Abstract
The 14-3-3 proteins are acidic, dimeric proteins that have been implicated in many eukaryotic cellular processes because of direct protein association with enzymes and other metabolic and regulatory proteins. 14-3-3 proteins are largely considered to be cytoplasmic, but a search for proteins that specifically interact with a plant 14-3-3 resulted in the isolation of a nuclear-encoded, thylakoid-targeted chloroplast precursor, the full-length Arabidopsis photosystem I N-subunit At pPSI-N (P.C. Sehnke, R.J. Ferl [1995] Plant Physiol 109: 1126). Using precursor truncations in the two-hybrid system, it was determined that the leader sequence is the site of PSI-N that associates with 14-3-3. This suggested the novel possibility that 14-3-3 would be found within chloroplasts. Immuno-electron microscopy of leaf tissue and western analysis of chloroplast fractions with monoclonal anti-14-3-3 antibodies localized 14-3-3 proteins to the chloroplast stroma and the stromal side of thylakoid membranes. Using peptide-generated, isoform-specific antibodies, GF14ν, GF14ε, GF14μ, and GF14υ were shown to be present in the chloroplast stromal extract. These isoforms represent two distinct phylogenetic 14-3-3 groupings. These data suggest a novel interorganellar role for these phylogenetically distinct 14-3-3 proteins.
The roles of 14-3-3 proteins are diverse, ranging from regulation of metabolic enzymes to participation in transcriptional complexes and phytotoxin receptors and partnerships with structural proteins (Aitken, 1996; Ferl, 1996; Sehnke and Ferl, 1996; Ku et al., 1998). All of these roles, however, include the common feature of direct physical association between the 14-3-3 protein and its target. The 14-3-3 proteins are dimeric, soluble proteins that can bind to other proteins via a phospho-Ser-mediated interaction (Muslin et al., 1996). The nature of the interacting domains within the 14-3-3 appears to vary depending upon the binding partners, however, it was observed early on that the central domains of the 14-3-3s (residues 171–213 of the human isoform η) contain the residues directing the phospho-Ser interaction with the enzyme Trp hydroxylase (Ichimura et al., 1995). Ichimura et al. (1995) called this region of the 14-3-3 the “box-1” domain, which is extremely well conserved in all species and isoforms of 14-3-3. In addition, this central region containing box 1 is essentially an autonomous domain capable of interacting with the target protein as an independent monomeric protein (Ichimura et al., 1995; Liu et al., 1996).
The three dimensional x-ray crystallographic structures of two 14-3-3 isoforms revealed a nine-helical, compartmentalized architecture with the C terminus externally exposed (Liu et al., 1995; Xiao et al., 1995). The entire C terminus is not completely visible in the model, however, the most terminal region forms an accessible “flap” over a hollow amphipathic core derived by the union of the monomers. Additionally, the extreme termini of the 14-3-3s are the most divergent regions of the proteins, suggesting that they may direct isoform-specific functions.
In an effort to identify functions for plant 14-3-3s that occur via the central box 1 and C-terminal regions, we screened an Arabidopsis-yeast two-hybrid cDNA library with an N-terminal truncation of 14-3-3 protein GF14-12 (de Vetten et al., 1992) as bait. This N-terminal truncation results in the removal of the first four helices, which constitute the dimerization domain (Wu et al., 1997a). This construct ensures that the box 1 and C-terminal domain are unencumbered by dimerization and are therefore available for direct interaction with potential targets. Of the three 14-3-3 interacting clones isolated, one was a full-length Arabidopsis cDNA homolog to the barley (Hordeum vulgare) precursor of the thylakoid lumenal localized photosystem I N-subunit (Hv pPSI-N) (Knoetzel and Simpson, 1993), which we called At pPSI-N (Sehnke and Ferl, 1995). The discovery of an interaction between 14-3-3 proteins and a thylakoid lumen protein presented two important questions that are addressed in the present paper. What is the nature of the interaction between 14-3-3s and PSI-N, and are 14-3-3s present inside of chloroplasts in spite of the fact that they lack any import signals?
MATERIALS AND METHODS
Materials
Subcloning was performed as described previously (Sambrook et al., 1989) with commercial enzymes. PCR was performed as per the manufacturer's instructions supplied with the enzyme (Promega, Madison, WI). All chemicals used were purchased commercially.
