Abstract
It is clear that innate immune system status is altered in numerous neurodegenerative diseases. Human genetic studies have demonstrated that triggering receptor expressed in myeloid cells 2 (TREM2) coding variants have a strong association with Alzheimer’s disease (AD) and other neurodegenerative diseases. To more thoroughly understand the impact of TREM2 in vivo, we studied the behavioral and cognitive functions of wild-type (WT) and Trem2−/− (KO) mice during basal conditions and brain function in the context of innate immune stimulation with peripherally administered lipopolysaccharide (LPS). Early markers of neuroinflammation preceded Aif1 and Trem2 upregulation that occurred at later stages (24–48 h post-LPS). We performed a transcriptomic study of these cohorts and found numerous transcripts and pathways that were altered in Trem2−/− mice both at baseline and 48 h after LPS challenge. Importantly, our transcriptome analysis revealed that our Trem2−/− mouse line (Velocigene allele) results in exaggerated Treml1 upregulation. In contrast, aberrantly high Treml1 expression was absent in the Trem2 knockout line generated by the Colonna lab and the Jackson Labs CRISPR/Cas9 Trem2 knockout line. Notably, removal of the floxed neomycin selection cassette ameliorated aberrant Treml1 expression, validating the artifactual nature of Treml1 expression in the original Trem2−/− Velocigene line. Clearly further studies are needed to decipher whether the Treml1 transcriptional artifact is functionally meaningful, but our data indicate that caution is warranted when interpreting functional studies with this particular line. Additionally, our results indicate that other Velocigene alleles or targeting strategies with strong heterologous promoters need to carefully consider downstream genes.
Introduction
Microglia are the central nervous system (CNS) resident innate immune cell population that act as sentinels and continually sample and survey the local milieu in the brain parenchyma (1–4). Microglia participate in both basal and pathological disease states, orchestrating aspects of processes including development, synaptogenesis, and inflammation (5,6). Notably, microglial reactivity has been demonstrated in numerous neurodegenerative disorders, including Alzheimer’s disease (AD), frontotemporal dementia (FTD), and Parkinson’s disease (PD), suggesting that they may play a pivotal role in the outcome of disease progression (7–13). Within the context of AD, many of the human genetic risk factors, including APOE, CLU, CD33, and TREM2, are genes that have the potential to alter innate immunity, making it critical to understand how these factors regulate microglial function (14–20).
Triggering receptor expressed on myeloid cell 2 (TREM2) is an immunoglobulin (Ig) superfamily cell surface receptor that is expressed by macrophages, dendritic cells, and osteoclasts in the periphery (21–24). Within the CNS, TREM2 is expressed almost exclusively on microglia and is one of the more highly expressed molecules that constitute the microglial sensome (25–27). TREM2 engagement results in intracellular signaling mediated by its association with TYRO protein tyrosine kinase binding protein (TYROBP), also known as DNAX-activating protein of 12 kDa (DAP12) (22,28,29). Several ligands for TREM2 have been revealed including lipopolysaccharide (LPS), bacteria, anionic and zwitterionic lipids, and, interestingly, both apoE and CLU have been identified as TREM2 ligands, potentially converging several human AD risk gene pathways together (30–34). The R47H allele as well as other TREM2 variants have been associated with increased risk of AD as well as FTD and PD (18–20, 35,36). Additionally, TREM2 or TYROBP loss of function mutations result in Nasu-Hakola disease (NHD), also known as polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL), characterized by microglial activation and dementia. These human genetic data indicate a critical role for TREM2 in neuropathological outcomes (18–20,37,38). TREM2 deficiency is associated with increased pro-inflammatory cytokine production following either in vitro and in vivo LPS stimulation and decreased bacterial clearance, suggesting an anti-inflammatory, protective function (24,39,40). Additionally, TREM2 has been implicated in shaping microglial phagocytosis of apoptotic neurons, microglial clustering around amyloid plaques, and microglial expansion and survival (34,41–46). Although data has emerged focusing on TREM2 during disease, it remains unclear whether a TREM2 deficiency actually impacts basal behavioral and cognitive function. Moreover, it is unknown how TREM2 impacts CNS transcriptomes under homeostatic conditions following systemic inflammation.
Here, we used a Trem2 knockout mouse model generated from the Velocigene ‘definitive null’ targeting strategy in which the entire coding region was replaced by selection cassette (lacZ-flox-human Ubiquitin C promoter-neomycin-flox), beginning at 16 bp upstream of the ATG start codon and ending at the TGA stop codon of Trem2. We demonstrate that TREM2 has no impact on behavioral and cognitive readouts as well as little to no impact on early CNS pro-inflammatory cytokine levels following peripheral LPS challenge, suggesting that TREM2 may be dispensable under basal conditions and at early time points after systemic inflammation. Following peripheral LPS challenge, we show that TREM2 is not significantly up-regulated until 48–72 h post challenge, well past the point of the initial wave of inflammation. Examination of the transcriptomes from WT and Trem2−/− mice revealed alterations in several genes 48 h after LPS challenge; however, the predominant transcript that was overwhelmingly elevated in Trem2−/− mice, Treml1, was also observed under basal conditions. Examination of Treml1 expression in the Trem2−/− line generated in the Colonna lab (24) revealed a trend for a slight increase in expression and a recently generated Trem2 CRISPR/Cas9 knockout allele had no significant change in Treml1 expression. Removal of the floxed neomycin cassette, driven by the human ubiquitin C promoter, abolished the Treml1 overexpression artifact in the Velocigene knockout line. These data suggest that the original Velocigene targeting of the Trem2 locus introduced an artifactual alteration in Treml1 expression that is of unknown consequence and caution must be taken while interpreting the role of Trem2 with this particular mouse model if the neomycin cassette remains intact.
Results
Trem2 deficiency does not alter behavior or cognition during basal conditions
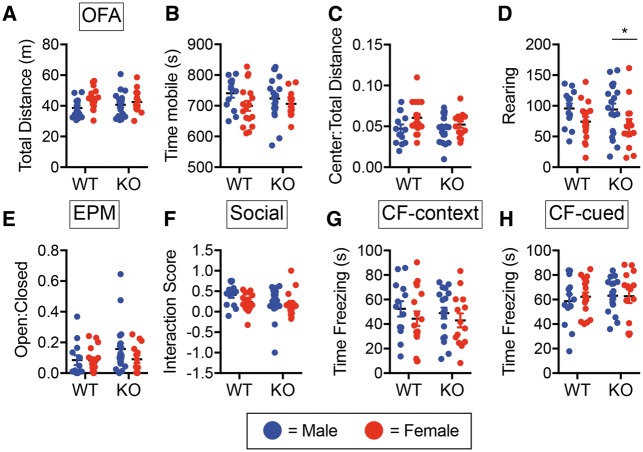
To determine if TREM2 deficiency has an impact on behavioral or cognitive abilities under basal conditions, 6-month-old male and female WT and Trem2−/− mice from the Velocigene allele were run through a series of behavioral and cognitive tests. Open field assay (OFA) was used to examine general activity/locomotion and anxiety but revealed no significant differences between the two genotypes in mobility (total distance traveled, time mobile) (Fig. 1A and B) or anxiety (center: total distance ratio, rearing) (Fig. 1C and D). Although there was a slight decrease in rearing between female versus male Trem2−/− mice, there were no significant genotype by sex effects. Use of the elevated plus maze (EPM) as another anxiety readout also showed no difference between WT and Trem2−/− mice in open: closed arm ratios (Fig. 1E), indicative of similar overall anxiety-like behavior. Additionally, three chamber social interaction behavioral testing to examine sociability in WT and Trem2−/− revealed comparable interaction scores (Fig. 1F). We next used conditioned fear (CF) behavioral testing to assess whether there were any alterations in cognitive function between 6-month old WT and Trem2−/− mice. Similar memory retention observed between WT and Trem2−/− mice in both the hippocampus-dependent context test and the hippocampus/amygdala-dependent cued test (Fig. 1G and H). Together these data show that Trem2 deficiency has no overt impact on behavioral and cognitive parameters at 6-months of age. Since there were no sex by genotype effects, the remainder of the study was conducted with male mice.
Figure 1.
