Abstract
Significant transcriptome alterations are detected in the brain of patients with amyotrophic lateral sclerosis (ALS), including carriers of the C9orf72 repeat expansion and C9orf72-negative sporadic cases. Recently, the expression of repetitive element transcripts has been associated with toxicity and, while increased repetitive element expression has been observed in several neurodegenerative diseases, little is known about their contribution to ALS. To assess whether aberrant expression of repetitive element sequences are observed in ALS, we analysed RNA sequencing data from C9orf72-positive and sporadic ALS cases, as well as healthy controls. Transcripts from multiple classes and subclasses of repetitive elements (LINEs, endogenous retroviruses, DNA transposons, simple repeats, etc.) were significantly increased in the frontal cortex of C9orf72 ALS patients. A large collection of patient samples, representing both C9orf72 positive and negative ALS, ALS/FTLD, and FTLD cases, was used to validate the levels of several repetitive element transcripts. These analyses confirmed that repetitive element expression was significantly increased in C9orf72-positive compared to C9orf72-negative or control cases. While previous studies suggest an important link between TDP-43 and repetitive element biology, our data indicate that TDP-43 pathology alone is insufficient to account for the observed changes in repetitive elements in ALS/FTLD. Instead, we found that repetitive element expression positively correlated with RNA polymerase II activity in postmortem brain, and pharmacologic modulation of RNA polymerase II activity altered repetitive element expression in vitro. We conclude that increased RNA polymerase II activity in ALS/FTLD may lead to increased repetitive element transcript expression, a novel pathological feature of ALS/FTLD.
Introduction
Amyotrophic lateral sclerosis (ALS), the most common motor neuron disease, is a devastating and fatal neurodegenerative disease characterized by progressive muscle weakness, atrophy and spasticity. Accumulating evidence suggests that ALS is a complex, multisystem disease involving various brain regions (1). On interest, a subset of ALS patients develop frontotemporal lobar degeneration (FTLD), a neurological condition affecting frontal and temporal lobes, and leading to cognitive and behavioral impairments (2,3). Furthermore, about 15% of the patients suffering from FTLD also develop motor neuron disease (4).
A pathological hallmark linking ALS and FTLD is the presence of cytoplasmic inclusions in neurons and glia. In 2006, Tar DNA binding protein 43 (TDP-43) was identified as the major component of the ubiquitin-positive inclusions observed in the brain and/or spinal cord of ALS patients (5). In fact, TDP-43-positive inclusions are observed in the majority of ALS patients and in the most frequent pathological subtype of FTLD (FTLD-TDP) (5–8).
While the majority of ALS and FTLD cases have an unknown etiology, a subset can be attributed to known gene mutations. A hexanucleotide GGGGCC repeat expansion [(G4C2)exp] in a non-coding region of chromosome 9 open reading frame gene (C9orf72), is the most common genetic abnormality in ALS/FTLD cases (c9ALS/FTLD) (9,10). While unaffected individuals typically carry up to 30 repeats, c9ALS/FTLD patients carry hundreds to thousands of (G4C2) repeats (11), accounting for approximately 6% of sporadic cases, as well as 25-40% of familial FTLD and ALS cases (9,10,12). In addition to TDP-43 pathology, the accumulation of C9orf72 sense and antisense transcripts containing (G4C2)exp repeats into nuclear RNA foci constitutes characteristic pathology associated with c9ALS/FTLD. Furthermore, repeat-containing C9orf72 transcripts are susceptible to repeat-associated non-ATG (RAN) translation resulting in dipeptide-repeat proteins collectively referred to as c9RAN proteins (13–16).
Recently, our analyses of RNA sequencing (RNAseq) data obtained from human brain tissues revealed widespread transcriptome changes in c9ALS and C9orf72-negative ALS cases compared to healthy controls, and highlighted divergence in mRNA expression, splicing and alternative polyadenylation between c9ALS and C9orf72-negative ALS (17). While these analyses were mostly focused on protein-coding RNAs, little is known about repetitive sequences which account for ∼45% of the human genome (18). While previously considered “junk DNA”, repetitive sequences are now recognized as a major source of genetic variation and genome evolution (18), and have been associated with diverse genetic disorders (19–22), some of which involve the central nervous system (23–29).
DNA transposons and retrotransposons constitute the two main classes of repetitive elements. In human genomes, DNA transposons lack the ability to move, whereas a subset of retrotransposons retain their ability to copy themselves through an RNA intermediate that is reverse transcribed and inserted into a new genomic location. Retrotransposons can be divided based on the presence (e.g. endogenous retroviruses) or absence (e.g. long interspersed nuclear elements or LINEs, and short interspersed nuclear elements or SINEs) of long terminal repeats (LTR). Among the non-LTR retrotransposons, only the LINE-1 class, together with the Alu and SVA SINE elements, are currently reported to be active in humans, and collectively represent approximately one third of the human genome (19). Retrotransposons are particularly active in neuronal progenitor cells, and have been proposed to create genomic diversity between neurons (30).
Recently, repetitive element biology has been associated with TDP-43 and neurodegenerative diseases. In particular, the human endogenous retrovirus HERV-K was found to accumulate in the brain of ALS patients, and the RNA levels of HERV-K polymerase positively correlated with the levels of transcripts encoding TDP-43 (TARDBP) (23). Recent studies also support TDP-43 as a non-transcriptional modulator of HERV-K protein accumulation (31). Moreover, analyses of pre-existing datasets on TDP-43 RNA binders showed that TDP-43 binds to RNA of diverse transposable elements, and reduced binding of these elements to TDP-43 was observed in the brain of FTLD patients (24). In more recent studies, a human TDP-43 Drosophila model recapitulating TDP-43 pathology seen in human disease, as well as locomotor abnormalities and early death, observed expression of diverse repetitive elements (32). Of interest, downregulation of one of these repetitive elements in the flies was able to reduce the human TDP-43-associated toxicity (32). Overall, these studies suggest an important link between TDP-43 and repetitive element biology.
In differentiated cells, diverse mechanisms are in place to silence the expression and/or mobility of repetitive elements. These include not only post-transcriptional regulation (e.g. RNA interference) but also various epigenetic mechanisms (33–35). Despite this tight repression, some degree of repetitive element accumulation or retrotransposition activity has been observed in patients with Schizophrenia, Ataxia telangiectasia, Creutzfeldt-Jakob disease, Fragile X, Rett syndrome, and ALS (23,25–29). In the present study, we examined repetitive element expression in post-mortem brain tissue from ALS patients carrying the C9orf72 (G4C2)exp repeat versus C9orf72-negative cases and healthy controls. We also examined whether observed changes in repetitive element expression extend to cases presenting FTLD phenotypes, either with or without comorbid ALS (including both carriers and non-carriers of the C9orf72 repeat expansion). Providing mechanistic insight, we identify a positive relationship between RNA polymerase II activity and repetitive element expression in ALS/FTLD postmortem brain, and demonstrate that pharmacologic inhibition of RNA polymerase II activity in cells leads to a reduction in repetitive element transcript abundance.
