Abstract
Nematodes are highly abundant animals, and many species have a parasitic lifestyle. Nematode parasites are important pathogens of humans and other animals, and there is considerable interest in understanding their molecular and genomic adaptations to nematode parasitism. This has been approached in three main ways: comparing the genomes of closely related parasitic and free-living taxa, comparing the gene expression of parasitic and free-living life cycle stages of parasitic nematode species, and analysing the molecules that parasitic nematodes excrete and secrete. To date, these studies show that many species of parasitic nematodes have genomes that have large gene families coding for proteases/peptidases, protease inhibitors, SCP/TAPS proteins and acetylcholinesterases, and in many cases there is evidence that these appear to be used by parasitic stages inside hosts, and are often secreted. Many parasitic nematodes have taxa-restricted gene families that also appear to be involved in parasitism, emphasizing that there is still much to be discovered about what it takes to be a parasitic nematode.
Keywords: nematode, parasite, genomics, evolution
There are many nematode parasites
Nematodes are the most abundant and speciose extant group of metazoans. Memorably, some 100 years ago, Nathan Cobb said of nematodes: ‘In short, if all the matter in the universe except the nematodes were swept away, our world would still be dimly recognizable … we should find its mountains, hills, vales, rivers, lakes, and oceans represented by a film of nematodes. The location of towns would be decipherable, since for … human beings there would be corresponding massing of certain nematodes. Trees would still stand in ghostly rows representing our streets and highways. The location of various plants and animals would still be decipherable…’ [1]. He made the point well, that nematodes are ubiquitous, if rather rarely seen. As Cobb also makes plain, they are also common parasites, infecting all manner of multicellular animals and plants, in all terrestrial and aquatic environments, such that being parasitized by nematodes is a normal feature of life.
Parasitic nematodes are diverse in their biology and life histories, and so, it can be difficult to make generalizations. However, by way of introduction, let us consider the most common nematode parasite of humans, Ascaris lumbricoides [2]. Adult male and female worms (females are about 30 cm long, 5 mm in diameter; males are a little smaller) live in the host gut where they reproduce. Females lay eggs—some 250 000 per day—and these pass out of the host in its faeces. Ascaris eggs have a resistant egg shell and so the eggs persist in the environment for considerable periods. Hosts become infected when they accidently ingest eggs. Once inside the host gut the eggs hatch to release a larva; this leaves the gut and migrates around the host body before returning to the gut where the worms then mature as adults and reproduce, and so the cycle continues. Other species of nematodes that parasitize vertebrates can live in other within-host sites, including blood vessels, the lymphatic system, and within the tissues. Parasitic nematodes have a myriad of different life cycles; for example, where larvae (rather than eggs) infect hosts; where worms are transmitted among hosts by arthropod vectors (in which case the parasite then lives in two different host species during its life) and where worms are transmitted when a host predates on an infected host. Nematodes parasitize vertebrates, invertebrates and plants, though studies are biased towards those that parasitize vertebrates—a bias that will persist in this article.
Particular challenges for parasitic nematodes, to which they are adapted, are surviving within the host and transmitting among hosts. For parasites of vertebrates an important challenge is surviving the host anti-parasite immune response. There are two general strategies that parasitic nematodes use. The first is to evade the host immune response by using a molecular disguise, by protecting themselves from host immune system effector molecules, or by living within certain within-host niches that are less immunologically exposed. The second, not mutually exclusive approach, is immunomodulation of the host, which appears to be widespread among nematodes [3]. To immunomodulate their hosts, nematodes release molecules into the host, and these interact with the host immune system to alter the host’s immune response to the parasite’s advantage. Though these immunomodulation phenomena are now well known, the detailed molecular mechanisms underlying them are currently largely unknown.
Parasitic nematodes cause immense harm to humans and other animals. More than 1.5 billion people are infected with gastrointestinal nematode parasites, with this concentrated in the young, poor of the developing world [4, 5], and as such the World Health Organization recognizes these infections as a neglected tropical disease [6]. The World Health Organization estimated that in 2012, worldwide intestinal nematode infection caused a disease burden, measured by the Years Lost due to Disability (YLD), of 5 million YLD. This loss is greater than for other infectious diseases—4 million for malaria, 4.5 million for HIV/AIDS [7].
Nematode parasitism of agricultural animals is also common and its control is necessary to maintain productivity and profitability [8, 9]. The production losses caused by parasitic nematode infection translates into economic costs: in the UK the estimated annual cost of nematode infection of sheep is €99 million. Across the European Union the annual sales of anthelmintic drugs to treat nematode infections total approximately €400 million [9], but it is likely that this is only a small fraction of the true costs [10].
Nematodes are diverse
Nematodes have a conservative morphology making traditional approaches to their taxonomy frustratingly hard, even for experts. Perhaps perversely, the nematodes are a highly diverse group of organisms. In molecular phylogenetic analyses nematodes often have considerably extended phylogenetic branches compared with other taxa, pointing to nematodes’ elevated molecular evolutionary rates [11, 12]. One consequence of this is that many nematode species (and other taxonomic groupings) have genes and gene families that are specific to them [13].
