Abstract
Smoking-associated DNA hypomethylation has been observed in blood cells and linked to lung cancer risk. However, its cause and mechanistic relationship to lung cancer remain unclear. We studied the association between tobacco smoking and epigenome-wide methylation in non-tumor lung (NTL) tissue from 237 lung cancer cases in the Environment And Genetics in Lung cancer Etiology study, using the Infinium HumanMethylation450 BeadChip. We identified seven smoking-associated hypomethylated CpGs (P < 1.0 × 10−7), which were replicated in NTL data from The Cancer Genome Atlas. Five of these loci were previously reported as hypomethylated in smokers’ blood, suggesting that blood-based biomarkers can reflect changes in the target tissue for these loci. Four CpGs border sequences carrying aryl hydrocarbon receptor binding sites and enhancer-specific histone modifications in primary alveolar epithelium and A549 lung adenocarcinoma cells. A549 cell exposure to cigarette smoke condensate increased these enhancer marks significantly and stimulated expression of predicted target xenobiotic response-related genes AHRR (P = 1.13 × 10−62) and CYP1B1 (P < 2.49 × 10−61). Expression of both genes was linked to smoking-related transversion mutations in lung tumors. Thus, smoking-associated hypomethylation may be a consequence of enhancer activation, revealing environmentally-induced regulatory elements implicated in lung carcinogenesis.
Introduction
Alterations in DNA methylation, an epigenetic modification, are common in human malignancies (1). Environmental factors such as tobacco smoke can modulate the establishment and maintenance of DNA methylation, and could thereby influence disease through complex mechanisms (2,3). Altered genome-wide DNA methylation patterns associated with tobacco smoking have been reported in studies of genomic DNA derived from blood (4–26), buccal cells (27), and lung macrophages (14). These observed DNA methylation changes, most of which are hypomethylation events, show variable permanence; some alterations persist for many years while others wane rapidly after smoking cessation (4,7,8,10,11,17,24–26,28,29). Blood-based DNA methylation changes can thus be used as biomarkers of tobacco smoke exposure and history (8,13,21). Importantly, an association between smoking-related hypomethylation in blood and lung cancer risk has been reported (30,31). The most consistently replicated smoking-related DNA methylation change across previous studies is at the cg05575921 CpG probe, located in the third intron of the aryl hydrocarbon receptor repressor gene (AHRR). An inverse correlation between smoking-associated methylation and AHRR expression in human lung tissue was previously reported for two other CpGs in the AHRR gene, one of which is very near cg05575921 (9). The mechanisms by which these hypomethylation events arise and how they might increase lung cancer risk remain unclear.
To address this issue, we profiled genome-wide DNA methylation in histologically normal lung tissues from 237 lung cancer patients in the Environment and Genetics in Lung cancer Etiology (EAGLE) study (32) and evaluated the association of DNA methylation with cigarette smoking status and other quantitative measures of tobacco smoking. We then sought to replicate our findings using 60 histologically normal lung samples from The Cancer Genome Atlas (TCGA) (33). To gain insight into the functional significance of observed DNA methylation changes, we integrated the DNA methylation data with epigenomic profiles of purified primary alveolar epithelial cells and the widely-studied A549 lung adenocarcinoma cell line and investigated the functional elements we identified.
Results
Identification of DNA methylation alterations associated with smoking
Our experimental data generation and analyses are summarized in a flow chart in Supplementary Material, Figure S1. We used the Illumina Infinium HumanMethylation450 BeadChip array to profile genome-wide DNA methylation in histologically normal lung tissues (distant from the tumor) from the Environment And Genetics in Lung cancer Etiology (EAGLE) study (32,34). We opted not to use tumor samples to minimize potential confounding effects due to the pronounced DNA methylation changes associated with the tumorigenic process (1,35). We included non-tumor lung (NTL) samples from 237 lung cancer patients (121 current cigarette smokers and 116 current nonsmokers of which 106 former and 10 never smokers, Supplementary Material, Table S1). Seventy-five percent of former smokers had quit smoking for ≥10 years before sample collection. Following removal of repetitive and SNP-containing probes and those on sex chromosomes, we examined the association of log-transformed DNA methylation (338,456 probes) with current cigarette smoking status using linear regression models adjusted for age, sex, body mass index, and smoking tobacco types other than conventional cigarettes. Adjusting for the top three principal components based on methylation distribution did not materially change the results, excluding the potential effect of hidden confounding factors. A quantile–quantile plot revealed little evidence for global inflation of the test statistics as compared to the expected distribution (λ = 1.108, after adjustment for age, sex and principal components, Supplementary Material, Fig. S2A). After Bonferroni correction, eight CpG probes were significantly hypomethylated in NTL tissues of current smokers (P < 0.05/338,456 = 1.48 × 10−7Table 1, Supplementary Materials, Table S2, Fig. S2B).
Table 1.
Association between tobacco smoking measures and methylation in the EAGLE and TCGA studies
| EAGLE |
TCGA (Validation) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probea | Chr | MapInfo (bp)b | UCSC gene | Current smoking status (yes vs. no, n = 237) P-valuec | Average DNA methylation beta value current smokers | Average DNA methylation beta value non-smokers | Average difference beta value, current vs. non-smokers | Average % difference beta value, current vs. non-smokers | Current smoking status (yes vs. no, n = 60) P-valuec | Average DNA methylation beta value current smokers | Average DNA methylation beta value non-smokers | Average difference beta value, current vs. non-smokers | Average % difference beta value, current vs. non-smokers |
| cg17113147 | 17 | 753545 | NXN | 2.24 × 10−11 | 0.83 | 0.88 | −0.05 | −5.33 | 0.0044 | 0.78 | 0.83 | −0.05 | −6.16 |
| cg05575921 | 5 | 373378 | AHRR | 2.41 × 10−10 | 0.71 | 0.77 | −0.07 | −8.79 | 0.0069 | 0.63 | 0.70 | −0.07 | −9.69 |
| cg07992500 | 2 | 37896583 | CDC42EP3 | 5.29 × 10−10 | 0.68 | 0.74 | −0.06 | −8.57 | 0.040 | 0.60 | 0.65 | −0.05 | −7.75 |
| cg14120703 | 9 | 139416102 | NOTCH1 | 1.11 × 10−9 | 0.51 | 0.57 | −0.06 | −10.34 | 0.0053 | 0.44 | 0.50 | −0.06 | −11.84 |
| cg11152412 | 15 | 74927688 | EDC3 | 1.98 × 10−8 | 0.17 | 0.19 | −0.03 | −13.49 | 0.00033 | 0.15 | 0.19 | −0.03 | −17.64 |
| cg03224163 | 7 | 139420300 | HIPK2 | 3.07 × 10−8 | 0.78 | 0.82 | −0.05 | −5.51 | 0.0052 | 0.68 | 0.72 | −0.04 | −5.07 |
| cg15912732 | 14 | 105255285 | AKT1 | 6.90 × 10−8 | 0.82 | 0.86 | −0.04 | −4.88 | 0.495 | 0.79 | 0.81 | −0.02 | −3.04 |
| cg13787850 | 9 | 102195951 | – | 1.07 × 10−7 | 0.18 | 0.23 | −0.04 | −19.00 | 0.0053 | 0.17 | 0.21 | −0.04 | −17.23 |
Probes in italics have been observed to be hypomethylated in smokers’ blood cell DNA.
hg19 coordinates.
Association was assessed using linear regression models adjusted for age, sex, body mass index, and non-cigarette smoking. P-values were two-sided for the analysis with EAGLE as the discovery sample, and one-sided for the analysis using TCGA as the replication sample.
In sensitivity analyses, excluding never-smokers, subjects who smoked other types of tobacco, subjects with prior chemotherapy, or additionally adjusting for histology and stage of paired tumor tissues yielded similar findings as in the primary analyses (Supplementary Material, Table S3). Methylation of all eight CpGs was associated with smoking duration (Supplementary Material, Table S2). While methylation was inversely associated with smoking duration and pack-years, increased DNA methylation was positively associated with years after smoking cessation (P < 0.05, Supplementary Material, Table S2). We also evaluated time to first cigarette after waking (TTFC) with DNA methylation of significant probes in normal lung tissues, since TTFC was recently found to be independently associated with lung cancer and is a marker of smoking dependence (36). With adjustment for current cigarette smoking status, those with TTFC 6–30 min, or ≤ 5 min showed significant hypomethylation of probe cg05575921 (P = 0.02 and 0.03 respectively) and probe cg13787850 (P = 0.01 and 0.005 respectively), but this association did not remain significant after adjusting for smoking duration. Since we only had 10 never smokers, we performed clustering analysis using the top 5,000 most variable CpG probes and then tested whether never smokers were enriched for some clusters. We did not find significant evidence showing that never smokers differed from former smokers in terms of methylation profiles, but for the eight CpG probes that are identified in our primary analysis, the effect sizes were stronger for the current vs. never smokers’ analysis (Supplementary Material, Table S4). Methylation of six intronic CpGs (cg17113147 (NXN), cg05575921 (AHRR), cg07992500 (CDC42EP3), cg14120703 (NOTCH1), cg11152412 (EDC3), and cg03224163 (HIPK2) was also associated with pack-years and years from quitting smoking (P < 0.05, Supplementary Material, Table S2). The association between smoking status and methylation of these six loci and one additional intergenic probe from the original eight loci (cg13787850 on chromosome 9) replicated in NTL from TCGA samples (37,38) (n = 60; 12 current smokers and 48 current nonsmokers, including 45 former smokers and 3 never smokers (Supplementary Material, Table S5) with single-sided P-values ≤ 0.05 (Table 1 and Supplementary Material, Table S2).
