Abstract
The recent generation of induced pluripotent stem cells (iPSCs) from a patient with Parkinson’s disease (PD) resulting from triplication of the α-synuclein (SNCA) gene locus allows unprecedented opportunities to explore its contribution to the molecular pathogenesis of PD. We used the double-nicking CRISPR/Cas9 system to conduct site-specific mutagenesis of SNCA in these cells, generating an isogenic iPSC line with normalized SNCA gene dosage. Comparative gene expression analysis of neuronal derivatives from these iPSCs revealed an ER stress phenotype, marked by induction of the IRE1α/XBP1 axis of the unfolded protein response (UPR) and culminating in terminal UPR activation. Neuropathological analysis of post-mortem brain tissue demonstrated that pIRE1α is expressed in PD brains within neurons containing elevated levels of α-synuclein or Lewy bodies. Having used this pair of isogenic iPSCs to define this phenotype, these cells can be further applied in UPR-targeted drug discovery towards the development of disease-modifying therapeutics.
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disease, which manifests primarily as a devastating movement disorder. α-synuclein, encoded by the SNCA gene, is one of the most commonly implicated molecular mediators of both genetic and sporadic PD. Multiplications of the SNCA gene locus, including duplications and triplications, have been shown to cause autosomal dominant PD in which gene dosage determines severity and latency (1–4). This suggests that 1) the pathologic properties of α-synuclein are not dependent on mutations that alter the α-synuclein protein product, 2) overexpression of wild-type α-synuclein is sufficient to cause disease in a dose-dependent manner and 3) overabundance of α-synuclein may be a common feature of PD wherein smaller increases in α-synuclein, due to increased expression or decreased clearance, may contribute to sporadic disease (5,6). Defining the mechanism by which overabundance of α-synuclein contributes to PD pathogenesis may thus provide insight into the molecular mechanisms underlying PD and provide novel targets for therapeutic intervention.
The recent generation of induced pluripotent stem cells (iPSCs) from a PD patient harboring a triplication of the SNCA gene locus has facilitated efforts to study the role of α-synuclein in PD pathogenesis (7). Whereas one allele maintains a single copy of SNCA, a triplication on the other allele results in a doubling of the total SNCA gene dosage (Fig. 1A). This model has been utilized in studies comprising healthy controls for phenotypic comparison to identify α-synuclein-related pathogenic changes (7–12). However, the triplicated locus is a large, 1.5 Mb region that includes 15 other putative or annotated genes (7). Thus, inferences drawn by comparison of these cells and their derivatives to healthy controls may be confounded by altered expression of any or all of the other genes in this region.
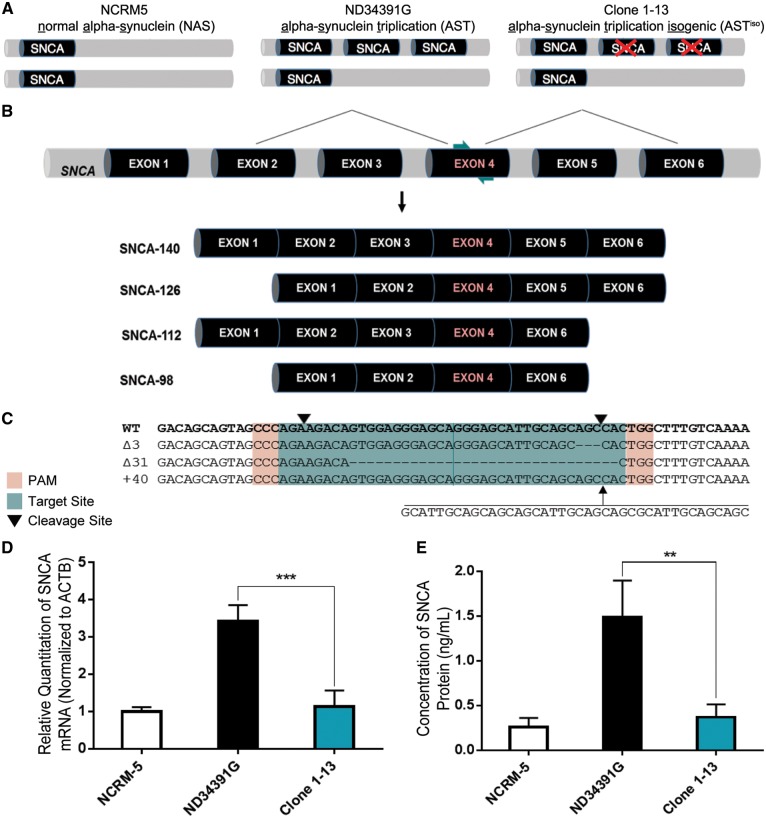
Figure 1.
Double Knockout of SNCA in iPSCs Derived from a PD Patient with SNCA Triplication via Double-Nicking CRISPR/Cas9. (A) Schematic representation of normal alpha synuclein (NAS), alpha-synuclein triplication (AST) and alpha-synuclein triplication isogenic (ASTiso) iPS lines. The triplication involves a large Mb region including the SNCA gene. (B) Position of sgRNAs (teal arrows) guiding Cas9 nickases to SNCA exon 4, common to all four α-synuclein transcript isoforms. (C) Wild-type, 3 bp deletion, 31 bp deletion, and 40 bp insertion alleles identified by sequencing of Clone 1-13. PAMs are highlighted in red, sgRNAs in teal and downward pointing black arrows represent cleavage sites. (D) qRT-PCR for total SNCA mRNA shows a normalization of α-synuclein transcript levels in Clone 1-13 iPSCs. Data are represented as mean ± SEM of biological triplicates. (E) ELISA shows a normalization of α-synuclein protein levels in Clone 1-13 iPSCs. Data are represented as mean ± SEM of biological triplicates. **P ≤ 0.01, ***P ≤ 0.001. See also Supplementary Material, Figs S1 and S2 and Table S4.
