Abstract
Iron–sulfur (Fe-S) clusters are ubiquitous cofactors essential to various cellular processes, including mitochondrial respiration, DNA repair, and iron homeostasis. A steadily increasing number of disorders are being associated with disrupted biogenesis of Fe–S clusters. Here, we conducted whole-exome sequencing of patients with optic atrophy and other neurological signs of mitochondriopathy and identified 17 individuals from 13 unrelated families with recessive mutations in FDXR, encoding the mitochondrial membrane-associated flavoprotein ferrodoxin reductase required for electron transport from NADPH to cytochrome P450. In vitro enzymatic assays in patient fibroblast cells showed deficient ferredoxin NADP reductase activity and mitochondrial dysfunction evidenced by low oxygen consumption rates (OCRs), complex activities, ATP production and increased reactive oxygen species (ROS). Such defects were rescued by overexpression of wild-type FDXR. Moreover, we found that mice carrying a spontaneous mutation allelic to the most common mutation found in patients displayed progressive gait abnormalities and vision loss, in addition to biochemical defects consistent with the major clinical features of the disease. Taken together, these data provide the first demonstration that germline, hypomorphic mutations in FDXR cause a novel mitochondriopathy and optic atrophy in humans.
Introduction
Iron–sulfur (Fe–S) clusters act as inorganic cofactors in many protein activities and are essential for multiple important biological processes, including electron transport in mitochondrial respiration, cofactor synthesis, substrate binding/activation, Fe-S storage, DNA repair, and gene regulation (1,2). Fe–S cluster biosynthesis is a tightly regulated process dependent upon the coordinated delivery of iron and sulfide, principally within the mitochondria of eukaryotic cells. Formation of Fe–S clusters involves the use of electrons from sulfur. Following the assembly of a Fe–S cluster on a scaffold protein, the cluster is transferred to an acceptor protein. A variety of human disorders have been associated with impaired Fe–S cluster biosynthesis, including neurodegenerative disorders (e.g. Friedreich ataxia and myopathy with lactic acidosis) (1). Iron homeostasis, which requires precise synthesis and localization of Fe–S clusters in mitochondria, is critical to ensure that there are sufficient, but not toxic levels of iron for cellular functions. Excessive levels of iron favor the formation of excess oxygen free radicals and consequent mitochondrial dysfunction. The optic nerves are especially vulnerable to such oxidative stress.
Ferredoxin-1 and -2 [FDX1 and FDX2 (a.k.a. FDX1L)], which are encoded by two homologous genes in mammals and expressed in mitochondria, serve as electron donors in Fe–S cluster biosynthesis (3,4). Mitochondrial membrane-associated ferredoxin reductase (FDXR) is a flavoprotein that initiates the mitochondrial electron transport chain reaction by transferring electrons from NADPH to the mitochondrial cytochrome P450 system via FDX1 or FDX2 (5). This pathway is also critical to steroid hormone biosynthesis (6). FDXR has been shown to be essential for cell viability and contributes to p53-mediated apoptosis through the generation of oxidative stress in mitochondria (7). FDX2, in particular, has been shown to act as an electron source in the electron transfer chain with NADPH and FDXR (8). Moreover, biallelic mutations in FDX2 have been identified in patients with metabolic myopathy who have deficient mitochondrial complexes I, II, and III and deficient aconitase, suggestive of mitochondrial Fe–S cluster-related defects (9).
We performed clinical exome sequencing in families with suspected mitochondriopathy, with the aim of identifying disease-causing genes. We report 17 individuals—from 13 independent families—with biallelic FDXR mutations, exhibiting a wide range of signs of mitochondriopathy. The affected individuals presented with neurological abnormalities, with most showing some degree of optic atrophy and neuropathy. Patient fibroblast cells showed deficient ferredoxin NADP reductase activity and mitochondrial dysfunction. Such defects were rescued by overexpression of wild-type FDXR. In a parallel analysis in the mouse model, we observed that mice with the Fdxr mutation allelic to the most common mutation among our patients recapitulated major aspects of the human phenotype, including progressive peripheral nerve impairments, decreased Fdxr enzyme activity, and mitochondrial dysfunction. Hence, we report a convergence of clinical, mouse model, and experimental findings suggesting that a biallelic mode of inheritance of mutant FDXR causes mitochondriopathy with optic atrophy.
Results
Clinical phenotypes associated with FDXR mutations
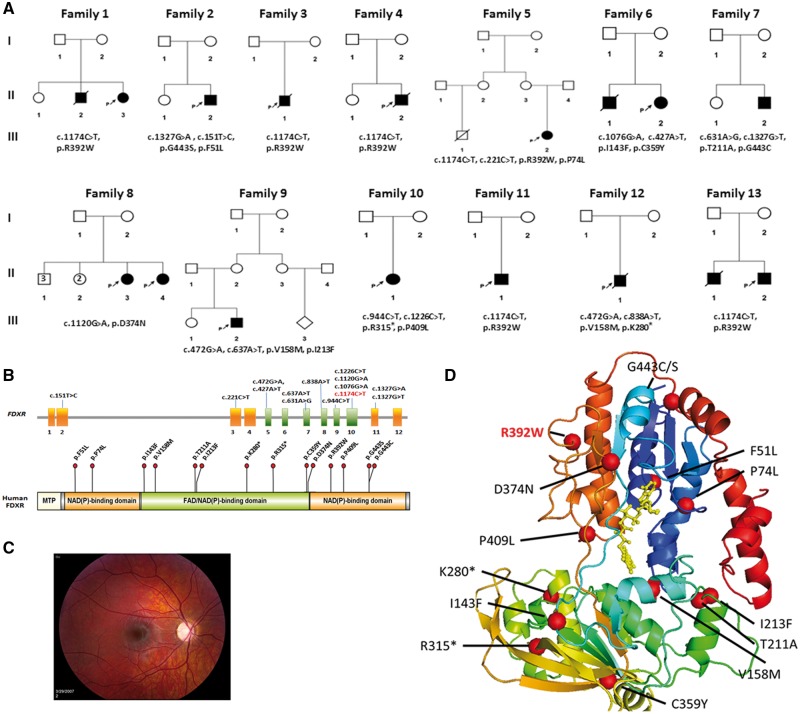
Among the 13 unrelated probands from diverse ethnic backgrounds with biallelic rare variants in FDXR (Fig. 1A), we identified 6 homozygotes and 7 compound heterozygotes (Table 1). As is common for mitochondrion-related diseases, patients presented with variable clinical phenotypes, consistent with mitochondriopathy. They were found to share the core clinical features of optic atrophy (Fig. 1C;Supplementary Material, Fig. S1A–C), ataxia, and hypotonia. Other common features in the cohort were neurological anomalies evidenced in MRI imaging, seizures, and global developmental delays. Other less common features included spasticity and respiratory failure. Several patients had abnormal movements and hearing loss. Six patients died in early age. The patients’ clinical features are summarized in Table 1 and additional detailed clinical information about the patients is provided in Supplementary Material, Note S1.
Figure 1.
Pedigree of affected families, eye phenotypes, and FDXR mutations. (A) Family pedigrees. Filled symbols represent affected individuals. Biallelic FDXR variants are indicated with each pedigree. (B) Schematic representation of FDXR and its predicted protein product. Exons are represented with boxes. The NAD(P) and FAD/NAD(P) binding domains are shown in orange and green, respectively, and the observed mutations are labelled. Figure was generated using Illustrator for Biological Sciences (IBS), available at http://ibs.biocuckoo.org/index.php as described previously (44). MTP: Mitochondrial Transit Peptide or mitochondrial targeting signals. (C) Retinal image of optic atrophy from a patient with a compound heterozygous FDXR mutation (p.F51L/p.G443S). (D) Patient mutations mapped to a three-dimensional FDXR structure, based on the homologous FDXR structure (starting from blue at the N-terminal region, and ending at red in the C-terminus) for Bos taurus complexed with FAD (gold). The residue position of missense variants and starting point of truncations are indicated with red spheres.
Table 1.
