Abstract
Known high-risk cutaneous malignant melanoma (CMM) genes account for melanoma risk in <40% of melanoma-prone families, suggesting the existence of additional high-risk genes or perhaps a polygenic mechanism involving multiple genetic modifiers. The goal of this study was to systematically characterize rare germline variants in 42 established melanoma genes among 144 CMM patients in 76 American CMM families without known mutations using data from whole-exome sequencing. We identified 68 rare (<0.1% in public and in-house control datasets) nonsynonymous variants in 25 genes. We technically validated all loss-of-function, inframe insertion/deletion, and missense variants predicted as deleterious, and followed them up in 1, 559 population-based CMM cases and 1, 633 controls. Several of these variants showed disease co-segregation within families. Of particular interest, a stopgain variant in TYR was present in five of six CMM cases/obligate gene carriers in one family and a single population-based CMM case. A start gain variant in the 5’UTR region of PLA2G6 and a missense variant in ATM were each seen in all three affected people in a single family, respectively. Results from rare variant burden tests showed that familial and population-based CMM patients tended to have higher frequencies of rare germline variants in albinism genes such as TYR, TYRP1, and OCA2 (P < 0.05). Our results suggest that rare nonsynonymous variants in low- or intermediate-risk CMM genes may influence familial CMM predisposition, warranting further investigation of both common and rare variants in genes affecting functionally important pathways (such as melanogenesis) in melanoma risk assessment.
Introduction
Cutaneous malignant melanoma (CMM) is an etiologically heterogeneous disease with genetic, host, and environmental factors, and their interactions contributing to its development (1). Approximately, 10% of CMM cases occur in a familial setting (2). CDKN2A and CDK4 are the two well-established high-risk genes for familial melanoma. Recently, BAP1, POT1, ACD, TERF2IP, and TERT were identified as potential high-risk melanoma susceptibility genes (3). However, these genes account for melanoma risk in less than 40% of melanoma-prone families, suggesting the existence of additional high-risk genes or perhaps a polygenic mechanism involving multiple genetic modifiers. In addition to high-risk genes, variants in intermediate-risk and low-risk genes also contribute to the predisposition of familial melanoma. For example, low-frequency and common variants in intermediate-risk melanoma genes such as MITF and MC1R have been associated with increased risk of melanoma in melanoma families (4–6). Further, genome-wide association studies (GWAS) have identified more than twenty low-risk melanoma susceptibility loci. Most of these loci involve genes and pathways that are known to play important roles in melanoma development such as pigmentation (MC1R, TYR, TYRP1, ASIP, HERC2/OCA2), nevi density (PLA2G6, MTAP/CDKN2A, CASP8, AGR3, FTO), cell cycle (CDKAL1 and CCND1), DNA repair (ATM and PARP1), and telomere length (TERT, OBCF1) (3). The role of these GWAS loci in the risk of familial melanoma has not yet been fully evaluated.
The advance in next-generation sequencing technology has made it possible to comprehensively characterize a large number of genes or gene panels in a large number of subjects. Using this approach, recent sequencing studies have demonstrated that germline variants, in particular loss-of-function (LOF) and pathogenic missense variants, in cancer predisposition genes occur with higher frequencies in cancer patients than healthy controls (7–10). The goal of this study was to systematically characterize rare germline variants in 42 established melanoma high-risk, intermediate-risk, and low-risk genes (Supplementary Material, Table S1) among melanoma patients in melanoma-prone families without identified mutations.
Results
The exome sequencing analysis included 144 melanoma cases from 76 families. We evaluated 42 known melanoma genes (Supplementary Material, Table S1), which included 32 established high-, intermediate-, and low-risk melanoma genes primarily based on recent reviews (3,11) and additional literature search, as well as 10 genes in cell-cycle regulation (BRCA1, BRCA2, CDK6, CDKN2B, PTEN, RB1, TP53) and telomere pathways (TERF1, TERF2, TINF2) that are strong candidates based on their functional closeness with known high-risk melanoma genes. Among these 42 genes, we identified 68 rare NS variants in 25 genes (Supplementary Material, Tables S2–S4), of which eight were LOF, two were inframe insertion/deletion, and 58 were missense variants. LOF variants, inframe changes, and 16 predicted deleterious missense variants, listed in Tables 1 and 2, were all technically validated by targeted sequencing.
Table 1.
Rare loss of function (LOF) variants in known melanoma genes identified in melanoma families
| Chr | Location | SNP ID | REF | VAR | Variant type | Protein change | Gene | Freq CMM with varianta | Family ID | #Pop case | #Pop ctlb | MAF in control datasets |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD NFE | Internal popc | Internal famc | ||||||||||||
| 7 | 124482937 | G | A | stop_gain | R232X | POT1 | 1/6 | A2 | 1 | 0 | 4.8 × 10−5 | 0 | 0 | |
| 9 | 12694335 | C | A | stop_gain | C113X | TYRP1 | 1/1 | B17 | 0 | 0 | 1.1 × 10−4 | 0 | 0 | |
| 9 | 12698595 | rs371562555 | C | T | stop_gain | Q285X | TYRP1 | 2/4 | FF2 | 0 | 0 | 2.7 × 10−5 | 0 | 0 |
| 9 | 12695535 | TAAG | frameshift | L136fs | TYRP1 | 1/1 | 110307 | 0 | 0 | 9.0 × 10−6 | 0 | 0 | ||
| 11 | 89017960 | rs62645917 | C | T | stop_gain | R402X | TYR | 5/6 | A2 | 1 | 0 | 4.8 × 10−5 | 0 | 0 |
| 13 | 48921999 | rs367654488 | C | A, T | stop_gain | S180X | RB1 | 2/6 | A2 | ND | ND | 8.0 × 10−5 | 0 | 0 |
| 14 | 24709627 | T | frameshift | K353fs | TINF2 | 2/3 | W | 0 | 0 | 0 | 0 | 0 | ||
| 22 | 38539385 | G | A | 5' UTR start_gain | PLA2G6 | 3/3 | D5 | ND | ND | 0 | 0 | 0 | ||
Chr, chromosome; REF, reference allele; VAR, variant allele; Freq, frequency; CMM, cutaneous malignant melanoma; Pop, population; ctl: control; MAF, minor allele frequency; NFE, non-Finish European; fam: family; ND, not determined.
