Abstract
Coiled-helix coiled-helix domain containing protein 10 (CHCHD10) and its paralogue CHCHD2 belong to a family of twin CX9C motif proteins, most of which localize to the intermembrane space of mitochondria. Dominant mutations in CHCHD10 cause amyotrophic lateral sclerosis (ALS)/frontotemporal dementia, and mutations in CHCHD2 have been associated with Parkinson’s disease, but the function of these proteins remains unknown. Here we show that the p.R15L CHCHD10 variant in ALS patient fibroblasts destabilizes the protein, leading to a defect in the assembly of Complex I, impaired cellular respiration, mitochondrial hyperfusion, an increase in the steady-state level of CHCHD2, and a severe proliferation defect on galactose, a substrate that forces cells to synthesize virtually all of their ATP aerobically. CHCHD10 and CHCHD2 appeared together in distinct foci by immunofluorescence analysis and could be quantitatively immunoprecipitated with antibodies against either protein. Blue native polyacrylamide gel electrophoresis analyses showed that both proteins migrated in a high molecular weight complex (220 kDa) in control cells, which was, however, absent in patient cells. CHCHD10 and CHCHD2 levels increased markedly in control cells in galactose medium, a response that was dampened in patient cells, and a new complex (40 kDa) appeared in both control and patient cells cultured in galactose. Re-entry of patient cells into the cell cycle, which occurred after prolonged culture in galactose, was associated with a marked increase in Complex I, and restoration of the oxygen consumption defect. Our results indicate that CHCHD10–CHCHD2 complexes are necessary for efficient mitochondrial respiration, and support a role for mitochondrial dysfunction in some patients with ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease and it is frequently associated with frontotemporal dementia (FTD). Mitochondrial dysfunction has often been suggested to underlie ALS pathology, but mutations in bona fide mitochondrial genes have rarely been reported in ALS cohorts. Recent reports of mutations in coiled-helix coiled-helix domain containing protein 10 (CHCHD10) in autosomal dominant familial ALS have changed this outlook. CHCHD10 mutations were first uncovered by whole exome sequencing in a family with motor neuron disease, ALS- and FTD-like symptoms, cerebellar ataxia and myopathy (1). Subsequent investigations uncovered autosomal dominant mutations in CHCHD10 in a clinically heterogeneous group of patients, ranging from sporadic and familial ALS (2–7), sporadic and familial FTD (2,8), autosomal dominant mitochondrial myopathy (9), spinal muscular atrophy (SMAJ) (10), Charcot–Marie–Tooth neuropathy (CMT2) (11), Alzheimer’s disease (5) and even Parkinson’s disease (PD) (1,5,12). Almost all of the reported mutations are missense substitutions in exon 2, with the exception of a stop-gain mutation (p.Q82X) suggesting a loss-of-function mechanism (2).
CHCHD10 is a member of the twin CX9C motif family of proteins, most of which localize to the mitochondrial intermembrane space (IMS). The CHCH domain forms a helix-turn-helix fold, stabilized by two disulphide bonds, which are formed upon import of these proteins into the IMS by the redox-regulated MIA40 pathway (13). CHCHD2, a paralogue of CHCHD10, arose by gene duplication from a common ancestor predating human speciation (14), and both proteins are conserved in all metazoans (15). The yeast Saccharomyces cerevisiae, contains a single orthologue, Mix17 (31% identity with CHCHD10, and 35% identity with CHCHD2) (16).
Recently, a missense mutation in CHCHD2 (c.182C>T, p.T61I) was shown to segregate with dominantly inherited late-onset PD in a large Japanese family and further screening revealed other PD patients with genetic variants in CHCHD2 (17). PD-associated CHCHD2 variants appear to be ethnic-specific, as variants found in Asian patients are not found in Caucasians and vice versa (18–20).
While mutations in CHCHD10 and CHCHD2 appear to be associated with a fairly wide spectrum of neurodegenerative disorders, the mechanisms of pathogenesis remain unclear. It has been reported that CHCHD10 is enriched at cristae junctions as part of the mitochondrial contact site and cristae organizing system (MICOS) complex (21), which is crucial for mitochondrial membrane architecture and cristae organization (22). Multiple different functions have been suggested for CHCHD2: a regulator of oxidative phosphorylation (OXPHOS) (23), a transcription factor that regulates the expression of the COX4I2 subunit during stress (24,25), an inhibitor of Bax oligomerization through its interaction with Bcl-xL (26), and finally a protein that sequesters SMAD4 to mitochondria, suppressing the activity of the TGFβ signalling pathway (27).
In this study, we have investigated the function of CHCHD10, and the mechanism of pathogenesis using fibroblasts derived from an ALS patient carrying the pathogenic p.R15L CHCHD10 variant.
Results
Reduced steady-state level of CHCHD10 in patient fibroblasts
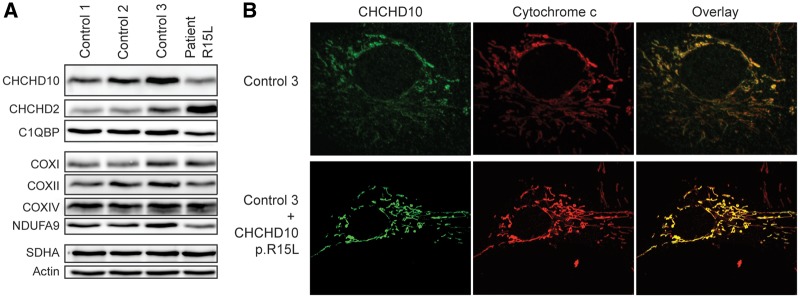
All reported mutations in CHCHD10 are heterozygous, suggesting autosomal dominant inheritance; however, whether this results from dominant negative effects, gain of function or haploinsufficiency is generally unknown. To begin to address this question we performed an immunoblot analysis on fibroblasts from an ALS patient, heterozygous for the c.44G>T (p.R15L) CHCHD10 variant. CHCHD10 was reduced to 57% of control levels in patient fibroblasts, whereas the level of its paralogue, CHCHD2, was increased 2.5-fold (Fig. 1A). Although both CHCHD10 and CHCHD2 have in silico predicted mitochondrial targeting sequences, these do not appear to be processed, as the mature forms of both proteins migrate on reducing gels at their predicted molecular masses (14, 16 kDa, respectively). Although the p.R15L mutation is contained in the putative targeting sequence, overexpression of the p.R15L variant in control cells showed that it localized to mitochondria without producing an obvious alteration in mitochondrial morphology (Fig. 1B). The steady-state level of the CHCHD10 mRNA in patient fibroblasts was not significantly different from control (data not shown), and DNA sequencing of an RT-PCR product indicated similar contributions from the wild-type and mutant alleles, as would be expected for a heterozygous missense mutation (Supplementary Material, Fig. S1). We conclude that the p.R15L variant targets correctly to mitochondria, but is unstable, leading to a loss of CHCHD10 function.
Figure 1.
Reduced steady-state level of CHCHD10 and localization of CHCHD10 p.R15L to mitochondria. (A) Whole cell extract from patient and control fibroblasts separated by SDS-PAGE and probed with antibodies against the indicated proteins. Actin was used as a loading control. (B) CHCHD10 p.R15L expression in control fibroblasts. Immunofluorescence analysis of control fibroblasts overexpressing mutant CHCHD10 p.R15L protein. Endogenous CHCHD10 staining is shown in green, the mitochondrial marker cytochrome c is shown in red.
