Abstract
Mitochondrial disorders affecting oxidative phosphorylation (OxPhos) are caused by mutations in both the nuclear and mitochondrial genomes. One promising candidate for treatment is the drug rapamycin, which has been shown to extend lifespan in multiple animal models, and which was previously shown to ameliorate mitochondrial disease in a knock-out mouse model lacking a nuclear-encoded gene specifying an OxPhos structural subunit (Ndufs4). In that model, relatively high-dose intraperitoneal rapamycin extended lifespan and improved markers of neurological disease, via an unknown mechanism. Here, we administered low-dose oral rapamycin to a knock-in (KI) mouse model of authentic mtDNA disease, specifically, progressive mtDNA depletion syndrome, resulting from a mutation in the mitochondrial nucleotide salvage enzyme thymidine kinase 2 (TK2). Importantly, low-dose oral rapamycin was sufficient to extend Tk2KI/KI mouse lifespan significantly, and did so in the absence of detectable improvements in mitochondrial dysfunction. We found no evidence that rapamycin increased survival by acting through canonical pathways, including mitochondrial autophagy. However, transcriptomics and metabolomics analyses uncovered systemic metabolic changes pointing to a potential ‘rapamycin metabolic signature.’ These changes also implied that rapamycin may have enabled the Tk2KI/KI mice to utilize alternative energy reserves, and possibly triggered indirect signaling events that modified mortality through developmental reprogramming. From a therapeutic standpoint, our results support the possibility that low-dose rapamycin, while not targeting the underlying mtDNA defect, could represent a crucial therapy for the treatment of mtDNA-driven, and some nuclear DNA-driven, mitochondrial diseases.
Introduction
Mitochondrial disease represents a diverse class of disorders caused by mutations affecting oxidative phosphorylation (OxPhos) (1). While presentation can vary greatly among disorders and among patients, common features are encephalopathy, myopathy, or both. Mutations have been identified in a wide range of genes encoded by both the nuclear and mitochondrial genome (mtDNA). Pathogenic mutations have been identified in genes specifying individual subunits of OxPhos complexes, assembly factors, and components of the mitochondrial genome maintenance and translational machinery. Identification of disease-causing nuclear mutations has led to the development of several targeted therapies (2–5). By contrast, clinically-applicable treatments aimed at reversing the mtDNA defects remain elusive.
One promising treatment candidate is rapamycin (also known as sirolimus), a drug that has been shown to extend lifespan in multiple animal models, including mice (6). Rapamycin inhibits the protein mechanistic target of rapamycin (mTOR), a master regulator of cell growth and metabolism that integrates numerous intracellular and extracellular nutrient signals to regulate a diverse array of processes, including autophagy, lipid synthesis, translation, mitochondrial biogenesis, and cell survival (7). Importantly, rapamycin has been shown to ameliorate morbidity and mortality in mouse models of several neurological diseases, most notably a model of mitochondrial disease caused by ablation of the nucleus-encoded gene specifying the Ndufs4 subunit of OxPhos complex I (8,9). Treated Ndufs4−/− mice showed improvements in neurological disease markers, as well as significant alterations in metabolism, most notably a shift away from glycolysis and towards amino acid metabolism that could be favorable in the setting of OxPhos dysfunction. However, the underlying mitochondrial dysfunction was not attenuated, and the mechanism of rapamycin‘s beneficial effect was not elucidated (8). There is independent cellular evidence that rapamycin could induce selective autophagy of mitochondria (‘mitophagy’) to degrade dysfunctional organelles (10), but this phenomenon remains to be demonstrated in mammals, and the applicability to disease is currently untested.
While mice have been engineered to express mutations in mtDNA (11–13), there are currently no available mice that model known pathogenic mutations in human mtDNA, due mainly to the difficulty of manipulating this genome in mammals (14,15). However, mice containing nuclear mutations that affect mtDNA replication and stability provide a good model of authentic mtDNA-driven disease phenotypes. One such mouse model expresses a homozygous knock-in mutation in the mitochondrial nucleotide salvage enzyme thymidine kinase 2 (Tk2)—specifically, p.His126Asn, corresponding to the pathogenic human TK2 variant p.His121Asn—that results in a progressive mtDNA depletion syndrome (MDS) (16). Similarly to human MDS patients, who typically present with a myopathy, occasionally with an encephalopathy (17), Tk2KI/KI (henceforth denoted TK2) mice exhibit a progressive severe depletion of mtDNA and concordant respiratory chain defects and disease (16). Newborn pups are comparable to littermates in size, but after the first week of life exhibit a failure to thrive and progressive motor dysfunction, culminating in a rapid decline phase prior to death at a median age of two weeks (16). Organs are differentially affected, with the most severe phenotype exhibited in brain (16). Brain mtDNA levels, while approximately normal at birth, decrease to ∼20% of WT levels by two weeks of age, concordant with the time frame of the rapid decline (18). Similarly, brain respiratory chain complex protein levels and activities are decreased dramatically. Skeletal muscle also exhibits mtDNA depletion and myofiber atrophy, but in contrast to human patients (17), retains normal respiratory function (16,19); the liver, kidney, and heart show only mild abnormalities (16).
In the present work, we treated TK2 mice with low-dose rapamycin to investigate this drug as a potential therapy for severe early-onset mtDNA disease.
Results
Rapamycin therapy extends mortality in TK2 mice
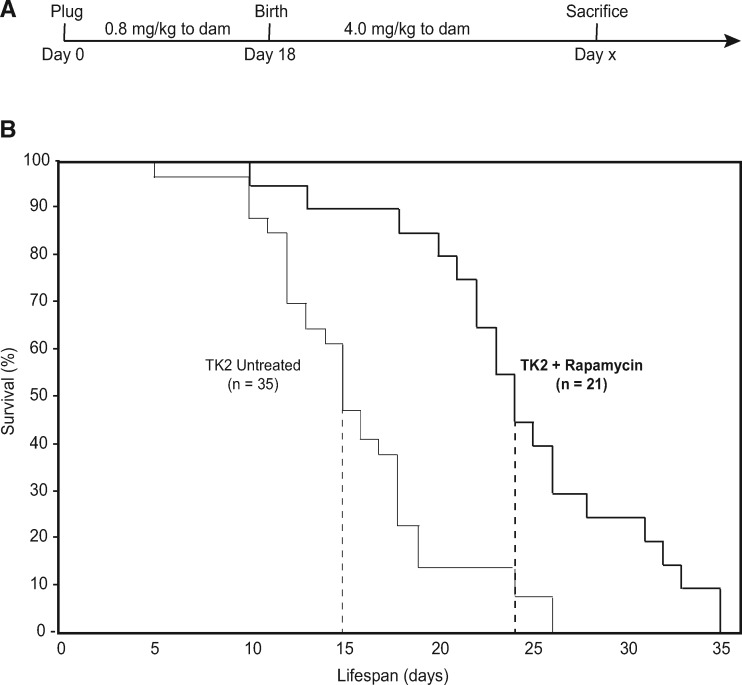
Because rapamycin has been shown previously to extend survival in the setting of nuclear-encoded mitochondrial disease (8), we hypothesized that it might also alter the disease severity and/or course in TK2 mice. As rapamycin has been administered successfully in utero in humans (20,21), and is known to cross the placenta (22,23), we administered it orally to dams, as described previously (24). Pilot studies in TK2 mice demonstrated that both pre- and postnatal treatments independently extended lifespan by several days, while combination pre- and postnatal therapy resulted in an even greater survival extension (data not shown). Accordingly, beginning at conception, dams were administered 0.8 mg/kg rapamycin, and dosage was increased to 4 mg/kg at delivery (Fig. 1A). This low-dosage regimen compares favorably with the range of reported values (25), and was selected to reduce the adverse effects of early and continuous rapamycin treatment.
Figure 1.
Rapamycin therapy extends lifespan in TK2 mice. (A) Rapamycin was delivered orally through the drinking water to dams, beginning at conception and continuing through the entire lifespan of the mice (no TK2 mice survived past weaning). Dose to dams was 0.8 mg/kg during gestation, and increased to 4 mg/kg upon delivery. (B) Kaplan-Meier curves representing survival of rapamycin-treated (thick lines) and untreated (thin lines) mice. WT littermates experienced no mortality during this period and are not shown. Dashed lines indicate 50% mortality. Median lifespan extension of treated vs untreated TK2 mice was ∼60%; maximum lifespan was ∼35%; log-rank test P-value = 3.4 × 10−5. n, total number of mice in each group.
Remarkably, rapamycin treatment extended significantly both the median (by ∼60%) and the maximum (by ∼35%) survival of TK2 mice (log-rank test, P = 3.4×10−5) (Fig. 1B). This extension of lifespan resulted in the longest-surviving TK2 animals surviving through weaning. No significant gender difference was observed in either median or maximum survival (Supplementary Material, Fig. S1A).
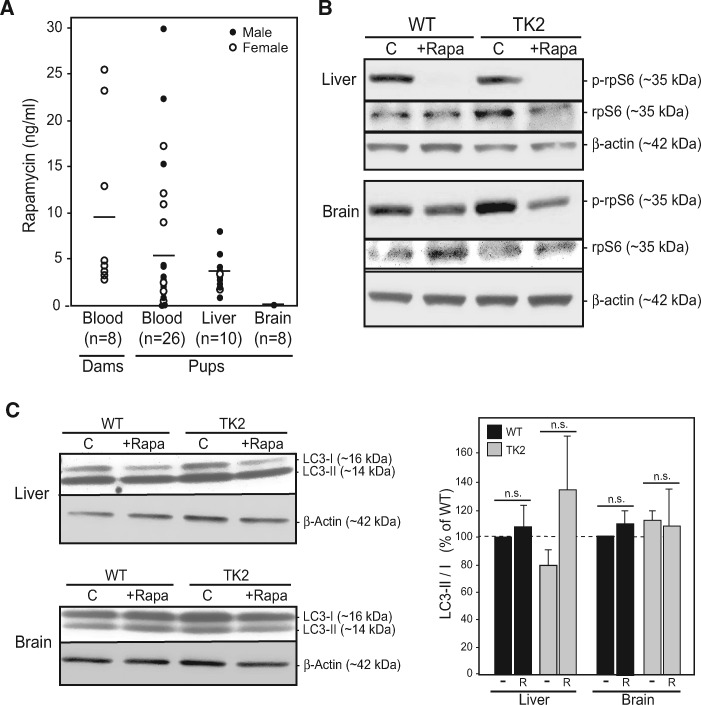
In order to establish a relationship between this enhanced survival and our rapamycin administration protocol, we measured steady-state rapamycin levels in these animals, both biochemically (by high performance liquid chromatography [HPLC]) and functionally [by monitoring the phosphorylation status of ribosomal protein S6 [rpS6], which is inhibited by rapamycin (6)]. HPLC values for male and female pups averaged ∼5 ng/ml in blood, ∼3 ng/g in liver, and were undetectable in brain (Fig. 2A). The steady-state levels in blood fell within the low end of reported doses in the literature (19). In agreement with the biochemistry, liver was responsive to rapamycin treatment (i.e. reduced phospho-rpS6 signal), whereas brain was not (Fig. 2B). The lack of a rapamycin signal in the brain has been observed by others (26,27), and is thought to be related to a dose-dependent permeability of the blood-brain barrier to rapamycin (28). Moreover, although rapamycin was clearly detected by analytical methods in both male and female livers (Fig. 2A), the phospho-rpS6 response was present in female (Fig. 2B), but not in male (Supplementary Material, Fig. S2), livers. This sexual dimorphism in rpS6 constitutive phosphorylation and in rapamycin response is also in agreement with previous reports (29,30). Taken together, these results show that despite gender differences in bioavailability and a low penetration into brain—the most vulnerable tissue in this disorder—low-dose rapamycin was sufficient to extend lifespan in TK2 mice of both genders.