Yeast Two-Hybrid Transformation and Library Screening
The coding region for residues 88–267 of the maize (Zea mays) Zm 14-3-3 GF14 12 (de Vetten et al., 1992) was subcloned into BamHI restricted yeast two-hybrid vector pGBT9 (CLONTECH Laboratories, Palo Alto, CA) as a BglII/BamHI cassette from pET15b GF14-12. pGBT9-monoGF14-12 was transformed in the yeast strain HF7C and used to screen a Matchmaker Arabidopsis cv Columbia 3-week-old vegetative tissue cDNA library using the manufacturer's protocol (CLONTECH Laboratories). Positively interacting clones, as indicated by growth on −His media and Lac Z gene reporter activity, were transferred to Escherichia coli strain HB101 and sequenced (model 373A sequencer, Applied Biosystems, Foster City, CA).
Interaction Domain Mapping Using Yeast Two-Hybrid System
PCR with the oligos (5′-AGGATCCCAGCATTAGCAGAAG-3′, 5′-TACCACTACAATGGATG-3′) and pGAD10 At pPSI-N as the template, was used to generate a BamHI cassette coding for the putative chloroplast and thylakoid targeting transit peptide (At pPSI-N residues 1–87). After restriction the cassette was subcloned into pGAD424 (CLONTECH Laboratories), yielding pGAD424-CTTD. A binding domain fusion construct, pGAD424-CTD, containing only the putative chloroplast transit peptide (residues 1–47) was produced by digestion of pGAD424-CTTD with BclI and BamHI and subcloned into BamHI-restricted pGAD424. A construct with the chloroplast transit peptide removed (retaining the putative thylakoid targeting domain and mature protein residues 49–171) was made by digestion of pGAD10 At pPSI-N with BclI and SalI and subcloning into BamHI/SalI-restricted pGAD424 yielding pGAD424 At iPSI-N (the intermediate form of the precursor). Finally, a construct (pGAD424-At mPSI-N) with the mature At PSI-N coding sequence (At pPSI-N residues 92–129) was created by sequential digestion of pGAD10 AT pPSI-N with XhoI, mung bean nuclease and PvuII, followed by subcloning into pGAD424 restricted with SmaI/PvuII. Yeast transformation and interaction screening were performed as described above.
SDS-PAGE and Immunoblotting Analysis
Samples of plant material or isolated organelles were analyzed for 14-3-3s using SDS-PAGE. Samples were combined with equal volumes of 2× SDS-PAGE sample loading buffer (50 mm Tris, 5% [v/v] β-mercaptoethanol, 2% [w/v] SDS, and 10% [v/v] glycerol, pH 6.8) prior to boiling for 90 s. Protein concentrations of both extract supernatants and column fractions were determined using Bradford analysis (Bradford, 1976). Samples of protein were loaded onto the gels along with molecular mass markers for western-immunoblotting analysis or with protein standards for Coomassie staining analysis. After electrophoresis, the gels were either stained with Coomassie Blue R-250 or transferred to nitrocellulose using a Minigel transfer system (Bio-Rad Laboratories, Hercules, CA) following the manufacturer's directions.
Coomassie-stained gels were destained with a methanol-acetic acid-water mixture until bands were visible. The blots were incubated in 5% (w/v) non-fat dry milk and 0.5% (v/v) Tween 20 in phosphate-buffered saline 7.6 (PSBT) overnight to block the remaining sites. Blots were washed with PBST and incubated with either anti-GF14 monoclonal antibody (Mab) containing ascites fluid (Lu et al., 1992) (1/3,000) or rabbit polyclonal anti-GF14 sera (1/10,000) produced by Bioworld (Dublin, OH) using conjugated peptides derived from the different At GF14 isoform cDNA sequences (P.C. Sehnke and R.J. Ferl, unpublished data). After washing, the blots were incubated with appropriate secondary antibody conjugate of horseradish peroxidase for 45 min. The blots were washed a final time and then processed with chemiluminescence kit (Amersham, Uppsala) prior to development on film. As controls to confirm antibody specificity, the peptides that were used to generate the respective antibodies were added to the diluted sera (at an approximately 20:1 molar ratio assuming specific antibody concentrations of 0.1% or 2 nm final peptide concentration) and allowed to complex for 1 h at room temperature prior to immunoblotting. Antibodies to the cytoplasmic marker protein BiP (a protein that associates with the endoplasmic reticulum luminal HSC70s) (Anderson et al., 1994) and the chloroplast stromal marker protein SecA (Yuan et al., 1994) were used to confirm cytoplasmic and chloroplast stromal sample integrity, respectively.