Hyperfusion of mitochondria but normal mitochondrial ultrastructure in patient fibroblasts
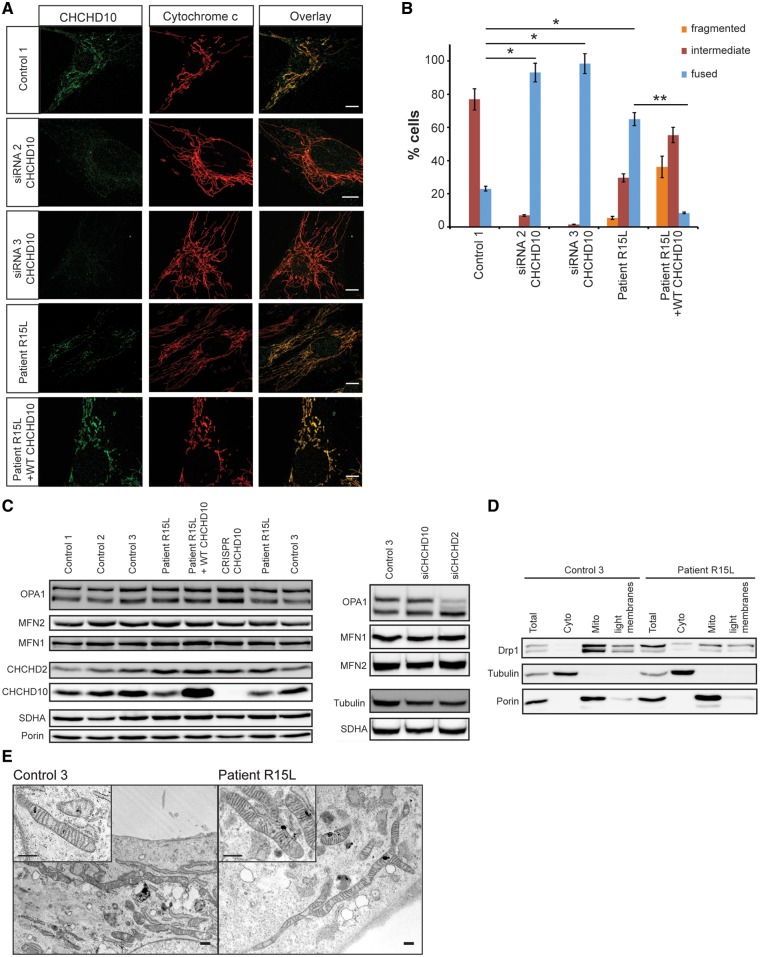
Immunofluorescence analysis revealed a striking mitochondrial hyperfusion phenotype in patient cells and in control fibroblasts in which CHCHD10 was depleted by siRNA (Fig. 2A and B); however, the steady-state levels of the major molecular components of the mitochondrial fusion machinery (OPA1 and MFN1, MFN2) were not significantly altered in patient or CHCHD10-depleted fibroblasts (Fig. 2C). Consistent with the hyperfusion phenotype, overall steady-state levels of Drp1, which is responsible for mitochondrial fission, were reduced, and a larger proportion appeared in the cytosolic fraction compared with control (Fig. 2D). Interestingly, depletion of CHCHD2 produced a shift towards short OPA1 isoforms (Fig. 2C), which was associated with increased mitochondrial fission (data not shown). Retrovirally mediated expression of a wild-type CHCHD10 cDNA (WT CHCHD10) rescued the mitochondrial hyperfusion in patient fibroblasts (from 65 to 9%). Transmission electron microscopy showed mitochondria with normal cristae organization in patient fibroblasts (Fig. 2E).
Figure 2.
Mitochondrial hyperfusion in patient fibroblasts without altered fusion machinery. (A and B) Immunofluorescence analysis of control and patient fibroblasts, control cells treated with siRNA against CHCHD10 and patient fibroblast stably overexpressing wild-type CHCHD10. (A) Endogenous CHCHD10 staining is shown in green, the mitochondrial marker cytochrome c in red. Scale bars: 10 μm. (B) Quantification analysis of mitochondrial network morphology. In each experimental condition described, 86 cells were analysed and the mitochondrial network organization was classified as fused, normal reticulum (intermediate) or fragmented, in three independent experiments. Data represent mean±SD and P-values were calculated using a two-tailed, unpaired t-test. P-values were calculated using a two-tailed, paired t-test *P < 0.05, **P < 0.01. (C) Immunoblot analysis of key players in mitochondrial fusion in patient or siRNA-depleted fibroblasts. Immunoblot analysis of MFN1, MFN2 and OPA1 levels in control and patient fibroblasts as well as fibroblasts where CHCHD10 (siRNA CHCHD10–2) or CHCHD2 (siRNA CHCHD2–2) was depleted by siRNA. SDHA, Porin and Tubulin were used as loading controls. (D) Analysis of DRP1 localization in control and patient fibroblasts by immunoblot analysis after cellular fractionation. Porin was used as a mitochondrial marker and tubulin as a cytosolic marker. (E) Analysis of the mitochondrial ultrastructure by transmission electron microscopy in control and patient fibroblasts. Scale bars: 500 nm.
Oxygen consumption and OXPHOS defect in patient fibroblasts
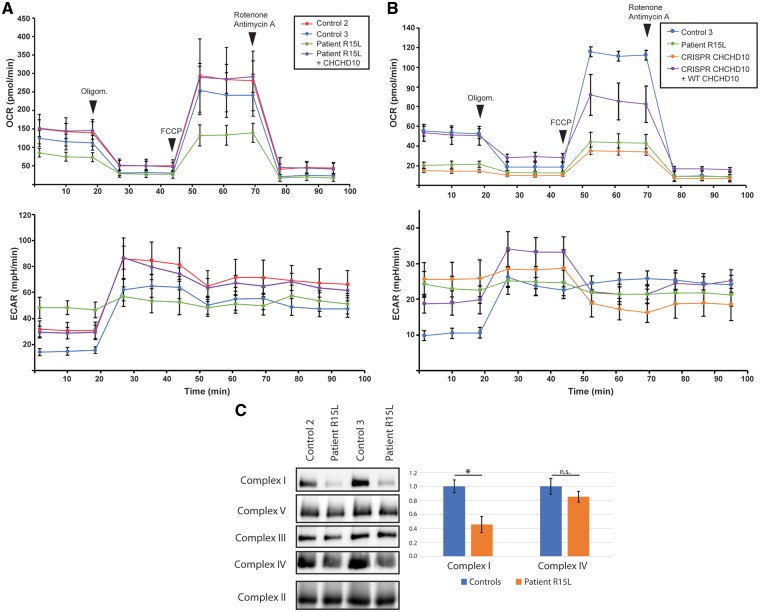
To determine if the p.R15L variant affected mitochondrial respiratory function, we measured oxygen consumption rates (OCR) using an Extracellular Flux Analyzer (Seahorse) in control and patient cells, as well as patient cells overexpressing wild-type CHCHD10. The OCR was measured in four steps: (i) basal OCR, (ii) after addition of oligomycin (1 μM) which inhibits ATP synthase, (iii) after uncoupling with cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (2 μM) and (iv) after addition of a mix of rotenone and antimycin A (0.5 μM), inhibitors of Complex I and III, respectively. Basal mitochondrial respiration of patient cell lines was reduced by 46 ± 12% (P <0.05) compared with controls (Fig. 3A) and the maximal respiratory capacity in response to uncoupling agent FCCP was reduced by 55 ± 12% (P <0.05). The extracellular acidification rate (ECAR) was increased about 2-fold in patient fibroblasts. This result was phenocopied with a CRISPR-Cas9-mediated CHCHD10 knockout cell line; however, not by the siRNA-mediated depletion of CHCHD10 (Fig. 3B;Supplementary Material, Fig. S2). Overexpression of CHCHD2 in control fibroblasts did not reduce OCR, arguing against role for increased CHCHD2 expression in disease pathogenesis (Supplementary Material, Fig. S3). Consistent with the rescue of the mitochondrial hyperfusion phenotype, overexpression of wild-type CHCHD10 in patient fibroblasts rescued the respiration defect (Fig. 3A and B).
