Synopsis
The mammalian gut microbiome plays a profound role in the physiology, metabolism, and overall health of its host. However, biologists have only a nascent understanding of the forces that drive inter-individual heterogeneity in gut microbial composition, especially the role of host social environment. Here we used 178 samples from 78 wild yellow baboons (Papio cynocephalus) living in two social groups to test how host social context, including group living, social interactions within groups, and transfer between social groups (e.g., dispersal) predict inter-individual variation in gut microbial alpha and beta diversity. We also tested whether social effects differed for prevalent “core” gut microbial taxa, which are thought to provide primary functions to hosts, versus rare “non-core” microbes, which may represent relatively transient environmental acquisitions. Confirming prior studies, we found that each social group harbored a distinct gut microbial community. These differences included both non-core and core gut microbial taxa, suggesting that these effects are not solely driven by recent gut microbial exposures. Within social groups, close grooming partners had more similar core microbiomes, but not non-core microbiomes, than individuals who rarely groomed each other, even controlling for kinship and diet similarity between grooming partners. Finally, in support of the idea that the gut microbiome can be altered by current social context, we found that the longer an immigrant male had lived in a given social group, the more closely his gut microbiome resembled the gut microbiomes of the group’s long-term residents. Together, these results reveal the importance of a host’s social context in shaping the gut microbiome and shed new light onto the microbiome-related consequences of male dispersal.
Introduction
Social animals are thought to acquire many of their resident bacteria from conspecifics, both through direct transmission from social partners and indirect transmission from shared environments (Lax etal. 2014; Powell etal. 2014; Tung etal. 2015). In support, several studies have shown that social organization and behavior shape an individual’s microbiome composition (e.g., White etal. 2010; Koch and Schmid-Hempel 2011; Meadow etal. 2013). These effects may be important to the evolution of animal social behavior because inter-individual variation in gut microbial composition is increasingly linked to variation in host health and fitness (Turnbaugh etal. 2009a; Huffnagle 2010; Heijtz etal. 2011; Koch and Schmid-Hempel 2011; Ezenwa etal. 2012; Forsythe and Kunze 2013; Bordenstein and Theis 2015). However, we still have only a limited understanding of how social organization and behavior affect patterns of microbial transmission between individuals in wild systems, and ultimately the composition and function of animal microbiomes.
To date, social organization and behavior are thought to influence two primary dimensions of microbiome composition: microbial alpha diversity, i.e., the number and distribution of bacterial taxa in an individual host, and beta diversity, i.e., differences in microbial community composition between hosts. In terms of alpha diversity, social partners have been proposed to serve as bacterial reservoirs, promoting microbial diversity within hosts and maintaining microbiome stability in the face of gains and losses of individual taxa (Lombardo 2008; Moeller etal. 2016a). In support, some studies have found that animals with high levels of social contact harbor more diverse gut microbiomes than animals who are less socially connected (Levin etal. 2016; Li etal. 2016b; Moeller etal. 2016b, although Levin etal. 2016 also found evidence of the opposite effect). Further, in bees, experimentally reducing an individual’s social contacts decreases their gut microbial diversity (Billiet etal. 2016). These effects may have important consequences for hosts: in free-living non-microbiome communities, high biodiversity is associated with greater community stability and productivity (e.g., Lehman etal. 2000; Tilman etal. 2006; Hooper etal. 2012a). In the microbiome, high alpha diversity is likewise proposed to promote long-term compositional and functional stability and resistance to invading pathogens (Dillon etal. 2005; Lozupone etal. 2012b). However, additional gut microbial taxa may also be largely functionally redundant, and the functional consequences of alpha diversity in animal microbiomes are the topic of considerable debate (Shade and Handelsman 2012; Moeller etal. 2016b).
In terms of beta diversity, socially mediated patterns of transmission are thought to promote microbiome community similarity among group members and social partners. Social group-specific microbiomes have been reported for several body sites and in a wide variety of taxonomic groups, including humans, non-human primates, carnivores, frogs, birds, and insects (Koch and Schmid-Hempel 2011; Degnan etal. 2012; McKenzie etal. 2012; Theis etal. 2012; Dunn etal. 2013; McCord etal. 2013; Song etal. 2013; Leclaire etal. 2014; Schloss etal. 2014; Gomez etal. 2015; Tung etal. 2015; Aivelo etal. 2016; Bennett etal. 2016; Levin etal. 2016; Whittaker etal. 2016). Such effects could be important because more similar microbial communities are presumed to have similar functional capacities and may provide similar “ecosystem services” to their hosts, including effects on digestion, immune responses, vitamin synthesis, or handling of plant secondary compounds (Costello etal. 2012; Delsuc etal. 2013; Ainsworth etal. 2015).
We tested the relationships between social behavior and gut microbial alpha and beta diversity in both “core” and “non-core” members of the gut microbiome. The presence and abundance of core and non-core gut microbial taxa are thought be shaped by different host and environmental factors. Core taxa are, by definition, found in the majority of hosts of a given species (Hamady and Knight 2009) and are thought to make major contributions to the gut microbiome’s normal functions (e.g., digestion and vitamin synthesis; Savage 1977; Walter and Ley 2011; Shade and Handelsman 2012; Zhang etal. 2016). The high prevalence of core taxa suggests that these microbes may be actively curated and retained by the host’s immune system (Hansen etal. 2010; Hooper etal. 2012b). Further, their abundance may be driven by interactions with other common microbial taxa (Stecher etal. 2010). In contrast, less prevalent, non-core taxa are proposed to often be transient, as they typically occur in a minority of hosts and are not consistently present in the same host over time (Martínez etal. 2013; Tinker and Ottesen 2016). Their dynamics may be shaped by patterns of microbial colonization from the environment, including conspecific hosts (Hanson etal. 2012). Hence, non-core microbes might be more likely to reflect recent social or external exposures.
To investigate this possibility, and to clarify the role of different social factors in gut microbiome composition, we performed 16S rRNA gene sequencing on 178 fecal samples (78 individuals) collected from baboons living in two social groups in a well-studied wild baboon (Papio cynocephalus) population living in the Amboseli ecosystem in Kenya. To do so, we took advantage of detailed data on the baboons’ demography, social relationships, and habitats collected by the Amboseli Baboon Research Project since 1971 (Alberts and Altmann 2012). Prior research on this population indicated that each social group harbored distinct gut microbiomes and that close grooming partners have more similar gut microbiomes than those who rarely groom each other (Tung etal. 2015).
Here, we expanded both the sample size and scope of our analyses to test three main hypotheses for both the core and non-core microbiome: (1) that sociality is linked to elevated gut microbial alpha diversity; (2) that increased social interaction promotes increased gut microbial similarity (beta diversity) between individuals; and (3) that the length of an immigrant male’s membership in his current social group predicts his microbiome similarity to long-term group residents. In all cases, we expected social effects on microbiome composition to be stronger in non-core than core gut microbial taxa. We predicted that baboons living in the larger social group and/or those who engaged in more grooming would have higher gut microbial diversity than individuals living in the smaller group or who were socially isolated. We also predicted that adult males, who disperse between social groups and encounter more diverse environments and social partners in the process, would exhibit higher gut microbial alpha diversity than adult females, who do not disperse. With respect to beta diversity, we expected that gut microbial similarity between individuals would be highest for members of the same social group and close grooming partners. Finally, we predicted that immigrant males that were members of their social group for a longer period of time would be more similar to other group residents than recent immigrants. Taken as a whole, our study improves our understanding of which aspects of microbiome community composition are most sensitive to a host’s social environment.