This diversity is also seen in various aspects of nematode genomes. Caenorhabditis elegans was the first nematode whose genome was sequenced, revealing a 100 Mb genome containing about 20 000 genes. Since then the genomes of other nematodes have also been sequenced, particularly those of parasitic species. There are currently about 25 nematode genome sequences published, with about 100 projects ongoing [14]. This work has already revealed substantial diversity in the size and content of nematode genomes. For example, sequenced nematode genomes now range from c. 20 Mb in Pratylenchus coffeae (a pest of bananas and other crops) coding for about 7000 genes, to 370 Mb in Haemonchus contortus (a gastrointestinal parasite of sheep) coding for c. 20 000 genes [15, 16]. This variation in genome size is due to changes in gene number, as well as changes in the size and number of introns, and in the extent of non-coding, intervening sequence, including repetitive sequence [16–19]. It is also interesting to note that even within the Caenorhabditis genus there are substantial differences in genome size and gene number. For example, genome size differs more than 2-fold ranging from 79 Mb in Caenorhabditis tropicalis to 190 Mb in Caenorhabditis brenneri, while estimates of gene number range from the low of C. elegans to c. 35 000 in Caenorhabditis sinica. Despite these substantial differences among nematode species, signals of the conservation of gene order across large nematode distances can still be seen [16, 18, 20–22].
Phylogenetic analysis of nematodes has shown that they have independently evolved parasitism on up to 18 different occasions [23, 24]. Consequently, many parasitic nematodes have close relatives that have a free-living lifestyle. Species that parasitize different hosts (vertebrates, invertebrates, plants) are also often in the same nematode clades, perhaps suggesting that there is some deep conservation of adaptations to a parasitic lifestyle independent of the type of host being parasitized.
Approaches to discovering the genetic and genomic basis of parasitism in nematodes
Parasitism is biologically fascinating, and so there is considerable interest in understanding the adaptations that underlie the parasitic lifestyle. This basic biological interest is, of course, supplemented by a desire to understand the biology of these parasites so as to find ways to control them and the harm that they cause.
Before the advent of genomics a whole series of physiological and molecular studies was aimed at investigating aspects of parasitic nematodes’ biology which, a priori, were likely to be different in parasitic nematodes, compared with free-living species. So, for example, the metabolism of many nematode parasites of the vertebrate gut was investigated, given the likely special gaseous, pH, etc. conditions in this environment. Similarly, because parasitic nematodes have to survive the host immune response there was considerable interest directed to understanding the structure and composition of parasitic nematodes’ surface cuticle and its interaction with the host immune response.
In pursuing the aim of understanding the genetic basis of nematode parasitism there is an underlying rationale that all such studies use, and it is useful to make this rationale explicit. It is this: parasitic nematodes have evolved from free-living ancestors, and as this has happened, parasites will have adapted existing traits of their free-living nematode ancestor, and evolved new traits, which together underlie the parasitic lifestyle. Therefore, comparing free-living nematodes with parasitic nematodes can be used to uncover the molecular basis of nematode parasitism. This is the first type of genomic study used to understand nematode parasites. A second approach, but one using the same fundamental rationale, is to compare, within a species, parasitic life cycle stages with free-living life cycle stages. Finally, the third approach that is used seeks to discover which molecules parasitic stages likely excrete/secrete [i.e. excretory/secretory (ES) material], with the rationale that many of these molecules will interact with the host in prosecuting a parasitic lifestyle.
Comparing the genomes of parasitic and free-living nematodes
This is potentially an enormously powerful approach to understand the genomic basis of nematode parasitism, but care must be taken in making the correct comparisons given the multiple, independent origins of nematode parasitism. Specifically, nematodes occur in five phylogenetic clades [23], with many clades containing both parasitic and free-living taxa. The most appropriate and powerful phylogenomic comparisons are those that compare taxa within the clades, rather than between clades. This is because parasitic versus free-living comparisons across different clades confounds differences in lifestyles with the different evolutionary history of those clades. In contrast, comparisons between taxa with different lifestyles within the same clade can reasonably be used to infer the genomic bases of those differences in lifestyle. Given the extensive genomic diversity among the nematodes, it is arguable that we currently have too few nematode genomes to make strong inferences about common genomic themes underlying parasitism across different nematode clades [14, 25]. Instead, more phylogenetically local, focused comparisons are probably more appropriate.
Because C. elegans was the first nematode genome to be sequenced, by necessity many studies of the genomes of parasitic nematodes have only been able to use C. elegans as a comparator species. However, important parasitic species are in the same clade—for example, the hookworms Necator spp. and Ancylostoma spp., parasites of livestock such as Haemonchus spp., together with common laboratory models such as Heligmosomoides polygyrus and Nippostrongylus brasiliensis [23].