Figure 3.
Mitochondrial oxygen consumption is impaired in patient fibroblasts as well as CRISPR-Cas9 CHCHD10 knockout cells. (A and B) Seahorse Bioscience XF24 Extracellular Flux Analyzer was used to measure the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) in control and (A) patient fibroblasts, and patient fibroblasts overexpressing wild-type CHCHD10. n = 3, data represent mean ± SD. (B) CRISPR-Cas9 knockout CHCHD10 cells, and CRISPR-Cas9 knockout CHCHD10 cells overexpressing wild-type CHCHD10. n = 5, data represent mean ± SD. (C) BN-PAGE analysis of patient and control fibroblasts shows a Complex I defect as revealed by subunit-specific antibodies against individual OXPHOS complexes. Quantification from five independent experiments, data represent mean±SEM. P-values were calculated using a two-tailed, paired t-test *P < 0.05.
To determine the steady-state levels of the OXPHOS complexes we performed a blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis. Patient fibroblasts showed about a 2-fold reduction in the level of fully assembled Complex I (Fig. 3C;Supplementary Material, Fig. S4), while the levels of the other OXPHOS complexes were not significantly altered. Consistent with this, the steady-state level of a Complex I subunit (NDUFA9) was reduced by immunoblot analysis (Fig. 1A).
CHCHD10 is required for growth in glucose-free medium
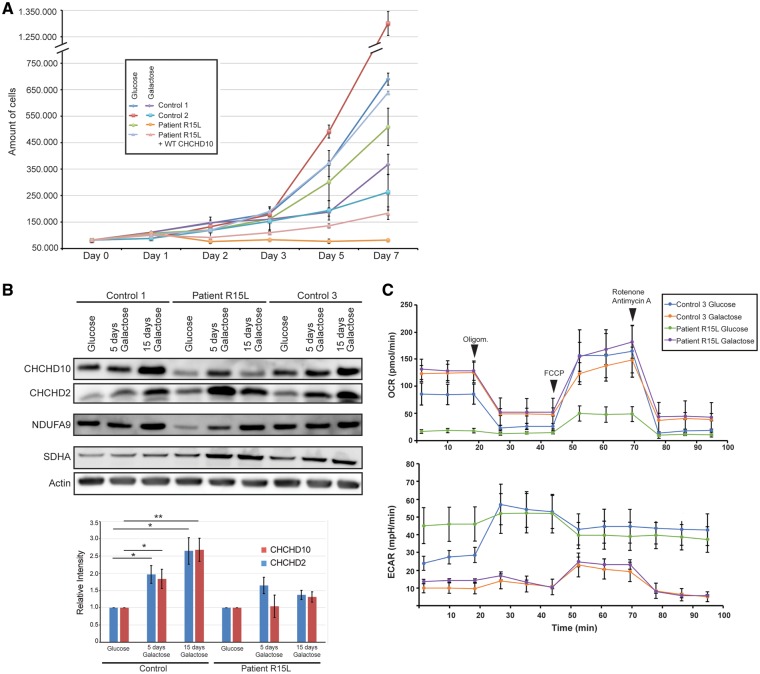
To interrogate mitochondrial respiratory function, we grew cells in medium containing a non-fermentable carbon source (in which glucose is replaced by galactose), forcing them to rely almost entirely on OXPHOS for ATP production. In general, cells with OXPHOS deficiencies grow more slowly than control cells in galactose medium or, if the deficiency is severe, undergo apoptosis. About 20% of the patient fibroblasts died within the first 24 h in galactose, and while the remaining cells stayed attached to the plate (Fig. 4A), they only started dividing after an adaptation period of ∼2 weeks (data not shown). Based on the severity of the Complex I assembly defect, this behaviour was unexpected, suggesting that other factors are at play.
Figure 4.
CHCHD10 is required for growth in glucose-free medium. (A) Growth curve for indicated cells grown in glucose- versus galactose-containing medium. (B) Immunoblot analysis and quantification of CHCHD10 and CHCHD2 levels in control and patient fibroblasts after subjecting the cells to galactose-containing medium for 5 and 15 days, respectively. Actin was used as a loading and normalization control. Graph represents the relative protein levels of CHCHD10 and CHCHD2 in control and patient cells grown in galactose medium compared with those grown in glucose medium (set to 1). Data represent mean ± SD (control, n = 6; patient, n = 3). P‐values were calculated using a two-tailed, paired t‐test *P < 0.05, **P < 0.01. (C) Seahorse Bioscience XF24 Extracellular Flux Analyzer was used to measure the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) in control and patient fibroblasts grown in either glucose or galactose-containing medium. n = 3, data represent mean ± SEM.
To further analyse the effects of galactose medium, we performed an immunofluorescence analysis of CHCHD10 in patient and control cells. CHCHD10 levels steadily increased in control, but not in the patient cells, over the time course of the experiment (Supplementary Material, Fig. S5). Consistent with these results, immunoblot analysis showed that CHCHD10 levels increased after 5 days in galactose medium, and continued to increase up to 15 days in control cells, whereas patient cells showed only a slight increase in CHCHD10 levels by Day 5, which, however, was not maintained by Day 15 (Fig. 4B). CHCHD2 levels were increased in both patient and control fibroblasts on Days 5 and 15. The changes in the levels of CHCHD10 in control cells were specific and did not reflect the small increase in mitochondrial content; however, there was a ∼1.5-fold increase in SDHA in patient cells after 15 days in galactose and strikingly a nearly 4-fold increase in NDUFA9, a proxy for Complex I assembly (Fig. 4B;Supplementary Material, Fig. S6A and B).
We next measured the changes in OCR in control and patient cells after 5 and 15 days in galactose medium. As expected, in control cells there was an increase in basal oxygen consumption over that observed in glucose medium, and a partial normalization of OCR in the patient cells after 5 days, which reached control levels after 15 days (Fig. 4C;Supplementary Material, Fig. S7).
