Abstract
Oculopharyngeal muscular dystrophy (OPMD) is a late onset disease caused by polyalanine expansion in the poly(A) binding protein nuclear 1 (PABPN1). Several mouse models have been generated to study OPMD; however, most of these models have employed transgenic overexpression of alanine-expanded PABPN1. These models do not recapitulate the OPMD patient genotype and PABPN1 overexpression could confound molecular phenotypes. We have developed a knock-in mouse model of OPMD (Pabpn1+/A17) that contains one alanine-expanded Pabpn1 allele under the control of the native promoter and one wild-type Pabpn1 allele. This mouse is the closest available genocopy of OPMD patients. We show that Pabpn1+/A17 mice have a mild myopathic phenotype in adult and aged animals. We examined early molecular and biochemical phenotypes associated with expressing native levels of A17-PABPN1 and detected shorter poly(A) tails, modest changes in poly(A) signal (PAS) usage, and evidence of mitochondrial damage in these mice. Recent studies have suggested that a loss of PABPN1 function could contribute to muscle pathology in OPMD. To investigate a loss of function model of pathology, we generated a heterozygous Pabpn1 knock-out mouse model (Pabpn1+/Δ). Like the Pabpn1+/A17 mice, Pabpn1+/Δ mice have mild histologic defects, shorter poly(A) tails, and evidence of mitochondrial damage. However, the phenotypes detected in Pabpn1+/Δ mice only partially overlap with those detected in Pabpn1+/A17 mice. These results suggest that loss of PABPN1 function could contribute to but may not completely explain the pathology detected in Pabpn1+/A17 mice.
Introduction
Oculopharyngeal muscular dystrophy (OPMD) is a late onset disease that causes ptosis, dysphagia, and loss of mobility due to progressive weakness of eyelid, pharyngeal, and proximal limb muscles. OPMD is not fatal but associated symptoms greatly affect quality of life. Dysphagia is often the prognostic symptom and the associated loss of nutrition and recurring aspiration pneumonia can be fatal (1,2). Treatments for OPMD involve surgical intervention to raise eyelids and loosen constrictor muscles of the throat to ease swallowing. No pharmacologic interventions are currently available.
The vast majority of OPMD patients have a dominant GCG expansion in the gene encoding the polyadenosine binding protein nuclear 1 (PABPN1) (3). Unlike other disease-causing expansion mutations, the dominant OPMD mutations are rather modest. Wild type PABPN1 contains a stretch of ten alanine residues that occurs just after the initial methionine. This alanine tract, which occurs in a domain with unknown function, is expanded to 11–18 alanine residues in patients with OPMD (3–5). PABPN1 is a ubiquitously expressed RNA-binding protein that is most well characterized as a regulator of polyadenosine (poly[A]) tail length and alternative polyadenylation (APA) as well as expression of antisense RNAs associated with gene promoters (6–11). Why a small expansion of alanine residues in a ubiquitously expressed RNA-binding protein causes a late-onset muscular dystrophy affecting specific muscle groups is not known.
The pathological hallmark identified in OPMD patient tissues is the presence of nuclear aggregates (12) that contain PABPN1 and other proteins including additional RNA-binding proteins and heat shock proteins (13–15). OPMD has been considered an aggregate disease akin to other triplet repeat diseases such as Huntington's disease. If toxic aggregates were the sole cause of pathology in OPMD, tissues that express the highest levels of the aggregate-prone protein would likely be most susceptible to pathology. However, muscle contains very low levels of PABPN1 relative to unaffected tissues (16), suggesting a more complex scenario that cannot be solely explained by aggregate-mediated toxicity. Consistent with a more complex mechanism underlying pathology, nuclear aggregates are found only in a small subset of myonuclei in OPMD patients (12,17,18) and have been detected in unaffected muscles and brain from an OPMD patient (19). Dynamic, non-pathogenic PABPN1 nuclear aggregates have also been detected in rat neurons (20,21), demonstrating that wild type PABPN1 can form aggregates. However, several studies link the presence of PABPN1 aggregates to cell death (22–25), supporting a role for aggregates contributing to pathology. Indeed, aggregation may deplete the functional PABPN1 pool. This model is supported by the fact that overexpression of wild type PABPN1 reverses apoptotic cell death caused by overexpression of alanine-expanded PABPN1 despite an increase in the number of nuclear aggregates (23).
Challenges exist for studying the functional consequences of PABPN1 mutation in OPMD patient muscle tissues. Patient tissues are difficult to obtain as OPMD is a rare muscle disease, affecting approximately 1:100,000 individuals (26), and muscle biopsy is invasive and painful. Furthermore, the late disease onset makes it difficult to perform systematic and controlled molecular studies with multiple time points and muscles in OPMD patient samples. Given the limited availability of patient samples, several model systems have been developed in which to study OPMD pathology including cell culture, Caenorhabditis elegans, Drosophila melanogaster, and mouse (15,19,25,27–32). However, these models have typically employed tissue-specific and/or constitutive promoters that lead to very high levels of expanded PABPN1 that do not account for the low expression of PABPN1 in muscle and thus complicate interpretation. Therefore, it is important to analyze the effects of endogenous levels of expanded PABPN1 in order understand how altered alanine-expanded PABPN1 causes muscle-specific pathology.
Many studies of OPMD pathology have been performed in mouse models. The most commonly used mouse model contains a transgenic expanded bovine PABPN1 allele under the control of the muscle-specific human skeletal actin promoter. These animals express 17 alanine-expanded PABPN1 at levels 10 to 30-fold over endogenous wild type PABPN1 levels (25,33). In these mice, approximately 5% of myonuclei contain intranuclear aggregates at three months of age and about 20% of myonuclei contain aggregates at 6 months of age. The high percentage of aggregate-positive nuclei and subsequent apoptotic myofiber death make this mouse an excellent model in which to study drug treatments that ameliorate aggregate formation and apoptosis. Indeed, several promising drugs have been tested in this model and results from some of these molecular and biochemical studies have been confirmed in OPMD patient samples (34–37).
Despite the utility of previous mouse models (22,24,25) in drug discovery, this model does not recapitulate the patient genotype and because PABPN1 has many important roles in RNA processing, overexpression of A17-PABPN1 may confound molecular data obtained from these studies. For example, in vitro experiments showed that wild type PABPN1 protein can form aggregates (38) and additional studies revealed that muscle-specific overexpression of wild type PABPN1 causes a functional phenotype in flies (30). While patient tissues would be the ideal system in which to define the early molecular events associated with OPMD pathology, they are limited in availability. Thus, a model system that closely mimics the genotype of OPMD patients is needed.
We have developed a novel mouse model of OPMD that represents the closest available genocopy of OPMD patients. This new OPMD model contains a conditional Pabpn1 knock-in allele (Pabpn1A17) at the native Pabpn1 locus, which encodes a 17 alanine-expanded PABPN1 protein (A17 PABPN1). Histologic analysis revealed that Pabpn1+/A17 mice have age-related and muscle-specific defects. We performed molecular and biochemical studies in young Pabpn1+/A17 mice and detected early-onset defects in polyadenylation and mitochondrial abnormalities. We also compared Pabpn1+/A17 mice to a heterozygous Pabpn1 knockout mouse and found similar but not identical histologic, proteomic, and molecular phenotypes. Taken together, these data suggest that loss of PABPN1 function could contribute to the muscle pathology detected in Pabpn1+/A17 mice.
Results
Generation of a new mouse model of OPMD
To generate the closest possible model to a patient genotype in autosomal dominant OPMD, we created a mouse model with a single (GCG)7 expanded Pabpn1 allele knocked in at the native Pabpn1 locus. As shown in Figure 1A, the targeted allele contains a cassette with murine Pabpn1 cDNA, an internal ribosome entry site (IRES), eGFP and a neomycin resistance gene (Neo) flanked by loxP sites followed by a 3' arm containing the (GCG)7 expansion. Following flippase and Cre-mediated recombination, the portion of the targeted allele between loxP sites is excised and the knock-in allele that encodes a 17-alanine expanded PABPN1 protein (A17 PABPN1) remains at the native Pabpn1 locus. To create mice that express A17 PABPN1 throughout development, mice containing the targeted allele were crossed with mice expressing Cre recombinase using the EIIa promoter (EIIA Cre mice), which is activated during early embryogenesis (39). Thus, resulting progeny (Pabpn1+/A17) contain a single wild type Pabpn1 allele and a knocked-in expanded Pabpn1A17 allele in all tissues, including germline tissues. These mice with the recombined allele were then crossed with each other to generate experimental animals that did not contain Cre recombinase. Mouse genotypes were determined by PCR on tail snips at weaning and in muscle tissue after tissue harvest (Supplementary Material, Fig. S1A and B).
Figure 1.
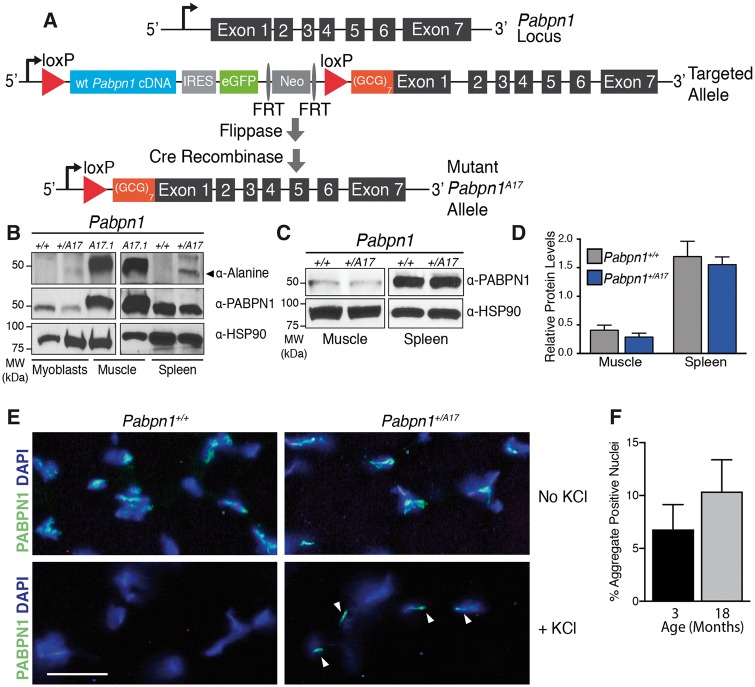
Generation of the Pabpn1+/A17 knock-in mouse. (A) Schematic of Pabpn1 knock-in allele. The wild type Pabpn1 locus is shown at the top. Before Cre-mediated recombination, the Targeted Allele contains murine wild type Pabpn1 cDNA sequence, an internal ribosome entry site (IRES), a gene encoding enhanced green fluorescent protein (eGFP) and a neomycin resistance gene (Neo) flanked by loxP sites. Downstream of the second loxP site is a start codon followed by a (GCG)7 stretch encoding the alanine expansion at the amino terminus of PABPN1. After flippase and Cre-mediated recombination, the Mutant Pabpn1A17 Allele contains the (GCG)7 expansion in Exon 1 upstream of the remaining Pabpn1 gene along with one remaining loxP site. (B) A17-PABPN1 was detected as a 50 kDa band in samples from 3-month-old male Pabpn1 mutant (+/A17) mice but not Pabpn1 wild type (+/+) mice using an α-Alanine antibody in lysates prepared from myoblasts and spleen isolated from Pabpn1+/A17mice. Lysate prepared from muscles overexpressing A17-PABPN1 (25) were used as a positive control (A17.1) for the α-Alanine antibody. Pabpn1+/+ and Pabpn1+/A17 samples were probed with α-PABPN1 antibody and α-HSP-90 antibody as controls. (C) Representative immunoblot using α-PABPN1 antibody in muscle and spleen lysates from 3-month-old male Pabpn1+/+ and Pabpn1+/A17mice demonstrating no change in total PABPN1 levels. Samples were probed with α-HSP-90 as a control. (D) Quantification of PABPN1 protein levels normalized to HSP-90 protein from muscle and spleen lysates from 3-month-old male Pabpn1+/+ and Pabpn1+/A17mice. Data are mean ± SEM of n = 3 animals per genotype. (E) Representative immunostaining for PABPN1 in rectus femoris muscle sections from 3-month-old male Pabpn1+/+ and Pabpn1+/A17mice with and without KCl incubation. Nuclei are labeled with DAPI. KCl-insoluble aggregates are marked with white arrowheads. Bar = 25 µm. (F) Quantification of percent KCl-insoluble aggregate nuclei in tibialis anterior muscle relative to total DAPI-stained nuclei from 3, and 18-month-old male Pabpn1+/A17mice. Data are mean ± SEM of n = 3 Pabpn1+/A17 animals per age.