One particularly useful genomic comparison between parasitic and free-living species involves six species within one sub-clade of nematodes. Specifically, this compared four obligate con-generic parasitic species (Strongyloides spp.), one facultatively parasitic species (Parastrongyloides trichosuri) and one free-living species (Rhabditophanes sp.) all of whom are closely phylogenetically related (Figure 1). Comparison of these species’ genomes was used to infer where genes and gene families arose, and where genes and gene families were lost, during the evolutionary history of the species and, crucially, as parasitism evolved (Figure 1). This analysis identified >1000 gene families that were evolutionarily acquired as the parasitic genera Parastrongyloides and Strongyloides evolved. Of these gene families, the two largest code for astacin-like zinc-metallopeptidases, and sperm-coating protein/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS proteins) (Pfam PF00188), which belong to the cysteine-rich secretory protein superfamily [26], but which are also known as cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins (CAP)-domain, Ancylostoma secreted protein (ASP) and venom allergen-like (VAL) proteins [27]). Specifically, the parasitic species Strongyloides ratti and Strongyloides stercoralis have 184 and 237 astacin-like metallopeptidase coding genes, compared with 36 in the free-living relative Rhabditophanes (C. elegans has 40). For SCP/TAPS proteins, there were 89 and 113 coding genes in S. ratti and S. stercoralis, respectively, compared with 12 in Rhabditophanes (C. elegans has 36) [19].
Figure 1.
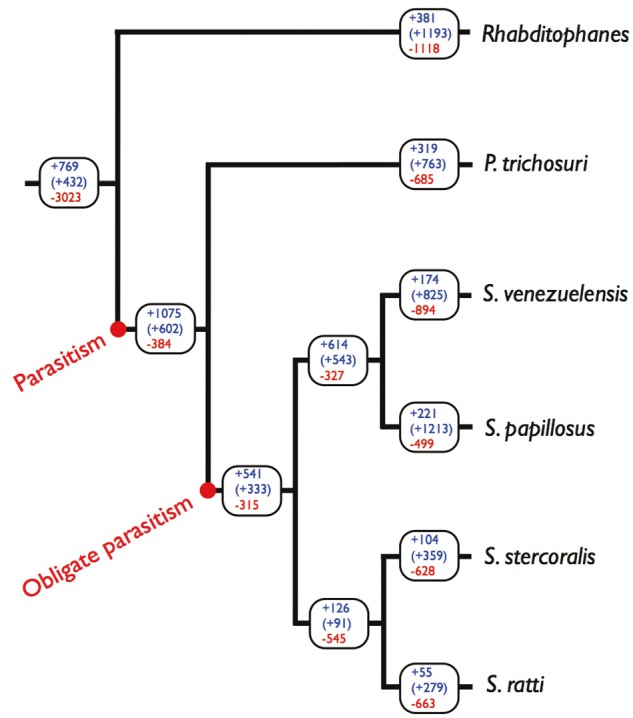
The phylogeny of Strongyloides and its relatives and the evolution of parasitism. A phylogeny of four species of Strongyloides, P. trichosuri and Rhabditophanes sp. Rhabditophanes is a free-living species. Parastrongyloides can have multiple free-living adult generations, but can also be parasitic, making it a facultative parasite. Strongyloides is an obligate parasitic species because, even though it has a free-living adult generation (as does Parastrongyloides), Strongyloides’ life cycle requires a parasitic adult generation every generation. Where parasitism and obligate parasitism are inferred to have arisen is shown. The boxes show, in descending vertical order, the number of (i) gene families originating on each branch (+, top in blue), (ii) gene families with at least one duplication event on each branch (+, middle in blue, in parentheses) and (iii) the number of gene families with at least one loss on each branch (-, bottom in red). The tree branch lengths do not show the relative distance among the taxa. Data from [19] . (A colour version of this figure is available online at: https://academic.oup.com/bfg)
The observed comparative expansion of astacin-like metallopeptidase coding genes in these parasitic taxa is consistent with similarly large gene families coding for peptidases and proteases in other parasitic nematode species [13]. For example, the Ascaris suum genome has 456 peptidase coding genes, with those coding for metallopeptidases and serine proteases predominating. The Toxocara canis genome has many similar genes too—165, 107 and 60 (total 332) coding for metallo, cysteine, and serine peptidases, respectively, together representing 89% of all the predicated peptidase coding genes in this species [28]. In the whipworm Trichuris suis, there are large numbers of genes coding for two classes of peptidases (116 for class S1 and 42 for class S8) [29]; Dictyocaulus viviparus has 478 protease coding genes [30]. Together, these results begin to suggest a common theme: that genomes of a range of parasitic nematodes have many genes coding for proteases or peptidases [25], though the specific class of proteases/peptidase coded for appears to not be particularly consistent among different taxa.