CHCHD10 immunoprecipitates CHCHD2 and C1QBP
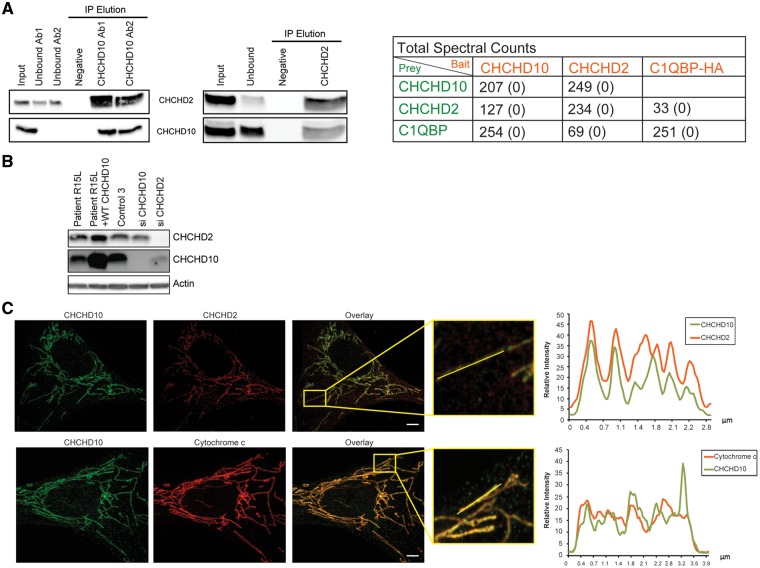
To characterize the CHCHD10 interactome we immunoprecipitated the endogenous protein and analysed it by liquid chromatography–tandem mass spectrometry (LC–MS/MS). This analysis identified CHCHD2 and C1QBP (p32), a multifunctional protein involved in mitochondrial translation (28), as major interactors of CHCHD10. Reciprocal immunoprecipitation of endogenous CHCHD2 also precipitated CHCHD10 and C1QBP (not shown in immunoblot) suggesting a robust interaction between these mitochondrial proteins (Fig. 5A;Supplementary Material, Tables S1 and S2).
Figure 5.
CHCHD10 co-immunoprecipitates with CHCHD2 and appears in distinct foci within mitochondria. (A) Co-immunoprecipitation experiment of CHCHD10 and CHCHD2 in 143B cells. Immunoprecipitation was performed with two different anti-CHCHD10 antibodies and an anti-CHCHD2 antibody, respectively. Stably overexpressing HA-tagged C1QBP was immunoprecipitated with anti-HA antibody. For negative controls no antibody was bound to the beads. Total spectral counts are listed for antibody-specific, versus negative controls (numbers in parentheses). (B) Immunoblot analysis of the CHCHD10 and CHCHD2 levels in patient and control fibroblasts, patient fibroblasts overexpressing wild-type CHCHD10 and control fibroblasts treated with siRNA against CHCHD10 (CHCHD10–2) and CHCHD2 (CHCHD2–2). Actin was used as a loading control. (C) Immunofluorescence analysis and line scan of the colocalization of CHCHD10 and CHCHD2. CHCHD10 is shown in green, CHCHD2 and the mitochondrial marker cytochrome c in red. Scale bar: 10 μm.
To further analyse the interdependence between CHCHD10 and CHCHD2 we performed siRNA-mediated depletion of each protein and analysed the levels of its potential partner by immunoblot analysis. Surprisingly, when CHCHD2 was depleted the levels of CHCHD10 decreased, whereas short-term depletion of CHCHD10 did not affect CHCHD2 levels (Fig. 5B). Immunofluorescence analysis of CHCHD2 and CHCHD10 using antibodies against the endogenous proteins showed a punctate pattern and a high degree of co-localization, consistent with the immunoprecipitation results (Fig. 5C).
CHCHD10 forms high molecular weight complexes with CHCHD2 and is not part of the MICOS complex
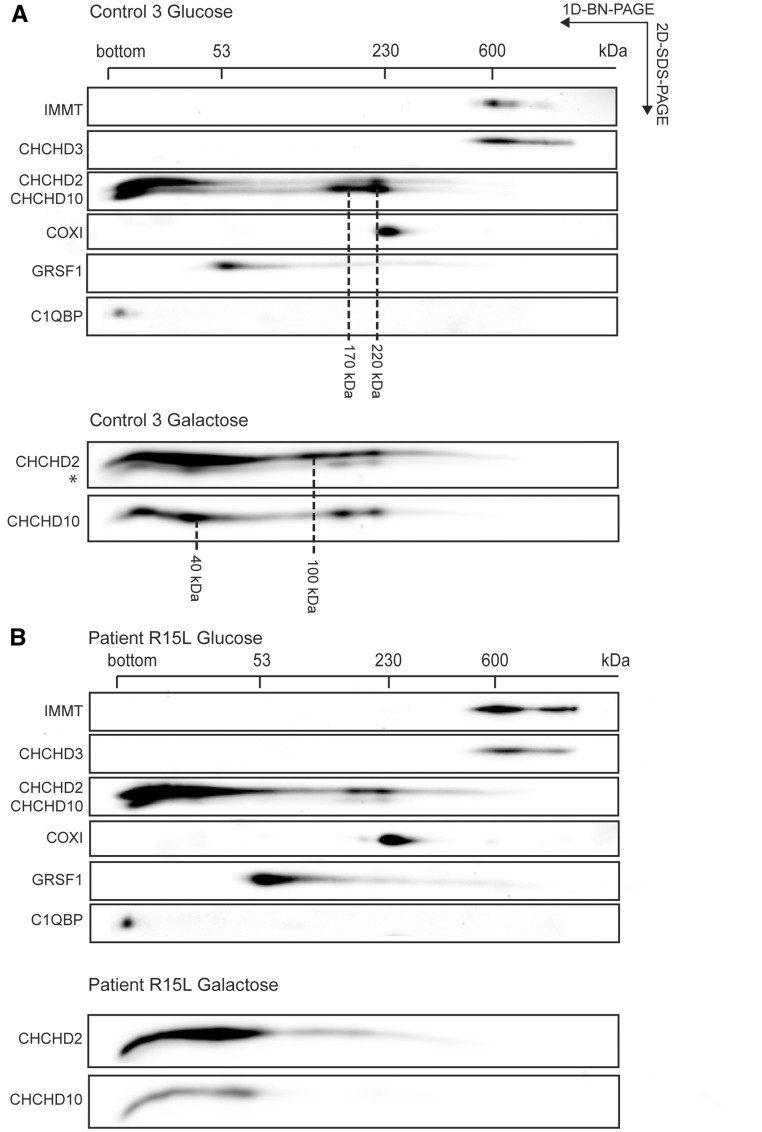
To investigate whether CHCHD10 forms stable complexes with CHCHD2 or C1QBP we performed two-dimensional PAGE (2D-PAGE) analysis (Fig. 6). In normal glucose-containing medium (Fig. 6A, top panel), CHCHD10 and CHCHD2 were present both in low molecular weight complexes [as either monomers (14 and 16 kDa, respectively), or dimers (not possible to distinguish between these possibilities on this gel), and in higher molecular weight complexes (CHCHD10: 170 and 220 kDa, CHCHD2: 220 kDa), whereas C1QBP ran only as a monomer (32 kDa)]. As expected, the level of CHCHD10 was reduced in patient mitochondria (Fig. 6B, top panel), but strikingly, it was virtually absent in the higher molecular weight complexes. Furthermore, the level of CHCHD2 appeared to compensate for the loss of CHCHD10 in the 170 kDa complex. Neither CHCHD10, nor CHCHD2 co-migrated with IMMT/Mic60 or CHCHD3/Mic19, key components of the MICOS complex, which itself was unaffected in patient fibroblasts.