Of the seven hypomethylated CpGs that replicated in TCGA data, five had been previously observed in the blood cell DNA of smokers, as indicated in italics in Table 1. Cg05575921 has been reported to be hypomethylated in virtually every published study of white blood cells (4–26); cg14120703 in two studies (17,25); cg11152412 in three studies (7,17,25); cg03224163 in one study (25); and cg13787850 in three studies (7,25,26). This suggests that there are key similarities in the effect of tobacco smoke on the DNA methylation status of specific loci in white blood cells and alveolar lung epithelium.
Investigation of epigenomic environment of replicated hypomethylated CpGs
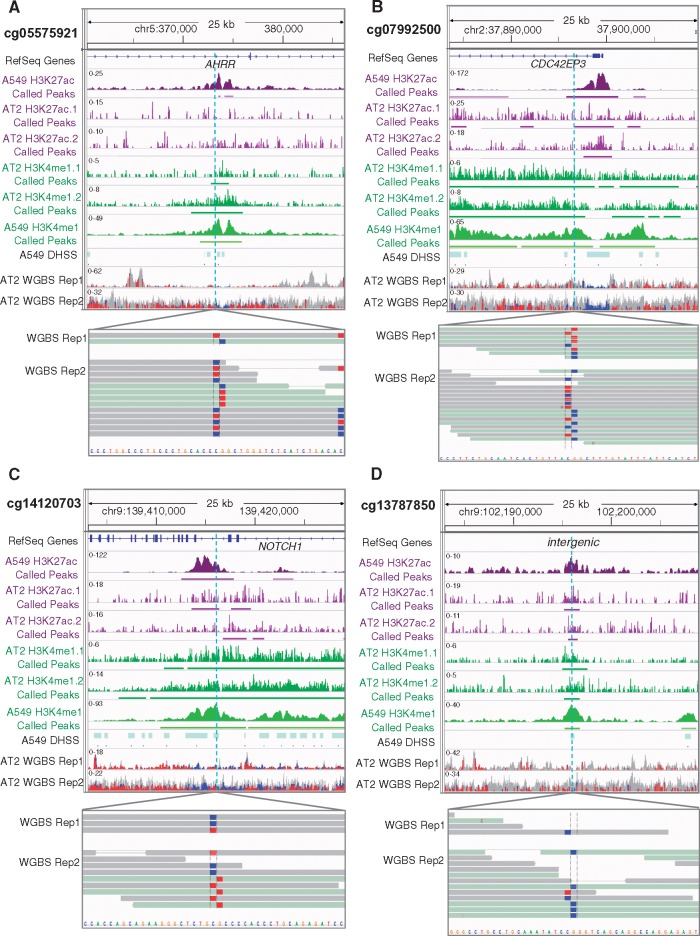
To gain insight into how smoke-induced hypomethylation of the CpG dinucleotides that were replicated in TCGA might relate to lung cancer, we investigated epigenetic marks in the region near each CpG in a cell type relevant to lung cancer risk: alveolar epithelial cells. Epigenetic patterns are highly cell type-specific; analyzing purified cells is important to avoid the confounding effects of mixed cell populations (39). In fact, a recent study demonstrated that changes in DNA methylation at a particular CpG in whole blood could be explained by tobacco smoking-associated induction of specific lymphocyte cell types rather than smoking-associated changes in DNA methylation levels (40). With these concerns in mind, we integrated our epigenome-wide data from lung tissue with epigenomic profiles obtained from purified primary human type 2 alveolar epithelial (AT2) cells acquired from remnant transplant lungs. AT2 cells are the suspected precursors of lung adenocarcinoma, the most common type of lung cancer (41,42). We used lungs from two never-smokers because isolation of primary alveolar epithelial cells from smokers is complicated by the frequent presence of inflammation. We generated whole genome bisulfite sequencing (WGBS) and chromatin immunoprecipitation-sequencing (ChIP-seq) data from these AT2 cells (∼85% pure (43); biological replicates) to examine the locations of each of the seven replicated hypomethylated CpGs. The WGBS data showed that cg05575921, cg07992500, and cg14120703 were partially methylated and lay adjacent to regions that are hypomethylated relative to the surrounding DNA (Fig. 1). The density of CpGs varied considerably per location, for example, the intergenic region showed only two hypomethylated CpGs (Fig. 1D). In addition, cg13787850 (Fig. 1) and cg11152412 (Supplementary Material, Fig. S3) were unmethylated (87–100%) and were located in very small hypomethylated patches in relatively CpG-poor areas. We next examined the regions around the seven CpGs for histone modifications typical of enhancers (44): histone 3 lysine 4 mono-methylation (H3K4me1, a mark of poised or active enhancers) and/or histone 3 lysine 27 acetylation (H3K27ac, often present when enhancers are active). We noted that cg05575921, cg07992500, cg14120703, and cg13787850 were in very clear enhancer peaks (Fig. 1). We chose to focus on those four CpGs showing hypomethylation and histone marks for poised or active enhancers: cg05575921, cg07992500, cg14120703, and cg13787850. The remaining three CpGs were either not hypomethylated or did not lie in enhancer marks (Supplementary Material, Fig. S3) and were not further investigated here.
Figure 1.
Four CpGs hypomethylated in smoker NTL are adjacent to unmethylated loci in AT2 cells and marked by regulatory histone marks in AT2 and A549 cells. Displayed from top to bottom for each CpG: the genomic location with RefSeq genes (if present; hg19 coordinates); H3K27ac (purple) & H3K4me1 (green) profiles in A549 cells and in two biological replicates of AT2 with called peaks underlined; DNase hypersensitive sites in A549; and two biological replicates of whole genome bisulfite sequencing (WGBS) with the CpG of interest magnified (blue= unmethylated; red = methylated; gray = no CpG present). A549 data were downloaded from ENCODE (45). The sequencing depth of AT2 cells is less than that of the A549 cell line owing to the difficulty in obtaining large numbers of primary human cells.
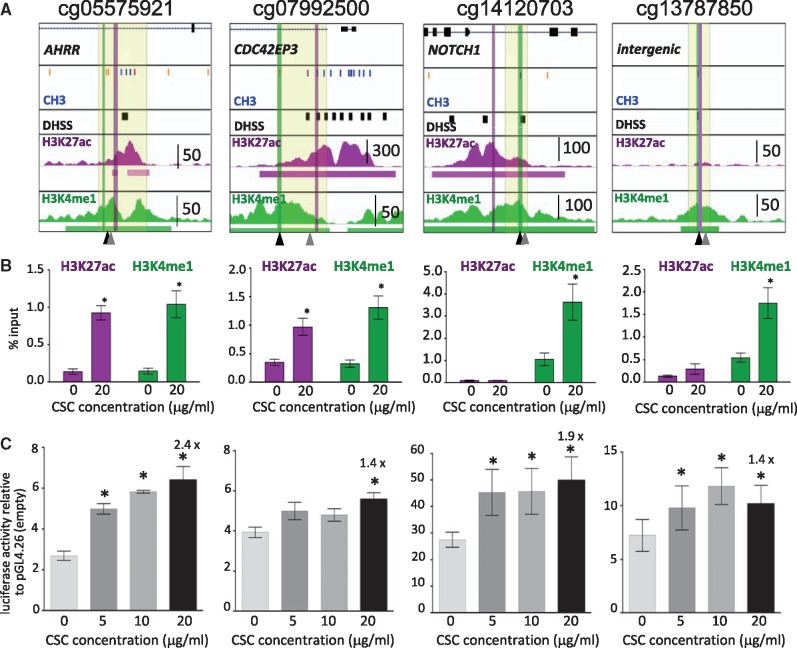
We also mined publicly available ChIP-seq data produced in the A549 lung adenocarcinoma cell line by the ENCODE consortium (45). A549 cells exhibited similar enhancer signatures and each region contained DNase-hypersensitive sites indicative of accessibility to transcription factors (TFs) (Fig. 1). Interestingly, analysis of these putative enhancer regions revealed the presence of a predicted aryl hydrocarbon receptor (AHR) binding motif (CACGCA) in all four regions (Fig. 2A). AHR is a ligand-activated helix-loop-helix transcription factor that is highly expressed in the alveolar lung epithelium (46) and is implicated in lung cancer (47). In response to xenobiotics, such as polycyclic aromatic hydrocarbons (PAHs) present in tobacco smoke, it heterodimerizes with the ARNT protein and induces expression of target genes (47–49), including cytochrome P450 enzymes (e.g. CYP1A1 or CYP1B1) that metabolize procarcinogens like PAH. The AHR/ARNT heterodimer also induces AHRR, which is a suppressor and feedback regulator of AHR activity (47,50,51).