Number of cases with the variant/number of cases sequenced in this family.
1, 559 population-based melanoma cases and 1, 633 matched controls from 3 cohort studies.
Internal population controls: 604 Caucasian healthy controls; internal family controls: ∼2, 000 exomes from ∼1, 000 cancer families (excluding melanoma or pancreatic cancer families).
Table 2.
Rare predicted deleterious inframe or missense variants in known melanoma genes identified in melanoma families
| Chr | Location | SNP ID | REF | VAR | Protein change | Gene | HGMD Class | HGMD Phen | METALR | #Pred dela | Freq CMM with variantb | Family ID | #Pop casec | #Pop ctlc |
MAF in control datasets |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD NFE | Internal popd | Internal famd | |||||||||||||||
| 5 | 1293893 | rs143148040 | G | A | P370S | TERT | D | 2 | 1/3 | W | 0 | 1 | 9.5 × 10−5 | 0 | 0 | ||
| 9 | 12704541 | rs199823942 | C | T | T366M | TYRP1 | D | 3 | 1/1 | EE106 | 0 | 0 | 1.2 × 10−4 | 0 | 0.00025 | ||
| 9 | 21971020 | GAC | CDKN2A | DM | Melanoma | 1/1 | EE107 | 0 | 0 | 2.8 × 10−5 | 0 | 0 | |||||
| 9 | 21971194 | C | T | G55D | CDKN2A | D | 5 | 1/4 | M | 0 | 0 | 0 | 0.0005 | 0 | |||
| 11 | 88911575 | rs145513733 | C | T | P152S | TYR | DM | Albinism oculocutaneous | T | 3 | 1/2 | B16 | 0 | 0 | 5.5 × 10−5 | 0 | 0 |
| 11 | 88961072 | rs61754388 | C | A | T373K | TYR | DM | Albinism oculocutaneous | D | 4 | 1/3 | B14 | 0 | 1 | 6.7 × 10−4 | 0 | 0 |
| 11 | 108199845 | rs370559102 | C | G | T2396S | ATM | DM? | Breast cancer | T | 2 | 2/3 | E2 | 2 | 0 | 2.8 × 10−4 | 0.0005 | 0.0005 |
| 11 | 108236072 | rs144636562 | A | G | N3003S | ATM | DM | Breast cancer | T | 6 | 3/3 | FE1 | 0 | 0 | 0 | 0 | 0 |
| 13 | 32907000 | rs56403624 | A | G | E462G | BRCA2 | DM? | Breast cancer | T | 2 | 3/4 | M | 3 | 0 | 4.4 × 10−4 | 0.001 | 0 |
| 13 | 32929222 | rs80358950 | A | C | K2411T | BRCA2 | D | 6 | 2/3 | X | ND | ND | 3.6 × 10−5 | 0 | 0 | ||
| 15 | 28096536 | C | T | C777Y | OCA2 | D | 5 | 1/2, 1/1 | B11, EE103 | ND | ND | 4.5 × 10−5 | 0 | 0 | |||
| 15 | 28259941 | rs142931246 | T | C | Y342C | OCA2 | DM | Albinism ocular | D | 7 | 1/3, 1/1 | E4, EE103 | ND | ND | 4.3 × 10−4 | 0.001 | 0 |
| 15 | 28326958 | C | G | Q21H | OCA2 | D | 2 | 1/1 | EE104 | ND | ND | 8.3 × 10−6 | 0 | 0 | |||
| 17 | 41197747 | C | A | C1847F | BRCA1 | D | 4 | 2/2 | EE105 | 0 | 0 | 6.7 × 10−5 | 0 | 0 | |||
| 17 | 41226369 | A | G | Y1552H | BRCA1 | D | 1 | 1/1 | Y | ND | ND | 9.0 × 10−6 | 0 | 0.00025 | |||
| 17 | 41244130 | rs80358337 | ACT | 1140_1140del | BRCA1 | 1/3 | D5 | 0 | 0 | 0 | 0 | 0 | |||||
| 17 | 41244252 | rs80357201 | G | A | P1099L | BRCA1 | DM? | Breast cancer | D | 4 | 1/3 | W | 0 | 3 | 2.4 × 10−4 | 0.0005 | 0 |
| 21 | 42749843 | A | G | D126G | MX2 | D | 7 | 2/3 | D5 | 2 | 0 | 9.5 × 10−5 | 0 | 0.00025 | |||
Chr, chromosome; REF, reference allele; VAR, variant allele; DM, disease-causing mutation; DM?, likely disease-causing mutation; Phen, phenotype; METALR, meta likelihood ratio; Pred, predicted; Del, deleterious; Freq, frequency; CMM, cutaneous malignant melanoma; Pop, population; ctl, control; MAF, minor allele frequency; NFE, non-Finish European; fam, family.
Number of algorithms predicted the variant as deleterious out of seven programs (SIFT, PolyPhen-2, Mutation Taster, Mutation Assessor, FATHMM, LRT, and Provean) evaluated.
Number of cases with the variant/number of cases sequenced in this family.
1, 559 population-based melanoma cases and 1, 633 matched controls from 3 cohort studies.
Internal population controls: 604 Caucasian healthy controls; internal family controls: ∼2, 000 exomes from ∼1, 000 cancer families (excluding melanoma or pancreatic cancer families).
