysis for our data and the LD structure of other populations was taken from the 1000 Genomes phase 3 data for comparison. The statistical power for associations was calculated by Quanto software (v.1.2.4) (52), using the previously reported effect size and effect allele frequency (EAF) from the present study at type 1 error of 0.01.
Bayesian Analysis and Generation of Credible Set
We used both genotyped and the imputed data from stage I samples and performed Bayesian quantitative trait association analysis under an additive model adjusted for first ten principal components, using SNPTEST v2.5.2 and expected method to take care of genotype uncertainty. For each SNP, we measured the Bayes factor which is the ratio between the probabilities of SNP under the alternative hypothesis that the SNP is associated with the phenotype and under the null hypothesis that it is not associated with the phenotype (53). The value of the Bayes factor is directly correlated with the strength of SNP-phenotype association. To identify the most probable causal SNP or set of SNPs in a genomic region, we calculated the posterior probability of each SNP as mentioned (54). The 95% credible set for a genomic region was constructed by ranking all the SNPs according to their Bayes factor value and accumulating the posterior probabilities of rank variants to attain 0.95 or more (55).
Tracking analysis
PMNS and PS are prospective cohort studies in which parents and children have been followed up at different stages, and demographic, anthropometric and biochemical data have been collected as described above (21,22). Data available at different ages reflects change in various environmental factors such as diet and lifestyle habits. Comparison of plasma B12 concentrations at different time points was performed using paired t-tests. Analysis of genetic associations of identified SNPs and GS (calculated from 8 independently associated SNPs) with B12 concentrations at different follow-up stages was performed using linear regression, adjusted for age and sex. Quantile based association analysis was also performed to compare between the lowest and highest GS.
Bioinformatics and Functional Analysis
We investigated the LD relationship between the SNPs in different populations using the 1000 Genomes phase 3 data. LD plots were generated using LDlink and regional association plots were drawn using LocusZoom (46,56,57). Bioinformatic analysis of transcription factor binding and functional prediction of regulatory elements was conducted using the UCSC genome browser, the JASPAR database and HaploReg (v4.1) (58–60). Based on bioinformatic predictions, we performed luciferase reporter assays in HepG2 cell line for promoter and enhancer activity by cloning the genomic region encompassing the new variants, rs3760775 and rs78060698 as well as the previously reported variant, rs3760776 in pGL4.10 and pGL4.23 vector respectively (Promega, USA). Primers were designed using Primer Blast and Primer3 to amplify specific regions and site-directed mutagenesis was performed if the amplified genomic region contained any other SNP (Supplementary Material, Table S5). The amplified genomic regions were cloned by digestion with Kpn1, Xho1 or Sac1 as required and ligated to the multiple cloning site of respective luciferase vectors. The clones were confirmed by Sanger sequencing and transfected with internal control vector pRL-TK in the molar ratio of 15:1 and cell lysate was collected 24 h after transfection. Luciferase activity was measured using PerkinElmer EnSpire Multimode Plate Reader.
