Abstract
The Mouse Genome Informatics (MGI) Resource supports basic, translational, and computational research by providing high-quality, integrated data on the genetics, genomics, and biology of the laboratory mouse. MGI serves a strategic role for the scientific community in facilitating biomedical, experimental, and computational studies investigating the genetics and processes of diseases and enabling the development and testing of new disease models and therapeutic interventions. This review describes the nexus of the body of growing genetic and biological data and the advances in computer technology in the late 1980s, including the World Wide Web, that together launched the beginnings of MGI. MGI develops and maintains a gold-standard resource that reflects the current state of knowledge, provides semantic and contextual data integration that fosters hypothesis testing, continually develops new and improved tools for searching and analysis, and partners with the scientific community to assure research data needs are met. Here we describe one slice of MGI relating to the development of community-wide large-scale mutagenesis and phenotyping projects and introduce ways to access and use these MGI data. References and links to additional MGI aspects are provided.
Keywords: database, genetics, genomics, human disease model, informatics, model organism, mouse, phenotypes
Introduction
The laboratory mouse is an essential model for understanding human biology, health, and disease. Among the key advantages of the mouse as an experimental tool are its characteristics of small size, short gestation period with multiple young, a short lifespan, and being a highly evolved mammal that is physiologically and genetically very similar to humans. In addition, the mouse is genetically the best-studied mammalian model, is experimentally accessible at all life stages, its complete genome has been sequenced, and plentiful technologies for precisely manipulating its genome make the mouse an exceptional animal for developing new living tools for scientific inquiry and human disease modeling. Further, the large number of inbred strains and special purpose strains that have been developed provide fertile ground for population studies and the potential for understanding multi-genic disease susceptibilities and testing therapeutics.
Many factors contribute to the strength of the mouse as a model system, including the sequencing of its genome (Mouse Genome Sequencing Consortium 2002) and subsequent whole genome sequencing of other mouse strains (c.f., Doran et al. 2016; Keane et al. 2011; Nikolskiy et al. 2015; Yalcin et al. 2012), discovery and analysis of single nucleotide polymorphisms (SNPs) and copy-number variation, the wide diversity of specialized strains (c.f., congenic, recombinant inbred, and humanized mice), and the Collaborative Cross (Collaborative Cross Consortium 2012; Morgan and Welsh 2015; Threadgill and Churchill 2012) and Diversity Outcross (Churchill et al. 2012; Morgan and Welsh 2015; Svenson et al. 2012) populations that model human populations, and the International Knockout Mouse Consortium (IKMC) (Austin et al. 2004; Auwerx et al. 2004; Bradley et al. 2012; International Mouse Knockout Consortium 2007) and International Mouse Phenotyping Consortium (IMPC) (Brown and Moore 2012a,b) that are revealing new functions of the genome. These and other important factors that make the mouse an exceptional model organism are beyond the scope of this review.
Rather, this review concentrates on some of the important inputs into the evolution of the mouse as a model genetic system. These include gene discovery, generation of defined mutations, and analysis of phenotypes that are core to the Mouse Genome Informatics (MGI) Resource (Mouse Genome Informatics 2016l). The continued advances in biological knowledge and computational technologies over the last 30 years, as well as the strong community input and participation, have together driven and shaped MGI into the international mouse knowledgebase it is today.
History
The Development of Mouse as an Experimental and Translational System
Mice have been with mankind for millennia, viewed as pests in early human grain stores, revered for bringing good luck in ancient Asia, and selectively bred for interesting physical attributes and docile characteristics by mouse fancier groups (Royer 2015). But the mouse did not ascend to a tractable, exceptional experimental system until after Cuénot’s demonstration that Mendelian genetic principles apply to mammals (Cuénot 1902).
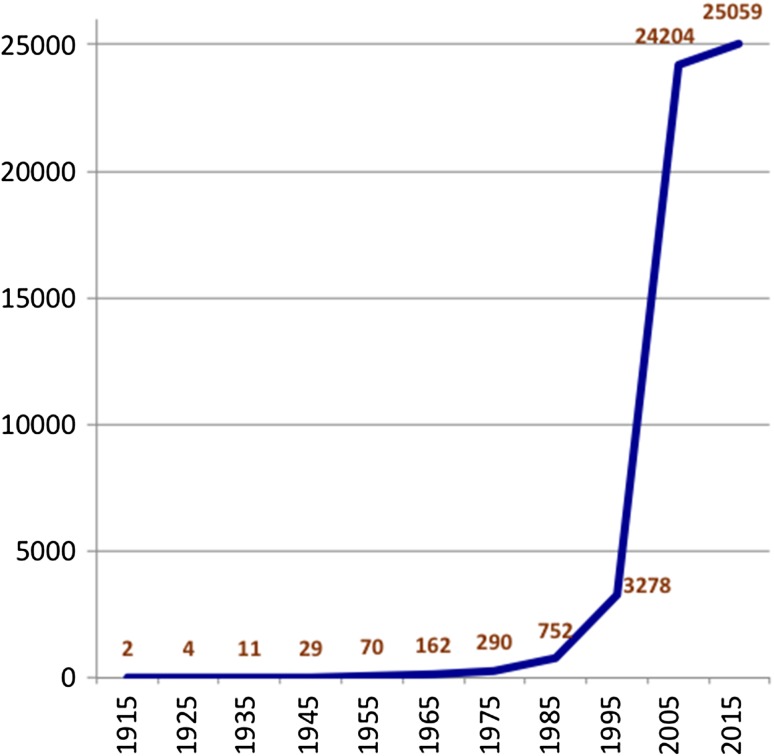
Once it was understood that the fundamental rules of inheritance were applicable, studies were undertaken on gene discovery via observations on segregation of visible traits. By 1945 a linkage map including 26 genes in 10 linkage groups, some with more than one allelic form, was published by the staff of the Roscoe B. Jackson Memorial Laboratory (1945). Gene discovery and mapping continued at a steady pace until the 1970s and 1980s when biochemical variants became tractable genetic markers and propelled a rapid expansion in gene discovery of many nonvisible, but physiologically important, molecular variants (c.f., Felder 1980; Hutton and Coleman 1969; Hutton and Roderick 1970; von Deimling et al. 1988). The advent of DNA technology, with the ability to detect polymorphisms based on restriction length polymorphisms (RFLPs) (c.f., Berman et al. 1986; Dandoy et al. 1985; Mock et al. 1987), further accelerated the density of the genetic markers on the mouse linkage map, from 752 known mapped genes in 1985 (Roderick and Davisson 1985) to a map of 7377 RFLP markers in 1996 (Dietrich et al. 1996). The Human Genome Project to sequence the human (and mouse) genomes (Collins et al. 1998) further accelerated the development of the mouse genome, its sequence map, and gene identification, which has settled in to a protein-coding gene number of approximately 20,000 to 25,000 genes in both human and mouse. Figure 1 illustrates the growth of mouse gene identification, with clear identification of the inflection points caused by the advent of molecular markers and the acceleration (to completion) of the mouse genome sequence in 2002 (Mouse Genome Sequencing Consortium 2002).
Figure 1.
The number of protein-coding genes identified in mice over time. The earliest genes discovered in mouse were in genetic segregation studies of morphological and physiological characters. A significant uptick can be seen when biochemical and molecular measurements became widespread in the 1980s, followed by the advent of molecular markers (e.g., cloned genes/gene fragments, RFLPs, SSLPs) in the 1990s, and finally the sequencing of the mouse genome, which was published by the Mouse Genome Sequencing Consortium in 2002. The number of protein-coding genes for mouse has consistently maintained at 20,000 to 25,000 for the last several years. The functional noncoding RNAs and myriad expanding identification of regulatory elements will continue to greatly expand the functional map of the mouse for the foreseeable future.
Important to the growth and development of the mouse as a model system were the network of researchers working on mice, who not only used standard publication methods to disseminate their work, but established many informal collaborations and open communication channels such as the Mouse News Letter and Chromosome Committees. Such cooperativeness is still observed through the International Committee for Standardized Genetic Nomenclature for Mice, the annual International Mammalian Genome Conferences, and the continued community contributions of data to the MGI resource.
More Data and Computer Advances Drive the Initiation and Expansion of MGI
The precursor of MGI, the “Encyclopedia of the Mouse Genome” software, was initiated in 1989 to develop an interactive tool to simultaneously visualize data from several investigator databases being maintained at The Jackson Laboratory. This was also a time when the Worldwide Web was not in common use and most biologists accessed a computer, if at all, through command line input into a central server. The first versions of this “encyclopedia” were distributed to scientists via postal mail on floppy disks (Richardson et al. 1995).
The genetic linkage map, having been carefully compiled from available data and constructed annually by personal efforts of many dedicated mouse geneticists over the years (M Lyon, R Meredith, S Hawkes, C Beechey, M Green, J Womack, TH Roderick, MT Davisson, Mouse News Letters 1965–1994), had reached a point of unsustainability. The last such map, produced by TH Roderick and MT Davisson in 1994, included nearly 800 genetically mapped loci.
The emergence of molecular biology and our ability to distinguish gene variants by restriction length polymorphisms (RFLPs) allowed the density of the genetic map of mouse markers to grow rapidly. Now, many, many strain variants could be identified and the map of the mouse quickly expanded. In 1992 a map of 317 simple sequence length polymorphism (SSLP) markers covering 99% of the genome was published (Dietrich et al. 1992), which grew to over 7000 markers by 1996 (Dietrich et al. 1996). Additional thousands of SSLP markers were added using interspecific backcross panels of mice that could be used to map virtually any newly identified gene that had a molecular probe with unprecedented speed (c.f., Avner et al. 1988; Copeland and Jenkins 1991; Reeves et al. 1991; Rhodes et al. 1998; Rowe et al. 1994; Watson et al. 1992). In 1993, a review of the status of gene-specific RFLP mapping in the mouse was published (Copeland et al. 1993a), along with a wall chart (Copeland et al. 1993b).
Fortuitously, information technology was also rapidly developing during this time. The growth and accessibility of the Worldwide Web allowed MGI to develop a web interface that was much easier to use than previous command line interfaces and provided the flexibility to create multiple views and combinations of information that were useful to the scientific community.
Ultimately, the direction and growth of MGI has been driven by biotechnology and major international projects from sequencing the genome (and now multiple mouse strain genomes), to large-scale mutagenesis programs, to genome manipulation technologies, developing mouse models of human disease and populations, and large-scale phenotyping efforts to understand gene function. Today, we can precisely and quickly create a new genetically engineered mouse with ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases), or CRISPR/Cas9 (Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9) technologies, and use exome or whole genome sequencing to determine the causative change in a spontaneous or induced phenotypic mutant or use comparative phenotyping to suggest a new mouse model for a human disease.
SOS: Communication Critical
Uniform application of authoritative nomenclatures, ontologies, and identifiers is essential to successful scientific discourse, to enabling experimental reproducibility, and to integrating and analyzing diverse data sets. The success of the MGI resources and our ability to provide high-quality integrated data that users can access across data types and data sets is critically dependent on development and application of these communication standards. Much of the work of MGI staff is devoted to critically assessing incoming data to resolve discrepancies in identifiers and harmonizing terminologies.
Common Language = Clear Communication
Language, and how it is used, can be precise or vague, with the same or similar thoughts expressed in very different words and contexts. For scientific communication, it becomes critical that precision is emphasized and meaning truly conveyed, as the enterprise of scientific advancement and the next discovery relies on the knowledge of what has been previously discovered. Thus, language used to describe scientific experiments, reagents, methods, outcomes and observations, and supported conclusions demands that words are chosen carefully to succinctly and accurately reflect the results. And, key to that communication is the use of standard vocabularies, nomenclatures, ontologies, and identifiers to uniquely define the elements of experimentation and their interrelationships. The result of failed communication through use of jargon or “common” laboratory terminology can be disastrous personally if one realizes the “gene” they thought they were studying is actually a different gene with extensive reported functional analysis already published. Use of conflicting terminology also causes significant confusion in the scientific literature when genes or other reagents are referred to by overlapping terms.