Trem2 deficiency does not alter behavioral or cognitive performance. Six-month-old male and female C57BL/6 N wild type (WT) or Trem2−/− (KO) mice were examined for behavioral alterations in the open field assay for mobility and anxiety (A–D), elevated plus maze for anxiety (E), and the 3 chamber social test of sociability (F). Contextual (G) and cued (H) cognitive memory performance was determined using conditioned fear assays. Males (blue) and females (red) had similar results. Shown are the averages ± S.E.M. for N = 13–19 animals/group/sex (WT female N = 17, WT male N = 13, KO female N = 14, KO male N = 19). There were no significant genotypes or sex by genotype differences noted in any of the assays (Two-way ANOVA, Fisher’s LSD).
TREM2 does not impact early CNS inflammation after systemic LPS challenge
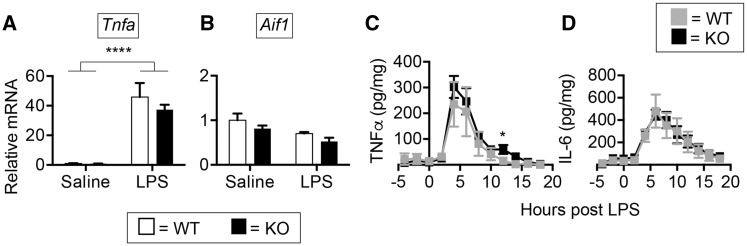
TREM2 has been shown to modulate pro-inflammatory cytokine production by microglia and macrophages after in vitro challenge with the gram negative bacterial mimetic, lipopolysaccharide (LPS) (24,40). Although Trem2 deficiency had no overt impact on behavioral/cognitive parameters during homeostatic conditions, it is possible that its effect is only revealed in adult animals within the context of inflammation. To examine if Trem2 expression impacts early CNS inflammation associated with systemic LPS challenge, WT and Trem2−/− mice were injected with 2 mg/kg LPS i.p. and PBS perfused brains were harvested and dissected 4 h after injection. RT-qPCR revealed a significant increase in hippocampal Tnfa transcript levels in both LPS challenged WT and Trem2−/− mice, but no effect of genotype on the magnitude of the response (Fig. 2A). Additionally, there was no difference in the expression of Aif1 (encoding the microglial marker IBA1) between WT and Trem2−/− mice, suggesting a lack of significant microglial activation at this time point for both genotypes (Fig. 2B). To further address any early neuroinflammatory changes that may occur in the absence of Trem2, in vivo microdialysis was conducted to monitor TNFα and IL-6 levels in the hippocampal interstitial fluid prior to and up to 12 h after peripheral LPS injection. Quantitative cytometric bead array analysis of TNFα levels revealed no significant differences in either basal levels (−4, −2, and 0 h before LPS injection) or early induction of TNFα protein in the CNS interstitial fluid of WT (grey line) or Trem2−/− (black line) mice (Fig. 2C). Between 12 and 14 h post challenge, there was a small but significant difference between WT and Trem2−/− TNFα levels in the hippocampus. No significant alterations based on genotype were observed in IL-6 levels at any of the times examined (Fig. 2D). These data suggested that TREM2 does not impact early CNS inflammation that is observed following systemic inflammation.
Figure 2.
TREM2 does not impact early CNS inflammation following a peripheral inflammatory challenge. Hippocampal tissues from PBS perfused male brains were isolated from C57BL/6 N wild type (WT) or Trem2−/− (KO) mice 4 h post i.p. challenge with 2 mg/kg LPS. RT-qPCR was conducted for (A) Tnfa and (B) Aif1 (encodes IBA1) levels. Shown are the averages ± S.E.M. for N = 4–7 mice per group. No significant differences based on genotype were noted. A main effect of LPS was noted for Tnfa, but no Trem2 genotype effect was observed (Two-way ANOVA, Tukey post-hoc, **P ≤ 0.01, ***P≤ 0.001). Hippocampal interstitial fluid (ISF) was collected from WT and Trem2−/− mice 4 h prior to and 18 h post i.p. injection with 2 mg/kg LPS using in vivo microdialysis. ISF fractions pooled in 2-h increments were measured for (C) TNFα and (D) IL-6. No significant Trem2 genotype differences were found with the exception of slight elevation of TNFα at the 12 h time point (Multiple t-test, Benjamini and Hochberg post hoc, *P ≤ 0.05). Shown are the averages ± S.E.M. for N = 3 mice per group.
Delayed microglial activation and CNS Trem2 induction after LPS challenge
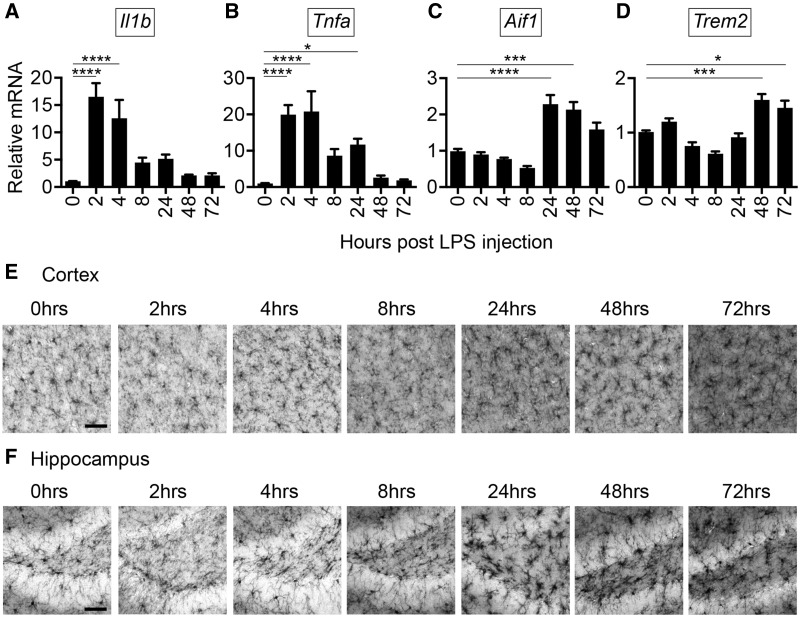
Although early TNFα production was evident, the absence of Aif1 induction at 4 h suggested that early time points might not address alterations in microglial function that could occur with Trem2 deficiency. To determine how CNS microglial activation is coordinated with inflammation and Trem2 expression following systemic LPS challenge, WT mice were injected with 2 mg/kg of LPS intraperitoneally (i.p.) and PBS perfused brains were harvested at 0, 2, 4, 8, 24, 48, and 72 h post injection. RT-qPCR demonstrated an early elevation in the expression of Il1b and Tnfa pro-inflammatory genes at 2 and 4 h followed by a drop in expression (Fig. 3A and B). Interestingly, Aif1 was not elevated during the early stage of robust inflammation (2 and 4 h post injection) and had a delayed induction observed at later time points (24 and 48 h post injection) (Fig. 3C). Tnfa levels were slightly but significantly still elevated at 24 h when increases in Aif1 were observed. Notably, Aif1 induction preceded induction of Trem2 gene expression, which was observed at 48 and 72 h post injection (Fig. 3D). IBA1 immunohistochemistry supported the delayed microglial activation identified by RT-qPCR after peripheral LPS stimulation, with microglia that appeared to look more activated after 24 h post challenge (Fig. 3E and F). The kinetics of altered IBA1 immunoreactivity was similar in both the cortex and hippocampus regions of the brain (Fig. 3E and F). Delayed Trem2 upregulation is suggestive of a potential anti-inflammatory role following systemic inflammation.
Figure 3.
Systemic inflammation induces CNS inflammation that precedes robust microglial activation and Trem2 induction. PBS perfused hemi brains from male C57BL/6 N mice were harvested at 0, 2, 4, 8, 24, 48, and 72 h post i.p. injection with 2 mg/kg LPS. RT-qPCR was conducted for (A) Il1b, (B) Tnfa, (C) Aif1 (encodes IBA1), and (D) Trem2. Il1b and Tnfa induction preceded Aif1 and Trem2 upregulation that was noted after the majority of inflammation subsided. Shown are the averages ± S.E.M. for N = 4–7 mice per group, N = 2 independent experiments. (One-way ANOVA, Tukey post-hoc, *P ≤ 0.05, ***P≤ 0.001, ****P ≤ 0.0001). IBA1 immunohistology was conducted on 50 μm sections from male mice harvested at 0, 2, 4, 8, 24, 48, and 72 h post i.p. injection with 2 mg/kg LPS and examined for reactivity in the (E) cortex and (F) hippocampus. Significant alterations in IBA1 reactivity was noted starting at 24 h post injection. Shown are representative images from N = 6–8 mice from N = 2 independent experiments. Scale bar = 50 μm.