Methods
Study subjects and sample collection
Since 1971, the Amboseli Baboon Research Project (ABRP) has collected continuous data on the demography, social interactions, and ranging patterns of hundreds of individual baboons in the Amboseli ecosystem in Kenya (Alberts and Altmann 2012). These data are collected by experienced field observers who visit each baboon social group 3–4 times per week, alternating between morning and afternoon sessions, year-round. All individuals are known and recognized by morphological characteristics.
Study subjects and fecal sampling
From 7 July to 8 August 2012, we collected fecal samples from the members of two baboon social groups, called “Mica’s” (n = 67 samples from 27 individuals) and “Viola’s” (n = 111 samples from 51 individuals) groups. These two groups occupied adjacent home ranges, with no home range overlap during the period of sample collection (Supplementary Fig. S1; Tung etal. 2015). Fecal samples from all group members were collected opportunistically within a few minutes of defecation. Samples were preserved in 95% ethanol and stored in the field in an evaporative cooling structure (approximate daily maximum temperature of 25 °C) until shipment to the US, where they were stored at −80 °C (Alberts and Altmann 2011). A total of 179 samples were collected from 79 individuals; 1 sample was removed during quality filtering of our sequencing data, yielding a final dataset of 178 samples from 78 individuals (Table 1; range = 1–5 samples per individual; median = 2 samples per individual).
Table 1.
Sample sizes for each social group and baboon age/sex classes
| Dataset | Number of samples | Number of samples in Mica’s group | Number of samples in Viola’s group | Number of individual hosts | Number of individual hosts in Mica’s group | Number of individual hosts in Viola’s group |
|---|---|---|---|---|---|---|
| All samples | 178 | 67 | 111 | 78 | 27 | 51 |
| Adult females | 57 | 22 | 35 | 30 | 11 | 19 |
| Adult males | 61 | 28 | 33 | 19 | 9 | 10 |
| Juveniles | 60 | 17 | 43 | 29 | 7 | 22 |
Profiling gut microbial composition
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from each fecal sample using the Powersoil DNA Isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA) (Turnbaugh etal. 2007; McInnes and Cutting 2010). Illumina libraries were prepared following Davenport etal. (2014). Specifically, we amplified a hypervariable section of the V4 region of the bacterial 16S rRNA gene via polymerase chain reaction using barcoded primers 515F and 806R (Caporaso etal. 2011; Degnan etal. 2012; Yatsunenko etal. 2012). Multiplexed libraries were single-end sequenced (102 bp per sequence) on the Illumina HiSeq 2000 platform at the University of California-Los Angeles Neuroscience Genomics Core, yielding 315,821,753 total raw sequencing reads.
Quality filtering and taxonomic assignment
Quality filtering and taxonomic assignments were conducted using the QIIME-based pipeline detailed in Supplementary Figure S2 (Caporaso etal. 2010). We rarefied the dataset to the sample with the lowest number of reads using the QIIME command single_rarefaction.py, yielding a rarefied OTU table of 151,166 reads per sample (26,907,548 reads total) and 16,583 OTUs (Supplementary Table S1). To differentiate the core and non-core gut microbiome, we split the rarefied OTU table into two tables following definitions used in previous studies: core OTUs were those present in ≥90% of samples, and non-core OTUs were present in <90% of samples (Ugland and Gray 1982; Qin etal. 2010; Li etal. 2013; Ainsworth etal. 2015). In addition to a non-core definition of <90% of samples, we re-ran the analyses with a non-core definition of taxa found in <50% of samples and found qualitatively similar results to those obtained using a 90% non-core cutoff, except where noted below (see Supplementary Results). We additionally repeated the analyses on the whole dataset without differentiating the core and non-core microbiomes, and found the results to be qualitatively similar to the core dataset (see Supplementary Results). Alpha and beta diversity metrics were calculated in QIIME.
Statistical analyses
Unless noted, all statistical tests were run in R (R Development Core Team 2014) and performed separately for the core and non-core datasets.
Testing H1: Sociality promotes gut microbial alpha diversity
We constructed linear mixed models using the lmekin function in the coxme package with the following fixed effects: the individual’s current social group, sex, grooming partner diversity, and age (Supplementary Table S2; see Supplementary Methods for information on how each of these were collected; Therneau 2015). We note that we did not test direct effects of group size (as opposed to group identity) because we only tested samples from two social groups. Kinship was incorporated in the random effect estimate to control for repeated sampling from some individuals and for relatedness in our study population (Supplementary Table S3). We used three measures of OTU alpha diversity as response variables to capture different aspects of diversity: OTU richness (i.e., the number of distinct OTUs in a sample), Shannon’s H (to account for evenness of OTU distribution), and Faith’s phylogenetic diversity (to test for a phylogenetic signature; Bates etal. 2015). The best-fitting models were identified using the log likelihood criterion.
Testing H2: Group living and social relationships within groups promote gut microbial community similarity
Gut microbial dissimilarity between individuals was estimated using weighted UniFrac (Lozupone and Knight 2005). Weighted UniFrac was chosen because it accounts for both differences in microbial abundance and evolutionary relationships between taxa (Lozupone and Knight 2005), although we found similar results when we repeated the analyses using unweighted UniFrac and Bray-Curtis beta diversity metrics (see Supplementary Results). To test whether members of the same social group had more similar gut microbiomes than members of different social groups, we performed PERMANOVA in the vegan package (Oksanen etal. 2012). Because some individuals were sampled more than others, and because samples from the same individual had similar community compositions (PERMANOVA; r2 = 0.64, P < 0.001; Supplementary Fig. S3), all analyses were conducted with one, randomly chosen sample per individual. We ran 1000 iterations of random subsampling to one sample per individual to check the robustness of the resulting r2 value to the samples included in our analysis. Because the r2 values varied little across random subsamples, we report the mean r2 value and associated permutation-based P values in the main text.
Baboon social groups contain maternal and paternal kin (Van Horn etal. 2007), so we ran partial Mantel tests to rule out kinship as a potential explanation for group level microbiome differences. We randomly subset the dataset to one sample per individual and ran 1000 iterations to produce a pseudo Mantel r and permutation-based P value for social effects on beta diversity, controlling for kinship.
To identify OTUs that differed significantly in abundance between social groups, we used linear discriminant effect size analysis (LEfSe; v.1) (Segata etal. 2011). We set the Kruskal–Wallis alpha level to 0.01 and the threshold on the logarithmic LDA scale to 3.0.
To test whether close grooming partners had more similar core and non-core microbiomes than individuals who rarely groomed each other, we ran partial Mantel tests on matrices of within-group beta diversity and grooming bond strength, controlling for kinship or diet for each social group using the vegan package in R (Supplementary Tables S4–S7; Oksanen etal. 2012).
Testing H3: Immigrant males who join a social group acquire their new group’s gut microbiome
We averaged the weighted UniFrac values between a sample from an immigrant male and samples from all other adult residents of the group who had been members of the social group for ≥1 year. We then ran linear mixed models with mean weighted UniFrac distance as the response variable, length of the immigrant male’s group membership as a fixed effect, and individual identity as a random effect.