Summary of clinical information
| Family | 1 |
2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II-3 | II-2 | II-2 | ||||||||||||
| Gender | F | M | M | M | M | F | F | M | M | F | M | M | M | |
| Genotype | p.R392W hom | p.R392W hom | p.F51L/ p.G443S | p.R392W hom | p.R392W hom | p.P74L/ p.R392W | p.I143F/ p.C359Y | p.T211A/ p.G443C | p.D374N hom | p.V158M/ p.I213F | p.R315*/ p.P409L | p.R392W hom | p.V158M/ p.K280* | p.R392W hom |
| Age examined | 1.6 yrs | d. 1.6 yrs | 14 yrs | 5.4 yrs; d. 6 yrs | 1.3 yrs; d. 2.3 yrs | 4 yrs | 1.8 yrs | 3 yrs | 20 yrs | 15.5 yrs | 4 yrs | 1.8 yrs | 2.5 yrs | 0.3 yrs |
| Microcephaly | No | No | No | Yes | Yes | Yes | Yes | No | No | No | No | |||
| Optic atrophy | Yes | Yes | Likely | Yes | Yes | Yes | No | Yes | No | Yes | Likely | Yes | Yes | Yes |
| Brain MRI | delayed myelina-tion | abnl CCa, Dgpb, dMBc, dCCAd | Normal | Normal | dCCe, abnl FLf | inCAg, deCWMh, deBGi, deGP&SNj | abnl BG &MP signalk | CAl, abnl PV signalm | Normal | swelling of the cerebellum | Normal | CAl | Normal | |
| Seizures | No | No | No | No | No | Yes, IS& Seizuren | No | No | No | Yes | No | |||
| EEG | Normal | Normal | Abnormal; dCFo | Abnormal; severe encephalopathy | Normal | Abnormal; abnl B&Sp | Normal | Abnormal; slow wave | Normal | |||||
| GDDq | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Regression | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | No | Yes |
| Hypotonia | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Yes | Yes | ||
| Spasticity | No | No | Yes | Yes | No | Yes | Yes | No | No | Yes | No | abnl MOr | Yes | Yes |
| Ataxia | Yes | MDs | No | Likely | No | Yes | Yes | |||||||
| Mitochodrial Morphology | abnl shapet, loss cristae | abnl shapeu | abnl shapev,w | Marginal loss of IV, normal, II-III | In#x | |||||||||
| Muscle ETC Results | I 51%, III 51%, II-III 15%, IV 48%; normal CS | Normal I-IV. | I-III 28%; CS high | Normal I -IV; CS high | ||||||||||
abnormal corpus callosum, abnl CC
diffusion of the globi palladi, dGP
diffusion of the midbrain, dMB
diffusion of the central and cortical atrophy, dCCA
decreased corpus callosum, dCC
chronic extra-axial collections in bilateral frontal convexities abnormal frontal lobe, abnl FL
Increased cerebellar atrophy, inCA
decreased cerebral white matter, deCWM
decreased basal ganglia, deBG
decreased signals globus palladia and substantia nigra, deGP&SN
abnormal basal ganglia and mesencephalic peduncles, abnl BG & MP signal
cerebral atrophy, CA
abnormal white matter periventricular signal, abnl PV signal
infantile spasms and seizures, IS&Seisure
diffuse cerebral function, dCF
chaotic background with spasm, abnl B&S
globe development delay, GDD
abnormal movements, abnl MO
Movement disorder, MD
abnormal mitochondrial morphology, abnl shape
abnormal shape, abnl shape
Elongated and abnormally shaped mitochondria, abnl shape
abnormal morphology, abnl shape
increased mitochondrial number, In#
FDXR mutations are evolutionarily conserved
The FDXR protein contains two NAD(P)-binding domains and one FAD/NAD(P)-binding domain (Fig. 1B). In Family 1, we identified a homozygous rare variant, c.1174 C > T (p.R392W), an amino acid change in an evolutionarily conserved residue of the FAD/NAD(P)-binding domain of FDXR (Supplementary Material, Fig. S2A and B). Multiple sequence alignment was used to analyze amino acid conservation (Supplementary Material, Note S2). The FDXR mutations identified in other patients are also in the FAD/NAD(P)-binding domain with a hotspot at p.R392W, which accounts for 11 (42.3%) out of the 26 mutant alleles (Fig. 1B). This alteration is a relatively common Latino allele with 0.19% minor allele frequency in this population (carrier rate 1/263; http://exac.broadinstitute.org/variant/17–72860036-G-A). Fourteen missense or nonsense FDXR mutations were identified in this study and eight of them (I143F, V158M, T211A, I213F, K280*, R315*, C359Y, D374N) clustered in the FAD binding site (Fig. 1B). Noteworthy, seven patients with homozygous R392W died at an early age, suggesting that this allele may have a more severe, detrimental impact on FDXR function. There were no biallelic truncating mutations identified in any patients and no homozygous loss-of-function alleles in gnomAD (http://gnomad.broadinstitute.org/), suggesting that complete loss of function of FDXR is likely incompatible with life.
Mutations of FDXR may cause loss of function due to protein instability
The FDX1/2 and FDXR proteins interact physically, and FDX1/2 mutations can affect binding to FDXR and/or cytochrome P450 (10). To investigate the possible functional consequences of FDXR mutations, we mapped all mutations to a crystal FDXR protein structure (Fig. 1D). P409L is a variant that appears to direct towards a carbonyl oxygen of the nucleotide of FADH (11). F51L, P74L, V158M are proximate to FADH. There is a possibility of the FADH and FDXR interaction. Although there is no experimental evidence of functional homo-dimerization of FDXR, two of the mutations, I143F and D374N, appear to lie at a putative interface between FDXR monomers (12). The remaining mutations, namely T211A, I213F, C359Y, R392W, G443C, and G443S, were buried, at least partially; these mutations could disrupt functionality by causing a destabilization of the protein structure (Fig. 1D). Examination of the energies of these mutations (Supplementary Material, Table S2) indicated that C359Y, G443C, and G443S are likely to disrupt the folding of FDXR. However, the modes of disruption of T211A, I213F, and R392W are unclear. There is no indication that these positions would be expected to disrupt a posttranslational modification site or protein interaction site. Previous results suggested that the mutation in the N-terminus could have significant effects on FDXR biochemistry by increasing the Kd of FDXR with cytochrome P450. Importantly, these mutations could also decrease cholesterol side chain cleavage activity (10). Together, the mutations in FDXR could impair protein functions through multiple mechanisms, including protein-protein interactions, protein stability and binding affinity to the substrates.
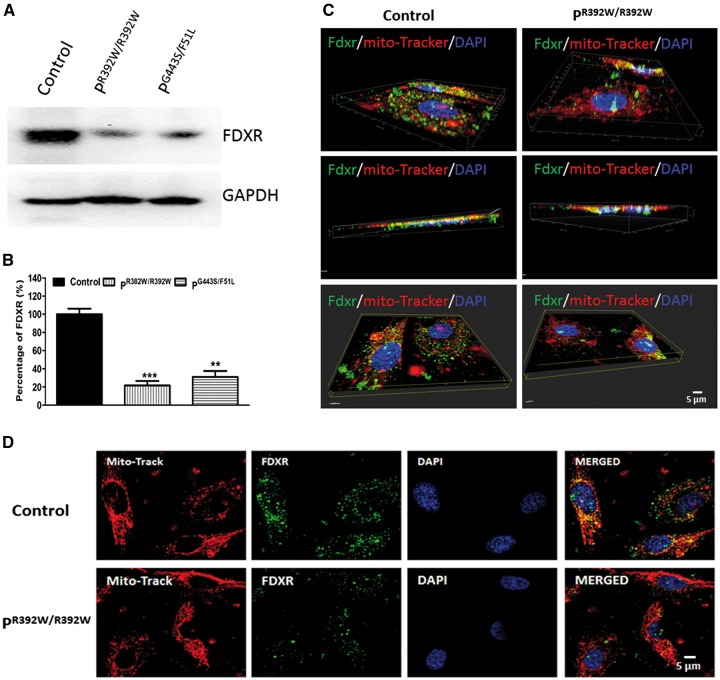
To investigate if the mutations of FDXR affect the protein’s stability, we tested the FDXR protein level in patient fibroblast cells. As shown in Figure 2A and B, the protein levels of FDXR in two of the patients’ fibroblast cell lines that were tested decreased dramatically. To confirm this finding, we examined the expression and localization of FDXR in control and patients’ fibroblasts by confocal microscopy. In this experiment, mitochondrial localization was demonstrated by a signal overlap between FDXR (Fig. 2C and D, green) and mitochondria (Fig. 2C and D, red). As shown in Figure 2C and D, we found that FDXR levels are significantly lower in the patient fibroblast cells with the homozygous mutations R392W. While we found that FDXR localization was mainly in mitochondria, FDXR was also detectable in nuclei and in the cytoplasm, in both control and patient fibroblasts. 3 D image analysis confirmed these observations (Fig. 2C and D, Supplementary Material, Movie S1). Previous electron microscopy and immunogold labeling studies report that FDXR is sublocalized to the mitochondrial matrix (13) where it initiates the electron-transport chain and ultimately leads to the conversion of cholesterol to pregnenolone in the first rate-limiting step of steroid hormone biosynthesis (13,14). The significance of the presence of FDXR in nuclei and the cytoplasm is yet to be elucidated. Mitochondrial Fe–S cluster biogenesis machinery proteins, namely NFS1 and IDS11, were also observed in the cytoplasm, though the functional relevance of their presence also remains to be elucidated (15,16).