The possible roles for such an array of proteases and peptidases in nematode parasites is not known, but there are two main ideas for the roles that they might play. First, they might be used by the parasite to digest components of the host, at least in part to acquire food. Strongyloides spp., as many other parasitic nematode species, undergoes a within-host migration before finally settling in the gut. This migration requires the breakdown of host tissue, which presumably these parasite-produced enzymes achieve. More generally, for many species of parasitic nematode, larval stages infect hosts by penetrating the host skin and then migrating through the host, and during these infection processes proteases, among other molecules, are used by the larvae to facilitate their migration. For Strongyloides adult parasitic stages, they live within and continuously burrow through the mucosa of the host’s small intestine [31]. Here the parasite is presumably digesting host tissue, which is then used as a food source by the parasite. In Strongyloides it is likely that the metallopeptidases are secreted by the parasite to achieve this. Other species of parasitic nematodes feed on the host in other ways, and the ability to digest host proteins would therefore seem to be a key component of living a parasitic lifestyle. The second potential role of proteases in the parasitic lifestyle concerns immunomodulation of their hosts. Here the idea is that these proteases act against, and disable, protein components of the host immune response, which thereby facilitates the parasites’ survival.
Turning to the second group of genes that are comparatively expanded in Strongyloides (compared with free-living species), the SCP/TAPS proteins. The expansion of this gene family in Strongyloides also chimes with analyses of genomes of other species [13]. For example, the hookworms Ancylostoma ceylanicum and Necator americanus have 432 and 137 of these genes [32, 33], and H. contortus 161, compared with 36 in the free-living species C. elegans (and as a further comparison, there are 33 in Pristionchus pacifiicus, which lives in close association with beetles, but does not appear to be a parasite [34]). This gene family is also reported to be expanded in another clade V nematode, D. viviparus [30].
There has been substantial interest in the SCP/TAPS proteins in parasitic nematodes, in large part because these molecules appear to be abundant in nematode parasites [27], with some of these proteins being immunodominant. Despite this notoriety, what these molecules do in parasitic nematodes remains far from clear. In vitro evidence is consistent with some of these molecules interacting with components of the host immune responses [35]. In H. polygyrus and Ancylostoma caninum SCP/TAPS molecules are on (and closely below) the parasite surface, as well as associated with the parasite’s gut and oesophagus. This is consistent with these molecules having a role in immunomodulating the host, but also consistent with interacting with the host more widely [27, 36]. More generally, related molecules occur in a wide variety of non-parasitic species, perhaps suggesting that the conserved features of these proteins are simply domains that can be usefully deployed in many settings [27].
Within the Strongyloides clade of nematodes, other gene families that expanded coincident with the evolution of parasitism include acetylcholinesterase-coding genes (33 and 34 in S. ratti and S. stercoralis, respectively, compared with three in Rhabditophanes; C. elegans has four), receptor-type protein tyrosine phosphatase-coding genes (83 and 75 in S. ratti and S. stercoralis, respectively, compared with 21 in Rhabditophanes; C. elegans has 59). The role of parasitic nematode-produced acetylcholinesterases in the parasitic lifestyle is also not fully understood. In some species of gastrointestinal parasitic nematodes these molecules are secreted into the host gut where they have been thought to play a role in modulating the activity of the gut—including its muscular contraction and secretion of mucus and other fluid—with this facilitating parasite survival [37]. However, more recently, evidence points to parasite acetylcholinesterases having an immunoregulatory role [38]. Careful assessment of this large family of genes in Strongyloides strongly suggests that, when translated, many of these gene products will be enzymatically inactive [39]. One possibility, therefore, is that this comparatively expanded gene family is a functionless family, though this then begs the question of why such a large family of putatively non-functional proteins became expanded and are maintained in a genome, and why they appear to be secreted by the parasitic stages of the life cycle. An alternative explanation is that these genes are coding for proteins with other functionalities that are completely unrelated to the acetylcholinesterase-like enzymatic function.
Importantly, in many of these genomic comparisons between parasitic and free-living species, gene families whose products are (i) hitherto unknown and (ii) unique to the taxa in which they are described have been found [13]. Given the diversity among nematode taxa, the existence of taxa-restricted gene families should be expected. Indeed, such taxa-restricted gene families should be as likely to occur among free-living species, as well as parasitic ones. Because of the taxa-restricted nature of these gene families, their possible role in parasitism is unclear, though this is clearly a research challenge for the future.
Comparing parasitic and free-living stages
This approach exploits the fact that many species of parasitic nematode have life cycles where some stages are parasitic and some are free-living (with these used for transmission among hosts). Comparing gene expression in parasitic and free-living life cycle stages might therefore identify the genes and proteins used specifically (or more) by the parasitic stages, which can then be used to infer what structures, metabolism, molecular processes, etc. underlie the parasitic lifestyle of the species concerned.
Interpreting results from such comparisons can be problematic, for two related reasons. First, these comparisons involve both different life cycle stages (for example, larvae and adults) and different lifestyles (free-living versus parasitic). Put formally, such comparisons therefore confound life cycle stage and lifestyle. This is a real problem because there can be considerable differences in the biology of different nematode life cycle stages, even when they are all living in the same environment. This can be seen using an example from the life cycle of the free-living nematode C. elegans. It has about 170 genes that code for collagen proteins, which are the principal component of its cuticle (as for all nematodes). Nematodes, including C. elegans, moult through four larval stages, before moulting into adult stages, and at each moult a new cuticle is formed. In C. elegans different groups of collagen coding genes are used by each life cycle stage. Therefore, transcriptomic comparison of different C. elegans life cycle stages would reveal life cycle stage-specific differences in the transcription of collagen coding genes. If these different life cycle stages had different lifestyles (such as free-living and parasitic), then we might wrongly infer that collagen molecules were an adaptation to the parasitic lifestyle. A second, related problem of comparing transcriptomes or proteomes between different nematode life cycle stages is that the results obtained depend on what stage is compared with what.