Results
Defining the core and non-core gut microbiome
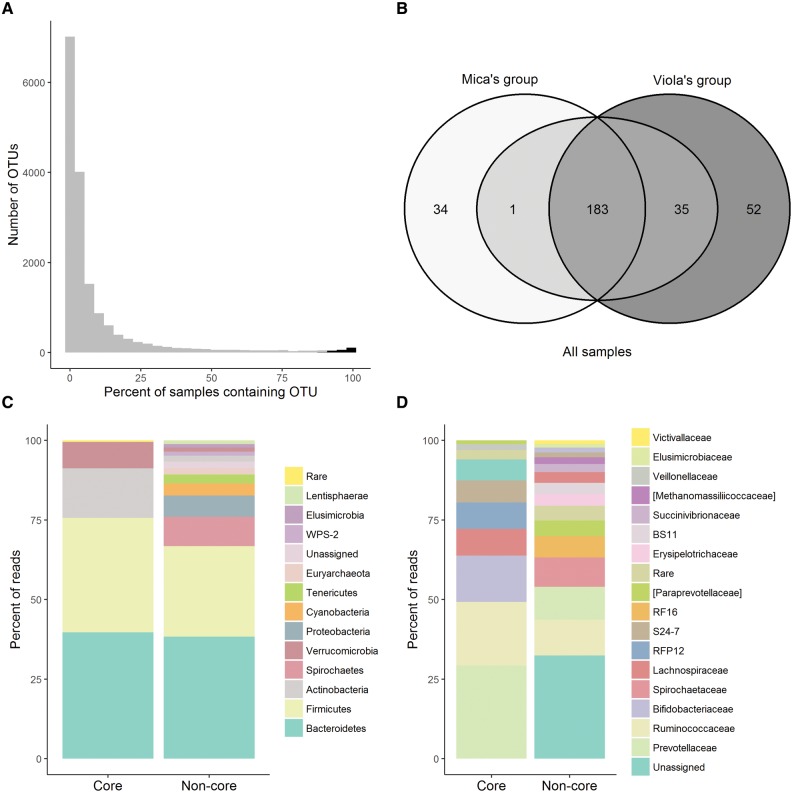
We identified 16,583 gut microbial OTUs in the 178 samples in our dataset. These OTUs exhibited a right-skewed distribution across samples such that the vast majority of OTUs (98.7%) were found in <10% of samples (Fig. 1A). Therefore, following previous studies (Ugland and Gray 1982; Qin etal. 2010; Li etal. 2013; Ainsworth etal. 2015), we defined “core” OTUs as those present in ≥90% of samples. The 219 OTUs that comprised this core occurred in 97.8% ± 3.0% (median ± SD) of samples and 98.7% of individuals (±1.9%; accounting for repeat sampling), and they comprised the majority of the sequencing reads in each sample (median ± SD = 62.0% ± 14.7%). The remaining 16,364 OTUs were classified as “non-core” OTUs. Each non-core taxon occurred in 2.8% ± 14.5% (median ± SD) of samples and 5.1% ± 18.3% of individual subjects. Only six phyla occurred in the core microbiome: Bacteroidetes (mean per sample abundance = 39.7%), Firmicutes (36.0%), Actinobacteria (15.6%), Verrucomicrobia (8.2%), Proteobacteria (0.4%), and Cyanobacteria (0.08%). In contrast, 29 phyla were represented in the non-core microbiome, including the six phyla also found in the core microbiome (Fig. 1C). Nineteen bacterial families were found in the core microbiome and 216 families in the non-core (Fig. 1D).
Fig. 1.
(A) Histogram of OTU prevalence in the 178 microbiome samples in this study. Core OTUs, shown in black (n = 218), were found in ≥90% of samples; the remaining OTUs were considered non-core OTUs (n = 16,364). (B) Venn diagram showing overlap in the number of core OTUs across the whole dataset (light gray), core OTUs in Mica’s group (white), and core OTUs in Viola’s group (dark gray). Numbers indicate overlap counts between datasets; e.g., 183 OTUs are found in ≥90% of the samples in the whole dataset, ≥90% of the samples in Mica’s group, and ≥90% of the samples in Viola’s group. The 219 core OTUs used in many of our analyses include 183 OTUs that are part of the core microbiome in both social groups, 1 OTU that is part of Mica’s core, but not Viola’s, and 35 OTUs that are part of Viola’s core, but not Mica’s. (C) Mean relative abundance of bacterial phyla represented by core and non-core OTUs across all samples. Rare phyla were those that comprised, on average, <1% of reads per sample. (D) Mean relative abundance of bacterial families represented by core and non-core OTUs across all samples. Rare families were those that comprised, on average, <1% of reads per sample. Bracketed taxa indicate taxon names proposed by the greengenes curators (DeSantis etal. 2006).
Group living, but not grooming partner diversity, predicted gut microbial alpha diversity
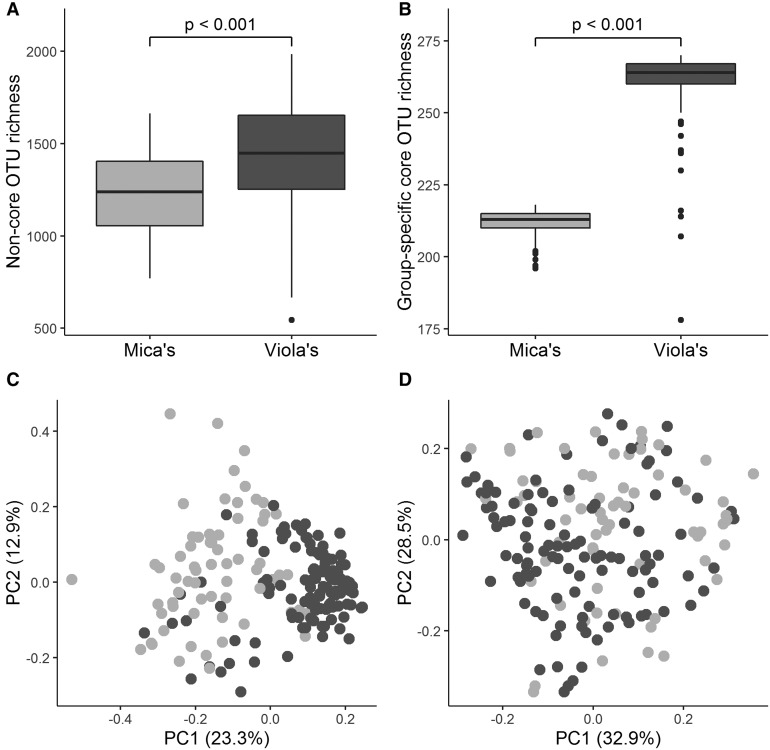
We expected gut microbial alpha diversity to be positively correlated with social group size and grooming partner diversity. With only two social groups, our ability to infer effects of group size is limited. However, we found that, for both the core and non-core microbiome, individuals living in the larger social group (Viola’s) exhibited higher gut microbial OTU richness than individuals in the smaller social group (Table 2 and Fig. 2A and B). Further, contrary to our expectations, the difference between the groups was more evident in the core gut microbiome than the non-core microbiome (Table 2 and Fig. 2A and B). Members of Viola’s group had 1448 ± 302 (median ± SD) non-core OTUs per sample, compared to 1238 ± 221 non-core OTUs in Mica’s group. This pattern was also apparent among the 219 core OTUs (Table 2; Viola’s group had 215 ± 8.5 (median ± SD) core OTUs per sample compared to 212 ± 7.1 in Mica’s group; Fig. 1B). Because core OTUs defined for the entire study population, by definition, minimize differences between the two social groups, we also repeated our analyses of between-group differences in the size of the core microbiome by defining group-specific core microbiomes (i.e., based on presence in ≥90% of members of each group, rather than the entire study population). We found that Viola’s group had a larger group-specific core microbiome than Mica’s group (Fig. 2B), with 270 group-specific core OTUs in Viola’s group, while Mica’s group only had 218 group-specific core OTUs (Fig. 1B).
Table 2.