To confirm that these mice express A17 PABPN1 in muscle and non-muscle tissue, we analyzed A17 PABPN1 expression by immunoblotting using an antibody specific to the alanine expansion (40) (Fig. 1B). As steady-state PABPN1 protein levels are low in muscle (41), we analyzed primary myoblasts and spleen tissue, which both have higher levels of PABPN1 than muscle tissue. We detected alanine-expanded PABPN1 in both myoblasts and spleen collected from Pabpn1+/A17 mice but not Pabpn1+/+mice. As a control, we included samples from an established transgenic OPMD mouse model (25), which shows high A17-PABPN1 levels as well as total PABPN1 levels consistent with the overexpression of transgenic alanine-expanded PABPN1 in this model. To assess whether total PABPN1 levels differ in the Pabpn1+/A17 mice compared to the Pabpn1+/+ mice, we analyzed total PABPN1 protein in muscle and spleen (Fig. 1C and D). This analysis revealed that, in contrast to the transgenic mouse model (25), PABPN1 protein levels are similar in Pabpn1+/+ and Pabpn1+/A17mice in both muscle and non-muscle tissue. We also detected comparable levels of Pabpn1 mRNA in muscle and non-muscle tissue from Pabpn1+/+ and Pabpn1+/A17mice when analyzed by quantitative reverse transcriptase PCR (qRT-PCR) (Supplementary Material, Fig. S1C). KCl-insoluble PABPN1 aggregates are a hallmark of OPMD pathology that are found in muscle sections from OPMD patients and animal models of OPMD (42). To determine whether muscle from the Pabpn1+/A17 mice show this hallmark of OPMD, we used immunofluorescence staining to detect PABPN1 (Fig. 1E) in KCl-insoluble aggregates in rectus femoris (RF) muscles from Pabpn1+/A17mice but not in Pabpn1+/+mice. To demonstrate that KCl-insoluble aggregates are detected in multiple muscles, we also use immunofluorescence staining to detect aggregates in tibialis anterior (TA) muscle (not shown). Quantification of immunostaining results from TA muscles revealed that, in Pabpn1+/A17mice, ∼7% of myonuclei contain KCl-insoluble PABPN1 aggregates at three months of age and ∼ 10% of myonuclei contain KCl-insoluble aggregates at 18 months of age (Fig. 1F), though the difference between the two ages was not statistically significant. Taken together, these results demonstrate that we have generated a new mouse model of OPMD.
To assess functional phenotypes in the Pabpn1+/A17mice, we employed standard assays including rotorod latency to fall and grip strength. Separate cohorts of male and female mice were tested at 6, 12, and 18 months of age. The Pabpn1+/A17mice did not have any significant defects as determined by these measurements (data not shown). However, the Pabpn1+/A17mice did display hind limb clasping starting at nine months of age (Supplementary Material, Fig. S2). This phenotype is consistent with a previously reported transgenic mouse model of OPMD that contains an expanded human PABPN1 gene under the control of the PABPN1 native promoter (19). To produce a more severe phenotype, we attempted to generate Pabpn1A17/A17 mice by crossing two Pabpn1+/A17mice. We used PCR primers designed to amplify differently sized products for the Pabpn1+ and Pabpn1A17 alleles to test for the presence of Pabpn1A17/A17 mice (Supplementary Material, Fig. S3A). Analysis of genotyping results revealed that only one of >40 pups tested positive for this genotype at weaning (Supplementary Material, Fig. S3B). This one mouse likely tested positive due to some tissue mosaicism, possibly due to uneven Cre expression during embryonic development, as the animal had no overt phenotype. We found that litters generated from these crosses were significantly smaller by ∼20% than crosses of Pabpn1+/A17mice with Pabpn1+/+mice, suggesting that Pabpn1A17/A17mice may be rarely generated or do not survive to weaning age (Supplementary Material, Fig. S3C).
Pabpn1+/A17 knock-in mice have smaller muscles than wild type littermates
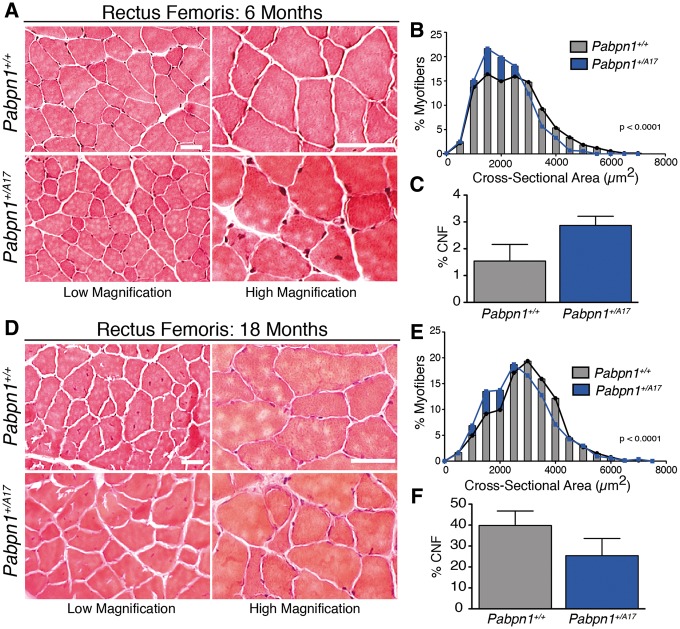
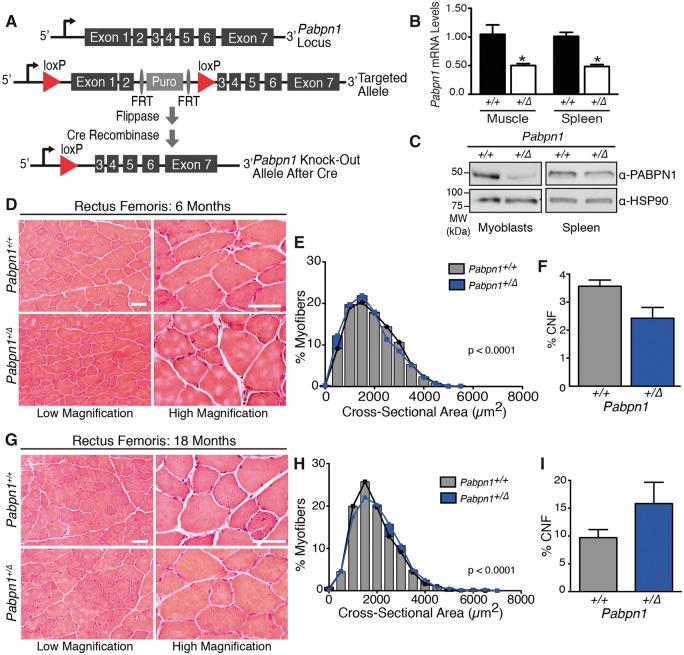
To evaluate muscle histopathology, we cross-sectioned representative proximal, rectus femoris (RF), and distal, tibialis anterior (TA), muscles from male Pabpn1+/A17and Pabpn1+/+ mice and stained with hematoxylin and eosin (H&E). Results from histologic analyses are summarized in Figure 2 (RF) and Supplementary Material, Figure S4 (TA). To take into consideration the late onset of OPMD, we analyzed muscles from animals at 6 and 18 months of age. No gross histologic defects were detected in Pabpn1+/A17mice aside from some small myofibers present in RF (Fig. 2A and D) and TA muscles (Supplementary Material, Fig. S4A and D). However, cross-sectional area (CSA) of Pabpn1+/A17myofibers was significantly smaller in both muscles at 6 months of age (Fig. 2B, Supplementary Material, Fig. S4B). In 18-month-old Pabpn1+/A17mice, RF myofiber CSA (Fig. 2E) was smaller, but TA myofiber CSA was larger (Supplementary Material, Fig. S4E). We quantified the percentage of central-nucleated fibers (CNF), a marker of myofiber regeneration (43), and detected a significant increase in CNF in TA muscles at 6 months of age in the Pabpn1+/A17mice (Supplementary Material, Fig. S4C) but no change in TA muscles at 18 months of age (Supplementary Material, Fig. S4F) or in RF muscles (Fig. 2C and F) at any age. These results demonstrate small but significant defects in muscles of Pabpn1+/A17mice compared to Pabpn1+/+mice that change with age in a muscle-specific manner.
Figure 2.
Smaller myofibers detected in rectus femoris muscles of Pabpn1+/A17mice (A) Histologic sections from rectus femoris (RF) muscles isolated from six-month-old male Pabpn1+/+ and Pabpn1+/A17mice shown at low and high magnification. Bar = 50 µm. (B) Significantly smaller myofiber cross-sectional area (CSA) was detected in six-month-old Pabpn1+/A17mice. Data are the frequency distribution of myofiber CSA. (C) No significant difference occurred in the percentage of centrally nucleated fibers (CNF) in six-month-old Pabpn1+/+ and Pabpn1+/A17mice. Data are mean ± SEM. (D) RF muscle sections from 18-month-old male Pabpn1+/+ and Pabpn1+/A17mice shown at low and high magnification. Bar = 50 µm. (E) Significantly smaller CSA detected in 18-month-old Pabpn1+/A17mice. Data are the frequency distribution of myofiber CSA. (F) No significant difference occurred in the percentage of CNF in 18-month-old male Pabpn1+/+ and Pabpn1+/A17mice. Data are mean ± SEM. In all cases, n = 3–5 mice per genotype.
Pabpn1+/A17mice have subtle early-onset RNA phenotypes
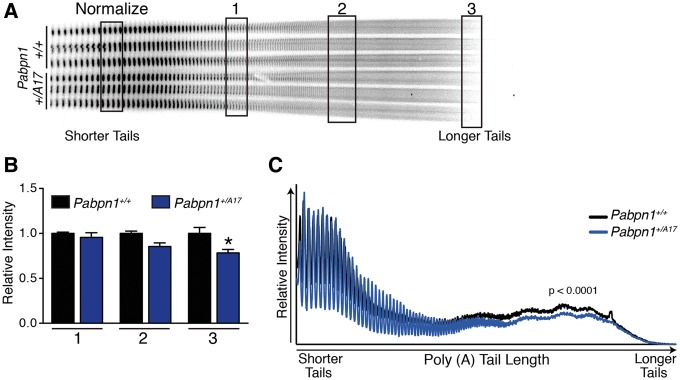
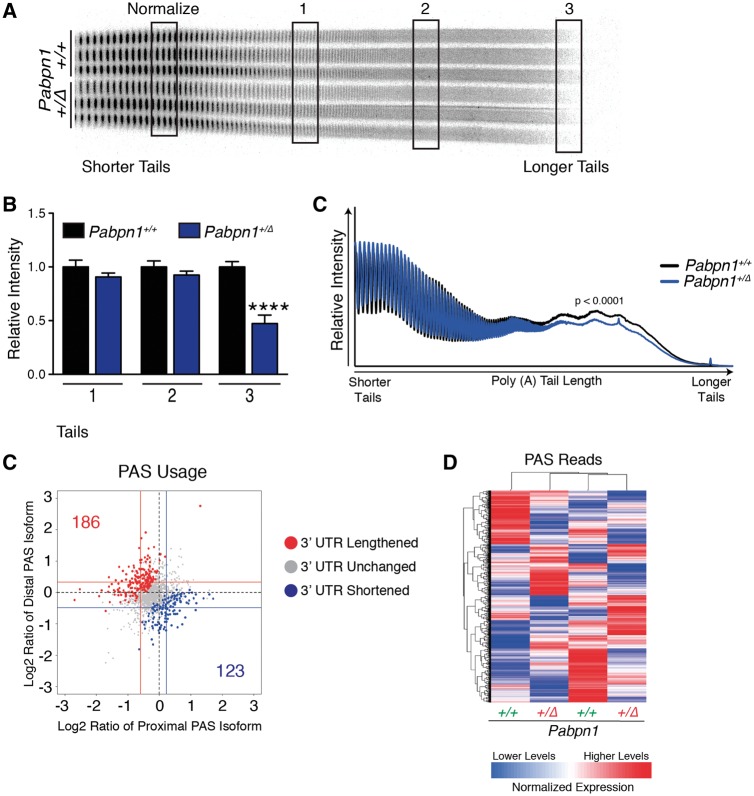
PABPN1 has multiple roles in RNA processing, but the most well-described function is enhancing the processivity of poly(A) polymerase to modulate the length of poly(A) tails added to RNAs (6–8). To determine whether polyadenylation is altered in Pabpn1+/A17mice prior to pathology, we measured bulk poly(A) tail length of total RNA isolated from RF muscles of three-month-old male Pabpn1+/+ and Pabpn1+/A17mice. We used an established assay where 3’ end radio labeled poly(A) tracts are resolved by polyacrylamide gel electrophoresis (41,42,44). As shown in Figure 3a, bulk poly(A) tail length is decreased in RNA from Pabpn1+/A17mice as compared to Pabpn1+/+ mice. Densitometry analysis at regions representing short, medium, and long poly(A) tails revealed that the longest poly(A) tails were decreased by ∼20% in Pabpn1+/A17mice relative to Pabpn1+/+ mice (Fig. 3B). Similarly, densitometry scans of each lane of poly(A) tails revealed a significant decrease in overall poly(A) tail length in Pabpn1+/A17mice (Fig. 3C). Taken together, these results indicate global poly(A) tail shortening in Pabpn1+/A17mice. This result demonstrates for the first time that expression of native levels of A17 PABPN1 affects polyadenylation, one of the major functions of PABPN1.