Immunotransmission Electron Microscopy
Arabidopsis tissue was prepared and analyzed with anti-14-3-3 Mabs as described previously (Bihn et al., 1997).
Chloroplast Isolation and Suborganellar Separation
Preparation of pea (Pisum sativum) chloroplasts, stromal extract, and isolated thylakoid membranes were as described previously (Cline, 1986). Isolation of chloroplasts from 21-d-old Arabidopsis plants was accomplished using a modified version of the procedure from Somerville et al. (1981). The chloroplasts were purified on a Percoll step gradient as described previously (Rensink et al., 1998). Stromal extract from Arabidopsis chloroplasts was obtained as described above. Protein concentrations were determined using Bradford analysis.
RESULTS AND DISCUSSION
Our preliminary finding of a plant 14-3-3 binding to the nuclear-encoded chloroplast precursor PSI-N in the yeast two-hybrid system (Sehnke and Ferl, 1995) and the reports of 14-3-3 involvement in mitochondrial import (Alam et al., 1994; Komiya et al., 1997), prompted our detailed exploration of the 14-3-3/PSI-N interaction. Potential domains of At pPSI-N were predicted by sequence identity with the delineated structural elements of Hv pPSI-N. Comparison of the amino acid sequence of At pPSI-N with Hv pPSI-N reveals conservation of the thylakoid peptidase cleavage site (AXA, where X can be any amino acid), separating the highly homologous mature proteins (85% identity) from the divergent NH2-terminal transit peptides (30% identity) (Fig. 1). Since both proteins utilize the ΔpH-dependent pathway (Nielsen et al., 1994; K. Cline, unpublished data) for which barley does not produce processing intermediates, differentiation of stromal and thylakoid targeting domains is, not surprisingly, less obvious. However the twin-Arg motif critical for thylakoid import of ΔpH- dependent precursors (Chaddock et al., 1995) is conserved in both C-terminal halves of the pPSI-N transit peptides (Fig. 1). In addition, an N-terminal truncation of At pPSIN (minus the first 40 residues) was competent in isolated thylakoid membrane import assays, but no longer for intact chloroplast import, which is again suggestive of a bipartite modular transit peptide (data not shown).
Figure 1.
Amino acid sequence of the 14-3-3 interacting chloroplast precursor protein At pPSI-N and comparison with barley PSI-N homolog. Amino acid identities between pPSI-N proteins are indicated by asterisks. Accession numbers for the PSI-N proteins are: Arabidopsis, U32176; barley, X66428. Transit peptide sequences are represented by lowercase characters, the thylakoid peptidase cleavage site in bold, and mature protein by uppercase characters. Identical residues between both proteins are indicated by asterisks. The characteristic twin Arg residues in the signal peptides critical for thylakoid import of precursors using the ΔpH pathway are boxed.