Figure 6.
CHCHD10 forms a complex with CHCHD2 not MICOS. (A and B) Two-dimensional BN-PAGE/SDS-PAGE electrophoresis analysis of mitochondria from control (A) and patient (B) fibroblasts immunoblotted for indicated proteins. The positions and molecular weight of identified CHCHD10/CHCHD2 containing complexes are indicated in (A). The monomeric molecular weights of CHCHD2 and CHCHD10 are 15.5 and 14.1 kDa, respectively. The bottom of the gel in the first dimension is indicated as ‘bottom’. (A and B bottom) Two-dimensional BN-PAGE/SDS-PAGE electrophoresis analysis of mitochondria from control (A) and patient (B) fibroblasts cultivated for 9 days in galactose-containing medium. *Lower band is a residual signal from a prior CHCHD10 detection. The positions of GRSF1, COX I, and the MICOS complex are indicated on top (in kDa).
To investigate whether these high molecular weight complexes would be influenced by growth in galactose medium we subjected cells grown for 9 days in galactose to 2D-PAGE analysis. CHCHD10 and CHCDH2 levels increased in control cells (Fig. 6a, bottom panel), confirming the immunoblot analysis (Fig. 4B), and we observed the appearance of additional complexes at 40 and 100 kDa, the appearance of CHCHD2 in the complex at 170 kDa, but no apparent change in the complex at 220 kDa. Patient fibroblasts (Fig. 6B, bottom panel) showed increased CHCHD2 levels in galactose, and a substantial shift into the complex at 40 kDa, but a further decrease in the higher molecular weight complexes containing either CHCHD2 or CHCHD10. These complexes are likely soluble in the IMS as CHCHD10 behaves like a soluble protein after alkaline carbonate extraction (Supplementary Material, Fig. S8).
Discussion
This study shows that two twin CX9C mitochondrial IMS proteins, CHCHD10 and CHCHD2, both of which have been implicated in neurodegenerative diseases, exist in complexes that support efficient mitochondrial respiration. Fibroblasts from an ALS patient with a mutation in CHCHD10 displayed a striking mitochondrial hyperfusion phenotype, a deficiency in both basal and maximal respiration, increased anaerobic glycolysis, and a severe growth defect on a non-fermentable carbon source, all strongly suggesting insufficient aerobic energy supply as the underlying disease mechanism.
Three lines of evidence, including immunoprecipitation, immunofluorescence and 2D-PAGE, place CHCHD10 and CHCHD2 in protein complexes of 40, 100, 170 and 220 kDa depending on culture conditions. In glucose-containing medium about half of the CHCHD10 in control cells was present in the 170 and 220 kDa complexes, which were essentially undetectable in patient fibroblasts. The appearance of the 40 kDa complex, which was only prominent in control and patient cells in galactose medium, correlated with the recovery of the oxygen consumption defect in the patient cells, and the re-entry into a proliferative phase. There appears to be some degree of functional interdependence between the two proteins as reduced levels of CHCHD10 in patient fibroblasts were associated with a marked increase in the levels of CHCHD2, perhaps as a compensatory mechanism, and the siRNA-mediated depletion of CHCHD2 resulted in decreased levels of CHCHD10, suggesting a role for CHCHD2 in CHCHD10 stability. Furthermore, similar increases were observed in the steady-state levels of both proteins in control cells growing in galactose. The immunoprecipitation data suggest that the CHCHD10–CHCHD2 complexes are largely hetero-oligomers. C1QBP (p32), a predominantly mitochondrial matrix protein that behaves as a monomer on second dimension gels, was the only other protein found to co-immunoprecipitate consistently in these experiments. We did not find any evidence for an association of CHCHD2 with cytochrome c oxidase, as had been previously reported (25). How might the interaction with matrix protein p32 occur? Although it is clear from our data that the majority of CHCHD10 runs on denaturing gels at its predicted (unprocessed) molecular mass, a recent investigation of the N-termini of mitochondrial proteins suggested that murine CHCHD10 is processed at amino acid 80 (full length is 142aa) (29). It is thus possible that some fraction of CHCHD10 engages the TIM machinery and is processed in the matrix; however, this would need to be verified experimentally.
Importantly, our study clearly shows that the CHCHD10–CHCHD2 complex does not associate with MICOS, a multi-subunit complex that determines the architecture of mitochondrial cristae in the inner mitochondrial membrane. Neither CHCHD10 nor CHCHD2 co-immunoprecipitated or co-migrated on second dimension Blue-Native gels with components of the MICOS complex, an observation that contrasts sharply with a recent report (21). We previously performed affinity purification/mass spectrometry studies with antibodies directed against the endogenous forms of two bona fide MICOS subunits (Mic60, Mic19) and did not recover either CHCHD10 or CHCHD2 in the immunoprecipitates (30). Consistent with our results, the MICOS complex was not destabilized in fibroblasts from our patient, and we found no evidence for a mitochondrial ultrastructural abnormality. Significantly, Mix17 the yeast orthologue of CHCHD10 and CHCHD2, does not constitute part of the yeast MICOS complex (31).
The oxygen consumption deficiency in patient cells, the growth defect of these cells in galactose medium, and the marked increase (3- to 4-fold) in CHCHD10 levels in control cells grown in galactose, suggest a key role for CHCHD10 in respiration, particularly under the stress conditions that are induced by forcing mitochondrial OXPHOS and increasing oxidative stress. In yeast, Mix17 is also required for normal oxygen consumption, and its steady-state level is reported to increase in response to replication stress (32). The level of CHCHD10 in patient cells was largely unresponsive to growth in galactose, despite the fact that these cells still carry one normal allele. However, there was a slow, progressive increase in oxygen consumption in the patient cells that correlated with an increase in mitochondrial volume, an even greater increase in a marker of Complex I assembly, and an increase in the level of CHCHD2—all likely adaptive responses to the low levels of CHCHD10. Whether other metabolic adaptations are also necessary for the recovery of respiration in patient cells remains to be determined.
Loss of CHCHD10 resulted in a hyperfused mitochondrial network, independent of significant changes in the levels of the proteins involved in mitochondrial fusion (MFN1, MFN2, OPA1). Mitochondrial fusion is a common response to nutrient starvation, increased oxidative stress etc. (33). It seems likely that stress induced by the loss of CHCHD10 results in reduced mitochondrial fission, owing to the repartitioning of DRP1 in the cell from the mitochondrial surface to the cytosol which we observed in the patient cells.
Overall, our data support a disease mechanism of haploinsufficiency, at least for the CHCHD10 p.R15L variant associated with ALS: (i) the steady-state level of CHCHD10 was reduced in patient fibroblasts, (ii) the respiration defect in patient cells (phenocopied in CHCHD10 knockout cells) could be rescued by expression of a wild-type CHCHD10 cDNA, and (iii) the defect in mitochondrial morphology was also rescued by expression of a wild-type CHCHD10 cDNA (iv) overexpression of the p.R15L variant in control cells did not produce an obvious phenotype. Our results differ from previous findings on the p.S59L variant in which mitochondria appeared fragmented, and mitochondrial cristae disorganized in patient fibroblasts, as well as HeLa cells overexpressing mutant cDNA (21), suggesting that some mutations in CHCHD10 could behave as dominant negatives or have a toxic gain of function.