Nomenclature Standards for Gene Identity, Allele Specification, Genomic Changes, Genotypes, and Strains
Nomenclature standards describing genomic and genetic entities and mutations exist today for well-studied species where there are large active research communities and genomes have been entirely sequenced (Table 1). For species where biological studies are sparse, or where sequencing has been done for comparative genomic data only, the standard is to follow the nomenclature of the most closely related, highly studied species (e.g., for primates, the human genetic nomenclature, for fish, the zebrafish nomenclature).
Table 1.
Nomenclature guidelines for human and major model species
| Species | Nomenclature guide URL | Email or contact URL |
|---|---|---|
| Human | http://www.genenames.org/about/guidelines1 | hgnc@genenames.org |
| Mouse | http://www.informatics.jax.org/nomen2 | nomen@jax.org |
| Rat | http://rgd.mcw.edu/nomen/nomen.shtml3 | http://rgd.mcw.edu/contact/index.shtml |
| Chicken | http://birdgenenames.org/cgnc/guidelines4 | agbase@igbb.msstate.edu |
| Xenopus | http://www.xenbase.org/gene/static/geneNomenclature.jsp5 | Joshua.Fortriede@cchmc.org |
| Zebrafish | https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines6 | nomenclature@zfin.org |
| Drosophila | http://flybase.org/wiki/FlyBase:Nomenclature7 | http://flybase.org/cgi-bin/mailto-fbhelp.html |
| C. elegans | http://wiki.wormbase.org/index.php/Nomenclature8 | genenames@wormbase.org |
| Arabidopsis | http://www.arabidopsis.org/portals/nomenclature/9 | curator@arabidopsis.org |
| Saccharomyces | http://www.yeastgenome.org/help/community/gene-registry10 | sgd-helpdesk@lists.stanford.edu |
1Human Gene Nomenclature Committee (HGNC). 2016. HGNC Guidelines. Available online (http://www.genenames.org/about/guidelines), accessed on December 1, 2016.
2Mouse Genome Informatics (MGI). 2016. Mouse Nomenclature Homepage. Available online (http://www.informatics.jax.org/nomen), accessed on December 1, 2016.
3Rat Genome Database (RGD). 2016. Rat Nomenclature Guidelines. Available online (http://rgd.mcw.edu/nomen/nomen.shtml), accessed on December 1, 2016.
4Chicken Gene Nomenclature Consortium. 2016. Gene Nomenclature Guidelines. Available online (http://birdgenenames.org/cgnc/guidelines), accessed on December 1, 2016.
5Xenbase. 2016. Gene Nomenclature Guidelines. Available online (http://www.xenbase.org/gene/static/geneNomenclature.jsp), accessed on December 1, 2016.
6Zebrafish Information Network (ZFIN). 2016. ZFIN Zebrafish Nomenclature Guidelines. Available online (https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines), accessed on December 1, 2016.
7FlyBase. 2016. FlyBase:Nomenclature. Available online (http://flybase.org/wiki/FlyBase:Nomenclature), accessed on December 1, 2016.
8WormBase. 2016. Nomenclature Homepage. Available online (http://wiki.wormbase.org/index.php/Nomenclature), accessed on December 1, 2016.
9The Arabadopsis Information Resource (tair). 2016. Nomenclature Homepage. Available online (http://www.arabidopsis.org/portals/nomenclature/), accessed on December 1, 2016.
10Saccharomyces Genome Database. 2016. SGD Help: Gene Registry. Available online (http://www.yeastgenome.org/help/community/gene-registry), accessed on December 1, 2016.
Gene identity for protein-coding genes of human and the major model organisms is largely established thanks to the Human Genome Sequencing Project. However, one must be aware that updates and changes to the sequence continue, as do revisions to annotations to the genome assemblies. Efforts, including those of the CCDs (Consensus Coding Sequences) (Farrell et al. 2014; NCBI CCDS Database 2016; Pruitt et al. 2009) to curate consensus cDNAs and analyze genomic regions that show differences in build algorithm results from NCBI and EBI genome assemblies continue to improve the quality of the sequenced genome of human, mouse, and other species. Tracking gene identities through common nomenclature and accession IDs is an effective way to ensure that the gene object that is being discussed is truly that gene. For the mouse, the International Committee on Standardized Genetic Nomenclature for Mice, an elected body of the International Mammalian Genome Society, establishes, revises, and maintains guidelines for mouse genetic nomenclature for genes, genetic markers, alleles, and mutations, for mouse strains, and for chromosome aberrations. Guidelines are available online (MGI 2016p) and assistance with nomenclature is available by emailing nomen@jax.org.
Ontologies for Gene Function, Sequence Features, Anatomy Terms, Phenotypes, and Disease Terminology
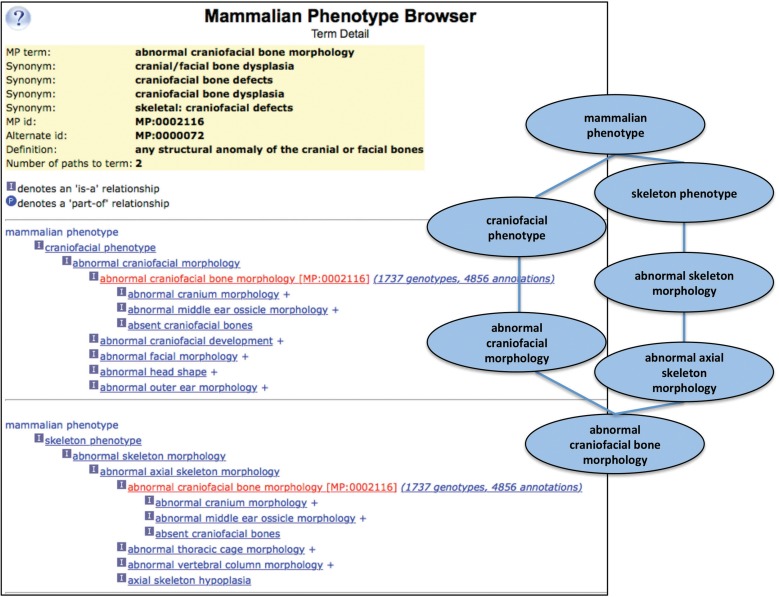
Where nomenclature provides standard vocabularies of discrete entities (genes, microRNAs, noncoding RNAs, strain designations, chromosomes, chromosome aberrations, mutagenesis methods, etc.), ontologies provide both terminology and relationships among term elements. The structure of these ontological relationships, a directed acyclic graph (DAG), is hierarchical in nature, but a single term can have multiple parent and/or child terms. For example, the anatomical structure “limb bone” is a bone; but is also part of the appendicular skeleton and part of the limb. By representing biological features and processes in a DAG structure, with various kinds of relationships among terms, the richness and complexity of biology can be better represented (Figure 2).
Figure 2.
The Mammalian Phenotype Ontology (MP) Browser example. At the left, the MP browser is shown displaying the detail for the term “abnormal craniofacial bone morphology.” At the top are the term name, common synonyms, the MP ID, and the definition. Below are two term paths shown as hierarchical trees, with paths listed as multiple sequential hierarchies. The “abnormal craniofacial bone morphology” term is followed by a link to MGI genotypes and annotations associated with that term. On the right is a graphical representation (as a DAG) for the abnormal craniofacial bone morphology term. The MP Browser is accessed using the pull-down “Search” menu from the MGI homepage and either following the Phenotypes submenu or the Vocabularies submenu to select the Mammalian Phenotype (MP) Browser (http://www.informatics.jax.org/searches/MP_form.shtml) (Mouse Genome Informatics (MGI) 2016b).
Within the MGI system, the following ontologies are used for data representation and annotation: the Gene Ontology (GO) (Ashburner et al. 2000; The Gene Ontology Consortium 2017), The Mammalian Phenotype Ontology (MP) (Smith and Eppig 2012, 2015), the Mouse Anatomy Ontologies (Hayamizu et al. 2013, 2015), and the Sequence Ontology (SO) (Eilbeck et al. 2005). The Human Phenotype Ontology (Köhler et al. 2014, 2017) is used to integrate human phenotypes into the Human-Mouse: Disease Connection portal (see Section Overview of the current MGI resources, below). The Disease Ontology (Kibbe et al. 2015; Schriml and Mitraka 2015) will be integrated into MGI in 2017. By curating and loading data using these standards, MGI produces the highest quality biological data representation and maximizes the robustness of searching the resource.
Mouse Mutagenesis in Large-scale
Systematic mutagenesis projects have been used effectively for many years in organisms such as worm, yeast, and Drosophila. Researchers studying organisms with larger genomes and more complex body-plans, such as zebrafish, rodents, and primates, have long envied having access to similar tools that would improve genetic accessibility and make possible various genomic manipulations. In the last 20 years, more of these precise genetic tools have been developed for mammals, and for the mouse most of all.
Phenotype-Driven (Forward) Mutagenesis: Large-scale ENU Mutagenesis
N-Ethyl-N-nitrosourea (ENU) was demonstrated to be an effective mutagen, and ENU dosage studies (Justice et al. 1999; Russell et al. 1982a,b) led to the development of several systematic experimental ENU protocols. These, in general, consisted of mutagenizing males of different inbred strains and generationally screening their progeny (using same strain or alternate strain matings) for dominant or recessive mutants via broad or specific phenotype testing. Several variations of mating schemes were developed, but in essence, mating of the mutagenized G0 male to wild-type females would detect dominant mutations (or uncover recessive mutations if the female carried a defined deletion) in the G1(F1) offspring. A further generation of mating, either backcrossing to the G0 father or intercrossing siblings, was required to detect recessive mutations (Barbaric and Dear 2007; Silver et al. 2007).
In the late 1990s, systematic, large-scale, chemical mutagenesis projects using ENU as the mutagen were proposed as a method for studying gene function and discovering new disease-related mutations (Brown and Nolan 1998; Hrabé de Angelis and Balling 1998). The first of these programs were developed in Germany (Hrabé de Angelis et al. 2000; Soewarto et al. 2000) and in the United Kingdom (Nolan et al. 2000a,b). Shortly thereafter ENU mutagenesis programs began in Australia (Nelms and Goodnow 2001) and at Japan’s RIKEN Center (Masuya et al. 2004). With the exception of the Australian program that concentrated on immunological phenotypes, these programs screened progeny of ENU-treated males using a wide range of standardized phenotype assays developed in their respective programs.
ENU mutagenesis projects in the United States and Canada were wide ranging but tended to focus on specific sets of phenotypes (Clark et al. 2004; Nadeau et al. 2001). Among the projects were neurological mutations at Northwestern University, The Jackson Laboratory, and the Tennessee Genome Consortium (Goldowitz et al. 2004); developmental/embryonic mutations at Baylor College of Medicine, Harvard Medical School, and the Sloan Kettering Institute (García-García et al. 2005; Herron et al. 2002); craniofacial mutations at the Stowers Institute (Sandell et al. 2011); cardiovascular and metabolic mutations at the Center for Modeling Human Disease, Toronto (Xie et al. 2007) and University of Pittsburgh (Yu et al. 2004); phenotypes associated with aging (e.g., cancer, obesity, Alzheimer’s) at the McLaughlin Institute and Case Western Reserve University; and a hypertension, obesity, sleep-centered program (Svenson et al. 2003) and reproductive genomics program (Lessard et al. 2004) at The Jackson Laboratory.
Sperm from ENU-treated males carry multiple mutations, and these may or may not result in obvious phenotypic changes in his immediate offspring. Thus, a new mutant discovered in an ENU screen is virtually certain to carry mutations unrelated to the novel phenotype identified. This fact is negative in that these additional mutations (so called “incidental” mutations) may have unknown input into the phenotypic change observed. With further generations of mating, many “incidental” mutations will be lost, as the mutation of interest remains selected for and may have unexpected effects on the variant phenotype that was initially identified. Conversely, frozen G0 sperm are a potential treasure of new mutations that can be identified via exome or genome sequencing as differing from the untreated G0 male, and their effect on the gene in which they occur can be predicted by analyzing sequence changes (Arnold et al. 2012; Bull et al. 2013; Moresco et al. 2013; Simon et al. 2015).
Today, there are nearly 4000 identified mouse genes with defined ENU mutations and described phenotypes. In addition, there are over 145,000 associated incidental mutations representing potential mutations of interest for functional or disease-related studies. These numbers will continue to grow as ENU remains a popular and accessible mutagen for projects generating single base pair mutations.