Figure 3.
Shortened poly(A) tails in Pabpn1+/A17 mice. (A) Poly(A) tail length was analyzed by polyacrylamide gel electrophoresis of 3’ end labeled poly(A) tracts from total RNA isolated from rectus femoris muscles of three-month-old male Pabpn1+/+ and Pabpn1+/A17 mice. n = 3 per genotype. *P < 0.05 (B) Quantification of regions representing short (1), medium (2), and long (3) poly(A) tails noted in (A) normalized to the total short poly(A) tails marked in (A) (Normalize) revealed fewer long poly(A) tails in Pabpn1+/A17mice. (C) Quantification of gel from (A) by plotting density along each lane revealed shorter poly(A) tails in Pabpn1+/A17mice.
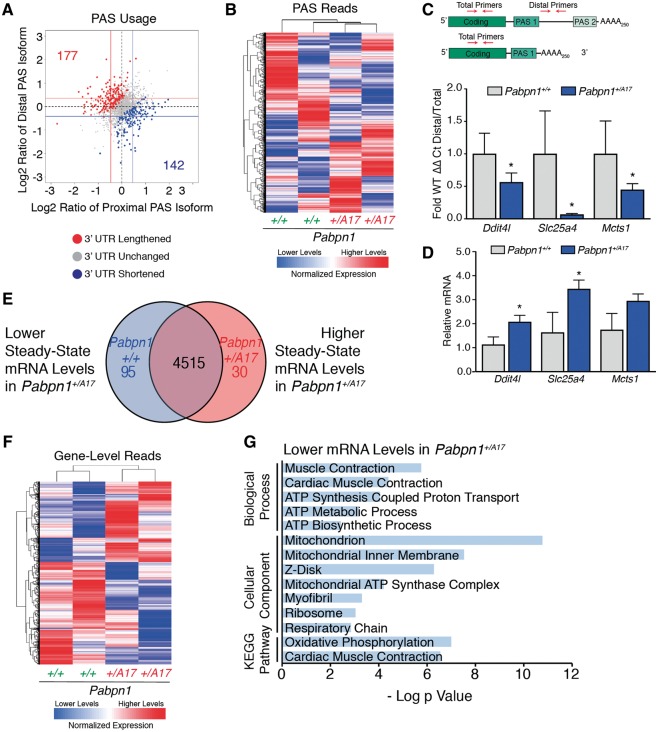
Another important function of PABPN1 in RNA processing is regulation of alternate polyadenylation signal (PAS) utilization, known as alternative polyadenylation (APA). APA can affect the coding region of the transcript or the length of the 3' untranslated region (UTR) depending on the cleavage site employed. Previous studies demonstrated that knockdown of PABPN1 in C2C12, HeLa and U2OS cell lines or transgenic overexpression of alanine-expanded PABPN1 in mouse muscles cause a global shift in PAS usage from distal to proximal PAS sites (9–11). To determine whether expression of native levels of A17 PABPN1 affects global PAS usage, we performed 3' region extraction and deep sequencing (3' READS) (45) on total RNA isolated from RF muscles of three-month-old male Pabpn1+/A17mice. As shown in Figure 4A, scatter plots representing the log ratio of distal or proximal PAS reads revealed no global shift toward either distal or proximal PAS utilization. However, a modest change in PAS usage (toward either distal or proximal PAS) in a small number of transcripts was detected (Fig. 4A), suggesting that native levels of A17 PABPN1 do affect PAS utilization. Heat maps of PAS reads (Fig. 4B) revealed that samples grouped by genotype, suggesting that changes detected in PAS usage were related to A17 PABPN1 expression rather than being stochastic or caused by some unknown variable. However, these data also indicate that the change in APA is very modest. The complete list of genes that show a change in PAS utilization compared to control can be found in Supplementary Material, Table S2.
Figure 4.
3' READS reveals modest changes in poly(A) signal usage in Pabpn1+/A17 mice. (A) Scatter plot depicting that few changes were detected in poly(A) sequence (PAS) selection by 3’ READS performed on RNA isolated from rectus femoris (RF) muscles from three-month-old male Pabpn1+/A17mice. Data are log2 ratio of distal to proximal PAS usage with increased distal PAS represented in red and increased proximal PAS represented in blue. n = 2 mice per genotype. (B) Sample clustering based on PAS usage as detected by 3'READS performed on RNA isolated from RF muscles of three-month-old mice showing that Pabpn1+/+ and Pabpn1+/A17 mice group by genotype. (C) Candidate genes validated for change in PAS selection and steady-state levels by qRT-PCR. Altered PAS selection was calculated as ΔΔCt of distal primer PCR product relative to total transcripts as detected by coding sequence primers and are reported in Pabpn1+/A17 mice as a fraction of wild type. (D) Steady-state levels of candidate genes as analyzed by qRT-PCR. All transcripts were normalized to Gapdh. *P < 0.05. (E) Venn diagram depicting changed steady-state mRNAs in RF from three-month-old Pabpn1+/A17. Of 4515 genes detected, 95 were depleted and 30 were increased. (F) Sample clustering showing that based on steady-state mRNA levels, Pabpn1+/+ and Pabpn1+/A17 mice group by genotype. (G) Histogram of the log P value of top GO terms and KEGG pathways for mRNAs decreased in Pabpn1+/A17mice revealing decreased levels of nuclear-encoded mitochondrial mRNAs.
To validate the 3’ READS data, we selected a subset of candidates to test for altered PAS usage using qRT-PCR primers targeting the coding sequence to amplify total transcript for each candidate and primers targeting the region between the proximal and distal PAS to amplify transcripts that were polyadenylated at the distal PAS (Fig. 4C, top). APA was detected as a shift in the ratio of distal PAS transcripts to the total transcript pool. We detected altered PAS usage in three candidate transcripts, Ddit4l, Slc25a4, and Mcts1 in Pabpn1+/A17mice as demonstrated by the altered ratio of transcripts utilizing the distal PAS to the total pool of transcripts (Fig. 4C, bottom). Two candidates had a corresponding change in steady-state levels when assayed by qRT-PCR and normalized to samples from Pabpn1+/+mice (Fig. 4D). Thus, expression of native levels of A17 PABPN1 has a modest effect on PABPN1 function in alternative polyadenylation, shortening and lengthening 3' UTRs of a subset of mRNAs as compared to Pabpn1+/+ mice.
The 3’ READS data also provide information about steady-state mRNA levels. As summarized in the Venn diagram in Figure 4E, of 4640 gene-level reads detected in Pabpn1+/A17and Pabpn1+/+ mice, 95 were depleted from Pabpn1+/A17mice and 30 were enriched in Pabpn1+/A17mice. A heat map of gene-level reads demonstrates that Pabpn1+/A17and Pabpn1+/+ mice group by genotype (Fig. 4F). We performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (46,47). This analysis revealed that genes with decreased steady-state mRNA levels in Pabpn1+/A17mice are enriched for terms involved in mitochondrial metabolism (Fig. 4G) suggesting there could be altered mitochondrial function in these mice.
Proteomic analysis reveals mitochondrial defects in Pabpn1+/A17mice
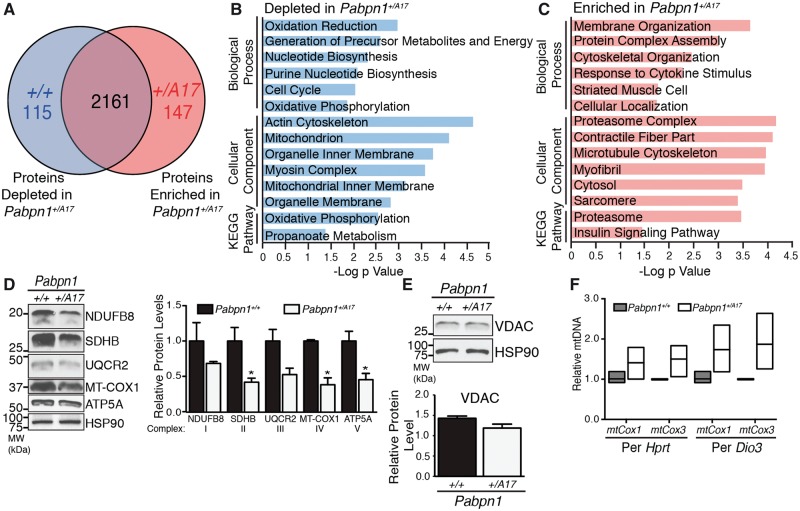
To determine whether the results obtained from the 3’ READS experiment were reflected in the proteome of Pabpn1+/A17mice, we analyzed global proteomic changes comparing RF muscle from three-month-old male Pabpn1+/+ and Pabpn1+/A17mice by mass spectrometry. Label-free quantification of proteins was performed by MaxQuant analysis of extracted ion currents (XIC). The average XIC across three biological repeats was calculated for Pabpn1+/A17mice and their corresponding Pabpn1+/+ littermates. To determine which proteins were altered, we calculated the log2 ratio of XIC values from Pabpn1+/A17relative to Pabpn1+/+mice. As represented in Figure 5A, of the 2423 proteins detected in all samples, 115 proteins were less abundant in Pabpn1+/A17mice and 147 proteins were more abundant in Pabpn1+/A17mice relative to the corresponding Pabpn1+/+mice. A summary of the total proteomic data for Pabpn1+/+ and Pabpn1+/A17mice is presented in Supplementary Material, Table S3. We performed GO and KEGG pathway analysis using DAVID. The results revealed that proteins that are depleted in Pabpn1+/A17 relative to Pabpn1+/+ are most significantly enriched for terms involving mitochondria and mitochondrial metabolism (Fig. 5B) while proteins that were more abundant in Pabpn1+/A17 relative to Pabpn1+/+ were most significantly enriched for terms involving cell structure and the proteasome complex (Fig. 5C).
Figure 5.
Proteomic analysis of Pabpn1+/A17 mice reveals altered mitochondrial proteins. (A) Diagram depicting number of total proteins detected in rectus femoris from three-month-old male Pabpn1+/+ and Pabpn1+/A17mice and those enriched or depleted in Pabpn1+/A17mice. (B) Histogram of the log P value of top GO terms and KEGG pathways for proteins depleted from Pabpn1+/A17mice revealing depletion of mitochondrial proteins. (C) Histogram of the log P value of top GO terms and KEGG pathways for proteins enriched in Pabpn1+/A17mice revealing increased levels of proteasome and cytoskeletal organization proteins. (D) Immunoblot (left) from homogenized rectus femoris muscles from three-month-old Pabpn1+/+ and Pabpn1+/A17mice showing decreased levels of electron transport chain proteins (NDUFB8, SDHB, UQCR2, MT-COX1, ATP5A) as quantified relative to total HSP-90 (right). (E) No change in total mitochondrial content as detected by immunoblot for VDAC (top) and quantified relative to HSP-90 (bottom) in muscles from Pabpn1+/+ and Pabpn1+/A17 mice. (F) Increased mitochondrial DNA (mtDNA) as detected by quantitative PCR of mtDNA markers mtCox1 and mtCox3 relative to nuclear genome markers Hprt and Dio3 in total genomic DNA from Pabpn1+/+ and Pabpn1+/A17 mice. All quantitative data are mean ± SEM from n = 3 animals per genotype. *P < 0.05.
Many of the mitochondrial proteins depleted in Pabpn1+/A17 mice are involved in the electron transport chain and oxidative phosphorylation (OXPHOS). To test for dysregulation of OXPHOS proteins in Pabpn1+/A17mice, we performed immunoblot analysis for electron transport chain complex subunits (Fig. 5D). While each complex of the electron transport chain contains multiple subunits, many of the subunits are labile when not incorporated into the complex. Thus, levels of labile subunits can be used as a proxy to quantify the entire complex. We detected significant decreases in SDHB (Complex II), COX1 (Complex IV), and ATP5A (Complex V) proteins in RF muscles from three-month-old male Pabpn1+/A17mice relative to Pabpn1+/+mice. Trends toward decreased levels of NDUFB8 (Complex I) and UQCR2 (Complex III) were also detected. To determine whether the decrease in OXPHOS proteins was due to a decrease in total mitochondrial content, we quantified total mitochondria by immunoblotting for the outer membrane voltage-dependent anion channel (VDAC) and by performing quantitative PCR (qPCR) to detect mitochondrial DNA (mtDNA). No change was detected in total mitochondrial content as assessed by immunoblotting for VDAC (Fig. 5E). However, Figure 5F shows that a trend toward increased levels of two mitochondrial genes, mtCox1 and mtCox3 (48), was detected when analyzed by qPCR and normalized to two nuclear genes, Hprt and Dio3, suggesting increased levels of mtDNA relative to nuclear DNA. Variability in samples from Pabpn1+/A17mice, potentially due to varying degrees of mitochondrial damage, likely contributed to the lack of statistical significance for these changes.