Figure 2.
CSC-induced changes in enhancer marks in A459 cells. (A) Each diagram displays RefSeq genes at top (if present; hg19 coordinates) and from top to bottom, ENCODE data in A549 cells: Illumina Infinium HumanMethylation450 BeadChip CpG methylation status (blue = unmethylated; orange/purple = partially methylated; the CpGs of interest are marked by black arrowheads at bottom); DNase hypersensitive sites (DHSS) as black boxes; H3K27ac and H3K4me1 profiles with called peaks underlined and peak height scale indicated. Locations of PCR primers used for ChIP assays of H3K27ac and H3K4me1 panel B are indicated by purple and green vertical lines respectively. The region highlighted in pale yellow was cloned into a basal promoter-containing luciferase gene reporter vector (pGL4.26, panel C). The positions of AHR binding sites are indicated by gray arrowheads at the bottom. (B) ChIP assays of H3K27ac and H3K4me1 in vehicle- or CSC-treated A549 cells expressed as % input. * indicates luciferase activity statistically significantly different from vehicle (P < 0.05 paired Student's t test). Bars represent mean ± SEM of samples assayed in three or more independent trials of A549 cells exposed to CSC for 48 h. (C) Luciferase reporter assays to test for CSC-regulated enhancer activity. The region highlighted in pale yellow in panel A was cloned into the basal promoter-containing luciferase vector pLG4.26. In each case we ensured that the DNase hypersensitive site closest to the hypomethylated CpG was included. The plasmids were transfected into A549 cells that were treated with vehicle or CSC at the indicated doses 24 h after transfection. A renilla expression plasmid was cotransfected for DNA quantity normalization. Luciferase activity was measured at 24 h after CSC exposure, and is indicated relative to empty vector. All expression levels were statistically significantly elevated relative to pGL4.26. Significant induction relative to vehicle is indicated by an asterisk and the fold induction is noted for 20 µg/ml CSC. Bars represent mean ± SEM of three or more independent experiments.
Functional investigation of putative regulatory elements
To investigate whether the four differentially methylated CpGs mark regulatory elements that can be activated by tobacco smoke, we exposed A549 cells to cigarette smoke condensate (CSC) for 48 h and performed targeted ChIP of H3K4me1 and H3K27ac (Fig. 2A and B). All four regions sustained significant increases in H3K4me1, indicating increased readiness for activity. In addition, the regions adjacent to cg05575921 and cg07992500 gained H3K27ac, indicative of regions actively engaged in regulating gene transcription. We note that enhancers can be activated in the absence of H3K27ac (52).
We further empirically verified the enhancer activity of the four regions in a reporter assay. We cloned regions spanning each CpG and nearby DNase hypersensitive sites (Fig. 2A) into a luciferase reporter vector containing a minimal promoter and transfected these plasmids into A549 cells. All four constructs exhibited statistically significantly elevated luciferase activity relative to the empty vector pGL4.26 (∼3–25-fold Fig. 2C) in the absence of CSC. Importantly, luciferase activity was significantly increased upon exposure to 20 µg/ml CSC for all four regions (Fig. 2C; fold increase over vehicle indicated). These observations indicate that the four regions can function as CSC-responsive enhancers even outside of their genomic context.
Identification of potential enhancer target genes
Enhancers are increasingly understood to regulate promoters of distant genes, and less than 10% are reported to interact with the nearest gene (53). Most target genes are located within 1 Mb from their enhancers (54–59). To identify candidate target genes of our four putative enhancers, we exposed A549 cells to CSC for 48 h and for two weeks. We confirmed the induction of CYP1A1, a gene encoding a cytochrome P450 enzyme that is known to be induced in response to CSC exposure (49) by targeted PCR (Supplementary Material, Fig. S4A). CYP1A1 was robustly induced at all CSC concentrations at 48 h and 2 weeks, so we proceeded to perform RNA-seq to identify genes that were differentially expressed in a 1Mb window flanking each CpG (Fig. 3A). AHRR was the only gene induced in the window surrounding cg05575921 (P = 3.20×10−7 and 1.13×10−62, at 48 h and two weeks, respectively, Bonferroni corrected for the number of genes in the 1 Mb flanking windows), indicating that the enhancer element, located in intron 3 of AHRR, is likely driving its nearest gene. This makes sense, given that AHRR is known to be induced by AHR in a regulatory feedback loop (47,50,51). Cg07992500 is located in the first intron/promoter region of CDC42EP3, but the expression of this gene was not regulated by CSC. Instead, CYP1B1, a member of the cytochrome P450 family located ∼400 kb upstream of cg07992500, was the only gene induced in the 1 Mb window around this CpG (48 h, P = 6.78×10−18; two weeks, P = 2.93×10−61). Cg14120703 is located in NOTCH1 intron 4, but we noted no significant changes in NOTCH1 expression. However, we observed smoking-associated downregulation of ENTPD2, located at the far end of the 1 Mb window, after 2 weeks of CSC exposure (P = 2.79 × 10−6). ENTPD2 is a member of the ecto-nucleoside triphosphate diphosphohydrolase family of proteins that hydrolyze 5’-triphosphates and may affect ATP and purine metabolism (60). These enzymes are implicated in immune responses related to cancer (61) and can be regulated by xenobiotics (62). Lastly, in the window surrounding cg13787850, we found that NR4A3, located ∼400 kb downstream, was significantly downregulated in response to CSC (48 h P =4.75 × 10−9; two weeks P = 1.34 × 10−25). NR4A3, previously called NOR1, encodes an orphan nuclear hormone receptor and is a known tumor suppressor (63). We validated CSC-regulated expression of AHRR, CYP1B1, ENTPD2, and NR4A3 in A549 cells by qRT-PCR (Supplementary Material, Fig. S4B).
Figure 3.
RNA-seq investigation of 1 Mb regions flanking the putative CSC-regulated enhancers. (A) Genes induced in a 1Mb window around each CpG in A549 cells treated for 48 h or 2 weeks with 20 µg/ml CSC. RNA-seq data were obtained from three independent treatments of A549 cells with CSC. (B) RNA-seq data from TCGA NTL samples (n = 100, Supplementary Material, Table S6) was analyzed to examine whether genes identified in A were associated with smoking history. NS= non-smoker; >15 and <15 indicates smoking cessation more than or less than 15 years ago. Fold change values between NS and current smoker were 4.58 for AHRR, 2.91 for CYP1B1, -0.40 for ENTPD2 and 0.87 for NR4A3. (C) TCGA NTL lung samples were used to examine the correlation between DNA methylation and RNA expression (n = 28, Supplementary Material, Table S8).
To validate these cell-based observations in lung tissue, we examined the relationship between smoking status and the expression of AHRR, CYP1B1, ENTPD2 and NR4A3 in TCGA NTL tissues (n = 100, Supplementary Material, Table S6) (Fig. 3B). AHRR and CYP1B1 were significantly elevated per smoking history (P = 1.53 ×10−13, and 3.48 ×10−10 respectively), while ENTPD2 was significantly downregulated (P = 0.021) (Supplementary Material, Table S7). These findings confirm the association between tobacco smoke exposure and altered regulation of the three genes in human lung tissue. In contrast, NR4A3 expression was not significantly associated with smoking (Fig. 3B and Supplementary Material, Table S7). In addition to these four target genes, we investigated whether other genes in the 1Mb window flanking each CpG exhibited smoking-associated expression in TGCA NTL. Only CYP1B1-AS1 (C2orf58), located in the 1Mb window flanking cg07992500 and encoding a CYP1B1-overlapping non-coding antisense RNA, was identified as an additional induced gene (P = 1.27 × 10−3, linear regression adjusted for age and sex and Bonferroni-corrected for the total number of genes in the windows around the four hypomethylated CpG probes) (Supplementary Material, Fig. S5). Extending the window to 2 Mb did not identify any additional genes whose expression significantly changed in relation to smoking exposure. We next examined the relationship between methylation of the four CpGs and expression of AHRR, CYP1B1, ENTPD2, and NR4A3, respectively, in TCGA NTL samples for which DNA methylation and RNA-seq data were available (n = 28, Supplementary Material, Table S8). As expected, expression of AHRR and CYP1B1 was significantly negatively correlated with methylation of the respective CpGs, while ENTPD2 expression was marginally correlated with methylation of cg14120703. In contrast, NR4A3 expression was not associated with cg13787850 methylation (Fig. 3C and Supplementary Material, Table S9). Thus, both our cell-based and the tissue-based expression analyses suggest that AHRR, CYP1B1, and ENTPD2 are likely the target genes regulated by the three out of four putative enhancers marked by smoking-associated CpGs. It is noteworthy that AHR binding sites (5’-CACGCA-3’) are predicted in the promoters of AHRR (chr5:420,905-421,071), CYP1B1 (chr2:38,304,144-38,304,788), and ENTPD2 (chr9:139,948,461-139,948,481) as well as in the putative enhancer elements that we identified (Fig. 2A), suggesting a common gene regulatory mechanism of these smoking-associated enhancers and their target genes.