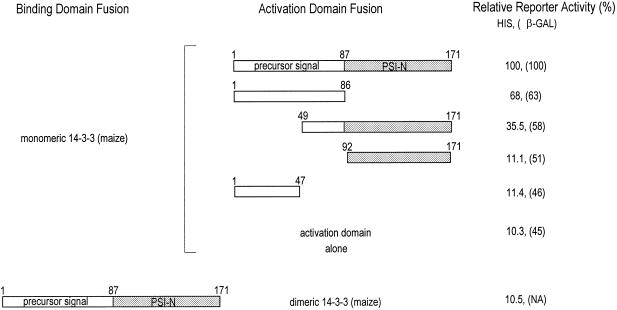
To determine the specific region of At pPSI-N that interacts with the 14-3-3 monomer, Gal4 fusion constructs encoding portions of At pPSI-N were transformed into yeast along with monomeric GF14-12 (Fig. 2). Interaction between the proteins was scored for both Lac Z and HIS reporter activity. The order of the strength of observed interaction was: full-length precursor (β-galactosidase activity was 24% of that of the p53/SV40 large T-antigen positive control supplied by manufacturer of the kit) > precursor transit peptide > carboxy-terminal half of the transit peptide containing the thylakoid targeted twin-Arg motif linked to At pPSI-N. The reporter activities for mature protein and the amino-terminal half of the transit peptide containing the putative stromal-targeting domain fusions were approximately equivalent to background for both reporters analyzed, indicating weak or no binding to the 14-3-3 fusion protein. This indicates that the 14-3-3s bind to the thylakoid targeting domain of the transit peptide. Since 14-3-3 proteins exist as dimers in vitro and are also thought to be dimers in vivo, we tested the ability of the At pPSI-N constructs to bind dimeric full-length maize 14-3-3, GF14-12. Surprisingly, no interaction above background was detectable (Fig. 2). In other two-hybrid experiments using full-length 14-3-3 proteins, we have observed that interactions through the 14-3-3 dimerization domains with endogenous or recombinant 14-3-3 proteins is strongly favored to the point of the exclusion of other interactions (Wu et al., 1997a). Full-length constructs are believed to maintain 14-3-3 fusion proteins as dimers and thereby limit or mask the interactions with library prey.
Figure 2.
Summary of interactions between At pPSI-N domains and 14-3-3 proteins using the two-hybrid system. Using the yeast two-hybrid protein-protein detection system, the domains important for 14-3-3/At PSI-N interaction were determined. At PSI-N GAL4 activation domain fusions and truncated Zm GF14-12 GAL4 binding domain fusions were transferred into yeast and assayed for reporter activity using β-galactosidase and His growth measurements. The results are summarized as relative units of activity corresponding to full-length At PSI-N/14-3-3 interaction. GAL4 activation domain was used as a negative control. Interactions between dimeric 14-3-3 and At PSI-N were also determined using a 14-3-3 GAL4 activation domain fusion, since full-length 14-3-3s possess transcriptional activation activity (Wu et al., 1997a).
Plant 14-3-3 proteins have been found in various tissues including leaves (de Vetten et al., 1992; Lu et al., 1992). While 14-3-3s are generally considered to be cytoplasmic, 14-3-3s have recently been identified within an organelle, the nucleus (Bihn et al., 1997; Todd et al., 1998). Furthermore, an early identification of plant 14-3-3s was made from external chloroplast membrane preparations, although the presence of internal chloroplast 14-3-3s was not demonstrated and cytosolic 14-3-3 contamination could not be ruled out as the source of the membrane-associated 14-3-3s (Hirsch et al., 1992). However, since the yeast two-hybrid 14-3-3/PSI-N data identified the putative thylakoid targeting domain of the PSI-N transit peptide as the 14-3-3 binding site, we speculated that 14-3-3s should be present within the chloroplast.