Trem2 mildly represses CNS inflammation under both basal and LPS-stimulated conditions
TREM2 has been suggested to impact several different aspects of microglial function including phagocytosis, inflammation, and survival (24,39–42,44,45). In order to examine the impact of TREM2 on CNS responses following systemic inflammation using an unbiased approach, RNAseq was conducted on hippocampal RNA isolated from PBS perfused saline or LPS injected WT or Trem2−/− mice 48 h post challenge when Trem2 is upregulated (Fig. 3D). RNAseq transcriptome data were analysed with false discovery rate (FDR) set at 5%.
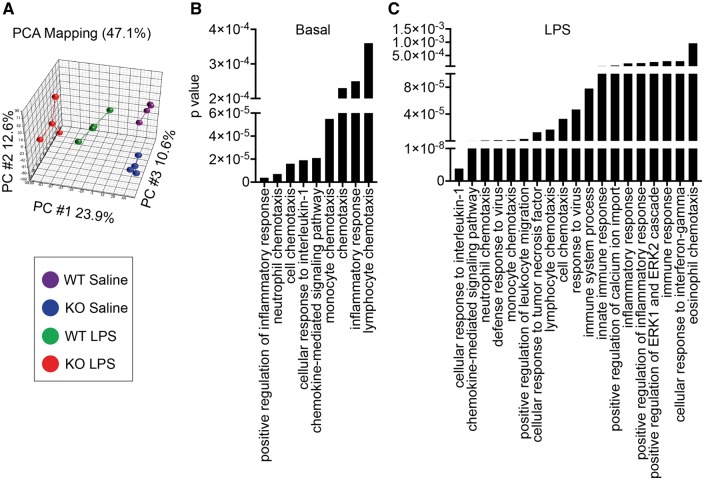
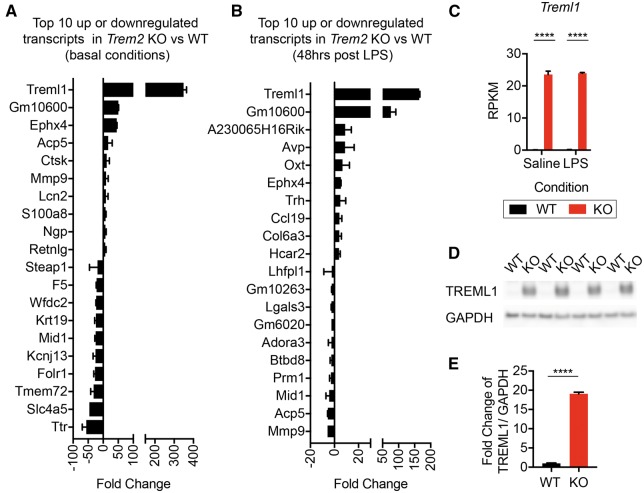
Examination of the CNS transcriptome in saline and LPS treated Trem2−/− and WT mice demonstrated some alterations in the gene expression patterns, and principle component analysis (PCA) revealed four distinct populations that were separated by both genotype and LPS treatment (Fig. 4A, Supplementary Material, Fig. S1). Gene ontology analysis revealed significant alterations in pathways involving inflammation and chemotaxis of immune cells including neutrophils, monocytes, and lymphocytes as well as regulation of inflammation in Trem2−/− mice relative to WT controls under basal conditions (Fig. 4B). After peripheral LPS challenge, pathways involved in cellular responses to pro-inflammatory cytokines, including IL-1 and TNF were significantly altered (Fig. 4C). Several transcripts were altered in Trem2−/− mice relative to WT mice during basal conditions including Treml1, Ctsk, Mmp9, Lcn2, and S100a8, while many additional transcripts were altered in LPS-treated Trem2−/− mice relative to WT including Treml1, Avp, Ccl19, Acp5, and Mmp9 (Fig. 5 and Supplementary Material, Tables S1 and S2).
Figure 4.
Trem2 deficiency alters CNS transcriptomes in both basal and inflammatory conditions. RNAseq was conducted on RNA extracted from PBS perfused hippocampal tissues from male C57BL/6 N wild type (WT) or Trem2−/− (KO) mice 48 h post i.p. challenge with saline or 2 mg/kg LPS. (A) Principal component analysis (PCA) of RNAseq datasets of hippocampal transcriptomes for WT and KO mice treated with LPS or saline is shown. Centroids for each group are indicated on the PCA plot. N = 3 animals per group. WT saline, KO saline, WT LPS, and KO LPS formed four non-overlapping groups on the PCA plot, indicating they have distinct transcriptomes. Gene ontology biological process (GO BP) pathway analysis demonstrating pathways that are significantly altered in Trem2−/− (KO) mice, relative to WT in (B) saline injected (basal levels) and (C) LPS challenged animals.
Figure 5.
Velocigene targeted Trem2 knockout mice have exaggerated levels of Treml1. The top 10 transcripts that were upregulated or downregulated in Trem2−/− relative to WT male hippocampal tissue at 48 h post i.p. injection of (A) saline or (B) 2 mg/kg LPS, as determined by RNAseq. Treml1 was increased 348.7 ± 15.6 and 165.6 ± 1.8 in Trem2−/− over WT controls in saline and LPS injected animals, respectively. Trem2 itself was excluded from the analysis of candidate transcripts. Shown are the averages ± S.E.M. from the N=3 animals per group. Data analysed by t-test with two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli with false discovery rate (FDR) set at 5%. (C) The RPKM values for Treml1, the most abundant transcript upregulated in both saline and LPS treated Trem2−/− mice, is shown for WT (black) and Trem2−/− (red) mice. Shown are the averages ± S.E.M. from the N=3 animals per group. (Two-way ANOVA, Tukey post-hoc, ****P≤ 0.0001). (D) Western blot analysis of TREML1 and GAPDH levels in both WT and Trem2−/− mice was assessed. Representative images from N=4 of the N=10 animals per group assayed are shown. (E) Fold change of TREML1/GAPDH ratios normalized to the average TREML1/GAPDH ratio of WT controls are shown as the averages ± S.E.M. for N=10 animals per group (Unpaired two-tailed t-test, ****P≤ 0.0001).
Trem2 deletion alters Treml1 expression depending on targeting construct
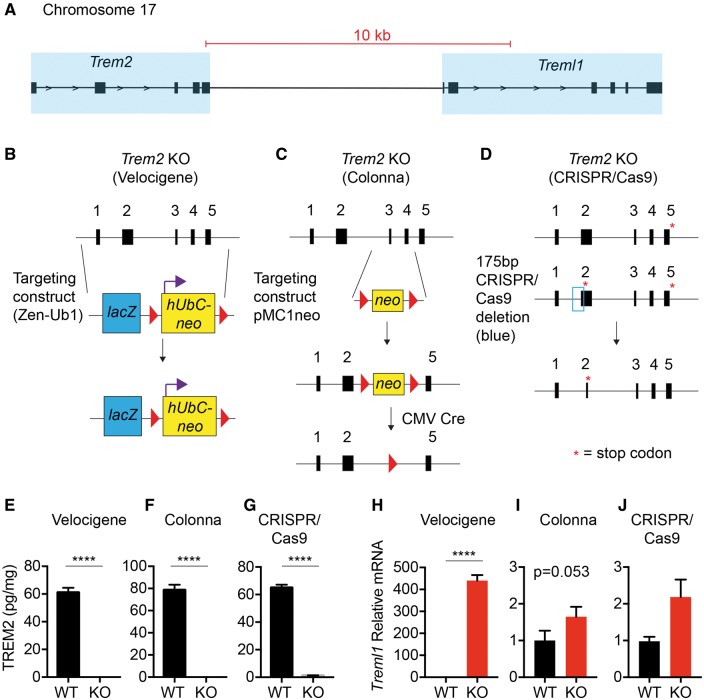
Under both basal and LPS-stimulated conditions, Treml1 was the top upregulated transcript in Trem2−/− mice, with a 349-fold change under basal conditions and a 165-fold change after LPS treatment (Fig. 5A and B and Supplementary Material, Tables S1 and S2). The deletion targeting strategy by Velocigene used a selection cassette (lacZ-flox-human Ubiquitin C promoter-neomycin-flox) to replace the coding sequence 16 bp upstream of the ATG start codon and ending at the TGA stop codon of Trem2. Interestingly, the Treml1 locus is directly downstream of Trem2, within ∼8, 000 bp of the 3’ end of Trem2. Examination of the raw RPKM values in Velocigene Trem2 targeted tissue by RNAseq demonstrated a significant increase in Treml1 expression based solely on genotype and not LPS treatment (Fig. 5C), suggesting an artifact of the targeting strategy itself. To examine whether TREML1 protein expression was also altered, we performed a western blot on hippocampal lysates from WT or Trem2−/− mice (Fig. 5D). TREML1 levels were significantly increased in Trem2−/− mice, with the TREML1/GAPDH ratio being 19 fold higher than WT controls (1.00 ±0.09 vs 19.07 ±0.42 WT vs Trem2−/− Ave ± S.E.M.) (Fig. 5D and E).