Methylation status of probes associated with expression in response to CSC exposure
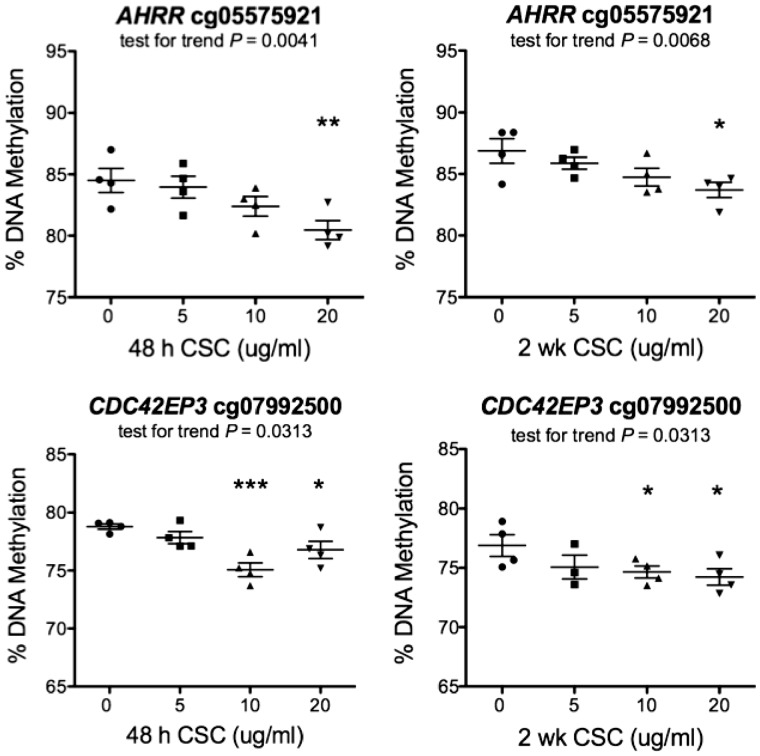
In TCGA LUAD data, the methylation status of cg05575921 and cg07992500 was inversely correlated with expression levels of the tobacco smoke-responsive genes AHRR and CYP1B1 respectively (Figs 2 and 3). We sought to determine whether methylation levels of either probe were responsive to CSC-exposure in vitro. To this end, we exposed A549 cells to 0, 5, 10, or 20 µg/ml CSC for 48 h or two weeks, and assessed the methylation of both probes by targeted sodium bisulfite pyrosequencing with the primers listed in Supplementary Material, Table S10. The methylation status of both probes changed in a dose-dependent and statistically significant manner at both time points (Fig. 4). Comparing DNA methylation levels at 0 and 20 µg/ml CSC, the methylation of cg05575921 diminished by 4.8% after a 48-h exposure (P = 0.009) and by ∼3.6% after a two-week exposure (P = 0.0177). For cg07992500, methylation diminished by 2.6% after 48 h of exposure (P = 0.0196), and 3.4% after two-weeks (P = 0.0297). These results suggest that loss of DNA methylation is an early event occurring during enhancer activation.
Figure 4.
Targeted sodium bisulfite pyrosequencing of cg05575921 and cg07992500 in CSC-treated A549 cells. A549 cells were exposed in quadruplicate to CSC in vitro for 48 h and 2 weeks at 0, 5, 10 or 20 µg/ml. DNA methylation was assessed by pyrosequencing of sodium bisulfite-treated DNA. All 4 plots showed a significant trend as indicated in the chart and a statistically significant reduction in DNA methylation between 0 and 20 µg/ml CSC (AHRR: P = 0.009 and 0.0177 at 48 h and 2 weeks respectively; CYP1B1: P = 0.0196 and 0.0297 at 48 h and 2 weeks, respectively (t-test, one-sided)).
A possible link between smoking-induced epigenetic events and lung cancer
Given the presence of AHR binding sites in all four putative enhancers, the presence of AHR binding sites in the promoters of AHRR, CYP1B1, and ENTPD2, and the dual role of AHR targets in detoxification and bioactivation of pro-carcinogens in tobacco smoke (47), we examined whether AHRR, CYP1B1, or ENTPD2 expression was associated with specific mutation signatures in adenocarcinoma tumors after adjusting for age and sex (TCGA, n = 407, Supplementary Material, Table S11). We found a positive association of AHRR expression and, to a lesser extent, CYP1B1 expression with smoking-associated transversion ratio (P = 1.14×10−7; and P = 2.04×10−4, respectively) and C > A mutations (P = 2.0 ×10−4 and P = 9.4×10−3, respectively) (Supplementary Material, Table S12), which remained significant after adjusting for smoking status (Supplementary Material, Table S13). Interestingly, while expression of these genes was positively associated with smoking-related mutations, it was negatively associated with C > T mutations, which have been found across many cancers and tend to increase with age (64). ENTPD2 expression, which was negatively associated with smoking, was marginally inversely associated with smoking-specific mutations (P = 1.45×10−3; for C > A mutations, P = 0.055) (Supplementary Material, Table S12). Thus, associations between expression of these genes and smoking-specific transversion mutations suggest one pathway linking smoking-induced epigenetic events to lung cancer.
Discussion
The relationship between DNA methylation alterations in blood and tobacco smoke exposure has been widely documented, but to date no epigenome-wide association study (EWAS) of tobacco smoking had been carried out in normal lung tissue. Using EAGLE samples, we identified seven smoking-associated hypomethylated CpGs that were replicated in lung tissue from TCGA. The number of significantly hypomethylated CpGs was smaller than in many blood-related studies, but was in line with expectations given the sample size and the need for Bonferroni correction for the number of functional probes on the HumanMethylation450 BeadChip. Analysis of larger numbers of samples in the future may uncover additional smoking-associated DNA methylation changes. We compared our results with those from a large blood DNA methylation study (7), which has 1793 blood samples in their discovery phase. This study identified 972 CpG probes with P < 10−7, of which 769 passed our quality control filters. Out of these 769 CpG probes, 578 (75.1%) have the same direction in their data and our data (P = 8 × 10−47 based on binomial test). In addition, 159 (out of 578) were replicated in our data with P < 0.05. These data suggest a reasonably high consistency between the two data sets even with a relatively small sample size in our study. Another factor potentially limiting the number of identified CpGs in our study might be that in the EAGLE population, only 10% of the current non-smokers were never smokers, and this limits the size of the DNA methylation effects that can be observed (Supplementary Material, Table S4).
Of the seven hypomethylated CpGs we identified, five had been previously reported in blood. Hypomethylation of cg05575921 in AHRR has been widely reported in whole blood, lymphoblasts, and pulmonary macrophages (6,7,10,11,13,14,16,21,28,30), and is inversely correlated with AHRR expression in lymphoblasts (14). Together with our observations in lung tissue, this suggests that hypomethylation of cg05575921 may be predictive of increased AHRR expression in multiple tissues. The nearby AHRR cg23576855 probe has been reported to be hypomethylated in 27 smokers’ lung tissue and to be inversely correlated with AHRR expression comparing five smokers with five non-smokers (9). Its location about 80 nts upstream of cg05575921 suggests it may mark the enhancer we identified, though interpretation of methylation data from this CpG is complicated by the fact that it carries a SNP (CpG to CpA) (9).
Besides cg05575921, four other CpGs, including two that mark putative enhancers in AT2 cells (cg14120703 and cg13787850), have been reported to be hypomethylated in blood in several studies (Table 1). We examined the epigenomic environment for these five common hypomethylated CpGs in the lymphoblastoid cell line GM12878 using epigenetic data from ENCODE (Supplementary Material, Fig. S6). Histone enhancer marks suggests that two CpGs might also mark the flanks of enhancers in GM12878 cells: cg05575921 and cg13787850. Thus, several smoking-associated CpG probes appear to be shared between blood and lung tissue, and might mark similar regulatory elements. This suggests that hypomethylation in blood DNA can be a good surrogate for hypomethylation at certain, although not all, CpGs in lung tissue.
Our observation that four of the hypomethylated CpGs flank regions that are unmethylated in primary human alveolar epithelial cells and that carry enhancer histone marks which are acutely regulated by CSC suggests that, at least in some cases, hypomethylation may be a byproduct of enhancer activation. Loss of DNA methylation has been well-documented to occur upon enhancer activation, as transcription factors interact with the enhancer element and presumably protect it from maintenance DNA methyltransferase activity (65,66). A recent study of disease-associated genetic variants supports the idea that differential occupancy of transcription factors affects DNA methylation (67). Loss of DNA methylation can thus be a powerful tool to identify regulatory elements and allow mechanistic dissection of the effects of environmental tobacco smoke; in the current investigation it led to the identification of four smoking-responsive regulatory elements. Our observation that DNA methylation is reduced at the two CpGs inversely associated with gene expression (cs05575921 and cg07992500) at 48 h following CSC exposure suggests that DNA methylation loss is a rapid event. The magnitude of the DNA methylation loss in vitro at 48 h and 2 weeks (<5%) is less than what we observed in EAGLE data (∼9–10%) but these two systems may not be fully comparable. Because A549 cells divide rapidly, the observed DNA methylation loss could be due to passive loss of the DNA methylation mark if it is not replaced following cell division. However, active demethylation is also a possibility and should be investigated.