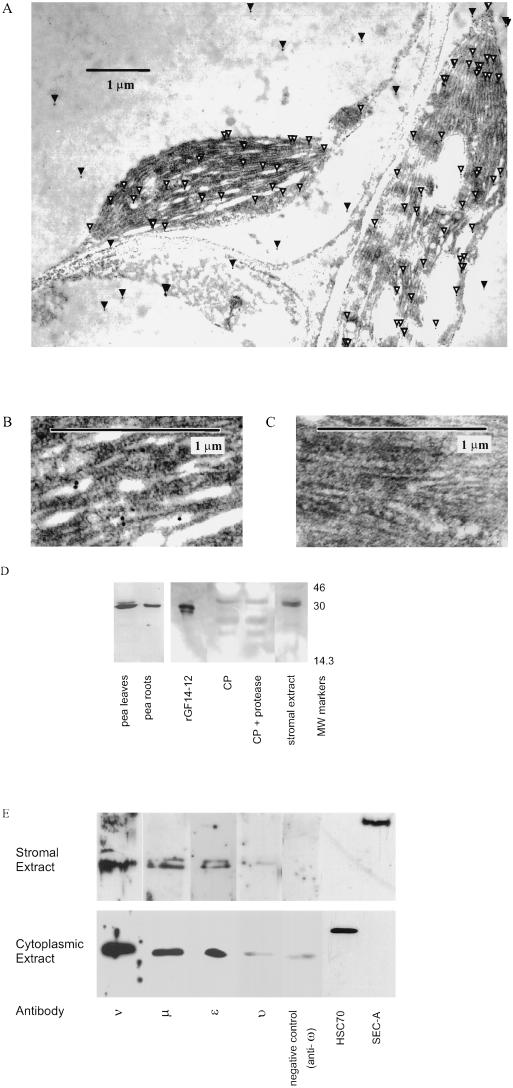
To investigate this theory, sectioned, embedded Arabidopsis leaves were treated with an anti-14-3-3 monoclonal antibody, Mab-15, which recognizes most of the Arabidopsis 14-3-3 isoforms. Localization was detected in the electron microscope by secondary antibodies linked to gold particles (Fig. 3A). Immunoreactive material was observed within the chloroplasts, in a higher concentration than that observed in the cytoplasm. Within the chloroplasts, gold particles were observed throughout the stroma and on thylakoid membranes (Fig. 3B). Control sections treated with an irrelevant monoclonal antibody did not demonstrate any immunoreactivity (Fig. 3C).
Figure 3.
Identification of 14-3-3 proteins within chloroplasts. A, Immunolocalization of the 14-3-3 proteins within the chloroplast using electron microscopy and anti-GF14 Mabs. Using a Mab that recognizes eight of the 10 Arabidopsis 14-3-3 isoforms, electron-dense gold-labeled particles representing 14-3-3s were found within the cytoplasm and the chloroplasts. White arrowheads indicate chloroplast 14-3-3s; black arrowheads indicate cytoplasmic 14-3-3s. B, Localization of 14-3-3 proteins within the chloroplast. Higher magnification of one of the chloroplasts shown in 3A indicates that gold particles representing 14-3-3 proteins were found within the chloroplast stroma and apparently on internal membranes. No concentration of gold particles was noted along the outer membrane. C, Control chloroplast . When an irrelevant monoclonal antibody directed against Dictyostelium spores was used as a control, no gold particles were detected. D, Western-blot analysis of pea chloroplasts and suborganellar fractions using anti-GF14 Mabs. Intact chloroplasts (CP), thermolysin-treated chloroplasts, and stromal extract from pea were analyzed for 14-3-3 content using SDS-PAGE and immunoblotting with anti-GF14 Mab. When possible, approximately equivalent amounts of protein (approximately 5 μg) were added to each lane as measured by Bradford analysis. Recombinant Zm GF14-12 (1 ng) was added as an antibody control. E, Molecular identification of specific 14-3-3 isoforms in Arabidopsis chloroplast-isolated stromal extract. Isoform-specific antibodies that recognize each of the Arabidopsis 14-3-3s were used to analyze Arabidopsis chloroplast stromal and cytoplasmic extracts electrophoresed with SDS-PAGE. Equivalent amounts of total protein (3 μg) from extracts were loaded in each lane. Transferred blots were incubated with antibody sera diluted 1/10,000, and immunogenic material was located using chemiluminescent detection. Positive western signals were detected for anti-ε, -ν, -υ, and -μ antibodies, while no signal was detected for the others, including anti-ω. To demonstrate antibody specificity, similar blots were incubated with diluted sera to which peptides that were used to generate the antibodies were added. These signals were abolished, indicating anti-14-3-3 specificity. Antibodies to the chloroplast stromal protein SecA and cytoplasmic protein BiP (a protein associated with the endoplasmic reticulum luminal HSC70s) were used to demonstrate the lack of cross-contamination between chloroplastic and cytoplasmic samples.