Examination of the genomic positioning of Trem2 and Treml1 revealed that the Treml1 is located less than 10 kb downstream of Trem2 on Chromosome 17 (Fig. 6A schematic). To determine if the disruption of the Trem2 gene using the Velocigene targeting strategy introduced a possible artifactual upregulation of Treml1 expression, other Trem2 knockout lines with different targeting strategies were obtained. We compared Trem2−/− (Velocigene) mice to Trem2−/− mice from the Colonna lab [targeting that replaced exons 3 and 4 with a neomycin cassette that was subsequently removed with CMV Cre (24)], along with Trem2−/− mice from CRISPR/Cas9 targeting that created a 175 bp deletion resulting in a stop codon at amino acid 17 (Fig. 6B–D). Using a sensitive MesoScale Discovery sandwich ELISA, we measured TREM2 protein levels and found that each deletion strategy resulted in ablation of TREM2 expression relative to controls (Fig. 6E–G). To assess whether the different targeting strategies differentially impacted Treml1 expression, RT-qPCR for Treml1 transcript levels was conducted from hippocampal RNA. In accordance with our RNAseq data (Fig. 5C), the Velocigene targeting of Trem2 resulted in a highly significant upregulation of Treml1 expression with a 440.2 ± 25.26 (avg ± S.E.M.) fold induction over controls (Fig. 6H). A trend (P = 0.053) toward a significant increased in Treml1 was observed in hippocampal Trem2−/− tissue derived from the Colonna deletion strategy; however, the magnitude of induction was only approximately two-fold and could be a true biological effect (Fig. 6I). Finally, although trending toward an increase, the CRISPR/Cas9 targeting of Trem2 by Jackson Labs did not induce any significant changes in Treml1 expression (Fig. 6J). Notably, Ephx4 transcripts were significantly increased in both Trem2 deficient animals but with much more upregulation in the Velocigene line compared with the CRISPR/Cas9 line. Additionally, S100a8 was only significantly altered in Velocigene animals (Supplementary Material, Fig. S2). This suggests that some, but not all, potential targets may be impacted by Treml1 overexpression whereas others may be a result of Trem2 deficiency itself. Together, these data indicate that these different targeting strategies have distinct outcomes on downstream Treml1 expression.
Figure 6.
Treml1 expression varies depending on the Trem2 knockout strategy. (A) Schematic of the relationship between Trem2 and Treml1 on Chromosome 17. Scale bar is indicated in red, transparent blue boxes indicate regions containing exons of the respective genes, and black arrows indicate direction of transcription. Trem2 targeting strategies for the (B) Velocigene, (C) Colonna, and (D) CRISPR/Cas9 approaches are depicted. TREM2 protein levels were determined by Meso Scale Discovery ELISA from Triton-X fractions of hippocampal tissues in (E) Velocigene, (F) Colonna, and (G) CRISPR/Cas9 derived Trem2−/− male mice, confirming that each line has no TREM2 (knockout verification). Shown are the averages ± S.E.M from N = 3–4 animals per group (Unpaired two-tailed t-test, ****P≤ 0.0001). (G–I) Hippocampal RNA was isolated from respective male wild-type (WT) and Trem2−/− mice (KO) for RT-qPCR analysis of Treml1 expression in (H) Trem2−/− mice created using the Velocigene strategy and their WT C57BL/6 N controls, (I) Trem2−/− mice from the Colonna targeting, and (J) Trem2−/− mice from CRISPR/Cas9 targeting and controls. Shown are the averages ± S.E.M. from N = 6–7 animals per group. (Unpaired two-tailed t-test, ****P≤ 0.0001).
Deletion of the floxed neomycin cassette eliminates aberrant Treml1 expression in Velocigene targeted Trem2−/− mice
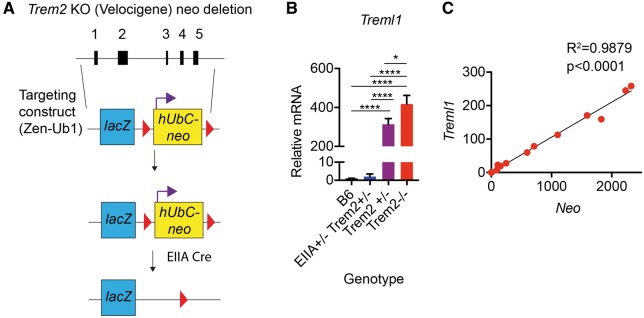
To examine if the human ubiquitin C promoter/neomycin cassette portion of the Velocigene Trem2−/− construct impacted transcription of the downstream Treml1 locus, we crossed Velocigene Trem2−/− mice with EIIA-Cre expressing animals in order to delete the floxed selection cassette (Fig. 7A). In contrast to Trem2+/− mice that retained the neo cassette, the resulting EIIA-Cre+/− Trem2+/− mice, which were negative for neomycin expression, no longer expressed aberrant levels of Treml1 (Fig. 7B). Since the EIIA-Cre mice results in different levels of mosaicism in neomycin deletion after the first cross (prior to establishing a germline colony), we were able to examine whether there was a correlation between neomycin and Treml1 expression. Hippocampal RT-qPCR revealed a strong positive correlation between neomycin and Treml1 expression with an R2 value of 0.9879 (P < 0.0001) (Fig. 7C). Resolution of Treml1 levels to wild-type levels through the excision of the human ubiquitin C promoter/neo cassette suggests that these residual elements in the Velocigene targeting construct are able to drive expression of downstream genes in the endogenous locus.
Figure 7.
Removal of the neomycin cassette from the Velocigene Trem2 knockout mice ameliorates aberrant Treml1 expression. (A) Schematic of Trem2−/− (Velocigene) male mice crossed with EIIA-Cre mice to remove the floxed neomycin cassette driven by the human Ubiquitin C promoter. The first cross of EIIA-Cre to Trem2−/− (Velocigene) mice creates a mosaic of neomycin deletion due to some inefficiencies in Cre activity at early embryonic stages. (B) RT-qPCR for Treml1 levels in hippocampal tissue from EIIA-Cre+/− Trem2+/− mice that were negative for neomycin, were compared with WT, Trem2+/−, and Trem2−/− Velocigene controls. EIIA-Cre+/− Trem2+/− mice that were negative for neomycin had significantly decreased levels of Treml1 relative to both Trem2+/−, and Trem2−/− Velocigene controls and were similar to WT. Shown are the averages ± S.E.M from N = 3–5 animals per group (One-way ANOVA, Tukey posthoc, *P <0.05, ****P≤ 0.0001). (C) RT-qPCR for Treml1 and neomycin levels in hippocampal tissue from EIIA Cre+/− Trem2+/− mice over a range of mosaicism was conducted. A highly significant, positive correlation between Treml1 and neomycin expression was observed. Shown is the correlation from N = 18 animals ranging in neomycin expression. (Linear regression, R2=0.9879, P < 0.0001).