Notwithstanding these caveats, comparative transcriptomic methods have been widely used, mainly focused on two within-host parasitic stages: (i) larval stages that infect hosts, and (ii) adult stages living and reproducing within hosts. The results are difficult to synthesize and summarize because of the diversity of comparisons made among many different parasitic taxa. In studies of infective larvae newly arrived within a host, increased transcription of genes whose products are involved in growth, metabolism, sensory processes, etc. are observed (for example, in D. viviparous and H. contortus [16, 30]), consistent with the parasites resuming growth and development once entry into a host has been achieved. The expression of genes coding for proteases, and protease inhibitors, is also often seen (for example, in Ancylostoma spp. and A. suum [22, 33, 40]) presumably reflecting the need of migratory larvae stages to digest host tissue as they migrate.
For parasitic adult stages, there is also a comparative increase in the expression of genes coding for a range of enzymes including proteases, peptidases, hydrolases, catalases (for example, in Necator americanus, H. contortus, T. canis and A. suum [16, 22, 28, 32]), and protease inhibitors. Also, consistent with adult stages growing and reproducing, there is a comparative increase in the abundance of transcripts of genes whose products are involved in DNA replication, cuticle formation, and spermatogenesis.
A more nuanced analysis of the hookworm A. ceylanicum compared parasitic stages at different times after infection, showing that in worms’ first few days within a host, SCP/TAPS protein coding transcripts are comparatively more abundant, but in more established worms, there was a comparative increase in the abundance of transcripts of genes whose products are likely involved in growth (for example, genes coding for cuticle components, and proteins that are able to bind cytoskeleton proteins [33]). Later still in infection the transcription profile alters towards genes coding for protein tyrosine phosphatases, serine/threonine kinases and C-lectins. These observations make the important, more general point that parasitic nematodes live a dynamic life where aspects of their biology change as the within-host environment alters. For example, hosts have changing physiological states (e.g. reproductive state, nutritional state, seasonal changes) as well as different infections and co-infections [41] with consequent effects on the host immune response, all of which changes the within-host environment to which parasitic nematodes are exposed.
For Strongyloides nematodes, an unusual feature of their life cycle largely overcomes the problem of confounding lifestyle with life cycle stage. This is because the Strongyloides life cycle has two adult generations—one parasitic (which is female only) and one free-living (which is dioecious). In this case, comparison of parasitic adult female worms with free-living adult female worms compares lifestyle, without life cycle stage being a confounding factor. Comparison of the transcriptome of these free-living and parasitic adults of S. ratti showed that the parasitic females had significantly more transcripts of astacin-metallopeptidase and SCP/TAPS coding genes—the same gene families that are comparatively enlarged in these genomes [19]. In addition to this, genes coding for transthyretin-like proteins, prolyl endopeptidases, aspartic peptidases, acetylcholinesterases, trypsin inhibitors also had comparatively greater expression in parasitic females [19]. Importantly, up to a third of all genes differentially expressed between these stages were hitherto unknown, hypothetical protein-coding genes, including some gene families that appear to be unique to these parasitic taxa [19], with similar phenomena reported in other species too [42].
For some larger parasitic adult worms, gene transcription in different regions of the worms’ bodies have been analysed [43]. For example, the whipworm Trichuris has a structurally modified anterior end (called the stichosome), and this anterior end lies embedded within the hosts’ mucosa, while the worms’ posterior end lies free in the host gut lumen into which worm eggs are then easily liberated. Transcriptional analysis of the stichosome (compared with the rest of the adult worm) of Trichuris suis shows comparatively greater expression of many genes coding for peptidases and for porins (along with many other genes), consistent with the idea that the stichosome's function is to allow the worm to burrow into host tissue [29, 44]. The transcriptional profile of the gut of two parasitic nematodes (H. contortus and T. canis [16, 28]) has also shown that genes coding for a range of protease/peptidases (and protease and peptidase inhibitors), and products involved in binding with and transporting a range of molecules, are comparatively upregulated, consistent with the a priori view of the function of the worm’s gut.
In summary, studies that have sought to discover the genes specifically used by within-host parasitic stages have shown that many different gene families are used. Often these genes belong to families that appear to be comparatively expanded in parasitic species, strengthening the inference that these genes have a special role in parasitism. Beyond this, many of the other genes specifically expressed by these adult stages are consistent with growth and reproduction, which is clearly the principal biological function of within-host adult parasitic stages.
Secreted molecules
Identifying the molecules that nematode parasites secrete into their hosts aims to understand the interactive interface between parasites and their hosts. Two approaches are used: proteomic analysis of ES molecules, and genomic analysis of genes whose products are predicted to be secreted.