Linear mixed models predicting variation in gut microbial alpha diversity in baboons (n = 178 samples from 78 individuals)
| Fixed Effects | Estimate | Standard Error | z | P | Direction of Effect | |
|---|---|---|---|---|---|---|
| Core | ||||||
| OTU richness | Social group | 0.556 | 0.169 | 3.29 | 0.001 | Viola’s > Mica’s |
| Age | 0.0226 | 0.0167 | 1.36 | 0.18 | — | |
| Shannon’s H | Social group | 0.221 | 0.0769 | 2.87 | 0.004 | Viola’s > Mica’s |
| Age | 0.0334 | 0.0077 | 4.34 | <0.001 | older > younger | |
| Faith’s PD | Social group | 0.224 | 0.062 | 3.62 | <0.001 | Viola’s > Mica’s |
| Age | 0.014 | 0.006 | 2.28 | 0.022 | older > younger | |
| Non-core | ||||||
| OTU richness | Social group | 227.8 | 48.86 | 4.66 | <0.001 | Viola’s > Mica’s |
| Age | 14.8 | 4.84 | 3.05 | 0.002 | older > younger | |
| Shannon’s H | Social group | 0.0911 | 0.127 | 0.72 | 0.47 | — |
| Age | 0.0326 | 0.0125 | 2.60 | 0.009 | older > younger | |
| Faith’s PD | Social group | 0.864 | 1.019 | 0.85 | 0.4 | — |
| Age | 0.344 | 0.101 | 3.39 | <0.001 | older > younger | |
Note: Models show fixed effects that were significant in at least one model. We also tested sex and grooming partner diversity as fixed effects, but these factors were never significant. Kinship between baboons was modeled as a random effect.
Fig. 2.
Boxplots showing differences in gut microbial OTU richness for (A) non-core and (B) the group-specific core gut microbial communities in each social group. Plots C and D show principal coordinates analyses of weighted UniFrac dissimilarities for (C) non-core and (D) core gut microbial communities. Mica’s group is shown in light gray and Viola’s group is shown in dark gray for each panel.
Contrary to our predictions, we found no evidence that individuals with more diverse grooming relationships had higher gut microbial alpha diversity. Indeed, there was no relationship between an individual’s grooming partner diversity and microbiome diversity for any measure of alpha diversity in either the core microbiome or the non-core microbiome (P > 0.28 for all linear mixed models).
Social effects on gut microbial beta diversity include the core microbiome
As in previous work in this population (Tung etal. 2015), we found that members of the same social group harbored more similar gut microbiomes than members of different social groups. Here, we observed that this effect extended to both the core and non-core microbiome. Social group membership explained 13.9% of the variance in gut microbial composition for the non-core microbiome (PERMANOVA of weighted UniFrac distances: non-core microbiome permuted r2 = 0.139, permuted P = 0.001; Fig. 2C), and 4.7% for the core microbiome (PERMANOVA of weighted UniFrac distances: core microbiome permuted r2 = 0.0477, permuted P = 0.007; Fig. 2D), even though core microbiome taxa, by definition, occurred in subjects from both groups. These group-level differences were not driven by kinship between members of the same social group. Gut microbial beta diversity between hosts was still correlated with group membership, even controlling for kinship (partial Mantel; core microbiome permuted r = 0.099, permuted P = 0.014; non-core microbiome permuted r = 0.396, permuted P = 0.001). Further, microbiome beta diversity between hosts was not correlated with kinship, controlling for group membership (partial Mantel; core microbiome permuted r = 0.004, permuted P = 0.55; non-core microbiome permuted r = 0.026, permuted P = 0.216).
Linear discriminant effect analysis (LEfSe) revealed several taxa that differed significantly in relative abundance between the two social groups. In the core microbiome, these differences were largely driven by OTUs from two genera (Bifidobacterium and Faecalibacterium) and two families (Coriobacteriaceae and RFP12) (Supplementary Fig. S4). Bifidobacterium also differed in relative abundance between social groups in the non-core microbiome, along with the genera Prevotella, YRC22, Coprococcus, Succinivibrio, and Treponema (Supplementary Fig. S5). When the non-core microbiome was defined more stringently, however, (50% instead of 90% threshold), non-core OTUs in Bifidobacterium did not differ in relative abundance between social groups (Supplementary Results).
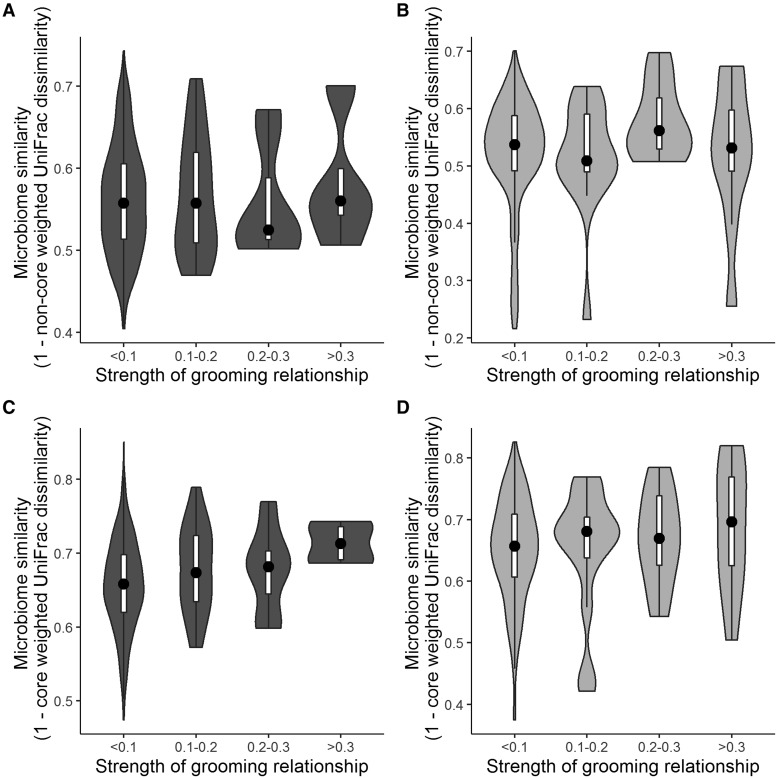
In Viola’s group, but not Mica’s, we found that close grooming partners had more similar core microbiomes than individuals who rarely groomed each other (Fig. 3). In Viola’s group, close grooming partners had more similar core gut microbiota and trended toward significance for non-core microbiota (partial Mantel tests controlling for kinship: core microbiome, r = 0.071, P = 0.009; non-core microbiome, r = 0.051, P = 0.0549; partial Mantel tests controlling for diet: core microbiome, r = 0.064, P = 0.047; non-core microbiome, r = 0.0597, P = 0.06). We did not find that grooming partners had more similar microbiomes in Mica’s group (partial Mantel tests controlling for kinship: core microbiome, r = 0.085, P = 0.11; non-core microbiome, r = 0.083, P = 0.12; partial Mantel tests controlling for diet: core microbiome, r = 0.0725, P = 0.18; non-core microbiome, r = 0.1298, P = 0.065). However, when we re-defined the non-core microbiome as taxa present in <50% of samples, grooming relationship strength significantly predicted gut microbial similarity for Mica’s group (Supplementary Results; partial Mantel tests controlling for kinship: r = 0.143, P = 0.024; partial Mantel tests controlling for diet: r = 0.154, P = 0.034). Further, the similar core microbiome effect sizes in both groups suggests that the lack of a significant relationship in Mica’s group may be due to lower statistical power (smaller sample size) than in Viola’s group. Subsetting Viola’s group to the same number of samples as Mica’s group no longer yielded significant grooming effects in Viola’s group (partial Mantel tests controlling for kinship on 1000 random subsets: core microbiome permuted r = 0.068, permuted P = 0.18). However, additional samples (e.g., repeated samples over time) would be needed to definitively distinguish between lack of power and lack of a true effect in Mica’s group.
Fig. 3.