To overcome this limitation, we applied the double-nicking clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system (13) to generate an isogenic iPSC line by introducing frameshifting mutations in two copies of SNCA in these cells. We then conducted a transcriptomic analysis of neurons derived from these iPSCs compared to the parental SNCA triplication line and identified an α-synuclein-induced ER stress phenotype, marked by activation of the IRE1α/XBP1 axis of the unfolded protein response (UPR), which was further confirmed by qRT-PCR and western blotting analysis. We further demonstrate that similar alterations in pIRE1α levels occur in post-mortem brain tissue from PD patients compared to controls, validating the clinical relevance of this cellular phenotype to the manifestation of PD pathology in patients. Taken together, these results highlight the IRE1α/XBP1 arm of the UPR as a mediator of α-synuclein-induced neuropathogenesis in PD, supporting further evaluation of this pathway as a candidate for therapeutic intervention. More generally, our results support the use of isogenic iPSC models as a platform to identify molecular contributors to PD pathology.
Results
Generation of isogenic SNCA triplication iPSCs
Cas9 nickase sgRNAs were designed to target SNCA exon 4, which is expressed in all four α-synuclein transcript isoforms (Fig. 1B), and tested for efficacy. Insertion/deletion (indel) mutations generated through imperfect non-homologous end joining (NHEJ) were detectable by SURVEYOR assay in HEK293T cells, as well as the neuroblastoma cell lines BE(2)-M17 and SH-SY5Y (Supplementary Material, Fig. S1A). Sequencing showed that the mutagenesis efficiency was 47% in HEK293T, 29% in BE(2)-M17 and 21% in SH-SY5Y cells (Supplementary Material, Fig. S1B). To confirm that these mutations were sufficient to alter SNCA gene expression, we analyzed total α-synuclein mRNA (Supplementary Material, Fig. S1C) and protein levels (Supplementary Material, Fig. S1D) by qRT-PCR and ELISA, respectively, and detected a reduction in α-synuclein levels in pooled, targeted populations of each cell type.
ND34391G iPSCs (obtained from the Coriell Institute, distributed through the NINDS Fibroblasts and iPSCs Collection), were derived from a PD patient carrying a triplication of a large 1.5 Mb region including SNCA and 10 additional genes (Fig. 1A), validated by copy number variant (CNV) quantification (14). After clonal expansion of Cas9 nickase-transfected iPSCs, four clones were identified as variants by high resolution melt (HRM) analysis. Sequencing (Fig. 1C) confirmed one clone as a double knockout, containing an out-of-frame 31 bp deletion and 40 bp insertion. It is worth noting that this clone also contains an in-frame 3 bp deletion, which is not predicted to affect gene expression from this copy of SNCA. We sequenced six putative off-target sites, and none of these sites were found to contain indels (Supplementary Material, Fig. S2A). The double knockout clone, Clone 1-13, was quality controlled by pluripotency immunostaining, karyotyping and hPSC ScoreCard analysis (Supplementary Material, Fig. S2B–D). Finally, we examined the expression of α-synuclein in NCRM-5 healthy control (normal α-synuclein = NAS), ND34391G α-synuclein triplication (AST) and Clone 1-13 isogenic (ASTiso) iPSCs (Fig. 1A), finding that α-synuclein mRNA (Fig. 1D) and protein (Fig. 1E) levels in the isogenic iPSC line were reduced to levels comparable to the healthy control iPSC line, with silencing of mRNA expression from the out-of-frame mutant alleles likely caused by nonsense mediated decay. In this context, isogenic refers to the fact that the parental SNCA triplication iPSC line and the double knockout clone possess identical genetic background, which includes overexpression of the other genes in the triplication locus, and selective reduction of SNCA gene dosage to achieve normal expression levels.
Characterisation of AST and ASTiso iPSC-derived neurons
NAS, AST and ASTiso iPSCs were differentiated into cortical neurons (14) (Supplementary Material, Fig. S3A) and consistent with previous reports (10), we observed phase-dense structures in the perinuclear compartment of the AST cells that were absent in the NAS neurons (Supplementary Material, Fig. S3B). These structures stained with an anti-SNCA antibody (Supplementary Material, Fig. S3B), further demonstrating that they contain α-synuclein protein, and that they may represent early stage Lewy-body like structures in these neurons.
Analysis of AST and ASTiso neurons by RT-PCR showed that they expressed comparable levels of neuronal marker genes (Supplementary Material, Fig. S3C) and showed similar morphology (Supplementary Material, Fig. S3D). A thorough transcriptomic analysis of the AST and ASTiso neurons and iPSCs was performed by RNA-seq. Analysis of expression of a number of markers of cell fate, including those enriched in neurons, neural progenitor cells, astrocytes, oligodendrocytes, microglia and pluripotent stem cells revealed that although there was a mixed population of cell types produced upon differentiation, these populations were highly similar between AST and ASTiso genotypes (Supplementary Material, Fig. S3E). This demonstrates that the triplication of the SNCA locus has relatively minor effects on the neuronal differentiation process, and results in highly similar cell populations to neurons derived from the ASTiso genotype that can be directly compared with each other.