Electrophoretic-mobility shift assay (EMSA) for rs78060698 alleles was conducted using nuclear extract (NE) from HepG2 cell line. Harvested HepG2 cells (∼108 cells) were incubated with 5 ml cell lysis buffer and buffer A (10 mM HEPES, pH 7.8; 1.5 mM MgCl2; 10 mM KCl; 0.5 mM DTT; and 1X Protease inhibitor) for 30 min on ice. Nuclei were collected by centrifugation at 2000 rpm after 10 strokes of Dounce homogenizer. The resulting nuclei were resuspended in 500 μl of buffer C (20 mM HEPES, pH 7.8; 25% (v/v) glycerol; 0.42 M NaCl; 1.5 mM MgCl2; 0.2 mM EDTA; 0.5 mM DTT; and 1X Protease inhibitor) and incubated for 1 h on ice. NE was collected by centrifugation at 20,000 g for 30 min at 4°C. Primers for EMSA were designed such that the SNP was positioned at the centre with a 15/16-base overhang on either side (Supplementary Material, Table S5). Double stranded (ds) oligonucleotides were obtained by incubating the forward and reverse oligonucleotides (50 μl of 50 pmol/μl each) at 95°C for 2 min and gradually cooling to 37°C. The oligo probes were radiolabeled by incubating 25 pmoles of ds oligo with 2.5 ul of γ32P-ATP and 10 units of T4 polynucleotidekinase (PNK) in 1X PNK buffer for 1 h at 37°C and unincorporated nucleotides were removed using a sephadex G-25 column. A binding reaction was set up with 5 μg of NE, 1 μl of labeled oligo, 1X Binding buffer (20mM HEPES, pH 7.9; 15% (v/v) ficol; 150 mM KCl; 1 mM EDTA; and 0.5 mM DTT), 0.5 μl of poly-dIdC (1 mg/ml) and 0.5 μl of yeast tRNA (1 mg/ml) in a total volume of 20 μl, on ice, for 1 h. For the super-shift assay, 1 μg of different antibodies (HNF1α (C19: sc-6547x), HNF4α (S20: sc-6557x) and HNF3β (H150: sc-20692); Santa Cruz Biotechnology) was added after 30 min incubation, followed by additional incubation for 90 min. The reaction mixtures were finally run on 6% TBE polyacraylamide gel with 0.5X TBE running buffer (pH 7.5) for about 8 h at 100V at 4°C. The resulting shift and super-shift bands were visualized by autoradiography using PMI Bio-Rad phosphoimager.
Data Availability
The summary association statistics from the genome-wide data presented in this study is available at http://www.ccmb.res.in/staff/chandak/data.html.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank the participants of all the cohorts for agreeing to join the study and field staff for their contributions in sample collection and community work. The help of Dr Seema Bhaskar, K Radha Mani and Inder Deo Mali, CSIR-Centre for Cellular and Molecular Biology, Hyderabad in genomic DNA isolation from blood samples and in managing the DNA samples is sincerely acknowledged. We acknowledge major contributions by S Rao, S Hirve, P Gupta, D S Bhat, H Lubree, S Rege, P Yajnik and the invaluable community work contributed by T Deokar, S Chaugule, A Bhalerao and V Solat from the KEM Hospital Research Centre, Pune. We are grateful to Professor Oluf Pedersen, Professor Niels Grarup and collaborators, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Denmark for providing anonymized genotype data for our replication study. We also thank Prof T Tanaka, Translational Gerontology Branch, NIA at Harbor Hospital, Baltimore, USA and acknowledge the contribution of the data from three studies, Sardinia, BLSA and InCHIANTI.
Conflict of Interest statement. None declared.
Funding
Council of Scientific and Industrial Research (CSIR), Ministry of Science and Technology, Government of India, India (XII Five-Year Plan titled “CARDIOMED”). Wellcome Trust, London, UK, Medical Research Council, London, UK and Department for International Development, UK. Parthenon Trust, Switzerland and ICICI Bank, Social Initiatives Group. Funding to pay the Open Access publication charges for this article was provided by Council of Scientific and Industrial Research (CSIR), Ministry of Science and Technology, Government of India, India.