Results
Repetitive element transcripts are enriched in the frontal cortex of c9ALS cases
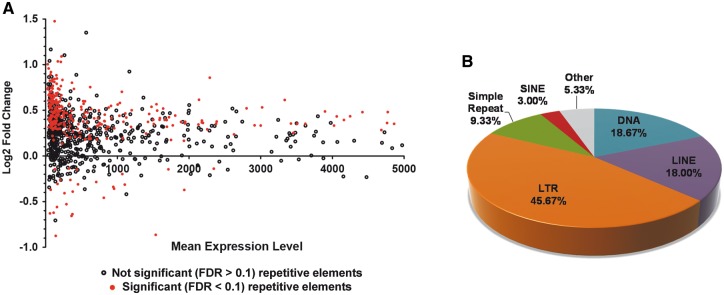
We have previously identified changes in gene expression, alternative polyadenylation and alternative splicing using RNAseq data from 8 c9ALS, 10 C9orf72-negative ALS (referred as sALS from now on), and 9 healthy controls (17). Here we analysed the same datasets to examine expression of repetitive element transcripts in frontal cortex and cerebellum. Using the HOMER program analyzeRepeats in tandem with DESeq for differential expression analyses (see Methods for more details), we identified transcripts belonging to multiple types of repetitive elements in the frontal cortex of c9ALS, sALS, and healthy cases (Table 1). Of note, each class of repetitive element sequences identified was represented in similar proportions for all three different study groups, with the most represented classes being SINEs, LINEs and LTRs (Table 1). When comparing the global population of repetitive elements among the three experimental groups, we found a significant increase in repetitive element expression in the frontal cortex, but not in the cerebellum, of c9ALS subjects compared to sALS (Fig. 1A). In particular a total of 300 different repetitive elements were significantly altered in c9ALS compared to sALS (FDR < 0.1), which are distributed across several classes (repetitive elements of the LTR class are the most abundant [45.67%], followed by DNA elements [18.67%] and LINEs [18%]; Fig. 1B). Of note, the majority of altered repetitive elements in c9ALS were increased (272, 90.7%) compared to a much lower proportion of downregulated elements (28, 9.3%). In addition, the majority of repetitive elements with altered expression belong to classes that are not necessarily the most representative classes typically found to be expressed in the frontal cortex (compare Table 1 and Fig. 1B). This data demonstrates that despite uniform distribution of repetitive element classes expressed in c9ALS, sALS and healthy control frontal cortices, a global event may lead to the observed changes in disease, which are not specific to a particular repetitive element class.
Table 1.
Proportion of repetitive element classes identified in the frontal cortex of the different study groups used for RNAseq
| % of c9ALS | % of sALS | % of Healthy | |
|---|---|---|---|
| SINE | 26.00 | 26.82 | 26.77 |
| rRNA | 27.83 | 24.85 | 25.28 |
| LINE | 18.45 | 18.53 | 18.27 |
| LTR | 9.74 | 10.36 | 10.10 |
| DNA | 5.66 | 6.36 | 6.00 |
| Low_Complexity | 4.68 | 5.48 | 5.43 |
| Simple_repeat | 4.05 | 4.83 | 4.73 |
| tRNA | 1.92 | 1.11 | 1.69 |
| Other | 1.68 | 1.66 | 1.74 |
Figure 1.
Repetitive element transcripts are enriched in the frontal cortex of c9ALS cases. Using RNAseq data from 9 healthy controls, 10 sALS and 8 c9ALS cases (17), we evaluated changes in the expression of repetitive elements. (A) MA plot showing the Log2 fold change and mean repetitive element expression levels in the human cortex tissue of c9ALS brains compared to sALS. Note the 300 significant changes are depicted in red (FDR < 0.1). (B) Main class distribution of the repetitive elements significantly altered in the frontal cortex of c9ALS vs. sALS cases.
Misregulation of repetitive elements in the frontal cortex extends to C9orf72 ALS cases with FTLD clinical diagnosis and non-ALS FTLD cases
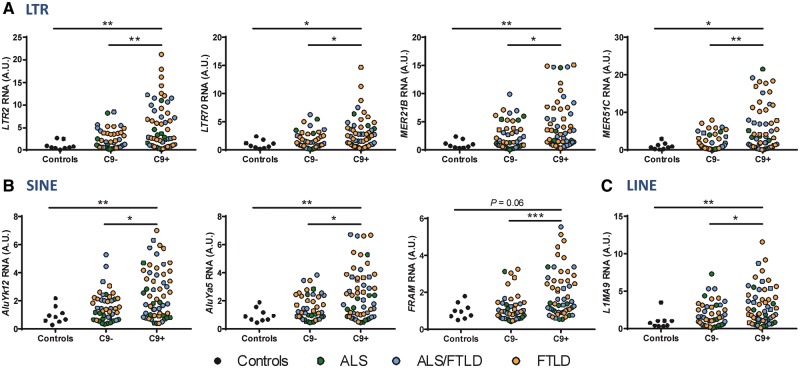
Quantitative RT-PCR (qRT-PCR) analyses were used to assay several repetitive element transcripts across a large collection of patient samples, representing not only ALS, but also ALS/FTLD and FTLD cases with (C9+) and without (C9-) the C9orf72 hexanucleotide repeat expansion (Supplementary Material, Table S1). Validations of several repetitive elements from each of three different repetitive element classes (LTR, SINE and LINE) are included in Figure 2. Note that in addition to validating some of the 300 altered repetitive elements with FDR < 0.1 (LTR2, MER51C, AluYk12, FRAM, L1MA9), we were also able to validate some repetitive elements presenting FDR > 0.1 (LTR70, MER51B, AluYa5). Furthermore, not only were the repetitive elements significantly increased in C9+ vs. C9-, but they were also significantly increased in C9+ with respect to healthy controls (Fig. 2). Of note, while qRT-PCR validation revealed a trend towards increased repetitive element expression in non-C9orf72 cases compared to controls, this did not reach statistical significance.
Figure 2.
Increased repetitive element accumulation in the frontal cortex of c9ALS/FTLD cases. A series of repetitive elements identified to be upregulated in c9ALS vs. sALS cases in our RNAseq analyses were validated in a large cohort of cases (Supplementary Material, Table S1) by qRT-PCR: 56 C9+, 46 C9-, 9 healthy controls. Validations were done for elements belonging to LTR (A), SINE (B) and LINE (C) repetitive element classes. Note, three of the repetitive elements shown (LTR70, MER21B, AluYa5) did not reach statistical significance from the RNAseq data (FDR > 0.1) but we observed significant changes by qRT-PCR. Statistical analyses were performed to compare all groups using a One-way ANOVA followed by Dunn’s multiple comparison test, *P < 0.05, **P < 0.01, ***P < 0.001.