Comparison of AST and ASTiso genotypes confirmed an approximately 90-fold overexpression of SNCA mRNA in AST neurons that was restored in ASTiso neurons (Supplementary Material, Fig. S4A). Further analysis of the other genes in the triplication showed that they show relatively subtle changes in expression, with the exception of SNCA-AS1 and CCSER1, which are upregulated in the AST neurons versus ASTiso neurons (Supplementary Material, Fig. S4C). One gene present in the triplicated region, MMRN1, was unable to be analysed due to very low expression levels. It is likely that the change in expression of the SNCA antisense transcript (SNCA-AS1) is due to a transcriptional feedback mechanism resulting from the SNCA mutations. The effect on CCSER1 expression is more unexpected, but it could be a potential downstream consequence of altered SNCA expression, since this gene was not mutated in the ASTiso line. These data suggest that the differences seen between AST and ASTiso genotypes are most likely due to the reduction in the level of SNCA mRNA after CRISPR/Cas9 mutagenesis rather than effects from other genes contained within the triplicated region.
Comparative transcriptomic analysis reveals an ER stress phenotype in SNCA triplication iPSC-derived neurons
We next performed differential expression analysis of our RNAseq data between different cell types and genotypes using DEseq2 (15). Cross correlations between the datasets (Fig. 2A) and principal component analysis (Fig. 2B) showed that the largest component of the variance (PC1, 74.38%) was due to differentiation stage. Differences between AST and ASTiso genotypes in neurons formed the next largest source of variation (PC2, 9.51%), followed by differences between these genotypes in iPSCs (PC3, 2.91%) (Fig. 2A and B). We applied a generalized linear model to test the effect of differentiation stage (iPSC vs. neuron), genotype (AST vs ASTiso) and interaction differentiation x genotype on gene expression. This identified 5368 genes that were differentially expressed between genotypes (Q < 0.05) (Supplementary Material, Fig. S4B). The number of differences in gene expression resulting from SNCA triplication are greater in neurons than iPSCs, and the genes responsible for these differences are not the same. The fact that iPSCs and neurons do not respond to SNCA triplication in the same way may provide a rationale as to why disease phenotypes manifest specifically in neurons, and indicates that the neuron-specific differentially expressed gene set may contain promising candidates for modulating such phenotypes.
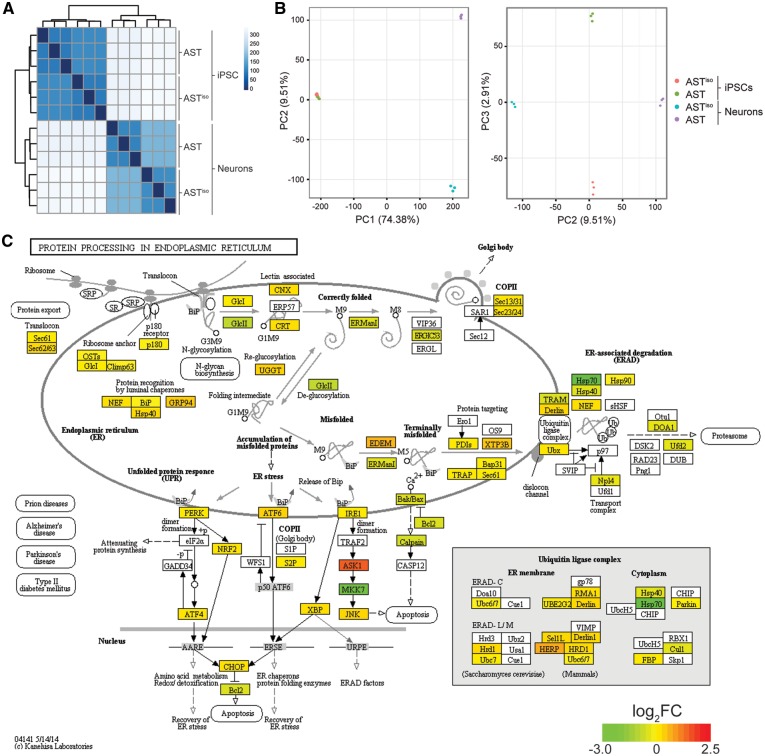
Figure 2.
Comparative transcriptomic analysis reveals an ER stress phenotype specific to SNCA triplication iPSC-derived neurons. (A) Cross-correlation and clustering analysis between datasets. Color denotes correlation coefficient. The primary source of variation is between iPSCs and iPSC-derived neurons, followed by AST and ASTiso genotypes in neurons. (B) Principal component analysis of differentially expressed genes. The first principal component (PC1) accounts for 74.38% of the variance, largely due to differentiation stage. PC2 accounts for the differences in AST and ASTiso genotypes in neurons (9.51% variance) and PC3 similarly accounts for genotype differences in iPSCs (2.91% variance). Cell types are indicated as follows: red = ASTiso iPSC, green = AST iPSC, blue = ASTiso neurons and purple = AST neurons. (C) Protein processing in the endoplasmic reticulum KEGG pathway diagram highlighting fold changes in gene expression between AST and ASTiso iPSC-derived neurons. Color indicates log2 gene expression change (red is higher in AST neurons). Data show biological triplicates, **Q < 0.01, ***Q < 0.01, ns = not significant. See also Supplementary Material, Figs S3–S5, Tables S1 and S2.