Discussion
Microglial activation is observed in numerous neuropathologies, from neurodegeneration to autoimmune disease, and may contribute to disease through the production of pro-inflammatory mediators (5,6). As a result, regulation of the progression and resolution of microglial responses can be critical to the outcome of disease. Several cell surface regulatory molecules on microglia, including CX3CR1 and CD200R1, are thought to modulate function (47,48). Although TREM2 is almost exclusively expressed on microglia and TREM2 variants are associated with neurological disease, the impact of TREM2 on CNS function is still under debate. Overall, we have demonstrated that the targeting strategy used to study the role of TREM2 on CNS sequelae in mouse models is pertinent in the interpretation of data. Using the Velocigene Trem2−/− mice, we have demonstrated that TREM2 does not impact basal behavioral or cognitive readouts in adult animals or result in large shifts in hippocampal ISF early inflammation following systemic LPS challenge. As TREM2 has been shown to impact microglial expansion during aging and has been shown to impact survival in both macrophages and microglia, it is possible that behavioral and cognitive readouts would be different in an aged cohort of Trem2−/− mice (44,45,49). Importantly, RNAseq analysis of CNS transcriptomes revealed exaggerated Treml1 expression that was over 300-fold in Trem2−/− (Velocigene) mice relative to controls during basal conditions. However, this large elevation in Treml1 expression was not observed in brain tissues derived from other Trem2 KO mice using two independent targeting strategies. Additionally, removal of the floxed neomycin cassette driven by the human ubiquitin C promoter in the Velocigene targeted Trem2 line completely prevented the Treml1 overexpression artifact, indicating that the promoter activity was sufficient to drive expression of a locus over 8 kb downstream. Because many Velocigene targeting strategies utilize this selection cassette, we suggest removal of the floxed neomycin cassette in order to circumvent transcription of neighboring genes. Due to this confound, it is therefore uncertain as to whether Treml1 is either compensating for or masking the impact of TREM2 deficiencies in these animals.
Although it is unknown if and how Treml1 impacts the overall response in Trem2 deficient animals, our RNAseq data demonstrated alterations in inflammatory pathways in the absence of Trem2. During basal conditions, increased levels of Ephx4, Mmp9, Ctsk, and S100a8 were within the top 10 induced genes. S100a8 and S100a9, the 11th most upregulated transcript in Trem2−/− mice in our data set, have been shown to play a role focal cerebral ischemia, with a deficiency in both transcripts resulting in diminished disease (50). Epoxide hydrolases are also involved in the regulation of neuroinflammation and inhibition of soluble epoxide hydrolases resulted in decreased pro-inflammatory cytokines (IL-1β and Il-6) within the context of seizures, and increased IL-10 secretion in the CNS following cardiac arrest (51,52). Additionally, both Mmp9 and Ctsk are also associated with microglial activation (53). Trem2−/− mice also show a significant upregulation of Lcn2, which we been shown to be a potent anti-inflammatory molecule in the context of peripheral LPS challenge (54). These changes may reflect a compensatory mechanism to offset the upregulation of the aforementioned transcripts that are likely to be more pro-inflammatory. Therefore, these data suggest that TREM2 may be playing an anti-inflammatory role in the CNS.
Recently, the role of TREM2 on microglial function has been under intense investigation in light of data associating TREM2 variants with an increased AD risk (18–20,55,56). Several studies using Trem2−/− mice from Velocigene targeting or Colonna lab targeting have revealed alterations in the clustering and morphology of plaque associated myeloid or microglial cells (34,46,57). However, there are conflicting results that remain regarding not only in the origin of the plaque-associated cells (i.e. peripherally derived TREM2+ myeloid cells vs TREM2+ microglia) but the role of TREM2 in amyloid pathology. Usage of the Velocigene Trem2−/− mice demonstrated slightly decreased amyloid pathology whereas a study using the Colonna derived Trem2−/− mice showed a slight increase in pathology in the hippocampus (34,57). Recently, it has been suggested that TREM2 functions in a distinct manner based on the stage of AD pathology, with an ability to mitigate early disease and exacerbate late disease (58). However, given our findings that there is a large discrepancy in the expression of Treml1 between the two model systems, this may play a significant role in shaping disease. Interestingly, TREML1 levels in the brain have recently been associated with decreased AD risk in humans (59), suggesting that upregulated expression may be protective in disease. Although soluble TREML1 can decrease LPS induced neutrophil inflammation suggesting an anti-inflammatory role with that cell type, to our knowledge nothing is currently known regarding the role of TREML1 and microglial function (60). Whether exaggerated Treml1, which is observed in the Velocigene Trem2−/− mice, is also playing a protective role when crossed to an amyloid model of disease is unknown. Therefore, caution must be taken when using the Velocigene Trem2−/− mice to define the role of TREM2 on microglial function until more is known regarding the nature of TREML1.
Materials and Methods
Animals
C57BL/6 N mice, Trem2−/− mice (KOMP line #10093 generated by Velocigene), and Trem2CRISPR-KO mice (Jackson Labs stock #027197) were housed under standard laboratory conditions in ventilated cages on 12-h light: dark cycles in a specific pathogen-free environment at Mayo Clinic. Trem2−/− mice generated by the Colonna lab (24) were also housed under standard laboratory conditions in ventilated cages on 12-h light: dark cycles in a specific pathogen-free environment. For LPS studies, animals were injected intraperitoneally (i.p.) with 2 mg/kg LPS (0111: B4; Sigma, Saint Louis, MO). Animal protocols were reviewed and approved by Mayo Clinic Institutional Animal Care and Use Committee.
Behavioral battery
To measure readouts of anxiety, mobility, sociability, and cognition, a behavioral battery consisting of open field assay (OFA), elevated plus maze (EPM), three chambered social interaction (3SI), and contextual and cued fear conditioning was performed on consecutive days, as previously described (61,62). All testing was conducted between 9: 00 A.M. and 5: 00 P.M. and animals were acclimated to the testing room for 1 h prior to the onset of testing. Prior to the initial test and in between each test animal, the behavioral apparatus was cleaned with 30% ethanol (or 30% isopropanol for cued fear conditioning) to remove any residual odors on the equipment.
Open field assay
To assess activity/anxiety in the open field assay (OFA), mice were placed in a 40x40x30 cm (WxLxH) black Plexiglass box with a brightly lit center. Fujinon overhead cameras recorded behavioral activity for 15 min and ANY-maze software was utilized for tracking (Stoelting Co.; Wood Dale, IL). An imaginary 13 × 13 cm center region in the box was digitally defined using ANY-maze software. Side mounted photobeams located 7.6 cm from the floor of the box were used to monitor rearing activity. Multiple readouts including total distance traveled, time mobile, center: total distance, and rearing were analysed.
Elevated plus maze
Elevated plus maze (EPM) is another test for assessing anxiety-like behavior. A four- armed (50 × 10 cm) maze is elevated 50 cm above the floor, with two of the arms enclosed by open top gray walls (35 × 15 cm) while the other two remain ‘open’. Mice were placed in the maze in the same orientation and their exploratory behavior was monitored for 5 min using an overheard camera and ANY-maze software. The ratio of time spent in open: closed arms was utilized as an anxiety measurement.
Three-chambered social interaction
To examine sociability, mice were placed in a three-chamber Plexiglass 40×40 cm box consisting of two 17 × 40 cm regions separated by 2 dividers forming a smaller 5×40 cm center region. Mice were able to move freely through a small 8×5 cm opening that was aligned in both dividers. Two mesh cylindrical containers were placed in the box, one in the corner of the ‘empty’ chamber and one in the corner of the ‘mouse’ chamber. The test mouse was placed in the box for 4 min to explore the surroundings and then removed and placed in a temporary holding cage. A same sex matched probe mouse was placed under the mesh container in the ‘mouse’ chamber compartment allowing for visual, olfactory, and auditory interactions between probe and test mice. After placement of the probe mouse, the test mouse was returned and allowed to explore the chambers undisturbed for 10 min. All behavior was tracked with and overhead camera and ANY-maze software. The interaction score was calculated as: (time spent near the mouse container- time spent near the empty container)/(time spent near the mouse container + time spent near the empty container).