Figure 2.
FDXR protein levels in patient fibroblasts are decreased relative to control cells. (A) Comparison of western blot results showed lower levels of FDXR expression in patient fibroblasts compared with control fibroblasts. (B) Quantification data of bands intensity was performed. Data are representative of three independent replications. (C) 3 D images evaluated by confocal microscopy using a Nikon AIR LUN-V Inverted confocal microscope and analyzed by Imaris software. (D) 2 D images evaluated by confocal microscopy using a Nikon AIR LUN-V Inverted confocal microscope and analyzed by Imaris software. The antibody we used was obtained from abcam (ab204310). The recombinant fragment corresponding to Human Ferredoxin Reductase is from aa 1 to aa 134.
FDXR mutations cause mitochondrial dysfunction in patient fibroblast cells
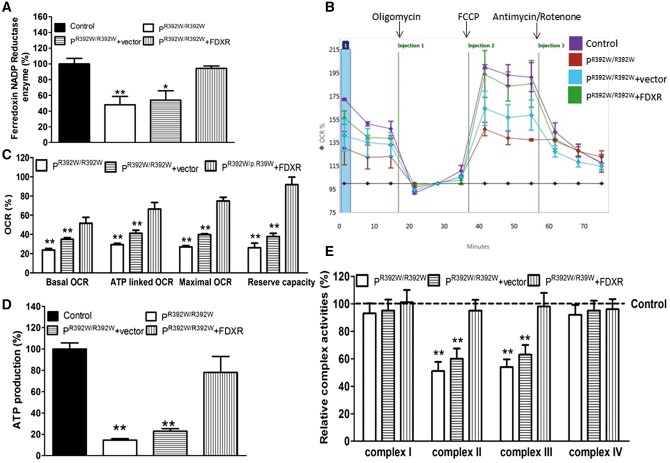
To investigate the functional consequences of patient mutations and to demonstrate causation, we assessed FDXR enzyme activity and mitochondrial functions in cultured fibroblasts from individual II-2 in Family 1 (PR392W/R392W) who is homozygous for the p.R392W mutation. To confirm causation, wild-type FDXR was overexpressed in the patient fibroblasts for complementation experiments (rescue) and expression was confirmed by immunofluorescence microscopy of tdTomato protein (Supplementary Material, Fig. S3A–C). As shown in Figure 3A, Ferredoxin NADP reductase activity in patient fibroblasts, non-transduced patient fibroblasts, and patient fibroblasts overexpressing FDXR (rescued) were 48%, 53%, and 94% respectively of the mean value measured for age and gender matched control fibroblasts. Basal OCR, a measure of cellular bioenergetics, in patient fibroblasts was 24% of that in control fibroblasts (100%, reference) (Fig. 3B and C). ATP production, an index of mitochondrial oxidative phosphorylation capacity, was significantly decreased in patient fibroblasts compared with that in control fibroblasts and rescued in patient fibroblasts transduced with FDXR (As shown in Fig. 3D). FDXR transduction of patient fibroblasts resulted in a partial rescue of basal OCR. Likewise, ATP-linked OCR, maximal OCR and reserve capacity OCR in patient fibroblasts were significantly lower than the rates observed for control fibroblasts and these reductions were also reversed in FDXR-transduced patient fibroblasts (Fig. 3C). Furthermore, complex II and III activities in patient fibroblasts were reduced to 51% and 54% of control levels, respectively (Fig. 3E). Complex II and III activities in FDXR-transduced patient fibroblasts were 95% and 98% of control levels, respectively. However, complex I and IV activities did not differ significantly among the various groups. Consistent with this observation, this patient’s deceased brother who is also homozygous for the p.R392W mutation, showed major deficiency in complexes II-III and mild reduction of complex I and IV activities in the muscle specimen. More importantly, the fibroblasts from the proband of family 2 (PG443S/F51L), a compound heterozygote, showed the similar mitochondrial dysfunction (Supplementary Material, Fig. S6A–D), suggesting that the mutations in FDXR not only impair the enzymatic activities, but also cause general mitochondrial dysfunctions.
Figure 3.
Mitochondrial dysfunction in patient fibroblasts. (A) Enzymatic assay of ferredoxin NADP reductase in fibroblasts (includes all products with a specification for this enzyme). Reductase activity was normalized relative to control fibroblasts. (B) An analysis of O2 consumption in indicated cell lines. OCRs were first measured for 1 × 104 cells/sample under basal conditions, and then measured repeatedly following the sequential addition of oligomycin (1.5 μM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) (4 μM), rotenone (1 μM), and antimycin A (1 μM) at the indicated times. (C) Basal, ATP-linked, maximal, and reserve capacity OCR in mutant and control cell lines. Basal OCR is OCR before the addition of oligomycin minus OCR after the addition of rotenone/antimycin A. ATP-linked OCR is OCR before the addition of oligomycin minus OCR after the addition of oligomycin. Maximal OCR is OCR after the addition of FCCP minus non-mitochondrial OCR. Reserve capacity OCR is the difference between maximal OCR after the addition of FCCP and basal OCR. (D) Measurement of cellular and mitochondrial ATP levels in fibroblasts by bioluminescence assay. Average ATP levels (6–7 replicates per cell line) are shown as a percentage of control levels. (E) Enzymatic activities of respiratory chain complexes I, II, III and IV. The calculations were based on three independent determinations. Data are represented as the means ± SEM; *P<0.05, **P <0.01, ***P <0.001 vs. control, as determined by Student’s t test.
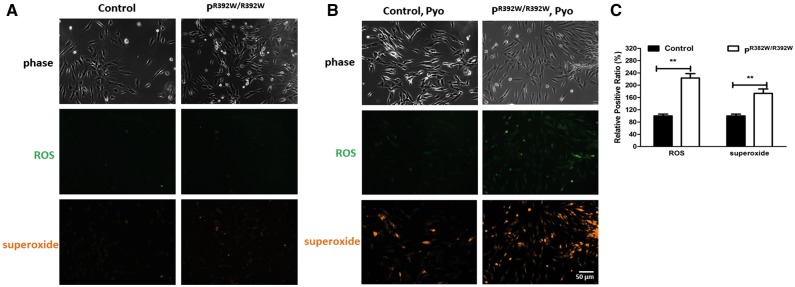
FDXR mutations cause increased ROS production in patient fibroblasts
Fe–S clusters are components of the mitochondrial complexes I and III. Mutations of FDXR could impair Fe-S cluster biosynthesis and result in decreased complex I and complex III activity. However, our results suggest that mutations of FDXR cause a broader impairment of mitochondrial function. Accumulating evidence suggests that FDXR plays an important role in ROS production (17). Therefore, we tested ROS production in patient cell lines. Our results showed that although superoxide production was altered in patient fibroblasts, reactive oxygen species (ROS) levels did not differ (Fig. 4A). However, when ROS production was induced by introduction of pyocyanin ROS, superoxide production levels increased significantly in patient fibroblasts compared with the control fibroblasts (Fig. 4B). This effect was blocked in the presence of a ROS production inhibitor (Supplementary Material, Fig. S5). This result is consistent with previously published data, which demonstrate that FDXR can influence ROS-mediated apoptosis in epithelial cancer cells (13,17). Altogether, these findings indicate that mutations in FDXR could have a broader effect on mitochondrial functions through the ROS pathway, contributing to more general mitochondrial dysfunction.