An unbiased Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on the differentially expressed genes in each of these sets identified “protein processing at the ER” as the pathway that was most highly enriched between AST and ASTiso genotypes (Fig. 2C). Interestingly, genes differentially expressed only between AST and ASTiso iPSC-derived neurons showed the strongest enrichment in this pathway, with a FDR corrected p-value of 5.99 × 10−8. Out of 140 total genes, 92 (66%) of the genes in this pathway were significantly differentially expressed (Fig. 2C), including components involved in the detection of unfolded proteins, and the response mediated through the UPR and ER-associated protein degradation (ERAD) pathways. The three branches of the UPR were all upregulated (Fig. 2C), with increased levels of PERK, ATF6 and IRE1α mRNA, and corresponding changes in their upstream activators and downstream effectors (Fig. 2C). The induction of the UPR by SNCA overexpression is likely to be a direct effect, since treatment of cultured human SH-SY5Y neurons with recombinant α-synuclein oligomers (although interestingly not fibres or monomers) can induce the UPR (16). It is also consistent with previous data that show 1) that UPR activation co-localises in neurons showing increased α-synuclein expression in human post-mortem brain samples from patients with PD (17), 2) that α-synuclein directly interacts with the ER chaperone BiP in cultured cells (18) and 3) that these proteins co-localise in mouse brains overexpressing truncated α-synuclein (18). Other KEGG pathways enriched in the differentially expressed gene set are listed in Supplementary Material, Table S1, and include spliceosomal (Q = 7.63 × 10−7) and lysosomal (Q = 1.96 × 10−3) components.
Further analysis of the genes that are differentially expressed between AST and ASTiso genotypes in iPS-derived neurons for enrichment of particular gene ontology (GO) categories highlighted numerous other biological processes that were affected by SNCA triplication (Supplementary Material, Table S2). These largely fall into three categories, namely cell death through both autophagy and apoptosis (e.g. autophagy Q = 1.90×10−8, apoptotic process Q = 1.69×10−5), alterations in cellular metabolism especially catabolic processes (e.g. cellular protein catabolic process Q = 1.61×10−5) and changes in gene expression particularly with respect to cell fate regulation (e.g. cell fate commitment Q = 8.64×10−9) (Supplementary Material, Table S2). These processes are likely to be downstream effects of α-synuclein aggregation, and have been implicated in Parkinson’s disease pathogenesis (19–21). Taken together with the ER stress response, they may form the basis for a diagnostic cellular phenotype underlying the early stages of disease progression.
We also attempted to overexpress SNCA in NAS neurons by the delivery of dCas9-VPR (14) and a sgRNA targeting the promoter of the SNCA gene to see whether we could recapitulate some of these effects in an orthologous system. We obtained an approximately 8-fold overexpression of SNCA at the RNA level in these neurons (Supplementary Material, Fig. S5A). Interestingly, we saw a corresponding 2-fold overexpression of IRE1α, suggesting that the UPR comprises a rapid, immediate response to SNCA overexpression, and further demonstrating that this effect is due to increased levels of α-synuclein rather than any other effects of the triplicated region (Supplementary Material, Fig. S5B). However, in contrast to the AST neurons, no effect on the level of PERK or ATF6 was observed. The greater specificity and lower magnitude of these effects is not unexpected, since these cells have only an 8-fold increase in SNCA RNA levels (Supplementary Material, Fig. S5A) as opposed to a nearly 90-fold change in AST neurons (Supplementary Material, Fig. S4A). Additionally, they have only experienced a short, transient exposure to increased SNCA expression, and therefore would only show the direct, immediate responses, not the indirect effects of chronic exposure to high levels of α-synuclein for longer periods of time.
UPR targets are activated in neural derivatives of SNCA triplication iPSCs
To further elucidate the role of the ER stress and unfolded protein response (UPR) pathways in the neuronal response to SNCA triplication, we examined expression of UPR pathway components in our RNAseq data, by qRT-PCR and western blotting (Fig. 3, Supplementary Material, Fig. S6). We focused on the IRE1α branch of the UPR (Fig. 3A), since this was most sensitive to the upregulation of SNCA levels in AST neurons and also upon transient upregulation of SNCA in NAS neurons (Supplementary Material, Fig. S5). IRE1α is a UPR master regulator which is itself activated by autophosphorylation, becoming pIRE1α, when its native suppressor, BiP, dissociates from its ER luminal domain to bind to unfolded proteins for which it has a greater affinity. Activated pIRE1α mediates the non-canonical splicing of a 26 nt intron from XBP1 mRNA, producing the functional XBP1(S) mRNA transcript that is capable of being translated into a transcription factor. XBP1(S) translocates to the nucleus to regulate UPR target genes in response to accumulated unfolded proteins (Fig. 3A). We found that IRE1α mRNA levels were upregulated 10-fold in AST neurons compared to NAS neurons, and partially rescued by reduction of α-synuclein levels in ASTiso neurons (Fig. 3B).
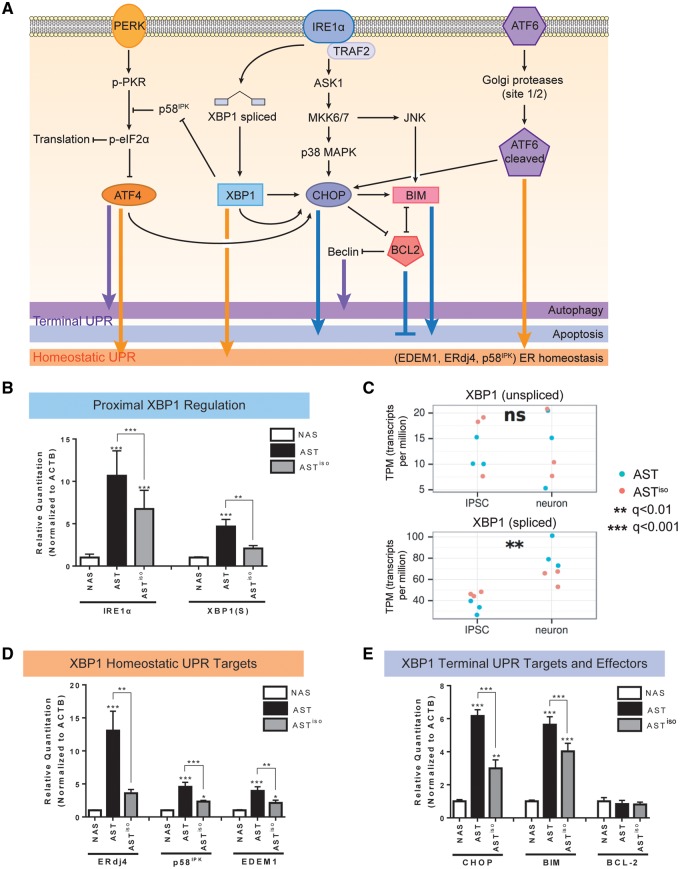
Figure 3.