Context and cued fear conditioning
To measure freezing as a learning/memory readout, mice were trained in a sound attenuated chamber with an overhead mounted camera and FreezeFrame tracking software (Actimetrics, Wilmette, IL) and a floor consisting of rods that were capable of inducing a mild electrical stimulus. Initial training of mice occurred by allowing animals to remain undisturbed for 2 min in the chamber while baseline freezing behavior was recorded. This was followed by a 30 s conditioned stimulus (CS) of an 80 dB white noise that was paired with an unconditioned stimulus (UC) of a mild foot shock (0.5 mA) for the final 2 s of the CS. Mice were undisturbed for the next consecutive minute and then received a second CS-US session followed by another 30 s of rest. Afterward, mice were removed from the chamber, returned to their home cage and left undisturbed overnight. The following day, mice were re-acclimated to the behavior room under the same conditions, returned to the test chamber, and freezing behavior was recorded for 5 min to examine contextual learning/memory (context test). Mice were then returned to their home cage, moved to a different room with dimmed lighting, and allowed to re-acclimate to these new conditions for a minimum of 1 h. To examine cued learning/memory (cued test) to the auditory CS independent of the previous contextual cues, several parameters of the environment were altered including both visual (i.e. changing from white to red ceiling lighting, dimming the anterior chamber, altering the test chamber itself by covering the floor with a white opaque plastic, changing the shape and spatial cues within the chamber, and altering the cages used to transport the animals to the test chamber) and olfactory (i.e. replacing 30% ethanol with 30% isopropanol to wipe down the test chamber, adding a vanilla extract scent to the test chamber and the anterior room). Mice were placed in the altered chamber for 3 min followed by initiation of the auditory CS for 3 min during which time freezing behavior was recorded. For both context and cued tests, baseline-freezing behavior obtained during the training period was subtracted from the overall freezing score to control for animal variability.
Tissue processing for RNAseq and RT-qPCR
Mice were deeply anesthetized with pentobarbital prior to cardiac perfusion with phosphate-buffered saline (PBS) to expunge blood components from the cerebrovasculature. For biochemical analysis, dissected hippocampal brain tissues were quickly frozen on dry ice and stored at −80 °C until further processing. Tissues were sonicated with 2–3 pulses in Tris buffered saline with EDTA (TBSE) (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA) with 1x protease and phosphatase inhibitors (Thermo Scientific, Waltham, MA). An aliquot of the tissue suspension was processed for RNA isolation using either the Aurum Total RNA mini kit (Biorad, Hercules, CA) or RNeasy Plus kit (Qiagen, Valencia, CA). Random-primed reverse transcription was performed according to manufacturer protocols (Invitrogen-Life Technologies, Grand Island, NY). cDNA was added to a reaction mix (10 μl final volume) containing 300 nM gene-specific primers and Universal SYBR green supermix (Biorad, Hercules, CA). All samples were run in triplicate and were analysed on a Quant Studio 7 Flex Real Time PCR instrument (Applied Biosystems - Life Technologies). Relative gene expression was normalized to GAPDH controls and assessed using the 2-ΔΔCT method. Primer sequences are as follows (5’ to 3’): Gapdh F: CTGCACCACCAACTGCTTAG, Gapdh R: ACAGTCTTCTGGGTGGCA GT, Aif1(Iba1) F: GGATTTGCAGGGAGGAAAAG, Aif1(Iba1) R: TGGGATCATCGAGGAATTG, Treml1 F: GTGATGGCCAGAAAGAAAGG, Treml1 R: TTCCTGGAACTCCAGTGCTC, Neo F: TGAATGAACTGCAGGACGAG, Neo R: AGTGACAACGTCGAGCACAG, Ephx4 F: TCATTGTTATTAACTTCCCACATC, Ephx4 R: AACAGCTGGGCAGGATGC, S100a8 F: CCTTTGTCAGCTCCGTCTTC, S100a8 R: CAAGGCCTTCTCCAGTTCAG
Meso-scale discovery ELISA
PBS perfused hippocampal brain tissues were isolated and briefly sonicated with 2–3 pulses in Tris buffered saline with EDTA (TBSE) (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA) with 1x protease and phosphatase inhibitors (Thermo Scientific, Waltham, MA). An aliquot was added to an equal amount of 2% Triton TBSE with 1x protease and phosphatase inhibitors for a final concentration of 1% Triton. Samples were lysed for 30 min on ice and then centrifuged at 14, 000 g for 15 min. Triton-X supernatants were stored at −80 °C until further use. For MSD ELISA, 96 well plates were coated with streptavidin (Meso Scale Discovery, Rockville, MD) and then washed and blocked in 0.5% BSA/PBST (0.05% Tween-20) overnight at 4 °C. Plates were incubated with 0.25 μg/ml biotinylated sheep anti-mouse TREM2 antibody (R&D Systems, Minneapolis, MN) for 1 h at RT. Following three washes with PBST, samples and standards diluted in 0.25% BSA/PBST supplemented with protease inhibitors (Roche, Indianapolis, IN) were incubated overnight at 4 °C. Plates were washed three times with PBST and incubated with 1 μg/ml rat monoclonal anti-mouse TREM2 (R&D Systems, Minneapolis, MN) for 1 h at RT. After 4 washes in PBST, 0.5µg/ml SULFO-TAG labeled anti-rat antibody was added and incubated for 1 h protected from light. Plates were washed 3 times in PBST and then once in PBS prior to being developed in Meso Scale Discovery Read buffer and read on the Meso Scale Discovery SECTOR Imager 2400 (Meso Scale Discovery, Rockville, MD).
Western blot
Hippocampal tissues from PBS perfused mice were sonicated with 2–3 pulses in Tris buffered saline with EDTA (TBSE) (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA) with 1x protease and phosphatase inhibitors (Thermo Scientific, Waltham, MA). An aliquot of the sample was incuvated with 2% Triton-X TBSE with 1x protease and phosphatase inhibitors for a final concentration of 1% Triton-X TBSE (TBSE-X). Samples were incubated for 1 h on ice and the centrifuged at 20, 000rcf at 4 °C for 15 min. Supernatants were examined for protein concentration using a BCA assay (ThermoScientific, Waltham, MA). 15 μg of protein was boiled for 5 min and resolved on a 10% polyacrylamide bis-tris gel and transferred to a nitrocellulose membrane. Membranes were blocked in 5% non-fat dry milk in PBS with 0.1% Tween 20 (PBS-T) (Sigma) and probed overnight at 4 °C with 1: 500 biotinylated goat anti-TREML1 (R&D Systems, Minneapolis, MN), followed by horseradish peroxidase (HRP) conjugated mouse anti-biotin secondary antibody. Membranes developed using supersignal west dura extended duration chemiluminescent substrate (Thermo Scientific) prior to film exposure. Membranes were stripped and re-probed with 1: 5000 mouse anti-GAPDH (clone 6C5, Advanced Immunochemicals, Long Beach, CA) overnight 4 °C, followed by goat anti-mouse HRP (Jackson Immunoresearch Laboratories). Membranes were washed and developed with enhanced chemiluminescent substrate (Thermo Scientific).
Immunohistochemistry
PBS perfused hemi-brains were drop fixed into 10% neutral buffered formalin (Fisher Scientific, Waltham, MA) overnight at 4 °C. Tissue was then transferred to 30% sucrose in PBS overnight at 4 °C. Coronal brain sections (50 μ) were cut on a freezing-sliding microtome and stored at −20 °C in cryprotectant until staining. Sections were washed in PBS to remove cryoprotectant and then blocked for endogenous peroxidase activity and permeabilized with 0.6% H2O2, 0.1% NaN3 in PBS-X (1X PBS containing 0.3% Triton-X) for 30 min at RT. Sections were blocked with 1% milk in PBS-X followed by incubation with rabbit anti-Iba1 at 1: 6000 (cat# 019–9741, Wako, Richmond, VA) in 0.5% milk PBS-X for 2 days at 4 °C. Sections were then incubated with the Vectastain anti-Rabbit IgG kit (Vector Labs, Burlingame, CA) overnight at 4 °C followed by ABC component for 4 h and developed using the DAB kit (Vector Labs) according to manufacturer’s instructions. Images were acquired using an Aperio XT Scanner (Aperio, Vista, CA) at a 20x magnification.