Eight rare LOF variants including five stopgain, two frameshift-insertion, and one 5’ UTR premature start gain variants were observed in six families (Table 1). The stop-gain variant in TYR (NM_000372: c.C1204T, p.R402X) was present in four of five sequenced melanoma cases and one obligate gene carrier in a single family; the single non-carrier case had the latest CMM onset in this family (Fig. 1). We sequenced this variant in all cases and unaffected members in this family with available DNA; 5 of 10 unaffected members had the variant (Fig. 1). The variant was not present in our in-house databases of non-melanoma familial cancer cases and population controls; it was observed in 6 out of 125 990 alleles (0.0048%) among non-Finnish Europeans (NFEs) in Genome Aggregation Database (gnomAD). In addition, among the 1559 population-based CMM cases and 1633 matched controls we sequenced, this variant was seen in one CMM case and none of the controls. This stopgain variant was classified as pathogenic in ClinVar and Human Gene Mutation Database (HGMD) and the associated phenotype is albinism. Interestingly, one of these cases (3020) and her unaffected brother (3021) also harbored a stopgain variant in POT1. This rare variant was also observed in one CMM case but zero controls in the population-based case-control studies (Table 1). Further, two of the CMM cases in this family (1001 and 1008) also harbored a rare stopgain variant in RB1 (Table 1). The variant was also present in their unaffected mother (2002) who was not related to the affected cousin (3020).
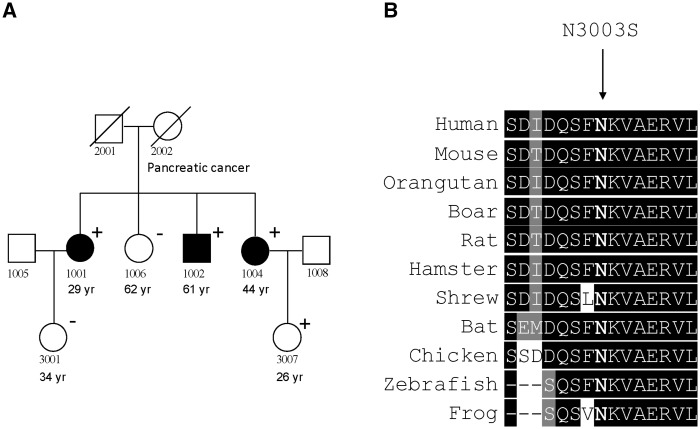
Figure 1.
The family (A2) with the TYR R402X variant. Black square or circle: melanoma cases; Square with diagonal stripes: obligate gene carriers. ‘+’: subjects with the TYR R402X variant; ‘−’: the non-carrier subject for R402X. Age under each subject denotes the age at diagnosis of first melanoma (for cases) or the age at exam/last status/death (for unaffected subjects).
Among the other five LOF variants, three are in TYRP1 (two stopgain and one frameshift), with each seen in a single separate family. Two variants were seen in families with only a single case available for sequencing. The third variant was observed in two of four sequenced cases in another family. All three variants were extremely rare (Table 1). The other two LOF variants occurred one each in PLA2G6 and TINF2. The 5′-untranslated region (5’-UTR) startgain variant in PLA2G6 occurred in a family with three affected people (mother and two children) and all three CMM cases had the variant. It was not present in any internal or external databases examined. The frameshift variant in TINF2 occurred in two of three affected people in one family. This variant was not seen in any internally sequenced case/control subjects or any external databases reviewed (Table 1).
Table 2 lists two inframe insertion/deletion variants and 16 missense variants predicted as deleterious based on an integrated Ensemble prediction score (Meta Likelihood ratio [LR]) or HGMD: four variants in BRCA1, three in OCA2, two in TYR, BRCA2, ATM, and CDKN2A, and one each in MX2, TYRP1, and TERT. Albinism and breast cancer are the most common disease traits associated with these variants. Two of these variants (one in OCA2 and one in MX2) were predicted as deleterious by all seven individual in-silico programs we evaluated (SIFT, PolyPhen-2, Mutation Taster, Mutation Assessor, FATHMM, LRT, and Provean). The OCA2 variant (rs142931246), which was seen in two independent CMM patients, was a known variant associated with Albinism. The MX2 variant was seen in two cases in a family with three affected people and in none of the three unaffected people sequenced in this family. This variant was also seen in two population-based CMM cases (1 PLCO and 1 AHS) but was very rare in internal/external control databases (Table 2).
Among the 18 variants (Table 2) that are likely to be deleterious, several variants showed disease co-segregation within families. In particular, one variant in ATM occurred in all three CMM cases and only one unaffected individual in a single family (Fig. 2A). This variant (NM_000051: c.A9008G, p.N3003S) was classified as a disease-causing mutation (DM) by HGMD and the associated phenotype was breast cancer. Although it was predicted as benign by Meta LR, it was predicted as deleterious by all individual in silico algorithms except FATHMM (Table 2). The variant is located in a highly conserved region (Fig. 2B) and is very rare in the population (absent in gnomAD-NFE). It was not seen in any sequenced population-based CMM cases or controls. Interestingly, the mother of the three CMM cases, who was deceased and did not have DNA available for sequencing, had pancreatic cancer, for which ATM is a susceptibility gene.
Figure 2.
The ATM missense variant (N3003S). (A) The family (FE1) with the ATM variant. Black square or circle: melanoma cases. ‘+’: Carriers for the ATM N3003S variant; ‘−’: non-carriers. Age under each subject denotes the age at diagnosis of first melanoma (for cases) or the age at exam/last status/death (for unaffected subjects). (B) Amino acid conservation among ATM homologs.
Since many of the 76 families had not previously been tested for mutations in CDKN2A, the best established high-risk CMM susceptibility gene, we examined this gene in all subjects and identified two rare variants in CDKN2A. The inframe insertion variant, identified as a Swedish founder mutation (12), was seen in a single family with only a single case available for sequencing and not seen in anyone else. The missense variant was seen in two independent families. In a large family with 13 affected people, the CDKN2A variant was present in only one sibship (three affected siblings and one unaffected daughter of one of the carrier cases) and absent in 22 other tested family members (4 CMM and 18 unaffected). The same variant was also observed in only one of three sequenced CMM cases in another family.