Smoking-associated increases in histone marks and luciferase activity and the presence of AHR binding sites suggest all four regions function as tobacco smoke-inducible enhancers. No induced genes were as yet identified for cg14120703 and cg13787850, and examination of a window expanded to 2 Mb around the elements did not yield any candidates (data not shown). ENTPD2 downregulation could be mediated by AHR, which has been known to cause gene repression (68,69). Repression could also be a consequence of transcription factor sequestration, a possible mechanism proposed for gene silencing by enhancers (70). Alternatively, the enhancer might trans-activate genes more distant than 2 Mb or genes on different chromosomes that in turn repress ENTPD2.
While the significance of cg14120703 and cg13787850 remains to be further investigated, the identification of AHRR and CYP1B1 as likely target genes provides clues to the link with lung cancer risk. Cytochrome P450 enzyme CYP1B1, which makes relatively inert and hydrophobic PAHs in tobacco smoke more hydrophilic for purposes of detoxification by excretion (‘biotransformation’), can also make these compounds more reactive and toxic in the process (‘bioactivation’), causing DNA adducts that commonly result in transversion mutations (71). Indeed, mutation signatures with high transversion mutation ratios are common in tobacco-related tumors (64), and both AHRR and CYP1B1 expression were positively associated with transversion mutations in TCGA.
Materials and Methods
Human tissue
The EAGLE study (32) included 2098 lung cancer cases and 2120 population controls enrolled in Italy between 2002 and 2005. The study was approved by local and NCI Institutional Review Boards, and all participants signed an informed consent form. Lung tissue samples were snap-frozen in liquid nitrogen within 20 min of surgical resection. Surgeons and pathologists ensured correct sampling of tissue from the tumor, the area adjacent to the tumor and an additional area distant from the tumor (1–5 cm). For the purpose of this study, we used the samples from an area distant from the tumors. Multiple samples were taken from each subject. At least one sample/subject was histologically confirmed to have no tumor nuclei. Remnant human transplant lungs were obtained in compliance with Institutional Review Board-approved protocols for the use of human source material in research (HS-07-00660) and processed to obtain type 2 alveolar epithelial cells (AT2) as described (43). AT2 Rep1 cells were obtained from a 62-year-old family-reported never-smoker Caucasian male who died of a cerebrovascular accident and intracerebral hemorrhage. AT2 Rep2 cells were obtained from a 25-year-old family-reported never-smoker Caucasian male who died of head trauma.
Primary human alveolar epithelial and A549 cell culture
Human lung tissue was processed as previously described (43) from non-smoker donors by inclusion of anti-EpCAM conjugated beads to select for epithelial cells. A549 lung adenocarcinoma cells were kindly provided by Dr. Zea Borok (USC Keck School of Medicine) and were mycoplasma-free. A549 and primary lung epithelial cells were respectively plated in RPMI 1640 and 50:50 DMEM high glucose media (Gibco # 21063, Grand Island, NY) DMEM-F12 (Sigma # D6421, St. Louis MO) supplemented with 10% fetal bovine serum, 2.0 mM of L-glutamine, 100 units/mL of penicillin, and 100 µg/mL of streptomycin. A549 cells were exposed to the indicated doses of cigarette smoke condensate (CSC) manufactured by Murty Pharmaceuticals (Lexington, KY; purchased from Fisher # NC9028647, Waltham, MA) and renewed every 2 days.
RNA and DNA isolation
EAGLE fresh frozen lung tissue samples remained frozen while approximately 30 mg was subsampled for DNA extraction into pre-chilled 2.0 ml microcentrifuge tubes. Lysates for DNA extraction were generated by incubating 30 mg of tissue in 1 ml of 0.2 mg/ml Proteinase K (Ambion) in DNA Lysis Buffer (10 mM Tris-Cl (pH 8.0), 0.1 M EDTA (pH 8.0), and 0.5% (w/v) SDS) for 24 h at 56 °C with shaking at 850 rpm in Thermomixer R (Eppendorf). DNA was extracted from the generated lysate using the QIAamp DNA Blood Maxi Kit (Qiagen) according to the manufacturer’s protocol. RNA and DNA were extracted from primary human epithelial cells with the Illustra TriplePrep Kit (GE LifeSciences # 28-9425-44, Piscataway, NJ) and from A549 cells with the AllPrep DNA/RNA/Protein kit (Qiagen #8004, Valencia, CA).
EAGLE Sample DNA methylation measurement and pre-processing
Bisulfite treatment and Illumina Infinium Human Methylation450 BeadChip assays were performed by the Southern California Genotyping Consortium at UCLA following Illumina’s protocols. This assay generates DNA methylation data for 485,511 methylation probes and 66 SNP probes for the purpose of data quality control. Raw methylated and unmethylated intensities were background corrected, and dye-bias equalized, to correct for technical variation in signal between arrays. For background correction, we applied a normal-exponential convolution, using the intensity of the Infinium I probes in the channel opposite their design to measure non-specific signal. Dye-bias equalization used a global scaling factor computed from the ratio of the average red and green fluorescing normalization control probes. Both methods were conducted using the methylumi package in Bioconductor version 2.11. For each probe, DNA methylation level was summarized as a β-value, estimated as the fraction of signal intensity obtained from the methylated beads over the total signal intensity. Probes with detection P-values of >0.05 were considered not significantly different from background noise and were labeled as missing. Methylation probes were excluded if any of the following criteria was met: on X/Y chromosome, annotated in repetitive genomic regions, annotated to harbor SNPs, or missing rate >5% (34). This method is described in FDb.InfiniumMethylation.hg19 package combined with UCSC common SNPs track v136 http://bioconductor.org/packages/release/data/annotation/html/FDb.InfiniumMethylation.hg19.html; date last accessed May 19, 2017 and http://bioconductor.org/packages/release/data/annotation/html/FDb.UCSC.snp135common.hg19.html; date last accessed May 19, 2017. After excluding probes on the X-/Y-chromosomes, 473,864 probes remained. After further excluding probes with genetic variants, 414,634 probes remained. After excluding probes in repetitive regions, 338,730 remained. After additionally excluding those with a missing rate of >5%, methylation for 338,456 autosomal probes remained. Because the β-values for the 66 SNP probes included in the array are expected to be similar in matched pair of normal and tumor tissues, we performed PCA using these 66 SNP probes to confirm the labeled pairs. We then performed PCA using the 5000 most variable methylation probes with var > 0.02 and found that the normal tissues were clustered together and well separated from the tumor tissues. We further excluded 5 normal tissues that were relatively close to the tumor cluster. From the remaining 239 normal tissue samples, we used 237 of them with smoking information available, including 121 current cigarette smokers and 116 current nonsmokers (106 former and 10 never smokers). Among the former smokers, 75% had quit for 10 or more years. The median (inter-quartile range) time since quitting smoking was 13 (11) years.
AT2 cell whole genome bisulfite sequencing
For whole genome bisulfite sequencing (WGBS) of AT2 cells, total DNA from 8×106 cells was isolated using the Illustra Triple Prep kit (GE Healthcare Life Sciences, Pittsburg, PA). Four μg of sample genomic DNA was sonicated using a Covaris S2 to an average molecular weight of 150 bp. Achievement of the desired size range was verified by Bioanalyzer (Agilent) analysis. Fragmented DNA was repaired to generate blunt ends using the END-It kit (Epicentre Biotechnologies, Madison, WI) according to manufacturer’s instructions. Following incubation, the treated DNA was purified using AmpureX beads from Agencourt. In general, magnetic beads were employed for all nucleic acid purifications in the following protocol. Following end repair, A-tailing was performed using the NEB dA-tailing module according to manufacturer’s instructions (New England Biolabs, Ipswich, MA). Adapters with a 3’ ‘T’ overhang were then ligated to the end-modified DNA. For whole genome bisulfite sequencing, modified Illumina paired-end (PE) adapters were used in which cytosine bases in the adapter are replaced with 5-methylcytosine bases. Ligation was carried out using ultrapure, rapid T4 ligase (Enzymatics, Beverly, MA) according to manufacturer’s instructions. The final product was then purified with magnetic beads to yield an adapter-ligation mix. Prior to bisulfite conversion, bacteriophage lambda DNA that had been through the same library preparation protocol described above to generate adapter-ligation mixes was combined with the genomic sample adapter ligation mix at 0.5% w/w. Adapter-ligation mixes were then bisulfite converted using the Zymo DNA Methylation Gold kit (Zymo Research, Orange, CA) according to the manufacturer’s recommendations. The final modified product was purified by magnetic beads and eluted in a final volume of 20 µl. Amplification of one-half the adapter-ligated library was performed using Kapa HiFi-U Ready Mix for the following protocol: 98° 2’, then six cycles of: 98° 30”, 65° 15”, 72° 60”, with a final 72° 10’ extension, in a 50 µl total volume reaction. Final library product was examined on the Agilent Bioanalyzer, then quantified using the Kapa Biosystems Library Quantification kit according to manufacturer’s instructions. Optimal concentrations to get the right cluster density were determined empirically but tended to be higher than for non-bisulfite libraries. Libraries were plated using the Illumina cBot and run on the Hi-Seq 2000 according to manufacturer’s instructions using HSCS v 1.5.15.1. Rep 1 underwent Paired End 100 cycling; rep 2 underwent Paired End 75 cycling. Image analysis and basecalling were carried out using RTA 1.13.48.0, deconvolution and fastq file generation were carried out using CASAVA_v1.7.1a5. Alignment to the genome was carried out using bsmap V 2.5. Aligned bam files were visualized using IGViewer V2.3.40 (Broad Institute, Cambridge MA) with alignments colored by Bisulfite mode “CG”, which colors each methylated C red, each unmethylated C in a CpG context blue, and each C not residing in a CpG gray. AT2 WGBS has been made publically available through GEO record GSE94986.