Transcript and protein expression validation of IRE1α/XBP1-induced UPR activation in SNCA triplication iPSC-derived neurons. (A) Schematic of signaling pathways downstream of the UPR. PERK, IRE1α and ATF6 branches are indicated, and the effects of downstream effectors on homeostatic, autophagic and apoptotic outcomes are indicated. (B, D, E) Validation of mRNA expression level differences in IRE1α/XBP1 pathway components by qRT-PCR. Levels of IRE1α and spliced XBP1 [XBP1(S)] (B), the homeostatic UPR targets ERdj4, p58IPK and EDEM1 (D), and the terminal UPR targets CHOP, BIM and BCL-2 (E) at the mRNA level were evaluated by qRT-PCR in NAS, AST and ASTiso iPSC-derived neurons. Data are represented as mean ± SEM of biological triplicates. (C) Gene expression from RNAseq data (in transcripts per million, TPM) of unspliced and spliced XBP1 transcripts across differentiation stage (iPSC or neuron) and genotype (blue = AST, red = ASTiso). *P ≤ 0.5, ** P ≤ 0.01, *** P ≤ 0.001, ns = not significant. See also Supplementary Material, Figs S5–S7 and Table S4.
Interestingly, expression of the spliced XBP1(S) isoform, but not the unspliced form, was found to be altered by SNCA triplication by both isoform-specific qRT-PCR (Fig. 3B) and RNAseq (Fig. 3C). This is consistent with the known role of splicing in regulation of signaling through XBP1 and resultant activation of the UPR. We further validated the expression of downstream XBP1(S) targets that are involved in both homeostatic and terminal UPR outputs, ERdj4, EDEM1, p58IPK and CHOP by qRT-PCR in AST neurons. In all cases, expression was elevated when compared to NAS neurons (Fig. 3D and E). For ERdj4, EDEM1, p58IPK and CHOP, expression was partially rescued by reduction of α-synuclein levels in ASTiso neurons. These results were confirmed by analysis of our RNAseq data as being upregulated in AST neurons when compared to the isogenic control (Supplementary Material, Fig. S6A). As CHOP is regarded as the major terminal UPR effector, indicating a commitment by the cell to undergo death, we explored whether the apoptotic targets of CHOP, BIM and BCL-2, were differentially expressed between NAS, AST and ASTiso neurons (Fig. 3E). Pro-apoptotic BIM was upregulated in AST neurons compared to NAS neurons, and partially rescued in ASTiso neurons (Fig. 3E). We did not observe significant differences in anti-apoptotic BCL-2 expression at the mRNA level (Fig. 3E). However, BCL-2 protein levels were substantially reduced in AST neurons compared to NAS neurons (Supplementary Material, Fig. S6B) and a small restoration of BCL-2 protein levels was observed in ASTiso neurons (Supplementary Material, Fig. S6B). Changes in Beclin-1 protein levels, a mediator of autophagy, were also observed, and were highest in NAS neurons, reduced in AST neurons, and partially restored in ASTiso neurons (Supplementary Material, Fig. S6B).
Interestingly, comparison of NAS neurons to ASTiso neurons highlights differences in gene expression of some components of this pathway. Given that α-synuclein expression is normalized in ASTiso neurons, these effects are probably due to altered expression of other genes within the triplication locus or other background genetic variability, and highlight the importance of generating an isogenic iPSC line to dissect such phenotypes.
We compared the expression of UPR genes in NAS and AST derivatives at the iPSC, intermediate neural stem cell (NSC), and neuron differentiation stages, finding that the expression levels of UPR components were generally indistinguishable at the iPSC stage with terminal UPR activation becoming progressively evident through differentiation to neurons. (Supplementary Material, Fig. S7). Taken together with the enrichment for autophagy (Q = 1.90×10−8) and apoptotic (Q = 1.69×10−5) pathways in genes differentially expressed between AST and ASTiso neurons, these results suggest that α-synuclein accumulation due to SNCA triplication activates an ER stress response in neurons, resulting in activation of the UPR and resulting in programmed cell death.