In vivo microdialysis
In vivo microdialysis of freely moving, awake mice was performed as described (63). Mice were provided acetaminophen 48 h before surgical probe implantation. Mice were anesthetized using 1.5–2.5% isoflurane prior to shaving the head and transecting the skin along the midline to expose the skull. A bore hole (0.75 mm) was made above the left hippocampus (coordinates bregma -3.1 mm, 2.5 mm lateral) using a small animal sterotaxic device. A second bore hole was made into the right, posterior skull quadrant for an anchoring bone screw. An AtmosL Guide Cannula (PEG-X, Eicom) was then stereotactically inserted into the hippocampus (12° angle, dura mater -1.2 mm) and cemented into place with dental cement. An AtmosLM Dummy probe (PED-X, Eicom) was inserted into the guide cannula and the wound was closed with surgical adhesive glue. Body temperature was maintained during the procedure using a heating blanket set at 37 °C. Mice were injected with antibiotic (ampicillin; 100 mg/kg, i.m.) and placed into a clean cage and provided with access to food and water ad libitum. Mice were allowed to awaken and recover overnight. To sample large molecules from the extracellular space, microdialysis MegaProbes (1000-kDa MWCO membrane, Eicom) were used with a peristaltic push-pull pump (SciPro). Microdialysis perfusion buffer was aCSF (1.3 mM CaCl2, 1.2 mM MgSO4, 3 mM KCl, 0.4 mM KH2PO4, 25 mM NaHCO3, 122 mM NaCl, pH 7.35) containing 0.15% BSA (Sigma) that was filtered through a 0.1 μM membrane. Mice were kept under constant light conditions for the remainder of the experiment. LPS (2 mg/kg) was injected (i.p.) 10 h post probe insertion. Microdialysis was performed at a flow rate of 1 μl/min and interstitial fluid (ISF) samples were collected hourly using a refrigerated fraction collector (SciPro).
Cytometric bead array
ISF samples were pooled into 2-h fractions and assayed for pro-inflammatory cytokines using a mouse inflammatory cytokine bead array kit according to manufacturer’s instructions (BD Bioscience, San Jose, CA). Samples were collected on a FACS Calibur (BD Bioscience, San Jose, CA) and analysed with FlowJo software (Tree Star, Inc).
Illumina RNA sequencing and pathway analysis
A total of 12 mRNA samples were sequenced at Mayo Clinic Genome Facility using NuGen Ovation library preparation and sequenced on an Illumina HiSeq 4000. Reads were mapped to the mouse genome mm10 and reads per kilobase per million mapped reads (RPKM) were generated for each transcript. RPKM values were log2 transformed and low abundance transcripts were removed from further analysis (log2 RPKM value less than -2 across all experimental groups were considered too lowly expressed). Principal component analysis (PCA) was performed using Partek Genomics Suite (Partek Inc., St. Louis, MO) with ANOVA and multiple comparison adjustment of p values using Benjamini–Hochberg-Yekutieli correction with false discovery rate (FDR) set at 5%.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
Mayo Foundation, GHR Foundation, Mayo Clinic Center for Individualized Medicine, Mayo Clinic Gerstner Family Career Development Award, Ed and Ethel Moore Alzheimer’s Disease Research Program of Florida Department of Health (6AZ06), NIH NS094137, AG047327 and AG049992 (JDF), The Robert and Clarice Smith and Abigail Van Buren Alzheimer’s Disease Research Program Fellowship, Mayo Clinic Program on Synaptic Biology and Memory, and NIH MH103632 (SK), and NIH AG027924, AG035355, and NS074969 (GB). Funding to pay the Open Access publication charges for this article was provided by the Mayo Foundation.
Supplementary Material
References
- 1. Nimmerjahn A., Kirchhoff F., Helmchen F. (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science, 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- 2. Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., Gan W.-B. (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci., 8, 752–758. [DOI] [PubMed] [Google Scholar]
- 3. Gyoneva S., Davalos D., Biswas D., Swanger S.A., Garnier-Amblard E., Loth F., Akassoglou K., Traynelis S.F. (2014) Systemic inflammation regulates microglial responses to tissue damage in vivo. Glia, 62, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heppner F.L., Skutella T., Hailer N.P., Haas D., Nitsch R. (1998) Activated microglial cells migrate towards sites of excitotoxic neuronal injury inside organotypic hippocampal slice cultures. Eur. J. Neurosci., 90, 3284–3290. [DOI] [PubMed] [Google Scholar]
- 5. Saijo K., Glass C.K. (2011) Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol., 11, 775–787. [DOI] [PubMed] [Google Scholar]
- 6. Nayak D., Roth T.L., McGavern D.B. (2014) Microglia development and function. Annu. Rev. Immunol., 32, 367–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imamura K., Hishikawa N., Sawada M., Nagatsu T., Yoshida M., Hashizume Y. (2003) Distribution of major histocompatibility complex class II-positive microglia and cytokine profile of Parkinson’s disease brains. Acta Neuropathol. (Berl.), 106, 518–526. [DOI] [PubMed] [Google Scholar]
- 8. McGeer P.L., Itagaki S., Boyes B.E., McGeer E.G. (1988) Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology, 38, 1285–1291. [DOI] [PubMed] [Google Scholar]
- 9. Ouchi Y., Yoshikawa E., Sekine Y., Futatsubashi M., Kanno T., Ogusu T., Torizuka T. (2005) Microglial activation and dopamine terminal loss in early Parkinson’s disease. Ann. Neurol., 57, 168–175. [DOI] [PubMed] [Google Scholar]
- 10. Perry V.H., Nicoll J.A.R., Holmes C. (2010) Microglia in neurodegenerative disease. Nat. Rev. Neurol., 6, 193–201. [DOI] [PubMed] [Google Scholar]
- 11. Heneka M.T., Carson M.J., El Khoury J., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M.. et al. (2015) Neuroinflammation in Alzheimer’s disease. Lancet Neurol., 14, 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cagnin A., Rossor M., Sampson E.L., Mackinnon T., Banati R.B. (2004) In vivo detection of microglial activation in frontotemporal dementia. Ann. Neurol., 56, 894–897. [DOI] [PubMed] [Google Scholar]
- 13. Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.-Y., Shang Y., Oldham M.C., Martens L.H., Gao F.. et al. (2016) Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell, 165, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu C.-C., Liu C.-C., Kanekiyo T., Xu H., Bu G. (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol., 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A.. et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet., 41, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert J.-C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B.. et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet., 41, 1094–1099. [DOI] [PubMed] [Google Scholar]
- 17. Naj A.C., Jun G., Beecham G.W., Wang L.-S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K.. et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet., 43, 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S.. et al. (2013) TREM2 variants in Alzheimer’s disease. N. Engl. J. Med., 368, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pottier C., Wallon D., Rousseau S., Rovelet-Lecrux A., Richard A.-C., Rollin-Sillaire A., Frebourg T., Campion D., Hannequin D. (2013) TREM2 R47H variant as a risk factor for early-onset Alzheimer’s disease. J. Alzheimers Dis., 35, 45–49. [DOI] [PubMed] [Google Scholar]
- 20. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J.. et al. (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med., 368, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daws M.R., Lanier L.L., Seaman W.E., Ryan J.C. (2001) Cloning and characterization of a novel mouse myeloid DAP12-associated receptor family. Eur. J. Immunol., 31, 783–791. [DOI] [PubMed] [Google Scholar]
- 22. Bouchon A., Hernández-Munain C., Cella M., Colonna M. (2001) A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med., 194, 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cella M., Buonsanti C., Strader C., Kondo T., Salmaggi A., Colonna M. (2003) Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J. Exp. Med., 198, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turnbull I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. (2006) Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol., 177, 3520–3524. [DOI] [PubMed] [Google Scholar]
- 25. Schmid C.D., Sautkulis L.N., Danielson P.E., Cooper J., Hasel K.W., Hilbush B.S., Sutcliffe J.G., Carson M.J. (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J. Neurochem., 83, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L-c., Means T.K., El Khoury J. (2013) The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci., 16, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N.. et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci., 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terme M., Tomasello E., Maruyama K., Crepineau F., Chaput N., Flament C., Marolleau J.-P., Angevin E., Wagner E.F., Salomon B.. et al. (2004) IL-4 confers NK stimulatory capacity to murine dendritic cells: a signaling pathway involving KARAP/DAP12-triggering receptor expressed on myeloid cell 2 molecules. J. Immunol., 172, 5957–5966. [DOI] [PubMed] [Google Scholar]