Gene-Driven (Reverse) Mutagenesis: Knocking Out Each of the Mouse Protein Coding Genes
As differentiated from forward mutagenesis, gene-driven or reverse mutagenesis targets specific genes for mutation whether or not there is any a priori knowledge about the phenotypic or physiological consequences. Attendees at a meeting at the Banbury Center at Cold Spring Harbor Laboratory in New York in September of 2003 endorsed creating an international resource of ES cell lines containing knockout alleles for each of the ≈20,000 to 25,000 mouse protein coding genes over a 5-year period using a common genetic background (C57BL/6N) and created using either gene trap or gene targeting techniques. Further, mutant ES cell lines (or mice created from them) would be made available to the entire research community as a resource for studying gene function and phenotype. The anticipated systematic phenotyping of these mutants would likewise be a coordinated international effort, with wide base-line data collected collaboratively, with future specialized phenotyping being done by individual research laboratories and specialists (Austin et al. 2004; Auwerx et al. 2004). This undertaking was not without controversy (see, e.g., Accili 2004), because there was little evidence that there would be demand for this large array of mutants; and although frozen mutant ES cells take up little space and expense, their usage would likely further fade with new technology developments; the systematic (but limited) phenotyping was not thought to yield enough substantive results to garner researcher interest; and a better investment could be made in providing access to less expensive facilities making tailored mutations for researchers (e.g., to study allelic series for specific genes).
Despite these concerns, in 2007 this ambitious plan took shape in the form of the IKMC (International Mouse Knockout Consortium et al. 2007), initially consisting of NIH’s Knockout Mouse Project, European Conditional Mouse Mutagenesis Program, and North American (Canadian) Conditional Mouse Mutagenesis Program. Later, Texas Institute for Genomic Medicine also joined the IKMC (Collins et al. 2007).
Five years later, Bradley et al. (2012) reviewed the progress of the IKMC efforts. Information about the IKMC is also available on the web (International Knockout Mouse Consortium 2016). Mouse ES cell lines with targeted or trapped alleles were available for 17,473 protein-coding genes, and nearly 1000 of these ES cell lines had been made into live mice for research. The EUCOMMtools project continued to expand the number of genes in the IKMC collection, as well as creating conditional-ready alleles for those genes where only deletion knockouts were created, and developing a set of cre-recombinase carrying knock-ins and transgenes to complement the IKMC conditional-ready alleles (Rosen et al. 2015). The next step in developing and utilizing the IKMC resources is now underway as the IMPC (see Phenotyping and functional discovery projects below).
Conditional Mutagenesis
Conditional mutagenesis is a powerful technique for interrogating gene function in a cell- or tissue-specific manner, potentially with a defined temporal component (Birling et al. 2009; Branda and Dymecki 2004; García-Otín and Guillou 2006; Nagy 2000). This technique is particularly useful to examine gene function temporally where a conventional gene knockout might result in embryonic lethality or to examine age-specific tissue expression. In principle, one mates a mouse carrying a “conditional-ready” allele, in which a recombinase recognition sequence has been inserted flanking the sequence of interest, to a mouse carrying an appropriate recombinase with a driver/promoter that directs its activation to a particular tissue at a particular developmental time. Offspring that inherit both the “conditional-ready” allele and the recombinase sequences will have a deletion of the sequence of interest, due to the activation of the recombinase protein and the resulting recombination between the two inserted recombinase sites (Nagy 2000; Sauer and Henderson 1988).
The cre-loxP system is the most commonly used recombinase system. In this case, the genomic region to be manipulated is flanked by loxP sites, unique 34-base pair sequences recognized by the cre protein. Expression of the cre protein results in recombination between the loxP sites and, depending on the experimental design, can cause deletion, inversion, or translocation of the loxP flanked sequence (Friedel et al. 2011; Gierut et al. 2014; Schnütgen et al. 2003). Variations that have been developed for conditional mutagenesis include use of non-cre recombinases, such as flp, dre, and ΦC31, and various inducible forms of recombinases that are activated exogenously, such as by administration of tamoxifen (Anastassiadis et al. 2010).
In evaluating which cre-bearing strain to use for experiments, the cre activity, tissue specificity, and expression timing of the cre driver must be known for the intended target tissue and any off-target sites (Heffner et al. 2012; Murray et al. 2012; Smedley et al. 2011). Without adequate knowledge of cre specificity, experimental results can be confounded and interpretation of observed phenotypes difficult due to cre excision in unanticipated tissues or at unexpected times.
A plethora of new conditional-ready mutations have been generated by the IKMC (Bradley et al. 2012) and are available to the scientific community. To take advantage of the conditional character of these constructs, appropriate cre recombinase mice are needed to mate with these mice. As of December 2016, over 2620 cre-containing transgenes and knock-in alleles have been documented in the MGI CrePortal (see Overview of the current MGI resources below).
Genome Editing: Endonuclease-Mediated Mutagenesis (Zinc Finger Nucleases, TALENS, CRISPR/Cas9)
Newest among the tools for creating specific mutations are endonuclease-mediated methods, referred to as gene editing or genome editing techniques. These have burst into the mouse toolkit in three waves of technical developments, and much work continues to improve the technology and fidelity of results and to push the flexibility of genome editing systems. An overview of the methodologies can be found in Carroll (2014), Doudna and Charpentier (2014), and Menke (2013).
ZFNs as targeting tools in mice were first wave, reported at the beginning of this decade (Carbery et al. 2010; Connelly et al. 2010; Meyer et al. 2010). ZFNs consist of a series of zinc finger domains fused to the cleavage domain of the FokI restriction enzyme. ZFNs are targeted by designing pairs to bind sequences adjacent to the endogenous target site and, once bound, the FokI domains attached to each of the ZNF pairs dimerize and create double-strand breaks in the target DNA. Nonhomologous end joining then occurs, usually resulting in small deletions. Repeated application of the same constructs can produce multiple unique mutations (alleles) in the same target sequence. The double-strand breaks can be repaired via homology directed repair to produce other types of mutations if DNA with homology to the target region is co-introduced with the ZFNs.
Quickly on its heels was the second wave, using TALENS, which had similar properties and method of action to the ZFNs. The distinct advantages of TALENS were a greater range of sequence specificities, higher efficiency, and better predictability of their action (Panda et al. 2013; Qiu et al. 2013). Thus, TALENs rapidly overtook ZFNs as the genome editing tool of choice.
The third wave of gene editing technology is CRISPR/Cas9 originating from a bacterial adaptive immune system. The CRISPR/Cas9 system, unlike the ZNFs and TALENS, does not require that unique endonuclease pairs be designed for each genomic target, thus making it a simpler and less time-consuming method (Wiles et al. 2015). In addition, its fidelity is higher, with the targeting to specific DNA sequences determined by the 5’ 20 nucleotide sequence of the synthetic single guide RNA, designed to be complementary to the genomic target sequence. And fewer off-target events are observed with CRISPR/Cas9 than with ZNFs or TALENS (Li et al. 2013; Miano et al. 2016; Shen et al. 2013; Singh et al. 2015; Tycko et al. 2016).
The IMPC has recently adopted use of CRISPR/Cas9 mutations in the mouse knockout phenotyping program, a move that will greatly reduce its reliance on the IKMC ES cell line mutations that require recovery of ES cell lines into mice and subsequent breeding removal of the critical exon via cre-excision, in the case of conditional-ready alleles or breeding to remove the neo-cassette in the case of deletion alleles (Bradley et al. 2012). This move will enable quick and direct generation of mice carrying single gene knockout mutations and accelerate the ability to produce the mouse cohorts needed for the IMPC’s high-throughput phenotyping pipeline (KL Svenson, The Jackson Laboratory, 2017, personal communication).
To emphasize the rapid adoption of gene editing technology in mice, a search of PubMed (Dec. 2016), lists over 1000 publications using ZNFs, TALENs, or CRISPR/Cas9 for genome editing in mice, with over 750 of these using the CRISPR/Cas9 technology. Further, the flexibility of the CRISPR/Cas9 system in its ability to create a variety of genomic changes in mice is illustrated by these “for example” sampling of outcomes: (1) to create conditional-ready alleles (“floxed” alleles) for use in creating mice for conditional mutagenesis experiments (Bishop et al. 2016; Lee and Lloyd 2014; Yang et al. 2013); (2) for developing humanized mice through gene replacement (Gennequin et al. 2013); (3) to model human disease (Lewis et al. 2016; Zhong et al. 2015); (4) to repair gene defects (Mianné et al. 2016; Nelson et al.; 2016; Wu et al. 2013; Yin et al. 2014); (5) to use in multiplexing to simultaneously create mutations in multiple genes (Wang et al. 2013; Zhou et al. 2014); and (6) to create large deletions, insertions, and chromosomal rearrangements (Boroviak et al. 2016; Maddalo et al. 2014; Zhang et al. 2015).
Phenotyping and Functional Discovery Projects: Large-scale Phenotyping
The discovery and creation of new mutations and variants in mouse have been accelerating thanks to the large-scale mutagenesis efforts using forward and reverse genetic technology and the continued development and improvement of new methods to create, validate, and preserve those mutants for the scientific community. Making these resources, however, is only the first step; the goal is to understand the function of genes and develop useful models for studying biology and disease. Thus, standardized, robust phenotyping must accompany our genetic knowledge and mutant mouse resources.
Efforts over the last 20 years have made significant progress on collecting systematic phenotypic data, developing standard protocols and phenotyping pipelines, and on making these data accessible online. Initially each project with significant throughput, whether performing ENU mutagenesis or studying knockouts, developed their own phenotyping methods, many of which contributed to the large-scale phenotyping projects that came later (c.f., Ayala et al. 2010; Brommage et al. 2014; Lessard et al. 2004; Li et al. 2015; Pack et al. 2007).
One of the earliest collaborative efforts attempted to establish globally standard phenotyping procedures and test phenotype assay robustness was the European Eumorphia project (Brown et al. 2006; Mallon et al. 2008). Baseline data from four inbred strains, C57BL/6J, C3HeB/FeJ, BALB/cByJ, and 129S2/SvPas (formerly 129/SvPas), was collected using identical protocols in multiple European laboratories and differences were investigated to determine if differences were attributable to procedure modifications, equipment differences or calibrations, technician error, lack of robustness of the test, etc. The Eumorphia progam was also responsible for the initial development of the Europhenome data repository (Mallon et al. 2008; Morgan et al. 2010) and the European Mouse Phenotyping Resource for Standardized Screens. European Mouse Disease Clinical program, with the Sanger Institute Mouse Genetics Program, continued the collaborative work of Eumorphia, developing protocols and phenotyping over 650 mutant mouse lines, largely from the growing IKMC mutant ES cell lines (Ayadi et al. 2012; Gates et al. 2011; Hrabé de Angelis et al. 2015).
In 2011, the IMPC was established, encompassing these groups and other participant worldwide (Brown and Moore 2012a,b). Its current members include the Medical Research Council, Harwell, UK; Wellcome Trust Sanger Institute, Cambridge, UK; Helmholtz-Zentrum Muenchen, Germany; PHENOMIN, Strasbourg, France; Australian Phenomics Network, Australia; RIKEN BioResource Center, Tsukuba, Japan; CNR Montorotondo, Italy; MARC Nanjing University, Nanjing, China; The Jackson Laboratory, Bar Harbor, USA; The Davis, Toronto; Charles River and CHORI Consortium, USA and Canada; Korea Mouse Phenotype Consoritum, South Korea; Baylor College of Medicine, Houston, USA; National Laboratory Animal Center, NARLabs, Taiwan; European Bioinformatics Institute, Hinxton, UK; Czech Centre for Phenogenomics, IMG, Czech Republic; and Universitat Autònoma de Barcelona, Spain (Koscielny et al. 2014). With a goal of systematically phenotyping cohorts of mice from each of the mutant lines produced from the IKMC resource within 10 years, the task ahead is significant. But, these baseline data will be invaluable for researchers looking for appropriate model systems and for developing more sophisticated tools for refining phenotypes and exploring functional biological networks. Phenotyping data are accessible on the IMPC website (International Mouse Phenotyping Consortium (IMPC) 2016; Ring et al. 2015), and some initial global studies are being published (c.f., Dickinson et al. 2016; White et al. 2013).