Previous studies have reported an increased accumulation of repetitive element transcripts with aging (36,37). Thus, we used a linear regression model to test whether disease duration (defined as the time from disease onset to death), age at death, C9orf72 genotype or disease subtype are associated with repetitive element accumulation in our cohort. We found that, neither disease duration nor age at death were associated with the accumulation of repetitive elements, suggesting that the increase in repetitive elements is not driven by either age at death or disease duration (Supplementary Material, Table S2). Instead, repetitive element transcript expression was strongly associated with C9orf72 genotype after correcting for disease duration, disease subtype and age at death (Supplementary Material, Table S2), confirming our observed results. Of interest, having an FTLD phenotype also appears to contribute to a larger accumulation of repetitive elements in the frontal cortex (Supplementary Material, Table S2). Overall, the data indicate that significantly increased repetitive expression is detected in C9orf72 repeat expansion carriers (including c9ALS, c9ALS/FTLD, and c9FTLD) in comparison to sporadic cases and controls; and that the observed accumulation of repetitive elements is not due to differences in age at death or disease duration of our patient cohort.
Accumulation of repetitive elements does not associate with TDP-43
Previous studies have associated TDP-43 biology with repetitive element expression. In one of these studies, the levels of TARDBP RNA positively correlated with RNA levels of the human endogenous retrovirus HERV-K (23). However, when measuring TARDBP RNA levels in the frontal cortex of our large cohort of human cases, we were unable to find significant positive correlations with repetitive element expression (Supplementary Material, Fig. S1). Moreover, another report has shown that TDP-43 binds repetitive elements and this binding is lost in cortical tissues of FTLD cases (24), and, more recently, TDP-43 has been shown to modulate repetitive element expression in a Drosophila model by disrupting siRNA-mediated gene silencing of repetitive elements (32). In order to determine whether the burden of TDP-43 pathology was associated with repetitive element expression, we used an immunoassay to measure the amount of phosphorylated TDP-43 (pTDP-43) in protein insoluble, urea soluble fractions from the frontal cortex of our large patient cohort. While ALS cases contained virtually no detectable amounts of pTDP-43, the presence of ALS/FTLD and FTLD cases in both non-C9orf72 and C9orf72 study groups allowed the detection of significant amounts of pTDP-43 compared to healthy controls (Supplementary Material, Fig. S2). When testing associations between pTDP-43 protein levels and repetitive element transcript expression we found a significant but mild positive correlation (Spearman’s r ∼0.2, Supplementary Material, Fig. S2). However, this correlation was lost when adjusting for disease duration, age at death, C9orf72 genotype, and disease subtype (data not shown). Overall, our data did not support the levels of pTDP-43 as a major factor in the regulation of repetitive element accumulation.
RNA polymerase II function in repetitive element expression
Previous reports have observed that changes in chromatin can reactivate HIV from latency (38–40). More recently, it has been proposed that the positive transcriptional elongation factor b (p-TEFb) can activate HIV from latency by promoting recruitment of RNA polymerase II (RNApol II) to HIV LTRs, leading to transcription elongation. P-TEFb is a cyclin-dependent kinase composed of different subunits, including CDK9. In HIV, RNApol II elongation is favored by p-TEFb-mediated phosphorylation of both the C-terminal domain of RPB1, which is the largest subunit of RNApol II, along with phosphorylation of the negative elongation factors NELF and DSIF (41–43). Of interest, DSIF is composed of SUPT4H1 and SUPT5H, and phosphorylation of SUPT5H transforms DSIF into a positive elongation factor. Thus, we aimed to explore the possibility that repetitive element expression may require RNApol II activation, through removal of either repressors or phosphorylation of associated factors.
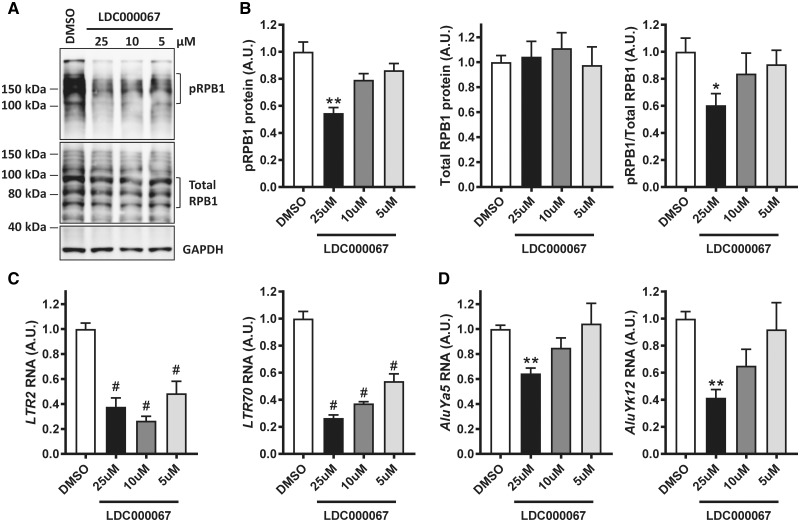
To determine if pharmacologic modulation of RNApol II activity impacts expression of repetitive elements, we treated human embryonic kidney (HEK293T) cells with an established inhibitor of CDK9 activity, LCD000067 (44), for 3 h at different doses. It has been previously shown that 10 μM of this compound for 1-3 h is sufficient to decrease CDK9 catalytic activity in vitro, as demonstrated by a decrease in phosphorylation of serine 2 in RPB1 (44). We observed a dose-dependent decrease in pRPB1 following exposure to the CDK9 inhibitor in HEK293T cells, while no significant change was detected in overall RPB1 protein (Fig. 3A and B) or RNA levels (POL2RA, Supplementary Material, Fig. S3). Moreover, the expression of diverse repetitive elements (that we observed to be altered in human tissues) was significantly decreased as a result of reduced pRPB1 levels following CDK9 inhibition (Fig. 3C and D), with a larger effect in repetitive elements from the LTR class (Fig. 3C). In addition, the levels of pRPB1 strongly correlated with repetitive element transcript expression in cells treated with different amounts of CDK9 inhibitor (Supplementary Material, Table S3), further supporting the role of RNA pol II in modulating the expression of repetitive element transcripts. Of interest, the levels of SUPT4H1 RNA (Supplementary Fig. S3) were also reduced in cells treated with the CDK9 inhibitor, suggesting that SUPT4H1 may modulate the expression of repetitive elements by RNApol II. This is of particular importance as SUPT4H1 has been involved in the transcription of long nucleotide repeats, including those in huntingtin (CAG repeats) (45) and C9orf72 (G4C2 repeats) (46). However, the levels of repetitive element transcripts and pRPB1 protein were unchanged following SUPT4H1 knockdown in HEK293T cells for 48 h (Supplementary Material, Fig. S4). Overall, the data demonstrate that transcription of repetitive elements does not require SUPT4H1 as it is for huntingtin (CAG repeats) (45) and C9orf72 (G4C2 repeats) expression (46).