Confirmation and further sublocalization of 14-3-3 proteins within the chloroplast was accomplished using the GF14 Mab-15 monoclonal antibody for western analysis of suborganellar fractions of well-characterized and protease-treated pea chloroplasts (Cline, 1986). Consistent with the immuno-electron microscopy results of Arabidopsis, 14-3-3s were present in intact pea chloroplasts and stroma (Fig. 3D). The strongest immunoreactive signal was present in the stroma as a doublet of approximately 30 to 32 kD. Treatment of the chloroplast preparation with protease did not alter the size of the 14-3-3 proteins detected, nor was the amount of 14-3-3 within the chloroplast significantly reduced by protease treatment. Given that 14-3-3s are known to be especially sensitive to proteases (Lu et al., 1994; data not shown), these data indicate that the majority of the chloroplastic 14-3-3s were internal to the chloroplast, and not attached to the outside of the chloroplast or inadvertently carried along with the preparation. The use of pea chloroplasts establishes the presence of chloroplastic 14-3-3s in a well-characterized chloroplast system and across species, but cannot address issues of whether there is any selectivity of which isoforms might be present within chloroplasts.
To determine which specific 14-3-3 isoforms are present in chloroplasts, we returned to Arabidopsis, for which the complete family of 10 14-3-3 isoforms has been described (Wu et al., 1997b). Isoform-specific antibodies were used in a western analysis of stromal extracts from isolated, protease-treated Arabidopsis chloroplasts (Fig. 3E). Surprisingly, multiple isoforms were found in the stromal extract. The relative levels of 14-3-3 isoforms detected in the stromal extract were At GF14ν ≫ At GF14ε > At GF14μ ≫ At GF14υ. Each isoform was itself present as a doublet on denatured SDS-PAGE, suggesting either cleavage or modification of the stromal 14-3-3s. While cDNA sequence analysis of the stromal-localized 14-3-3s did not reveal any obvious transit peptide/cleavage sites, phosphorylation of plant 14-3-3s has been reported (Lu et al., 1994). Further experiments are necessary to fully understand the apparent modification of isoforms in the chloroplasts.
For comparison of cytoplasmic levels of the chloroplast-identified 14-3-3 stromal extracts, cytoplasmic supernatant fractions were analyzed with the 14-3-3 isoform-specific antibodies. Cytoplasmic levels of the isoforms were different from that of the chloroplast stromal material, again indicating potential selectivity of the 14-3-3 isoforms present in the chloroplast. In fact, At GF14ω which was present at low levels in the leaf cytoplasm, appeared to be excluded from the chloroplast stromal fraction. Antibodies to cytoplasmic (BiP) and chloroplast (SecA) marker proteins were used to demonstrate the integrity of the stromal and cytoplasmic fractions used in these studies. No cytoplasmic contamination, as indicated by BiP, was detected in the stromal extract.
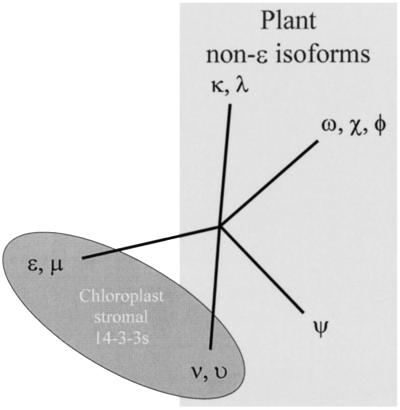
Phylogenetic analysis of the 14-3-3s detected in the chloroplast stromal extracts revealed several important features (Fig. 4). First, the four chloroplast stromal 14-3-3s are from two distinct classes, the ε-like and the non-ε-like isoforms. This was surprising, as the ε and non-ε distinction is thought to represent the earliest divergence within plant species (Wu et al., 1997b). The second feature is that the ν and υ isoforms are from their own distinctive branch of the non-ε forms. This clustering suggests that function may be reflected in a phylogenetic manner, such that similar phylogeny relates to similar function or location. The ε-like isoforms ε and μ may represent an ancestoral or constant function, as all ε-like isoforms (animals, plants, etc.) are anciently diverged from the other isoforms of their own species.
Figure 4.
Phylogenetic similarity of chloroplast 14-3-3s. Amino acid sequence comparison of the 10 Arabidopsis 14-3-3 isoforms using the phylogenetics analysis program PHYLIP 3.5 distinguished five distinct groups, one ε-like and four non-ε (Wu et al., 1997b). Placement on the phylogenetic tree of the 14-3-3s detected in the chloroplast stromal extract was confined to two divergent and distinct groups, one ε-like and one non-ε-like. Isoforms ε and μ are ε-like forms; ν and υ are clearly non-ε forms, but represent a unique cluster among the non-ε isoforms. This suggests the possibility that chloroplast-specific 14-3-3s may be a distinct evolutionary lineage. However, the presence of ε-like forms in the chloroplast stroma and elsewhere further suggests that certain chloroplast 14-3-3 duties require a general form (ε) and a specialized form (ν or υ), perhaps in the form of a heterodimer.