Figure 2.
Rapamycin treatment regimen. (A) Rapamycin concentrations in whole blood (ng/ml), liver (ng/g), and brain (ng/g) of the indicated mice was measured by HPLC at time of sacrifice. Dots represent individual animals, with total number indicated. Lines represent average value. (B) Representative western blots illustrating phosphorylation of rpS6 at Ser240/244 in untreated vs rapamycin-treated WT and TK2 livers and brains from female mice, compared with total rpS6 and β-actin. (C) Representative western blot of LC3-I and LC3-II in liver and brain from female mice; quantitation LC3-II/LC3-I at right; liver n = 7; brain n = 5). Quantitation of all experiments is at right. -, untreated; R, treated with rapamycin; n.s., not significant.
Rapamycin therapy does not affect morbidity in TK2 mice
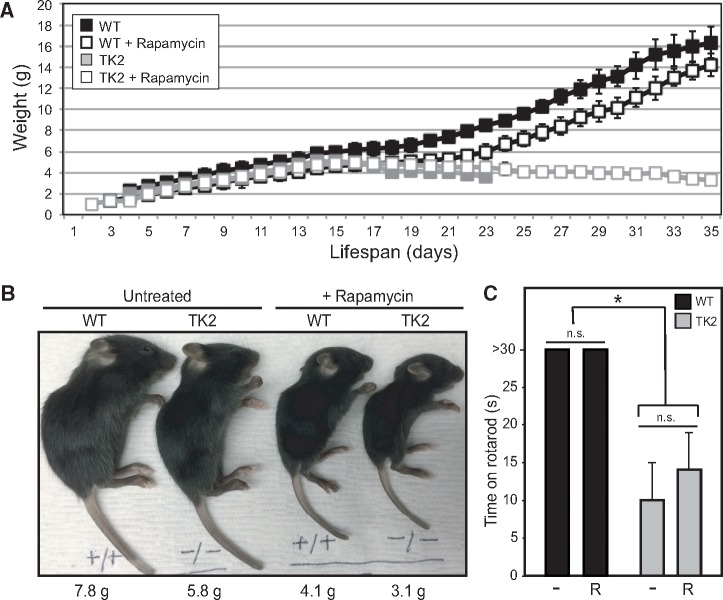
We next asked whether this increase in survival corresponded to an improvement in morbidity. A notable characteristic of TK2 mice is their failure to thrive, which is evident within one week of birth by a decreased weight gain relative to wild-type (WT) littermates. This is followed approximately one week later by rapid weight decline, and subsequent death within 2–3 days. Considering that rapamycin therapy, in and of itself, is known to cause decreased weight (25), we monitored pup weight throughout the lifespan of both treated and untreated litters. In fact, all treated mice exhibited decreased weight compared with untreated mice, with the effect most pronounced in treated TK2 animals (Fig. 3A and B;Supplementary Material, Data File S1). Segregation of the data by gender showed that this trend was present in both male and female mice (Supplementary Material, Fig. S1B). Some of this weight loss might be attributable to depleted fat stores, as necropsy of mice sacrificed after overt disease manifestation revealed decreased adiposity in treated animals of both genotypes, consistent with the literature (31,32), and again, most severe in treated TK2 mice (Supplementary Material, Fig. S3). Gross morphology and histology of the brain, heart, liver, kidney, spleen, and skeletal muscle were notable only for hyposplenism in treated WT and TK2 mice and muscle atrophy in TK2 mice (Supplementary Material, Fig. S4). Given this finding, and previous studies in mice and flies demonstrating the adverse effects of rapamycin on muscle mitochondrial gene transcripts and functions (33,34), we assayed respiratory chain activity by staining muscle for cytochrome c oxidase (COX) activity. While deficient in many TK2 patients (19), COX activity appeared normal in our TK2 mice and was not adversely affected by rapamycin treatment (Supplementary Material, Fig. S4). Assessment of motor function using a rotating rod assay of motor function (e.g. coordination and balance) revealed severe deficiencies in TK2 mice compared with WT littermates that was unaffected by rapamycin treatment (Fig. 3C). There was no change, either by underlying disease or by rapamycin therapy, in liver lipid droplet content as measured by Oil-Red-O staining (Supplementary Material, Fig. S4). Similarly, there was no evidence of increased leukocytic infiltration (data not shown) or apoptosis (by TUNEL staining) in the brain, liver, or muscle of treated or untreated WT or TK2 animals (Supplementary Material, Fig. S4).
Figure 3.
Rapamycin does not improve TK2 mouse fitness. (A) Pup weight of rapamycin-treated and untreated litters was measured daily, and indicated for untreated WT (black filled squares), rapamycin-treated WT (black open squares), untreated TK2 (gray filled squares), and rapamycin-treated TK2 (gray open squares) mice. Error bars represent standard deviation, when applicable. (B) Representative image of WT and TK2-treated and untreated mice at 15 days, with weights indicated. (C) Motor function was assessed using a rotating rod assay by recording length of time mice could remain on the rod, up to 30 s. Average values are shown, with error bars indicating standard deviation and asterisk indicating significance compared with untreated WT at the 95% confidence interval by Welch’s P-value; n = 7 for untreated WT and TK2 groups, n = 9 for rapamycin-treated WT and TK2 mice. Other notation as in Figure 2.
Rapamycin does not improve molecular markers of mtDNA disease
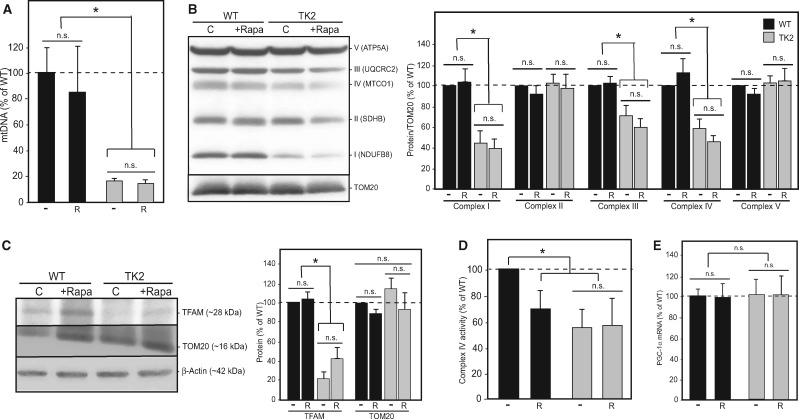
We were particularly interested in the effect of rapamycin therapy on mtDNA disease as measured by molecular indicators of mitochondrial dysfunction. We compared treated and untreated animals of both genotypes, again sacrificing animals after overt disease manifestation but before end-stage. As the brain is the organ most affected in our TK2 mouse model, we first focused on this tissue. Depletion of brain mtDNA was unaltered by treatment, as assessed both by quantitative PCR of mtDNA levels (Fig. 4A) and by measurement of mitochondrial transcription factor A (TFAM), a stoichiometric protein marker of mtDNA content (Fig. 4C). Similarly, there was no change in the decreased protein levels of respiratory chain subunits (specifically subunits of complexes I, III and IV) (Fig. 4B) or in complex IV activity (Fig. 4D), whether normalized to total mitochondrial content (Fig. 4) or to complex II (data not shown). Finally, there was no significant difference in mitochondrial content between WT and TK2 brains from treated and untreated animals, as demonstrated by unaltered total cellular levels of TOM20, an mtDNA-independent protein marker of total mitochondrial mass (Fig. 4C), and by unchanged RNA expression of the mitochondrial biogenesis master regulator PGC-1α (34) (Fig. 4E).
Figure 4.
Rapamycin does not affect mtDNA disease in the brains of treated TK2 mice. (A) Quantitation of brain mtDNA levels [relative to nuclear DNA (Gapdh)] by qPCR in 15- to 18-day-old pups (n = 11, n = 12, n = 10, and n = 13 for untreated WT, rapamycin-treated WT, untreated TK2, and rapamycin-treated TK2 mice, respectively). (B) Representative western blot and quantitation (n = 6) of representative subunits from respiratory complexes I–V, normalized to TOM20. (C) Representative western blot and quantitation (n = 6) for TFAM, TOM20 and β-actin; TFAM was quantitated relative to TOM20, and TOM20 relative to β-actin (n = 6). (D) Complex IV activity was measured in isolated mitochondria from brain tissue using a Seahorse XFe24 flux analyzer and the standard electron flow assay; values were normalized to total mitochondrial content (n = 5). (E) Quantitation of PGC-1α signal by qRT-PCR (n = 5). For all graphs, values are shown as percentages relative to untreated WT, with error bars indicating standard deviation, and significance tested by Welch’s t-test at 95% confidence. Other notation as in Figure 3.
The lack of improvement in brain markers of mtDNA disease was not surprising in light of the fact that rapamycin was not detected in the brains of these low-dose treated animals. However, it was quite surprising in the context of the significant survival benefit seen after therapy. We therefore decided to assess the effects of rapamycin in the liver, because despite being a relatively unaffected organ in TK2 disease (16), it is the first organ exposed to drug from the gut in oral rapamycin therapy (with clearly detectable levels in our animals) and plays a key role in drug metabolism (35). Consistent with the results seen in the brain, the liver exhibited no change in mtDNA levels as a result of rapamycin therapy (Supplementary Material, Fig. S5A). There was also no significant change in liver expression of respiratory chain subunits after treatment (Supplementary Material, Fig. S5B) [with the exception of the decreased expression of complex II in rapamycin-treated TK2 animals], and COX activity was normal in the setting of TK2 disease (Supplementary Material, Fig. S5D), as seen previously (16). There were trends toward decreased liver mitochondrial mass (Supplementary Material, Fig. S5C) and biogenesis (Supplementary Material, Fig. S5E) following rapamycin therapy, which is consistent with the known inhibitory effect of rapamycin on mitochondrial function and biogenesis (36). Despite the decrease in mitochondrial mass, mtDNA content was preserved so as to maintain a constant level of total cellular mtDNA (Supplementary Material, Fig. S5C), implying that rapamycin does not affect mtDNA copy control (37).
Taken together, these findings indicate that rapamycin therapy dosed at the lowest end of the reported therapeutic range is nevertheless sufficient to enhance survival of mtDNA disease mice significantly, but does so without any apparent amelioration of the underlying mitochondrial dysfunction.