To assess the overall genetic burden due to rare nonsynonymous (NS) variants in these melanoma genes in the families, we conducted a rare variant burden test by comparing familial cases to 604 population controls sequenced and analysed at CGR, NCI, using similar approaches (see Methods). Familial CMM cases showed higher frequencies of rare variants for OCA2 (P = 0.032) and TYR (P = 0.033). Burden test results are shown in Table 3 for genes with P < 0.2. We also conducted burden tests for the six genes (ATM, CDKN2A, CDKN2B, TINF2, TYRP1, and TYR) sequenced in 795 cases and 807 controls from the PLCO and AHS studies. CMM cases had an increased burden of carrying rare NS variants in TYRP1 (P = 0.024), TYR (P = 0.032), and TINF2 (P = 0.037), based on SKAT-O test. In general, results were similar when using the CAST test (Supplementary Material, Table S5) or when we restricted the analysis to variants called by both the Torrent Variant Caller and GATK (data not shown).
Table 3.
Burden test comparing familial cases to population controls
| Gene | #Cases | #Families | #Controls | P |
|---|---|---|---|---|
| OCA2 | 8 | 5 | 11 | 0.032 |
| TYR | 6 | 3 | 6 | 0.033 |
| RB1 | 3 | 2 | 3 | 0.084 |
| CDKAL1 | 4 | 2 | 5 | 0.091 |
| HERC2 | 14 | 11 | 39 | 0.093 |
| BRCA1 | 6 | 5 | 13 | 0.124 |
| MITF | 3 | 3 | 5 | 0.125 |
| TYRP1 | 5 | 4 | 10 | 0.143 |
| CDKN2A | 2 | 2 | 3 | 0.177 |
| TINF2 | 2 | 1 | 3 | 0.184 |
| BRCA2 | 6 | 5 | 17 | 0.194 |
| BAP1 | 2 | 2 | 3 | 0.196 |
Discussion
In this study, we investigated the role of rare germline variants in known CMM genes in melanoma-prone families without known mutations. We identified a number of rare LOF and predicted deleterious missense variants that were enriched in CMM cases, some demonstrating co-segregation with disease within families, such as variants in TYR, PLA2G6, and ATM. Several variants also showed evidence for enrichment in population-based sporadic CMM cases. Further, compared with their corresponding controls, familial CMM cases had an increased burden of rare germline variants in TYR and OCA2 and sporadic cases had an increased burden for TYR, TYRP1, and TINF2. In particular, CMM cases seemed to have a higher frequency of rare variants in albinism associated genes. Given the rarity (all LOF and predicted pathogenic variants have minor allele frequency < 0.07% in the general population), co-segregation for some variants, predicted damaging effect, functional relevance (all these are known CMM genes), and increased genetic burden for several genes in familial and/or sporadic CMM cases, our findings suggest that rare germline variants in several of these genes are likely to contribute to CMM susceptibility in high-risk families.
TYR encodes tyrosinase, the key enzyme catalyzes the conversion of tyrosine to melanin. Mutations in TYR account for 46% of cases of albinism in European populations (13), and non-pathologic polymorphisms have been associated with skin pigmentation variation. The stopgain variant (R402X) found in one of the most informative families examined was classified as pathogenic by ClinVar and HGMD for albinism, an autosomal recessive disorder. It is possible that TYR may function similarly to some other cancer susceptibility genes for which disease manifestation differs depending on bi-allelic (e.g. albinism) or mono-allelic (e.g. melanoma) inheritance. The pathogenic R402X variant has been reported in albino patients either in a homozygous or compound heterozygous pattern (14,15). This variant is located next to a common polymorphism (R402Q) that has been associated with hypopigmentation, increased sun sensitivity, and the risk for developing basal cell carcinoma and CMM (16–19). R402Q has not been associated with albinism (20), although it did have deficient enzyme activity (21). Interestingly, four of the five cases in the family with the TYR R402X variant had R402Q, including the case who did not carry R402X. Unlike R402Q, which is common among Caucasians (27% in gnomAD-NFEs), R402X is very rare (0.0048% in gnomAD-NFEs). Further, the R402X variant was also seen in one population-based sporadic CMM case we evaluated. In the family that harbored the variant, all but one CMM case had the variant and the single non-carrier case had the latest age onset (52 years). However, 5 out of 10 unaffected family members sequenced also carried the R402X variant suggesting incomplete penetrance. Nevertheless, the analysis of whole exome sequencing data in this family found only one rare nonsynonymous variant that was shared by all CMM cases but the variant allele is five times more common (0.02% in gnomAD-NFEs) compared with R402X and the gene function has not been shown to be relevant to melanoma. Although we cannot rule out the possibility of non-coding regulatory variants as the major susceptibility mechanism, our exome findings suggest that the TYR R402X variant is the most likely candidate in this family. It is also possible that the effect is further modified by R402Q since the two cases with only one of the TYR variants had the oldest ages at onset. In addition, a subset of cases in this family also harbored rare LOF variants in RB1 and POT1, raising the possibility of a potentially more complicated underlying susceptibility mechanism in this family, although the role of LOF variants in RB1 and POT1 is less certain given the lack of co-segregation.
Interestingly, both LOF and predicted deleterious missense variants seemed to be enriched for albinism genes such as TYR, TYRP1, and OCA2, which encode proteins that are essential to normal pigmentation and production of melanin (22). All these variants were very rare in the general population. Some of these variants have been shown to cause protein instability and reduced tyrosinase activity (23). In particular, burden tests for both familial and sporadic case-control comparisons were significant for TYR. Results from a similar burden test for rare variants in all five albinism genes examined (SLC45A2, TYRP1, TYR, OCA2, SLC45A5) indicate that these variants were enriched in familial CMM cases (P = 0.01). These results suggest that rare heterozygous variants in melanogenesis genes may play an important role in CMM susceptibility through partially impairing pigmentation.