Figure 3.
Inhibition of CDK9 leads to a decrease in RNApol II activity and repetitive element expression. HEK293T cells were treated with DMSO control and the indicated amounts of LDC000067, a CDK9 inhibitor, for a total of 3 h. (A) Immunoblots showing the levels of phosphorylated RNApol II largest subunit (pRPB1), total RPB1 and GAPDH, in response to different doses of CDK9 inhibitor. (B) Quantification of immunoblots from 2 independent experiments in which each condition was performed in duplicates, and in which pRPB1 and total RPB1 protein levels were normalized to GAPDH or as pRPB1/Total RPB1 ratio. (C, D) qRT-PCR analyses show a significant response to CDK9 inhibition of repetitive elements belonging to the LTR (C) and SINE (D) classes. Statistical analyses were performed to compare all groups using a One-way ANOVA followed by Bonforreoni’s multiple comparison test, *P < 0.05, **P < 0.01, #P < 0.0001.
Increased RNApol II activity in the human frontal cortex of ALS/FTLD cases
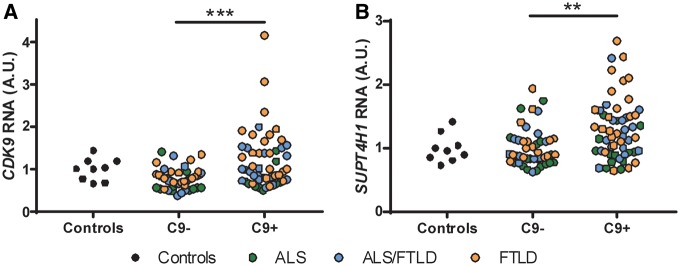
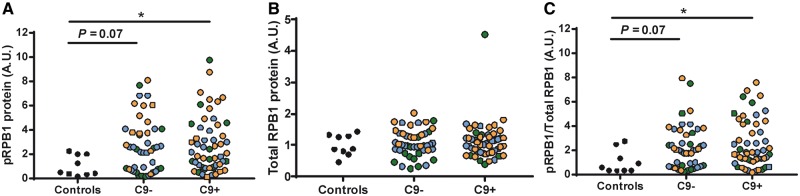
To determine whether RNApol II activity may be increased in the frontal cortex of ALS/FTLD cases, we first measured the RNA levels of CDK9, SUPT4H1 and SUPT5H RNA. Interestingly, in our large cohort of cases, we found a significant increase in the RNA levels of CDK9 (Fig. 4A) and SUPT4H1 RNA (Fig. 4B), but not in SUPT5H RNA (Supplementary Material, Fig. S5), in the C9orf72 cases. Moreover, both CDK9 and SUPT4H1 RNA levels strongly correlated with repetitive element expression (Table 2). Overall, the data suggest that elements that regulate RNApol II activity are involved in expression of repetitive elements. While CDK9 and SUPT4H1 RNA levels may not accurately reflect the proportion of active CDK9 and SUPT4H1 proteins promoting RNApol II transcription and elongation, we measured the levels of total and phosphorylated RPB1 (pRPB1) protein in the frontal cortex of our large cohort of cases for which we had sufficient tissue available (Supplementary Material, Table S1). The levels of pRPB1 protein (Fig. 5A and C), but not that of total RPB1 (Fig. 5B), were significantly increased in C9orf72 cases, suggesting increased RNApol II activity. More importantly, the levels of pRPB1 positively correlated with the transcript levels of all repetitive elements analysed (Table 3). Overall, our data suggest that increased RNApol II activity is involved in transcription of repetitive elements in the human frontal cortex of ALS/FTLD cases by a mechanism that is independent of C9orf72 genotype or a particular disease subtype.
Figure 4.
CDK9 and SUPT4H RNA levels are elevated in C9orf72-positive cases. The levels of CDK9 (A) and SUPT4H1 (B) RNA were assessed by qRT-PCR as described in methods for our large cohort of cases (Supplementary Material, Table S1). Statistical analyses were performed to compare all groups using a One-way ANOVA followed by Dunn’s multiple comparison test, **P < 0.01, ***P < 0.001. Of note, results from a subsequent linear regression model were also significant (C9+ vs. C9-: SUPT4H, P = 0.0288; CDK9, P = 0.0059), while correcting for disease duration, disease subtype, and age at death.
Table 2.
Significant associations were found between repetitive element transcript levels and CDK9 or SUPT4H1 RNA, measured by qRT-PCR, in the human frontal cortex
|
CDK9 RNA |
SUPT4H1 RNA |
||||
|---|---|---|---|---|---|
| ID | Class | Spearman's r (95% CI) | P (two-tailed) | Spearman's r (95% CI) | P (two-tailed) |
| LTR2 | LTR | 0.7261 (0.6180–0.8073) | <0.0001 | 0.8022 (0.7201–0.8622) | <0.0001 |
| LTR70 | LTR | 0.7395 (0.6355–0.8171) | <0.0001 | 0.8059 (0.7263–0.8641) | <0.0001 |
| MER21B | LTR | 0.6680 (0.5429–0.7641) | <0.0001 | 0.7464 (0.6473–0.8207) | <0.0001 |
| MER51C | LTR | 0.7577 (0.6595–0.8305) | <0.0001 | 0.8120 (0.7343–0.8688) | <0.0001 |
| AluYa5 | SINE | 0.7665 (0.6712–0.8369) | <0.0001 | 0.8311 (0.7597–0.8826) | <0.0001 |
| AluYk12 | SINE | 0.7366 (0.6316–0.8150) | <0.0001 | 0.7935 (0.7089–0.8557) | <0.0001 |
| FRAM | SINE | 0.7909 (0.7039–0.8545) | <0.0001 | 0.8431 (0.7759–0.8914) | <0.0001 |
| L1MA9 | LINE | 0.6379 (0.5048–0.7413) | <0.0001 | 0.7135 (0.6044–0.7963) | <0.0001 |
Spearman’s r correlation coefficients, 95% confidence intervals (CIs), and P values are presented. Note a P ≤ 0.005 was considered significant after Bonferroni correction.
Figure 5.