We speculate that the focal expression of CHCHD10–CHCHD2 complexes creates a mitochondrial platform or microenvironment that is necessary for efficient organization and function of the mitochondrial OXPHOS system, which is especially required in times of transient increases in demand for aerobically produced ATP, a characteristic of neurons and muscle cells. Further studies will be required to determine the precise molecular function of these proteins.
Materials and Methods
Human studies
The research studies on cell lines were approved by the institutional review board of the Montreal Neurological Institute, McGill University.
Cell lines and media
A primary fibroblast culture was established from a previously published heterozygous carrier of c.44G>T (p.R15L) (patient DNA #8807) diagnosed with sporadic ALS at age 54 (involving his upper limb) who remains alive 14 years later (5). The p.R15L substitution, has been reported in ALS patients in several independent studies (3–5), including its segregation with disease in large ALS family (3). Control (Montreal Children’s Hospital cell repository; Montreal; Canada; MCH58, 64, 65) and patient fibroblasts were immortalized by transduction with retroviral vectors expressing the HPV-16 E7 gene and the catalytic component of human telomerase (htert) as described previously (34). Fibroblasts and 143B cells were grown in 4.5 g/l glucose DMEM supplemented with 10% fetal bovine serum (FBS), at 37 °C in an atmosphere of 5% CO2. Cells stably overexpressing CHCHD10 in patient cells were engineered using retroviral vectors as previously described (35). For carbon source-dependent experiments, cells were grown in DMEM with 10% dialysed FBS supplemented with either 4.5 g/l glucose or 4.5 g/l galactose.
siRNA transfection
siRNA duplex constructs were used for transient depletion of CHCHD10 (ambion: CHCHD10-1: s53405; CHCHD10-2: s196436; CHCHD10-3: s226550) or CHCHD2 (ambion: CHCHD2-1: s27540; CHCHD2-2: s27541) in control fibroblasts. siRNA duplexes were transiently transfected into cells using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s specifications. The transfection was repeated on Day 3 and the cells were harvested on Day 6 for analysis.
Generation of stable cell lines over-expressing CHCHD10
CHCHD10 was amplified through PCR with Taq-polymerase with 7-deaza GTP/nucleotide mix (NEB) using cDNA from control fibroblasts as a template, and cloned into pBABE-Puro using Gateway Cloning Technology (Invitrogen). Retroviral constructs were transfected into the Phoenix packaging cell line using the HBS/Ca3(PO4)2 method. Control and patient fibroblasts were infected 48 h later by exposure to virus-containing medium in the presence of 4 μg/ml of polybrene as described previously (36).
Generation of CRISPR-Cas9 CHCHD10 knockout cell line
The sgRNA oligomers for CHCHD10 targeting exon 2 were annealed and inserted into plasmid pSpCas9(BB)-2A-Puro (PX459) V2.0 from Addgene (62988) via combined ligation (T7 ligase NEB) and restriction enzyme (BpiI, NEB) reaction.
Sequence for sgRNA targeting exon 2 of CHCHD10:
5′-caccGAAGCCGCAGCGCGGCCTCC-3′
3′-CTTCGGCGTCGCGCCGGAGGcaaa-5′
The plasmid (1.5 μg) was transfected into a fibroblasts cell line with jetPRIME reagent. Twenty-four hours after transfection puromycin was added for 2 days at a concentration of 5 μg/mL. Cells were than expanded and single clones were selected and analysed by immunoblot and sequencing for positive CHCHD10 knockout cell lines.
Mitochondrial isolation
Fibroblasts or 143B cells were rinsed twice with phosphate buffered saline (PBS), resuspended in ice-cold 250 mM sucrose/10 mM Tris–HCl (pH 7.4), and homogenized with 10 passes of a pre-chilled, zero-clearance homogenizer (Kimble/Kontes). A post-nuclear supernatant was obtained by centrifugation of the samples twice for 10 min at 600g. Mitochondria were pelleted by centrifugation for 10 min at 10 000g and washed once in the same buffer. Protein concentration was determined using the Bradford assay.
Mitochondrial fractionation
Control and patient fibroblasts were rinsed twice in PBS, resuspended in ice‐cold 250 mM sucrose/10 mM Tris–HCl (pH 7.4). Ten percent of the suspension was lysed in 1.5% DDM in PBS and centrifuged for 15 min at 20 000g, and the supernatant was considered as the total fraction. The rest (90%) of the cells were homogenized in ice‐cold 250 mM sucrose/10 mM Tris–HCl (pH 7.4) with 10 passes of a pre‐chilled, zero‐clearance homogenizer (Kimble/Kontes). A post‐nuclear supernatant was obtained by centrifugation of the samples three times for 10 min at 600g. Mitochondria were pelleted by centrifugation for 10 min at 10 000g, washed twice in the same buffer, and lysed in 1.5% DDM in PBS. The lysate was considered as crude mitochondrial fraction. The remaining supernatant was cleared by three further rounds of centrifugation at 10 000g for 10 min. This supernatant was then centrifuged at 100 000g for 1 h. The pellet was washed two times with PBS, lysed in 1.5% DDM in PBS, and considered as light membranes. The supernatant was adjusted to 1.5% DDM and considered as the cytosol fraction. Protein concentration was determined by Bradford assay.
Alkaline carbonate extraction
Mitochondria isolated from control and patient fibroblasts were extracted with 100 mM alkaline carbonate at pH 11.5 as previously described (35), centrifuged at 20 000g and the pellet and supernatant fractions analysed by SDS-PAGE.
Denaturing, native and 2D-PAGE
For SDS-PAGE, cells were extracted with 1.5% n-dodecyl-d-maltoside (DDM) in PBS, after which 20 μg of protein was run on denaturing polyacrylamide gels. BN-PAGE was used to separate individual OXPHOS complexes. Isolated mitochondria were solubilized with 1% DDM, and 20 μg of solubilized samples were run in the first dimension on 6–15% or 8–15% polyacrylamide gradient gels as previously described (37). Two-dimensional (2D)-BN-PAGE/SDS-PAGE was carried out as detailed previously (38). The separated proteins were transferred to a nitrocellulose membrane and immunoblot analysis was performed with the indicated antibodies.
Oxygen consumption measurements using the SeaHorse apparatus
Cells were seeded the day before measurement in a 24-well culture plate at 4 × 104 cells/well. Culture media was removed the following day and replaced with XF Assay Media from SeaHorse Bioscience (Billerica, MA, USA) containing 4.5 g/l glucose or galactose and incubated at 37 °C. OCR and the ECAR were measured using the SeaHorse XF24 Extracellular Analyzer over a 100-min time period. The assay consisted of three basal rate measurements followed by sequential injections of oligomycin (1 μM), FCCP (2 μM) and rotenone (0.5 μM) + antimycin (0.5 μM). Three rate measurements were performed after each injection. Error bars represent mean ± SD of four to five independent experiments and the P-values were calculated using a t-test.
Immunofluorescence analyses
Immunofluorescence analyses were performed by fixation of the cells grown on coverslips with 4% formaldehyde in PBS at room temperature for 20 min, permeabilization in 0.1% Triton X‐100 in PBS, blocking with 5% bovine serum albumin (BSA) in PBS, followed by incubation with primary antibodies in 5% BSA in PBS, for 1 h at RT. The appropriate anti‐species secondary antibodies coupled to Alexa fluorochromes (Invitrogen) (1:1000) were further used for 30 min at RT. Coverslips were mounted onto slides using fluorescence mounting medium (Dako).