Among the 26 LOF or predicted deleterious variants identified in our families, two occurred in all affected people within their respective families with three or more cases sequenced. The startgain variant in the 5’-UTR region of PLA2G6 was carried by all three cases in the family, although the cases were all siblings and therefore the expected sharing of genetic information among them is high. Common variants in PLA2G6 have been associated with nevi count and melanoma risk in previous association studies (24). The impact of this rare variant is unclear, although sequence changes in the 5’-UTR region are known to have great impact on mRNA stability and protein expression levels (25). Another variant that was seen in all three sequenced CMM cases was a missense variant in ATM, a gene that plays central roles in DNA damage repair and cell cycle control. Mutations in this gene are associated with ataxia telangiectasia (A-T), breast cancer, and pancreatic cancer (26–28). Interestingly, the mother of the three CMM cases who carried the ATM variant had pancreatic cancer. The variant was only seen in one of the three unaffected people we sequenced, and this carrier was young (26 years) when examined. The missense variant (N3003S) is located in the kinase domain of ATM, where sequence residues are well conserved among different species. In an evaluation of the functional impact of known ATM missense mutations identified in A-T or breast cancer patients, Scott et al. found that three mutants (V2716A, R2849P and G2867R) altered normal ATM function by a dominant interfering mechanism (29) and these mutations are also located in the kinase domain. In contrast, four other ATM missense variants, which are located upstream of the kinase domain, did not have an adverse effect on kinase activity or impact cell survival, highlighting the importance of the kinase domain in maintaining normal ATM function.
In our analysis, several patients harbored multiple deleterious/likely deleterious rare variants of known CMM genes. Among the cases who harbored the TYR R402X variant, one case (the youngest one) had a stopgain variant in POT1 and another two cases had a stopgain variant in RB1. The patient with the POT1 stopgain variant had the lowest POT1 RNA expression level in the Epstein-Barr-virus-transformed lymphoblastoid cell line among 50 CMM patients in our families with RNASeq data [Reads Per Kilobase of transcript per Million mapped reads (RPKM) = 785 for the carrier versus the average value of 1138 among all examined 50 cases], suggesting that this variant may affect POT1 expression. In the family with the PLA2G6 5’-UTR variant, two of the three cases also carried a missense variant in MX2 that was predicted as deleterious by all in silico programs evaluated. Finally, a patient from a family with only one case sequenced had two missense variants of OCA2, both predicted as deleterious by most prediction algorithms. Although the exact mechanisms remain unclear, it is possible that multiple common/rare variants in known and unknown genes are responsible for CMM susceptibility in some high-risk families, which is consistent with a more complicated genetic predisposition mechanism that has been increasingly recognized in the field.
Our study has several limitations. First, our analyses did not have sufficient statistical power to conduct individual rare variant association tests. Second, the unequivocal proof of the functionality of these rare variants in CMM development has not been obtained. In addition, most variants did not show full co-segregation with disease. Among the few variants showing evidence for disease co-segregation, each was only observed in a single family, making it challenging to determine causality. Nevertheless, our study is the first systematic investigation of rare germline variants in multiple melanoma genes in high-risk CMM families. The strengths of the study include the evaluation of co-segregation with disease for many of these variants in the larger families with unaffected people available for sequencing, the inclusion of two internal control datasets consisting of a large number of subjects from cancer families and population controls from PLCO and ACS to control for platform-specific artifacts in addition to investigating publicly available databases, and the assembly of a substantial number of population-based cases and their matched controls from multiple cohort studies to follow up on the top variants/genes identified in our families. We technically validated all LOF and predicted deleterious missense variants using targeted sequencing. In addition to the evaluation of individual rare variants, we also conducted a gene-level burden test to compare the cumulative frequency of rare variants in each of these genes in CMM cases and controls. For six of these genes, we also conducted the burden test in population-based cases and controls. Our study identified a number of potentially disease-related variants in known melanoma genes, some of which may have implications for risk assessment that require further replication and functional characterization.
Materials and Methods
Study population
The details of this family study have been previously described (6,30). All family members who were willing to participate in the study provided written informed consent under an NCI IRB approved protocol. All diagnoses of melanoma were confirmed by histologic review of pathologic materials/reports or medical records. All study participants were of European ancestry.
The present exome sequencing analysis included 144 melanoma cases from 76 families (40 families with 1 case, 11 families with 2 cases, 20 families with 3 cases, 3 families with 4 cases, and 2 families with 5 cases sequenced). Sixty-five families (29 families with 1 case and all families with ≥2 cases sequenced) included at least two first-degree relatives with a history of melanoma. The remaining 11 families included single high-risk melanoma patients with early age at diagnosis before 30 years (n = 9) or multiple primary melanomas (n = 2).
We conducted targeted sequencing of top genes/variants in 1559 population-based melanoma cases and 1633 controls including 1278 Prostate, Lung, Colorectal and Ovarian Screening Trial (PLCO), 324 Agriculture Health Study (AHS), and 1590 Harvard cohort studies [Nurses’ Health Study (NHS) and Health Professional’s Follow-up Study (HPFS)] (see the ‘Targeted sequencing of top genes/variants in population-based melanoma cases and controls’ section below).
The dbGAP accession number for sequencing data included in this study is phs001177.v2.
Whole exome sequencing and bioinformatics analysis
Whole exome sequencing (WES) was performed at the Cancer Genomics Research Laboratory, National Cancer Institute (CGR, NCI), as previously described (31,32). Briefly, 1.1 µg of genomic DNA was extracted by standard methods from whole blood. SeqCAP EZ Human Exome Library v3.0 (Roche NimbleGen, Madison, WI) was utilized for exome sequence capture. Supplementary Material, Table S6 shows the start and end positions of genomic regions for each gene covered by the capture probes. The captured DNA was then subject to paired-end sequencing utilizing the Illumina HiSeq2000 sequencer for 2 X 100-bp sequencing of paired-ends (Illumina, San Diego, CA). Exome sequencing was performed to a sufficient depth to achieve a minimum coverage of 15 reads in at least 80% of the coding sequence from the UCSC hg19 transcripts database.