The levels of phosphorylated RPB1 protein are elevated in C9orf72-positive cases. The levels of phosphorylated RPB1 (pRPB1, (A) and total RPB1 (B) proteins were assessed by immunoblotting as described in methods for our large cohort of cases (Supplementary Material, Table S1) and normalized to GAPDH protein levels. Note the levels of pRPB1 are also presented normalized to total RPB1 levels (C). Statistical analyses were performed to compare all groups using a One-way ANOVA followed by Dunn’s multiple comparison test, *P < 0.05.
Table 3.
Repetitive element transcript levels significantly associate with protein levels of phosphorylated RPB1 (pRPB1), in the human frontal cortex
| pRPB1/Total RPB1 protein |
|||
|---|---|---|---|
| ID | Class | Spearman's r (95% CI) | P (two-tailed) |
| LTR2 | LTR | 0.5548 (0.3972–0.6805) | <0.0001 |
| LTR70 | LTR | 0.5689 (0.4170–0.6899) | <0.0001 |
| MER21B | LTR | 0.5088 (0.3446–0.6428) | <0.0001 |
| MER51C | LTR | 0.4976 (0.3305–0.6345) | <0.0001 |
| AluYa5 | SINE | 0.5120 (0.3466–0.6465) | <0.0001 |
| AluYk12 | SINE | 0.4996 (0.3319–0.6367) | <0.0001 |
| FRAM | SINE | 0.4030 (0.2188–0.5595) | <0.0001 |
| L1MA9 | LINE | 0.4931 (0.3261–0.6303) | <0.0001 |
Spearman’s r correlation coefficients, 95% confidence intervals (CIs), and P values are presented. Note a P ≤ 0.005 was considered significant after Bonferroni correction.
Discussion
In the present study, we performed new transcriptome analyses in a cohort of c9ALS, sALS and healthy control cases in which poly-A RNAseq was previously performed (17). Specifically, we geared our studies towards determining if repetitive element expression is altered in ALS. Previous studies have implicated altered expression of repetitive elements in ALS, as is the case of the human endogenous retrovirus-K (HERV-K) (23,47), and possibly extending to FTLD cases presenting TDP-43 pathology (24). Our bioinformatics analyses uncovered a significant upregulation of repetitive element transcript expression in c9ALS compared to sALS cases. In validating these changes using a larger cohort of patients that included ALS, ALS/FTLD and FTLD cases, we were able to validate not only those repetitive elements with a more stringent cut-off criteria (FDR < 0.1), but also some changes that did not meet the established cut-off. This apparent discrepancy can be easily explained by the fact that the RNAseq was done in libraries containing poly-adenylated (poly-A) RNA (mRNA), while the qRT-PCR validations were performed using cDNA that was made from total RNA, using random primers. Therefore, as the majority of repetitive elements are unlikely to contain a poly-A tail, our measurement of repetitive elements from the RNAseq data is likely uncovering only a small proportion of all repetitive elements altered in disease.
RNA polymerase II (RNApol II) has been involved in HIV reactivation from latency, through transcription elongation of its LTRs (48). Here we aimed to evaluate whether RNApol II activity is involved in the elevated expression of repetitive elements observed in ALS/FTLD. To do so, we performed cell culture studies aimed to decrease RNApol II activity in HEK293T cells. Despite being a non-neuronal line, inhibition of CDK9 was sufficient to decrease RNApol II activity in HEK293T cells, as assessed by phosphorylation of RPB1 (the largest subunit of RNApol II). Most importantly, this decrease in RNApol II activity significantly correlated with reduced repetitive transcript expression, while other non-repetitive element transcripts (e.g. GAPDH and RPLP0 used as endogenous controls) were unaltered under these conditions.
In the frontal cortex, altered repetitive element expression was most pronounced in cases containing the C9orf72 repeat expansion compared to non-C9orf72 cases. While qRT-PCR validation revealed a trend towards increased repetitive element expression in non-C9orf72 cases compared to controls, this did not reach statistical significance. When evaluating the levels of RNApol II activity and other transcription elongation factors in our large cohort of patients, we found increased pRPB1 in C9orf72 cases compared to healthy controls. In addition, the RNA levels of transcription factors CDK9 and SUPT4H1 were significantly upregulated in C9orf72 compared to non-C9orf72 cases. Moreover, the levels of pRPB1 protein, as well as of CDK9 and SUPT4H1 RNA, correlated with repetitive transcript expression in frontal cortex tissues, further supporting the idea that repetitive element expression in disease results from increased activity of RNApol II.
While SUPT4H1 RNA levels were elevated in the frontal cortex of c9ALS/FTLD cases, downregulation of SUPT4H1 did not alter repetitive element expression in vitro. This is of particular interest as SUPT4H1 has been shown to modulate expression of huntingtin (CAG repeats) (45) and C9orf72 (G4C2)exp repeats (46). Overall, this is not all surprising as repeats within repetitive element transcripts are not as homogenous as for the GGGGCC hexanucleotide repeat in the C9orf72 gene or the CAG trinucleotide repeat in huntingtin. Furthermore, our studies demonstrate that downregulation of SUPT4H1 does not affect RNApol II activity, suggesting that targeting SUPT4H1 for treatment of diseases with nucleotide repeat expansions in the C9orf72 and huntingtin genes will not alter transcription of other transcripts. While this particular approach for c9ALS/FTLD may not target repetitive element expression by directly affecting RNApol II activity, it is possible that reduction of C9orf72 repeat expansion transcript and its associated pathology would still have an effect on repetitive transcript expression, as we observed a strong association between the presence of the C9orf72 repeat expansion and repetitive element expression.
In ALS/FTLD cases containing the C9orf72 repeat expansion, the presence of protein pathology known as c9RAN proteins are characteristic features (13–16). The c9RAN proteins are dipeptide repeats that are translated from the G4C2 repeat expansion by a yet unknown mechanism. A total of 5 different dipeptide-repeat proteins can be generated from the sense (poly-GA, poly-GP, poly-PR) or antisense (poly-GP, poly-PR, poly-PA) strands containing the repeat expansion. While all inclusions are most abundant in brain areas including the frontal cortex and cerebellum, there are differences in the specific distribution of each particular dipeptide-repeat protein (49). The relative contribution of each particular dipeptide-repeat protein to disease pathogenesis remains largely unknown, but an increased number of recent and ongoing studies in this area will aid in deciphering the role of these dipeptide-repeat proteins in neurodegeneration. However, an intriguing link between dipeptide-repeat proteins and heterochromatin has recently been uncovered. As repetitive elements are known to be silenced in the nucleus through histone modifications and DNA methylation (50), the association of nuclear dipeptide-repeat protein inclusions with H3K9me2, a marker of heterochromatin (49), suggests that they may play a role in gene expression. Therefore, it is possible that more widespread changes involving chromatin may take place in C9orf72 cases compared to non-C9orf72 cases, which could then explain the increased expression of repetitive elements in C9orf72 cases. However, whether altered epigenetic modifications significantly affect repetitive element accumulation in ALS/FTLD remains to be elucidated.