Rapamycin does not act through canonical pathways
Previous studies have implicated several downstream targets of rapamycin in ameliorating neurological (including mitochondrial) disease (7,10,38). Given the dramatic mortality improvement seen in our TK2 mice with rapamycin treatment, albeit in the absence of an obvious change in morbidity, we sought to understand the mechanism of action by investigating candidate pathways associated with rapamycin treatment. Again, we focused our analysis on the brain, as we hypothesized that the most affected organ ought to manifest treatment-induced changes. Similarly, we analysed liver, because this organ is highly responsive to oral rapamycin therapy (35).
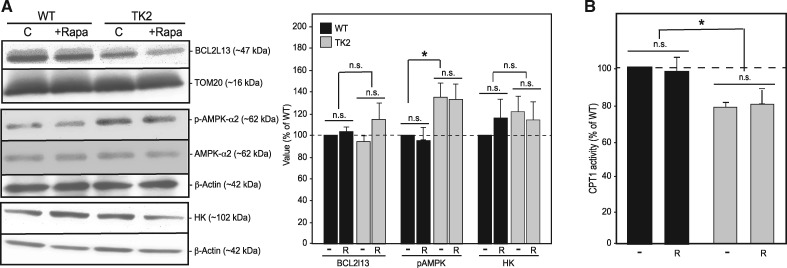
Perhaps the best-known target of rapamycin is the appropriately named protein complex mechanistic target of rapamycin (mTOR), which serves as a master regulator for numerous cellular processes, including translation, lipid metabolism, mitochondrial biogenesis, and autophagy (39). Rapamycin binds the immunophilin protein FKBP12 to inhibit mTOR, thereby de-repressing autophagy and inducing the conjugation of phosphatidylethanolamine to the cytosolic form of the autophagic membrane protein light chain protein 3 (LC3-I) to form the autophagosome-bound form (LC3-II); elevated LC3-II relative to LC3-I indicates autophagic induction (40). The LC3-II/I ratio was measured in all four treatment groups, and was found to be statistically unchanged in the brains of both WT and TK2 rapamycin-treated animals (Fig. 2C), though trending upwards in the livers of treated animals. A marker of mitochondrial-specific autophagy (‘mitophagy’), BCL2L13 (41), was unchanged in the brain (Fig. 5A) and was elevated in TK2 liver (Supplementary Material, Fig. S6A), but these values were unchanged in response to rapamycin treatment (Supplementary Material, Fig. S6A). These data, taken together with the tissue-dependent detection of rapamycin in our treated mice (Fig. 2), agree with previous studies showing that rapamycin induces autophagy in treated tissues, although this does not translate into an increase in mitochondrial-specific degradation by mitophagy (42).
Figure 5.
Canonical pathways of rapamycin are not perturbed in TK2 brains. (A) Representative western blots and quantitation for LC3-II/LC3-I (n = 4), BCL2L13 normalized to TOM20 (n = 6), pAMPK normalized to β-actin (n = 7), and hexokinase (HK) normalized to β-actin (n = 8). (B) Assay of CPT1 activity (n = 4 for all groups). For all graphs, values are shown as percentages relative to untreated WT, with error bars indicating standard error; *P < 0.05. Other notation as in Figure 3.
Next, we hypothesized that rapamycin might provide a survival benefit in TK2 mice by upregulating OxPhos-independent energy pathways to maintain cellular ATP levels (8). A primary candidate pathway was glycolysis, where glucose can be oxidized to pyruvate and ultimately converted to lactate to bypass mitochondrial oxidative phosphorylation. We therefore measured cellular levels of hexokinase (HK), the first enzyme in the brain glycolytic pathway, which has been shown to be differentially regulated in response to both mitochondrial disease and rapamycin treatment (8). Brain HK was unchanged in both untreated and treated animals (Fig. 5A). In contrast, the liver homologue, glucokinase (GK), which is also differentially regulated in response to insulin signaling (43), trended upwards in TK animals and downwards following rapamycin treatment (Supplementary Material, Fig. S6A).
Another candidate metabolic pathway for ATP generation was β-oxidation of fatty acids. Although this pathway is partially mitochondrial, it does not involve mtDNA-encoded subunits of the respiratory chain and thus would likely be spared in the setting of TK2 disease. Beta-oxidation requires the importation of CoA-acylated long chain fatty acids into the mitochondrion via the carnitine shuttle, whose activity can be approximated by measuring the activity of the rate-limiting enzyme carnitine palmitoyltransferase I (CPT1) (44). Surprisingly, measurement of CPT1 activity in the brain revealed a slight decrease in activity in untreated TK2 mice, which was unaltered by rapamycin treatment (Fig. 5B). Liver CPT1 activity in untreated TK2 animals did not differ significantly from WT, and showed no change with rapamycin treatment (Supplementary Material, Fig. S6B).
Finally, we asked if rapamycin could be extending lifespan by altering cellular energy homeostasis. In response to elevated cellular AMP/ATP ratios, the regulatory kinase AMPK is activated by phosphorylation at Thr-172 (45,46) and proceeds to inhibit anabolic processes and activate catabolic processes through downstream effectors, including PGC-1α and mTORC1 (47), in efforts to increase ATP production and restore homeostasis. We hypothesized that in TK2 disease, tissues deficient in mitochondrial respiration might exhibit elevated AMPK activation. Additionally, since AMPK acts upstream of mTORC1 (7), we considered the possibility that rapamycin might be acting by interdicting AMPK signaling. However, while Thr172-pAMPK levels were elevated significantly in TK2 animals compared with WT, they were unaltered by rapamycin treatment (Fig. 5A). In the liver, by contrast, Thr172-pAMPK levels were decreased in rapamycin-treated WT animals, as predicted (Supplementary Material, Fig. S6A).
Taken together, we found little evidence that the effect of rapamycin on survival in TK2 mice could be ascribed to pathways most typically associated with this molecule [notably, this conclusion was unchanged upon re-analysis of the data segregated by gender, despite sex differences in rpS6 phosphorylation (data not shown)]. However, there was evidence of altered metabolic homeostasis in response to treatment, particularly in the liver.
Unbiased approaches reveal potential mediators of the rapamycin effect
Because the investigation of candidate pathways did not reveal an obvious mechanism for rapamycin‘s role in the survival extension in TK2 mice, we embarked upon an unbiased approach that might elucidate putative mechanisms. We investigated the brain and liver RNA expression and metabolite profiles of treated and untreated WT and TK2 mice sacrificed after disease manifestation. Metabolomics analysis was conducted in plasma as well, because of the traditional role of certain plasma metabolite levels in diagnosing mitochondrial disease (48,49).
RNA expression profiles of brain and liver were analysed by sequencing the RNA transcriptome (‘RNA-Seq’) of three biological replicates for each of the four biological groups: untreated WT, rapamycin-treated WT, untreated TK2, and rapamycin-treated TK2 (raw read counts in Supplementary Material, Data File S2). We then compared these animals in three ways: untreated WT vs untreated TK2; untreated WT vs treated WT; and untreated TK2 vs treated TK2. Genes of interest varying significantly within each comparison were identified using two complementary packages for differentially-expressed gene (DEG) analysis, available in Bioconductor (www.bioconductor.org): DESeq2 (50,51) and edgeR (52–54). The DEGs were then used to identify differentially regulated biological functions via overrepresentation analysis (ORA) based on the GO-Slim biological process annotations from the Protein Analysis THrough Evolution Relationships (PANTHER) gene ontology classification system (55–57). As both platforms are commonly employed in analysing RNA-Seq data, and have previously been shown to differ in their selection of DEGs [DESeq2 is considered more conservative in DEG selection through control for false-positives, whereas edgeR is optimized for <12 biological replicates (58)], we employed both packages to increase the sensitivity of our analysis (59,60). Differences in output from the two platforms were therefore not surprising, although consistency between analyses was used as stronger evidence of differential regulation.
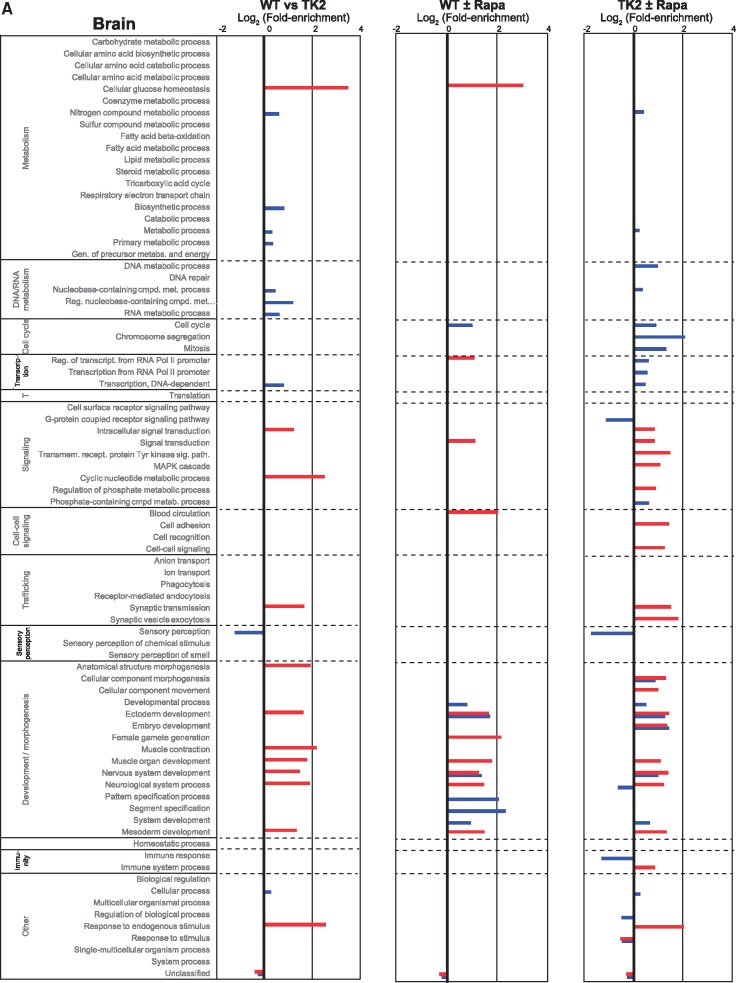
We first performed overrepresentation analysis on the brain with the three comparisons noted above (Fig. 6A). Comparison of untreated WT to untreated TK2 animals (Fig. 6A, Column 1) showed that, beyond the expected overrepresentation in nucleotide pathways, differences fell primarily in neurological and myological developmental pathways (e.g. GO categories for ectoderm and mesoderm development, as well as for muscle organ and nervous system development). These results likely represent hitherto unknown effects of TK2 disease on developmental processes. Interestingly, comparison of both untreated WT to treated WT (Fig. 6A, Column 2) and untreated TK2 to treated TK2 (Fig. 6A, Column 3) yielded a similar result. In addition, the last comparison also showed differences in intracellular and cell-cell signaling, cell cycle, and transcriptional processes (e.g. GO categories for intracellular signaling mediated by the MAPK cascade, cyclic nucleotides, and phosphate-containing compounds). Thus, rapamycin apparently influenced systemic signaling events and developmental pathways in the brain, possibly modifying disease.
Figure 6.