Figure 4.
Increased ROS production in patient fibroblasts. (A) Profiling of ROS formation in fibroblasts loaded with ROS/superoxide detection reagents and untreated by pyocyanin or NAC. (B) Profiling of ROS formation by fluorescence microscopy in fibroblasts loaded with ROS/superoxide detection reagents and treated with pyocyanin. General oxidative stress levels were monitored in the green channel, while superoxide production was detected in the orange channel. (C) Quantification data of the relative positive signal ratio were performed after the cells were treated with pyocyanin. Data collected from 10 images for each group).
Mitochondrial dysfunction is recapitulated in FdxrR389Q/R389Q mice
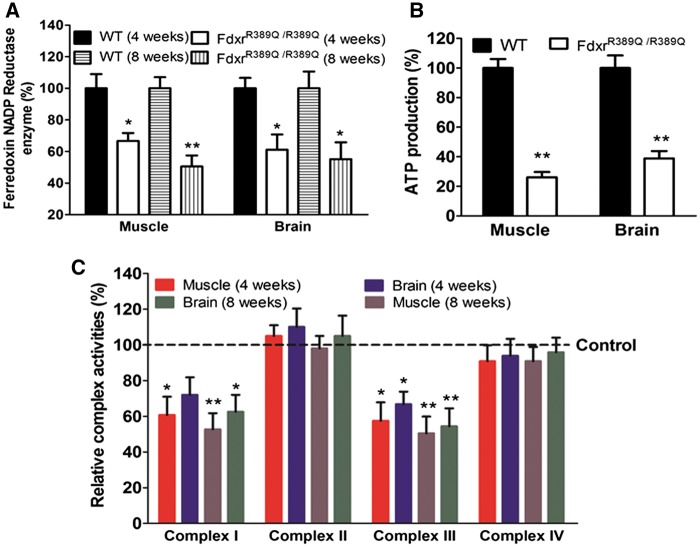
Taking advantage of a newly discovered mouse strain carrying a spontaneous mutation in Fdxr, we further examined the role of FDXR in peripheral neuropathy, mitochondrial function, and optic atrophy. These mice were previously reported to have a recessive gait phenotype and were shown by exome sequencing to harbor a homozygous missense mutation in Fdxr that results in an R389Q change in the encoded protein (corresponding to human FDXR p.R392Q and allelic to the common patient mutation p.R392W) (18). Using comprehensive gait analyses, we found that this mutation causes progressive impairment of posterior gait dynamics (Supplementary Material, Movie S2). To test FDXR and mitochondrial function in this mouse model, we conducted FDXR enzyme activity assays using tissues from homozygous FdxrR389Q/R389Q mice and WT littermate controls (Fig. 5A). Consistent with the human enzyme assays, we observed significantly reduced FDXR activity levels, relative to WT levels in both muscle (33 and 49%, respectively) and brain (39 and 45%, respectively). Similar results were observed in other tissues, including the liver and heart (Supplementary Material, Fig. S7A) which showed impaired complex I and complex III activities (Fig. 5C;Supplementary Material, Fig. S7C), and decreased ATP production (Fig. 5B;Supplementary Material, Fig. S7B).
Figure 5.
Mitochondrial function in mouse muscle and brain tissues. (A) Results of the Ferredoxin NADP reductase activity assay (all enzyme-specific products) in mouse tissues. Relative activity levels were normalized relative to levels in WT mice. (B) Quantitative bioluminescence assay of cellular and mitochondrial ATP levels. (C) Activities of respiratory chain complexes I, II, III and IV in mitochondria isolated from three mutant mouse tissues and three WT mouse tissues. The calculations were based on four independent determinations.
Visual acuity defects are associated with loss of retinal ganglion cells (RGCs) and abnormal myelination of optic nerve in FdxrR389Q/R389Q mice
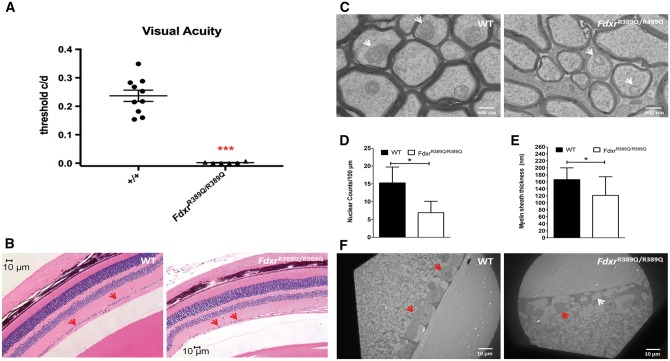
In addition to ataxia, the majority of the patients with FDXR mutations in this study had vision impairment or loss and optic atrophy (Table 1 and Supplementary Material, Note S1). To test if FdxrR389Q/R389Q mice recapitulate these clinical features, we tested visual acuity using functional optokinetics in adult mice (15–17 weeks of age). The optokinetic response scores from FdxrR389Q/R389Q mice were significantly reduced as compared with WT controls (Fig. 6A). We then performed a histopathological assessment of the retina. We counted the retinal ganglion cell (RGC) layer nuclei in H&E stained, paraffin sections of eyes of 2-month-old and 6-month-old WT and FdxrR389Q/R389Q mice. No significant difference in the number of RGC layer nuclei between WT and mutant mice at the age of 2 months old was found (Supplementary Material, Fig. S8A). However, a significant reduction in the RGC layer nuclear number was observed in 6-month-old FdxrR389Q/R389Q mice, with a 54% loss of the RGC layer nuclei (Fig. 6B) as compared with WT mice. Transmission electron microscopy (TEM) of retina from 6-month-old mice confirmed RGC loss in FdxrR389Q/R389Q mice, and revealed abnormal cytoplasm, shrunken nuclei, and abnormal chromatin aggregates in remaining RGCs (Fig. 6F).
Figure 6.
Visual acuity defects associated with loss of retinal ganglion cells (RGCs) and abnormal myelination of optic nerve in 6-month-old mice with p.R389Q/R389Q mutations. (A) Ability to track directionality of visual cues (black and white lines) in real time via closed circuit camera monitoring was assessed. Frequency thresholds were measured by systematically increasing the spatial frequency of the gratings (narrowing of stripes) at 100% contrast until animals could no longer track (45) and optokinetic scores were calculated. FdxrR389Q/R389Q showed significantly impaired visual acuity compared with age and sex-matched controls. (B) H&E-stained sagittal sections through the optic disc of eyes showed that the number of retinal ganglion cell (RGC) layer nuclei was reduced in H&E sections of eyes from 6-month-old p.R389Q mice as compared with WT control mice. Nuclei (excluding the flat and smaller endothelial nuclei) were counted in the RGC layer in H&E sections (D). Nuclear number in the RGC layer of p.R389Q mice decreased significantly (B and D). (C) Transmission electron microscopy (TEM) showed a significant decrease in the thickness of the myelin sheath of RGC axons in 6-month-old p.R389Q mice. Compared with WT RGC axons, RGC axons of p.R389Q mice lost their myelin sheath (C); the reduction in the thickness of the myelin sheath of p.R392Q RGC axons was statistically significant (E). Thickness of the myelin sheath of the RGC axons in the optic nerve was also measured using ImageJ software (E). (F) TEM study of the 6-month-old retina indicated that the p.R389Q mice had a clear reduction in the number of their RGCs; the remaining RGCs also showed an empty cytoplasm, shrinkage of the nuclei, and aggregation of chromatin inside the nuclei (F, arrow).