While previous studies have observed a relationship between repetitive element expression and TDP-43 biology (24,32), we were unable to establish a link between the levels of either TARDBP RNA or pTDP-43 protein and repetitive element expression. Given that our initial identification of repetitive element accumulation in ALS cases was derived from RNAseq data of frontal cortex tissues, a region we know contains very minimal or no TDP-43 pathology in ALS, this finding is not surprising. Moreover, we observed no significant differences in pTDP-43 burden between non-C9orf72 and C9orf72 cases that could explain changes in repetitive element accumulation between these two groups. Of interest, while we found a very mild although significant association of pTDP-43 protein levels with repetitive element expression, this association was lost when adjusting for disease duration, age at death, C9orf72 genotype and disease subtype. Still, the presence of an FTLD phenotype was associated with repetitive element expression in the frontal cortex (Supplementary Material, Table S2), suggesting certain features characteristic of FTLD cases may account for repetitive element accumulation. As one of the obvious pathological features distinguishing the FTLD subtype is the presence of TDP-43 pathology in the frontal cortex, although our current findings do not support a major role for pTDP-43 burden in contributing to the aberrant regulation of repetitive element expression, additional investigation into the relationship between TDP-43 and repetitive element biology in disease is warranted.
Overall, our studies suggest that repetitive element expression is a key feature of ALS/FTLD, which strongly associates to a C9orf72 genotype and FTLD disease subtype. Moreover, our data suggest that repetitive element expression is mediated by increased transcription through RNApol II in frontal cortex tissues. Given that repetitive element transcripts have been shown to accumulate in neurodegenerative diseases (22,26,27), and expression of repetitive elements has been linked to toxicity (32,47), regulation of repetitive elements may occupy a more active and mechanistic role in the pathogenesis of aging-related diseases than previously considered. Therefore, future studies should be aimed to evaluate both the significance and specific contribution of key repetitive elements to the pathophysiology of ALS/FTLD.
Materials and Methods
Patient sample consents and approvals
Written informed consent was given by all participants or authorized family members. Then, participant and family information was gathered, autopsies were performed and postmortem analyses were conducted. All protocols were approved by the Mayo Clinic Institutional Review Board and Ethics Committee.
Clinical, genetic and pathological assessments
ALS/FTLD patients in this study were independently ascertained by trained neurologists as having ALS and/or FTLD upon neurological and pathological examinations. The presence or absence of a C9orf72 repeat expansion was determined using a previously described repeat-primed polymerase chain reaction method (9). A description of the cases used in this study is included in Supplementary Material, Table S1.
Bioinformatics analyses
Paired-end 100 bp reads were aligned to the hg19 human reference genome using Tophat v.2.0.6 (51). Repetitive element expression levels were produced with the software Homer (v.4.8) (52) and the hg19 repeatmasker annotation (53). The steps to obtain repetitive elements expression levels in Homer were as follows: Construct tag directories that capture primary alignments [makeTagDirectory -keepOne -flip -sspe], obtain raw counts of repetitive elements with the requirement that the repetitive element not be located within an exon [analyzeRepeats hg19 repeats -condenseGenes -noAdj -noexon], then get raw gene-counts from the same tag directory [analyzeRepeats rna hg19 -condenseGenes -count exons -strand - -noAd]. After obtaining the expression levels we then moved our analyses to DESeq for normalization and differential expression (54). To normalize the RNAseq libraries, we used the gene-counts as input into DESeq (v1.26.0), we then extracted the size factors for the RNAseq libraries from the gene-counts, and applied the size factors to their respective sample for the repetitive element counts during DESeq’s normalization step. The DESeq dispersion estimates were calculated using options “per-condition”, “gene-est-only”, “local”. Repetitive element significance was determined if FDR <10%. The RNAseq data used in our analyses is publicly available at the NCBI Gene Expression Omnibus under accession number GSE67196. Detailed information on library preparation and sequencing can be obtained from Prudencio et al. 2015 (17).
Cell culture, antibodies, primers and reagents
HEK293T cells were used in experiments aimed to alter RNApol II activity. Knock-down experiments were performed using siLentfect (Bio-Rad) following the manufacturer’s protocol for a total of 48 h. For immunoblot analyses the following antibodies were used: pRPB1 (1:1000, Phospho-Rpb1 CTD Ser2/5, Cell Signaling #4735), total RPB1 (1:1000, Rpb1 NTD, D8L4Y, Rabbit mAb, Cell Signaling #14958), GAPDH (1:10000, Meridian Life Science #H86504M). The CDK9 inhibitor, LDC000067 compound (44), was purchased from Selleck Chemicals (#S7461). Each experiment involving HEK293T cells was performed in triplicates per condition, with each experiment performed a minimum of two times. Then cells were harvested for both RNA and protein. RNA extraction from HEK293T cells is explained in “RNA extraction and quantitative RT-PCR” section. A list of qRT-PCR primers used in this study is provided in Supplementary Material, Table S4. Protein lysates from cells were prepared by lysing the cells with sonication in co-IP buffer (50 mM Tris–HCl, pH 7.4, 300 mM NaCl, 1% Triton X-100, 5 mM EDTA) supplemented with 10% SDS and 1% of protease and phosphatase inhibitors. Concentration of protein lysates was measured using bicinchonic acid assays (BCA), and 20 μg of heat-denatured protein was analysed by immunoblotting.
RNA extraction and quantitative RT-PCR
RNA was extracted from frozen human postmortem samples of frontal cortex using the RNAeasy Plus Mini Kit (QIAGEN, Germantown, MD, USA), as previously described (17). RNA integrity (RIN) was verified on an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and 500 ng of RNA (RIN ≥ 7.0) was used for reverse transcription polymerase chain reaction (RT-PCR). RNA extracted from HEK293T cells was performed using the Direct-zol RNA Miniprep (Zymo Research, Orange, CA). A total of 1,500 ng were used for RT-PCR. For human and cell extracted RNA, the RT-PCR reaction was performed using the High Capacity cDNA Transcription Kit with random primers (Applied Biosystems, Foster City, CA, USA), as per manufacturer’s instructions. Following standard protocols, quantitative real-time PCRs (qRT-PCR) were conducted using SYBR GreenER qPCR SuperMix (Invitrogen, Carlsbad, CA, USA) for all samples in triplicates. All primer pairs used are listed in Supplementary Material, Table S4. qRT-PCRs were run in an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative quantification was determined using the ΔΔCt method and normalized to the endogenous controls RPLP0 and GAPDH. Note the relative transcript levels were normalized to that of the healthy controls (mean set to 1).