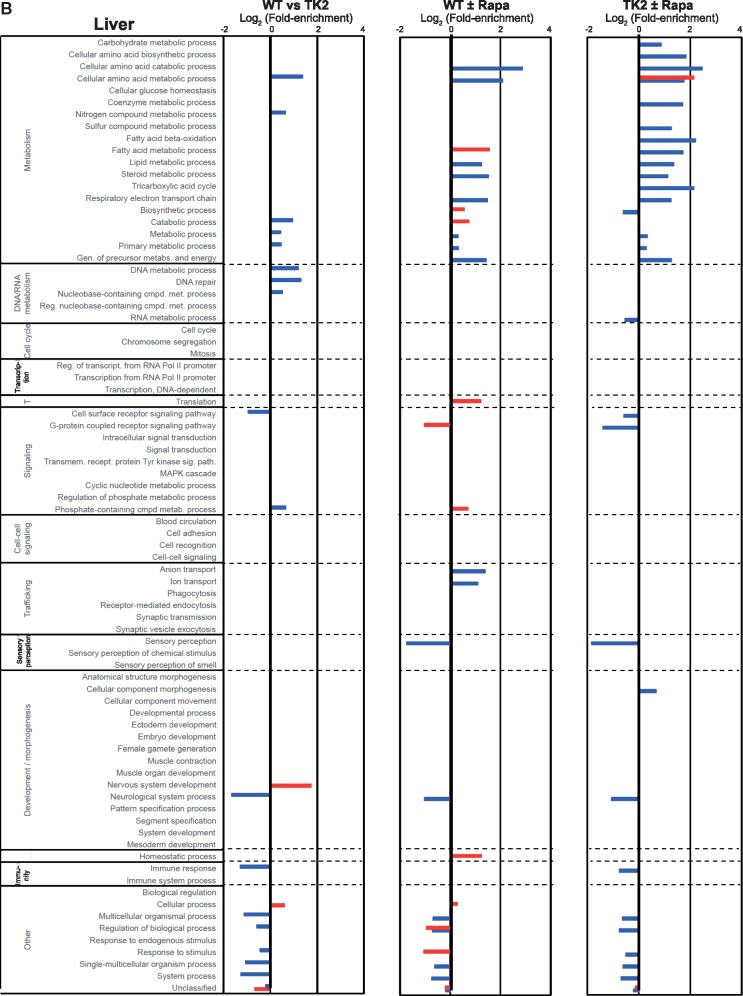
RNA-Seq analysis. Overrepresentation analysis based on the PANTHER GO-Slim biological process gene ontology categorization of differentially expressed genes identified by the DESeq2 (blue bars) and edgeR (red bars) algorithms for the indicated comparisons (3 biological replicates for each condition). Bars denote log2 fold-enrichment of the indicated GO category, representing relative likelihood that the process is (positive values) or is not (negative values) an important source of the difference between groups. (A) Brain, (B) Liver.
The expression pattern in liver (Fig. 6B) contrasted dramatically with that in the brain. When comparing treated and untreated livers, both in WT and TK2 mice (Fig. 6B, Columns 2 and 3), differences from both platforms were seen primarily in metabolic, not developmental, pathways. In particular, there were alterations in amino acid, fatty acid, lipid, steroid, and TCA metabolism (e.g. GO categories for cellular amino acid metabolism, fatty acid metabolism, fatty acid β-oxidation, and respiratory electron transport chain). On the other hand, few changes were seen when comparing untreated WT to untreated TK2 animals (Fig. 6B, Column 1), including primarily nucleotide metabolism, and thus those changes likely reflected underlying disease.
In addition to the overrepresentation analysis, which examines the transcriptome in a global fashion, we assessed the effects of rapamycin on the differential expression of specific genes that are markers for cellular processes of potential interest, focusing on mediators of metabolism, oxidative stress, apoptosis, and epigenetics (summarized in Supplementary Material, Data File S3). We found evidence in the liver supporting the role of fatty acid metabolism/transport (29 of 69 genes were differentially regulated in one or both treatment comparisons), β-oxidation (4 of 9 genes), and especially ketogenesis (5 of 6 genes), whereas fatty acid biosynthesis was unaffected (0 of 9 genes). There was little evidence of rapamycin-mediated effects on adipokine signaling (14 of 19 genes unchanged), oxidative stress (38 of 42 genes unchanged), apoptosis (11 of 14 genes unchanged), or epigenetics (150 of 166 genes unchanged). The latter two categories were also unaffected in the brain (15 of 15 genes and 146 of 166 genes unchanged, respectively; see Supplementary Material, Data File S3). Finally, to validate the RNA-Seq results, we conducted side-by-side comparison of the RNA-Seq and qRT-PCR expression values for five representative genes: acetyl-CoA carboxylase α (Acaca), diacyglycerol O-acyltransferase 2 (Dgat2), fatty acid synthase (Fasn), lipoprotein lipase (Lpl); and the mitochondrial master regulator PGC-1α (Ppargc1a). The qRT-PCR results for these five genes were essentially congruent with the expression values for these same genes obtained by RNA-Seq analysis (Supplementary Material, Fig. S7).
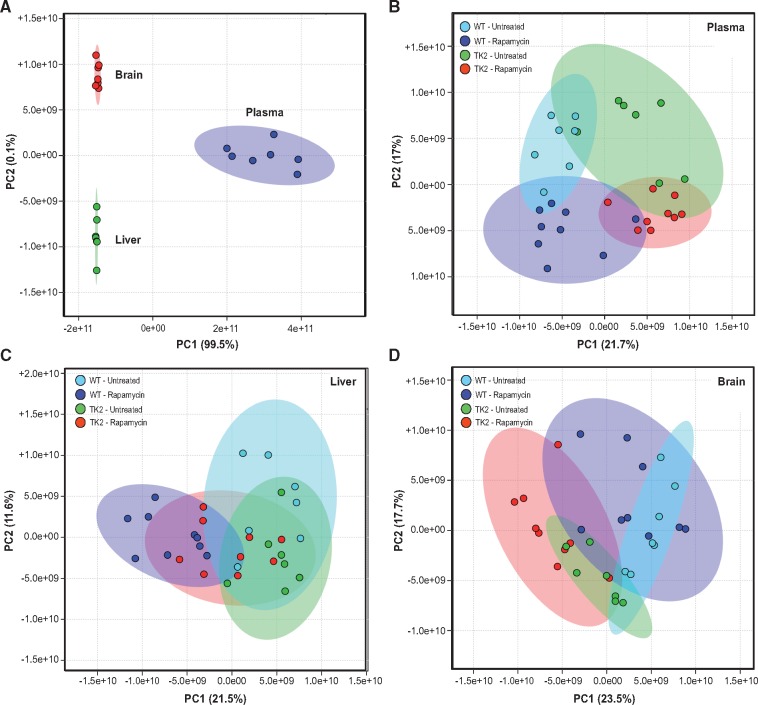
The metabolomics analysis included a panel of ∼130 assigned compounds from a total of >2000 detected peaks (3) (Supplementary Material, Data File S4). We analysed seven untreated TK2 animals plus 7 age-, gender- and treatment-matched WT littermates; and 9 treated TK2 animals plus 9 age-, gender- and treatment-matched WT littermates. Data analysis was performed using two complementary approaches (61). First, we conducted unsupervised analysis by principal component analysis (PCA) to assess the effect size of genotype and treatment, i.e. our ability to separate the 32 mice into the four known biological groups based solely on their metabolite values. Next, we expanded on the results of the unsupervised analysis by dividing the samples into non-overlapping datasets and conducting univariate analyses to identify differentially expressed metabolites.
After validating the ability of PCA to distinguish among the metabolic profiles of the three tissues analysed (Fig. 7A), we performed PCA on each tissue, using the 25–30 most significantly altered metabolites, as identified by ANOVA (analysis was also performed by hierarchical clustering, and yielded similar results [Supplementary Material, Fig. S8]). Plasma showed the greatest segregation of the 32 animals, forming four relatively distinct groups based on genotype and treatment (Fig. 7B). Liver segregated primarily based on common treatment, indicating a larger effect on liver metabolites of rapamycin than that of TK2 disease (Fig. 7D). We therefore conducted univariate analysis in liver and plasma to compare untreated vs treated WT mice, as well as untreated vs treated TK2 mice. Treatment-induced metabolite changes that were common to both genotypes (i.e. altered both in the WT ± rapa and TK2 ± rapa analyses) were deemed worthy of inclusion in a ‘rapamycin signature’ (Table 1; Supplementary Material, Fig. S9). The signature in liver (compounds in Table 1 highlighted in bold) included 15 compounds that were primarily intermediates of amino acid and carbohydrate metabolism, but also included compounds associated with nucleotide and fatty acid metabolism and with cofactor biosynthesis. The signature in plasma included nine compounds, namely intermediates of amino acid and carbohydrate metabolism, as well as β-oxidation and collagen biosynthesis. Notably, three metabolites (marked with asterisks in Table 1) were present, and altered in the same direction, in the signatures of both tissues: the amino acid metabolic intermediates 4-acetamidobutanoic acid (a GABA derivative; increased after rapa), trans-4-hydroxyproline (a hydroxylated form of the imino acid proline; increased after rapa), and L-pipecolic acid (an imino acid generated during lysine degradation; decreased after rapa). Of note, there were no significant changes seen in liver or plasma levels of glucose, lactate, pyruvate, and β-hydroxybutyrate (Supplementary Material, Data File S4).
Figure 7.
Unsupervised analysis of metabolomics data by principle component analysis. PCA was performed on all 32 mice in the 4 treatment groups: untreated WT, rapamycin-treated WT, untreated TK2, and rapamycin-treated TK2. (A) Positive control for PCA using ∼55 identified compounds common to the brain, liver and plasma of untreated WT mice (n = 7). (B–D) PCA performed on all 32 mice using the 25–30 most significantly altered compounds in plasma (B), liver (C), and brain (D). Colored ellipses represent 95% confidence range for the indicated biological group.
Table 1.