Violin plots showing the relationship between the strength of grooming relationships and the gut microbial communities. Black dots represent median values and white rectangles represent the first and third quartiles of the data. Rotated kernel density plots representing the underlying data are shown on each side. Stronger bonds predict more similar gut microbiotas in (C) the core microbiome in Viola’s group but not in (A) the non-core microbiome in Viola’s group, (B) the non-core microbiome in Mica’s group, or (D) the core microbiome in Mica’s group.
Longer male residency increases gut microbiome similarity to other group members
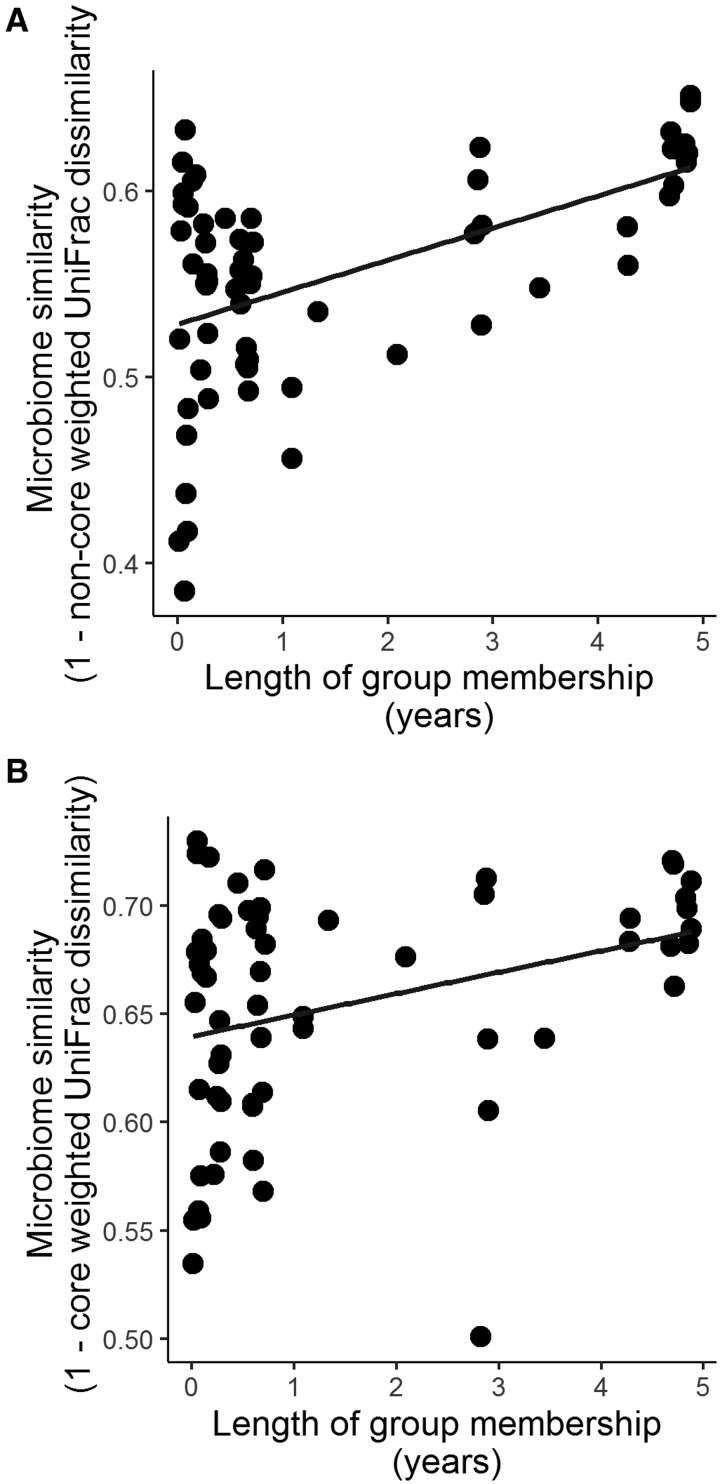
Immigrant males who had lived in their current social group longer had core and non-core microbiota that were more similar to other long-term adult group residents than males with shorter group residency times (Table 3 and Fig. 4). If these effects were solely due to dietary shifts when males moved between groups, we would expect microbiome convergence to occur relatively quickly, over a period of a few days (David etal. 2014). Instead, our results suggest that this process occurs over a more extended time period (months to years). Immigrant males may acquire some microbes from group members via physical contact. Consistent with this hypothesis, we found that immigrant males who had been in the group longer engaged in more frequent grooming interactions than males who had recently immigrated to the group (linear model; β = 0.05063, P = 0.001). Figure 4 appears to show that males who had been group members for less than a year had greater variance in similarity to long-term residents than adult males who had been members for over year. However, we found no statistical evidence for this pattern (Bartlett’s Test for differences in variance; P > 0.5 for both the core and non-core microbiome), and, when we subset the data to immigrant males who had been group members for <1 year, we did not find that individuals with greater social integration had more similar microbiomes to the rest of the group than those who were less socially integrated. Future work that uses a longitudinal study design would have more power to detect such a relationship.
Table 3.
Best supported linear mixed models (based on the log likelihood criterion) predicting gut microbial similarity between immigrant males (n = 61 samples from 19 individuals) and long-term, adult group residents (n = 78 samples from 38 individuals)
| Fixed effects | Estimate | Standard error | DF | t | P | Direction of effect | |
|---|---|---|---|---|---|---|---|
| Core | |||||||
| Weighted UniFrac | consecutive years in group | −0.0281 | 0.0106 | 59 | −2.64 | 0.0106 | ↑ time ↓dissimilarity |
| Non-core | |||||||
| Weighted UniFrac | consecutive years in group | −0.0126 | 0.00306 | 9.39 | −4.12 | 0.0024 | ↑ time ↓dissimilarity |
Note: Subject identity was modeled as a random effect
Fig. 4.
The longer an immigrant male has lived in his new social group, the more similar his gut microbiome composition is to those of his new group members for both (A) the non-core microbiome and (B) the core microbiome. The Y-axis represents the average pairwise gut microbial similarity (1—weighted UniFrac dissimilarity) between a given sample from an immigrant male and the adult members of his current social group.
Finally, we also found that, compared to females, males had more diverse core gut microbiomes based on Shannon’s H (linear mixed model; z = 2.07, P = 0.039), and more diverse non-core gut microbiomes based on Faith’s PD (linear mixed model; z = 2.97, P = 0.003). While there are many physiological and behavioral differences between male and female baboons, these results are consistent with the idea that sex-differences in dispersal lead to higher gut microbial alpha diversity in males than in females. However, this result should be treated with caution as we did not observe sex differences in all three measures of alpha diversity; we found no differences in gut microbial richness between males and females (linear mixed models; z = −0.30, P = 0.7 for core OTU richness and z = 1.66, P = 0.096 for non-core OTU richness). Further, we did not find statistically significant sex differences in microbial alpha diversity in either the non-core 50% analysis or the whole microbiome (Supplementary Results).
Discussion
Social effects occur in both core and non-core gut microbial taxa
The processes that shape gut microbial presence and abundance are thought to differ for core and non-core gut microbial taxa. Core taxa may be acquired early in life and, because they make substantial contributions to basic gut microbial functions (Walter and Ley 2011; Shade and Handelsman 2012; Zhang etal. 2016), they may be actively retained and managed by hosts (Hansen etal. 2010; Franzosa etal. 2015; Hooper etal. 2012b). In contrast, non-core taxa do not occur consistently between hosts, or even in the same host over time, and their dynamics are thought to reflect recent environmental and social transmission events (Martínez etal. 2013; Tinker and Ottesen 2016). If true, social signatures on the gut microbiome should be stronger in non-core versus core taxa. However, we found that social interactions predict microbiome composition for both core and non-core taxa, and we detected stronger effects in the core microbiome than the non-core microbiome in some cases.