Human postmortem sample preparation and immunoblotting procedure
Human brain tissue samples were homogenized in 10x volume of homogenate buffer (50mM Tris base [pH 8], 274mM NaCl, 5mM KCl, 5mM EDTA, 1% Triton-X-100, 1% SDS, protease inhibitor cocktail, 1mM PMSF, phosphatase inhibitor cocktail II and III), followed by sonication. Samples were centrifuged at 16,000 g for 15 min (4 °C) to remove cellular debris, and a standard BCA protein assay performed on the supernatant. A total of 30 µg of protein from each sample was diluted in dH2O, 2x Tris-glycine SDS sample buffer (Life Technologies), and 5% beta-mercaptoethanol (Sigma Aldrich), and heat-denatured for 5 min at 95 °C. Samples were run on 10% SDS-PAGE Tris-glycine gels (Life Technologies), and transferred to PVDF membrane (Millipore). Membranes were blocked in 5% non-fat dry milk in TBS/0.1% Triton-X-100, and incubated overnight in primary antibody diluted in 5% milk in TBS/0.1% Triton-X-100 rocking at 4 °C (anti-pRBP1 [1:1000, Cell Signaling Technology, Danvers MA]; anti-RBP1 [1:1000, Cell Signaling Technology, Danvers MA]; anti-GAPDH [1:10000, Meridian Life Science, Memphis TN]). Membranes were incubated in HRP-conjugated secondary antibodies (1:5000; Jackson ImmunoResearch Laboratories, West Grove PA) for 1 h at room temperature, and detected by ECL (PerkinElmer). Bands were quantified using Scion Image by analysing pixel density, and protein levels were normalized to GAPDH to control for protein loading. Note the relative protein levels were normalized to that of the healthy controls (mean set to 1).
Immunoassay analysis of insoluble pTDP-43 protein in human frontal cortex tissues
Brain homogenates were prepared as previously described (55). In brief, approximately 50 mg of frontal cortex tissue was homogenized in cold RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, protease and phosphatase inhibitors) and sonicated on ice. Homogenates were cleared by centrifugation at 100,000 g for 30 min at 4 °C. The supernatant was collected and the resulting pellet was resuspended in RIPA buffer and, re-sonicated and re-centrifuged to prevent carry-over of soluble material. The RIPA-insoluble pellet was then extracted using 7 M urea buffer, sonicated and centrifuged at 100,000 g for 30 min at 22 °C. The protein concentration of the urea-soluble supernatant was determined by Bradford assay. pTDP-43 levels in this fraction were evaluated using a sandwich immunoassay that utilizes MSD electrochemiluminescence detection technology. A mouse monoclonal antibody that detects TDP-43 phosphorylated at serines 409 and 410 (1:500, #CAC-TIP-PTD-M01, Cosmo Bio U.S.A.) was used as the capture antibody. The detection antibody was a Sulfo-tagged rabbit polyclonal C-terminal TDP-43 antibody (2 µg/ml; 12892-1-AP, Proteintech). Fractions were diluted in TBS and 35 µg of protein per well were tested in duplicate wells. Serial dilutions of recombinant pTDP-43 in TBS were used to prepare the standard curve. Response values corresponding to the intensity of emitted light upon electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120 were acquired, and pTDP-43 concentrations were interpolated using the standard curve.
Statistics
Unless otherwise indicated, one-way ANOVA followed by Dunn’s multiple comparison test was used to compare all study groups. We considered the difference between comparisons to be significant when P < 0.05 (*P < 0.05; **P < 0.01; ***P < 0.001, #P < 0.0001). Spearman r correlations were performed where indicated, and P ≤ 0.005 was considered significant after Bonferroni correction. To assess whether age at death is associated with repetitive element transcript expression, CDK9 and SUPT4H1 RNA, we used a generalized linear regression model in R, also adjusting for C9orf72 genotype, disease subtype and disease duration. We considered a significant association when P < 0.05, after adjusting within a group using the Bonferroni correction.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to all patients who donated tissues.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) [R01NS063964 (C.D.L.); R21NS089979 (K.B.B., T.F.G.); R35NS097273 (L.P.); R21NS084528 (L.P.); P01NS084974 (L.P., K.B.B., R.R.); P01NS099114 (L.P.); R01NS088689 (L.P.); R01NS093865 (L.P.); R35NS097261 (R.R.); R01NS080882 (R.R.)]; Department of Defense (ALSRP AL130125, L.P.); Mayo Clinic Foundation (L.P.); Mayo Clinic Center for Individualized Medicine (K.B.B., T.F.G., L.P.); Amyotrophic Lateral Sclerosis Association (K.B.B., T.F.G., L.P., M.P.); Robert Packard Center for ALS Research at Johns Hopkins (L.P.), Target ALS (L.P.); Association for Frontotemporal Degeneration (L.P.); Muscular Dystrophy Association (#416137, T.F.G.); Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Clinical Research Fellowship (U54 NS092091, M.V.B.). CReATe is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS). CReATe is funded through collaboration between NCATS and the NINDS. Funding to pay the Open Access publication charges for this article was provided by Mayo Clinic Foundation.