Details of the bioinformatics analysis pipeline used in this study have been previously described (31–33). Variant discovery and genotype calling were performed globally using three variant callers (UnifiedGenotyper and HaplotypeCaller modules from GATK and FreeBayes [v9.9.2]). We included all target regions as well as 250 bp flanking region on each side. An Ensemble variant calling pipeline (v0.2.2) was then implemented to integrate analysis results from the above three callers. Subsequently, the Ensemble variant calling pipeline applies a Support Vector Machine (SVM) learning algorithm to identify an optimal decision boundary based on the variant calling results out of the multiple variant callers, to produce a more balanced decision between false positives and true positives. In addition, insertions and deletions were left-aligned at both post-alignment (BAM) and post-variant-calling (VCF) levels using GATK’s LeftAlignIndels and LeftAlignVariants modules, respectively.
Annotation of each variant locus was made via a custom software pipeline based on public data from ANNOVAR, dbNSFP, SnpEff, and SnpSift integrated using a CGR in-house script, including Ensembl, refGene, and UCSC KnownGene databases, the ESP6500 dataset from University of Washington’s Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), dbNSFP—database of human nonsynonymous SNPs and function predictions (https://sites.google.com/site/jpopgen/dbNSFP), the Molecular Signatures Database—MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp), the National Center for Biotechnology Information ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and dbSNP databases build 137 (34), the 1000 Genomes Project (35), the Exome Aggregation Consortium (ExAC) database (36) (http://exac.broadinstitute.org), the Human Gene Mutation Database (HGMD) (37), and others. We also used the recently released Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/), which provides aggregate data on 123, 136 exome and 15, 496 whole-genome sequences from unrelated individuals sequenced as part of various disease-specific and population genetic studies, for allele frequency estimation in the general population (36).
Exome sequencing of population controls
Data from 604 population controls from two cohort studies (Cancer Prevention Study [CPS]-II, n = 224; PLCO, n = 378) (32) were available for inclusion in the current study to evaluate genetic burden for known melanoma genes. The sequencing/analysis methods for the population control samples followed the same ensemble calling process as was used for the familial CMM patients. However, the SeqCAP EZ Human Exome Library v3.0 + UTR (Roche NimbleGen, Madison, WI) was utilized for exome sequence capture. Variant calling for the population controls was done together with that for the in-house database (CGR, NCI) of cancer-prone families that included melanoma families.
Variant characterization
Variants in the 42 examined genes were included for further evaluation if they 1) passed the quality control (QC) filter in the in-house bioinformatics pipeline; 2) were called by at least two of the three variant callers; 3) had an allele frequency <0.1% in the 1000 Genomes Project (overall and European sample) and ESP6500 (overall and European sample); 4) were present in ≤2 families from an in-house database (CGR, NCI) of ∼2000 exomes in ∼1000 cancer-prone families (excluding melanoma-prone or pancreatic cancer families); and 5) were classified as nonsynonymous (NS) including frameshift, stopgain, inframe deletion or insertion, or NS substitutions (missense). Although we primarily focused on exonic variants, we included intronic variants if they were in splicing regions or impacted ‘start’ or ‘stop’ codons or transcription factor binding sites as potential NS variants. Frameshift and stopgain variants were defined as loss-of-function (LOF) variants and coded as deleterious. To classify missense variants as predicted deleterious, we used an ensemble prediction score [Meta Likelihood ratio (LR)] that incorporates results from nine in silico algorithms (SIFT, PolyPhen-2, GERP ++, Mutation Taster, Mutation Assessor, FATHMM, LRT, SiPhy, and PhyloP) and allele frequency. This ensemble score achieved the highest discriminative power compared with 18 deleterious scoring methods and also showed low false positive prediction rate for benign yet rare NS variants (38). Missense variants classified as disease-causing mutation (DM) or likely disease-causing (DM?) in HGMD were also considered as predicted deleterious variants.
Variant validation by targeted sequencing
LOF variants, inframe deletions/insertions, and all predicted deleterious missense variants were technically validated using Sanger sequencing or Ampliseq at CGR. For technical validation using Ampliseq, a targeted, multiplexed PCR primer panel was designed using the Ion AmpliSeq Designer v4.4.4 (Life Technologies, Carlsbad, CA, USA). Average amplicon size of the panel was 244 bp. Sample DNA (30 ng) was amplified using this custom AmpliSeq primer panel, and sequencing libraries were prepared following the manufacturer’s Ion AmpliSeq Library Preparation protocol (Life Technologies), using Ion Xpress Barcode Adapters. Individual sample libraries were pooled, then templated and sequenced on the Ion Torrent PGM Sequencer using Ion PGM Hi-Q Chef chemistry. Base calling and alignment were performed using Torrent Suite 4.4. Variant calling was done separately with GATK and Torrent Variant Caller.
Targeted sequencing of top genes/variants in population-based melanoma cases and controls
We sequenced the majority of LOF variants and a subset of predicted deleterious NS variants in 1559 population-based melanoma cases and 1633 controls including 1, 278 PLCO, 324 AHS, and 1590 Harvard cohort studies (NHS and HPFS) using a custom-designed AmpliSeq panel. We also sequenced the entire exonic regions for six of these genes: ATM, CDKN2A, CDKN2B, TINF2, TYRP1, and TYR in 795 cases and 807 controls from the PLCO and AHS studies. Deep sequence coverage was generated at each locus of interest for each sample. Genotypes were determined independently for each sample based on the ratio of base calls in those sequence reads at a given locus.