Univariate analysis of liver and plasma metabolites in rapamycin-treated vs untreated mice
| Liver | |||
|---|---|---|---|
| Metabolite | WT ± rapa(Fold change) | TK2 ± rapa(Fold change) | Pathway |
| L-glutamine | 1.3 | Amino acid metabolism | |
| L-methionine | 1.5 | ||
| N-acetyl-l-alanine | 1.3 | ||
| 5-oxo-l-proline | 1.5 | 1.6 | |
| Trans-4-hydroxyproline (∗) | 1.2 | 1.6 | |
| L-glutamic acid | 1.5 | 1.5 | |
| 4-acetamidobutanoic acid (∗) | 1.4 | 1.4 | |
| Ketoleucine | 1.7 | 1.4 | |
| 2-hydroxyglutarate | 1.4 | 1.4 | |
| Taurine | 0.7 | 0.8 | |
| S-(5'-adenosyl)-l-homocysteine | 0.6 | 0.6 | |
| L-pipecolic acid (∗) | 0.6 | 0.4 | |
| Guanidinoacetate | 0.6 | ||
| Hypotaurine | 0.3 | ||
| N-acetyl-d-galactosamine | 1.4 | 1.9 | Carbohydrate metabolism |
| Melibiose | 0.4 | ||
| D-(+)-raffinose | 0.4 | ||
| Palatinose | 0.4 | ||
| D-ribose | 0.6 | 0.6 | |
| Nicotinamide | 0.7 | Cofactor metabolism | |
| 6-carboxyhexanoate | 1.5 | ||
| 4-pyridoxate | 0.6 | ||
| 3-(4-hydroxyphenyl)lactate | 0.7 | ||
| Omega-hydroxydodecanoic acid | 1.5 | Fatty acid/lipid metabolism | |
| Ethylmalonic acid | 0.9 | 0.6 | |
| Glyoxylic acid | 1.6 | ||
| Myo-inositol | 0.8 | ||
| N-methyl-l-glutamate | 0.5 | Methane metabolism | |
| Uridine | 1.4 | Nucleotide metabolism | |
| Deoxycytidine | 2.3 | ||
| 5'-methylthioadenosine | 2.0 | ||
| Cytidine | 1.6 | ||
| Uracil | 0.7 | 0.7 | |
| Xanthine | 0.7 | ||
| Inosine | 0.7 | ||
| Adenosine | 0.6 | ||
| 2-hydroxybutyric acid | 0.8 | Oxidative stress | |
| Alpha-ketoglutaric acid | 2.0 | 2.0 | TCA cycle |
| L-fucose | 0.7 | 0.7 | |
| Plasma | |||
| Alpha-ketobutyrate | 1.4 | Amino acid metabolism | |
| N-acetylserine | 1.3 | ||
| 4-acetamidobutanoic acid (∗) | 1.5 | 1.7 | |
| Trans-4-hydroxyproline (∗) | 1.3 | 1.6 | |
| N-acetyl-l-alanine | 1.3 | 1.4 | |
| O-acetyl-l-serine | 1.3 | 1.3 | |
| L-pipecolic acid (∗) | 0.7 | 0.7 | |
| Ketoleucine | 0.8 | ||
| L-lysine | 0.7 | ||
| L-tryptophan | 0.6 | ||
| Phenylpyruvate | 0.3 | ||
| Propanoate | 0.8 | 0.8 | β-oxidation |
| Gluconic acid | 1.3 | Carbohydrate metabolism | |
| N-acetylneuraminic acid | 1.6 | 1.7 | |
| L-arabitol | 1.3 | 1.3 | |
| 4-hydroxybenzoate | 0.7 | Cofactor metabolism | |
| Pyridoxal | 0.7 | 0.7 | |
| β-Glycerophosphoric acid | 0.6 | Glycerolipid metabolism | |
| Creatinine | 1.2 | Muscle breakdown | |
| 2-amino-2-methylpropanoate | 1.4 | Nucleotide metabolism | |
| Inosine | 0.6 | ||
| Xanthine | 0.7 | ||
| Ophthalmic acid | 4.2 | Oxidative stress | |
Numbers indicate fold change of significantly altered metabolites (p>0.05); upregulated values (treated/untreated) shown in red, downregulated in blue. Metabolites are also shown as colored data points in Supplementary Material, Fig. S9. Blank boxes denote no significant change. Compounds highlighted in bold were altered significantly in both the WT±rapa and TK2±rapa columns. Asterisks indicate the compounds identified in both liver and plasma.
PCA in the brain separated the 32 mice into relatively distinct groups based on genotype, but with almost no segregation based on treatment, indicating that rapamycin had little effect on the metabolite signature of this tissue (Fig. 7C;Supplementary Material, Fig. S8A). As a result, we pooled the data from all WT and all TK2 mice and conducted univariate analysis to compare the metabolic profiles of the two genotypes. Significant changes were seen in intermediates of the TCA cycle and in nucleotide, amino acid, and fatty acid metabolism, as well as in markers of oxidative stress and cofactor biosynthesis (Supplementary Material, Table S1). The results in brain, while not pointing to a treatment effect of rapamycin, may nevertheless provide key insights for future studies into the pathogenesis of TK2 disease.
Discussion
This work presents the first evidence of the therapeutic benefit of rapamycin in a mouse model of mtDNA-driven mitochondrial disease. Administration of low-dose oral rapamycin to TK2 mice throughout life extended median and maximum survival by 60 and 35%, respectively. Our results complement previous studies of rapamycin therapy used to treat a mouse model of mitochondrial disease due to ablation of a nuclear-encoded OxPhos subunit gene, Ndufs4 (8). Importantly, we expand here on that finding by demonstrating the clinical efficacy of a significantly lower dose, which is relevant to patient applications because of the importance of minimizing side effects when administering rapamycin long term (62).
Remarkably, although this dosage was sufficient to extend lifespan dramatically, it produced no detectable improvement in fitness or in molecular measures of disease; in none of our assays did rapamycin reverse TK2 phenotypes to more closely resemble those in untreated WT animals, as all examined performance markers were unimproved by rapamycin. Specifically, the brains of TK2 animals exhibited a severe and progressive mtDNA depletion and respiratory chain dysfunction that was unaltered by therapy. Additionally, there were no apparent changes in the brain—the most affected organ in this mouse—in canonical rapamycin-associated pathways, including macroautophagy, mitophagy, mitochondrial biogenesis, lipid metabolism, or glycolytic energy production. This was not surprising, given that rapamycin at the dose used here could not be detected in the brain. However, in the liver, which retains relatively preserved mtDNA levels and no significant respiratory chain defects (16), there was a clear response to rapamycin treatment, including inhibition of translation and inhibition of mitochondrial biogenesis. We note that there were no differences observed between male and female mice with the exception of phospho-rpS6 [as has been reported previously (16)], and which likely relates to differences in constitutive rpS6 phosphorylation that become more apparent at low doses, such as those employed here). Because we saw no evidence of the absence of either mitophagy or altered mtDNA depletion, it is unlikely that selective degradation of dysfunctional or mtDNA-depleted mitochondria in the liver explains the lifespan extension. Similarly, we found little evidence in support of an immune- or apoptosis-mediated rescue mechanism.
The results of targeted analyses of brain and liver eliminated possible mechanisms by which rapamycin exerts its effects, but shed no light on the actual mechanism(s) involved. We considered the possibility that our inability to deduce the mechanism of rapamycin rescue was due to omission of other disease-critical target tissues. However, we considered this possibility to be unlikely, as previous studies of TK2 mice indicate that brain is the most severely affected tissue (16) and ought to demonstrate phenotypes associated with rescue. Furthermore, the other tissue demonstrating degenerative changes in TK2 disease histologically was skeletal muscle, which did not manifest respiratory deficiency, either with or without treatment, and showed no improvement in the degenerative phenotype upon treatment. We therefore turned to unbiased hypothesis-generating approaches, namely metabolomics and transcriptomics of both liver and brain, and uncovered several interesting consequences of rapamycin treatment in both tissues (even though low-dose rapamycin does not enter the brain).
Metabolomics analysis of brain revealed a high degree of similarity between genotype-matched brains, regardless of treatment. Combining this finding with the biochemical evidence showing unaltered hexokinase expression and AMPK activation with treatment, and with the transcriptomics analysis showing no difference in key metabolic processes, such as amino acid, lipid, and glucose metabolism, it seems unlikely that rapamycin extended TK2 survival by rescuing brain function through metabolic reprogramming. However, transcriptomics demonstrated significant differences between untreated WT and TK2 animals in neural developmental processes and in nucleic acid metabolism in the brain, which were also similarly perturbed when comparing treated and untreated mice. Altered developmental processes included cell-cell and intracellular signaling processes and cell cycle and transcriptional pathways. While rapamycin is known to inhibit cell cycle (7), the evidence from our metabolomics, HPLC, and expression analyses all imply that the drug did not penetrate this organ. It is therefore possible that the effects seen in the brains of rapamycin-treated animals represent systemic signaling changes induced by the drug.
Another surprising finding from the untargeted analysis came from the liver, which showed profound effects of rapamycin treatment. Transcriptomics revealed alterations in metabolic processes between untreated WT and TK2 animals, namely amino acid, nitrogen, and nucleic acid metabolism. There were even more marked metabolic pathway changes after rapamycin treatment, including fatty acid, lipid, steroid, and TCA metabolic gene expression, but notably not in oxidative stress pathway expression. Metabolomics similarly showed that rapamycin treatment had a larger effect on metabolic profile than did TK2 disease, and identified treatment-induced changes in amino acid, carbohydrate, fatty acid, cofactor, and nucleic acid metabolism, but not in key markers of mitochondrial function, such as glucose, lactate, pyruvate, and β-hydroxybutyrate. These results, taken together with the altered liver glycolytic and energy homeostasis markers, agree with previous studies finding that rapamycin exerts a significant metabolic effect on the liver in the setting of mitochondrial disease (8,35). This is also consistent with changes seen in plasma, including alterations in amino acid, carbohydrate, β-oxidation, and lipid metabolites. It is possible that systemic energy homeostasis, which is differentially regulated by rapamycin, is a key mediator of TK2 mouse survival through downstream signals on affected tissues. A targeted lipidomics analysis might shed light on these questions.
Of note, the results of the metabolomics analysis were concordant with those of transcriptomics. Specifically, both analyses identified amino acid, carbohydrate, and fatty acid metabolism as being perturbed by rapamycin treatment in liver, whereas these were not perturbed by rapamycin in the brain. By combining the information derived from both transcriptomics and metabolomics, we identified tissue-specific and systemic changes that may constitute a ‘rapamycin signature,’ thereby providing insight into potential mechanisms of TK2 rescue as well as putative future biomarkers of disease and treatment. Further work will be required to determine if this signature can be validated in humans treated with rapamycin.
Perhaps the most striking finding in this work was the demonstration of lifespan extension in the face of a failure to reverse the underlying phenotype of mitochondrial disease in general, and severe mtDNA depletion in particular. This result is all the more remarkable in the specific mouse model studied here, in which rapamycin apparently was unable to reach the most severely affected tissue, namely brain.
We note that this finding contrasts with the mitigation of neurological disease markers and progressive weight decline seen in rapamycin-treated Ndufs4−/− mice (8). It is possible that this discrepancy is due to differences in the disease model employed or to differences in the mode of rapamycin administration [our study administered ∼0.8 mg/kg orally, compared with 8 mg/kg given intraperitoneally (8)]. Supporting this conclusion, follow-up studies by the same group detected a striking decrease in weight of up to ∼50% in their Ndufs4−/− model after modifying dose and mode of administration, which agrees with numerous other literature reports (25). Variability in rapamycin’s effects on disease morbidity, including weight loss, represents a crucial revelation that requires further exploration prior to considering rapamycin therapy in human mitochondrial disease patients.
Our results thus support the possibility that rapamycin treatment may circumvent the OxPhos defect by tapping into alternative energy reserves, such as amino acids and lipids, a view supported by decreased weight and adiposity of all treated animals described in the literature (31,32). It is equally possible, however, that rapamycin exerts its effects indirectly, by inducing systemic signaling events through third-party molecules to modulate disease. One possible outcome of such signaling could be rapamycin-mediated developmental delay, consistent with the retardation in growth irrespective of the animals' genetic status and with the alteration of brain transcriptional pathways associated with development. In either case, from a therapeutic standpoint, our results support the possibility that even though the TK2 mice are destined to die by OxPhos deficiency, this outcome can be postponed significantly while other therapies [e.g. nucleoside supplementation in TK2 disease (63)] are being developed or employed, through administration of low-dose rapamycin, either alone or in combination with other treatment modalities.