The fact that increased mtDNA level does not correspond with an increased mitochondrial protein marker level suggests an increase in mtDNA replication, which can indicate mitochondrial dysfunction related to age (49). To further test for aberrant mitochondrial function in Pabpn1+/A17mice, we tested for defects in electron transport chain complex activity. We stained sectioned RF and TA muscles from three-month-old male mice for succinate dehydrogenase activity (SDH, Complex II) using nitro blue tetrazolium assay. We detected a significant decrease in SDH positive fibers in RF muscles from Pabpn1+/A17mice (Supplementary Material, Fig. S5) but did not detect any change in SDH positive fibers in TA muscles from Pabpn1+/A17mice. This result suggests that functional mitochondrial defects are muscle-specific and are more prevalent in proximal limb muscle than distal limb muscle. To further assess mitochondrial function, we performed in vitro assays of NADH:ubiquinone oxidoreductase (Complex I) activity using homogenize RF muscle from 3, 12, and 18-month-old Pabpn1+/+ and Pabpn1+/A17mice. We detected a trend toward decreased Complex I activity in RF muscle homogenate from 3-month-old male mice and a statistically significant decrease in 12-month-old male Pabpn1+/A17mice (Supplementary Material, Fig. S5C). These results suggest that early-onset mitochondrial defects are exacerbated with age. These results support the proteomic and immunoblot data and indicate functional mitochondrial defects in Pabpn1+/A17mice.
Comparison with Pabpn1 knock-out suggests loss of function may contribute to but does not fully explain pathology
Recent studies have suggested that a loss of PABPN1 function could contribute to phenotypes associated with OPMD (16,23,33,41,50). To determine whether a loss of PABPN1 function contributes to the phenotypes detected in Pabpn1+/A17mice, we generated Pabpn1 knock-out mice for comparison. As shown in Figure 6A, the Pabpn1 targeted allele contains regions homologous to exons 1 and 2 of the Pabpn1 gene and a puromycin resistance gene within two FRT sites flanked by loxP sites. Following flippase and Cre-mediated recombination, exons 1 and 2 are excised resulting in a null Pabpn1 allele with no transcriptional start site. To generate mice with Pabpn1 knocked out from early embryogenesis, mice containing the targeted allele were crossed with EIIA Cre mice to generate Pabpn1+/Δmice. Mouse genotype was determined by PCR of tail snips taken at weaning and by PCR of muscle samples after tissue harvest (Supplementary Material, Fig. S6A and B). When Pabpn1+/Δmice were crossed with Pabpn1+/Δmice to generate homozygous Pabpn1 knock-out mice, none were detected at weaning (Supplementary Material, Fig. S6C). Furthermore, quantification of total pups produced by crossing Pabpn1+/Δmice to Pabpn1+/Δmice revealed that litters produced by these crosses were significantly smaller than those generated by crossing Pabpn1+/Δmice to Pabpn1+/+ (Supplementary Material, Fig. S6D). These data show that Pabpn1Δ/Δmice may not be viable and suggest that Pabpn1 may be an essential gene. All subsequent studies to explore the functional consequences of decreased PABPN1 were carried out using Pabpn1+/Δmice. Analysis of qRT-PCR and immunoblot data confirmed that muscle and non-muscle tissue from Pabpn1+/Δmice show a ∼50% decrease in steady-state Pabpn1 mRNA and PABPN1 protein when compared to Pabpn1+/+ littermates (Fig. 6B and C), validating this as a model to assess the consequence of decreased levels of PABPN1.
Figure 6.
Age-related hypertrophy in Pabpn1+/Δ knock-out mice. (A) Schematic of Pabpn1 knock-out allele. Before Cre recombinase-mediated recombination, the targeted allele contains a puromycin resistance gene flanked by FRT elements between exons 2 and 3 of the Pabpn1 locus. The FRT-flanked puromycin resistance gene and exons 1 and 2 of Pabpn1 are surrounded by loxP sites. After flippase and Cre-mediated recombination, exons 1 and 2 of Pabpn1 are excised resulting in a null allele with no start codon. (B) Steady-state Pabpn1 mRNA levels were decreased by ∼50% in muscle and spleen from 3-month-old male Pabpn1+/Δmice as detected by qRT-PCR. Data are mean fold change ± SEM from n = 3 mice per genotype. *P < 0.05. (C) Steady-state PABPN1 protein levels were decreased by half in 3-month-old male Pabpn1+/Δmice as detected by immunoblot using an α-PABPN1 antibody (41). HSP-90 was used as a loading control. (D) Histologic sections from rectus femoris (RF) muscles isolated from six-month-old male Pabpn1+/+ and Pabpn1+/Δ mice shown at low and high magnification. Bar = 50 µm. (E) Significantly smaller myofiber cross-sectional area (CSA) was detected in RF muscles from six-month-old Pabpn1+/Δ mice. Data are the frequency distribution of myofiber CSA. (F) No significant difference detected in the percentage of centrally nucleated fibers (CNF) from RF sections from six-month-old male Pabpn1+/+ and Pabpn1+/Δ mice. Data are mean ± SEM. (G) Histologic sections from RF muscle isolated from 18-month-old male Pabpn1+/+ and Pabpn1+/Δ mice shown at low and high magnification. Bar = 50 µm. (H) Significantly larger CSA was detected in RF muscles from 18-month-old male Pabpn1+/Δ mice. Data are the frequency distribution of myofiber CSA. (I) No significant difference was detected in the percentage of centrally nucleated fibers (CNF) from RF sections from 18-month-old male Pabpn1+/+ and Pabpn1+/Δ mice. Data are mean ± SEM. In all cases, n = 5 mice per genotype.
To test for muscle histopathology associated with decreased PABPN1 levels, we analyzed H&E stained sections of RF and TA muscles from male Pabpn1+/+and Pabpn1+/Δ mice at 6 and 18 months of age. In six-month-old Pabpn1+/Δmice, we detected some small myofibers in both RF and TA muscles but no other obvious defects (Fig. 6D and G and Supplementary Material, Fig. S7A and D). Frequency distribution of myofiber CSA of both RF and TA from six-month-old Pabpn1+/+and Pabpn1+/Δ mice (Fig. 6E and Supplementary Material, Fig. S7B) revealed significant shifts toward smaller myofibers in Pabpn1+/Δmice in both muscles. These results are consistent with defects detected in six-month-old Pabpn1+/A17 mice (Fig. 2 and Supplementary Material, Fig. S4). In 18-month-old Pabpn1+/Δmice, CSA was significantly increased in both RF (Fig. 6G and H) and TA (Supplementary Material, Fig. S7D and E) myofibers indicating age-related hypertrophy. In contrast, this age-related hypertrophy was detected only in TA muscles from 18-month-old Pabpn1+/A17 mice. CNF were not significantly increased in RF or TA muscles (Fig. 6F and I, Supplementary Material, Fig. S7C and F) of Pabpn1+/Δmice relative to Pabpn1+/+ mice at 6 months of age whereas CNF were increased in TA muscles of Pabpn1+/A17 mice at this age (Supplementary Material, Fig. S4C). These results indicate defects in Pabpn1+/Δ muscles are similar to those detected in Pabpn1+/A17 muscles in adult animals. However, aged Pabpn1+/Δmice do not recapitulate all histologic defects detected in aged Pabpn1+/A17 mice.
To determine whether Pabpn1+/Δ mice had similar molecular phenotypes to Pabpn1+/A17 mice, we analyzed RF muscles isolated from three-month-old male Pabpn1+/Δand Pabpn1+/+ mice. Quantification of poly(A) tail length revealed a ∼ 50% decrease in the longest poly(A) tails in the Pabpn1+/Δ mice (Fig. 7A and B), which is comparable to the 50% decrease in longest poly(A) tails caused by Pabpn1 knockdown in primary myoblasts (41). Densitometry analysis of each lane indicates a global decrease in bulk poly(A) tail length in RNA isolated from RF muscles of Pabpn1+/Δ mice (Fig. 7C). This result is similar to the global poly(A) tail shortening detected in primary myoblasts after Pabpn1 knockdown (41), and to the poly(A) tail shortening detected in Pabpn1+/A17 mice (Fig. 3). To determine whether loss of 50% of PABPN1 affects APA, we performed 3’ READS analysis comparing Pabpn1+/Δand Pabpn1+/+ mice. Scatter plots of distal to proximal PAS reads shown in Figure 7C revealed very little change in PAS usage with 50% PABPN1 depletion. Cluster analysis of PAS reads revealed no grouping of Pabpn1+/Δand Pabpn1+/+ samples by genotype (Fig. 7D), indicating no significant change in PAS usage on a global scale. These data indicate that PABPN1 depletion and expression of native levels of A17 PABPN1 both lead to shorter poly(A) tails but 50% depletion of PABPN1 does not cause a global shift in PAS utilization at least in young mice.
Figure 7.
Decreased poly(A) tail length but no change in PAS usage in Pabpn1+/Δ mice. (A) Poly(A) tail length was analyzed by polyacrylamide gel electrophoresis of 3’ end labeled poly(A) tracts from total RNA isolated from rectus femoris muscles of three-month-old male Pabpn1+/+ and Pabpn1+/Δ mice. n = 3 per genotype. (B) Quantification of regions representing short (1), medium (2), and long (3) poly(A) tails noted in (A) normalized to the total short poly(A) tails marked in (A) (Normalize) revealed fewer long poly(A) tails in Pabpn1+/A17mice. (C) Quantification of gel from (A) by plotting density along each lane revealed shorter poly(A) tails in Pabpn1+/Δmice. ****P < 0.0001 (D) Scatter plot depicting few changes were detected in poly(A) sequence (PAS) selection as detected by 3'READS. Data are log2 ratio of distal to proximal PAS usage with increased distal PAS represented in red and increased proximal PAS represented in blue. (E) Sample clustering based on PAS usage as detected by 3'READS showing that Pabpn1+/+ and Pabpn1+/Δ mice do not group by genotype. n = 2 mice per genotype.
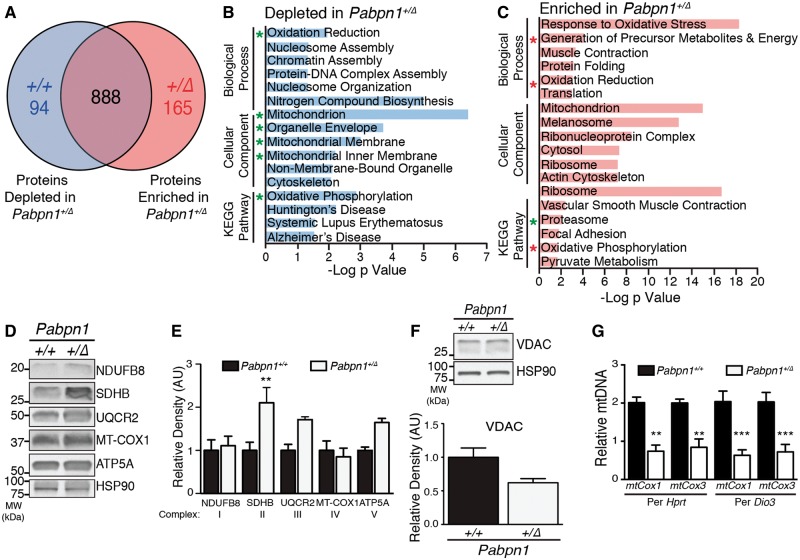
To explore whether the proteomic changes we identified in Pabpn1+/A17 mice are also detected in Pabpn1+/Δ mice, we used global proteomic analysis of RF muscles comparing three-month-old Pabpn1+/Δto Pabpn1+/+ mice (Fig. 8A). We detected a total of 1147 proteins with 94 proteins depleted from and 165 proteins enriched in Pabpn1+/Δmice relative to Pabpn1+/+. Analysis of GO terms and KEGG pathways revealed that several terms related to mitochondrial metabolism and the proteasome complex were altered in RF muscles from Pabpn1+/Δ mice relative to Pabpn1+/+ mice (Fig. 8B and C), which was similar to changes detected in Pabpn1+/A17 mice (compare to Fig. 5B and C). To determine whether mitochondrial dysregulation occurs in RF muscles from Pabpn1+/Δmice, we examined levels of electron transport chain complex subunits by immunoblotting and examining total mitochondrial content in Pabpn1+/Δand Pabpn1+/+ mice. Representative immunoblot and densitometric quantification revealed that, in contrast to Pabpn1+/A17mice, RF muscles from Pabpn1+/Δ had significantly increased steady-state levels of SDHB while levels of UQCR2 and ATP5A trended upward but this increase was not statistically significant (Fig. 8D and E). Analysis of SDH activity revealed that no significant change was detected in the number of SDH positive myofibers in TA or RF (Supplementary Material, Fig. S8). However, consistent with data showing increased levels of electron transport chain complex proteins (Fig. 8D and E), a trend toward an increase in SDH positive myofibers was detected in RF muscles. These results contrast with the data showing decreased SDH positive myofibers in RF muscles of Pabpn1+/A17 mice (Supplementary Material, Fig. S5). No significant change in total mitochondrial content was detected by immunoblot analysis of VDAC levels (Fig. 8F), though levels trended downward. Analysis of qPCR data revealed decreased levels of mtCox1 and mtCox3 relative to nuclear DNA markers (Fig. 8G), indicating decreased mtDNA content whereas Pabpn1+/A17 mice showed increased mtDNA content (compare to Fig. 6C). These data suggest that different biochemical consequences result from similar molecular phenotypes detected in Pabpn1+/Δ mice and Pabpn1+/A17 mice.