The presence of divergent isoforms within the chloroplast suggests the potential of specialized heterodimerization among these 14-3-3s. It was interesting that one monomer of the heterodimeric mitochondrial import stimulating factor (MSF) is the mammalian 14-3-3ε form, which is then partnered with δ or ζ, non-ε forms. One further recent piece of evidence that supports the theory of ε and non-ε isoform heterodimers is the identification of both ε-like and non-ε isoforms in the nuclei of HeLa cells as cruciform-binding proteins (Todd et al., 1998). It is tempting to speculate that the pairing of an ε form with a non-ε isoform will be a common factor for 14-3-3 heterodimers. This model is not without biophysical merit, as the association of plant ε isoforms with non-ε isoforms has been demonstrated to be dynamic in nature (Wu et al., 1997a). This could serve to allow increased flexibility for function from a limited isoform pool. The common-link ε isoforms may also provide some type of basal requirement for the 14-3-3 dimers. A more complete understanding of the actual physical makeup and distribution of heterodimers is required to fully answer this question of significance.
The interaction between PSI-N and 14-3-3s broadens the paradigm for 14-3-3/target associations. The PSI-N leader domain does not contain the canonical 14-3-3 phospho-Ser interaction sequence. However, recent data analyzing the interaction between 14-3-3 and ExoS clearly demonstrate that the canonical 14-3-3 binding site need not be present in the target protein (Masters et al., 1999). The PSI-N/14-3-3 interaction extends the growing realization that 14-3-3s can interact with non-canonical targets, and mutagenesis of the PSI-N interaction domain will reveal the amino acids required for this type of interaction.
The ability of a plant 14-3-3 to bind to a nuclear-encoded chloroplast precursor (Sehnke and Ferl, 1995) suggested the heretofore-undiscovered presence of 14-3-3s within chloroplasts. The only other example of 14-3-3s binding to import precursor proteins is MSF, a collection of 14-3-3s that (in animals, at least) acts to mediate mitochondrial precursor import on the external surface of the organelle (Komiya et al., 1997). MSF releases the precursor to import receptors on the mitochondrial outer membrane surface and, after ATP hydrolysis, is released back into the cytosol to recruit additional precursors. However, no reports of intra-mitochondrial 14-3-3s exist, further suggesting that MSF is a cytosolic factor. Therefore, the presence of 14-3-3s within chloroplasts represents a unique organellar localization for 14-3-3, especially since 14-3-3s have no import or precursor sequences to suggest their own direct import. This interaction also suggests a functional role for 14-3-3 during import. However, it would be premature to assign or limit the stromal 14-3-3s discovered here to roles involving solely import. The discovery of pPSI-N/14-3-3 interactions should allow for future functional characterization studies and further our understanding of 14-3-3 involvement in diverse cellular processes.
ACKNOWLEDGMENTS
We are grateful to the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) DNA Sequencing Core Laboratory for sequencing the yeast two-hybrid subclones and the original At pPSI-N cDNA clone. We also thank Dr. Charles Guy for providing the BiP monoclonal antibody used in this study.
Footnotes
This work was supported by a grant from the U.S. Department of Agriculture, National Research Initiative (grant no. 97–35304–4942). This article is Florida Agricultural Experiment Station Journal Series No. R–07231.