Materials and Methods
Treatment and analysis of TK2 knock-in mice
The homozygous recessive Tk2-H126N knock-in model of mtDNA Depletion Syndrome (MDS) was generated and characterized as reported previously (16). All experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committee of the Columbia University Medical Center and were consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The mouse colony was housed and bred in accordance with international standards, including a 12-h/12-h light/dark cycle.
To generate homozygous Tk2 mice, heterozygous Tk2+/KI mice were crossed, and plugs were checked daily to mark date of conception. Upon detection of a plug, each litter was designated for rapamycin or control (vehicle) treatment to generate a ‘timed mating’ of vehicle- and rapamycin-treated TK2 pups and WT littermates for comparison. Rapamycin treatment was initiated by dissolving rapamycin (LC Laboratories, #R-5000) in 100% ethanol (Sigma) to a concentration of 8 mg/ml. This rapamycin stock was then diluted in sterilized drinking water (24) and administered to the pregnant dam at a final concentration of 8 µg/ml. Because mice are expected to consume approximately 10% of their body mass in water daily, the anticipated resulting dose to the dam was 0.8 mg/kg. Water containing rapamycin or ethanol vehicle was replaced every 48 h. At delivery, the rapamycin dose administered to the dam was increased to 40 µg/ml (dilution of a stock of 40 µg/ml). The resulting target dose delivered to the dam was 4 mg/kg, and blood concentrations were verified by HPLC to be ∼10 ng/ml. All untreated control litters were given water with equivalent concentrations (0.1%) of vehicle alone (100% ethanol).
To generate Kaplan-Meier curves, animals were monitored daily and sacrificed humanely immediately upon exhibiting extreme distress: inability to remain upright, labored breathing, or loss of >10% body weight within 24 h. All deaths were recorded in rapamycin-treated WT litters regardless of genotype, and no WT animal deaths were observed which could not be explained by extenuating circumstance.
All analyses of TK2 mouse phenotypes were performed on mouse ‘tetrads’ consisting of at least one untreated TK2 and one untreated WT littermate, and at least one rapamycin-treated TK2 pup and one rapamycin-treated WT littermate, all of the same age and gender-matched when possible. Mice were sacrificed when the untreated TK2 pups stopped gaining weight and reached the weight plateau, at ∼p15–18.
Necropsy and histological analysis of mouse tissues
For necropsy, mouse weight and general appearance were recorded at the time of sacrifice. Subdermal dorsal adipose tissue was inspected by making a dorsal incision immediately below the level of the scapula, then anteriorly peeling back dermal tissue to expose the skull, and photo-documenting adipose tissue. Brain was subsequently removed and weighed, sliced laterally, and placed in ∼20 volumes of 4% paraformaldehyde (PFA) in 1x phosphate-buffered solution (PBS) pH 7.6 at 4 °C for >48 h. Next, a ventral incision was made at the base of the diaphragm and along the sternum, and dermal tissue was removed to photo-document thoracic musculature and adiposity. General appearance of the thoracic and peritoneal cavities was recorded, along with appearance of bowels and stomach. We then removed and weighed the liver, spleen, kidneys, heart, and quadriceps muscle. Half of the muscle samples were frozen immediately by submerging in dry ice-chilled 2-methylbutane and stored at −80 °C. All remaining organs were fixed for >24 h in ∼20 volumes of 4% PFA/1× PBS at 4 °C. After 24 h, one-half of each liver was transferred to 20% sucrose for 24 h prior to freeze-embedding for Oil-Red-O staining. Remaining samples (except brain) were transferred to 70% ethanol/1× PBS for up to 1 week, until embedding and sectioning. All histology and specialized assays were performed at the Columbia University Medical Center‘s Molecular Pathology Shared Resource, as follows: brain, heart, liver, spleen, kidney, and muscle tissues for organ histology were paraffin-embedded, sectioned at 5 µm, and stained with hematoxylin and eosin (H&E); TUNEL staining was performed on PFA-fixed, paraffin-embedded brain, liver, and muscle sections; Oil-Red-O staining was performed on PFA/sucrose-equilibrated frozen liver sections; COX/SDH staining was performed on 5-µm flash-frozen muscle sections as described previously (64). Pathological analyses of histological samples were performed blinded as to sample identity.
Measurement of rapamycin concentrations
All rapamycin measurements in whole blood and tissues were performed as described (6).
mtDNA quantitation
Tissue levels of mtDNA were quantitated by quantitative real-time PCR using an ABI PRISM 7000 sequence detection system, as described previously (65), with the following modifications. The intercalating dye SYBR® Green was used as a fluorescent probe, and separate reactions were performed on each sample in triplicate using mtDNA-specific primers for murine COX I (Mtco1) (5'-TGCTAGCCGCAGGCATTACT-3'; 5'-CGGGATCAAAGAAAGTTGTGTTT-3') or nuclear DNA-specific primers for murine glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (5'-ACAGTCCATGCCATCACTGCC-3'; 5'-GCCTGCTTCACCACCTTCTTG-3'). Technical replicates were averaged and normalized to Gapdh. Ratios of mtDNA/nDNA were reported as a percentage of levels in untreated WT mice. Selected samples analysed in parallel by both qPCR and Southern blot demonstrated agreement between the two methods.
Rotating rod test
A rotating rod (adapted rotarod) performance test was used to assess and quantitate locomotor activity in mouse pups at p15 (i.e. following disease onset in TK2 mice, but prior to end-stage). Due to the age and exceptionally small size of both WT and mutant mice after rapamycin treatment, the traditional assay was adapted. A ∼1-cm diameter plexiglass 10-ml serological pipette (Sigma-Aldrich) was coated with one layer of Nalgene™ Super Versi-Dry™ Surface Protector (ThermoFischer Scientific), with the padded side facing outward to enhance traction. Mouse pups were placed upright in the center of the rod, which was held 5 cm above a cushioned surface, and rotated at 1 rotation/s for 30 s, or until the pup was unable to maintain grip and slipped to the cushioned surface. Each rotation event was filmed, and total time maintained on the rod (up to 30 s) was recorded.
Molecular analysis of mouse tissues
TK2 and littermate controls were sacrificed at early stage disease (p15–18), and ∼0.3 g tissue were immediately homogenized on ice in an isotonic and viscous mitochondrial extraction buffer (70 mM sucrose, 210 mM mannitol, 5 mM HEPES, and 1 mM EDTA, pH 7.2) using 15 strokes in a glass Dounce tissue grinder with a Teflon® pestle (Thomas Scientific). Aliquots of total lysate were saved for mtDNA and western blot analysis. To enrich for mitochondria, the remaining homogenate was centrifuged at 900×g for 10 min at 4 °C. Supernatant was transferred and re-centrifuged at 900×g for 10 min at 4 °C, followed by a final transfer and re-centrifugation at 4000×g for 10 min at 4 °C. Finally, supernatant was removed and discarded, and the resulting mitochondrial-enriched pellet was re-suspended in mitochondrial assay solution (70 mM sucrose, 220 mM mannitol, 5 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, and 0.2% fatty acid-free BSA, pH 7.4) to a concentration of approximately 10 µg/µl. The resulting mitochondrial enrichment sample was used to assess respiratory chain and CPT1 enzymatic activities, and for western blot analysis.
Respiratory chain activity
Respiratory chain activity was assessed in enriched mitochondrial fractions from brain or liver according to the electron flow assay using a Seahorse XFe24 Flux Analyzer (Agilent Technologies, Santa Clara, CA), as described (66). Brain and liver respiratory chain activity was performed in parallel on age-matched ‘tetrads’ as described above, with five technical replicates per animal. Each technical replicate contained either 5 µg (brain) or 50 µg (liver) of enriched mitochondrial fraction.
Western blotting
Western blotting was performed as described previously (4), with the following specifications, For all western blotting, 10–25 µg of whole tissue extract or mitochondrial enrichment fraction were prepared as described above, electrophoresed through a Novex™ 4–20% Tris-Glycine mini gel (ThermoFischer Scientific), transferred to Immun-Blot™ PVDF membranes (Biorad, Hercules, CA), and probed overnight. Antibodies included: AMPKα (#2532, Cell Signaling, Danvers, MA), phospho-AMPKα (Thr172) (40H9) (#2535, Cell Signaling), BCL2L13 (16612-1-AP, Proteintech, Chicago, IL, USA), β-actin monoclonal AC-15 clone (A5441, Sigma-Aldrich), glucokinase (ab37796, abcam, Cambridge, U.K.), hexokinase (AB3279, EMD Millipore, Darmstadt, Germany), LC3B (ab48394, abcam), phospho-S6 Ribosomal Protein (Ser240/244) (#2215, Cell Signaling), S6 Ribosomal Protein (54D2) (#2317, Cell Signaling), TFAM (ab131607, abcam), TOM20 FL-145 (SC-11415, Santa Cruz Biotechnology, Dallas, TX), and Total Rodent OXPHOS antibody cocktail (ab110413, abcam). Signal was detected using SuperSignal® West Dura Extended Duration Substrate (34076, Thermo Scientific) and a KwikQuant Digital Western System (Kindle Biosciences LLC, Greenwich, CT). Quantification of proteins was carried out using NIH ImageJ 1.46r software (Rasband, W.S., ImageJ, U.S. National Institutes of Health, Bethesda, MD, http://imagej.nih.gov/ij/). Average gray value after lane background subtraction was normalized to loading control (β-actin for whole-tissue lysates, TOM20 for mitochondrial enrichment), and reported as a percentage of untreated WT signal.
RNA expression analysis by quantitative reverse transcription
Total tissue RNA was isolated by TRIzol® (ThermoFischer Scientific) extraction followed by treatment with RNAse-free DNAse (Qiagen) and subsequent purification with a RNeasy kit (Qiagen). For expression analysis by quantitative reverse-transcriptase PCR (qRT-PCR), cDNA synthesis and qPCR analysis was conducted as described previously (18) with specific murine primers and Taqman® probes (Applied Biosystems, Invitrogen) for mouse Acaca (Mm01304257_m1), Dgat2 (Mm00499536_m1), Fasn (Mm00662319_m1), Lpl (#Mm00434764_m1), Ppargc1a (PGC-1α; #Mm01208835_m1), and mouse Gapdh endogenous control [VIC®/MGB probe #4352339E (all probes from ThermoFischer Scientific)].
RNA transcriptome analysis by RNA-seq
RNA transcriptome analysis was performed on brain and liver for four sample groups (untreated WT, rapamycin-treated WT, untreated TK2, and rapamycin-treated TK2) each with 3 biological replicates. Tissues were isolated from 15-day-old female TK2 pups and age- and treatment-matched WT littermate controls. RNA was extracted as above, and quality control was performed using an Agilent Bioanalyzer 2100 (Agilent Technologies) through the Columbia University Medical Center’s Molecular Pathology Shared Resource; subsequent analysis was limited to samples with RNA integrity numbers of 8.0–10.0.
RNA sequencing was performed at the Columbia University Sulzberger Genome Center. In brief, we used poly-A pull-down to enrich mRNAs from total RNA samples (∼200 ng per sample) and proceeded with library preparation using the TruSeq RNA Prep kit (Illumina). Libraries were sequenced using an Illumina HiSeq2500/HiSeq4000. Samples were multiplexed in each lane, yielding targeted number (30 million) of single-end 100-bp reads per sample, as a fraction of 280–400 million reads for the whole lane.