Figure 8.
Mitochondrial defects detected in Pabpn1+/Δ mice. (A) Diagram depicting number of total proteins detected in rectus femoris muscles from three-month-old male Pabpn1+/+ and Pabpn1+/Δ mice and those enriched or depleted in Pabpn1+/Δ mice. (B) Histogram of the log P value of top GO terms and KEGG pathways for proteins depleted from Pabpn1+/Δ mice revealing decreased levels of mitochondrial proteins. Groups that are also depleted in Pabpn1+/A17 mice are marked with a green asterisk. (C) Histogram of the log p value of top GO terms and KEGG pathways for proteins enriched in Pabpn1+/Δmice revealing increased levels of mitochondrial and proteasome proteins. Groups that are depleted rather than enriched in Pabpn1+/A17 mice are marked with a red asterisk. (D) Increased levels of electron transport chain proteins (SDHB, UQCR2, ATP5A) as detected by immunoblot (left) and quantified relative to total HSP-90 (E) in Pabpn1+/Δmice. (F) No change in total mitochondrial content as detected by immunoblot for VDAC (top) and quantified relative to HSP-90 (bottom) in Pabpn1+/+ and Pabpn1+/Δmice. (G) Decreased mitochondrial DNA (mtDNA) as detected by quantitative PCR of mtDNA markers mtCox1 and mtCox3 relative to nuclear genome markers Hprt and Dio3 in total genomic DNA fromPabpn1+/+ and Pabpn1+/Δmice. All quantitative data are mean ± SEM from n = 3 animals per genotype. *P < 0.05.
Discussion
We have generated a mouse model that is the closest available genocopy to OPMD patients. This mouse model stands out in comparison to other models in that the expanded Pabpn1 allele is present at the native locus and thus the model does not rely on transgenic expression of alanine-expanded PABPN1 and does not overexpress alanine-expanded PABPN1. In Pabpn1+/A17 mice, we detected small but significant muscle-specific and age-related muscle histopathology. We also demonstrated that expression of native levels of A17-PABPN1 causes an overall decrease in poly(A) tail length. Although we did not detect a global shift in PAS, we detected modest changes in APA in a subset of RNAs. We also demonstrated that mitochondrial defects occur at the RNA and protein level. Comparing Pabpn1+/Δ and Pabpn1+/A17 mice revealed a partial overlap in histologic and molecular phenotypes, supporting a model where a partial loss of PABPN1 function contributes to but does not fully explain muscle pathology.
These novel mouse models allow analysis of mammalian PABPN1 function in vivo. We were unable to generate homozygous Pabpn1Δ/Δ mice, which suggests that Pabpn1 may be an essential gene in mice. We also found that we were unable to generate homozygous Pabpn1A17/A17 mice in Mendelian ratios, suggesting that A17-PABPN1 may not be able to perform essential PABPN1 functions. However, because aggregate-mediated PABPN1 sequestration or cell death may be exacerbated by increased A17-PABPN1 doses, it is unclear whether decreased PABPN1 function, PABPN1 aggregation, or some combination of aggregation and loss of function underlie this lethality. Considering that Ala13 homozygous OPMD patients have been reported (51), perhaps a mouse model containing Pabpn1A13/A13 would be a useful system in which to study the effects of expanded PABPN1 dosage.
To assess muscle histopathology in these newly generated mouse models, we performed histologic analysis on both proximal (RF) and distal (TA) limb muscles from six-month-old (adult) and 18-month-old (aged) mice. We detected atrophy in RF muscles of adult and aged Pabpn1+/A17 mice along with small and central nucleated fibers (CNF), though no significant change in total CNF was detected. These results are consistent with proximal limb involvement reported in OPMD patients (52,53). We also detected atrophy in TA muscles of adult Pabpn1+/A17 mice but detected hypertrophy in TA muscles of aged Pabpn1+/A17 mice. Age-related hypertrophy may be a result of a compensatory myogenic response, which is supported by the finding of increased CNF in TA muscles of adult Pabpn1+/A17 mice. Age-related hypertrophy has also been detected in other mouse models of muscle disease such as the mdx model of Duchenne muscular dystrophy (54,55). Atrophy was also detected in RF and TA muscles from adult Pabpn1+/Δ mice. However, in aged Pabpn1+/Δ mice, hypertrophy was detected in both RF and TA muscles. These data indicate phenotypic overlap in adult Pabpn1+/A17 and Pabpn1+/Δ mice that diverges with age in proximal (RF) but not distal (TA) muscles.
To examine molecular events that could precede pathology in Pabpn1+/A17 and Pabpn1+/Δ mice, we analyzed RF muscle isolated from three-month-old animals. Decreased bulk poly(A) tail length detected in muscles from three-month-old Pabpn1+/A17 and Pabpn1+/Δ mice is an intriguing result that suggests that alanine-expanded PABPN1 is not fully functional in this canonical role for PABPN1. Previous work has shown that Pabpn1 knockdown in cell culture results in a similar decrease in poly(A) tail length (41), so this result could be due to depletion of the total pool of PABPN1 in Pabpn1+/Δ mice. Our analysis shows that steady state levels of PABPN1 protein are comparable in muscles from Pabpn1+/A17 mice and Pabpn1+/+mice, suggesting that the poly(A) tail defect is caused by loss of PABPN1 function rather than PABPN1 protein. However, the decrease in bulk poly(A) tail length in the Pabpn1+/A17 mice could result from sequestration of PABPN1 protein in aggregates with no impact on total protein levels detected. This possibility of loss of PABPN1 function due to sequestration complicates studies to assess the function of alanine-expanded PABPN1.
We did not detect a global shift in alternative polyadenylation signal (PAS) usage (APA) in muscles from Pabpn1+/A17 mice. Previous reports linking PABPN1 to global changes in APA with a shift toward more proximal PAS usage were performed in cell culture or animal models that either had Pabpn1 knocked down or transgenic alanine-expanded PABPN1 overexpressed (9–11). Though these studies point to an important function of PABPN1 in regulating PAS selection, our results suggest that this function is only modestly disrupted by expressing native levels of alanine-expanded PABPN1 in young Pabpn1+/A17 mice. Also notable is no discernable regulation of antisense transcripts from gene promoters in Pabpn1+/A17 mice, which was reported previously with Pabpn1 knockdown in cell culture (10). Therefore, aberrant PAS utilization on a global scale may not be an early driver of pathology associated with OPMD. Although small changes in PAS utilization were detected in Pabpn1+/A17 mice, even fewer were detected in Pabpn1+/Δ mice. A previous study of mouse TA muscle with viral-delivered shRNA-mediated acute Pabpn1 knockdown demonstrated altered PAS utilization in transcripts corresponding to a subset of proteasome genes (50) so perhaps in the case of some RNAs, 50% loss of PABPN1 is sufficient to alter PAS usage. However, in this model, while the overall knockdown achieved was ∼50%, comparable to our genetic model, the viral delivery could have led to heterogeneity in the amount of PABPN1 knockdown achieved in different muscle fibers. Because it has been shown in humans that steady-state PABPN1 mRNA levels decrease with age (34), perhaps the effect of alanine-expanded PABPN1 on PAS utilization becomes more relevant to OPMD pathology over time. Further work is needed to determine whether changes in PAS utilization increase with age in Pabpn1+/A17 or Pabpn1+/Δ mice.
Quantification of steady-state mRNA levels from the 3’READS experiment as well as validated proteomic data showed that mitochondria are impaired in Pabpn1+/A17 mice. These results are consistent with those reported previously in a study implicating mitochondrial dysfunction in Drosophila and mouse models of OPMD that overexpress alanine-expanded PABPN1 as well as in OPMD patient tissues (35). In those studies, decreased poly(A) tail length of key nuclear-encoded mitochondrial mRNAs was also reported. We detected altered PAS usage and increased levels of Ddit4l mRNA, which is a regulator of mTOR signaling that may contribute to impaired mitochondrial function. Elevated levels of Ddit4l have been reported in cases of disuse atrophy (56,57) so decreased myofiber CSA detected in RF of adult and aged mice could be a result of increased Ddit4l and decreased mTOR signaling. We detected altered levels of nuclear-encoded mitochondrial mRNAs and protein but did not detect any change in levels of VDAC protein, a marker of total mitochondrial content. We did detect increased mitochondrial genome copy number in Pabpn1+/A17 mice, which is associated with aging in skeletal muscle (49). Increased mitochondrial DNA detected in Pabpn1+/A17 mice suggests that premature mitochondrial aging may be an early-onset symptom of OPMD.
Interestingly, we detected different effects on OXPHOS protein levels and mtDNA content in Pabpn1+/A17 mice and Pabpn1+/Δ mice. Previous reports of mitochondrial defects have been attributed to shortened poly(A) tails in key nuclear-encoded mitochondrial RNAs (35). However, the fact that both Pabpn1+/A17 mice and Pabpn1+/Δ mice had shortened bulk poly(A) tails but different mitochondrial phenotypes suggests that another mechanism may contribute to mitochondrial defects. A recent report demonstrated that transgenic expression of alanine-expanded PABPN1 causes accumulation of misspliced TNNT3 mRNA due to TNNT3 mRNA sequestration in aggregates away from SC35-containing splicing speckles in samples from a cell culture model of OPMD as well as samples from OPMD patients (58). Perhaps other aberrant splicing events in nuclear-encoded mitochondrial genes could occur in Pabpn1+/A17 mice and contribute to aberrant mitochondrial function.
While patient tissues are the ideal system in which to study mechanisms of OPMD pathology, they present a challenge in that their limited availability, combined with the late onset nature of the disease, makes controlled studies of symptomatic and pre-symptomatic individuals difficult. This mouse model, which is the closest available genocopy to OPMD patients, can be used to perform the types of studies that are hindered by limited patient tissue availability. Critically, comparison of molecular and biochemical data generated in Pabpn1+/A17 mice recapitulate some of the major results obtained from OPMD patient tissue studies. For example, the validated 3’ READS and proteomics data supporting mitochondrial dysfunction in young Pabpn1+/A17 mice agree with proteomic data reported from OPMD patient biopsies showing mitochondrial proteins as a major category of dysregulated proteins (35). Other microarray studies have shown altered levels of mRNAs encoding mitochondrial proteins in OPMD patient tissues (34) and defects in mitochondrial morphology have been reported in several patient case studies (59–62). With the availability of these mice, future studies can determine the mechanistic details of mitochondrial defects.
In addition to our study, several additional reports suggest that loss of PABPN1 function could contribute to muscle pathology in OPMD (23,33,41,50). If alanine expansion in PABPN1 interferes with normal PABPN1 function(s), overexpression of the wildtype PABPN1 should ameliorate deleterious effects. Indeed, overexpression of wildtype PABPN1 is protective against depletion of the X-linked inhibitor of apoptosis and cell death in mice overexpressing A17-PABPN1 (23) and recent studies of potential gene therapies for OPMD revealed that expression of exogenous wild type Pabpn1 reversed the myofiber cross-sectional area defect in muscles from the A17.1 mouse (63). Furthermore, over-expression of wildtype PABPN1 prevents functional defects in pharyngeal muscles associated with aging in mice (33). In fact, loss of protein function may be more relevant than previously appreciated in other aggregate-associated diseases. For example, loss of APP/Aβ protein function may contribute to Alzheimer’s disease pathology (64) and loss of huntingtin protein during development is associated with age-related neurodegeneration in a mouse model (65). Future work is needed to elucidate the complicated interplay between toxicity, cell death, and loss of protein function in aggregate-associated proteinopathies. These studies would benefit from use of model systems such as the one described here that do not employ transgenic overexpression of pathogenic proteins.