RGC axons in the optic nerve were also studied with TEM. No difference was found in the 2-month-old WT and FdxrR389Q/R389Q mouse RGC axons (Supplementary Material, Fig. S8B). However, a significant decrease in the thickness of the myelin membrane of RGC axons was found in 6-month-old FdxrR389Q/R389Q mice (Fig. 6C), indicating that the FdxrR389Q/R389Q mice had compromised myelination of RGC axons, probably related to defects in oligodendrocyte function. Also, mitochondria inside RGC axons of the FdxrR389Q/R389Q mice had structural damage, including cristae loss and the appearance of “ghost” mitochondria (Fig. 6C), which were consistent with the findings in the patient fibroblasts indicating that mutations of FDXR had broader effects on mitochondrial structure and functions.
Discussion
Synthesis of iron-sulfur (Fe–S) cluster scaffold proteins requires a coordinated delivery of iron, sulfur, and electrons. Although the iron donor for Fe–S clusters remains elusive, two Fe–S cluster assembly proteins, IscA (19) and frataxin (20), have been proposed as possible iron chaperones delivering iron for Fe–S cluster assembly in IscU (21). The electrons required for Fe–S cluster assembly in IscU are delivered by ferredoxin, which is reduced by ferredoxin reductase using NADPH as the electron donor (8,22). In humans, there are two ferredoxins: FDX1 and FDX2 (8). Both FDX1 and FDX2 appear to be important for Fe–S cluster biogenesis (4,8). Nevertheless, deletion of FDX1 in Hela cells did not elicit any defects in Fe–S cluster biogenesis (3), and mutation of FDX2 has been associated with a mitochondrial muscle myopathy, which is characterized by severely impaired activities of Fe–S proteins in mitochondria (9). Furthermore, while FDX1 is exclusively expressed in adrenal gland, FDX2 is ubiquitously found in virtually all tissues (3). Thus, FDX1 may participate in the synthesis of bile acid, vitamin D, and steroid hormones (8,22–24), while FDX2 is likely involved in Fe–S cluster biogenesis (3,22). Humans have one ferredoxin reductase (FDXR) (14), which is a FAD-dependent enzyme that reduces FDX1 and FDX2 using NADPH as the electron donor (8,22). Biallelic mutations in the FDX2 gene (FDX1L) have been identified in patients with metabolic myopathy with deficient mitochondrial complexes I, II, and III and aconitase, suggesting mitochondrial Fe–S-related defects (9). Depletion of FDXR diminishes Fe–S cluster assembly and causes mitochondrial iron overload (8), indicating that FDXR has a crucial role in Fe–S cluster biogenesis in human cells.
Here, we identified biallelic rare variants in FDXR by clinical exome sequencing in 17 affected individuals from 13 independent families who share a wide range of presentations consistent with a mitochondrial disorder, and optic atrophy was observed in the vast majority of these patients. We performed genetic complementation experiments by transfecting the wildtype FDXR gene in patient-derived fibroblast cells to support the pathogenesis. Mechanistically, mutations of FDXR may disrupt the function domains of the enzyme based on 3-D crystal model. Our data also suggest some mutations may affect the protein conformation and/or the protein stability. Abnormal ROS production has been identified in this study and in many mitochondrial diseases, which could be secondary to mitochondrial respiratory chain defects and may play very important roles in the pathogenesis of FDXR mutations. A mouse model carrying a mutation allelic to a common mutation found in our patients showed progressive impairments in enzyme activity and mitochondrial function and recapitulated the major human phenotypes. Mouse model provided a great opportunity to study the pathogenesis of Fdxr mutations. In this study, we were also able to examine the pathogenesis of vision loss. The vision loss seen in FdxrR389Q/R389Q mice is progress- and aging-dependent, suggesting degenerative process. This is further supported by aging-dependent RGC loss and dysmorphic mitochondria in optic nerve. The clinical presentations, mouse models, and experimental findings suggest a biallelic mode of inheritance of FDXR mutations causing mitochondriopathy with optic atrophy.
Mitochondrial diseases often present with myopathy, optic atrophy, and pigmentary retinopathy in addition to common central nervous system symptoms such as encephalopathy, global developmental delays, and seizure (25). Eye abnormalities, ataxia, and hypotonia exhibited by our patients (Table 1) are consistent with Fe–S disease clinical features (1). Our initial phenotypic analysis of FdxrR389Q/R389Q mice revealed that they share many congruent neurological features with human patients, but there were some discrepancies, including differences in complex activities that could be explained by species-related differences. Alternatively, the most common mutation in humans is p.R392W, rather than p.R392Q. Since tryptophan (W) is a much more radical change from arginine than glutamine (Q), this could explain differences between the mouse and human phenotypes. To further refine the mouse model and provide more complete recapitulation of clinical features, future efforts will be directed towards the generation and characterization of mouse models carrying patient specific mutations. CRISPR/Cas9 genome editing technology to generate knock-in and knockout mouse lines with the same amino acid changes as the hot spot mutation (p.R392W) in humans would provide additional value since the phenotypes of the “stiff” mouse are slightly different from human subjects.
This study is the very first to investigate the clear fact that biallelic FDXR mutations cause human disease, and provides us a unique opportunity to understand of the biology of the FDXR pathway. This is a unique opportunity to characterize the molecular mechanisms behind this new disorder. Since FDXR occupies a crucial upstream position at the top of the iron-sulfur cluster biogenesis pathway, with the potential to influence numerous downstream proteins in an epistatic manner, this study provides us the opportunity to understand the major biochemical consequences of mutations in this pathway. Some FDXR variants, such as p.R392W, appear quite frequently in our patients, suggesting that these particular mutations could be the tip of the iceberg for a large, previously unidentified population of patients afflicted by FDXR-mediated diseases. Furthermore, given the fact that FDXR functions as both a mitochondrial protein and a protein involved in Fe–S cluster biogenesis, this study permitted us to compare and contrast the distinct yet overlapping phenotypes of these two major classes of disorders (mitochondrial disorders versus Fe–S cluster biogenesis disorders), which will be critical in the development of sound medical management related to these pathways. The combination of human subjects and mouse model allowed us to clearly elucidate these biochemical pathways and will help in the development of precise, targeted therapies for FDXR-related disorders, as well as Fe–S cluster disorders in general.
In conclusion, we identified a novel disease-causing gene FDXR associated with mitochondrial diseases. The biallelic FDXR mutations cause optic atrophy and neuropathy. Because of its upstream role in the Fe–S cluster biogenesis pathway, FDXR mutations can also affect many pathways. In addition, FDXR is required for the cholesterol side-chain cleavage to pregnenolone, a metabolic intermediate of many steroid hormones, raising the possibility that cholesterol pathway could be a potential therapeutic target for the treatment of this condition. Further characterization of the existing models will provide insights for the function of FDXR and the molecular pathology associated with disruption of this protein function. This knowledge will contribute to the existing framework for understanding mitochondriopathy and improve clinical intervention of these diseases.
Materials and Methods
Clinical information
Patients with optic atrophy, encephalopathy and/or peripheral neuropathy were recruited for clinical whole exome sequencing. They were evaluated by a clinical geneticist, and the clinical features are summarized in Table 1 and Supplementary Material, Note S1. Patients have diverse ethnic backgrounds (Hispanic, Asian, Iranian, and Caucasian ethnic background). For patients who underwent additional procedures, informed consent was obtained in accordance with the Human Subjects Committee, Cincinnati Children’s Hospital Medical Center (CCHMC).
Whole-exome sequencing
For patients 1, 2, and 5, whole exome sequencing was performed on the parent-proband trio at Ambry Genetics (Aliso Viejo, CA). Exome library preparation, sequencing, bioinformatics, and data analyses were performed as previously described (26). In brief, exome enrichment was performed using the IDT xGen Exome Research Panel V1.0 (Integrated DNA Technologies; Coralville, IA) and sequenced using paired-end, 100- or 150-cycle chemistry on the HiSeq or NextSeq system (Illumina; San Diego, CA). Data were annotated with the Ambry Variant Analyzer tool, and stepwise filtering included the removal of common alterations, intergenic and 3′/5′ untranslated region variants, intronic variants outside ±2, and synonymous variants unlikely to have splicing effects. Variants were then filtered based on family history and possible inheritance models.