There has been relatively limited proteomic analysis of parasitic nematodes. Of particular note is the analysis of the ES of H. polygyrus, showing that this contained >400 ES-specific proteins. SCP/TAPS proteins dominated the ES, but they also contained proteases, lysozymes, apyrases and acetylcholinesterases [25]. The ES of the hookworm A. caninum also contained similar groups of proteins, but lectins and galectins too [42]. Given that different species of parasitic nematodes have different within-host niches, and so different biology, the molecules they secrete to interact with their host are likely to vary in respect of their different biology. Proteomic analyses of the H. contortus ES has focused on those proteins that are immunologically recognized by hosts [45]. A wide variety of molecules were identified including, once again, proteases. However, notably, the most immunogenic molecules in these ES was a transthyretin-like protein [45]. In S. ratti the secreted proteomes of parasitic and free-living females, and infective larvae were compared, finding a range of proteins specifically secreted by the parasitic stage, including proteases, prolyl endopeptidases and acetylcholinesterases [46].
Proteomic analyses are the most direct way of characterizing parasites’ ES. However, many studies have used a transcriptomic approach where ES molecules are computationally predicted based on the presence of signal peptides and the absence of trans-membrane domains—a so-called secretome—though, of course, this approach does not only identify those proteins specifically secreted outside of the worm. While this is a widespread approach, there is a worrying empirical discordance between protein and transcript abundance, probably because of post-translational processes which decouple the abundance of mRNA transcripts from the quantity of protein present [47–49].
Many of the results from analyses of predicted secretome overlap with results obtained from the two other approaches (above) used to investigate nematode parasitism. The predicted secretome can be large—in N. americanus it is a third of the entire proteome—and genes whose expression was comparatively greater in parasitic stages are more likely to be predicted secretory molecules (N. americanus, S. ratti) [19, 32].
In many species, proteases and peptidases are predicted to be secreted (A. suum, T. canis, T. suis, S. ratti [19, 22, 28, 29]); in N. americanus, over half (325 of 592) of all proteases fall into this category, and 19% of the 478 of D. viviparus [30]. Some SCP/TAPS proteins are also predicted to be secreted (in A. suum, T. canis, S. ratti, D. viviparus). But, species also differ in their predicted secretome. For example, in A. suum, o-linked glycosylated proteins are the single largest group of predicted secreted proteins, while other species have a diversity of molecules often implicated in immunomodulatory roles [22].
The genomic basis of nematode parasitism and prospects for the future
While impressive advances have been made in interrogating nematode genomes, these studies are still essentially at an early stage, especially given the diversity that exists among the nematodes. Notwithstanding this, it appears that the genomes of many parasitic nematodes are abundantly resourced with some particular gene families—those coding for proteases/peptidases, protease inhibitors, SCP/TAPS proteins, acetylcholinesterases, etc.—and in many cases these genes appear to be used especially during the within-host parasitic stages, and are often secreted [19, 46]. The apparent preponderance of proteases/peptidases and protease inhibitors makes the, perhaps obvious, point that parasitic nematodes make their living by managing and manipulating host proteins and that, similarly, hosts seek to interact with parasite proteins (which parasites, in turn, aim to resist). The apparent widespread occurrence of SCP/TAPS proteins in parasitic nematodes is fascinating, but it remains frustrating that we have such a limited understanding of what these molecules might do.
One consistent, if somewhat opaque, finding in many of these studies is that there are often taxa-restricted genes and gene families, and that these often have many of the hallmarks of being involved in the parasitic lifestyle. Discovering what the products of these genes do is clearly an important task to be able to fully understand the biology of nematode parasitism. This also makes the more general point that different groups of nematodes’ genomes are specialized in different ways for a parasitic lifestyle, and in the molecules they use and secrete as they interact with the host. While this diversity of function might be daunting, it might also give good purchase on separating out nematode-wide, from taxa-restricted, aspects of nematode parasitism.
Over the next few years it can be envisaged that further great strides will be made in interrogating the genomes of parasitic nematodes. However, beyond genomic analyses, functional manipulations of genomes will be needed to rigorously test hypotheses concerning the function of particular genes and their products. Despite the enormous success of transgenesis, RNAi analysis and genome editing etc. in C. elegans, these methods have proved much harder to establish with parasitic species. For example, it appears that parasitic nematodes are generally refractory to RNAi [50], whereas C. elegans is, by lucky chance, particularly susceptible (and indeed more susceptible than most other Caenorhabditis spp. [51]). Clearly, for parasitic nematodes the use of such approaches is considerably trickier because of the need for these parasites to be maintained in (usually vertebrate) hosts. However, progress is being made, particularly with Strongyloides spp., where transgenesis is possible [52] (and in the related species, P. trichosuri [53]), and where the first evidence showing the possible potential of CRISPR has now been obtained [52]. Using these and other techniques we will eventually reveal how nematodes live their parasitic lives, and so we will answer a fundamentally fascinating question, but will also be able to consider how to manipulate and control these organisms so as to alleviate human and animal suffering.