Stained cells were imaged using a 60× or a 100× objective lenses (NA1.4) on an Olympus IX81 inverted microscope with appropriate lasers using an Andor/Yokogawa spinning disk system (CSU-X), with a sCMOS camera. Mitochondrial network morphology was classified in a blinded manner as fused, normal reticulum (intermediate) or fragmented. For each condition, 86 cells were analysed. Experiments were done three times independently. Error bars represent mean ± SD and P-values were calculated using a t-test.
Analysis of mitochondrial ultrastructure by transmission electron microscopy
Cells grown on NuncTM Lab-TekTM Chamber Slide System (Thermofisher) were washed in 0.1 M Na cacodylate washing buffer (Electron Microscopy Sciences) and fixed with 2.5% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M Na cacodylate buffer overnight at 4 °C. Cells were then washed three times in 0.1 M Na cacodylate washing buffer for a total of 1 h, incubated in 1% osmium tetroxide (Mecalab) for 1 h at 4 °C, and washed with ddH2O three times for 10 min. Then, dehydration in a graded series of ethanol/deionized water solutions from 30 to 90% for 8 min each, and 100% twice for 10 min each, was performed. The cells were then infiltrated with a 1:1 and 3:1 Epon 812 (Mecalab): ethanol mixture, each for 30 min, followed by 100% Epon 812 for 1 h. Cells were embedded in the culture wells with new 100% Epon 812 and polymerized overnight in an oven at 60 °C. Polymerized blocks were trimmed and 100 nm ultrathin sections were cut with an Ultracut E ultramicrotome (Reichert Jung) and transferred onto 200-mesh Cu grids (Electron Microscopy Sciences). Sections were post-stained for 8 min with 4% aqueous uranyl acetate (Electron Microscopy Sciences) and 5 min with Reynold’s lead citrate (Fisher Scientific). Samples were imaged with a FEI Tecnai-12 transmission electron microscope (FEI Company) operating at an accelerating voltage of 120 kV equipped with an XR-80C AMT, 8 megapixel CCD camera.
Immunoprecipitation
Mitochondria (200 μg) isolated from 143B cells were pelleted, rinsed once with PBS and extracted in 200 μl of lysis buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% DDM] (Sigma), and complete protease inhibitors (Roche) on ice for 30 min. The extract was centrifuged at 20 000g at 4 °C for 20 min, and the supernatant was pre‐cleared overnight with non-coated Dynabeads Protein A (Invitrogen) to reduce non-specific protein binding to the beads. Binding of indicated antibodies to Dynabeads Protein A (Invitrogen) was performed overnight. Antibodies were then cross-linked to the beads using 20 mM dimethyl pimelimidate (Sigma). The immunoprecipitation reaction was performed overnight at 4 °C. Samples were eluted using 0.1 M glycine pH 2.5/0.5% DDM, trichloroacetic acid precipitated and analysed by mass spectrometry on an Orbitrap (Thermo Scientific) at the Institute de Recherches Cliniques de Montreal. The false discovery rate is <5% with a Mascot score of 50.
Antibodies
Antibodies directed against the following proteins were used in this study: CHCHD10 (Sigma, HPA003440; Proteintech, 25671-1-AP), CHCHD2 (Sigma, HPA027407; Proteintech, 66302-1-Ig) SDHA (Abcam, ab14715), ATP5A1 (Abcam, ab14748), NDUFA9 (Abcam, ab14713), COXI (Abcam, ab14705) and COXII (Abcam, ab110258), COXIV (Abcam, ab110261), IMMT/MIC60 (Proteintech, 10179-1-AP), CHCHD3/MIC19 (Proteintech, 25625-1-AP), TOMM20 (Santa Cruz, sc‐11415), Core2 (Abcam, ab14745), Actin (Abcam, ab3280), cytochrome c (BioSciences, 556432), GRSF1 (Sigma, HPA036984), C1QBP (GC1qR, Abcam, ab24733), anti-HA (Abcam, ab130275), Porin (Calbiochem, 529534), Tubulin (Invitrogen, 32-2500), OPA1 (BD BioSciences, 612606), MFN1 (Cell Signaling, 14739S), MFN2 (Cell Signaling, 11925S), DRP1 (BD Biosciences, 611113).
Cell growth assay
For immortalized fibroblasts, 82 000 cells were seeded in 6-well culture dishes at Day 0. For the galactose experiments, cells were switched to galactose-containing medium on Day 1. After 1, 2, 3, 5 and 7 days of culture, cells were trypsinized, homogenized, and counted using a Bio-Rad TC10 automated cell counter. Experiments were done in independent triplicates. Error bars represent mean ± SD and P-values were calculated using a t-test.
Statistical analysis
All data are reported as mean ± SD or ±SEM as indicated in the figure legend. Statistical significance was determined using Student’s two-tailed, unpaired and paired t-tests. P-values <0.05 were considered statistically significant and labelled as follows: *P < 0.05, and **P < 0.01.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Hana Antonicka, Julien Prudent, and Heidi McBride for fruitful discussions and critique of the manuscript, and Philip McGoldrick (ALS Double Play Christopher Chiu Fellow) and Raphael Schneider (Clinical Research Fellowship from ALS Canada) for establishing the patient fibroblast culture.
Conflict of Interest statement. None declared.
Funding
This work was supported by grants from the Canadian Institutes of Health Research (366577), Parkinson Canada, and the Muscular Dystrophy Association (480233) to E.A.S., and by the James Hunter ALS Initiative (J.R., L.Z.). I.S. was supported by a scholarship from the Deutscher Akademischer Auslands Dienst (DAAD) (German Exchange Service).