ChIP-seq of AT2 cells
Human AT2 cells underwent ChIP-seq using a modified version of the Sigma ChIP-it kit Protocol (Sigma). Specifically, 2 million human AT2 were used per ChIP-seq of H3K27Ac (Cat # 39135, ActiveMotif) and H3K4me1 (Cat # 39297, ActiveMotif). All cells were crosslinked with 3.75% formaldehyde at 25° for 5 min with gentle nutation. Fixation was quenched with 2.5mM glycine and samples were frozen at -80° before further processing. Each sample underwent nuclei fractionation using cell lysis buffer prepared per manufacturer’s recommendations. Upon nuclei isolation, cells were sonicated into fragments (average size 300bp) using a Bioruptor2000 (Diagenode) with 30-s pulses for a total of 30 min of sonication (15 on, 15 off). StaphSeq (Cat # S6576, Sigma) was used for antibody binding. All ChIPs were verified for enrichment by site-specific PCR prior to sequencing using the active region of PGDH for enhancer marks. All ChIP-seq samples underwent library preparation and sequencing at the USC Epigenomic Core, where they were multiplexed using adapter barcoding and underwent single-end 50bp sequencing using the IlluminaHiSeq2000. Reads with a quality score > 20 were aligned to the hg19 human genome build using Bowtie2 (72). Alignment metrics are provided in Supplementary Material, Table S14. For A549 cells, Bowtie-mapped ChIP-seq and DNAse Hypersensitivity datasets were downloaded from the UCSC Genome browser (https://genome.ucsc.edu; date last accessed May 19, 2017).
Targeted ChIP in A549 cells
ChIP for H3K4me1 (Diagenode #pAb-037-050, Denville, NJ) and H3K27Ac (Active Motif #39133, Carslbad, CA) were performed as previously described with DNA sonicated to an average length of 200–800 bp (43). Enrichment was normalized to % input DNA for each CSC exposure group after subtracting non-specific binding determined using pre-immune IgG (Santa Cruz #sc-2027, Santa Cruz, CA).
ChIP-seq peak calling and data visualization
SICERv1.1 peak calling was performed using a window size of 200 bp and gap size of 200 bp (73). Input DNA was used for background normalization. ChIP-seq reads from AT2 cells were observed at or near read saturation. The Integrative Genomics Viewer v1.5 was used to visually inspect and graph peak quality (74). Respective window sizes for H3K27ac ChIP-seq and H3K4me1 ChIP-seq were set to: 50bp and 100bp, while gap sizes were set to 50bp and 200bp, respectively (FDR cutoff = 5 × 10−4).
Quantitative polymerase chain reactions
For expression studies, mRNA was reverse transcribed from RNA using the iScript cDNA Synthesis kit (Biorad #170-8891 Hercules, CA). Quantitative PCR and reverse transcription(RT)-PCR were respectively performed with ChIP elutes and template cDNA using iQ SYBR Green Supermix (BioRad # 170-8882) and primers listed in Supplementary Material, Table S15 and S16. All PCR reactions were analyzed using a DNA engine Opticon (MJ Research, Waltham, MA).
Luciferase reporter gene assays
Putative enhancer regions spanning each hypomethylated CpG and closest A549 DNase HSS were PCR amplified from normal human gDNA (Promega) and inserted upstream of the minimal promoter contained within the pGL4.26-luciferase construct (Promega). All constructs were verified by sequencing. Primers used in amplification of each locus are provided in Supplementary Material, Table S17. A549 cells were transiently transfected with one of the constructs under study, pmaxGFP (constitutive transfection efficiency control vector, Lonza/Amaxa, # VSC-1001, Walkersville, MD), and pRL-CMV (constitutive Renilla control vector, Promega) with Fugene HD (Promega) according to the manufacturer’s instructions. Cells were exposed to the indicated doses of CSC 24 h post-transfection. Cells were harvested 48 h post-treatment and assayed for luciferase activity with the Dual-Luciferase Reporter Assay System™ (Promega, #1960) according to the manufacturer’s instructions. Raw luciferase values were normalized to Renillla luciferase activity and pGL4.26 ‘empty’ vector activity. Data are expressed as the mean ± S.E.M. of three independent biological replicates assayed in duplicate. Significance was assessed via student's t-test.
RNA-seq analysis of A549 cells
Library construction was performed at the USC Epigenome Center at the Norris Comprehensive Cancer Center from total cell RNA extracted from vehicle or CSC-exposed A549 cells as described previously (43) with modifications. Briefly, total cell RNA was DNase I digested and then subjected to ribosomal RNA depletion with the Ribominus™ Eukaryote v2 kit (Life Technologies, # A15020, Grand Island, NY). Libraries were constructed with the TruSeq RNA Sample Prep Kit v2 (llumina # RS-122-2001) and underwent Illumina HiSeq 2000 paired-end sequencing (2 × 75 bp) according to the manufacturer’s instructions. Sequence reads were filtered such that >90% of each read had a quality score > 20. Reads that passed filter were aligned to hg19 with TopHat2 v2.0.7 and corrected for GC-bias (75). Alignment metrics for BAM files were assessed with Picard Tools (https://broadinstitute.github.io/picard/; date last accessed May 19, 2017) and Samtools (http://samtools.sourceforge.net; date last accessed May 19, 2017), and are provided in Supplementary Material, Table S18. Reads that mapped to ribosomal RNA or microRNA were excluded from analysis, and genes that did not have at least 10 counts per million (CPM > 10) mapped reads in at least two samples were also filtered prior to differential expression testing performed in edgeR (70). Genes that were more than 2-fold different between treatment groups (absolute log2 ≥1) and had a Benjamini-Hochberg FDR < 0.05 were considered differentially expressed.
Targeted DNA methylation assessment in A549 cells via sodium bisulfite pyrosequencing
DNA was extracted from CSC or vehicle-exposed A549 cells after the indicated exposure periods with the Qiagen DNeasy Blood and Tissue Kit (Qiagen, #69504) according to the manufacturer’s instructions. Two micrograms of DNA from each sample were subjected to bisulfite treatment with the EpiTect Fast Bisulfite Conversion Kit (Qiagen, # 59802). Bisulfite-converted DNA was PCR amplified with the Pyromark PCR Kit (Qiagen # 978703) and the primers listed in Supplementary Material, Table S10. Biotin-labeled amplicons were pyrosequenced and analyzed on a Qiagen Pyromark Q96 ID with PyroMark Gold Q96 Reagents (Qiagen #972804) and the analysis software included with the instrument (v 1.0.9) according to the manufacturer’s instructions.
Statistical analyses
EWAS of DNA methylation and smoking in EAGLE. We evaluated the association between current cigarette smoking and methylation in 237 NTL tissues (Supplementary Material, Table S1). For the methylation of each probe, a linear regression analysis was conducted using log-transformed methylation as the response variable, current smoking status as the main exposure, adjusting for age, sex, body mass index, non-cigarette smoking and the top three principal components based on methylation with the goal to remove potential confounding factors. The significance level for the discovery stage in EAGLE was set at P < 1.48 × 10−7 (0.05/338,456 probes), after Bonferroni-correction for multiple comparisons.
We conducted a series of sensitivity analyses, using similar models as we did in the primary analyses (Supplementary Material, Table S2). First, we excluded never smokers to compare only current smokers to former smokers, leaving 227 samples (121 current and 106 former smokers). Second, we additionally adjusted for histology or stage of the tumor to verify whether the association between smoking and methylation in normal lung tissues was confounded by the status of the paired tumor tissues. Third, we excluded subjects who smoked non-cigarettes (including cigarillos, cigars, or pipes, n = 39) to control for the effect of other tobacco products on methylation, leaving 198 samples (102 current smokers and 96 non-current smokers, including 86 former and 10 never smokers). Fourth, we excluded seven subjects who underwent chemotherapy before sample collection to minimize the potential molecular alterations caused by chemotherapy, leaving 230 samples (118 current smokers and 102 former smokers). These sensitivity analyses did not appreciably change the results.