Analysis of post-mortem tissue confirms elevation of pIRE1 in PD patient brains
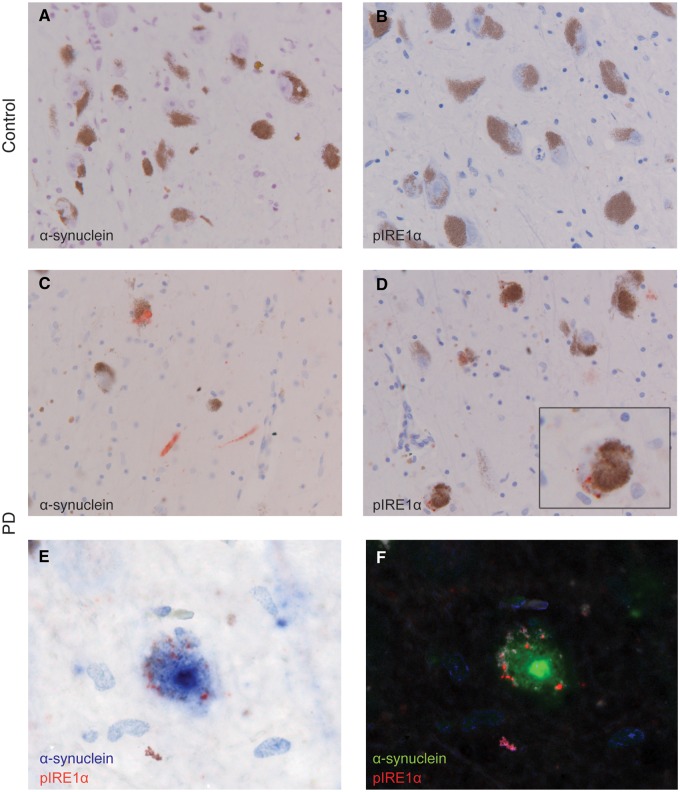
To evaluate the relevance of the findings from iPSC-derived neurons to the pathogenesis of PD in patients, we conducted a neuropathological analysis of α-synuclein and pIRE1α levels in PD versus control post-mortem brain tissue. We performed immunohistochemistry for pIRE1α on formalin-fixed paraffin-embedded brain tissue from a small cohort of PD (n = 5) and control cases (n = 6) (Fig. 4). No α-synuclein immunoreactivity was detected in controls (Fig. 4A), although α-synuclein was detectable in PD cases (Fig. 4C). Likewise, immunoreactivity for pIRE1α was found only in PD cases (Fig. 4D) and was absent in controls (Fig. 4B). In PD cases immunoreactivity for pIRE1α was observed as a punctate or granular staining in neuromelanin-containing neurons in the substantia nigra (Fig. 4D). Two cases were further investigated for the colocalization of pIRE1α with α-synuclein. Immunoreactivity for pIRE1α was observed in 5–20% of neurons staining for α-synuclein but the subcellular localizations of pIRE1α and α-synuclein were distinct. Neurons in PD cases positive for pIRE1α contained neuromelanin and increased cytoplasm-localized immunoreactivity for α-synuclein and/or α-synuclein positive Lewy bodies (Fig. 4E and F). These results support the conclusion that overexpressed α-synuclein induces activation of the IRE1α/XBP1 arm of the UPR, confirms that this effect is evident in patient-derived dopaminergic neurons, and emphasizes the utility of iPSC-based screening platforms for identifying molecular alterations that manifest in PD patients.
Figure 4.
Immunohistochemical detection of α-synuclein and pIRE1α in substantia nigra pars compacta of representative PD and control cases. (A,B) Immunoreactivity for α-synuclein and pIRE-1α is absent in the control case. (C) α-synuclein positive structures, Lewy neurites and bodies, (red) are observed in the PD case. (D) pIRE1α immunoreactive granules (red) are observed in neurons in the substantia nigra of the PD case. Inset shows a higher magnification of a single neuron. (E) Double-immunohistochemistry for pIRE1α (red) and α-synuclein (blue) shows double-labelling of a melanin-containing dopaminergic neuron. (F) Double-staining deconvoluted with spectral imaging showing α-synuclein (green), pIRE1α (red), haematoxylin (blue) and neuromelanin (white). Scale bar: 50 µm in A-D; 20 µm in the inset (D) and E, F., n = 5 PD cases, n = 6 controls. See also Supplementary Material, Table S3.
Discussion
Patient-specific iPSCs have revolutionized efforts to elucidate disease-relevant molecular and cellular phenotypes. However, like other disease models, there are limitations in exploiting iPSC-based models to evaluate complex human diseases. Foremost among these is that when comparing iPSC derivatives of healthy controls to those of patients, there exists an inherent difference in the genetic background between individuals. An important development in the use of iPSCs for modeling human disease was the development of isogenic iPSCs, differing exclusively at disease-relevant loci, using for example site-specific zinc finger nucleases to introduce the SNCA A53T PD-associated mutation to healthy control cells, and conversely to correct the same mutation in iPSCs derived from a PD patient who carried this mutation (22).
The development of iPSCs from a PD patient with a triplication of the SNCA gene locus provides an unprecedented means of evaluating the molecular consequences of endogenous α-synuclein overexpression (7). Alongside their characterizations of the aforementioned SNCA A53T isogenic iPSC pair, Chung et al. compared SNCA triplication iPSC-derived cortical neurons to those derived from wild-type human embryonic stem cells (12) and showed that SNCA triplication neurons closely phenocopy the A53T mutant. However, this comparison was performed in cells of different genetic backgrounds. Additionally, the triplication involves a large, 1.61–2.04 Mb region, containing 16 genes (Supplementary Material, Fig. S4C). Some of these genes may participate in fundamental cellular processes that could further affect neuronal physiology and contribute to PD pathogenesis, including CEB1, HERC3, FAM13A1 and KIAA1680. CEB1 and HERC3 are E3 ubiquitin ligases that mediate ISGylation (23) and vesicle trafficking (24), FAM13A1 is a Rho GTPase activating protein identified as a candidate gene involved in Alzheimer’s disease (25) and KIAA1680 is part of a family of genes, some of which are mediators of neuronal migration (26). Here, we build on the initial observations described above, conducting a detailed analysis of α-synuclein-induced ER stress and UPR activation in neuronal derivatives of SNCA triplication iPSCs compared to an isogenic line with normalized SNCA gene dosage. We have therefore been able to delineate those cellular perturbations caused specifically by elevated SNCA gene dosage, without the interference of variable genetic background or the overexpression of other genes in the triplication locus.