Overview of the Current MGI Resources
The mission of the MGI (Mouse Genome Informatics 2016l) resource is to provide integrated genetic, genomic, and biological data about the laboratory mouse to facilitate the study of human health and disease. To fulfill this mission MGI must maintain a dynamic, evolving system that continues to grow and make directional changes as biological science changes and the computing tools supporting its infrastructure advance.
MGI is a collection of integrated data resources and tools that can all be accessed via the central MGI homepage (Mouse Genome Informatics 2016l). Data originate from high-throughput data projects, direct investigator or collaborative group data submissions, and through curation of the scientific literature. Semantic and contextual integration is achieved through automated and semiautomated methods, coupled with expert curation and extensive quality controls. By promoting integration throughout, MGI has been able to consistently develop and maintain a unified genome feature catalog (Zhu et al. 2015); enforce nomenclature standards for genes, genome features, alleles, strains, and other mouse entities; and develop and apply ontologies. These unify metadata and vocabulary terminology across data sources, adding value and enabling robust data searching and superior accessibility to users.
This review cannot thoroughly cover all aspects of MGI data content or address all possible use cases for MGI. Below, we discuss a few of the common access methods for MGI content. Table 2 provides a high-level look at more of the primary types of data found in MGI, and the value added that MGI provides in integrating and curating these data. Table 3 provides a snapshot of MGI statistics that are updated weekly on the MGI website (Mouse Genome Informatics 2016l). An expanded list of statistics can be found linked on the MGI homepage (Mouse Genome Informatics 2016o). Users are encouraged to explore MGI, review the tutorial and informational items on the MGI homepage (Mouse Genome Informatics 2016l), and contact MGI User Support at mgi-help@jax.org or use the “Contact Us” link in the navy blue navigation bar at the top of MGI webpages for assistance with using MGI or with questions about MGI content or methods. In addition, readers should consult two recent reviews that cover other aspects of MGI data and access methods (Eppig et al. 2015c, 2017) and a retrospective on MGI (Eppig et al. 2015b).
Table 2.
Overview of types of data found in MGI
| Data type | Description and URL link |
|---|---|
| Unified nonredundant genome feature catalog | MGI’s fjoin algorithm (Richardson 2006) is used to computationally compare genome assembly predictions from NCBI (NCBI Resource Coordinators 2016) and Ensembl (Aken et al. 2017) and from gene models curated by Havana/VEGA (Harrow et al. 2014). Results are integrated with MGI’s genome features. Curators from MGI and these groups collaboratively analyze and resolve conflicting data on an ongoing basis. The process of creating the unified non-redundant genome feature catalog is described in Zhu et al. (2015). Also see MGI Gene Query (http://www.informatics.jax.org/marker, Mouse Genome Informatics (MGI) 2016j) and the downloadable Marker Coordinates report (Mouse Genome Informatics (MGI) 2016m). |
| Strain-specific genome features | MGI captures instances of genes that are not present in the C57BL/6J reference sequence genome, but are found in other mouse strains (Eppig et al. 2012). A classical example is Ren2 (renin 2, Piccini et al. 1982). A number of inbred strains carry this duplicated gene, but it is absent in C57BL/6J (see MGI’s Ren2 gene page (Mouse Genome Informatics (MGI) 2016i). |
| Associating genes to sequences | MGI, NCBI, Ensembl, and Havana/VEGA co-curate the mapping of nucleotide sequences to their corresponding genes; for protein sequences the co-curation effort includes MGI, UniProt KB, and Protein Ontology groups. See NCBI’s information for CCDS (NCBI CCDS Database 2016). |
| Nomenclature for mouse (genes, genome features, strains, chromosome aberrations) | MGI maintains the website for the International Committee on Standardized Genetic Nomenclature for Mice and Rats online (Mouse Genome Informatics (MGI) 2016p). MGI implements the committee’s guidelines in assigning new or revised nomenclature. Nomenclature is coordinated with HGNC (Human Gene Nomenclature Committee, Gray et al. 2015) and RGD (Rat Genome Database, Shimoyama et al. 2015) to maximize co-assignment of orthologous gene names. |
| Mouse-Human Orthologs | MGI currently uses a hybrid orthology representation, where ortholog calls from NCBI’s Homologene (NCBI Resource Coordinators 2016) and HGNC’s Comparison of Ortholog Prediction (HCOP, Eyre et al. 2007) are compared by a rule-based algorithm to maximize human-mouse ortholog accuracy. These results are used widely in MGI where human-mouse data are compared (Dolan et al. 2015). |
| Gene function data—annotations to mouse genes and gene products | MGI is the authoritative source for Gene Ontology (GO) annotations for mouse (The Gene Ontology Consortium 2017). This dataset is available from the MGI website and downloadable from the server, and is provided to the Gene Ontology Consortium website for display with GO data from other species. MGI staff curate GO terms to mouse genes/gene products and integrate efforts from UniProt KB/GOA (The UniProt Consortium 2017). See the Gene Function Topic link (Mouse Genome Informatics (MGI) 2016i) and the GO download report (Mouse Genome Informatics (MGI) 2016k). |
| Gene Expression data | Data for endogenous gene expression during mouse development is emphasized, for both wild-type and mutant genotypes, and covering a wide range of expression assay types. Supporting images are included. Integration with genomic and phenotype data in MGI provides powerful query capability. (Ringwald and Eppig 2011). Key references for how best to use the expression data web interfaces include Finger et al. (2015) and Smith et al. (2015). See also the Gene Expression Topic link (Mouse Genome Informatics (MGI) 2016e). |
| Gene Expression Literature Index | For rapid user access, gene expression data for mouse development is indexed by curators to capture the genes, ages, and assay types described for each scientific publication. Users can view a high-level overview and rapidly access data details. See the Gene Expression Literature search (Mouse Genome Informatics (MGI) 2016f). |
| Recombinase/cre data | MGI maintains data on the recombinase containing transgenes and knock-in alleles used for conditional mutagenesis, capturing both the descriptions of those alleles, the genotypes used in experimentation, and the results (www.creportal.org, CrePortal 2016). Important for researchers is the integration of data on the site specificity for cre activity, as off-target expression can significantly affect experimental interpretation (Heffner et al. 2012). |
| Genetic variants and mutations | MGI maintains a comprehensive catalog of mutant alleles, including those spontaneously occurring, induced, or genetically engineered. Naturally occurring variants, including SNPs and QTL also are captured (Bult et al. 2013; Eppig et al. 2007, 2012, 2015a). Search forms for mutant alleles (Mouse Genome Informatics (MGI) 2016a) and SNPs (Mouse Genome Informatics (MGI) 2016r) allow flexible searching for a variety of parameters. |
| Phenotypes | Phenotypes are associated with the genotypes (allelic composition and strain background) in which they were observed. MGI gathers phenotypic data from large-scale programs such as Europhenome, German Mouse Clinic, Sanger Institute Mouse Genetics Program, the IMPC (and others), as well as incorporating researchers’ data submissions, and through curation of scientific publications (Bello et al. 2015; Bult et al. 2016; Eppig et al. 2015a, 2017). Phenotype data in can be found in MGI by choosing the Phenotypes and Mutant Alleles topic area on the MGI Homepage (Mouse Genome Informatics (MGI) 2016q). |
| Mouse strains available worldwide, the International Mouse Strain Resource (IMSR) | MGI maintains a composite mouse strains catalog, the International Mouse Strain Resource (www.findmice.org, International Mouse Strain Resource (IMSR) 2016) that consolidates the holdings of mouse resource repositories worldwide, including mice available in various states (live, cryopreserved embryos and sperm, ES cell lines) (Eppig et al. 2015b). Data are released weekly to update repository holdings as they change. The website allows searching by strain name, gene or allele symbol or name, as well as by strain type (e.g., inbred strain, congenic strain), allele type (e.g., insertion, targeted mutation), specific repository or geographic region. More than 38,600 mouse resources and nearly 200,000 ES cell lines from 20 repositories and consortia, representing 47 individual repositories are included. |
| Tumor models | The tumor model data offering within the MGI Resource (Mouse Models of Human Cancer: The Mouse Tumor Biology Database (MTB) 2016) includes data on mouse models of cancer from studies on spontaneous and induced tumors in genetically defined mice (inbred or genetically modified), accompanied by the tumor frequencies observed, diagnoses and metastases, germline and somatic genome information, and pathology reports and images. Data on patient derived xenograft (PDX) mice include diagnoses, images, and, genomic properties of the tumors (Bult et al. 2015). |
| Human disease models | The Human-Mouse: Disease Connection (HMDC) displays integrated data curated in MGI on mouse genotypes and phenotypes and the human diseases that are modeled with the associations between human genes and human diseases from OMIM and human phenotypes from Human Phenotype Ontology. A grid view displays human and mouse orthologs with their comparative phenotype and disease associations, allowing users to view direct orthologous gene disease models, as well as examining potential mouse models and potential human causative gene associations. (Bello and Eppig 2016; Bello et al. 2015; Eppig et al. 2015d, 2017; www.diseasemodel.org, Human-Mouse: Disease Connection (HMDC) 2016). |
Table 3.
MGI content summary December 2016*
| MGI data type (number of mouse…) | Number |
|---|---|
| Genes with protein sequences | 24,193 |
| Genes/genome features with nucleotide sequences | 48,663 |
| Genes with human orthologs | 17,091 |
| Genes with rat orthologs | 18,513 |
| Genes with Gene Ontology (GO) annotations | 24,218 |
| Gene Ontology (GO) annotations (Total) | 301,832 |
| Mutant alleles in mice | 50,035 |
| Genes with targeted mutations | 16,841 |
| Recombinase (cre) transgenes and knock-ins | 2626 |
| QTL | 5672 |
| Genotypes with Mammalian Phenotype Ontology (MP) annotations | 59,133 |
| Mammalian Phenotype Ontology (MP) annotations (Total) | 306,913 |
| Models (genotypes) associated with human diseases | 5128 |
| Genes with expression assay results | 14,333 |
| Expression assay results (Total) | 76,990 |
| Tumor frequency records | 71,000 |
| Tumor Images | >5800 |
| Clinical, pathological, expression, and genomic data from PDX models | 450 |
| References in the MGI bibliography | 231,595 |
*For additional MGI statistics, see Mouse Genome Informatics (MGI) 2016o.
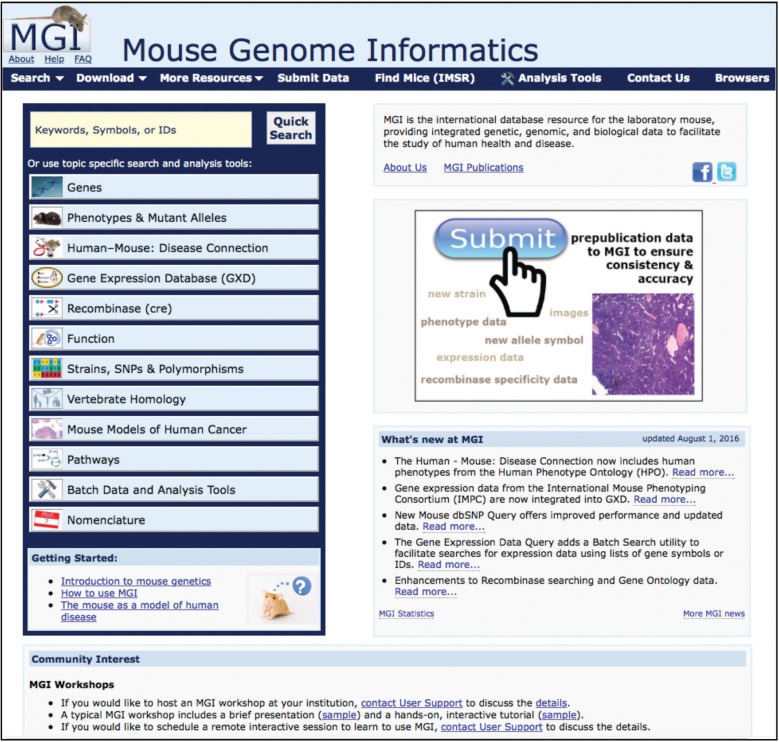
Guide to the MGI Homepage (Mouse Genome Informatics (MGI) 2016l)
The MGI homepage (Figure 3) is a jumping-off platform to the various data sections in MGI. In addition links are provided to “About” pages, “Help,” “FAQ,” tutorials and introductory materials, MGI publications, as well as to “What’s new” in MGI and various community news items.