- 29. Painter M.M., Atagi Y., Liu C.-C., Rademakers R., Xu H., Fryer J.D., Bu G. (2015) TREM2 in CNS homeostasis and neurodegenerative disease. Mol. Neurodegener., 10, 43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atagi Y., Liu C.-C., Painter M.M., Chen X.-F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S.S., Xu H.. et al. (2015) Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem., 290, 26043–26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jendresen C., Årskog V., Daws M.R., Nilsson L.N.G. (2017) The Alzheimer’s disease risk factors apolipoprotein E and TREM2 are linked in a receptor signaling pathway. J. Neuroinflammation, 14, 59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeh F.L., Wang Y., Tom I., Gonzalez L.C., Sheng M. (2016) TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron, 91, 328–340. [DOI] [PubMed] [Google Scholar]
- 33. Daws M.R., Sullam P.M., Niemi E.C., Chen T.T., Tchao N.K., Seaman W.E. (2003) Pattern recognition by TREM-2: binding of anionic ligands. J. Immunol., 171, 594–599. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H.. et al. (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell, 160, 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rayaprolu S., Mullen B., Baker M., Lynch T., Finger E., Seeley W.W., Hatanpaa K.J., Lomen-Hoerth C., Kertesz A., Bigio E.H.. et al. (2013) TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Mol. Neurodegener., 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thelen M., Razquin C., Hernández I., Gorostidi A., Sánchez-Valle R., Ortega-Cubero S., Wolfsgruber S., Drichel D., Fliessbach K., Duenkel T.. et al. (2014) Investigation of the role of rare TREM2 variants in frontotemporal dementia subtypes. Neurobiol. Aging, 35, 2657.e13–2619. [DOI] [PubMed] [Google Scholar]
- 37. Paloneva J., Autti T., Raininko R., Partanen J., Salonen O., Puranen M., Hakola P., Haltia M. (2001) CNS manifestations of Nasu-Hakola disease: a frontal dementia with bone cysts. Neurology, 56, 1552–1558. [DOI] [PubMed] [Google Scholar]
- 38. Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A.. et al. (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet., 71, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gawish R., Martins R., Bohm B., Wimberger T., Sharif O., Lakovits K., Schmidt M., Knapp S. (2015) Triggering receptor expressed on myeloid cells-2 fine-tunes inflammatory responses in murine Gram-negative sepsis. FASEB J., 29, 1247–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng H., Liu C.-C., Atagi Y., Chen X.-F., Jia L., Yang L., He W., Zhang X., Kang S.S., Rosenberry T.L.. et al. (2016) Opposing roles of the triggering receptor expressed on myeloid cells 2 and triggering receptor expressed on myeloid cells-like transcript 2 in microglia activation. Neurobiol. Aging, 42, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsieh C.L., Koike M., Spusta S.C., Niemi E.C., Yenari M., Nakamura M.C., Seaman W.E. (2009) A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem., 109, 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takahashi K., Rochford C.D.P., Neumann H. (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med., 201, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kawabori M., Kacimi R., Kauppinen T., Calosing C., Kim J.Y., Hsieh C.L., Nakamura M.C., Yenari M.A. (2015) Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J. Neurosci., 35, 3384–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poliani P.L., Wang Y., Fontana E., Robinette M.L., Yamanishi Y., Gilfillan S., Colonna M. (2015) TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest., 125, 2161–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng H., Jia L., Liu C.-C., Rong Z., Zhong L., Yang L., Chen X.-F., Fryer J.D., Wang X., Zhang Y.-W.. et al. (2017) TREM2 promotes microglial survival by activating Wnt/β-catenin pathway. J. Neurosci., 37, 1772–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ulrich J.D., Finn M.B., Wang Y., Shen A., Mahan T.E., Jiang H., Stewart F.R., Piccio L., Colonna M., Holtzman D.M.. et al. (2014) Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener., 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., Lamb B.T. (2010) Regulation of tau pathology by the microglial fractalkine receptor. Neuron, 68, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lyons A., Downer E.J., Crotty S., Nolan Y.M., Mills K.H.G., Lynch M.A. (2007) CD200 ligand receptor interaction modulates microglial activation in vivo and in vitro: a role for IL-4. J. Neurosci., 27, 8309–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu K., Byers D.E., Jin X., Agapov E., Alexander-Brett J., Patel A.C., Cella M., Gilfilan S., Colonna M., Kober D.L.. et al. (2015) TREM-2 promotes macrophage survival and lung disease after respiratory viral infection. J. Exp. Med., 212, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ziegler G., Prinz V., Albrecht M.W., Harhausen D., Khojasteh U., Nacken W., Endres M., Dirnagl U., Nietfeld W., Trendelenburg G.. et al. (2009) Mrp-8 and -14 mediate CNS injury in focal cerebral ischemia. Biochim. Biophys. Acta, 1792, 1198–1204. [DOI] [PubMed] [Google Scholar]
- 51. Wang J., Fujiyoshi T., Kosaka Y., Raybuck J.D., Lattal K.M., Ikeda M., Herson P.S., Koerner I.P. (2013) Inhibition of soluble epoxide hydrolase after cardiac arrest/cardiopulmonary resuscitation induces a neuroprotective phenotype in activated microglia and improves neuronal survival. J. Cereb. Blood Flow Metab., 33, 1574–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hung Y.-W., Hung S.-W., Wu Y.-C., Wong L.-K., Lai M.-T., Shih Y.-H., Lee T.-S., Lin Y.-Y. (2015) Soluble epoxide hydrolase activity regulates inflammatory responses and seizure generation in two mouse models of temporal lobe epilepsy. Brain. Behav. Immun., 43, 118–129. [DOI] [PubMed] [Google Scholar]
- 53. Lively S., Schlichter L.C. (2013) The microglial activation state regulates migration and roles of matrix-dissolving enzymes for invasion. J. Neuroinflammation, 10, 75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kang S.S. et al. (2017) Lipocalin-2 protects the brain during inflammatory conditions. Mol. Psychiatry, doi: 10.1038/mp.2016.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Efthymiou A.G., Goate A.M. (2017) Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener., 12, 43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jay T.R., von Saucken V.E., Landreth G.E. (2017) TREM2 in Neurodegenerative Diseases. Mol. Neurodegener., 12, 56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jay T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L.. et al. (2015) TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med., 212, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jay T.R., Hirsch A.M., Broihier M.L., Miller C.M., Neilson L.E., Ransohoff R.M., Lamb B.T., Landreth G.E. (2017) Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J. Neurosci., 37, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carrasquillo M.M. et al. (2017) A candidate regulatory variant at the TREM gene cluster associates with decreased Alzheimer’s disease risk and increased TREML1 and TREM2 brain gene expression. Alzheimers Dement., 13, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Derive M., Bouazza Y., Sennoun N., Marchionni S., Quigley L., Washington V., Massin F., Max J.-P., Ford J., Alauzet C.. et al. (2012) Soluble TREM-like transcript-1 regulates leukocyte activation and controls microbial sepsis. J. Immunol., 188, 5585–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kang S.S., Kurti A., Fair D.A., Fryer J.D. (2014) Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. J. Neuroinflammation, 11, 156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cook C., Kang S.S., Carlomagno Y., Lin W.-L., Yue M., Kurti A., Shinohara M., Jansen-West K., Perkerson E., Castanedes-Casey M.. et al. (2015) Tau deposition drives neuropathological, inflammatory and behavioral abnormalities independently of neuronal loss in a novel mouse model. Hum. Mol. Genet., 24, 6198–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ulrich J.D., Burchett J.M., Restivo J.L., Schuler D.R., Verghese P.B., Mahan T.E., Landreth G.E., Castellano J.M., Jiang H., Cirrito J.R.. et al. (2013) In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol. Neurodegener., 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.