References
- 1. Bannwarth S., Ait-El-Mkadem S., Chaussenot A., Genin E.C., Lacas-Gervais S., Fragaki K., Berg-Alonso L., Kageyama Y., Serre V., Moore D.G.. et al. (2014) A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain, 137, 2329–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dols-Icardo O., Nebot I., Gorostidi A., Ortega-Cubero S., Hernandez I., Rojas-Garcia R., Garcia-Redondo A., Povedano M., Llado A., Alvarez V.. et al. (2015) Analysis of the CHCHD10 gene in patients with frontotemporal dementia and amyotrophic lateral sclerosis from Spain. Brain, 138, e400.. [DOI] [PubMed] [Google Scholar]
- 3. Johnson J.O., Glynn S.M., Gibbs J.R., Nalls M.A., Sabatelli M., Restagno G., Drory V.E., Chio A., Rogaeva E., Traynor B.J. (2014) Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain, 137, e311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muller K., Andersen P.M., Hubers A., Marroquin N., Volk A.E., Danzer K.M., Meitinger T., Ludolph A.C., Strom T.M., Weishaupt J.H. (2014) Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain, 137, e309.. [DOI] [PubMed] [Google Scholar]
- 5. Zhang M., Xi Z., Zinman L., Bruni A.C., Maletta R.G., Curcio S.A., Rainero I., Rubino E., Pinessi L., Nacmias B.. et al. (2015) Mutation analysis of CHCHD10 in different neurodegenerative diseases. Brain, 138, e380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chio A., Mora G., Sabatelli M., Caponnetto C., Traynor B.J., Johnson J.O., Nalls M.A., Calvo A., Moglia C., Borghero G.. et al. (2015) CHCH10 mutations in an Italian cohort of familial and sporadic amyotrophic lateral sclerosis patients. Neurobiol. Aging, 36, 1767 e1763–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ronchi D., Riboldi G., Del Bo R., Ticozzi N., Scarlato M., Galimberti D., Corti S., Silani V., Bresolin N., Comi G.P. (2015) CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain, 138, e372.. [DOI] [PubMed] [Google Scholar]
- 8. Jiao B., Xiao T., Hou L., Gu X., Zhou Y., Zhou L., Tang B., Xu J., Shen L. (2016) High prevalence of CHCHD10 mutation in patients with frontotemporal dementia from China. Brain, 139, e21.. [DOI] [PubMed] [Google Scholar]
- 9. Ajroud-Driss S., Fecto F., Ajroud K., Lalani I., Calvo S.E., Mootha V.K., Deng H.X., Siddique N., Tahmoush A.J., Heiman-Patterson T.D.. et al. (2015) Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics, 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Penttila S., Jokela M., Hackman P., Maija Saukkonen A., Toivanen J., Udd B. (2012) Autosomal dominant late-onset spinal motor neuronopathy is linked to a new locus on chromosome 22q11.2-q13.2. Eur. J. Hum. Genet., 20, 1193–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Auranen M., Ylikallio E., Shcherbii M., Paetau A., Kiuru-Enari S., Toppila J.P., Tyynismaa H. (2015) CHCHD10 variant p.(Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol. Genet., 1, e1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ikeda A., Matsushima T., Daida K., Nakajima S., Conedera S., Li Y., Yoshino H., Oyama G., Funayama M., Nishioka K.. et al. (2017) A novel mutation of CHCHD2 p.R8H in a sporadic case of Parkinson’s disease. Parkinsonism Relat. Disord., 34, 66–68. [DOI] [PubMed] [Google Scholar]
- 13. Becker T., Bottinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci., 37, 85–91. [DOI] [PubMed] [Google Scholar]
- 14. Cavallaro G. (2010) Genome-wide analysis of eukaryotic twin CX9C proteins. Mol. Biosyst., 6, 2459–2470. [DOI] [PubMed] [Google Scholar]
- 15. Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J.M., Riemer J. (2009) Systematic analysis of the twin cx(9)c protein family. J. Mol. Biol., 393, 356–368. [DOI] [PubMed] [Google Scholar]
- 16. Pfanner N., van der Laan M., Amati P., Capaldi R.A., Caudy A.A., Chacinska A., Darshi M., Deckers M., Hoppins S., Icho T.. et al. (2014) Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol., 204, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Z.D., Saw W.T., Tan E.K. (2017) Mitochondrial CHCHD-containing proteins: physiologic functions and link with neurodegenerative diseases. Mol. Neurobiol., 54, 5534–5546. [DOI] [PubMed] [Google Scholar]
- 18. Jansen I.E., Bras J.M., Lesage S., Schulte C., Gibbs J.R., Nalls M.A., Brice A., Wood N.W., Morris H., Hardy J.A.. et al. (2015) CHCHD2 and Parkinson’s disease. Lancet Neurol., 14, 678–679. [DOI] [PubMed] [Google Scholar]
- 19. Puschmann A., Dickson D.W., Englund E., Wszolek Z.K., Ross O.A. (2015) CHCHD2 and Parkinson’s disease. Lancet Neurol., 14, 679.. [DOI] [PubMed] [Google Scholar]
- 20. Zhang M., Xi Z., Fang S., Ghani M., Sato C., Moreno D., Liang Y., Lang A.E., Rogaeva E. (2016) Mutation analysis of CHCHD2 in Canadian patients with familial Parkinson’s disease. Neurobiol. Aging, 38, 217.e7–217.e8 [DOI] [PubMed] [Google Scholar]
- 21. Genin E.C., Plutino M., Bannwarth S., Villa E., Cisneros-Barroso E., Roy M., Ortega-Vila B., Fragaki K., Lespinasse F., Pinero-Martos E.. et al. (2016) CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol. Med., 8, 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rampelt H., Zerbes R.M., van der Laan M., Pfanner N. (2016) Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta, 1864, 737–746. [DOI] [PubMed] [Google Scholar]
- 23. Baughman J.M., Nilsson R., Gohil V.M., Arlow D.H., Gauhar Z., Mootha V.K., Dermitzakis E.T. (2009) A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet., 5, e1000590.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aras S., Pak O., Sommer N., Finley R. Jr., Huttemann M., Weissmann N., Grossman L.I. (2013) Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res., 41, 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aras S., Bai M., Lee I., Springett R., Huttemann M., Grossman L.I. (2015) MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion, 20, 43–51. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y., Clegg H.V., Leslie P.L., Di J., Tollini L.A., He Y., Kim T.-H., Jin A., Graves L.M., Zheng J., Zhang Y. (2015) CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ., 22, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu L., Gomez-Duran A., Saretzki G., Jin S., Tilgner K., Melguizo-Sanchis D., Anyfantis G., Al-Aama J., Vallier L., Chinnery P., Lako M., Armstrong L. (2016) The mitochondrial protein CHCHD2 primes the differentiation potential of human induced pluripotent stem cells to neuroectodermal lineages. J. Cell Biol., 215, 187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yagi M., Uchiumi T., Takazaki S., Okuno B., Nomura M., Yoshida S., Kanki T., Kang D. (2012) p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res., 40, 9717–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calvo S.E., Julien O., Clauser K.R., Shen H., Kamer K.J., Wells J.A., Mootha V.K. (2017) Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell Proteomics, 16, 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janer A., Prudent J., Paupe V., Fahiminiya S., Majewski J., Sgarioto N., Des Rosiers C., Forest A., Lin Z.Y., Gingras A.C.. et al. (2016) SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med., 8, 1019–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harner M., Korner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J., 30, 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C.. et al. (2012) Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol., 14, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aasen T., Raya A., Barrero M.J., Garreta E., Consiglio A., Gonzalez F., Vassena R., Bilic J., Pekarik V., Tiscornia G.. et al. (2008) Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol., 26, 1276–1284. [DOI] [PubMed] [Google Scholar]
- 34. Lochmuller H., Johns T., Shoubridge E.A. (1999) Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp. Cell Res., 248, 186–193. [DOI] [PubMed] [Google Scholar]
- 35. Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmuller H., Chevrette M., Kaufman B.A., Horvath R.. et al. (2009) Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet., 41, 833–837. [DOI] [PubMed] [Google Scholar]
- 36. Pear W.S., Scott M.L., Nolan G.P. (1997) Generation of high-titer, helper-free retroviruses by transient transfection. Methods Mol. Med., 7, 41–57. [DOI] [PubMed] [Google Scholar]
- 37. Leary S.C. (2012) Blue native polyacrylamide gel electrophoresis: a powerful diagnostic tool for the detection of assembly defects in the enzyme complexes of oxidative phosphorylation. Methods Mol. Biol., 837, 195–206. [DOI] [PubMed] [Google Scholar]
- 38. Antonicka H., Ogilvie I., Taivassalo T., Anitori R.P., Haller R.G., Vissing J., Kennaway N.G., Shoubridge E.A. (2003) Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem., 278, 43081–43088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.