References
- 1. Geser F., Brandmeir N.J., Kwong L.K., Martinez-Lage M., Elman L., McCluskey L., Xie S.X., Lee V.M., Trojanowski J.Q. (2008) Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch. Neurol., 65, 636–641. [DOI] [PubMed] [Google Scholar]
- 2. Lomen-Hoerth C., Murphy J., Langmore S., Kramer J.H., Olney R.K., Miller B. (2003) Are amyotrophic lateral sclerosis patients cognitively normal? Neurology, 60, 1094–1097. [DOI] [PubMed] [Google Scholar]
- 3. Seltman R.E., Matthews B.R. (2012) Frontotemporal lobar degeneration: epidemiology, pathology, diagnosis and management. CNS Drugs, 26, 841–870. [DOI] [PubMed] [Google Scholar]
- 4. Cruts M., Gijselinck I., Van Langenhove T., van der Zee J., Van Broeckhoven C. (2013) Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci., 36, 450–459. [DOI] [PubMed] [Google Scholar]
- 5. Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M.. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. [DOI] [PubMed] [Google Scholar]
- 6. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y.. et al. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun., 351, 602–611. [DOI] [PubMed] [Google Scholar]
- 7. Davidson Y., Kelley T., Mackenzie I.R., Pickering-Brown S., Du Plessis D., Neary D., Snowden J.S., Mann D.M. (2007) Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol., 113, 521–533. [DOI] [PubMed] [Google Scholar]
- 8. Dickson D.W., Josephs K.A., Amador-Ortiz C. (2007) TDP-43 in differential diagnosis of motor neuron disorders. Acta Neuropathol., 114, 71–79. [DOI] [PubMed] [Google Scholar]
- 9. DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.. et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron, 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L.. et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Blitterswijk M., DeJesus-Hernandez M., Niemantsverdriet E., Murray M.E., Heckman M.G., Diehl N.N., Brown P.H., Baker M.C., Finch N.A., Bauer P.O.. et al. (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol., 12, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Majounie E., Renton A.E., Mok K., Dopper E.G., Waite A., Rollinson S., Chio A., Restagno G., Nicolaou N., Simon-Sanchez J.. et al. (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol., 11, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W. 3rd, Rademakers R.. et al. (2013) Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron, 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mori K., Arzberger T., Grasser F.A., Gijselinck I., May S., Rentzsch K., Weng S.M., Schludi M.H., van der Zee J., Cruts M.. et al. (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol., 126, 881–893. [DOI] [PubMed] [Google Scholar]
- 15. Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C.. et al. (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science, 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- 16. Gendron T.F., Bieniek K.F., Zhang Y.J., Jansen-West K., Ash P.E., Caulfield T., Daughrity L., Dunmore J.H., Castanedes-Casey M., Chew J.. et al. (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol., 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P.. et al. (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci., 18, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodega B., Orlando V. (2014) Repetitive elements dynamics in cell identity programming, maintenance and disease. Curr. Opin. Cell Biol., 31, 67–73. [DOI] [PubMed] [Google Scholar]
- 19. Cordaux R., Batzer M.A. (2009) The impact of retrotransposons on human genome evolution. Nature Rev. Genet., 10, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H. Jr. (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet., 73, 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ostertag E.M., Kazazian H.H. Jr. (2001) Biology of mammalian L1 retrotransposons. Annu. Rev. Genet., 35, 501–538. [DOI] [PubMed] [Google Scholar]
- 22. Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A.. et al. (2011) DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature, 471, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Douville R., Liu J., Rothstein J., Nath A. (2011) Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol., 69, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W., Jin Y., Prazak L., Hammell M., Dubnau J. (2012) Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One, 7, e44099.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeong B.H., Lee Y.J., Carp R.I., Kim Y.S. (2010) The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt-Jakob disease. J. Clin. Virol., 47, 136–142. [DOI] [PubMed] [Google Scholar]
- 26. Muotri A.R., Marchetto M.C., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H. (2010) L1 retrotransposition in neurons is modulated by MeCP2. Nature, 468, 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan H., Qurashi A., Poidevin M., Nelson D.L., Li H., Jin P. (2012) Retrotransposon activation contributes to fragile X premutation rCGG-mediated neurodegeneration. Hum. Mol. Genet., 21, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bundo M., Toyoshima M., Okada Y., Akamatsu W., Ueda J., Nemoto-Miyauchi T., Sunaga F., Toritsuka M., Ikawa D., Kakita A.. et al. (2014) Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron, 81, 306–313. [DOI] [PubMed] [Google Scholar]
- 29. Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., Gage F.H. (2011) Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U.S.A, 108, 20382–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erwin J.A., Marchetto M.C., Gage F.H. (2014) Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci., 15, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manghera M., Ferguson-Parry J., Douville R.N. (2016) TDP-43 regulates endogenous retrovirus-K viral protein accumulation. Neurobiol. Dis., 94, 226–236. [DOI] [PubMed] [Google Scholar]
- 32. Krug L., Chatterjee N., Borges-Monroy R., Hearn S., Liao W.W., Morrill K., Prazak L., Rozhkov N., Theodorou D., Hammell M.. et al. (2017) Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet., 13, e1006635.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slotkin R.K., Martienssen R. (2007) Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet., 8, 272–285. [DOI] [PubMed] [Google Scholar]
- 34. Whitelaw E., Martin D.I. (2001) Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet., 27, 361–365. [DOI] [PubMed] [Google Scholar]
- 35. Garcia-Perez J.L., Morell M., Scheys J.O., Kulpa D.A., Morell S., Carter C.C., Hammer G.D., Collins K.L., O'Shea K.S., Menendez P.. et al. (2010) Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature, 466, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li W., Prazak L., Chatterjee N., Gruninger S., Krug L., Theodorou D., Dubnau J. (2013) Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci., 16, 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maxwell P.H. (2016) What might retrotransposons teach us about aging? Curr. Genet., 62, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheridan P.L., Mayall T.P., Verdin E., Jones K.A. (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev., 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Lint C., Emiliani S., Ott M., Verdin E. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. embo J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 40. Blazkova J., Trejbalova K., Gondois-Rey F., Halfon P., Philibert P., Guiguen A., Verdin E., Olive D., Van Lint C., Hejnar J.. et al. (2009) CpG methylation controls reactivation of HIV from latency. PLoS Pathog., 5, e1000554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parada C.A., Roeder R.G. (1999) A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. embo J., 18, 3688–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parada C.A., Roeder R.G. (1996) Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature, 384, 375–378. [DOI] [PubMed] [Google Scholar]
- 43. Pan X.Y., Zhao W., Wang C.Y., Lin J., Zeng X.Y., Ren R.X., Wang K., Xun T.R., Shai Y., Liu S.W. (2016) Heat shock protein 90 facilitates latent HIV reactivation through maintaining the function of positive transcriptional elongation factor b (p-TEFb) under proteasome inhibition. J. Biol. Chem., 291, 26177–26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Albert T.K., Rigault C., Eickhoff J., Baumgart K., Antrecht C., Klebl B., Mittler G., Meisterernst M. (2014) Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. Br. J. Pharmacol., 171, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu C.R., Chang C.R., Chern Y., Wang T.H., Hsieh W.C., Shen W.C., Chang C.Y., Chu I.C., Deng N., Cohen S.N.. et al. (2012) Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell, 148, 690–701. [DOI] [PubMed] [Google Scholar]
- 46. Kramer N.J., Carlomagno Y., Zhang Y.J., Almeida S., Cook C.N., Gendron T.F., Prudencio M., Van Blitterswijk M., Belzil V., Couthouis J.. et al. (2016) Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science, 353, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W., Lee M.H., Henderson L., Tyagi R., Bachani M., Steiner J., Campanac E., Hoffman D.A., von Geldern G., Johnson K.. et al. (2015) Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med., 7, 307ra153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kao S.Y., Calman A.F., Luciw P.A., Peterlin B.M. (1987) Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature, 330, 489–493. [DOI] [PubMed] [Google Scholar]
- 49. Schludi M.H., May S., Grasser F.A., Rentzsch K., Kremmer E., Kupper C., Klopstock T., Arzberger T., Edbauer D. (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol., 130, 537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goodier J.L. (2016) Restricting retrotransposons: a review. Mob. DNA, 7, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smit A.F.A., Hubley R., Green P. (2013-2015) RepeatMarker Open 4.0.
- 54. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gendron T.F., van Blitterswijk M., Bieniek K.F., Daughrity L.M., Jiang J., Rush B.K., Pedraza O., Lucas J.A., Murray M.E., Desaro P.. et al. (2015) Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol., 130, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.