There are several possible explanations for this finding. First, group-living and social interactions may predict gut microbial composition for both core and non-core taxa because of microbe-microbe interactions. Specifically, because microbes within a community interact, they likely promote or decrease each other’s relative abundances in ways that are independent of microbial transmission. Hence even if transmission exerts stronger effects on non-core than core taxa, there may be ripple effects that influence the abundance of core microbiome taxa. Such ripple effects might be caused by competitive and mutualistic interactions between resident taxa (Ley etal. 2006; Coyte etal. 2015), as well as indirect interactions, such as when microbes alter the gut environment to make it more conducive for related taxa to thrive (Stecher etal. 2010). For instance, using a mouse infection model, Stecher etal. (2010) found that closely related bacterial phylotypes were more likely to co-occur in the same host than less related phylotypes. Mice with high levels of Lactobacilli were more likely to be successfully colonized by experimentally introduced Lactobacillus reuteri than mice with low abundances of Lactobacilli. This “like will to like” phenomenon, in which closely related taxa co-occur and promote related taxa, has been found in environmental microbes (Chaffron etal. 2010) and in human gut microbes including Bifidobacterium spp. and Proteobacteria, both of which occur in our dataset (Lozupone etal. 2012a).
A second explanation is that socially mediated transmission is likely not restricted to non-core taxa, but also exerts strong effects on the abundance of core microbes. It is well known that physical contact between individuals shapes the core microbiome early in life (e.g., Ley etal. 2005; Walke etal. 2011; Sanders etal. 2014); hence individuals may continue to acquire core microbes from conspecifics throughout life. In support, Billiet etal. (2016) found that limiting contact with nestmates or colony material in adult bumblebees led to a significant drop in the abundance of certain core taxa. Further, Li etal. (2016a) suggest that pikas acquire core gut microbial taxa in adulthood via coprophagy. Although the baboons in our study are not coprophagic, physical contact between group members may lead to the transmission of core gut microbes (Song etal. 2013), and future work should explore if mechanisms of social transmission differ between terrestrial hosts, who presumably have more contact with fecal material, and their closely related arboreal relatives. Indeed group members are proposed to serve as reservoirs for core microbes, and it may be advantageous for a host to access a social reservoir of core microbes to recover after an illness or to adapt to local circumstances (Lombardo 2008; Moeller etal. 2016a).
Finally, other aspects of group living, besides social transmission, may influence the abundance of core and non-core gut microbial taxa—at least at the social group level. Specifically, Mica’s and Viola’s groups had only nominal home range overlap in the year prior to sampling (Supplementary Fig. S1; Tung etal. 2015). Hence, the members of each social group may have been colonized by distinct sources of environmentally transmitted microbes. Other studies have found group- or site-specific microbes in species with geographically close but non-overlapping territories (Leclaire etal. 2014; Maurice etal. 2015; Bennett etal. 2016). For instance, in one study of wild pikas, a substantial portion of the core gut microbes harbored by individuals were also common in local environmental samples (Li etal. 2016a). However, this mechanism cannot explain within social group effects, such as those linked to grooming relationships, because members of the same social group experience very similar environmental exposures, and controlling for habitat use does not remove the effects of grooming on gut microbial similarity (Tung etal. 2015).
Regardless of the underlying explanation for why social effects extend to both the core and non-core microbiome, social structuring in the core microbiome could have functional consequences for hosts. For example, the genus Bifidobacterium, which was socially structured in baboon core and non-core microbiomes, colonizes the gut early in life and plays an important role in processing complex carbohydrates and producing vitamins (Pokusaeva etal. 2011; Turroni etal. 2014). Faecalibacterium, which was socially structured in the core microbiome, is one of the most common genera in the human microbiome and can indicate a disease state when present at low levels (Sokol etal. 2008; Miquel etal. 2013). Finally, the genera Prevotella, Succinivibrio, and Treponema, which were structured in the non-core microbiome, are associated with high-fiber human diets (Schnorr etal. 2014). Treponema, which was more abundant in the larger, more diverse baboon social group, has been proposed to be an indicator of high gut microbial diversity, perhaps indicating a healthy gut community (Schnorr etal. 2014). As these genera differ in abundance between social groups, future work in this study system could test if differences in individual health between social groups are correlated with the relative abundance of certain taxa.
Host social behavior and gut microbial alpha diversity
A growing number of studies propose that social partners serve as reservoirs of gut microbial diversity, and individuals with more social partners should exhibit higher gut microbial diversity than socially isolated animals (Lombardo 2008; Levin etal. 2016; Li etal. 2016b; Moeller etal. 2016b). In the baboons in our study, we found that the members of the larger social group exhibited higher gut microbial alpha diversity; however, individuals with the highest grooming partner diversity did not have the most diverse gut microbiomes. Although we cannot draw strong conclusions based on only two social groups, a possible explanation for our results is that indirect transmission of microbes from environmental sources may be more important in shaping baboon gut microbial alpha diversity than direct transmission via physical contact between hosts. For instance, the social group with more members (Viola’s) also occupied a larger home range than the group with fewer members (Supplementary Fig. S1; Tung etal. 2015). Larger home ranges may put baboons into contact with more diverse microbes, especially if microbial populations are spatially heterogeneous, and if larger home ranges contain more diverse resources, substrates, and microbial communities. However, testing this hypothesis would require repeating these analyses with three or more social groups. To date, no studies have tested the relationship between home range area and gut microbial alpha diversity; but, previous research has shown that home range size predicts intestinal parasite diversity and abundance (Nunn and Dokey 2006; Bordes etal. 2009).
Regardless of the mechanism, social effects on gut microbial alpha diversity may have functional consequences for mammalian hosts. Some papers have proposed that diverse microbiomes are more stable and “healthier” than less diverse microbiomes (Dillon etal. 2005; Lozupone etal. 2012b). In free-living communities, biodiversity stabilizes ecosystems such that more diverse communities experience less stochasticity (Tilman etal. 2006; de Mazancourt etal. 2013), greater stability against perturbations (Eisenhauer etal. 2012), and increased productivity (Lehman etal. 2000; Venail and Vives 2013). Alternatively, alpha diversity may be functionally redundant, such that multiple unrelated taxa can fulfill the same role (Shade and Handelsman 2012), or have potentially negative consequences, such as Chiyo and colleagues’ finding that elephants that had greater gut Escherichia coli haplotype diversity also were more likely to harbor pathogenic strains (Chiyo etal. 2014). Further studies are necessary to demonstrate if differences in gut microbial communities have functional consequences for their hosts. Taken together, our results suggest that, if greater core microbial diversity is both biologically significant and beneficial, higher gut microbial alpha diversity may constitute a benefit of living in a large social group with a large home range
Dispersal and the local microbiome
In baboons and many other animals, the consequences of dispersal can range from higher risk of predation and difficulty finding food in unfamiliar habitats, to new reproductive opportunities and improved social status (Alberts and Altmann 1995; Bonte etal. 2012). Our results suggest a novel consequence of dispersal: changes in gut microbial composition. To our knowledge, ours is the first study to show that residence time in a social group predicts similarity of an immigrant animal’s microbiome to those of other long-term group residents. There are several potential routes by which dispersing males may acquire a local microbiome, including changes in diet, microbial exposures from the environment, and microbial colonization from the members of their new social group. In our population, dietary shifts are unlikely to be the sole mechanism by which dispersal alters the gut microbiome. Dietary shifts in gut microbiome composition tend to occur rapidly, over hours or days (Turnbaugh etal. 2009b; Fernando etal. 2010; David etal. 2014), whereas our data suggest that males’ microbiomes continue to converge with their new social group years after emigration. Thus, direct and indirect transmission are probably important in explaining our results, especially since males who have been resident in a social group longer groom more with others. These interactions create potential routes for direct transmission. In addition, we found that, by some metrics, immigrant males had more diverse microbiomes than adult females, who do not leave their natal groups. While hormonal or dietary differences between males and females may also contribute to male-female differences, the hypothesis that dispersal contributes to diversity in the gut microbiome will be important to test in the future, by comparing males with different dispersal histories over a similar time frame.