We used RTA (Illumina) for base calling and bcl2fastq2 (version 2.17) for converting BCL to fastq format, coupled with adaptor trimming. We mapped the reads to the mouse reference genome (UCSC/mm10) using STAR (2.5.2 b) and featureCounts (v1.5.0-p3).
Differentially expressed gene (DEG) analysis was conducted in parallel, using two Bioconductor packages, DESeq2 and edgeR, as described previously (51,67). In brief, raw gene counts were filtered according to 1 count per million in at least two samples, normalized using the empirical Bayes method, and DEGs were identified using the Benjamini-Hochberg correction for multiple testing, with a false discovery rate (FDR) <0.05 according to a threshold log2-transformed fold-change of 1.2. In the brain, DESeq2 identified a total of 725, 407, and 1334 DEGs for the untreated WT vs TK2, rapa-treated vs untreated WT, and rapa-treated vs untreated TK2 groups, respectively; for the brain, DESeq2 identified 1477, 902, and 1924 DEGs. For the same comparisons, EdgeR identified 489, 670, and 1530 DEGs in the brain and 167, 1733, and 210 DEGs in the liver.
Metabolomics analysis
Metabolomics analysis was performed as described previously (3) on 32 animals representing four distinct sample groups: seven untreated TK2 mice together with seven gender-matched WT littermates (also untreated), and nine rapamycin-treated TK2 mice together with nine gender-matched littermates (also rapamycin-treated). All animals were sacrificed when 15 days old. Blood was collected immediately following sacrifice and anticoagulated with sterile ethylenediamine tetraacetic acid (EDTA) (ThermoFischer Scientific), pH 8.0 to a concentration of 100 µM, then centrifuged at 2000×g for 15 min at 4 °C. Supernatant (plasma) was transferred to a new tube and stored at −80 °C until further analysis. Intact liver and brain were isolated, divided into two equal sections (for brain, divided into left and right hemispheres) and immediately frozen and stored at −80 °C until further analysis.
Metabolite extraction of plasma was performed as described previously (68). Briefly, 25 μl plasma was mixed with 250 μl ice-cold acetonitrile: methanol (75%/ 25% v/v). The solution was vortexed for 15 s, placed on ice for 30 min and then centrifuged for 20 min at 4 °C at maximum speed (i.e. ∼16, 000×g). Supernatant (100 μl) was transferred to an LC-MS glass vial. For brain and liver, samples were cut from frozen tissue sections and ∼ 50–100 mg was placed in 2 ml safe-lock Eppendorf tube and 1.5 ml dry ice-cooled methanol: H2O (80%/20% v/v) was added. Samples were then homogenized in a Qiagen TissueLyzer II (2 × 1 min at 30 Hz) and then centrifuged for 20 min at 4 °C at maximum speed. An aliquot of the supernatant (750 μl) was transferred to a new 2-ml tube and 1250 μl water were added. After vortexing, samples were frozen at −80 °C and lyophilized overnight. The remaining pellet was dissolved in a volume of acetonitrile: H2O (70%/30% v/v) proportional to 3 times the initial mass of the sample and transferred to an LC-MS vial; for example, a 50-mg tissue sample was extracted and dissolved into 150 μl of 70% acetonitrile. LC-MS data collection and analysis were performed as described previously (68). Briefly, negative mode data were collected using a Q Exactive Plus Orbitrap Mass Spectrometer coupled to a Dionex Ultimate 3000 UHPLC system (ThermoFisher Scientific) with a Xbridge Amide HILIC 100×2.1 mm, 2.5-μm particle size column (Waters, 186006091). Mobile phase A was 20 mM ammonium acetate, 0.25% ammonium hydroxide pH 9; mobile phase B was 100% acetonitrile. The flow rate was 220 µl/min and the elution gradient was from 85% B to 2% B over 11 min followed by re-equilibration to 85% B. Full scan data were collected in a range of 70–1000 m/z, with the resolution set at 140 000, the AGC target at 3E6, and the maximum injection time at 400 ms. Progenesis QI software (Waters) was used to perform peak picking, peak identification, deconvolution, and intensity integration and normalization. Using metabolite reference standards purchased from IROA Technologies (Bolton, MA), an in-house metabolite retention time library of 520 reference standards was established. Metabolite identification in Progenesis was based on mass tolerance of 5 ppm and retention time tolerance of 0.3 min. For each tissue, of the >2000 peaks detected, ∼100 compounds were identified and assigned.
For each tissue, we analysed data using several complementary approaches, via the comprehensive, web browser-based bioinformatics tool MetaboAnalyst 3.0, designed for metabolomics data processing, statistical analysis, and functional interpretation (69). Analysis was performed as described previously (70), with the following specifications. In brief, we first performed unsupervised exploratory analysis using both principle component analysis (PCA) and hierarchical clustering. Normalized peak intensities were variance-filtered and mean-centered, then analysis was conducted in each tissue using the 25–30 most significantly altered metabolites as identified by parametric ANOVA (25 compounds for brain, 30 for liver and plasma). Results of the PCA were reported as the score plot for the two principle components explaining the greatest variance, with 95% confidence regions indicated for each biological group. Hierarchical clustering was performed using Euclidian distance measures and the Ward clustering algorithm, and results were displayed as a dendrogram with heatmap displaying auto-scaled features in red/yellow. Validations for unsupervised analyses were performed with all ∼100 identified compounds from the seven untreated WT samples from each tissue.
We next divided the tissues into non-overlapping datasets, based on the results of the clustering analysis, to conduct univariate analysis. For the brain, there was one comparison: pooled untreated and rapamycin-treated WT vs pooled untreated and rapamycin-treated TK2 mice. For liver and plasma, we compared untreated to rapamycin-treated WT mice, and also untreated to rapamycin-treated TK2 mice. To confirm Gaussian distribution of data, we generated normal probability plots for randomly selected metabolites from each sample group (genotype + treatment combination). We then calculated fold-difference and adjusted P-values using Welch’s t-test adjusted for multiple testing using the Benjamini-Hochberg approach at a FDR, Q, of 10% (71). Metabolites of interest were defined as those with a fold change of at least 20% between groups and significant at a 95% confidence level, with P-values adjusted by the Benjamini-Hochberg method for a 10% false discovery rate (72).
Statistics
Unless otherwise indicated, all quantitations, including those of Western blots and qPCR data, are expressed as average values of at least three biological replicates, normalized to untreated WT and expressed as a % of WT, with error bars representing standard deviation. Significance was established using a two-sided Welch’s t-test, and a P-value of α < 0.05 was considered to be statistically significant. To determine significance of survival data represented by the Kaplan–Meier curve, the log-rank test was used to compare the survival of rapamycin-treated vs vehicle-treated mice, and a log-rank statistic <0.05 was considered to be statistically significant. Because statistical analysis is integral to the RNA-expression and metabolite profile analyses, all relevant testing is listed under the section for metabolomics analysis.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Drs Brad Siegmund, John Summerville, Kurenai Tanji, and Katherine Xu for assistance with interpretation of the histology, Dr. Xiaoyun Sun for assistance with transcriptomics analyses, Drs Ali Naini and Jiesheng Lu for help with Southern blotting, and Drs Orhan Akman, Estela Area-Gomez, Robert Gilkerson, Vernice Jackson-Lewis, Delfina Larrea, Carlos Lopez-Gomez, Marta Pera, Liza Pon, and Serge Przedborski for helpful advice and comments.
Conflict of Interest statement. None declared.
Funding
Marriott Mitochondrial Disorders Clinical Research Network (M.H., V.M., E.A.S., and R.S.), U.S. National Institutes of Health (P01-HD080642 to E.A.S. and M.H.; F30-NS093798 to S.S.; P30-AG013319-20 and 5U01-AG022307-09 to M.J.), US Department of Defense (W911F-15-1-0169 to E.A.S.), and Arturo Estopinan TK2 Research Fund (to M.H.). V.M. is an investigator of the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the U.S. National Institutes of Health and by the Marriott Mitochondrial Disorders Clinical Research Network.