Figure 3.
MGI homepage (www.informatics.jax.org) (Mouse Genome Informatics (MGI) 2016l) The MGI Homepage is the gateway to data, tools, news, and release notes. The major sections of the Homepage allow users to (1) do a “Quick Search” to broadly sweep their area of interest, (2) jump to topic area pages, (3) visit the tutorial and introduction areas, (4) use the navy blue navigation bar to find specific search forms or tools, and (5) read news about MGI and follow informational links for MGI statistics, publications, about pages, help documents, and FAQs.
Accessing MGI data via the “Quick Search”
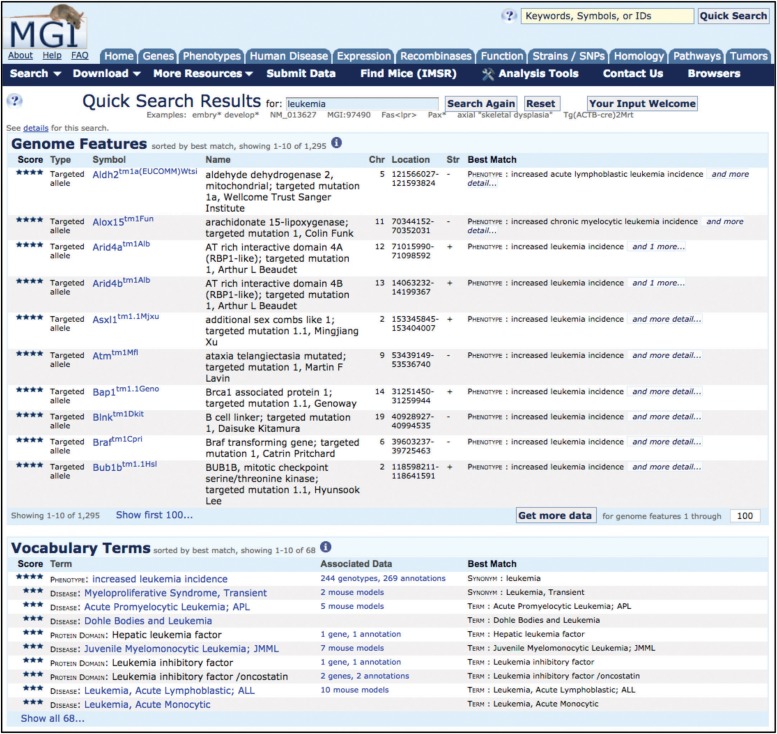
Analysis of web logs for the MGI website clearly shows that the majority of users use the “Quick Search” over all other search tools that MGI provides (Figure 4). The Quick Search is accessible on virtually all MGI web pages and can initiate your exploration of MGI, or quickly allow the user to jump to a new data search of interest from any MGI page being viewed.
Figure 4.
Quick Search results page example. The Quick Search tool is found at the top left of the MGI homepage and on other MGI pages in the upper right corner. In this example, leukemia was entered as the search term, resulting in 1295 results in the Genome Features section and 68 results in the Vocabulary Terms section. From the Quick Search Results page, the symbols in the Genome Features section link to the relevant MGI Gene Detail page or Mutant Allele Detail page. In the Vocabulary Terms section, the term links to the relevant term page: for phenotype terms, to the Mammalian Phenotype Ontology Browser; for disease terms, to the MGI Human Disease and Mouse Model Detail Page; and for a protein family term to the MGI Protein Superfamily Detail page. The Associated Data column links to the underlying annotations and brings the user to the relevant annotation detail page.
For new users, the Quick Search provides a fast entry point into a MGI topical area without the need to be familiar with the MGI query forms that provide more precise results with multi-parameter searches (e.g., leukemia). The Quick Search results are broad, including all genes and alleles where the term entered is included in the current or former nomenclature or associated to the gene via annotations. In this example, 1295 genome features were returned, including genome features with annotations to a phenotype term that “contains” leukemia, such as Alox15tm1Fun, as well as genome features whose names included “leukemia,” such as Pmv54, polytropic murine leukemia virus 54. In addition, 68 hits to leukemia appear in the vocabulary term section, which shows annotations to leukemia in MGI disease terms, phenotype terms, protein domain terms, and functional (GO) terms.
For experienced users the Quick Search can quickly identify your data of choice by using a simple, but narrowly defined term (e.g., “Bloom Syndrome” using quotes for “exact match” or “Bmp6”). Each of these searches retrieves only six genome features and three vocabulary term hits (data as of Dec. 30, 2016). Keep in mind that many such simple terms may return a large number of results, for example, the term “anemia” returns 1067 genome feature matches and 129 vocabulary matches (as of 30 Dec. 2016), but can be considerably reduced by using the more specific term “macrocytic anemia,” with quotes, which returned 48 genome feature matches and 3 vocabulary terms.
Using Topic-Specific Searching and Tools
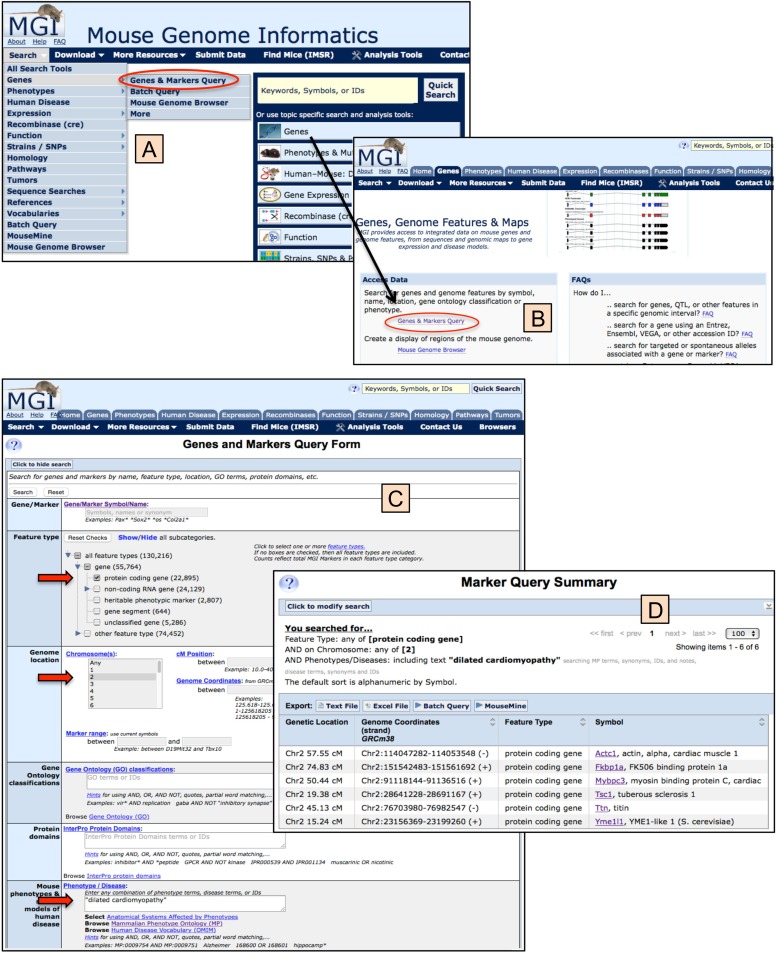
On the MGI homepage, in the left column, are a series of buttons providing access to topic areas within MGI, including Genes; Phenotypes and Mutant Alleles; Human-Mouse: Disease Connection; Gene Expression Database (GXD); Recombinase (cre); Function; Strains; Strains, SNPs and Polymorphisms; Vertebrate Homology; Mouse Models of Human Cancer; Pathways; Batch Data and Analysis Tools; and Nomenclature. Clicking on any of these buttons takes the user to a subsection of MGI optimized for information and searching for that particular topic. Each search form within a topic behaves in a similar manner and takes advantage of the integration of MGI data. Figure 5 shows a topic-specific search form and results for the genes and genome feature area of MGI. Note that within each topical area and almost all MGI pages, the header section of the page continues to provide full access to different sections of the database: to the Quick Search at the upper right; via the topical area tabs across the top of the page; and via pull-down menus available in the navy blue navigation bar.
Figure 5.
Genes and markers query form and results example. Panels A and B show two methods for accessing the Genes and Markers Query Form and illustrate the general principle for accessing other Query Forms within MGI. Beginning on the MGI homepage (www.informatics.jax.org) (Mouse Genome Informatics (MGI) 2016l), use either the pull-down “Search” (panel A) navigating to the Genes submenu and the Genes & Marker Query (circled) or click on the “Genes” topical area button that leads to the Genes, Genome Features & Maps subpage (panel B), where the Genes & Markers Query (circled) also can be selected. On the Genes and Markers Query Form (Mouse Genome Informatics (MGI) 2016j), panel C users may specify as one or more search parameters as desired. In this case, the following parameters were chosen: Feature type = protein coding gene AND Genome location = Chromosome 2 AND Mouse phenotype term = “dilated cardiomyopathy” (enclosed in parentheses for an exact match against the two-word term). The Results page is shown in panel D, where six genes satisfied the query. Note the “You searched for…” feature at the top, which tells the user what parameters were used, and the “Export” utilities for downloading or forwarding the results for additional analysis.
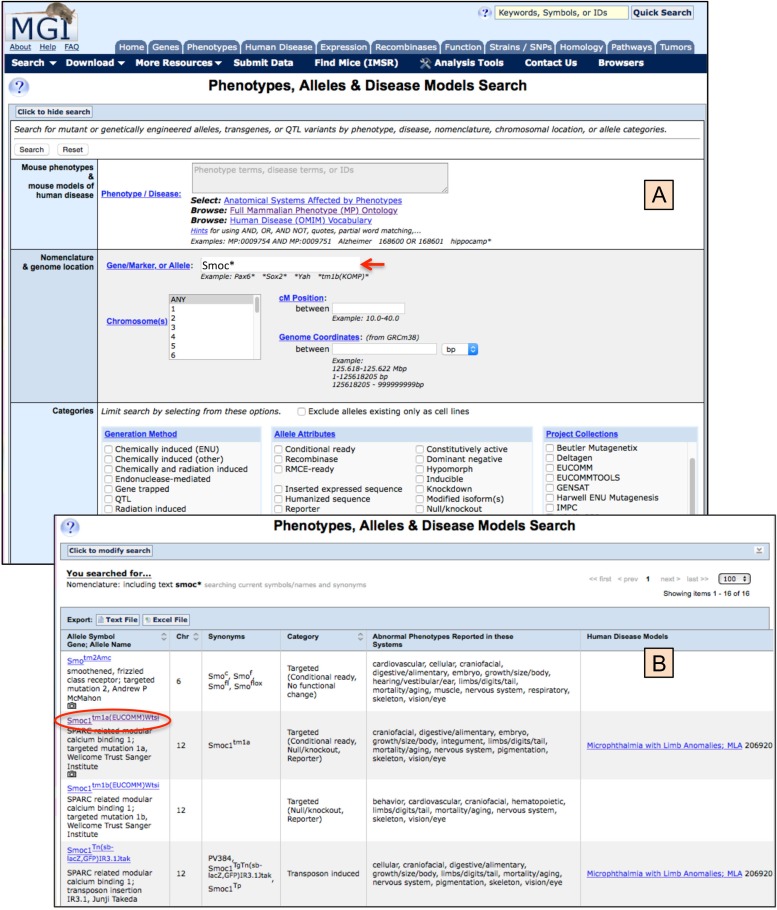
Figure 6 illustrates access to phenotype and mutant allele data. The search form for Phenotypes and Mutant Alleles, with an example summary results is shown in Figure 6.1, with further access to finer data levels shown in Figure 6.2. While this figure does not show all the possible data details that can be accessed, it does illustrate the principle that one is able to drill down to multiple levels of finer data detail. The reader is referred to the “How to use MGI” link (http://www.informatics.jax.org/mgihome/other/homepage_usingMGI.shtml) on the MGI Homepage beneath the listing of topical areas for additional navigation methods (Mouse Genome Informatics (MGI) 2016g).