Statistical analyses
To test whether the cumulative frequency of rare NS variants was increased in familial CMM cases compared with 604 population controls that were sequenced using the same platform and were analysed together with cancer cases, we performed a gene-level test of association for each of the 42 melanoma genes. For each gene, we counted the number, N, of cases carrying a rare NS variant that was included in both SeqCAP EZ Human Exome Library v3.0 (used for familial CMM patients) and SeqCAP EZ Human Exome Library v3.0 + UTR (used for population controls). We then calculated the probability of observing N variants under the null hypothesis using a randomization test. We started by creating a list of the 1208 haplotypes from the 604 controls sequenced with our cases. In the rare scenario when a control had two or more rare variants, each haplotype was assumed to carry at least one rare variant. We then used these haplotypes to perform 1000 iterations of a two-step randomization procedure. For each iteration, we first randomly generated Identical-By-Descent (IBD) patterns for the family cases using the laws of Mendelian Inheritance. We then assigned each of the founder chromosomes in the families to carry a haplotype randomly selected from the list of control haplotypes. After 1000 iterations, our p-value was the proportion of iterations where the number of family members carrying a rare variant was at least N.
We also conducted a rare variant burden test for the six genes (ATM, CDKN2A, CDKN2B, TINF2, TYRP1, and TYR), for which we sequenced the entire coding region, in 795 cases and 807 controls from the PLCO and AHS studies using the SKAT-O statistic (39), which is a linear combination of the burden test (aimed to test effect size of variants with the same direction in cases and in controls) and variance component test (aimed to test effect size of variants with different directions in cases and controls). We also used another burden test statistics, CAST, which makes a strong assumption that all rare variants in a set are causal and associated with a trait with the same direction and magnitude of effect, to compare results. We included all NS variants with minor allele frequency less than 0.1% in ESP and 1000 Genomes, regardless of their predicted functions. We also conducted a sensitivity analysis by only including variants that were called by both the Torrent Variant Caller and GATK to ensure the accuracy of the burden test. All statistical analyses were conducted using R (version 3.3.3) and all P values were two-sided.
Web Resources
1000 Genomes Project, http://www.internationalgenomes.org ANNOVAR, http://annovar.openbioinformatics.org/ ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/ dbNSFP, http://varianttools.sourceforge.net/Annotation/DbNSFP dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/ Ensemble, http://www.ensemble.org/ ExAC, http://exac.broadinstitute.org Exome Sequencing Project, http://evs.gs.washington.edu/EVS/ FreeBayes, https://github.com/ekg/freebayes GATK, https://software.broadinstitute.org/gatk/ Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/ HGMD, http://www.hgmd.cf.ac.uk/ac/index.php MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp RefGene, http://refgene.com/ SnpEff, http://snpeff.sourceforge.net/ SnpSift, http://snpeff.sourceforge.net/SnpSift.html UCSC Genome Browser, http://genome.ucsc.edu
All websites were last accessed on September 29, 2017.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are indebted to the participating families, whose generosity and cooperation have made this study possible. We acknowledge the contributions to this work that were made by Virginia Pichler, Deborah Zametkin, Mary Fraser, and Barbara Rogers. We thank the NCI DCEG Cancer Sequencing Working Group: Lynn R. Goldin, Mary L. McMaster, Neil E. Caporaso, Bari Ballew, Sharon Savage, Mark H. Greene, Allan Hildesheim, Nan Hu, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, and Guoqin Yu and members of the NCI DCEG Cancer Genomics Research Laboratory: Sarah Bass, Joseph Boland, Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Sally Larson, Kerry Lashley, Hyo Jung Lee, Michelle Manning, Jason Mitchell, David Roberson, Mingyi Wang. We are indebted to the participants in the NHS and HPFS for their dedication to this research. We thank the following state cancer registries for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington, and Wyoming.
Conflict of Interest statement. None declared.
Funding
Intramural Research Program of the NIH, NCI, DCEG. This work was also supported in part by NIH R01 CA49449, P01 CA87969, UM1 CA186107, and UM1 CA167552.
References
- 1. Tucker M.A., Goldstein A.M. (2003) Melanoma etiology: where are we? Oncogene, 22, 3042–3052. [DOI] [PubMed] [Google Scholar]
- 2. Goldstein A.M., Tucker M.A. (2001) Genetic epidemiology of cutaneous melanoma: a global perspective. Arch. Dermatol., 137, 1493–1496. [DOI] [PubMed] [Google Scholar]
- 3. Read J., Wadt K.A., Hayward N.K. (2016) Melanoma genetics. J. Med. Genet., 53, 1–14. [DOI] [PubMed] [Google Scholar]
- 4. Yokoyama S., Woods S.L., Boyle G.M., Aoude L.G., MacGregor S., Zismann V., Gartside M., Cust A.E., Haq R., Harland M.. et al. (2011) A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature, 480, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Demenais F., Mohamdi H., Chaudru V., Goldstein A.M., Newton Bishop J.A., Bishop D.T., Kanetsky P.A., Hayward N.K., Gillanders E., Elder D.E.. et al. (2010) Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J. Natl. Cancer Inst., 102, 1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldstein A.M., Landi M.T., Tsang S., Fraser M.C., Munroe D.J., Tucker M.A. (2005) Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol. Biomarkers Prev., 14, 2208–2212. [DOI] [PubMed] [Google Scholar]
- 7. Ramus S.J., Song H., Dicks E., Tyrer J.P., Rosenthal A.N., Intermaggio M.P., Fraser L., Gentry-Maharaj A., Hayward J., Philpott S.. et al. (2015) Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J. Natl. Cancer Inst., 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grant R.C., Selander I., Connor A.A., Selvarajah S., Borgida A., Briollais L., Petersen G.M., Lerner-Ellis J., Holter S., Gallinger S. (2015) Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology, 148, 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J., Walsh M.F., Wu G., Edmonson M.N., Gruber T.A., Easton J., Hedges D., Ma X., Zhou X., Yergeau D.A.. et al. (2015) Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med., 373, 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu C., Xie M., Wendl M.C., Wang J., McLellan M.D., Leiserson M.D., Huang K.L., Wyczalkowski M.A., Jayasinghe R., Banerjee T.. et al. (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun., 6, 10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Law M.H., Macgregor S., Hayward N.K. (2012) Melanoma genetics: recent findings take us beyond well-traveled pathways. J. Invest. Dermatol., 132, 1763–1774. [DOI] [PubMed] [Google Scholar]