References
- 1. O'Leary F., Samman S. (2010) Vitamin B12 in health and disease. Nutrients, 2, 299–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Selhub J. (1999) Homocysteine metabolism. Annu. Rev. Nutr., 19, 217–246. [DOI] [PubMed] [Google Scholar]
- 3. Takahashi-Iniguez T., Garcia-Hernandez E., Arreguin-Espinosa R., Flores M.E. (2012) Role of vitamin B12 on methylmalonyl-CoA mutase activity. J. Zhejiang Univ. Sci. B, 13, 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen N.C., Yang F., Capecci L.M., Gu Z., Schafer A.I., Durante W., Yang X.F., Wang H. (2010) Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J., 24, 2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albert M.J., Mathan V.I., Baker S.J. (1980) Vitamin B12 synthesis by human small intestinal bacteria. Nature, 283, 781–782. [DOI] [PubMed] [Google Scholar]
- 6. Nielsen M.J., Rasmussen M.R., Andersen C.B., Nexo E., Moestrup S.K. (2012) Vitamin B12 transport from food to the body's cells–a sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol., 9, 345–354. [DOI] [PubMed] [Google Scholar]
- 7. Baik H.W., Russell R.M. (1999) Vitamin B12 deficiency in the elderly. Annu. Rev. Nutr., 19, 357–377. [DOI] [PubMed] [Google Scholar]
- 8. Toh B.H., van Driel I.R., Gleeson P.A. (1997) Pernicious anemia. N. Engl J. Med., 337, 1441–1448. [DOI] [PubMed] [Google Scholar]
- 9. Refsum H., Yajnik C.S., Gadkari M., Schneede J., Vollset S.E., Orning L., Guttormsen A.B., Joglekar A., Sayyad M.G., Ulvik A., et al. (2001) Hyperhomocysteinemia and elevated methylmalonic acid indicate a high prevalence of cobalamin deficiency in Asian Indians. Am. J. Clin. Nutr., 74, 233–241. [DOI] [PubMed] [Google Scholar]
- 10. Shobha V., Tarey S.D., Singh R.G., Shetty P., Unni U.S., Srinivasan K., Kurpad A.V. (2011) Vitamin B(1)(2) deficiency & levels of metabolites in an apparently normal urban south Indian elderly population. Indian J. Med. Res., 134, 432–439. [PMC free article] [PubMed] [Google Scholar]
- 11. Yajnik C.S., Deshpande S.S., Lubree H.G., Naik S.S., Bhat D.S., Uradey B.S., Deshpande J.A., Rege S.S., Refsum H., Yudkin J.S. (2006) Vitamin B12 deficiency and hyperhomocysteinemia in rural and urban Indians. J. Assoc. Physicians India, 54, 775–782. [PubMed] [Google Scholar]
- 12. Yajnik C.S., Deshpande S.S., Jackson A.A., Refsum H., Rao S., Fisher D.J., Bhat D.S., Naik S.S., Coyaji K.J., Joglekar C.V., et al. (2008) Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: the Pune Maternal Nutrition Study. Diabetologia, 51, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yajnik C.S., Chandak G.R., Joglekar C., Katre P., Bhat D.S., Singh S.N., Janipalli C.S., Refsum H., Krishnaveni G., Veena S., et al. (2014) Maternal homocysteine in pregnancy and offspring birthweight: epidemiological associations and Mendelian randomization analysis. Int. J. Epidemiol., 43, 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krishnaveni G.V., Hill J.C., Veena S.R., Bhat D.S., Wills A.K., Karat C.L., Yajnik C.S., Fall C.H. (2009) Low plasma vitamin B12 in pregnancy is associated with gestational ‘diabesity‘ and later diabetes. Diabetologia, 52, 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nilsson S.E., Read S., Berg S., Johansson B. (2009) Heritabilities for fifteen routine biochemical values: findings in 215 Swedish twin pairs 82 years of age or older. Scand. J. Clin. Lab. Invest., 69, 562–569. [DOI] [PubMed] [Google Scholar]
- 16. Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. (2008) Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet., 40, 1160–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M.G., Usala G., Lai S., Mulas A., Corsi A.M., Vestrini A., et al. (2009) Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet., 84, 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hazra A., Kraft P., Lazarus R., Chen C., Chanock S.J., Jacques P., Selhub J., Hunter D.J. (2009) Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum. Mol. Genet., 18, 4677–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin X., Lu D., Gao Y., Tao S., Yang X., Feng J., Tan A., Zhang H., Hu Y., Qin X., et al. (2012) Genome-wide association study identifies novel loci associated with serum level of vitamin B12 in Chinese men. Hum. Mol. Genet., 21, 2610–2617. [DOI] [PubMed] [Google Scholar]