References
- 1. Alston C.L., Rocha M.C., Lax N.Z., Turnbull D.M., Taylor R.W. (2017) The genetics and pathology of mitochondrial disease. J. Pathol., 241, 236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrari M., Jain I.H., Goldberger O., Rezoagli E., Thoonen R., Chen K.-H., Sosnovik D.E., Scherrer-Crosbie M., Mootha V.K., Zapol W.M. (2017) Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl. Acad. Sci. USA, 114, E4241–E4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jain I.H., Zazzeron L., Goli R., Alexa K., Schatzman-Bone S., Dhillon H., Goldberger O., Peng J., Shalem O., Sanjana N.E.. et al. (2016) Hypoxia as a therapy for mitochondrial disease. Science, 352, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garone C., Garcia-Diaz B., Emmanuele V., Lopez L.C., Tadesse S., Akman H.O., Tanji K., Quinzii C.M., Hirano M. (2014) Deoxypyrimidine monophosphate bypass therapy for thymidine kinase 2 deficiency. EMBO Mol. Med., 6, 1016–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halter J.P., Michael W., Schupbach M., Mandel H., Casali C., Orchard K., Collin M., Valcarcel D., Rovelli A., Filosto M.. et al. (2015) Allogeneic haematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Brain, 138, 2847–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrison D.E., Strong R., Sharp Z.D., Nelson J.F., Astle C.M., Flurkey K., Nadon N.L., Wilkinson J.E., Frenkel K., Carter C.S.. et al. (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature, 460, 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaur A., Sharma S. (2017) Mammalian target of rapamycin (mTOR) as a potential therapeutic target in various diseases. Inflammopharmacology, 25, 293–312. [DOI] [PubMed] [Google Scholar]
- 8. Johnson S.C., Yanos M.E., Kayser E.B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V.Z., Gagnidze A.. et al. (2013) mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science, 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kruse S.E., Watt W.C., Marcinek D.J., Kapur R.P., Schenkman K.A., Palmiter R.D. (2008) Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab., 7, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dai Y., Zheng K., Clark J., Swerdlow R.H., Pulst S.M., Sutton J.P., Shinobu L.A., Simon D.K. (2014) Rapamycin drives selection against a pathogenic heteroplasmic mitochondrial DNA mutation. Hum. Mol. Genet., 23, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kauppila J.H., Baines H.L., Bratic A., Simard M.L., Freyer C., Mourier A., Stamp C., Filograna R., Larsson N.G., Greaves L.C.. et al. (2016) A phenotype-driven approach to generate mouse models with pathogenic mtDNA mutations causing mitochondrial disease. Cell Rep., 16, 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan W., Waymire K.G., Narula N., Li P., Rocher C., Coskun P.E., Vannan M.A., Narula J., Macgregor G.R., Wallace D.C. (2008) A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science, 319, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inoue K., Nakada K., Ogura A., Isobe K., Goto Y., Nonaka I., Hayashi J.I. (2000) Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat. Genet., 26, 176–181. [DOI] [PubMed] [Google Scholar]
- 14. Gammage P.A., Gaude E., Van Haute L., Rebelo-Guiomar P., Jackson C.B., Rorbach J., Pekalski M.L., Robinson A.J., Charpentier M., Concordet J.-P.. et al. (2016) Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucl. Acids Res., 44, 7804–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reddy P., Ocampo A., Suzuki K., Luo J., Bacman S.R., Williams S.L., Sugawara A., Okamura D., Tsunekawa Y., Wu J.. et al. (2015) Selective elimination of mitochondrial mutations in the germline by genome editing. Cell, 161, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akman H.O., Dorado B., López L.C., García-Cazorla Á., Vilà M.R., Tanabe L.M., Dauer W.T., Bonilla E., Tanji K., Hirano M. (2008) Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum. Mol. Genet., 17, 2433–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Hattab A.W., Craigen W.J., Scaglia F. (2017) Mitochondrial DNA maintenance defects. Biochim. Biophys. Acta, 1863, 1539–1555. [DOI] [PubMed] [Google Scholar]
- 18. Dorado B., Area E., Akman H.O., Hirano M. (2011) Onset and organ specificity of Tk2 deficiency depends on Tk1 down-regulation and transcriptional compensation. Hum. Mol. Genet., 20, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chanprasert S., Wang J., Weng S.W., Enns G.M., Boue D.R., Wong B.L., Mendell J.R., Perry D.A., Sahenk Z., Craigen W.J.. et al. (2013) Molecular and clinical characterization of the myopathic form of mitochondrial DNA depletion syndrome caused by mutations in the thymidine kinase (TK2) gene. Mol. Genet. Metab., 110, 153–161. [DOI] [PubMed] [Google Scholar]
- 20. Faehling M., Wienhausen-Wilke V., Fallscheer S., Trinajstic-Schulz B., Weber J., Leschke M. (2015) Long-term stable lung function and second uncomplicated pregnancy on sirolimus in lymphangioleiomyomatosis (LAM). Sarcoidosis Vasc. Diffuse Lung Dis., 32, 259–264. [PubMed] [Google Scholar]
- 21. Framarino dei Malatesta M., Corona L.E., De Luca L., Rocca B., Manzia T.M., Orlando G., Tisone G., Iaria G. (2011) Successful pregnancy in a living-related kidney transplant recipient who received sirolimus throughout the whole gestation. Transplantation, 91, e69–e71. [DOI] [PubMed] [Google Scholar]
- 22. Anderl S., Freeland M., Kwiatkowski D.J., Goto J. (2011) Therapeutic value of prenatal rapamycin treatment in a mouse brain model of tuberous sclerosis complex. Hum. Mol. Genet., 20, 4597–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villa-Cuesta E., Holmbeck M.A., Rand D.M. (2014) Rapamycin increases mitochondrial efficiency by mtDNA-dependent reprogramming of mitochondrial metabolism in Drosophila. J. Cell Sci., 127, 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komarova E.A., Antoch M.P., Novototskaya L.R., Chernova O.B., Paszkiewicz G., Leontieva O.V., Blagosklonny M.V., Gudkov A.V. (2012) Rapamycin extends lifespan and delays tumorigenesis in heterozygous p53+/- mice. Aging, 4, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson S.C., Yanos M.E., Bitto A., Castanza A., Gagnidze A., Gonzalez B., Gupta K., Hui J., Jarvie C., Johnson B.M.. et al. (2015) Dose-dependent effects of mTOR inhibition on weight and mitochondrial disease in mice. Front. Genet., 6, 247.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y., Bokov A., Gelfond J., Soto V., Ikeno Y., Hubbard G., Diaz V., Sloane L., Maslin K., Treaster S.. et al. (2014) Rapamycin extends life and health in C57BL/6 mice. J. Gerontol. A, 69, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadaczek P., Beyer J., Kells A., Narrow W., Bowers W., Federoff H.J., Forsayeth J., Bankiewicz K.S., Borlongan C.V. (2011) Evaluation of an AAV2-based rapamycin-regulated glial cell line-derived neurotrophic factor (GDNF) expression vector system. PLOS One, 6, e27728.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banerjee S., Gianino S.M., Gao F., Christians U., Gutmann D.H. (2011) Interpreting mammalian target of rapamycin and cell growth inhibition in a genetically engineered mouse model of Nf1-deficient astrocytes. Mol. Cancer Ther., 10, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leontieva O.V., Paszkiewicz G.M., Blagosklonny M.V. (2012) Mechanistic or mammalian target of rapamycin (mTOR) may determine robustness in young male mice at the cost of accelerated aging. Aging, 4, 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drake J.C., Peelor F.F. 3rd, Biela L.M., Watkins M.K., Miller R.A., Hamilton K.L., Miller B.F. (2013) Assessment of mitochondrial biogenesis and mTORC1 signaling during chronic rapamycin feeding in male and female mice. J. Gerontol. A Biol. Sci. Med. Sci., 68, 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cai H., Dong L., Liu F. (2016) Recent advances in adipose mTOR signaling and function: Therapeutic prospects. Trends Pharmacol. Sci., 37, 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houde V.P., Brûlé S., Festuccia W.T., Blanchard P.-G., Bellmann K., Deshaies Y., Marette A. (2010) Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes, 59, 1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye L., Widlund A.L., Sims C.A., Lamming D.W., Guan Y., Davis J.G., Sabatini D.M., Harrison D.E., Vang O., Baur J.A. (2013) Rapamycin doses sufficient to extend lifespan do not compromise muscle mitochondrial content or endurance. Aging, 5, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. (2007) mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature, 450, 736–740. [DOI] [PubMed] [Google Scholar]
- 35. Fok W.C., Bokov A., Gelfond J., Yu Z., Zhang Y., Doderer M., Chen Y., Javors M., Wood W.H., Zhang Y.. et al. (2014) Combined treatment of rapamycin and dietary restriction has a larger effect on the transcriptome and metabolome of liver. Aging Cell, 13, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schieke S.M., Phillips D., McCoy J.P. Jr., Aponte A.M., Shen R.F., Balaban R.S., Finkel T. (2006) The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem., 281, 27643–27652. [DOI] [PubMed] [Google Scholar]
- 37. Tang Y., Schon E.A., Wilichowski E., Vazquez-Memije M.E., Davidson E., King M.P. (2000) Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol. Biol. Cell, 11, 1471–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palavra F., Robalo C., Reis F. (2017) Recent advances and challenges of mTOR inhibitors use in the treatment of patients with tuberous sclerosis complex. Oxid. Med. Cell Longev., 2017, 9820181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laplante M., Sabatini D.M. (2013) Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci., 126, 1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A.. et al. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy, 4, 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murakawa T., Yamaguchi O., Hashimoto A., Hikoso S., Takeda T., Oka T., Yasui H., Ueda H., Akazawa Y., Nakayama H.. et al. (2015) Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun., 6, 7527.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morán M., Delmiro A., Blázquez A., Ugalde C., Arenas J., Martín M.A. (2014) Bulk autophagy, but not mitophagy, is increased in cellular model of mitochondrial disease. Biochim. Biophys. Acta, 1842, 1059–1070. [DOI] [PubMed] [Google Scholar]
- 43. Matschinsky F.M. (2005) Glucokinase, glucose homeostasis, and diabetes mellitus. Curr. Diabetes Rep., 5, 171–176. [DOI] [PubMed] [Google Scholar]
- 44. Dobrzyn P., Dobrzyn A., Miyazaki M., Cohen P., Asilmaz E., Hardie D.G., Friedman J.M., Ntambi J.M. (2004) Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc. Natl. Acad. Sci. USA, 101, 6409–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carling D. (2017) AMPK signalling in health and disease. Curr. Opin. Cell Biol., 45, 31–37. [DOI] [PubMed] [Google Scholar]
- 46. Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., Hardie D.G. (1996) Characterization of the AMP-activated protein kinase kinase from rat liver and identification of Threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem., 271, 27879–27887. [DOI] [PubMed] [Google Scholar]
- 47. Jeon S.M. (2016) Regulation and function of AMPK in physiology and diseases. Exp. Mol. Med., 48, e245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rodenburg R.J.T. (2011) Biochemical diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis., 34, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim C., Potluri P., Khalil A., Gaut D., McManus M., Compton S., Wallace D.C., Yadava N. N. (2017) An X-chromosome linked mouse model (Ndufa1S55A) for systemic partial Complex I deficiency for studying predisposition to neurodegeneration and other diseases. Neurochem. Int., pii: S0197-0186(17)30060-8. [DOI] [PubMed] [Google Scholar]
- 50. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson M.D., Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol., 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCarthy D.J., Chen Y., Smyth G.K. (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucl. Acids Res., 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robinson M.D., McCarthy D.J., Smyth G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res., 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc., 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. (2017) PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res., 45, D183–d189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schurch N.J., Schofield P., Gierlinski M., Cole C., Sherstnev A., Singh V., Wrobel N., Gharbi K., Simpson G.G., Owen-Hughes T.. et al. (2016) How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA, 22, 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yendrek C.R., Ainsworth E.A., Thimmapuram J. (2012) The bench scientist's guide to statistical analysis of RNA-Seq data. BMC Res. Notes, 5, 506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X.. et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol., 17, 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vinaixa M., Samino S., Saez I., Duran J., Guinovart J.J., Yanes O. (2012) A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites, 2, 775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson S.C., Kaeberlein M. (2016) Rapamycin in aging and disease: maximizing efficacy while minimizing side effects. Oncotarget, 7, 44876–44878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lopez-Gomez C., Levy R.J., Sanchez-Quintero M.J., Juanola-Falgarona M., Barca E., Garcia-Diaz B., Tadesse S., Garone C., Hirano M. (2017) Deoxycytidine and deoxythymidine treatment for thymidine kinase 2 deficiency. Ann. Neurol., 81, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ross J.M. (2011) Visualization of mitochondrial respiratory function using cytochrome C oxidase/succinate dehydrogenase (COX/SDH) double-labeling histochemistry. J. Vis. Exp., e3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Timken M.D., Swango K.L., Orrego C., Buoncristiani M.R. (2005) A duplex real-time qPCR assay for the quantification of human nuclear and mitochondrial DNA in forensic samples: implications for quantifying DNA in degraded samples. J. Forensic Sci., 50, 1044–1060. [PubMed] [Google Scholar]
- 66. Rogers G.W., Brand M.D., Petrosyan S., Ashok D., Elorza A.A., Ferrick D.A., Murphy A.N., Kowaltowski A.J. (2011) High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLOS One, 6, e21746.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen Y., McCarthy D., Robinson M., Smyth G.K. (2011) EdgeR: differential expression analysis of digital gene expression data. User‘s Guide. [http://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf; date last accessed June, 2017].
- 68. Titov D.V., Cracan V., Goodman R.P., Peng J., Grabarek Z., Mootha V.K. (2016) Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science, 352, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia J., Psychogios N., Young N., Wishart D.S. (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucl. Acids Res., 37, W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xia J., Wishart D.S. (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protocols, 6, 743–760. [DOI] [PubMed] [Google Scholar]
- 71. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B, 57, 289–300. [Google Scholar]
- 72. Noble W.S. (2009) How does multiple testing correction work?. Nat. Biotechnol., 27, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.