For patients 4 and 6–9, whole-exome sequencing was performed at Baylor College of Medicine. The sequencing was performed as described by Yang and colleagues (27). A minimum of 20× coverage was required for variant calling. About 95% of single nucleotide variants and 88.2–95.0% of indels could be identified.
For patients 10–14, sequencing was performed on exon targets isolated by capture with the Clinical Research Exome kit (Agilent Technologies; Santa Clara, CA) at GeneDx. Genomic DNA was extracted from whole blood samples obtained from affected children and their parents. The supporting sequencing technology and variant interpretation protocol employed are described elsewhere (28).
Generation of stable FDXR overexpressing cell line
Fibroblasts derived from the proband in Family 1 and a control were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fisher Scientific; Grand Island, NY) supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. WT human FDXR cDNA was obtained from Addgene (pTRE-FDXR WT, transcript variant 3, NM_001258012.3). The full-length cDNA was cloned into the PLVX-IRES-tdTomato lentiviral expression vector (Clontech, Takara Bio Inc.; Shiga, Japan) with an In-Fusion Cloning kit (Clontech) as described previously (25). Briefly, the FDXR PCR fragment and 15-bp vector sequence were fused with linearized vector by In-Fusion Enzyme. The construct was packaged into viral particles in HEK293T cells and lentiviruses were concentrated by ultracentrifugation (CCHMC Vector Core). Transduction of the patient’s fibroblast cell line was performed according to established methods (25). Fibroblasts were seeded in 6-well plates; 24 h later, concentrated lentivirus (titered at 5.44 × 108) was added to the cells with 4 μg/ml polybrene. Transduced cells were sorted by flow cytometry with a fluorescein isocyanate (FITC) filter, and tdTomato-positive cells were selected for FDXR overexpression. Phase contrast and immunofluorescence was evaluated by conventional fluorescence microscopy (Zeiss Axiophot; Oberkochen, Germany).
Mouse strain
All procedures involving mice were approved by The Jackson Laboratory’s Institutional Animal Care and Use Committee and performed in accordance with the National Institutes of Health guidelines for the care and use of animals in research. The strain used for this study was B6; 129 S-Fdxrm1J Otop3m1J/GrsrJ (The Jackson Laboratory, Stock #026096). This strain was previously shown to exhibit an abnormal, recessive gait phenotype, and harbors a missense (autosomal recessive) mutation in the Fdxr gene resulting in a p.R389Q change in the Fdxr protein (18). Protein alignment also shows that this amino acid change is located at the same location as mutation R392W in the human ortholog. Although these mice also carry a linked homozygous variant of uncertain clinical significance in the 5’ untranslated region of Otop3, it is likely not contributing to the phenotype because mice with a targeted deletion of the Otop3 gene show increased startle reflex and oligodactyly (MGI: 5450617).
Enzymatic assay of ferredoxin NADP reductase
Ferredoxin NADP reductase activity was measured as described previously (29). Briefly, a reaction buffer containing 50 mM Tris-HCl (pH 7.8 at 25 °C), 200 µM NADPH, 75 µM cytochrome C-oxidized, and 10 µM ferredoxin was prepared and incubated at room temperature (RT) for 30 min. Cells were grown to stationary phase in DMEM supplemented with 10% fetal bovine serum, harvested, and lysed in standard RIPA lysis buffer. Similarly, mouse tissues (muscle, brain, liver, heart, and kidney) from 4-week-old mice were minced into 1–2-mm3 pieces in ice-cold isolation buffer [10 mM EDTA/0.05% trypsin in phosphate buffered saline (PBS)], incubated for 30 min on ice, and centrifuged at 300 ×g for 5 min. The harvested pellets were homogenized in ice-cold PBS containing 10 mM EDTA and protease inhibitor using a GentleMACS dissociator. The homogenized samples were filtered with 70 µm filters and centrifuged (4 °C, 5 min at 800 ×g). Finally, human cell or mouse tissue lysates were added to the reaction buffer, mixed, and spectrophotometric measurements were taken at A550nm. Relative enzyme activity was calculated after normalization relative to control samples.
Measurements of mitochondrial respiration
OCRs in fibroblast cell lines were measured with a XF-96 extracellular flux analyzer (Seahorse Biosciences; North Billerica, MA) (30,31). The XF96 analyzer creates a transient, 7 µl-chamber in specialized microplates that allows determination of oxygen and proton concentrations in real time. The cultured cells were transferred to an XF96 analyzer assay plate at a density of 1 × 104 cells/well and allowed to grow overnight. The measured parameters included basal respiration, ATP production, and maximal respiratory capacity. To allow comparison across experiments, OCR and acidification rate data are expressed in pmol/min and mpH/min, respectively, normalized to cell protein in individual wells determined by Bradford protein assays (Bio-Rad; Hercules, CA). OCRs were determined under normal conditions and after the addition of oligomycin (1.5 µM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) (0.5 µM), rotenone (1 µM), and antimycin A (1 µM).
ATP levels
ATP was measured using the ATP Bioluminescence Assay Kit CLS II, according to the manufacturer’s instructions (Roche Diagnostics; Basel, Switzerland) (32). Briefly, the assay buffer and substrate were equilibrated to RT, and the buffer was mixed with the substrate (assay reagent) to obtain a homogeneous solution. After a 30-min equilibration of the cell plates to RT, 100 µl of assay reagent was added to each well with 2 × 104 fibroblasts, and the content was mixed for 2 min on an orbital shaker to induce cell lysis. After a 10-min incubation at RT, the luminescence was read on a Synergy H1 microplate reader (BioTek Instruments; Winooski, VT) (33,34). ATP production in mouse tissues was measured similarly.
Mitochondrial respiratory chain complex I–IV activity assay
The enzymatic activities of complexes I, II, III, and IV were assayed as described previously (35,36). Complex I (NADH dehydrogenase) activity was determined by the rotenone-sensitive NADH oxidation at 340 nm, using the coenzyme Q analog 2, 3-dimethyl-5-methyl 6-n-decyl-1, 4-benzomethyluinone as an electron acceptor. The activity of complex II (succinate dehydrogenase) was analyzed by tracking the secondary reduction of 2, 6 dichlorophenolindophenol by ubiquinone-2 at 600 nm. Complex III (cytochrome bc1 complex) activity was determined by measuring the reduction of cytochrome C at 550 nm with reduced decylubiquinone. Complex IV (cytochrome c oxidase) activity was measured by monitoring the oxidation of reduced cytochrome C as a decrease in absorbance at 550 nm. Citrate synthase activity was analyzed by measuring the reduction of 5, 5′-dithiobis-2-nitrobenzoic acid at 412 nm in the presence of acetyl-CoA and oxaloacetate. Complex activities were normalized relative to citrate synthase activity.
Visual acuity
Analysis of visual acuity was performed by the Mouse Neurobehavioral Phenotyping Core at The Jackson Laboratory using the OptoMotry system (CerebralMechanics Inc.). Untrained, unrestrained mice were placed on an elevated platform in the center of an arena surrounded by a cylinder with a 3 D sine wave grid projection. The directionality of the subject’s head in response to variable frequency rotation of the cylinder was measured and an optokinetic (OKT) response score was calculated by the accompanying instrumentation software. A total of 41 mice (15 FdxrR389Q/R389Q, 15 Fdxr+/R389Q, 11 Fdxr+/+) were tested at 15–17 weeks of age. Technicians were blind to genotypes during testing. Graphing and statistical analyses were performed using Prism 7 (GraphPad Software; La Jolla, CA). Ordinary one-way ANOVA tests with a Dunnett’s post hoc test (to adjust for multiple comparisons) were used to compare each genotype (homozygous and heterozygous) to WT. No significant differences between wild type and heterozygous mice were found; therefore, unpaired, nonparametric t-tests (Mann Whitney U test) were ultimately used to demonstrate significance between WT and FdxrR389Q/R389Q. P values of less than 0.05 were considered significant [<0.05 (*), <0.01 (**) and <0.001 (***)].