OPMD is often characterized as a disease of aggregate-mediated toxicity, though new data are emerging to argue that this model is too simplistic to explain the pathology that occurs in a subset of muscles in OPMD patients. Several studies performed in a well-established mouse model that overexpresses alanine-expanded PABPN1 demonstrate that aggregate formation leads to severe muscle phenotypes and that removal of aggregates reverses those phenotypes (22,24,25). However, a different study reported that soluble expanded PABPN1 was more toxic to cells than the aggregated form (66) and still another reported that overexpression of wild type PABPN1 ameliorated phenotypes related to alanine-expanded PABPN1 overexpression (23). Data from our newly-created Pabpn1+/A17 and Pabpn1+/Δ mice indicate some molecular and histologic overlap but also divergence in biochemical phenotypes. The fact that phenotypes in Pabpn1+/A17 and Pabpn1+/Δ mice do not overlap completely hint at a more complex model of pathology in which defects due to loss of function and PABPN1 aggregation are not mutually exclusive. With the creation of this mouse model, a new tool is available for future studies such as determining tissue-specific requirements for PABPN1, assessing the function of alanine-expanded PABPN1 in muscle, and exploring whether native levels of alanine-expanded PABPN1 form aggregates in other tissues. These studies will help to determine how alanine-expanded PABPN1 expression causes pathology in skeletal muscle and may help to identify new targets for the treatment of OPMD.
Materials and Methods
Generation of Pabpn1+/A17 knock-in mice
The conditional Pabpn1+/A17 knock-in mouse line was generated by Ozgene Pty Ltd (Bentley WA, Australia) (Fig. 1A). A targeting construct was cloned to contain the (GCG)7 expansion at the 5' end. Upstream of the expansion mutation, the targeting construct contained a wild type murine Pabpn1 cDNA sequence, an internal ribosome entry site (IRES), a gene encoding enhanced green fluorescent protein (eGFP) and a neomycin resistance gene (Neo) flanked by flippase recognition target (FRT) sites. The wild type Pabpn1 cDNA, IRES, eGFP, and Neo were flanked by loxP sites. The entire targeting construct was flanked at each end with regions of homology to the 5' end of the Pabpn1 coding sequence and the region immediately upstream of the Pabpn1 coding sequence. Targeting constructs were electroporated into a C57BL/6 ES cell line, Bruce4 (67). Homologous recombinant ES cell clones were identified by Southern hybridization and injected into blastocysts. Male chimeric mice were obtained and crossed to C57BL/6J females to establish heterozygous offspring. Mice heterozygous for the targeted allele were crossed with mice expressing flippase to remove the Neo gene and subsequently crossed with mice containing the Cre recombinase gene under the control of the EIIa promoter (The Jackson Laboratory, Bar Harbor, ME) to generate mice containing Pabpn1+/A17 in all cells including the germline (Fig. 1A) (39). Pabpn1+/A17 knock-in mice were genotyped by PCR using primers designed to detect the presence of the loxP site (A17 Allele primers) and the presence of the eGFP gene (Floxed Allele primers) found in the floxed allele (Supplementary Material, Fig. S1). Animals that generated a PCR product for the loxP site but not for the eGFP gene were considered positive for the Pabpn1A17 allele. Initial crosses were tested for the presence of the Cre recombinase gene using Cre primers designed by the Jackson Laboratory (oIMR1084 and oIMR1085). To determine whether knock-in mice were homozygous or heterozygous for Pabpn1A17, primers were designed to amplify differently sized products for the Pabpn1+ and Pabpn1A17 alleles (WT/A17 primers, Supplementary Material, Fig. S3A). Animals that generated products for both Pabpn1+ and Pabpn1A17 alleles were considered to be heterozygous knock-ins. Sequences for PCR primers used for genotyping can be found in Supplementary Material, Table S1. Experiments were performed using male mice at 3, 6, 9, or 18 months of age. All animal experiments were performed in accordance with approved guidelines and ethical approval from Emory University’s Institutional Animal Care and Use Committee and in compliance with the National Institutes of Health.
Generation of conditional Pabpn1 knock-out mouse
Generation of the Pabpn1 conditional knockout allele was performed using standard methods (Fig. 3A) (68). The targeting construct contained regions of homology to exons one and two of the Pabpn1 gene, a puromycin resistance gene (Puro) flanked by FRT sequences between two loxP sites. Targeting constructs were cloned into the pFlexible vector (68), which was linearized by KpnI digest and electroporated into HZ1.1 mouse embryonic stem cells (C57BL/6 background) at the Emory University School of Medicine Transgenic Mouse and Gene Targeting Core Facility. Puromycin resistant clones were screened by Southern blot to identify clones with proper construct integration. Selected clones were expanded, injected into blastocysts, and implanted into C57Bl/6 mice. Mice heterozygous for the targeted allele were crossed with mice expressing flippase to remove the Neo gene and subsequently crossed with mice containing the Cre recombinase gene under the control of the EIIa promoter (Jackson Laboratories) to generate mice with one Pabpn1 allele knocked out in all cells including the germline (Fig. 3A). Heterozygous Pabpn1 knock-out mice were genotyped by PCR using a set of three primers (WT/Floxed Allele and Knockout Allele primers) designed to amplify differently-sized products from the wild type, floxed, and knock-out alleles. Animals that were positive for the knockout allele and wild type allele but not the floxed allele were considered heterozygous Pabpn1 knock-out animals. Sequences for PCR primers used for genotyping can be found in Supplementary Material, Table S1. Experiments were performed using mice at 3, 6, or 18 months of age.
Real-time PCR analysis
For analysis of steady-state mRNA levels, RNA was isolated from rectus femoris and tibialis anterior muscles, or non-muscle tissue from three-month-old mice using TRIzol (ThermoFisher Scientific) according to the manufacturer's instructions. From isolated RNA, cDNA was synthesized using the M-MLV reverse transcriptase kit (Invitrogen) and Rnasin plus (Promega). For analysis of mitochondrial and nuclear DNA markers, total DNA was isolated from muscles using phenol chloroform: isoamyl alcohol (25:24:1, ThermoFisher Scientific) according to the manufacturer's instructions. For quantitative PCR analysis, approximately 10 ng of cDNA or genomic DNA samples were mixed with appropriate primers (Supplementary Material, Table S1) and SYBR Select Master Mix (Applied Biosciences). All samples were analyzed using the comparative Ct method (69) on an Applied Biosciences Step One Plus Real Time PCR System. For analysis of alternative polyadenylation, primers were designed to amplify a region within the coding sequence of the mRNA (Total) or the region between the distal and proximal PAS (Distal). PAS usage was quantified by calculating ΔΔCt as Distal-Total and fold change was reported as a ratio of Pabpn1+/+ controls. For analysis of cDNA, all samples were normalized to 18S rRNA or Gapdh. For analysis of mtDNA, samples were normalized to two nuclear reference genes, Hprt and Dio3.
Immunoblotting
From three-month-old mice, rectus femoris and tibialis anterior muscles were either homogenized or primary mouse cells were isolated and cultured, and lysed by standard methods (41). Samples were separated by SDS-PAGE on Mini-PROTEAN TGX Any kD polyacrylamide gels (BioRad), and transferred to 0.2 µm nitrocellulose membrane (BioRad). Membranes were blocked in either 5% bovine serum albumin (BSA) in Tris-buffered saline pH 7.4, with 0.1% Tween-20 (TBS-T) to be probed with α-VDAC antibodies or with 5% nonfat milk in TBS-T for all other immunoblots. The following primary antibodies were used: α-PABPN1 (1:8000) (41), α-Alanine expansion (1:2000) (40), α-HSP90 (1:5000, Santa Cruz Biotechnology #sc-13119), α-OXPHOS antibody cocktail (1:4000, Abcam #ab110413), and α-VDAC (1:4000, Abcam #15895). In all cases, following primary antibody incubation, immunoblots were incubated in the appropriate horseradish peroxidase conjugated secondary α-IgG antibodies (1:4000, Jackson ImmunoResearch Laboratories #115-036-062, #711-036-152).
Preparation of muscle tissue for histologic analysis
Tibialis anterior and rectus femoris muscles from six or eighteen-month-old mice were dissected and flash frozen in cryo-freezing medium (Sakura OCT-TissueTek). Muscles were cut into 14 µm serial cross-sections using a cryostat (Leica Biosystems) and sections were mounted on Superfrost Plus slides (Fisher Scientific). Sectioned muscles were either immediately stained with hematoxylin and eosin (H&E) or frozen for later analysis.
Succinate dehydrogenase activity assay
Sectioned tibialis anterior and rectus femoris muscles from three-month-old mice were stained for succinate dehydrogenase activity as described (70). Briefly, muscle sections were incubated in staining solution (1.5 mM nitroblue tetrazolium, 130 mM sodium succinate, 0.2 mM phenazine methosulfate, 1 mM sodium azide in PBS) at 37˚C for 20 min, washed in PBS, dehydrated in ethanol and xylene, and mounted with Permount mounting medium (Fisher Scientific).
Complex I activity assay
Activity assay kits (Cayman Chemical) were used to quantify mitochondrial electron transport chain Complex I activity in homogenized rectus femoris muscle according to the manufacturer’s instructions. Briefly, whole RF muscles isolated from 3, 12, and 18-month-old male mice were homogenized on ice in muscle homogenization buffer (71) using a Dounce homogenizer and insoluble debris was removed by centrifugation. Complex I activity was measured as oxidation of NADH by measuring a decrease in Absorbance at 340 nm using a BioTek Neo plate reader.
Histologic analyses
For analysis of H&E and SDH-stained slides, stained sections were imaged in the same anatomic region across all genotypes and ages. Exact localization within the muscle was determined using blood vessels and nerves as landmarks. For analysis of myofiber cross-sectional area (CSA), images were obtained from four separate sections representing the proximal, middle, and distal sections of each muscle for each biological replicate. CSA was measured by manually outlining individual myofibers using ImageJ (NIH). For each biological replicate, the CSA of 400–500 myofibers was quantified. For quantification of central nucleated fibers (CNF), the number of myofibers with one or more centrally-located nuclei was quantified and reported as a percentage of total myofibers analyzed. For SDH analysis, images were obtained for 6–8 separate sections representing the proximal, middle, and distal portion of the muscle. SDH positive myofibers were counted using ImageJ and calculated as a percentage of total myofibers for 500–700 myofibers analyzed per biological replicate.
Immunostaining
For detection of PABPN1, rectus femoris or tibialis anterior muscle sections from three-month-old mice were permeabilized in 0.3% Triton x-100 in PBS at room temperature (15 min), incubated in blocking buffer (0.3% BSA, 0.1% Triton x-100 in PBS) at room temperature (1–2 h) and incubated in primary α-PABPN1 diluted 1:1000 in 0.5x blocking buffer in PBS at 4 ˚C (overnight). Slides were washed in 0.5x blocking buffer and incubated in FITC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories #711-096-152) diluted 1:500 in 0.5x blocking buffer at room temperature (1 h). Slides were washed in 0.5× blocking buffer and incubated in 4'6-diamidino-2-phenylindole (DAPI, 1 µg/ml in PBS) to visualize nuclei, and mounted with Vectashield anti-fade mounting medium (Vector Laboratories). To remove soluble proteins, slides were incubated at room temperature in 1 M KCl in HPEM buffer (30 mM HEPES, 65 mM PIPES, 10 mM EDTA, 2 mM MgCl2, pH 6.9) for 1 h after permeabilization and before blocking.
Quantification of PABPN1 aggregates
KCl-insoluble aggregates were quantified as percentage of aggregates positive for PABPN1 by immunostaining in KCl-treated sections relative to total DAPI-stained nuclei. For each genotype and age, ≥ 12 sections for n = 3 animals were quantified.
Determination of bulk poly(A) tail length
Analysis of poly(A) tails from bulk RNA was performed as described (41). Briefly, 10 µg of RNA isolated from rectus femoris muscles of three-month-old male mice was labeled with [32P]-cytidine 3',5' bisphosphate using T4 RNA ligase and non-poly(A) RNA was digested using RNase A and T1. Labeled RNA was separated on a 7% polyacrylamide gel containing 8 M urea, 90 mM tris-borate, and 2 mM EDTA and visualized by autoradiography. Poly(A) tail length was quantified by densitometric analysis of gels using ImageJ. Different areas on images of poly(A) tail gels were chosen to represent short, medium, and long poly(A) tails. Integrated density was quantified and normalized to the integrated density of a region representing very short poly(A) tails that is not expected to be affected by loss of PABPN1 function. Entire lanes were also scanned and data for each genotype was binned by poly(A) tail length using GraphPad Prism 7.0. Statistical significance was determined using one-way ANOVA. Changes in bulk poly(A) tail length were detected as a decrease in the density in regions representing longer poly(A) tails. Statistical significance was determined by non-parametric t-test using GraphPad Prism 7.0.