We tested the association between cumulative smoking measures (smoking pack-years, duration, and time after smoking cessation) with methylation of significant probes identified in the discovery stage in normal lung tissues. We conducted linear regression analyses with log-transformed methylation as the response variable and smoking variables as the main exposure, adjusting for the same covariates as in the primary analyses. For analysis of TTFC, we additionally adjusted for current cigarette smoking status or smoking duration. For the analysis of smoking pack-years, duration, and TTFC, never smokers were excluded, leaving 223 subjects for the analysis. For the analysis of smoking cessation, we only included former smokers (n = 91 with information). Regression coefficient and P value were calculated for methylation alteration of each probe per 10 pack-years, per 10 years’ smoking duration, or per 10 years after quitting smoking. P < 0.05 with the same direction as in the primary analyses (positive for smoking pack-years and duration and negative for years after smoking cessation) was considered significant for the analyses of cumulative smoking measures.
Replication of DNA methylation and smoking using TCGA NTL data. We used linear regression adjusting for age and sex. A significant epigenome-wide association identified in EAGLE lung data was considered replicated if the association was found in the TCGA samples with the same direction and one-sides P-value < 0.05.
A549 CSC-treated vs. vehicle treated analysis of gene expression in 1 Mb window flanking CpGs. Differentially expressed genes were tested using the edgeR package in Bioconductor (76) (TMM normalization). Genes within a 1 Mb window flanking each of the four CpGs showing gene expression changes of >2 fold and a Bonferroni-corrected P value <0.05 (corrected for the number of genes within the 1 Mb flanking windows) were considered significantly differentially expressed.
Validation of significant smoking vs. expression genes in TCGA and examination of smoking vs. expression in 1 Mb windows flanking CpGs. Both Human Methylation450 BeadChip data (level 3) and RNA-seq data (level-3) from 100 NTL samples obtained from lung adenocarcinoma (LUAD) and squamous cell lung cancer (LUSC) patients was downloaded from the TCGA Data Portal website (http://cancergenome.nih.gov/; date last accessed May 19, 2017). RNA-seq expression value (rsem or RNA-seq by expected maximization) was log2 transformed and normalized by quantile normalization. Samples were categorized into 4 groups by smoking status: Smokers (n = 21); Current reformed smokers for > 15 years (n = 24); Current reformed smokers for ≤15 years (n = 48); and Non-smokers (n = 7). Linear regression analysis was performed to examine associations between smoking status and AHRR, CYP1B1, ENTPD2 and NR4A3 gene expression, and models were adjusted for age and gender of the patient. To investigate all genes within a 1Mb region on either side of the four methylation probes in TCGA we use R package TxDb.Hsapiens.UCSC.hg19.knownGene: Annotation package for TxDb objects(s) R package version 3.0.0 [https://bioconductor.org/packages/release/data/annotation/html/TxDb.Hsapiens.UCSC.hg19.knownGene.html; date last accessed May 19, 2017] with average normalized log2-transformed expression (log2(1 + rsem)) ≥ 3, testing under the linear regression model; P values are corrected with Bonferroni method, adjusting for the number of genes in the 1 Mb windows. Associations between gene expression and smoking-status with a Bonferroni-corrected P value < 0.05 were considered significant.
TCGA NTL expression vs. DNA methylation. Based on 28 TCGA NTL samples, we calculated the β-coefficient and log2 transformed expression values (rsem) for the associations between DNA methylation and the expression level of four genes (AHRR, CYP1B1, ENTPD2, and NR4A3), using “Pearson’s product moment correlation coefficient” method in R 3.0.0.
TCGA gene expression-mutation association. We tested for the associations between the total number of non-synonymous point mutations, and the expression level of three genes in lung adenocarcinoma tissues. Because the data show non-normality, we performed quantile normalization for the expression data so that each genomic feature had a standard normal distribution. The association was assessed by linear regression, adjusted for sex, age of diagnosis, and stage. Analysis was based on 407 subjects with non-missing genomic/clinical data, including 91 current smokers and 316 current non-smokers.
Data Access
ChIP-seq and WGBS data for AT2 cells can be accessed at the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/; date last accessed May 19, 2017) under accession number GSE94986; the RNA sequencing data of A459 under the GEO accession number GSE69770; and the methylation data under the GEO accession code GSE52401.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are extremely grateful to the EAGLE participants and the large number of collaborators (listed in https://dceg.cancer.gov/research/cancer-types/lung/environment-genetics-lung-cancer-etiology-eagle; date last accessed May 19, 2017) that made the EAGLE study possible.
Conflict of Interest statement. None of the funding agencies compromised the authors’ freedom to design, conduct, interpret and publish the results of the study. The authors declare that they have no actual or potential competing financial interests.
Funding
Intramural Research Program of National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics; National Institutes of Health grants R01 HL114094 (to IALO and ZB), R37HL062569-13 (to ZB), and the Norris Comprehensive Cancer Center core grant (NCI P30CA01408); National Cancer Institute contract HHSN261201300361P to IALO; National Institute of Environmental Health Sciences training grant T32ES013678 (to TRS); the USC Provost’s Postdoctoral Scholar Research Grant (to TRS); The American Cancer Society/Canary Foundation postdoctoral fellowship # PFTED-10-207-01-SIED (to CNM); the Thomas G. Labrecque Foundation, the L. K. Whittier Foundation, the Hastings Foundation and generous donations from Conya and Wallace Pembroke. ZB is supported by the Ralph Edgington Chair in Medicine. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/systems/; date last accessed May 19, 2017).
References
- 1. Baylin S.B., Jones P.A. (2011) A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer, 11, 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feil R., Fraga M.F. (2011) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet., 13, 97–109. [DOI] [PubMed] [Google Scholar]
- 3. Jones P.A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet., 13, 484–492. [DOI] [PubMed] [Google Scholar]
- 4. Wan E.S., Qiu W., Baccarelli A., Carey V.J., Bacherman H., Rennard S.I., Agusti A., Anderson W., Lomas D.A., Demeo D.L. (2012) Cigarette smoking behaviors and time, since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet., 21, 3073–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. (2011) Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet., 88, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joubert B.R., Håberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun Ø., Cupul-Uicab L.A.. et al. (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect., 120, 1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeilinger S., Kühnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A.. et al. (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One, 8, e63812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shenker N.S., Ueland P.M., Polidoro S., van Veldhoven K., Ricceri F., Brown R., Flanagan J.M., Vineis P. (2013) DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology, 24, 712–716. [DOI] [PubMed] [Google Scholar]
- 9. Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. (2013) Epigenome-wide association study in the European prospective investigation into cancer and nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet., 22, 843–851. [DOI] [PubMed] [Google Scholar]
- 10. Flanagan J.M., Brook M.N., Orr N., Tomczyk K., Coulson P., Fletcher O., Jones M.E., Schoemaker M.J., Ashworth A., Swerdlow A.. et al. (2015) Temporal stability and determinants of white blood cell DNA methylation in the breakthrough generations study. Cancer Epidemiol. Biomarkers Prev., 24, 221–229. [DOI] [PubMed] [Google Scholar]
- 11. Lee K.W., Richmond R., Hu P., French L., Shin J., Bourdon C., Reischl E., Waldenberger M., Zeilinger S., Gaunt T.. et al. (2015) Prenatal exposure to maternal cigarette smoking and DNA methylation: epigenome-wide association in a discovery sample of adolescents and replication in an independent cohort at birth through 17 years of age. Environ. Health Perspect., 123, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Markunas C.A., Xu Z., Harlid S., Wade P.A., Lie R.T., Taylor J.A., Wilcox A.J. (2014) Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ. Health Perspect., 122, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao X., Jia M., Zhang Y., Breitling L.P., Brenner H. (2015) DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin. Epigenetics, 7, 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. (2012) Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet., 159B, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elliott H.R., Tillin T., McArdle W.L., Ho K., Duggirala A., Frayling T.M., Davey Smith G., Hughes A.D., Chaturvedi N., Relton C.L. (2014) Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics, 6, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dogan M.V., Shields B., Cutrona C., Gao L., Gibbons F.X., Simons R., Monick M., Brody G.H., Tan K., Beach S.R.. et al. (2014) The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics, 15, 151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guida F., Sandanger T.M., Castagné R., Campanella G., Polidoro S., Palli D., Krogh V., Tumino R., Sacerdote C., Panico S.. et al. (2015) Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum. Mol. Genet., 24, 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zaghlool S.B., Al-Shafai M., Al Muftah W.A., Kumar P., Falchi M., Suhre K. (2015) Association of DNA methylation with age, gender, and smoking in an Arab population. Clin. Epigenetics, 7, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harlid S., Xu Z., Panduri V., Sandler D.P., Taylor J.A. (2014) CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the Sister Study. Environ. Health Perspect., 122, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsaprouni L.G., Yang T.P., Bell J., Dick K.J., Kanoni S., Nisbet J., Viñuela A., Grundberg E., Nelson C.P., Meduri E.. et al. (2014) Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics, 9, 1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Philibert R.A., Beach S.R., Lei M.K., Brody G.H. (2013) Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin. Epigenetics, 5, 19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun Y.V., Smith A.K., Conneely K.N., Chang Q., Li W., Lazarus A., Smith J.A., Almli L.M., Binder E.B., Klengel T.. et al. (2013) Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum. Genet., 132, 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Besingi W., Johansson A. (2014) Smoke-related DNA methylation changes in the etiology of human disease. Hum. Mol. Genet., 23, 2290–2297. [DOI] [PubMed] [Google Scholar]