- 12. Borg A., Johannsson U., Johannsson O., Hakansson S., Westerdahl J., Masback A., Olsson H., Ingvar C. (1996) Novel germline p16 mutation in familial malignant melanoma in southern Sweden. Cancer Res., 56, 2497–2500. [PubMed] [Google Scholar]
- 13. Sturm R.A., Duffy D.L. (2012) Human pigmentation genes under environmental selection. Genome Biol., 13, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gershoni-Baruch R., Rosenmann A., Droetto S., Holmes S., Tripathi R.K., Spritz R.A. (1994) Mutations of the tyrosinase gene in patients with oculocutaneous albinism from various ethnic groups in Israel. Am. J. Hum. Genet., 54, 586–594. [PMC free article] [PubMed] [Google Scholar]
- 15. Hutton S.M., Spritz R.A. (2008) Comprehensive analysis of oculocutaneous albinism among non-Hispanic caucasians shows that OCA1 is the most prevalent OCA type. J. Invest. Dermatol., 128, 2442–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Council M.L., Gardner J.M., Helms C., Liu Y., Cornelius L.A., Bowcock A.M. (2009) Contribution of genetic factors for melanoma susceptibility in sporadic US melanoma patients. Exp. Dermatol., 18, 485–487. [DOI] [PubMed] [Google Scholar]
- 17. Nan H., Kraft P., Qureshi A.A., Guo Q., Chen C., Hankinson S.E., Hu F.B., Thomas G., Hoover R.N., Chanock S.. et al. (2009) Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol., 129, 2250–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nan H., Kraft P., Hunter D.J., Han J. (2009) Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int. J. Cancer, 125, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bishop D.T., Demenais F., Iles M.M., Harland M., Taylor J.C., Corda E., Randerson-Moor J., Aitken J.F., Avril M.F., Azizi E.. et al. (2009) Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet., 41, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oetting W.S., Pietsch J., Brott M.J., Savage S., Fryer J.P., Summers C.G., King R.A. (2009) The R402Q tyrosinase variant does not cause autosomal recessive ocular albinism. Am. J. Med. Genet. A, 149A, 466–469. [DOI] [PubMed] [Google Scholar]
- 21. Jagirdar K., Smit D.J., Ainger S.A., Lee K.J., Brown D.L., Chapman B., Zhen Zhao Z., Montgomery G.W., Martin N.G., Stow J.L.. et al. (2014) Molecular analysis of common polymorphisms within the human Tyrosinase locus and genetic association with pigmentation traits. Pigment Cell Melanoma Res., 27, 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montoliu L., Gronskov K., Wei A.H., Martinez-Garcia M., Fernandez A., Arveiler B., Morice-Picard F., Riazuddin S., Suzuki T., Ahmed Z.M.. et al. (2014) Increasing the complexity: new genes and new types of albinism. Pigment Cell Melanoma Res., 27, 11–18. [DOI] [PubMed] [Google Scholar]
- 23. Dolinska M.B., Kus N.J., Farney S.K., Wingfield P.T., Brooks B.P., Sergeev Y.V. (2017) Oculocutaneous albinism type 1: link between mutations, tyrosinase conformational stability, and enzymatic activity. Pigment Cell Melanoma Res., 30, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falchi M., Bataille V., Hayward N.K., Duffy D.L., Bishop J.A., Pastinen T., Cervino A., Zhao Z.Z., Deloukas P., Soranzo N.. et al. (2009) Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet., 41, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dvir S., Velten L., Sharon E., Zeevi D., Carey L.B., Weinberger A., Segal E. (2013) Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. U. S. A, 110, E2792–E2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S.. et al. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science, 268, 1749–1753. [DOI] [PubMed] [Google Scholar]
- 27. Roberts N.J., Jiao Y., Yu J., Kopelovich L., Petersen G.M., Bondy M.L., Gallinger S., Schwartz A.G., Syngal S., Cote M.L.. et al. (2012) ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov., 2, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Renwick A., Thompson D., Seal S., Kelly P., Chagtai T., Ahmed M., North B., Jayatilake H., Barfoot R., Spanova K.. et al. (2006) ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet., 38, 873–875. [DOI] [PubMed] [Google Scholar]
- 29. Scott S.P., Bendix R., Chen P., Clark R., Dork T., Lavin M.F. (2002) Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc. Natl. Acad. Sci. U. S. A, 99, 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goldstein A.M., Struewing J.P., Chidambaram A., Fraser M.C., Tucker M.A. (2000) Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J. Natl. Cancer Inst., 92, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 31. Shi J., Yang X.R., Ballew B., Rotunno M., Calista D., Fargnoli M.C., Ghiorzo P., Bressac-de Paillerets B., Nagore E., Avril M.F.. et al. (2014) Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet., 46, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X.R., Rotunno M., Xiao Y., Ingvar C., Helgadottir H., Pastorino L., van Doorn R., Bennett H., Graham C., Sampson J.N.. et al. (2016) Multiple rare variants in high-risk pancreatic cancer-related genes may increase risk for pancreatic cancer in a subset of patients with and without germline CDKN2A mutations. Hum. Genet., 135, 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pathak A., Pemov A., McMaster M.L., Dewan R., Ravichandran S., Pak E., Dutra A., Lee H.J., Vogt A., Zhang X.. et al. (2015) Juvenile myelomonocytic leukemia due to a germline CBL Y371C mutation: 35-year follow-up of a large family. Hum. Genet., 134, 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Durbin R.M., Altshuler D.L., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Collins F.S., De La Vega F.M., Donnelly P.. et al. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60, 706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet., 133, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. (2015) Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet., 24, 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee S., Wu M.C., Lin X. (2012) Optimal tests for rare variant effects in sequencing association studies. Biostatistics, 13, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.