3' region extraction and deep sequencing (3' READS)
3' READS analysis was performed on total RNA isolated from rectus femoris muscles of three-month-old mice as described (45,72). Briefly, 1 µg of input RNA was used for each sample, and poly(A)+ RNA was selected using oligo d(T)25 magnetic beads (NEB), followed by on-bead fragmentation using RNase III (NEB). Poly(A)+ RNA fragments were then selected using a chimeric oligo containing 15 regular dTs and five locked nucleic acid dTs conjugated on streptavidin beads, followed by RNase H (NEB) digestion. RNA fragments with ligated with 5’ and 3’ adapters were reverse-transcribed, followed by PCR (15×) to obtain cDNA libraries for sequencing on the Illumina platform. Processing of 3’READS data was carried out as previously described (45). Briefly, reads were mapped to the mouse genome using bowtie 2 (73). Reads with ≥ 2 unaligned Ts at the 5’ end were used to identify PASs. PASs located within 24 nt from each other were clustered together.
APA analysis
Differential expression of APA isoforms was carried out with Fisher’s exact test. Significant events were those with P < 0.05 and delta relative abundance >5%. Relative expression of two most abundant 3’UTR APA isoforms, e.g., proximal and distal PASs, was calculated by log2 (Distal PAS/Proximal PAS).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of whole muscle tissue
Rectus femoris muscles were dissected from three-month-old mice and analyzed by LC-MS/MS as described (74). Muscles were homogenized in urea lysis buffer (8M urea, 100 mM NaHPO4) with Pierce 1× HALT protease and phosphatase inhibitor cocktail (ThermoFisher) on a Bullet Blender (Next Advance) at 4 ˚C according to the manufacturer's instructions. Samples were then sonicated (Sonic Dismembrator, Fisher Scientific) and protein concentration was measured by bicinchoninic assay (BCA) (75). Protein homogenate was diluted in 50 mM NH4HCO and treated with 1 mM dithiothreitol (30 min at room temperature) and 5 mM iodoacetamide (20 min at room temperature protected from light). Proteins were digested for 2 h in 1:100 lysyl endopeptidase at room temperature and overnight in 1:50 trypsin (Promega) at room temperature. Digested peptides were removed from buffers on a Sep-Pak C18 column (Waters) and dried under vacuum. Dried peptides (2 µg) were resuspended in loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile) with 0.2 pmol of isotope-labeled peptide calibrants (Life Technologies #88321). Resuspended peptides were separated on a C18 column (25 × 75 µm internal diameter, New Objective) by a NanoAcquity UPLC (Waters) and monitored on an Orbitrap Fusion Tribrid Mass Spectrometer with Electron Transfer Dissociation (Thermo Scientific). Separation was performed over a 120-min gradient (400 nl/min) of 3–80% buffer B (0.1% formic acid 0.5% DMSO in acetonitrile) into buffer A (0.1% formic acid 5% DMSO in water). The mass spectrometer cycle was programmed to collect one full MS scan followed by MS/MS scans. MS scans were collected at m/z 200 at a resolution of 70,000 in profile mode and MS/MS scans were collected at 200 m/z at a resolution of 17,500. Dynamic exclusion was set to exclude previously sequenced precursor ions for 30 s within a 10 ppm window. Also excluded were precursor ions with +1 or ≥ +6 charge states.
Label-free protein quantification (LFQ)
LFQ was performed as previously described (76,77). Raw sample data were analyzed using MaxQuant v1.5.3.30 with Thermo Foundation 2.0 for RAW file reading capability. The search engine Andromeda, integrated into MaxQuant, was used to search a concatenated target-decoy Uniprot mouse reference protein database (retrieved August 2015; 54,489 target sequences), plus 245 contaminant proteins from the common repository of adventitious proteins (cRAP) built into MaxQuant. Methionine oxidation (+15.9949 Da), asparagine and glutamine deamidation (+0.9840 Da), and protein N-terminal acetylation (+42.0106 Da) were variable modifications (up to 5 allowed per peptide); cysteine was assigned a fixed carbamidomethyl modification (+57.0215 Da). Only fully tryptic peptides were considered with up to two miscleavages in the database search. A precursor mass tolerance of ±20 ppm was applied prior to mass accuracy calibration and ±4.5 ppm after internal MaxQuant calibration. Other search settings included a maximum peptide mass of 6000 Da, a minimum peptide length of 6 residues, and 0.6 Da tolerance for low resolution (ITMS) MS/MS scans. Co-fragmented peptide search was enabled to deconvolute multiplex spectra. The false discovery rate (FDR) for peptide spectral matches, and site decoy fraction were set to 1 percent. Quantification settings were as follows: requantify with a second peak finding attempt after protein identification has completed; match MS1 peaks between runs; a 0.7 min retention time match window was used after an alignment function was found with a 20-min RT search space. Quantitation of proteins was performed using the LFQ algorithm built into MaxQuant, which normalizes for technical variations across samples. LFQ minimum ratio count allowed was 1. The quantitation method only considered razor plus unique peptides for protein level quantitation. The full list of parameters used for MaxQuant are available upon request as a parameter.XML file. To determine changes in steady-state protein levels between muscle samples, the log2 ratio of XIC from Pabpn1+/A17 and Pabpn1+/Δ samples relative to XIC of the wild type littermates was compared. Proteins with a log2 ratio of ≥ 0.5 were considered to be enriched in Pabpn1+/A17 or Pabpn1+/Δ while proteins with a log2 ratio of ≤ −0.5 were considered to be depleted in Pabpn1+/A17 knock-in or Pabpn1+/Δ knock-out samples. Gene ontology (GO) term and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment was determined using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (46,47).
Microscopy
All stained slides were viewed using an Axioplan microscope with either a 0.5 NA 10x Plan-Neoflaur or a 0.5 NA 20x Plan-Neoflaur objective lens (Carl Zeiss MicroImaging, Inc.). Digital images were obtained with a charge-couple device camera (Carl Zeiss MicroImaging, Inc.) and Scion Image 1.63 (Scion Corp., NIH). All images were assembled and equally processed for color levels, brightness, and contrast as needed using Photoshop CC 2015 (Adobe).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5 or GraphPad Prism 7. For analysis of frequency distributions of myofiber CSA, Kruskal-Wallace test plus Dunn's post-test was performed. In all other cases, statistical significance was determined by either Student's t-test or one-way analysis of variance (ANOVA) unless otherwise noted. In all cases, samples with P < 0.05 were considered to be statistically significant.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared
Funding
National Institutes of Health [5F32AR068207 to KEV, 1F31AR069994 to BLP, GM084089 to BT and NS069234, NS84870, and AR061987 to AHC and GKP], Muscular Dystrophy Association [255856 to AB, 157523 to LHA, 276125 to GKP, and 422006 to AHC], Mouse Transgenic and Gene Targeting Core (TMF), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Proteomic analysis was performed by the Emory University School of Medicine Integrated Proteomics Core. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Supplementary Material
References
- 1. Victor M., Hayes R., Adams R.D. (1962) Oculopharyngeal muscular dystrophy. A familial disease of late life characterized by dysphagia and progressive ptosis of the evelids. N. Engl. J. Med., 267, 1267–1272. [DOI] [PubMed] [Google Scholar]
- 2. Youssof S. (2016) The relationship between physical symptoms and health-related quality of life in oculopharyngeal muscular dystrophy. Muscle Nerve, 53, 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brais B., Bouchard J.P., Xie Y.G., Rochefort D.L., Chretien N., Tome F.M., Lafreniere R.G., Rommens J.M., Uyama E., Nohira O.. et al. (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet., 18, 164–167. [DOI] [PubMed] [Google Scholar]
- 4. Richard P., Trollet C., Gidaro T., Demay L., Brochier G., Malfatti E., Tom F.M., Fardeau M., Lafor P., Romero N.. et al. (2015) PABPN1 (GCN)11 as a Dominant Allele in Oculopharyngeal Muscular Dystrophy -Consequences in Clinical Diagnosis and Genetic Counselling. J. Neuromuscul. Dis., 2, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jouan L., Rocheford D., Szuto A., Carney E., David K., Dion P.A., Rouleau G.A. (2014) An 18 alanine repeat in a severe form of oculopharyngeal muscular dystrophy. Can. J. Neurol. Sci., 41, 508–511. [DOI] [PubMed] [Google Scholar]
- 6. Kerwitz Y., Kuhn U., Lilie H., Knoth A., Scheuermann T., Friedrich H., Schwarz E., Wahle E. (2003) Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. embo. J., 22, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhn U., Gundel M., Knoth A., Kerwitz Y., Rudel S., Wahle E. (2009) Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem., 284, 22803–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wahle E. (1995) Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem., 270, 2800–2808. [DOI] [PubMed] [Google Scholar]
- 9. Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kühn U., Menzies F.M., Oude Vrielink J.A., Bos A.J., Drost J., Rooijers K.. et al. (2012) The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell, 149, 538–553. [DOI] [PubMed] [Google Scholar]
- 10. Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., Park J.Y., Gunderson S.I., Kalsotra A., Manley J.L.. et al. (2015) Systematic profiling of poly(A)+ transcripts modulated by core 3' end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet., 11, e1005166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Klerk E., Venema A., Anvar S.Y., Goeman J.J., Hu O., Trollet C., Dickson G., den Dunnen J.T., van der Maarel S.M., Raz V.. et al. (2012) Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res., 40, 9089–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tome F.M., Fardeau M. (1980) Nuclear inclusions in oculopharyngeal dystrophy. Acta Neuropathol., 49, 85–87. [DOI] [PubMed] [Google Scholar]
- 13. Fan X., Messaed C., Dion P., Laganiere J., Brais B., Karpati G., Rouleau G.A. (2003) HnRNP A1 and A/B interaction with PABPN1 in oculopharyngeal muscular dystrophy. Can. J. Neurol. Sci., 30, 244–251. [DOI] [PubMed] [Google Scholar]
- 14. Corbeil-Girard L.P., Klein A.F., Sasseville A.M., Lavoie H., Dicaire M.J., Saint-Denis A., Page M., Duranceau A., Codere F., Bouchard J.P.. et al. (2005) PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol. Dis., 18, 551–567. [DOI] [PubMed] [Google Scholar]
- 15. Tavanez J.P., Bengoechea R., Berciano M.T., Lafarga M., Carmo-Fonseca M., Enguita F.J. (2009) Hsp70 chaperones and type I PRMTs are sequestered at intranuclear inclusions caused by polyalanine expansions in PABPN1. PLoS One, 4, e6418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Apponi L.H., Corbett A.H., Pavlath G.K. (2013) Control of mRNA stability contributes to low levels of nuclear poly(A) binding protein 1 (PABPN1) in skeletal muscle. Skelet. Muscle, 3, 23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gidaro T., Negroni E., Perie S., Mirabella M., Laine J., Lacau St Guily J., Butler-Browne G., Mouly V., Trollet C. (2013) Atrophy, fibrosis, and increased PAX7-positive cells in pharyngeal muscles of oculopharyngeal muscular dystrophy patients. J. Neuropathol. Exp. Neurol., 72, 234–243. [DOI] [PubMed] [Google Scholar]
- 18. Uyama E., Tsukahara T., Goto K., Kurano Y., Ogawa M., Kim Y.J., Uchino M., Arahata K. (2000) Nuclear accumulation of expanded PABP2 gene product in oculopharyngeal muscular dystrophy. Muscle Nerve, 23, 1549–1554. [DOI] [PubMed] [Google Scholar]
- 19. Dion P., Shanmugam V., Gaspar C., Messaed C., Meijer I., Toulouse A., Laganiere J., Roussel J., Rochefort D., Laganiere S.. et al. (2005) Transgenic expression of an expanded (GCG)13 repeat PABPN1 leads to weakness and coordination defects in mice. Neurobiol. Dis., 18, 528–536. [DOI] [PubMed] [Google Scholar]
- 20. Berciano M.T., Villagra N.T., Ojeda J.L., Navascues J., Gomes A., Lafarga M., Carmo-Fonseca M. (2004) Oculopharyngeal muscular dystrophy-like nuclear inclusions are present in normal magnocellular neurosecretory neurons of the hypothalamus. Hum. Mol. Genet., 13, 829–838. [DOI] [PubMed] [Google Scholar]