Figure 6.1 and 6.2.
Phenotypes, alleles, and disease models search form and results example. For navigation to the Search Form, see the Genes and Markers access method illustrated in Figure 5, panels A and B. The same overall method utilizing the Search pull-down menu or the topical area buttons on the MGI homepage are used throughout the MGI system. Figure 6.1 shows the phenotypes, alleles, and disease model search form (panel A) where, in this example, only a single search parameter was entered into the gene/marker, or allele field. The entry, Smoc*, uses a wildcard and will return all alleles beginning with Smoc…, as well as any synonyms or allele names containing a term beginning Smoc… Additional parameters may be specified in the search, including phenotype and disease terms, genome location, allele generation methods, allele attributes, and/or alleles that were created as part of large projects. There also is an option to exclude alleles if they only exist in cell lines. Panel B show the results of this search, with 4 of the 16 alleles returned shown here (see upper right of the screen for the allele count). For each allele, its symbol, name, synonyms, chromosome assignment, category of mutant generation, and attributes, systems showing abnormal phenotypes and human disease models are provided. Links are provided from the allele symbol (circled) to the Allele Detail Page (Figure 6.2) and from the human disease to the Disease Ontology browser.
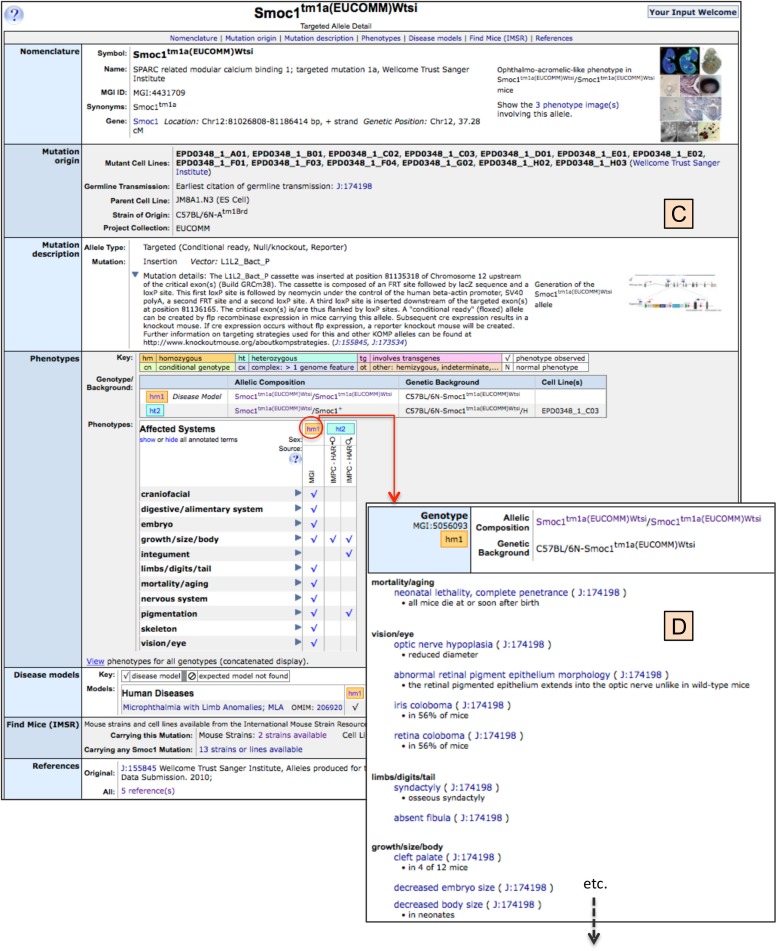
Figure 6.2.
In panel C the Smoc1tm1a(EUCOMM)Wtsi allele detail page is shown, containing data on the nomenclature and location of the mutant allele, cell lines that contain this mutation, if it has been germline transmitted, its strain of origin, and the project collection that created it. This is followed by a brief description of the mutation itself and its molecular specificity, as known. Images (with additional links to more images and image captions and references) are provided for phenotypic images as well as mutation/vector images, if available. The Phenotypes section of this page shows various genotypes that have been studied with this allele, in this case there are both homozygous and heterozygous genotypes on a C57BL/6N genetic background. The full genotype is always presented, given that genetic background can have significant effect on the phenotypic presentation of mutant genes. A table specifying the anatomical systems in which phenotype was detected is presented, along with the source project, and sex-specific phenotypes, if applicable. It should be noted for each anatomical system, a toggle opens to reveal finer phenotypic detail. Towards the bottom of this detail page are the disease models section showing disease association for this allele, the Find Mice (IMSR) section showing mice available from repositories for the specific mutation being viewed or for all mutations in this gene and linking the user directly to IMSR for information and further access to holding repositories, and finally, a link to all references describing this mutation. Panel D illustrates one of the many links from this Allele Detail Page. Here the link is shown from the genotype “hm1” (homozygous Smoc1tm1a(EUCOMM)Wtsi allele on C57BL/6N) to the finer detail of the phenotypes observed in this genotype. Within panel D, links can be seen from the Mammalian Phenotype Ontology term (e.g., neonatal lethality, complete penetrance) to the Mammalian Phenotype Ontology page (see Figure 2) and to the reference from which this phenotypic data came (e.g., J:174198).
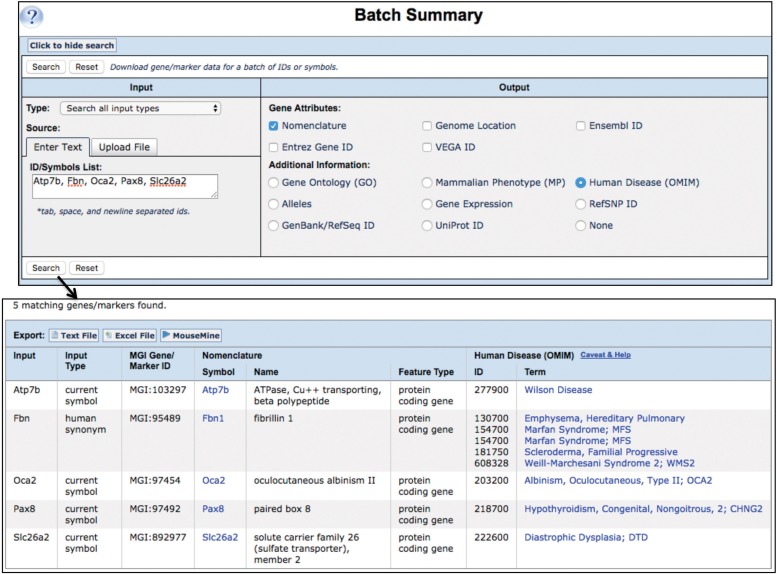
Batch Data and Analysis Tools
Many of the search results pages that are returned from a query include the ability to download the results you are viewing in either text or Excel format, or to forward your results for further analysis to the MGI Batch Query or MouseMine tools. In addition, over 75 data files are generated weekly and available for download, either from the Batch Data and Analysis Tools topical area page (Mouse Genome Informatics (MGI) 2016c) or using the pull-down menu labeled “Download” from the navy blue navigation bar at the top of MGI web pages.
The Batch Query (Bult et al. 2008, 2010; Eppig et al. 2015c, 2017) allows the user to upload a file or enter a set of IDs or gene symbols and selectively retrieve other database IDs (making it an excellent ID translator) as well as selected gene attributes such as phenotypes, human disease, function (GO annotations), expression data, or RefSNP, UniProt, or RefSeq IDs. (Figure 7).
Figure 7.
MGI batch query and results example. The Batch Query (Mouse Genome Informatics (MGI) 2016d) is a quick way to translate IDs between databases and to pull selected data from MGI for further analysis. For navigation to the Batch Query Form, follow the method shown in Figure 5, panels A and B, and use the Search pull-down menu or the topical area buttons on the MGI homepage. In this Batch Query example, a list of gene symbols was entered into the Input box at the upper left: Atp7b, Fbn, Oca2, Pax8, Slc26a2. Note that various IDs can be entered (IDs from MGI, Ensembl, GenBank, UniProt, etc.), or a file of gene symbols or IDs, as well. In the Output box at the upper right, Nomenclature, Genome Location and Human Disease (OMIM) were selected. The results returned are a tabular display of all data matching the request and can be downloaded as text or Excel files or forwarded to MouseMine (Motenko et al. 2015) for further analysis. Links in the Nomenclature Symbol column take the user to the Gene Detail page for that gene; and links in the Disease (OMIM) Term column take the user to the MGI Human Disease and Mouse Model Detail page.
MouseMine (Motenko et al. 2015), a mouse-specific instance using InterMine software (Smith et al. 2012), offers great flexibility for building datasets that are not possible to retrieve using the regular MGI web pages. This tool is particularly useful for bioinformaticians or advanced users for mining data with very specific parameters. Mousemine provides a number of common “prepared” queries as well as the ability for users to build unique queries against MGI data. The functionality available includes uploading, manipulating, and saving results lists, as well as downloading data or forwarding data to Galaxy (Afgan et al. 2016). MouseMine also includes utilities for doing enrichment analyses of your results. The web service API for MGI also resides within MouseMine (Kalderimis et al. 2014).
The Human-Mouse: Disease Connection (HMDC)
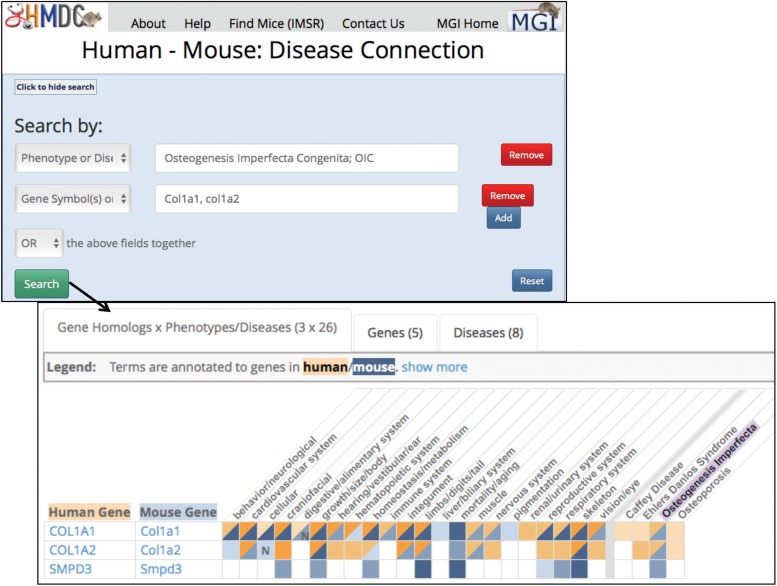
HMDC (Human-Mouse: Disease Connection 2016) supports translational studies by providing access to published and potential mouse models of human disease and supports the discovery of candidate genes and comparisons of phenotypes in mouse models and human patients. HMDC combines the phenotype and disease model data in MGI with integrated human data from OMIM (Online Mendelian Inheritance in Man) (Amberger et al. 2015) and human phenotype data from Human Phenotype Ontology (Köhler et al. 2014, 2017). The web interface allows users to search (using mouse or human data) by gene symbols, names, or IDs; phenotype or disease names or IDs; and genome locations. The query can use one or multiple of these parameters and will accept a file of gene symbols or IDs as well. Figure 8 shows an example search and the result summary returned.
Figure 8.
Human-Mouse Disease Connection (www.diseasemodel.org) (Human-Mouse: Disease Connection (HMDC) 2016) example. For navigation to the Human-Mouse Disease Connection page, follow the method shown in Figure 5, panels A and B, and use the Search pull-down menu or the topical area buttons on the MGI Homepage. In this figure, the top panel shows the HMDC homepage with its facile search form. Users select what they wish to search by from a pull-down list that includes: Gene symbol or ID; Gene name; Phenotype or Disease ID; Phenotype or Disease term; Genome location; or Gene File upload. Once a category and value are entered, the user may choose to add additional search parameters. In this example, the Phenotype/Disease term Osteogenesis Imperfecta Congenita; OIC was selected and an additional search for Gene symbols was selected and entered as: Col1a1, col1a2. Finally the selection was made to “or” these fields together. The lower panel shows the resulting grid display where human and mouse orthologs are shown in rows and phenotypes and disease shown in columns. Blue indicates mouse data; orange indicates human data. The highlighted Osteogenesis Imperfecta column shows both human COL1A1 and COL1A2 and mouse Col1a1 and Col1a2 are associated to the disease. In addition, mice mutant for Smpd3 have been used to model Osteogenesis Imperfecta, suggesting that the human orthologous gene might be a candidate gene for a patient without COL1A1 or COL1A2 mutations. Note also, that in the column for Caffey Disease that there is an association to human COL1A1, but no mouse model has shown this association. In this case, a researcher might want to look at (or engineer) a Col1a1 mutation to create a potential model for Caffey Disease.