As yet, we do not know whether changes in the gut microbiome during dispersal have consequences for hosts, although it may be advantageous for immigrants to develop a “local microbiome” (i.e., one specific to the geographic region). Alberdi etal. (2016) proposed that a plastic gut microbiome may help vertebrate hosts adjust more quickly to changing environmental conditions. Research on humans shows that gut microbial composition correlates with the likelihood of developing gut-related illnesses when traveling (Youmans etal. 2015), which suggests that developing a local microbiome may help hosts adjust to local diets. Finally, some have proposed that a local microbiome can modulate susceptibility to local parasites (Koch and Schmid-Hempel 2012).
Prior research on the disease-related consequences of dispersal have tended to consider effects on the group itself, rather than the individuals who themselves transfer. For instance, social groups may minimize disease risk by excluding immigrants that display signs of illness or refusing to accept immigrants until after a “waiting period” that would reveal whether the immigrant was sick (Freeland 1976). However, because group members greatly outnumber dispersers, the social group should arguably have stronger effects on the microbiomes of immigrants than vice versa. Previous work on chimpanzees suggests that individuals who move between social groups maintain gut microbiome signatures from both groups (Degnan etal. 2012), consistent with our finding that dispersers acquire the local microbiome. One way to test this question in future studies would be to use longitudinal data to track a single disperser’s microbiome, along with the microbiomes of individuals in the group that he immigrates into. Leveraging longitudinal data in species that disperse between social groups repeatedly throughout their adult lives, such as baboons, translates to a series of natural experiments that can provide insight into long-term social structuring of the microbiome. Understanding how social context modulates the gut microbiome over time, and the consequences of such effects, is a key area to pursue in future behavioral ecology research.
Supplementary Material
Acknowledgments
We thank D. Jansen and E. Miller for help with statistical analyses. We thank the Kenya Wildlife Services, Institute of Primate Research, National Museums of Kenya, National Council for Science and Technology, members of the Amboseli-Longido pastoralist communities, Tortilis Camp, Ker and Downey Safaris, Air Kenya, and Safarilink for their cooperation and assistance in Kenya. A number of people contributed to the long-term data collection over the years, and we are grateful to all of them for their dedication and contributions. Particular thanks go to the Amboseli Baboon Project long-term field team (R.S. Mututua, S. Sayialel, and J.K. Warutere), and to V. Somen and T. Wango for their assistance in Nairobi. We are grateful to Karl Pinc for his contributions to the development of Babase, the Baboon Project database. We also thank the database technicians, particularly D. Onderdonk, C. Markham, T. Fenn, N. Learn, L. Maryott, P. Onyango and J. Gordon. This research was approved by the IACUC at Princeton University, the University of Notre Dame, and Duke University and adhered to all the laws and guidelines of Kenya.
Funding
This work was supported by the National Science Foundation and the National Institutes of Health, especially the National Institutes of Aging; in the past decade, in particular, the Amboseli Baboon Project acknowledges support from IOS 1053461, IBN 9985910, IBN 0322613, IBN 0322781, BCS 0323553, BCS 0323596, DEB 0846286, DEB 0846532, IOS 0919200, R01 AG034513, R21 AG049936, and P01 AG031719. This paper was also directly supported by IOS 1638630. We also thank Duke University, Princeton University, the University of Notre Dame, the Chicago Zoological Society, the Max Planck Institute for Demography, the L.S.B. Leakey Foundation, and the National Geographic Society for support at various times over the years.
Supplementary data
Supplementary data available at ICB online.
Data archiving
Sample metadata and supplements are available in the Dryad Data Repository (doi:10.5061/dryad.nh044). Raw sequencing data are deposited in NCBI’s Short Read Archive (BioProject PRJNA388566).
References
- Ainsworth T, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamino JL, Spalding HL, Smith C, et al. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aivelo T, Laakkonen J, Jernvall J.. 2016. Population and individual level dynamics of intestinal microbiota of a small primate. Appl Environ Microbiol 82:3537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP.. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation?. Trends Ecol Evol 31:689–99. [DOI] [PubMed] [Google Scholar]
- Alberts SC, Altmann J.. 1995. Balancing costs and opportunities: dispersal in male baboons. Am Nat 145:279–306. [Google Scholar]
- Alberts SC, Altmann J.. 2011. Monitoring guide for the Amboseli baboon research project: protocols for long-term monitoring and data collection. (http://amboselibaboons.nd.edu/assets/211484/abrp_monitoring_guide_sep2016.pdf).
- Alberts SC, Altmann J.. 2012. The Amboseli baboon research project: 40 years of continuity and change In: Kappeler PM, Watts DP, editors. Long-term field studies of primates. Berlin: Springer; p. 261–287. [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S.. 2015. Fitting linear mixed-effects models using lme4. 67:48. [Google Scholar]
- Bennett G, Malone M, Sauther ML, Cuozzo FP, White B, Nelson KE, Stumpf RM, Knight R, Leigh SR, Amato KR.. 2016. Host age, social group, and habitat type influence the gut microbiota of wild ring-tailed lemurs (Lemur catta). Am J Primatol 78:883–92. [DOI] [PubMed] [Google Scholar]
- Billiet A, Meeus I, Van Nieuwerburgh F, Deforce D, Wäckers F, Smagghe G.. 2016. Colony contact contributes to the diversity of gut bacteria in bumblebees (Bombus terrestris). Insect Sci 24:270–277. [DOI] [PubMed] [Google Scholar]
- Bonte D, Van Dyck H, Bullock JM, Coulon A, Delgado M, Gibbs M, Lehouck V, Matthysen E, Mustin K, Saastamoinen M, et al. 2012. Costs of dispersal. Biol Rev Camb Philos Soc 87:290–312. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Theis KR.. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13:e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes F, Morand S, Kelt DA, Van Vuren DH.. 2009. Home range and parasite diversity in mammals. Am Nat 173:467–74. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R.. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, von Mering C.. 2010. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res 20:947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyo PI, Grieneisen LE, Wittemyer G, Moss CJ, Lee PC, Douglas-Hamilton I, Archie EA.. 2014. The influence of social structure, habitat, and host traits on the transmission of Escherichia coli in wild elephants. PLoS One 9:e93408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA.. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR.. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350:663. [DOI] [PubMed] [Google Scholar]
- Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y.. 2014. Seasonal variation in human gut microbiome composition. PLoS One 9:e90731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mazancourt C, Isbell F, Larocque A, Berendse F, De Luca E, Grace JB, Haegeman B, Wayne Polley H, Roscher C, Schmid B, et al. 2013. Predicting ecosystem stability from community composition and biodiversity. Ecol Lett 16:617–25. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H.. 2012. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci U S A 109:13034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R.. 2013. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol 23:1301–17. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Vennard CT, Buckling A, Charnley AK.. 2005. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–8. [Google Scholar]
- Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL.. 2013. Home life: factors structuring the bacterial diversity found within and between homes. PLoS One 8:e64133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer N, Scheu S, Jousset A.. 2012. Bacterial diversity stabilizes community productivity. PLoS One 7:e34517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB.. 2012. Animal behavior and the microbiome. Science 338:198–9. [DOI] [PubMed] [Google Scholar]
- Fernando SC, Purvis HT, Najar FZ, Sukharnikov LO, Krehbiel CR, Nagaraja TG, Roe BA, DeSilva U.. 2010. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol 76:7482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe P, Kunze WA.. 2013. Voices from within: gut microbes and the CNS. Cell Mol Life Sci 70:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C.. 2015. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A 112:E2930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland WJ. 1976. Pathogens and the evolution of primate sociality. Biotropica 8:12–24. [Google Scholar]