- 21. Villagra N.T., Bengoechea R., Vaque J.P., Llorca J., Berciano M.T., Lafarga M. (2008) Nuclear compartmentalization and dynamics of the poly(A)-binding protein nuclear 1 (PABPN1) inclusions in supraoptic neurons under physiological and osmotic stress conditions. Mol. Cell. Neurosci., 37, 622–633. [DOI] [PubMed] [Google Scholar]
- 22. Davies J.E., Rose C., Sarkar S., Rubinsztein D.C. (2010) Cystamine suppresses polyalanine toxicity in a mouse model of oculopharyngeal muscular dystrophy. Sci. Transl. Med., 2, 34ra40.. [DOI] [PubMed] [Google Scholar]
- 23. Davies J.E., Sarkar S., Rubinsztein D.C. (2008) Wild-type PABPN1 is anti-apoptotic and reduces toxicity of the oculopharyngeal muscular dystrophy mutation. Hum. Mol. Genet., 17, 1097–1108. [DOI] [PubMed] [Google Scholar]
- 24. Davies J.E., Sarkar S., Rubinsztein D.C. (2006) Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum. Mol. Genet., 15, 23–31. [DOI] [PubMed] [Google Scholar]
- 25. Davies J.E., Wang L., Garcia-Oroz L., Cook L.J., Vacher C., O'Donovan D.G., Rubinsztein D.C. (2005) Doxycycline attenuates and delays toxicity of the oculopharyngeal muscular dystrophy mutation in transgenic mice. Nat. Med., 11, 672–677. [DOI] [PubMed] [Google Scholar]
- 26. Brais B., Xie Y.G., Sanson M., Morgan K., Weissenbach J., Korczyn A.D., Blumen S.C., Fardeau M., Tome F.M., Bouchard J.P.. et al. (1995) The oculopharyngeal muscular dystrophy locus maps to the region of the cardiac alpha and beta myosin heavy chain genes on chromosome 14q11.2-q13. Hum. Mol. Genet., 4, 429–434. [DOI] [PubMed] [Google Scholar]
- 27. Wang Q., Bag J. (2006) Ectopic expression of a polyalanine expansion mutant of poly(A)-binding protein N1 in muscle cells in culture inhibits myogenesis. Biochem. Biophys. Res. Commun., 340, 815–822. [DOI] [PubMed] [Google Scholar]
- 28. Raz V., Routledge S., Venema A., Buijze H., van der Wal E., Anvar S., Straasheijm K.R., Klooster R., Antoniou M., van der Maarel S.M. (2011) Modeling oculopharyngeal muscular dystrophy in myotube cultures reveals reduced accumulation of soluble mutant PABPN1 protein. Am. J. Pathol., 179, 1988–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Catoire H., Pasco M.Y., Abu-Baker A., Holbert S., Tourette C., Brais B., Rouleau G.A., Parker J.A., Neri C. (2008) Sirtuin inhibition protects from the polyalanine muscular dystrophy protein PABPN1. Hum. Mol. Genet., 17, 2108–2117. [DOI] [PubMed] [Google Scholar]
- 30. Chartier A., Benoit B., Simonelig M. (2006) A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. embo J., 25, 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hino H., Araki K., Uyama E., Takeya M., Araki M., Yoshinobu K., Miike K., Kawazoe Y., Maeda Y., Uchino M.. et al. (2004) Myopathy phenotype in transgenic mice expressing mutated PABPN1 as a model of oculopharyngeal muscular dystrophy. Hum. Mol. Genet., 13, 181–190. [DOI] [PubMed] [Google Scholar]
- 32. Mankodi A., Wheeler T.M., Shetty R., Salceies K.M., Becher M.W., Thornton C.A. (2012) Progressive myopathy in an inducible mouse model of oculopharyngeal muscular dystrophy. Neurobiol. Dis., 45, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Randolph M.E., Luo Q., Ho J., Vest K.E., Sokoloff A.J., Pavlath G.K. (2014) Ageing and muscular dystrophy differentially affect murine pharyngeal muscles in a region-dependent manner. J. Physiol., 592, 5301–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anvar S.Y., Raz Y., Verway N., van der Sluijs B., Venema A., Goeman J.J., Vissing J., van der Maarel S.M., t Hoen P.A., van Engelen B.G.. et al. (2013) A decline in PABPN1 induces progressive muscle weakness in oculopharyngeal muscle dystrophy and in muscle aging. Aging (Albany NY), 5, 412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chartier A., Klein P., Pierson S., Barbezier N., Gidaro T., Casas F., Carberry S., Dowling P., Maynadier L., Bellec M.. et al. (2015) Mitochondrial dysfunction reveals the role of mRNA poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLoS Genet., 11, e1005092.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anvar S.Y., t., Hoen P.A., Venema A., van der Sluijs B., van Engelen B., Snoeck M., Vissing J., Trollet C., Dickson G., Chartier A.. et al. (2011) Deregulation of the ubiquitin-proteasome system is the predominant molecular pathology in OPMD animal models and patients. Skelet. Muscle, 1, 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trollet C., Anvar S.Y., Venema A., Hargreaves I.P., Foster K., Vignaud A., Ferry A., Negroni E., Hourde C., Baraibar M.A.. et al. (2010) Molecular and phenotypic characterization of a mouse model of oculopharyngeal muscular dystrophy reveals severe muscular atrophy restricted to fast glycolytic fibres. Hum. Mol. Genet., 19, 2191–2207. [DOI] [PubMed] [Google Scholar]
- 38. Scheuermann T., Schulz B., Blume A., Wahle E., Rudolph R., Schwarz E. (2003) Trinucleotide expansions leading to an extended poly-L-alanine segment in the poly (A) binding protein PABPN1 cause fibril formation. Protein Sci., 12, 2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U S A, 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vest K.E., Apponi L.H., Banerjee A., Pavlath G.K., Corbett A.H. (2015) An antibody to detect alanine-expanded pabpn1: a new tool to study oculopharyngeal muscular dystrophy. J. Neuromuscul. Dis., 2, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Apponi L.H., Leung S.W., Williams K.R., Valentini S.R., Corbett A.H., Pavlath G.K. (2010) Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet., 19, 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Banerjee A., Apponi L.H., Pavlath G.K., Corbett A.H. (2013) PABPN1: molecular function and muscle disease. febs J., 280, 4230–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cooper R.N., Tajbakhsh S., Mouly V., Cossu G., Buckingham M., Butler-Browne G.S. (1999) In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell. Sci., 112, 2895–2901. [DOI] [PubMed] [Google Scholar]
- 44. Chekanova J.A., Shaw R.J., Belostotsky D.A. (2001) Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol., 11, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 45. Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B. (2013) Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat. Methods, 10, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang da W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 47. Huang da W., Sherman B.T., Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res., 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho Y., Hazen B.C., Gandra P.G., Ward S.R., Schenk S., Russell A.P., Kralli A. (2016) Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. faseb J., 30, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barrientos A., Casademont J., Cardellach F., Estivill X., Urbano-Marquez A., Nunes V. (1997) Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Brain Res. Mol. Brain. Res., 52, 284–289. [DOI] [PubMed] [Google Scholar]
- 50. Riaz M., Raz Y., van Putten M., Paniagua-Soriano G., Krom Y.D., Florea B.I., Raz V. (2016) PABPN1-Dependent mRNA Processing Induces Muscle Wasting. PLoS Genet., 12, e1006031.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richard P., Trollet C., Stojkovic T., de Becdelievre A., Perie S., Pouget J., Eymard B.. Neurologists of French Neuromuscular Reference Centers, C. and Filnemus. (2017) Correlation between PABPN1 genotype and disease severity in oculopharyngeal muscular dystrophy. Neurology, 88, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Der Sluijs B.M., Hoefsloot L.H., Padberg G.W., Van Der Maarel S.M., Van Engelen B.G. (2003) Oculopharyngeal muscular dystrophy with limb girdle weakness as major complaint. J. Neurol., 250, 1307–1312. [DOI] [PubMed] [Google Scholar]
- 53. Youssof S., Schrader R., Bear D., Morrison L. (2015) Hip flexion weakness is associated with impaired mobility in oculopharyngeal muscular dystrophy: a retrospective study with implications for trial design. Neuromuscul. Disord., 25, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duddy W., Duguez S., Johnston H., Cohen T.V., Phadke A., Gordish-Dressman H., Nagaraju K., Gnocchi V., Low S., Partridge T. (2015) Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet. Muscle, 5, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanabe Y., Esaki K., Nomura T. (1986) Skeletal muscle pathology in X chromosome-linked muscular dystrophy (mdx) mouse. Acta Neuropathol., 69, 91–95. [DOI] [PubMed] [Google Scholar]
- 56. Kelleher A.R., Pereira S.L., Jefferson L.S., Kimball S.R. (2015) REDD2 expression in rat skeletal muscle correlates with nutrient-induced activation of mTORC1: responses to aging, immobilization, and remobilization. Am. J. Physiol. Endocrinol. Metab., 308, E122–E129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miyazaki M., Esser K.A. (2009) REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am. J. Physiol. Cell. Physiol., 296, C583–C592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klein P., Oloko M., Roth F., Montel V., Malerba A., Jarmin S., Gidaro T., Popplewell L., Perie S., Lacau St Guily J.. et al. (2016) Nuclear poly(A)-binding protein aggregates misplace a pre-mRNA outside of SC35 speckle causing its abnormal splicing. Nucleic Acids Res., 44, 10929–10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gambelli S., Malandrini A., Ginanneschi F., Berti G., Cardaioli E., De Stefano R., Franci M., Salvadori C., Mari F., Bruttini M.. et al. (2004) Mitochondrial abnormalities in genetically assessed oculopharyngeal muscular dystrophy. Eur. Neurol., 51, 144–147. [DOI] [PubMed] [Google Scholar]
- 60. Sadeh M., Pauzner R., Blatt I., Mouallem M., Farfel Z. (1993) Mitochondrial abnormalities in oculopharyngeal muscular dystrophy. Muscle Nerve, 16, 982–983. [PubMed] [Google Scholar]
- 61. Schroder J.M., Krabbe B., Weis J. (1995) Oculopharyngeal muscular dystrophy: clinical and morphological follow-up study reveals mitochondrial alterations and unique nuclear inclusions in a severe autosomal recessive type. Neuropathol. Appl. Neurobiol., 21, 68–73. [DOI] [PubMed] [Google Scholar]
- 62. Wong K.T., Dick D., Anderson J.R. (1996) Mitochondrial abnormalities in oculopharyngeal muscular dystrophy. Neuromuscul. Disord., 6, 163–166. [DOI] [PubMed] [Google Scholar]
- 63. Malerba A., Klein P., Bachtarzi H., Jarmin S.A., Cordova G., Ferry A., Strings V., Espinoza M.P., Mamchaoui K., Blumen S.C.. et al. (2017) PABPN1 gene therapy for oculopharyngeal muscular dystrophy. Nat. Commun., 8, 14848.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kepp K.P. (2016) Alzheimer's disease due to loss of function: A new synthesis of the available data. Prog. Neurobiol., 143, 36–60. [DOI] [PubMed] [Google Scholar]
- 65. Arteaga-Bracho E.E., Gulinello M., Winchester M.L., Pichamoorthy N., Petronglo J.R., Zambrano A.D., Inocencio J., De Jesus C.D., Louie J.O., Gokhan S.. et al. (2016) Postnatal and adult consequences of loss of huntingtin during development: Implications for Huntington's disease. Neurobiol. Dis., 96, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Messaed C., Dion P.A., Abu-Baker A., Rochefort D., Laganiere J., Brais B., Rouleau G.A. (2007) Soluble expanded PABPN1 promotes cell death in oculopharyngeal muscular dystrophy. Neurobiol. Dis., 26, 546–557. [DOI] [PubMed] [Google Scholar]
- 67. Kontgen F., Suss G., Stewart C., Steinmetz M., Bluethmann H. (1993) Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int. Immunol., 5, 957–964. [DOI] [PubMed] [Google Scholar]
- 68. van der Weyden L., Adams D.J., Harris L.W., Tannahill D., Arends M.J., Bradley A. (2005) Null and conditional semaphorin 3B alleles using a flexible puroDeltatk loxP/FRT vector. Genesis, 41, 171–178. [DOI] [PubMed] [Google Scholar]
- 69. Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 70. Ross J.M. (2011) Visualization of mitochondrial respiratory function using cytochrome c oxidase/succinate dehydrogenase (COX/SDH) double-labeling histochemistry. J. Vis. Exp., 23, e3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc., 7, 1235–1246. [DOI] [PubMed] [Google Scholar]
- 72. Zheng D., Liu X., Tian B. (2016) 3'READS+, a sensitive and accurate method for 3' end sequencing of polyadenylated RNA. RNA. 22, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dammer E.B., Lee A.K., Duong D.M., Gearing M., Lah J.J., Levey A.I., Seyfried N.T. (2015) Quantitative phosphoproteomics of Alzheimer's disease reveals cross-talk between kinases and small heat shock proteins. Proteomics. 15, 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem., 150, 76–85. [DOI] [PubMed] [Google Scholar]
- 76. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res., 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.