The Recombinase (cre) Portal
The CrePortal (CrePortal 2016) facilitates identification of the most suitable cre mouse lines for conditional mutagenesis experiments. It describes over 2620 recombinase containing transgenes and knock-in alleles with detailed molecular information and tissue- and age-specific cre activity. Data on cre-expressing mice have been integrated from individual laboratories and large-scale programs including the NIH Neuroscience Blueprint Cre Driver Network (Tsien 2016), the Allen Institute for Brain Science (Madisen et al. 2010), the Pleiades Promoter project (Portales-Casamar et al. 2010), the JAX Cre Resources project (Heffner et al. 2012), GENSAT (Gerfen et al. 2013), and EUCOMMtools (Friedel et al. 2011). Cre activity data are annotated using the Mouse Anatomy ontologies with images of intended and off-target activity. The CrePortal can be searched by the anatomical structure in which recombinase activity was assayed and/or by the driver used to activate the cre recombinase. The search summary provides a list of drivers, the recombinase-containing allele symbols, associated gene and allele name, allele synonym, a list of tissues in which recombinase activity was detected or not detected, the inducible agent if required, links to references, and links to the International Mouse Strain Resource (IMSR) (International Mouse Strain Resource 2016) for locating those strains available through public repositories. Filters allow users to refine search results. Cre allele symbols are linked to the MGI (Mouse Genome Informatics (MGI) 2016l) phenotypic data pages that provide a more detailed cre activity summary and phenotypic information for genotypes involving the cre transgenes and knock-ins. We encourage you to explore the CrePortal and submit your laboratory’s cre line observations for inclusion in the CrePortal at the MGI online submission forms (Mouse Genome Informatics 2016n). A step-by-step tutorial is available linked from the Recombinase (cre) topic area page.
The IMSR
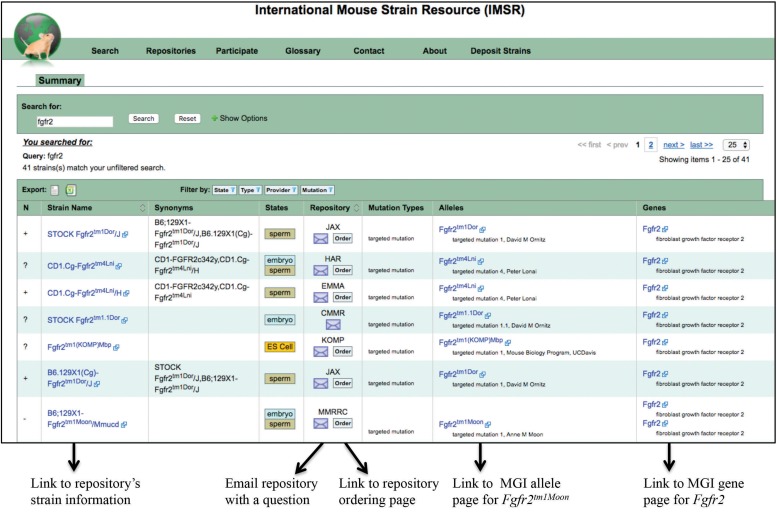
The IMSR (Eppig et al. 2015a; International Mouse Strain Resource 2016) facilitates access to mouse resources and mouse models of human disease that are used for basic and translational research. The unique and pivotal role of IMSR is in unifying information about mouse resource holdings worldwide, including inbred, mutant, and genetically engineered mice maintained as breeding stocks, cryopreserved embryos and gametes, and ES (embryonic stem) cell lines. At the IMSR website (International Mouse Strain Resource 2016) users can search for mouse resources, locate strains for their research, learn details about a strain, order mice from a repository and contact a repository for questions, and link to phenotype and disease model descriptions. (Figure 9).
Figure 9.
International Mouse Strain Resource (www.findmice.org) (International Mouse Strain Resource (IMSR) 2016) example. The link to the IMSR search is found on the navy blue navigation bar appearing near the top of all MGI pages. Here, the results page for an IMSR search for Fgfr2 is shown. The results of this search show 41 strains returned, with each strain row providing links from the strain name to the repository’s strain page, links to email the repository or go to its ordering page, and links to information in MGI about the mutant allele carried and to the gene page. Note that new searches can be initiated from this same page by replacing this gene symbol with other gene symbols or strain designations. A new search including additional options (e.g., strain state (ES cell, embryo, live, etc.), strain type (coisogenic strain, congenic strain, etc.), specific repository of choice, or specific mutations type of choice) can be accessed from the IMSR homepage or by choosing “show options” next to the search box.
Mouse Models of Human Cancer: The Mouse Tumor Biology Database (MTB)
MTB (Mouse Models of Human Cancer: The Mouse Tumor Biology Database 2016) is a semi-independent resource under the MGI umbrella of resources. Developed initially under a National Cancer Institute contract and in conjunction with the Mouse Models for Human Cancer Consortium that was initiated in 1998 (Marks 2009), its goal was and continues to be to provide a comprehensive guide to use of the mouse in understanding the genetic basis of human cancer and informing investigations of novel targets for therapeutic intervention. As are other MGI resources, MTB is an expertly curated resource, utilizing nomenclature and vocabulary standards, and includes data curated from the scientific literature, investigator-submitted data, as well as data from large-scale research efforts, and curated metadata from community resources such as Gene Expression Omnibus (Barrett et al. 2013) and Expression Atlas (Petryszak et al. 2016). MTB includes data about spontaneous and induced tumors in inbred strains and genetically modified mouse strains, as well as data on PDX mouse studies (patient derived xenograft created by implanting patient tumors into immunodeficient or humanized mouse hosts) (Bult et al. 2015).
MTB contains data from spontaneous or endogenously induced tumors from genetically defined mice including tumor classification, incidence and latency, tumor associated QTL, pathology reports, images and genetic changes in the tumor (somatic), and background strain (germline) genomes. The PDX resource enables searches based on tumor type, cancer diagnosis, and genomic properties of the engrafted tumors. The MTB (Mouse Models of Human Cancer: The Mouse Tumor Biology Database 2016) provides online query tools to facilitate cohesive searches and visualization of these varied data, thus enabling the identification of novel mouse models of human cancer and potential therapeutic treatments.
Summary and Future…
The Encyclopedia of the Mouse precursor project of what would become MGI was originally funded by NIH in 1989 as molecular biology was a rising star, and the audacious possibility of sequencing the human genome was being debated and planned (Robert 1989). The initial effort for MGI was to unify many nascent databases (reflecting the first steps in computerizing genetic data) and provide a visual interface that connected various data sources. The mouse/mammalian research community propelled MGI’s advancement by participating in the genome sequencing revolution and developing large-scale mutagenesis and phenotyping projects to enhance studies of gene function. Through many iterations and transformative changes, MGI has succeeded in its goal of providing an integrated gold-standard knowledgebase for basic researchers, translational investigators, and computational biologists.
The National Human Genome Research Institute has released plans to reduce biomedical resource funding for model organism databases in the coming years (see articles by Kaiser (2016) in Science and Hayden (2016) in Nature). In response MGI and the other primary model organisms in the National Human Genome Research Institute’s portfolio have formed a new Alliance for Genome Resources (AGR). Members include MGI, FlyBase (Gramates et al. 2017), Saccharomyces Genome Database (Engel et al. 2016), WormBase (Harris et al. 2014), the Zebrafish Information Network (Howe et al. 2017), Rat Genome Database (Shimoyama et al. 2015), and The Gene Ontology Consortium (The Gene Ontology Consortium 2017).
The immediate goals of the AGR are to study the content, relationships, and operating definitions for data in each resource to determine commonalities and differences that affect integration and cross-referencing between systems and to develop a web portal that allows users to traverse multiple model organism data in a shared system. Ultimately a shared infrastructure for these data will emerge for the common types of data. Species-specific data types and methods will need to be maintained so as not to compromise the functionality and data quality that each model organism database provides to its unique user communities. An announcement of the AGR appears on the Genetics Society of America website (Genetics Society of America (GSA) 2016).
While AGR members have ongoing collaborations that have lasted for many years, for example, in developing GO, in striving for common nomenclatures for gene orthologs, in adopting common tools such as JBrowse (Skinner et al. 2009), and participating in each other’s Advisory Boards, there remain many significant challenges and differences to overcome including curation policies, data acquisition infrastructure, and paradigms for user interactions for searching and visualization of data. MGI’s involvement in AGR will undoubtedly mean more change, but that is not a new paradigm; change is the name of the game in keeping MGI in tune with the ever-evolving science.
Acknowledgments
I gratefully acknowledge the support and dedication of the entire MGI team, former and present. Current MGI members include co-principal investigators Judith A. Blake, Carol J. Bult, James A. Kadin, Martin Ringwald, Joel E. Richardson, Cynthia L. Smith; informatics analysts and curators: Anna Anagnostopoulos, Dale A. Begley, Susan M. Bello, Richard M. Baldarelli, Karen Christie, Harold J. Drabkin, Jacqueline H. Finger, Paul Hale, Terry F. Hayamizu, David P. Hill, Michelle N. Knowlton, Debra M. Krupke, Monica McAndrews, Ingeborg J. McCright, Li Ni, Hiroaki Onda, Jill M. Recla, Deborah J. Reed, Beverly Richards-Smith, Dmitry Sitnikov, Constance M. Smith, Monika Tomczuk, Laurens Wilming, Jinxia Xu, Yunxia Zhu; software developers: Jonathan S. Beal, Olin Blodgett, Jeffrey W. Campbell, Lori E. Corbani, Mary E. Dolan, Peter Frost, Sharon C. Giannatto, Lucie Hutchins, Jill R. Lewis, David B. Miers, Howie Motenko, Steven B. Neuhauser, Kevin R. Stone; user support staff: MeiYee Law, David R. Shaw; and administrative support staff: Nancy E. Butler, Janice E. Ormsby.
Funding
This work was made possible by NIH grants HG000330 and HG002273 from the National Human Genome Research Institute, grant HD062499 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, grant CA089713 from the National Cancer Institute, and grant OD011190 from the Office of the Director, Division of Comparative Medicine.