LITERATURE CITED
- Aitken A. 14-3-3 and its possible role in coordinating multiple signaling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- Alam R, Hachiya N, Sakaguchi M, Kawabata S, Iwanaga S, Kitajima M, Mihara K, Omura T. cDNA cloning and characterization of mitochondrial import stimulation factor (MSF) purified from rat liver cytosol. J Biochem (Tokyo) 1994;116:416–425. doi: 10.1093/oxfordjournals.jbchem.a124541. [DOI] [PubMed] [Google Scholar]
- Anderson JV, Li Q-B, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994;104:1359–1370. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihn EA, Paul AL, Wang SW, Erdos GW, Ferl RJ. Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J. 1997;12:1439–1445. doi: 10.1046/j.1365-313x.1997.12061439.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chaddock AM, Mant A, Karnauchov I, Brink S, Herrmann RG, Klösgen RB, Robinson C. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Import of proteins into chloroplasts: membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J Biol Chem. 1986;261:14804–14810. [PubMed] [Google Scholar]
- de Vetten NC, Lu G, Ferl RJ. A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell. 1992;4:1295–1307. doi: 10.1105/tpc.4.10.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 proteins and signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Aitken A, Bertsch U, Soll J. A plant homologue to mammalian brain 14-3-3 protein and protein kinase C inhibitor. FEBS Lett. 1992;296:222–224. doi: 10.1016/0014-5793(92)80384-s. [DOI] [PubMed] [Google Scholar]
- Ichimura T, Uchiyama J, Kunihiro O, Ito M, Horigome T, Omata S, Shinkai F, Kaji H, Isobe T. Identification of the site of interaction of the 14-3-3 protein with phosphorylated tryptophan hydroxylase. J Biol Chem. 1995;270:28515–28518. doi: 10.1074/jbc.270.48.28515. [DOI] [PubMed] [Google Scholar]
- Knoetzel J, Simpson DJ. The primary structure of a cDNA for PsaN, encoding an extrinsic lumenal polypeptide of barley photosystem I. Plant Mol Biol. 1993;22:337–345. doi: 10.1007/BF00014940. [DOI] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Schatz G, Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku NO, Liao J, Omary MB. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 1998;17:1892–906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- Liu YC, Elly C, Yoshida H, Bonnefoy-Berard N, Altman A. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. J Biol Chem. 1996;271:14591–14595. doi: 10.1074/jbc.271.24.14591. [DOI] [PubMed] [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA. 1992;89:11490–11494. doi: 10.1073/pnas.89.23.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Sehnke PC, Ferl RJ. Phosphorylation and calcium binding properties of an Arabidopsis GF14 brain protein homolog. Plant Cell. 1994;6:501–510. doi: 10.1105/tpc.6.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of pseudomonas aeruginosa [in process citation] Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Nielsen VS, Mant A, Knoetzel J, Møller BL, Robinson C. Import of barley photosystem I subunit N into the thylakoid lumen is mediated by a bipartite presequence lacking an intermeditate processing site. J Biol Chem. 1994;269:3762–3766. [PubMed] [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P. Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 1998;118:691–699. doi: 10.1104/pp.118.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sehnke PC, Ferl RJ. Nucleotide sequence of an Arabidopsis thaliana cDNA clone (accession no. U32176) encoding the complete precursor for a homolog to the barley extrinsic thylakoid lumenal polypeptide PSI-N. Plant Physiol. 1995;109:1126. [Google Scholar]
- Sehnke PC, Ferl RJ. Plant metabolism: enzyme regulation by 14-3-3 proteins. Curr Biol. 1996;6:1403–1405. doi: 10.1016/s0960-9822(96)00742-7. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Somerville SC, Ogren WL. Isolation of photosynthetically active protoplasts and chloroplasts from Arabidopsis thaliana. Plant Sci Lett. 1981;21:89–96. [Google Scholar]
- Todd A, Cossons N, Aitken A, Price G, Zannis-Hadjopoulos M. Human cruciform binding protein belongs to the 14-3-3 family. Biochemistry. 1998;37:14317–14325. doi: 10.1021/bi980768k. [DOI] [PubMed] [Google Scholar]
- Wu K, Lu G, Sehnke P, Ferl RJ. The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch Biochem Biophys. 1997a;339:2–8. doi: 10.1006/abbi.1996.9841. [DOI] [PubMed] [Google Scholar]
- Wu K, Rooney MF, Ferl RJ. The Arabidopsis 14-3-3 multigene family. Plant Physiol. 1997b;114:1421–1431. doi: 10.1104/pp.114.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- Yuan J, Henry R, McCaffery M, Cline K. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science. 1994;266:796–798. doi: 10.1126/science.7973633. [DOI] [PubMed] [Google Scholar]