Our studies analysed the effect of SNCA triplication in iPS-derived cortical neurons, and showed that the observed effect on IRE1α activation could also be recapitulated in post-mortem samples of the substantia nigra, suggesting that these processes are more general across different brain regions. Although dopaminergic neurons of the substantia nigra are typically thought to be the most sensitive to neurodegeneration in PD patients, patients also experience a wide range of non-motor symptoms that are not treated by dopamine replacement. Progressive cognitive impairment leading to dementia is one of the most common and important non-motor symptoms of PD (cumulative prevalence ∼80%). Our results demonstrate that there are cellular consequences of SNCA overexpression on cortical neurons, which may underlie such cognitive phenotypes, and which may act more ubiquitously in other neuronal subtypes across the brain.
We demonstrate that SNCA triplication induces the IRE1α/XBP1 arm of the UPR, a finding that stands in agreement with previous literature supporting a role for α-synuclein-induced ER stress and UPR activation in the pathogenesis of PD (16–18). Past studies have demonstrated the need for careful evaluation of UPR-based therapies for neurodegenerative diseases, since it is challenging to accurately predict whether the outcome of altering levels or activity of UPR components will be protective or pathogenic (27). Our study supports further investigation into whether and in which way altering the UPR to target PD-associated α-synucleinopathy might be beneficial. The development of mechanism-based therapies with this goal will require careful evaluation of the contributions of downstream effectors of UPR activation (28,29). An understanding of the differential actions of the UPR branches in different brain regions, neuronal subtypes, and stages of disease will be required towards this end. Importantly, the isogenic iPSCs we generated can enhance the utility of SNCA triplication iPSCs as a platform for drug screening efforts to elucidate such outcomes towards developing a safe and efficacious disease-modifying therapy for α-synucleinopathy-related features of PD.
Materials and Methods
Targeted genome editing to generate isogenic iPSCs and neuronal differentiation
SNCA exon 4-targeting double-nicking CRISPRs were designed based on the principles described in Ran et al. 2013, and cloned into pSpCas9n(BB)-2 A-Puro (PX462) V2.0 (a gift from Feng Zhang, Addgene #62987). Plasmids were Neon transfected into single cell suspensions of AST iPSCs which were then seeded onto mouse embryonic fibroblast (MEF) feeders for expansion. After four days, individual clones were manually isolated and transferred to feeder-free culture on Matrigel-coated 96-well plates. Clones were expanded from 96- to 6-well plates and collected for analysis and cryopreservation. HRM analysis, SURVEYOR assay, and Sanger sequencing were used to identify a double knockout clone. Absence of off-target mutagenesis was confirmed by Sanger sequencing of the top six putative off-target sites. NAS, AST and ASTiso iPSCs were differentiated into neurons via a neural stem cell intermediate, as previously described (14).
Analysis of α-synuclein and UPR component expression in iPSC-derived neurons
For RNA-based analyses, NAS, AST and ASTiso iPSCs, NSCs, and neurons were lysed using Qiazol lysis reagent and RNA was purified using RNeasy spin columns. RNA was quality controlled before RNA sequencing, and results were validated by qRT-PCR. For protein-based analyses, NAS, AST and ASTiso iPSCs, NSCs and neurons were lysed using cell extraction buffer (FNN0011, Life Technologies), supplemented with 1X Halt Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific). Findings from RNA-seq and qRT-PCR were followed up with ELISA and western blotting analysis. Statistical significance was determined by one-way ANOVA using SPSS (IBM).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to acknowledge members of Professor Matthew Wood’s laboratory for helpful discussions and the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (grant 090532/Z/09/Z) for generation of the sequencing data. The SP-dCas9-VPR and pSpCas9n(BB)-2A-Puro vectors were obtained from Addgene (#63798 and #62987) and were gifts from George Church and Feng Zhang respectively.
Conflict of Interest statement. None declared.