- Gomez A, Petrzelkova K, Yeoman CJ, Vlckova K, Mrázek J, Koppova I, Carbonero F, Ulanov A, Modry D, Todd A, et al. 2015. Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol Ecol 24:2551–65. [DOI] [PubMed] [Google Scholar]
- Hamady M, Knight R.. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Gulati A, Sartor RB.. 2010. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol 26:564–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB.. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. [DOI] [PubMed] [Google Scholar]
- Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S.. 2011. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108:3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DU, Adair EC, Cardinale BJ, Byrnes JEK, Hungate BA, Matulich KL, Gonzalez A, Duffy JE, Gamfeldt L, O’Connor MI.. 2012a. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 486:105–8. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ.. 2012b. Interactions between the microbiota and the immune system. Science 336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB. 2010. The microbiota and allergies/asthma. PLoS Pathog 6:e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P.. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A 108:19288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P.. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett 15:1095–103. [DOI] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclaire S, Nielsen JF, Drea CM.. 2014. Bacterial communities in meerkat anal scent secretions vary with host sex, age, and group membership. Behav Ecol 25:996–1004. [Google Scholar]
- Lehman C, Xa L, Tilman D, Steven DG.. 2000. Biodiversity, stability, and productivity in competitive communities. Am Nat 156:534–52. [DOI] [PubMed] [Google Scholar]
- Levin II, Zonana DM, Fosdick BK, Song SJ, Knight R, Safran RJ.. 2016. Stress response, gut microbial diversity and sexual signals correlate with social interactions. Biol Lett 12:pii:20160352 published online (doi: 10.1098/rsbl.2016.0352). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI.. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI.. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–48. [DOI] [PubMed] [Google Scholar]
- Li H, Li T, Yao M, Li J, Zhang S, Wirth S, Cao W, Lin Q, Li X.. 2016a. Pika gut may select for rare but diverse environmental bacteria. Front Microbiol 7:1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qu J, Li T, Li J, Lin Q, Li X.. 2016b. Pika population density is associated with composition and diversity of gut microbiota. Front Microbiol 7:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Bihan M, Methé BA.. 2013. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One 8:e63139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MP. 2008. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav Ecol Sociobiol 62:479–97. [Google Scholar]
- Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R.. 2012a. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res 22:1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R.. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R.. 2012b. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Muller CE, Walter J.. 2013. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One 8:e69621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Cl Knowles S, Ladau J, Pollard KS, Fenton A, Pedersen AB, Turnbaugh PJ.. 2015. Marked seasonal variation in the wild mouse gut microbiota. ISME J 9:2423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord AI, Chapman CA, Weny G, Tumukunde A, Hyeroba D, Klotz K, Koblings AS, Mbora DNM, Cregger M, White BA, et al. 2013. Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. Am J Primatol 76:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes P, Cutting M.. 2010. Manual of Procedures for Human Microbiome Project, Core Microbiome Sampling, Protocol A, HMP Protocol # 07-001, Version Number: 11.0.
- McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL.. 2012. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J 6:588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow JF, Bateman AC, Herkert KM, O’Connor TK, Green JL.. 2013. Significant changes in the skin microbiome mediated by the sport of roller derby. PeerJ 1:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, Thomas M, Wells JM, Langella P.. 2013. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 16:255–61. [DOI] [PubMed] [Google Scholar]
- Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, Ochman H.. 2016a. Cospeciation of gut microbiota with hominids. Science 353:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H.. 2016b. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv 2:e1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn CL, Dokey AT-W.. 2006. Ranging patterns and parasitism in primates. Biol Lett 2:351–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H.. 2012. vegan: Community ecology package. R package version 2.0-5.
- Pokusaeva K, Fitzgerald GF, van Sinderen D.. 2011. Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6:285–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JE, Martinson VG, Urban-Mead K, Moran NA.. 2014. Routes of acquisition of the gut microbiota of Apis mellifera. Appl Environ Microbiol 80:7378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2014. R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE.. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23:1268–83. [DOI] [PubMed] [Google Scholar]
- Savage DC. 1977. Microbial ecology of gastrointestinal tract. Annu Rev Microbiol 31:107–33. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Iverson KD, Petrosino JF, Schloss SJ.. 2014. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome 2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5:3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60–R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Handelsman J.. 2012. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol 14:4–12. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, et al. 2013. Cohabiting family members share microbiota with one another and with their dogs. eLife 2:e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, et al. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6:e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis KR, Schmidt TM, Holekamp KE.. 2012. Evidence for a bacterial mechanism for group-specific social odors among hyenas. Sci Rep 2:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. 2015. coxme: mixed effects Cox models. R package version 2.2-5 Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Tilman D, Reich PB, Knops JMH.. 2006. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441:629–32. [DOI] [PubMed] [Google Scholar]
- Tinker KA, Ottesen EA.. 2016. The core gut microbiome of the American cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl Environ Microbiol 82:6603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, Archie EA.. 2015. Social networks predict gut microbiome composition in wild baboons. eLife 4:e05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. 2009a. A core gut microbiome in obese and lean twins. Nature 457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI.. 2007. The human microbiome project. Nature 449:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI.. 2009b. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Ventura M, Buttó LF, Duranti S, O’Toole PW, Motherway MOC, van Sinderen D.. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugland KI, Gray JS.. 1982. Lognormal distributions and the concept of community equilibrium. Oikos 39:171–8. [Google Scholar]
- Van Horn R, Buchan J, Altmann J, Alberts S.. 2007. Divided destinies: group choice by female savannah baboons during social group fission. Behav Ecol Sociobiol 61:1823–37. [Google Scholar]
- Venail PA, Vives MJ.. 2013. Phylogenetic distance and species richness interactively affect the productivity of bacterial communities. Ecology 94:2529–36. [DOI] [PubMed] [Google Scholar]
- Walke JB, Harris RN, Reinert LK, Rollins-Smith LA, Woodhams DC.. 2011. Social immunity in amphibians: evidence for vertical transmission of innate defenses. Biotropica 43:396–400. [Google Scholar]
- Walter J, Ley R.. 2011. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol 65:411–29. [DOI] [PubMed] [Google Scholar]
- White J, Mirleau P, Danchin E, Mulard H, Hatch SA, Heeb P, Wagner RH.. 2010. Sexually transmitted bacteria affect female cloacal assemblages in a wild bird. Ecol Lett 13:1515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DJ, Gerlach NM, Slowinski SP, Corcoran KP, Winters AD, Soini HA, Novotny MV, Ketterson ED, Theis KR.. 2016. Social environment has a primary influence on the microbial and odor profiles of a chemically signaling songbird. Front Ecol Evol 4:90. [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans BP, Ajami NJ, Jiang Z-D, Campbell F, Wadsworth WD, Petrosino JF, DuPont HL, Highlander SK.. 2015. Characterization of the human gut microbiome during travelers’ diarrhea. Gut Microbes 6:110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Derrien M, Levenez F, Brazeilles R, Ballal SA, Kim J, Degivry M-C, Quere G, Garault P, van Hylckama Vlieg JET, et al. 2016. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J 10:2235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.