- 24. Shah S., McRae A.F., Marioni R.E., Harris S.E., Gibson J., Henders A.K., Redmond P., Cox S.R., Pattie A., Corley J.. et al. (2014) Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res., 24, 1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S.. et al. (2016) Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet., 5, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ambatipudi S., Cuenin C., Hernandez-Vargas H., Ghantous A., Le Calvez-Kelm F., Kaaks R., Barrdahl M., Boeing H., Aleksandrova K., Trichopoulou A.. et al. (2016) Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics, 8, 599–618. [DOI] [PubMed] [Google Scholar]
- 27. Teschendorff A.E., Yang Z., Wong A., Pipinikas C.P., Jiao Y., Jones A., Anjum S., Hardy R., Salvesen H.B., Thirlwell C.. et al. (2015) Correlation of smoking-associated DNA methylation changes in buccal cells With DNA methylation changes in epithelial cancer. JAMA Oncol., 1, 476–485. [DOI] [PubMed] [Google Scholar]
- 28. Novakovic B., Ryan J., Pereira N., Boughton B., Craig J.M., Saffery R. (2014) Postnatal stability, tissue, and time specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics, 9, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee M.K., Hong Y., Kim S.Y., London S.J., Kim W.J. (2016) DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin. Epigenetics, 8, 103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fasanelli F., Baglietto L., Ponzi E., Guida F., Campanella G., Johansson M., Grankvist K., Johansson M., Assumma M.B., Naccarati A.. et al. (2015) Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun., 6, 10192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baglietto L., Ponzi E., Haycock P., Hodge A., Bianca Assumma M., Jung C.H., Chung J., Fasanelli F., Guida F., Campanella G.. et al. (2017) DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int. J. Cancer, 140, 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landi M.T., Consonni D., Rotunno M., Bergen A.W., Goldstein A.M., Lubin J.H., Goldin L., Alavanja M., Morgan G., Subar A.F.. et al. (2008) Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health, 8, 203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang J.T., Lee Y.M., Huang R.S. (2015) The impact of the Cancer Genome Atlas on lung cancer. Transl. Res., 166, 568–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi J., Marconett C.N., Duan J., Hyland P.L., Li P., Wang Z., Wheeler W., Zhou B., Campan M., Lee D.S.. et al. (2014) Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat. Commun., 5, 3365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M., Siegmund K.D., Koss M.N., Hagen J.A., Lam W.L.. et al. (2012) Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome. Res., 22, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu F., Wacholder S., Kovalchik S., Panagiotou O.A., Reyes-Guzman C., Freedman N.D., De Matteis S., Consonni D., Bertazzi P.A., Bergen A.W.. et al. (2014) Time to smoke first morning cigarette and lung cancer in a case-control study. J. Natl. Cancer Inst., 106, dju118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. (2012) Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. (2014) Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jaffe A.E., Irizarry R.A. (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol., 15, R31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bauer M., Linsel G., Fink B., Offenberg K., Hahn A.M., Sack U., Knaack H., Eszlinger M., Herberth G. (2015) A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin. Epigenetics, 7, 81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rowbotham S.P., Kim C.F. (2014) Diverse cells at the origin of lung adenocarcinoma. Proc. Natl. Acad. Sci. USA, 111, 4745–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Desai T.J., Brownfield D.G., Krasnow M.A. (2014) Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature, 507, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marconett C.N., Zhou B., Rieger M.E., Selamat S.A., Dubourd M., Fang X., Lynch S.K., Stueve T.R., Siegmund K.D., Berman B.P.. et al. (2013) Integrated transcriptomic and epigenomic analysis of primary human lung epithelial cell differentiation. PLoS Genet., 9, e1003513.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shlyueva D., Stampfel G., Stark A. (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet., 15, 272–286. [DOI] [PubMed] [Google Scholar]
- 45. Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G.. et al. (2013) ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res., 41, D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marconett C.N., Zhou B., Siegmund K.D., Borok Z., Laird-Offringa I.A. (2014) Transcriptomic profiling of primary alveolar epithelial cell differentiation in human and rat. Genomics Data, 2, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsay J.J., Tchou-Wong K.M., Greenberg A.K., Pass H., Rom W.N. (2013) Aryl hydrocarbon receptor and lung cancer. Anticancer Res., 33, 1247–1256. [PMC free article] [PubMed] [Google Scholar]
- 48. Baba T., Mimura J., Gradin K., Kuroiwa A., Watanabe T., Matsuda Y., Inazawa J., Sogawa K., Fujii-Kuriyama Y. (2001) Structure and expression of the Ah receptor repressor gene. J. Biol. Chem., 276, 33101–33110. [DOI] [PubMed] [Google Scholar]
- 49. Whitlock J.P. (1999) Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol., 39, 103–125. [DOI] [PubMed] [Google Scholar]
- 50. Lee J.S., Kim E.Y., Nomaru K., Iwata H. (2011) Molecular and functional characterization of Aryl hydrocarbon receptor repressor from the chicken (Gallus gallus): interspecies similarities and differences. Toxicol. Sci., 119, 319–334. [DOI] [PubMed] [Google Scholar]
- 51. Haarmann-Stemmann T., Bothe H., Kohli A., Sydlik U., Abel J., Fritsche E. (2007) Analysis of the transcriptional regulation and molecular function of the aryl hydrocarbon receptor repressor in human cell lines. Drug Metab. Dispos., 35, 2262–2269. [DOI] [PubMed] [Google Scholar]
- 52. Zhu Y., Sun L., Chen Z., Whitaker J.W., Wang T., Wang W. (2013) Predicting enhancer transcription and activity from chromatin modifications. Nucleic Acids Res., 41, 10032–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanyal A., Lajoie B.R., Jain G., Dekker J. (2012) The long-range interaction landscape of gene promoters. Nature, 489, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fraser P., Bickmore W. (2007) Nuclear organization of the genome and the potential for gene regulation. Nature, 447, 413–417. [DOI] [PubMed] [Google Scholar]
- 55. Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F.. et al. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature, 457, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Steensel B., Dekker J. (2010) Genomics tools for unraveling chromosome architecture. Nat. Biotechnol., 28, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bickmore W.A., van Steensel B. (2013) Genome architecture: domain organization of interphase chromosomes. Cell, 152, 1270–1284. [DOI] [PubMed] [Google Scholar]
- 58. Dekker J., Mirny L. (2016) The 3D genome as moderator of chromosomal communication. Cell, 164, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rowley M.J., Corces V.G. (2016) Capturing native interactions: intrinsic methods to study chromatin conformation. Mol. Syst. Biol., 12, 897.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schetinger M.R., Morsch V.M., Bonan C.D., Wyse A.T. (2007) NTPDase and 5’-nucleotidase activities in physiological and disease conditions: new perspectives for human health. Biofactors, 31, 77–98. [DOI] [PubMed] [Google Scholar]
- 61. Kumar V. (2013) Adenosine as an endogenous immunoregulator in cancer pathogenesis: where to go. Purinergic Signal, 9, 145–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wood E., Broekman M.J., Kirley T.L., Diani-Moore S., Tickner M., Drosopoulos J.H., Islam N., Park J.I., Marcus A.J., Rifkind A.B. (2002) Cell-type specificity of ectonucleotidase expression and upregulation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch. Biochem. Biophys., 407, 49–62. [DOI] [PubMed] [Google Scholar]
- 63. Safe S., Jin U.H., Morpurgo B., Abudayyeh A., Singh M., Tjalkens R.B. (2016) Nuclear receptor 4A (NR4A) family - orphans no more. J. Steroid Biochem. Mol. Biol., 157, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L.. et al. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu J., Watts J.A., Pope S.D., Gadue P., Kamps M., Plath K., Zaret K.S., Smale S.T. (2009) Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev., 23, 2824–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aran D., Sabato S., Hellman A. (2013) DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol., 14, R21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J.. et al. (2017) Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet., 49, 131–138. [DOI] [PubMed] [Google Scholar]
- 68. Marlowe J.L., Knudsen E.S., Schwemberger S., Puga A. (2004) The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem., 279, 29013–29022. [DOI] [PubMed] [Google Scholar]
- 69. Huai W., Zhao R., Song H., Zhao J., Zhang L., Zhang L., Gao C., Han L., Zhao W. (2014) Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat. Commun., 5, 4738.. [DOI] [PubMed] [Google Scholar]
- 70. Kolovos P., Knoch T.A., Grosveld F.G., Cook P.R., Papantonis A. (2012) Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin, 5, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luch A. (2005) Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer, 5, 113–125. [DOI] [PubMed] [Google Scholar]
- 72. Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics, 25, 1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011) Integrative genomics viewer [letter]. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Robinson M.D., McCarthy D.J., Smyth G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.