Funding
Cure Parkinson’s Trust and John Fell OUP Research Fund to S.M.H.-A., the John Fell OUP Research Fund to M.J.A.W. A.R.B, Wellcome Trust ISSF and contributions from the Departments of Pathology, Biochemistry, DPAG and Pharmacology. BBSRC, Institute Strategic Programme Grant [BB/J004669/1] (W.H.), University of North Carolina at Chapel Hill School of Medicine MD-PhD Program and NIH Oxford-Cambridge Scholars Program (S.M.H.-A.), ISAO, ZonMW, Alzheimer Nederland, and a Stellar grant from Janssen Pharmaceuticals (W.S.). The James Martin Stem Cell Facility (S.A.C., R.F.) receives financial support from the Wellcome Trust [WTISSF121302], the Oxford Martin School [LC0910-004] and EU IMI [StemBANCC]; the research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115439, resources of which are composed of financial contribution from the European Union' Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' in kind contribution. This publication reflects only the authors' views, and neither the IMI JU nor EFPIA nor the European Commission are liable for any use that may be made of the information contained therein. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R. (2003) Alpha-synuclein locus triplication causes Parkinson's disease. Science, 302, 841.. [DOI] [PubMed] [Google Scholar]
- 2. Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M.. et al. (2004) Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet, 364, 1167–1169. [DOI] [PubMed] [Google Scholar]
- 3. Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. (2004) Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet, 364, 1169–1171. [DOI] [PubMed] [Google Scholar]
- 4. Farrer M., Kachergus J., Forno L., Lincoln S., Wang D.S., Hulihan M., Maraganore D., Gwinn-Hardy K., Wszolek Z., Dickson D.. et al. (2004) Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol., 55, 174–179. [DOI] [PubMed] [Google Scholar]
- 5. Singleton A.C. J. (2007) Dawson T. (ed.), In Parkinson's Disease: Genetics and Pathogenesis. Informa Healthcare USA, Inc, New York, NY, in press., pp. 19–33. [Google Scholar]
- 6. McCormack A.L., Di Monte D.A. (2009) Enhanced alpha-synuclein expression in human neurodegenerative diseases: pathogenetic and therapeutic implications. Curr. Protein Pept. Sci., 10, 476–482. [DOI] [PubMed] [Google Scholar]
- 7. Devine M.J., Ryten M., Vodicka P., Thomson A.J., Burdon T., Houlden H., Cavaleri F., Nagano M., Drummond N.J., Taanman J.W.. et al. (2011) Parkinson's disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat. Commun., 2, 440.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flierl A., Oliveira L.M., Falomir-Lockhart L.J., Mak S.K., Hesley J., Soldner F., Arndt-Jovin D.J., Jaenisch R., Langston J.W., Jovin T.M.. et al. (2014) Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an alpha-synuclein gene triplication. PloS One, 9, e112413.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reyes J.F., Olsson T.T., Lamberts J.T., Devine M.J., Kunath T., Brundin P. (2015) A cell culture model for monitoring alpha-synuclein cell-to-cell transfer. Neurobiol. Dis., 77, 266–275. [DOI] [PubMed] [Google Scholar]
- 10. Byers B., Cord B., Nguyen H.N., Schüle B., Fenno L., Lee P.C., Deisseroth K., Langston J.W., Pera R.R., Palmer T.D., Chin W.-C. (2011) SNCA triplication Parkinson's patient's iPSC-derived DA neurons accumulate alpha-synuclein and are susceptible to oxidative stress. PloS One, 6, e26159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliveira L.M., Falomir-Lockhart L.J., Botelho M.G., Lin K.H., Wales P., Koch J.C., Gerhardt E., Taschenberger H., Outeiro T.F., Lingor P. (2015) Elevated alpha-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson's patient-derived induced pluripotent stem cells. 6, e1994. [DOI] [PMC free article] [PubMed]
- 12. Chung C.Y., Khurana V., Auluck P.K., Tardiff D.F., Mazzulli J.R., Soldner F., Baru V., Lou Y., Freyzon Y., Cho S.. et al. (2013) Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science, 342, 983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y.. et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heman-Ackah S.M., Bassett A.R., Wood M.J. (2016) Precision Modulation of Neurodegenerative Disease-Related Gene Expression in Human iPSC-Derived Neurons. Sci. Rep., 6, 28420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castillo-Carranza D.L., Zhang Y., Guerrero-Munoz M.J., Kayed R., Rincon-Limas D.E., Fernandez-Funez P. (2012) Differential activation of the ER stress factor XBP1 by oligomeric assemblies. Neurochem. Res., 37, 1707–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoozemans J.J., van Haastert E.S., Eikelenboom P., de Vos R.A., Rozemuller J.M., Scheper W. (2007) Activation of the unfolded protein response in Parkinson's disease. Biochem. Biophys. Res. Commun., 354, 707–711. [DOI] [PubMed] [Google Scholar]
- 18. Bellucci A., Navarria L., Zaltieri M., Falarti E., Bodei S., Sigala S., Battistin L., Spillantini M., Missale C., Spano P. (2011) Induction of the unfolded protein response by alpha-synuclein in experimental models of Parkinson's disease. J. Neurochem., 116, 588–605. [DOI] [PubMed] [Google Scholar]
- 19. Venderova K., Park D.S. (2012) Programmed cell death in Parkinson's disease. Cold Spring Harb. Perspect. Med., 2, a009365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lynch-Day M.A., Mao K., Wang K., Zhao M., Klionsky D.J. (2012) The role of autophagy in Parkinson's disease. Cold Spring Harb. Perspect. Med., 2, a009365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Griffith H.R., den Hollander J.A., Okonkwo O.C., O'Brien T., Watts R.L., Marson D.C. (2008) Brain metabolism differs in Alzheimer's disease and Parkinson's disease dementia. Alzheimers Dement., 4, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S.. et al. (2011) Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell, 146, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong J.J., Pung Y.F., Sze N.S., Chin K.C. (2006) HERC5 is an IFN-induced HECT- type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl Acad. Sci. USA, 103, 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cruz C., Ventura F., Bartrons R., Rosa J.L. (2001) HERC3 binding to and regulation by ubiquitin. FEBS Lett., 488, 74–80. [DOI] [PubMed] [Google Scholar]
- 25. Miller J.A., Woltjer R.L., Goodenbour J.M., Horvath S., Geschwind D.H. (2013) Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med., 5, 48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paracchini S., Thomas A., Castro S., Lai C., Paramasivam M., Wang Y., Keating B.J., Taylor J.M., Hacking D.F., Scerri T.. et al. (2006) The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum. Mol. Genet., 15, 1659–1666. [DOI] [PubMed] [Google Scholar]
- 27. Valdes P., Mercado G., Vidal R.L., Molina C., Parsons G., Court F.A., Martinez A., Galleguillos D., Armentano D., Schneider B.L.. et al. (2014) Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc. Natl Acad. Sci. USA, 111, 6804–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim I., Xu W., Reed J.C. (2008) Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov., 7, 1013–1030. [DOI] [PubMed] [Google Scholar]
- 29. Chen Y., Brandizzi F. (2013) IRE1: ER stress sensor and cell fate executor. Trends Cell Biol., 23, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.