Cell images
Fibroblasts were seeded in 6-well plates; 24 h later, concentrated lentivirus (titered at 2.44 × 108) was added together with 4 μg/ml polybrene. Images were captured by conventional fluorescence microscopy after transduction of PLVX-FDXR-tdTomato lenti-virus for 48 h. Transduced cells were sorted by flow cytometry with a FITC filter, and tdTomato-positive cells were selected for FDXR overexpression. Patient fibroblasts were sorted by FACS after 24 h in culture.
Confocal microscopy of immunofluorescence
Fibroblasts were cultured in DMEM containing 200 mM MitoTracker Red (Molecular Probes, Thermofisher Scientific; Eugene, OR) for 30 min at 37 °C. After washing with PBS, fibroblasts were fixed in 4% paraformaldehyde in PBS for 20 min. They were incubated in 0.3% Triton X-100 for 30 min, and treated with 50 mM NH4Cl in PBS for 15 min, followed by a PBS wash. The sections were blocked for 1 h at room temperature (RT) in blocking buffer (10% goat serum and 0.4% Triton X-100 in PBS) and then incubated with rabbit anti-Fdxr antibody (1: 100) overnight at 4 °C. After three 10-min washes in PBS, goat anti-rabbit secondary antibody conjugated with Alexa Fluor® 488 (1: 1500) was applied to the sections for 2 h at RT. Sections were mounted with VECTASHIELD medium containing DAPI (Vector H1200, Vector Laboratories, Burlingame, CA). Cell images were obtained via confocal microscopy using a Nikon A1R LUN-V Inverted confocal microscope (Nikon Instruments Inc.; Melville, NY) and analyzed with Imaris software (Bitplane; South Windsor, CT).
Detection of fluorescent ROS and superoxide production
We measured ROS generation levels in fibroblasts with the Cellular ROS/Superoxide Detection Assay Kit (ab139476, Abcam; Cambridge, UK). Seeded cells were placed directly onto glass slides or polystyrene tissue culture plates to ensure 50–70% confluency. After a media change, cells were treated by 30 min with the ROS inhibitor N-acetyl-L-cysteine. Pyocyanin was used to induce ROS production; controls included vehicle alone (no inhibitor and no inducer of ROS) and vehicle with inhibitor. ROS production was observed with oxidative stress detection reagent (green) and superoxide detection reagent (orange) for 30 min to 1 h. Afterwards, cells were washed twice with 1× wash buffer. Cell images were captured with a fluorescence microscope using standard excitation/emission filter sets (green 490/525 nm and orange 550/620 nm).
H&E staining and electron microscopy
Eyes were fixed in 4% paraformaldehyde for 24 h at RT and then embedded in paraffin. Sagittal sections of the eye (6–8 μm) were cut through the optic nerve head and then stained with H&E. Images were obtained under light microscopy (BX63; Olympus Corporation; Center Valley, PA). Central (250 µm from the edge of the optic disc) and peripheral (100 µm from the ora serrata) segments of the retina were photographed. Nuclei in the retinal ganglion cell layer (excluding the small and flat nuclei of the endothelial cells) were counted on the photographed images. Length of the retina was measured using an ImageJ program.
For electron microscopy, mice were anesthetized and perfused with 0.9% NaCl and EM fixative (4% paraformaldehyde, 2.5% glutaraldehyde, PBS, pH 7.4–7.6). Tissues were postfixed in EM fixative buffer, washed in 0.1 M sodium cacodylate buffer (EMS; Hatfield, PA), and postfixed in 1% osmium tetroxide (EMS) for 1 h at 4 °C. Samples were washed in 0.1 M sodium cacodylate buffer and dehydrated through a graded ethanol series. Samples were embedded in LX-112 (Ladd Research Industries; Williston, VT). Tissue blocks were sectioned (thickness, 0.5–1 μm) and stained with toluidine blue for light microscopy. Blocks were trimmed and cut into 90-nm-thick sections with an ultramicrotome (Leica EM UC7; Buffalo Grove, IL). The ultra-thin sections were counterstained with uranyl acetate 2% (EMS) and lead citrate. All images were taken with an 80-kV transmission electron microscope (Hitachi, H-7650, V01.07; Tokyo, Japan). Measurement of the thickness of myelin membrane in RGC axons was performed using ImageJ software (National Institutes of Health).
Western blotting
Cell lysates from mouse tissues were harvested as described above. Approximately, 50 μg of protein was mixed with 2× reducing sample buffer (Sigma-Aldrich; St. Louis, MO). The samples were heated to 95 °C for 5 min before being separated on a 10% SDS-PAGE gel (21). The gel was electrophoretically transferred onto PVDF membranes (Invitrogen, Thermo Fisher Scientific; Carlsbad, CA) at 80 V for 2 h. The membrane was blocked for 1 h in 10% milk prior to overnight incubation in primary antibody FDXR (Abcam) at 1: 100 dilution at 4 °C. This overnight incubation was then followed by a 1-h incubation at 37 °C with horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich). ECL plus detection reagent (GE Healthcare; Chicago, IL) was used for visualization of bands, and quantification analysis of band intensity was performed in ImageJ software (version 1.45 s). GAPDH at a 1: 1000 dilution (Ambion, Thermo Fisher Scientific; Foster City, CA) was used as a loading control.
3D protein structure
The structure of FDXR was built from the homologous structure of FDXR for Bos taurus complexed with FAD (PDB: 1CJC) (22) and complexed with FDX1 and FDXR (PDB: 1E6E) (37). The homology model of FDXR was built with the ROSETTA homology modeling module (38) with sequence alignments generated in TCOFFEE (39,40). The energy calculations for internal structure destabilization and protein-protein binding were done in the FoldX program (41). Potential phosphorylation and modification sites in FDXR were identified in Eukaryotic Linear Motif (42) to identify potential motifs, and checked in the experimental database of Phosphosite (43). The images were generated in the PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, LLC; New York, NY).
Supplementary Material
Acknowledgements
The authors thank the patients and their families for participating in our study. The authors also thank Geogianne M. Ciraolo and Tilat Rizvi (Cincinnati Children’s Hospital) for their help with electron microscopy and Jesse Slone for critical reading this manuscript.
Conflict of Interest statement. The authors declare no conflict of interest statement. D.N.S., A.C., K.L.H and S.T. are full time employees of Ambry Genetics; M.T.C, K.G.M, R.W., F.M., J.K.R., for GeneDx. Exome Sequencing is a commercially available test. The Department of Molecular & Human Genetics at Baylor College of Medicine derives revenue from clinical genetic testing performed at Baylor Genetics.
Funding
Cincinnati Children’s Hospital Research Foundation, National Eye Institute (1R01EY026609–01 awarded to TH), Jackson Laboratory, and NIH Office of Research Infrastructure Programs (R24 OD021325–01) awarded to L.G.R. Funding to pay the Open Access publication charges for this article was provided by Ambry Genetics Inc.
Supplementary Material
Supplementary Material is available at HMG online.