- 20. Grarup N., Sulem P., Sandholt C.H., Thorleifsson G., Ahluwalia T.S., Steinthorsdottir V., Bjarnason H., Gudbjartsson D.F., Magnusson O.T., Sparso T., et al. (2013) Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. PLoS Genet., 9, e1003530.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao S., Yajnik C.S., Kanade A., Fall C.H., Margetts B.M., Jackson A.A., Shier R., Joshi S., Rege S., Lubree H., et al. (2001) Intake of micronutrient-rich foods in rural Indian mothers is associated with the size of their babies at birth: Pune Maternal Nutrition Study. J. Nutr., 131, 1217–1224. [DOI] [PubMed] [Google Scholar]
- 22. Krishnaveni G.V., Veena S.R., Hill J.C., Karat S.C., Fall C.H. (2015) Cohort profile: Mysore parthenon birth cohort. Int. J. Epidemiol., 44, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Potdar R.D., Sahariah S.A., Gandhi M., Kehoe S.H., Brown N., Sane H., Dayama M., Jha S., Lawande A., Coakley P.J., et al. (2014) Improving women's diet quality preconceptionally and during gestation: effects on birth weight and prevalence of low birth weight–a randomized controlled efficacy trial in India (Mumbai Maternal Nutrition Project). Am. J. Clin. Nutr., 100, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mollicone R., Reguigne I., Fletcher A., Aziz A., Rustam M., Weston B.W., Kelly R.J., Lowe J.B., Oriol R. (1994) Molecular basis for plasma alpha(1,3)-fucosyltransferase gene deficiency (FUT6). J. Biol. Chem., 269, 12662–12671. [PubMed] [Google Scholar]
- 25. Adeyemo A., Rotimi C. (2010) Genetic Variants Associated with Complex Human Diseases Show Wide Variation across Multiple Populations. Public Health Genomics, 13, 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kato N. (2012) Ethnic diversity in type 2 diabetes genetics between East Asians and Europeans. J. Diabetes Invest., 3, 349–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCarthy M.I. (2008) Casting a wider net for diabetes susceptibility genes. Nat. Genet., 40, 1039–1040. [DOI] [PubMed] [Google Scholar]
- 28. Marigorta U.M., Navarro A. (2013) High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants. Plos Genet., 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Higai K., Miyazaki N., Azuma Y., Matsumoto K. (2008) Transcriptional regulation of the fucosyltransferase VI gene in hepatocellular carcinoma cells. Glycoconj. J., 25, 225–235. [DOI] [PubMed] [Google Scholar]
- 30. Lauc G., Essafi A., Huffman J.E., Hayward C., Knezevic A., Kattla J.J., Polasek O., Gornik O., Vitart V., Abrahams J.L., et al. (2010) Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet., 6, e1001256.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma B., Simala-Grant J.L., Taylor D.E. (2006) Fucosylation in prokaryotes and eukaryotes. Glycobiology, 16, 158R–184R. [DOI] [PubMed] [Google Scholar]
- 32. Becker D.J., Lowe J.B. (2003) Fucose: biosynthesis and biological function in mammals. Glycobiology, 13, 41R–53R. [DOI] [PubMed] [Google Scholar]
- 33. Pickard J.M., Maurice C.F., Kinnebrew M.A., Abt M.C., Schenten D., Golovkina T.V., Bogatyrev S.R., Ismagilov R.F., Pamer E.G., Turnbaugh P.J., et al. (2014) Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature, 514, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koropatkin N.M., Cameron E.A., Martens E.C. (2012) How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol., 10, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., et al. (2014) Human genetics shape the gut microbiome. Cell, 159, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Degnan P.H., Taga M.E., Goodman A.L. (2014) Vitamin B12 as a modulator of gut microbial ecology. Cell Metab., 20, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersen C.B., Madsen M., Storm T., Moestrup S.K., Andersen G.R. (2010) Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes. Nature, 464, 445–448. [DOI] [PubMed] [Google Scholar]