Key Points
Nematodes are ubiquitous and abundant animals and many species are parasites.
Nematodes have evolved parasitism on multiple different occasions.
Phylogenetically appropriate comparisons of parasitic and free-living nematodes can reveal nematodes’ genomic adaptations to parasitism.
Different parasitic nematodes have taxa-specific adaptations to parasitism, though the comparative expansion of genes and gene families coding for proteases and SCP/TAPS proteins appears to be a common theme among them.
Parasitic nematodes’ secreted molecules are the key interface between host and parasite, and discovering these molecules will be instrumental in understanding how nematodes are adapted to their parasitic lifestyle.
Acknowledgements
I would like to thank James Cotton for supplying data for the figure, Rebecca Cole and Vicky Hunt for useful discussions, and anonymous referees for helpful comments.
Funding
This work was supported by a grant from the Wellcome Trust.
Mark Viney studies the biology of nematodes, particularly parasitic species, seeking to understand how the environment controls their development, and how they are adapted to their parasitic lifestyle; nematode.bio.bris.ac.uk.
References
- 1. Cobb N. Nematodes and their relationships In: Yearbook United States Department of Agriculture. Washington DC: United States Department of Agriculture; 1914, pp. 457–90. [Google Scholar]
- 2. De Silva NR, Brooker S, Hotez PJ, et al. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol 2003;19:547–51. [DOI] [PubMed] [Google Scholar]
- 3. Maizels RM, Balic A, Gomez-Escobar N, et al. Helminth parasites – masters of regulation. Immunol Rev 2004;201:89–116. [DOI] [PubMed] [Google Scholar]
- 4. Savioli l, Albonico M.. Soil-transmitted helminthiasis. Nat Rev Microbiol 2004;2:618–9. [DOI] [PubMed] [Google Scholar]
- 5. Pullan RL, Smith JL, Jasrasaria R, et al. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 2014;7:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotez PJ, Kamath A.. Neglected tropical diseases in sub-saharan Africa: review of their prevalence, distribution, and disease burden. PLoS NTD 2009;3:e412.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. WHO. WHO Health Statistics and Information Systems, Estimates for 2000-2012, Disease Burden. who.int/healthinfo/global_burden_disease/estimates/en/index2.html.
- 8. Nieuwhof GJ, Bishop SC.. Costs of the major endemic diseases of sheep in Great Britain and the potential benefits of reduction in disease impact. Anim Sci 2005;81:23–9. [Google Scholar]
- 9. Selzer PM. Antiparasitic and Antibacterial Drug Discovery – From Molecular Targets to Drug Candidates. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA,2009. [Google Scholar]
- 10. Charlier J, Höglund J, von Samson-Himmelstjerna G, et al. Gastrointestinal nematode infections in adult dairy cattle: impact on production, diagnosis and control. Vet Parasitol 2009;164:70–9. [DOI] [PubMed] [Google Scholar]
- 11. Aguinaldo AMA, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 1997;387:489–93. [DOI] [PubMed] [Google Scholar]
- 12. De Ley PA. Quick tour of nematode diversity and the backbone of nematode phylogeny (January 25, 2006). In: WormBook The C. elegans Research Community. doi/10.1895/wormbook.1.41.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 13. Parkinson J, Mitreva M, Whitton C, et al. A transcriptomic analysis of the phylum Nematoda. Nat Genet 2004;36:259–1267. [DOI] [PubMed] [Google Scholar]
- 14. Blaxter ML, Koutsovoulos G.. The evolution of parasitism in Nematode. Parasitology 2015;142:S26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burke M, Scholl EH, Bird DM, et al. The plant parasite Pratylenchus coffeae carries a minimal nematode genome. Nematology 2015;17:621–37. [Google Scholar]
- 16. Laing R, Kikuchi T, Martinelli A, et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol 2013;14:R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dieterich C, Clifton SW, Schuster LN, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 2008;40:1193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitreva M, Jasmer DP, Zarlenga DS, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet 2011;43:228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hunt VL, Tsai IJ, Coghlan A, et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet 2016;48:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guiliano DB, Hall N, Jones SJ, et al. Conservation of long-range synteny and microsynteny between the genomes of two distantly related nematodes. Genome Biol 2002;3:research005710057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghedin E, Wang S, Spiro D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 2007;317:756–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jex AR, Liu S, Young ND, et al. Ascaris draft genome. Nature 2011;479:529–33. [DOI] [PubMed] [Google Scholar]
- 23. Blaxter ML, DeLey P, Garey JR, et al. A molecular evolutionary framework for the phylum Nematoda. Nature 1998;392:71–5. [DOI] [PubMed] [Google Scholar]