References
- 1. Beilschmidt L.K., Puccio H.M. (2014) Mammalian Fe-S cluster biogenesis and its implication in disease. Biochimie, 100, 48–60. [DOI] [PubMed] [Google Scholar]
- 2. Johnson D.C., Dean D.R., Smith A.D., Johnson M.K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem., 74, 247–281. [DOI] [PubMed] [Google Scholar]
- 3. Sheftel A.D., Stehling O., Pierik A.J., Elsasser H.P., Muhlenhoff U., Webert H., Hobler A., Hannemann F., Bernhardt R., Lill R. (2010) Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl Acad. Sci. USA, 107, 11775–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cai K., Tonelli M., Frederick R.O., Markley J.L. (2017) Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry, 56, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanukoglu I., Jefcoate C.R. (1980) Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin. J. Biol. Chem., 255, 3057–3061. [DOI] [PubMed] [Google Scholar]
- 6. Hanukoglu I. (1992) Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J. Steroid Biochem. Mol. Biol., 43, 779–804. [DOI] [PubMed] [Google Scholar]
- 7. Hwang P.M., Bunz F., Yu J., Rago C., Chan T.A., Murphy M.P., Kelso G.F., Smith R.A., Kinzler K.W., Vogelstein B. (2001) Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat. Med., 7, 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi Y., Ghosh M., Kovtunovych G., Crooks D.R., Rouault T.A. (2012) Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta, 1823, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spiegel R., Saada A., Halvardson J., Soiferman D., Shaag A., Edvardson S., Horovitz Y., Khayat M., Shalev S.A., Feuk L.. et al. (2014) Deleterious mutation in FDX1L gene is associated with a novel mitochondrial muscle myopathy. Eur. J. Hum. Genet., 22, 902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coghlan V.M., Vickery L.E. (1991) Site-specific mutations in human ferredoxin that affect binding to ferredoxin reductase and cytochrome P450scc. J. Biol. Chem., 266, 18606–18612. [PubMed] [Google Scholar]
- 11. Ziegler G.A., Vonrhein C., Hanukoglu I., Schulz G.E. (1999) The structure of adrenodoxin reductase of mitochondrial P450 systems: electron transfer for steroid biosynthesis. J. Mol. Biol., 289, 981–990. [DOI] [PubMed] [Google Scholar]
- 12. Novotna J., Jandova A., Janouskova M., Laurova L., Skoda V. (1976) [Levels of DNA and RNA in the carcinomatous tissue of the collum uteri before and after application of Cytembena (author's transl)]. Cesk. Gynekol., 41, 733–734. [PubMed] [Google Scholar]
- 13. Bhaduri A., Ungewickell A., Boxer L.D., Lopez-Pajares V., Zarnegar B.J., Khavari P.A. (2015) Network analysis identifies mitochondrial regulation of epidermal differentiation by MPZL3 and FDXR. Dev. Cell, 35, 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solish S.B., Picado-Leonard J., Morel Y., Kuhn R.W., Mohandas T.K., Hanukoglu I., Miller W.L. (1988) Human adrenodoxin reductase: two mRNAs encoded by a single gene on chromosome 17cen—-q25 are expressed in steroidogenic tissues. Proc. Natl Acad. Sci. USA, 85, 7104–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Land T., Rouault T.A. (1998) Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell, 2, 807–815. [DOI] [PubMed] [Google Scholar]
- 16. Shi Y., Ghosh M.C., Tong W.H., Rouault T.A. (2009) Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet., 18, 3014–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu G., Chen X. (2002) The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene, 21, 7195–7204. [DOI] [PubMed] [Google Scholar]
- 18. Fairfield H., Srivastava A., Ananda G., Liu R., Kircher M., Lakshminarayana A., Harris B.S., Karst S.Y., Dionne L.A., Kane C.C.. et al. (2015) Exome sequencing reveals pathogenic mutations in 91 strains of mice with Mendelian disorders. Genome Res., 25, 948–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu J., Bitoun J.P., Tan G., Wang W., Min W., Ding H. (2010) Iron-binding activity of human iron-sulfur cluster assembly protein hIscA1. Biochem. J., 428, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gakh O., Ranatunga W., Smith DYt., Ahlgren E.C., Al-Karadaghi S., Thompson J.R., Isaya G. (2016) Architecture of the human mitochondrial iron-sulfur cluster assembly machinery. J. Biol. Chem., 291, 21296–21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Bitoun J.P., Ding H. (2006) Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J. Biol. Chem., 281, 27956–27963. [DOI] [PubMed] [Google Scholar]
- 22. Webert H., Freibert S.A., Gallo A., Heidenreich T., Linne U., Amlacher S., Hurt E., Muhlenhoff U., Banci L., Lill R. (2014) Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun., 5, 5013.. [DOI] [PubMed] [Google Scholar]
- 23. Vickery L.E. (1997) Molecular recognition and electron transfer in mitochondrial steroid hydroxylase systems. Steroids, 62, 124–127. [DOI] [PubMed] [Google Scholar]
- 24. Griffin A., Parajes S., Weger M., Zaucker A., Taylor A.E., O'Neil D.M., Muller F., Krone N. (2016) Ferredoxin 1b (Fdx1b) is the essential mitochondrial redox partner for cortisol biosynthesis in zebrafish. Endocrinology, 157, 1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simon M., Richard E.M., Wang X., Shahzad M., Huang V.H., Qaiser T.A., Potluri P., Mahl S.E., Davila A., Nazli S.. et al. (2015) Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet., 11, e1005097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C.. et al. (2015) Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med., 17, 578–586. [DOI] [PubMed] [Google Scholar]
- 27. Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C.. et al. (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M.. et al. (2015) Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet., 97, 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zanetti G., Curti B. (1981) Interactions between ferredoxin-nadp+ reductase and ferredoxin at different reduction levels of the two proteins. FEBS Lett., 129, 201–204. [DOI] [PubMed] [Google Scholar]
- 30. Kang E., Wang X., Tippner-Hedges R., Ma H., Folmes C.D., Gutierrez N.M., Lee Y., Van Dyken C., Ahmed R., Li Y.. et al. (2016) Age-related accumulation of somatic mitochondrial DNA mutations in adult-derived human iPSCs. Cell Stem Cell, 18, 625–636. [DOI] [PubMed] [Google Scholar]
- 31. Kang E., Wu J., Gutierrez N.M., Koski A., Tippner-Hedges R., Agaronyan K., Platero-Luengo A., Martinez-Redondo P., Ma H., Lee Y.. et al. (2016) Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature, 540, 270–275. [DOI] [PubMed] [Google Scholar]
- 32. Tajiri N., Borlongan C.V., Kaneko Y. (2016) Cyclosporine A treatment abrogates ischemia-induced neuronal cell death by preserving mitochondrial integrity through upregulation of the Parkinson's disease-associated protein DJ-1. CNS Neurosci. Ther., 22, 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gong S., Peng Y., Jiang P., Wang M., Fan M., Wang X., Zhou H., Li H., Yan Q., Huang T.. et al. (2014) A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res., 42, 8039–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M.. et al. (2016) The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet., 25, 584–596. [DOI] [PubMed] [Google Scholar]
- 35. Leslie N., Wang X., Peng Y., Valencia C.A., Khuchua Z., Hata J., Witte D., Huang T., Bove K.E. (2016) Neonatal multiorgan failure due to ACAD9 mutation and complex I deficiency with mitochondrial hyperplasia in liver, cardiac myocytes, skeletal muscle, and renal tubules. Hum. Pathol., 49, 27–32. [DOI] [PubMed] [Google Scholar]
- 36. Tang S., Le P.K., Tse S., Wallace D.C., Huang T., Nollen E.A.A. (2009) Heterozygous mutation of Opa1 in Drosophila shortens lifespan mediated through increased reactive oxygen species production. PLoS One, 4, e4492.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muller J.J., Lapko A., Bourenkov G., Ruckpaul K., Heinemann U. (2001) Adrenodoxin reductase-adrenodoxin complex structure suggests electron transfer path in steroid biosynthesis. J. Biol. Chem., 276, 2786–2789. [DOI] [PubMed] [Google Scholar]
- 38. Kim D.E., Chivian D., Baker D. (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res., 32, W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Erb I., Notredame C. (2016) How should we measure proportionality on relative gene expression data? Theory Biosci., 135, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Catuara-Solarz S., Espinosa-Carrasco J., Erb I., Langohr K., Gonzalez J.R., Notredame C., Dierssen M. (2016) Combined treatment with environmental enrichment and (-)-epigallocatechin-3-gallate ameliorates learning deficits and hippocampal alterations in a mouse model of Down syndrome. eNeuro, 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schymkowitz J.W., Rousseau F., Martins I.C., Ferkinghoff-Borg J., Stricher F., Serrano L. (2005) Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl Acad. Sci. USA, 102, 10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dinkel H., Van Roey K., Michael S., Kumar M., Uyar B., Altenberg B., Milchevskaya V., Schneider M., Kühn H., Behrendt A.. et al. (2016) ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res., 44, D294–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J., Ning Y., Hedley W., Saunders B., Chen Y., Tindill N., Hannay T., Subramaniam S. (2002) The molecule pages database. Nature, 420, 716–717. [DOI] [PubMed] [Google Scholar]
- 44. Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y.. et al. (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics, 31, 3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prusky G.T., Alam N.M., Beekman S., Douglas R.M. (2004) Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest. Ophthalmol. Vis. Sci., 45, 4611–4616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.