- 38. Burger R.L., Schneider R.J., Mehlman C.S., Allen R.H. (1975) Human plasma R-type vitamin B12-binding proteins. II. The role of transcobalamin I, transcobalamin III, and the normal granulocyte vitamin B12-binding protein in the plasma transport of vitamin B12. J. Biol. Chem., 250, 7707–7713. [PubMed] [Google Scholar]
- 39. Yammani R.R., Seetharam S., Seetharam B. (2001) Identification and characterization of two distinct ligand binding regions of cubilin. J. Biol. Chem., 276, 44777–44784. [DOI] [PubMed] [Google Scholar]
- 40. Roman T.S., Marvelle A.F., Fogarty M.P., Vadlamudi S., Gonzalez A.J., Buchkovich M.L., Huyghe J.R., Fuchsberger C., Jackson A.U., Wu Y., et al. (2015) Multiple Hepatic Regulatory Variants at the GALNT2 GWAS Locus Associated with High-Density Lipoprotein Cholesterol. Am. J. Hum. Genet., 97, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He H.L., Li W., Liyanarachchi S., Srinivas M., Wang Y.Q., Akagi K., Wang Y., Wu D.Y., Wang Q.B., Jin V., et al. (2015) Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc. Natl Acad. Sci. USA, 112, 6128–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Iari R.C.S., Lupien M., Markowitz S., Scacheri P.C. (2014) Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res., 24, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yajnik C.S., Joglekar C.V., Pandit A.N., Bavdekar A.R., Bapat S.A., Bhave S.A., Leary S.D., Fall C.H. (2003) Higher offspring birth weight predicts the metabolic syndrome in mothers but not fathers 8 years after delivery: the Pune Children's Study. Diabetes, 52, 2090–2096. [DOI] [PubMed] [Google Scholar]
- 44. Yajnik C.S., Joglekar C.V., Lubree H.G., Rege S.S., Naik S.S., Bhat D.S., Uradey B., Raut K.N., Shetty P., Yudkin J.S. (2008) Adiposity, inflammation and hyperglycaemia in rural and urban Indian men: Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) Study. Diabetologia, 51, 39–46. [DOI] [PubMed] [Google Scholar]
- 45. Kehoe S.H., Chopra H., Sahariah S.A., Bhat D., Munshi R.P., Panchal F., Young S., Brown N., Tarwande D., Gandhi M., et al. (2015) Effects of a food-based intervention on markers of micronutrient status among Indian women of low socio-economic status. Br. J. Nutr., 113, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marchini J., Howie B., Myers S., McVean G., Donnelly P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. [DOI] [PubMed] [Google Scholar]
- 49. Yang J., Zaitlen N.A., Goddard M.E., Visscher P.M., Price A.L. (2014) Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet., 46, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., et al. (2010) A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 52. Gauderman W.J. (2002) Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med., 21, 35–50. [DOI] [PubMed] [Google Scholar]
- 53. Stephens M., Balding D.J. (2009) Bayesian statistical methods for genetic association studies. Nat. Rev. Genet., 10, 681–690. [DOI] [PubMed] [Google Scholar]
- 54. Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., Morris A., et al. (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet., 44, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gaulton K.J., Ferreira T., Lee Y., Raimondo A., Magi R., Reschen M.E., Mahajan A., Locke A., Rayner N.W., Robertson N., et al. (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet., 47, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Machiela M.J., Chanock S.J. (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res., 42, D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary association statistics from the genome-wide data presented in this study is available at http://www.ccmb.res.in/staff/chandak/data.html.