- 24. Weinstein SB, Kuris AM.. Independent origins of parasitism in Animalia. Biol Lett 2016;12:20160324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zarowiecki M, Berriman M.. What helminth genomes have taught us about parasite evolution. Parasitology 2015;142:S85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cantacessi C, Hofmann A, Young ND, et al. Insights into SCP/TAPS proteins of liver flukes based on large-scale bioinformatic analyses of sequence datasets. PLoS One 2012;7:e31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hewitson JP, Harcus Y, Murray J, et al. Proteomic analysis of secretory products from the model gastrointestinal nematode Heligmosomoides polygyrus reveals dominance of Venom Allergen-Like (VAL) proteins. J Proteomics 2011;74:1573–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu X-Q, Korhonen PK, Cai H, et al. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat Commun 2015;6:6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jex AR, Nejsum P, Schwarz EM, et al. Genome and transcriptome of the porcine whipworm Trichuris suis. Nat Genet 2011;46:701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McNulty SN, Strübe C, Rosa BA, et al. Dictyocaulus viviparus genome, variome and transcriptome elucidate lungworm biology and support future intervention. Sci Rep 2016;6:20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Viney ME, Kikuchi T.. Strongyloides ratti and S. venezuelensis – rodent models of Strongyloides infection. Parasitology 2017;144:285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang YT, Gao X, Rosa BA, et al. Genome of the human hookworm Necator americanus. Nat Genet 2014;46:261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwarz EM, Hu Y, Antoshechkin I, et al. The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection-specific gene families. Nat Genet 2015;47:416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sommer RJ. Pristionchus pacificus (August 14, 2006). In: WormBook The C. elegans Research Community, WormBook. doi/10.1895/wormbook.1.102.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 35. Del Valle A, Forbes BF, Harrison LM, et al. Isolation and molecular cloning of a secreted hookworm platelet inhibitor from adult Ancylostoma caninum. Mol Biochem Parasitol 2003;129:167–77. [DOI] [PubMed] [Google Scholar]
- 36. Zhan B, Liu Y, Badamchian M, et al. Molecular characterisation of the Ancylostoma-secreted protein family from the adult stage of Ancylostoma caninum. Int J Parasitol 2003;33:897–907. [DOI] [PubMed] [Google Scholar]
- 37. Selkirk ME, Lazari O, Matthews JB.. Functional genomics of nematode acetylcholinesterases. Parasitology 2005;131:S3–S18. [DOI] [PubMed] [Google Scholar]
- 38. Vaux R, Schnoeller C, Berkachy R, et al. Modulation of the immune response by nematode secreted acetylcholinesterase revealed by heterologous expression in Trypanosoma musculi. PLoS Path 2016;12:e1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hunt VL, Tsai IJ, Selkirk ME, et al. The genome of Strongyloides spp. gives insights into protein families with a putative role in nematode parasitism. Parasitology 2017;144:343–58. [DOI] [PubMed] [Google Scholar]
- 40. Mitreva M, McCarter JP, Arasu P, et al. Investigating hookworm genomes by comparative analysis of two Ancylostoma species. BMC Genomics 2005;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Viney ME, Graham AL.. Patterns and processes in parasite co-infection in animals and humans. Adv Parasitol 2013;82:321–69. [DOI] [PubMed] [Google Scholar]
- 42. Mulvenna J, Hamilton B, Nagaraj SH, et al. Proteomic analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol Cell Proteomics 2009;8:109–21. [DOI] [PubMed] [Google Scholar]
- 43. Yin Y, Martin J, Abubucker S, et al. Intestinal transcriptomes of nematodes: comparison of the parasites Ascaris suum and Haemonchus contortus with the free-living Caenorhabditis elegans. PLoS NTD 2008;2:e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Foth BJ, Tsai IJ, Reid AJ, et al. Whipworm genome and dual-species transcriptome anlayses provide molecular insights into an intimate host-parasite interaction. Nat Genet 2014;46:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yatsuda AP, Krijgsveld J, Cornelissen AWCA, et al. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J Biol Chem 2003;9:16941–1651. [DOI] [PubMed] [Google Scholar]
- 46. Soblik H, Younis AE, Mitreva M, et al. Life cycle stage-resolved proteomic analysis of the excretome/secretome from Strongyloides ratti – identification of stage-specific proteases. Mol Cell Proteomics 2011;10:M111.010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Foss EJ, Radulovic D, Shaffer SA, et al. Genetic basis of proteome variation in yeast. Nat Genet 2007;39:1369–75. [DOI] [PubMed] [Google Scholar]
- 48. Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 2011;7:e1001393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haider S, Pal R.. Integrated analysis of transcriptomic and proteomic data. Curr Genomics 2013;14:91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Selkirk Me, Huang SC, Knox DP, et al. The development of RNA interference (RNAi) in gastrointestinal nematodes. Parasitology 2012;139:605–12. [DOI] [PubMed] [Google Scholar]
- 51. Winston WM, Sutherlin M, Wright AJ, et al. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci USA 2007;104:10565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lok JB, Shao H, Massey HC, et al. Transgenesis in Strongyloides and related parasitic nematodes: historical perspectives, current functional genomic applications and progress towards gene disruption and editing. Parasitology 2016;144:327–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grant WN, Skinner SJ, Newton-Howes J, et al. Heritable transgenesis of Parastrongyloides trichosuri: a nematode parasite of mammals. Int J Parasitol 2006;36:475–83. [DOI] [PubMed] [Google Scholar]