References
- Accili D. 2004. A note of caution on the Knockout Mouse Project. Nat Genet 36:1132. [DOI] [PubMed] [Google Scholar]
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Juettemann T, Keenan S, Laird MR, Lavidas I, Maurel T, McLaren W, Moore B, Murphy DN, Nag R, Newman V, Nuhn M, Ong CK, Parker A, Patricio M, Riat HS, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Wilder SP, Zadissa A, Kostadima M, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Cunningham F, Yates A, Zerbino DR, Flicek P. 2017. Ensembl 2017. Nucleic Acids Res 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. 2010. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol 477:109–123. [DOI] [PubMed] [Google Scholar]
- Arnold CN, Barnes MJ, Berger M, Blasius AL, Brandl K, Croker B, Crozat K, Du X, Eidenschenk C, Georgel P, Hoebe K, Huang H, Jiang Z, Krebs P, La Vine D, Li X, Lyon S, Moresco EM, Murray AR, Popkin DL, Rutschmann S, Siggs OM, Smart NG, Sun L, Tabeta K, Webster V, Tomisato W, Won S, Xia Y, Xiao N, Beutler B. 2012ENU-induced phenovariance in mice: Inferences from 587 mutations. BMC Res Notes 5:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KCK, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melcher H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. 2004. The Knockout Mouse Project. Nat Genet 36:921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis MH, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Müller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, Rosen B, Rosenthal N, Skarnes B, Stewart AF, Thornton J, Tocchini-Valentini G, Wagner E, Wahli W, Wurst W. 2004. The European dimension for the mouse genome mutagenesis program. Nat Genet 36:925–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avner P, Amar L, Dandolo L, Guénet JL. 1988. Genetic analysis of the mouse using interspecific crosses. Trends Genet 4:18–23. [DOI] [PubMed] [Google Scholar]
- Ayadi A, Birling MC, Bottomley J, Bussell J, Fuchs H, Fray M, Gailus-Durner V, Greenaway S, Houghton R, Karp N, Leblanc S, Lengger C, Maier H, Mallon AM, Marschall S, Melvin D, Morgan H, Pavlovic G, Ryder E, Skarnes WC, Selloum M, Ramirez-Solis R, Sorg T, Teboul L, Vasseur L, Walling A, Weaver T, Wells S, White JK, Bradley A, Adams DJ, Steel KP, Hrabě de Angelis M, Brown SD, Herault Y. 2012. Mouse large-scale phenotyping initiatives: Overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome 23:600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, Wasserman DH, McGuinness OP NIH Mouse Metabolic Phenotyping Center Consortium . 2010. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech 3:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric I, Dear TN. 2007. Optimizing screening and mating strategies for phenotype-driven recessive N-ethyl-N-nitrosourea screens in mice. J Am Assoc Lab Anim Sci 46:44–49. [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. 2013. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello SM, Eppig JT MGI Software Group . 2016. Inferring gene-to-phenotype and gene-to-disease relationships at Mouse Genome Informatics: Challenges and solutions. J Biomed Semant 7:14. [Google Scholar]
- Bello SM, Smith CL, Eppig JT. 2015. Allele, phenotype and disease data at Mouse Genome Informatics: Improving access and analysis. Mamm Genome 26:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JW, Rocha AJ, Basch R. 1986. Restriction length polymorphism in the variable region of the Tcr locus linked to histocompatibility antigen H-8 on murine chromosome 14. Immunogenetics 24:328–330. [DOI] [PubMed] [Google Scholar]
- Birling MC, Gofflot F, Warot X. 2009. Site-specific recombinases for manipulation of the mouse genome. Methods Mol Biol 561:245–263. [DOI] [PubMed] [Google Scholar]
- Bishop KA, Harrington A, Kouranova E, Weinstein EJ, Rosen CJ, Cui X, Liaw L. 2016. Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3 (Bethesda) 6:2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak K, Doe B, Banerjee R, Yang F, Bradley A. 2016. Chromosome engineering in zygotes with CRISPR/Cas9. Genesis 54:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Bürger A, Bult CJ, Bushell W, Collins FS, Desaintes C, Doe B, Economides A, Eppig JT, Finnell RH, Fletcher C, Fray M, Frendewey D, Friedel RH, Grosveld FG, Hansen J, Hérault Y, Hicks G, Hörlein A, Houghton R, Hrabé de Angelis M, Huylebroeck D, Iyer V, de Jong PJ, Kadin JA, Kaloff C, Kennedy K, Koutsourakis M, Kent Lloyd KC, Marschall S, Mason J, McKerlie C, McLeod MP, von Melchner H, Moore M, Mujica AO, Nagy A, Nefedov M, Nutter LM, Pavlovic G, Peterson JL, Pollock J, Ramirez-Solis R, Rancourt DE, Raspa M, Remacle JE, Ringwald M, Rosen B, Rosenthal N, Rossant J, Ruiz Noppinger P, Ryder E, Schick JZ, Schnütgen F, Schofield P, Seisenberger C, Selloum M, Simpson EM, Skarnes WC, Smedley D, Stanford WL, Francis Stewart A, Stone K, Swan K, Tadepally H, Teboul L, Tocchini-Valentini GP, Valenzuela D, West AP, Yamamura KI, Yoshinaga Y, Wurst W. 2012. The mammalian gene function resource: The international knockout mouse consortium. Mamm Genome 23:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. 2004. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6:7–28. [DOI] [PubMed] [Google Scholar]
- Brommage R, Liu J, Hansen GM, Kirkpatrick LL, Potter DG, Sands AT, Zambrowicz B, Powell DR, Vogel P. 2014. High-throughput screening of mouse gene knockouts identifies established and novel skeletal phenotypes. Bone Res 28:14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Lad HV, Green ED, Gkoutos G, Gates H, Hrabé de Angelis M, Consortium TE. 2006. EUMORPHIA and the European mouse phenotyping resource for standardised screens (EMPReSS) In: Hrabé de Angelis M, Chambon P, Brown S, eds. Standards of Mouse Model Phenotyping. Weinheim: Wiley-VCH; p 309–320. [Google Scholar]
- Brown SD, Moore MW. 2012. a. The International Mouse Phenotyping Consortium: Past and future perspectives on mouse phenotyping. Mamm Genome 23:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. 2012. b. Towards an encyclopaedia of mammalian gene function: The International Mouse Phenotyping Consortium. Dis Model Mech 5:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Nolan PM. 1998. Mouse mutagenesis-systematic studies of mammalian gene function. Hum Mol Genet 7:1627–1633. [DOI] [PubMed] [Google Scholar]
- Bull KR, Rimmer AJ, Siggs OM, Miosge LA, Roots CM, Enders A, Bertram EM, Crockford TL, Whittle B, Potter PK, Simon MM, Mallon AM, Brown SD, Beutler B, Goodnow CC, Lunter G, Cornall RJ. 2013. Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS Genet 9:e1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE Mouse Genome Database Group . 2013. The mouse genome database: Genotypes, phenotypes, and models of human disease. Nucleic Acids Res 41:D885–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE Mouse Genome Database Group . 2016. The Mouse Genome Database 2016. Nucleic Acids Res 44:D840–D847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA Mouse Genome Database Group . 2008. The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Res 36:D724–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT Mouse Genome Database Group . 2010. The Mouse Genome Database: Enhancements and updates. Nucleic Acids Res 38:D586–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Krupke DM, Begley DA, Richardson JE, Neuhauser SB, Sundberg JP, Eppig JT. 2015. Mouse Tumor Biology (MTB): A database of mouse models for human cancer. Nucleic Acids Res 43:D818–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. 2010. Targeted genome modification in mice using zinc-finger nucleases. Genetics 186:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. 2014. Genome engineering with targetable nucleases. Annu Rev Biochem 83:409–439. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Gatti DM, Munger SC, Svenson KL. 2012. The diversity outbred mouse population. Mamm Genome 23:713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT, Goldowitz D, Takahashi JS, Vitaterna MH, Siepka SM, Peters LL, Frankel WN, Carlson GA, Rossant J, Nadeau JH, Justice MJ. 2004. Implementing large-scale ENU mutagenesis screens in North America. Genetica 122:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Finnell RH, Rossant J, Wurst W. 2007. A new partner for the international knockout mouse consortium. Cell 129:235. [DOI] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. 1998. New goals for the U.S. Human Genome Project: 1998–2003. Science 282:682–689. [DOI] [PubMed] [Google Scholar]
- Connelly JP, Barker JC, Pruett-Miller S, Porteus MH. 2010. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol Ther 18:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Gilbert DJ, Jenkins NA, Nadeau JH, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Steen RG, Lincoln SE, Weaver A, Joyce DC, Merchant M, Wessel M, Katz H, Stein LD, Reeve MP, Daly MJ, Dredge RD, Marquis A, Goodman N, Lander ES. 1993. b. Genome maps IV 1993. Wall chart. Science 262:67–82. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. 1991. Development and applications of a molecular genetic linage map of the mouse genome. Trends Genet 7:113–118. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, Stein LD, Nadeau JH, Lander ES. 1993. a. A genetic linkage map of the mouse: Current applications and future prospects. Science 262:57–66. [DOI] [PubMed] [Google Scholar]
- CrePortal 2016. CrePortal Homepage. Available online (www.creportal.org), accessed on December 1, 2016.
- Cuénot L. 1902. La loi de Mendel et l’hérédité de la pigmentation chez les souris. Arch Zool Exp Gen Ser 3:27–30. [Google Scholar]
- Dandoy F, De Maeyer E, Bonhomme F, Guenet JL, De Maeyer-Guignard J. 1985. Segregation of restriction fragment length polymorphism in an interspecies cross of laboratory and wild mice indicates tight linkage of the murine IFN-beta gene to the murine IFN-alpha genes. J Virol 56:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center , MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bućan M, Murray SA. 2016. High-throughput discovery of novel developmental phenotypes Nature 537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC, Lander ES. 1992. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O’Connor TJ. 1996. A comprehensive genetic map of the mouse. Nature 380:149–152. [DOI] [PubMed] [Google Scholar]
- Dolan ME, Baldarelli RM, Bello SM, Ni L, McAndrews MS, Bult CJ, Kadin JA, Richardson JE, Ringwald M, Eppig JT, Blake JA. 2015. Orthology for comparative genomics in the Mouse Genome Database. Mamm Genome 26:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran AG, Wong K, Flint J, Adams DJ, Hunter KW, Keane TM. 2016. Deep genome sequencing and variation analysis of 13 inbred mouse strains defines candidate phenotypic alleles, private variation and homozygous truncating mutations. Genome Biol 17:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E Genome editing . 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. [DOI] [PubMed] [Google Scholar]
- Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. 2005. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol 6:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Weng S, Binkley G, Paskov K, Song G, Cherry JM. 2016. From one to many: Expanding the Saccharomyces cerevisiae reference genome panel. Database (Oxford) Mar 17:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE members of the Mouse Genome Database Group . 2007. The Mouse Genome Database (MGD): New features facilitating a model system. Nucleic Acids Res 35:D630–D637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE Mouse Genome Database Group . 2012. The Mouse Genome Database (MGD): Comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res 40:D881–D886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE The Mouse Genome Database Group . 2015. a. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res 43:D726–D736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Motenko H, Richardson JE, Richards-Smith B, Smith CL. 2015. b. The International Mouse Strain Resource (IMSR): Cataloging worldwide mouse and ES cell line resources. Mamm Genome 26:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Richardson JE, Kadin JA, Ringwald M, Blake JA, Bult CJ. 2015. c. Mouse Genome Informatics (MGI): Reflecting on 25 years. Mamm Genome 26:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Richardson JE, Kadin JA, Smith CL, Blake JA, Bult CJ and the MGD Team . 2015. d. Mouse Genome Database: From sequence to phenotypes and disease models. Genesis 53:458–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Smith CL, Blake JA, Ringwald M, Kadin JA, Richardson JE, Bult CJ. 2017. Mouse Genome Informatics (MGI): Resources for mining mouse genetic, genomic, and biological data in support of primary and translational research. Methods Mol Biol 1488:47–73. [DOI] [PubMed] [Google Scholar]
- Eyre TA, Wright MW, Lush MJ, Bruford EA. 2007. HCOP: A searchable database of human orthology predictions. Brief Bioinform 8:2–5. [DOI] [PubMed] [Google Scholar]
- Farrell CM, O’Leary NA, Harte RA, Loveland JE, Wilming LG, Wallin C, Diekhans M, Barrell D, Searle SM, Aken B, Hiatt SM, Frankish A, Suner MM, Rajput B, Steward CA, Brown GR, Bennett R, Murphy M, Wu W, Kay MP, Hart J, Rajan J, Weber J, Snow C, Riddick LD, Hunt T, Webb D, Thomas M, Tamez P, Rangwala SH, McGarvey KM, Pujar S, Shkeda A, Mudge JM, Gonzalez JM, Gilbert JG, Trevanion SJ, Baertsch R, Harrow JL, Hubbard T, Ostell JM, Haussler D, Pruitt KD. 2014. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res 42:D865–D872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder MR. 1980. Biochemical and developmental genetics of isozymes in the mouse, Mus musculus. Isozymes Curr Top Biol Med Res 4:1–68. [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Xu J, Eppig JT, Kadin JA, Richardson JE, Ringwald M. 2015. The mouse gene expression database: New features and how to use them effectively. Genesis 53:510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel RH, Wurst W, Wefers B, Kühn R. 2011. Generating conditional knockout mice. Methods Mol Biol 693:205–231. [DOI] [PubMed] [Google Scholar]
- García-García MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, Huangfu D, Rakeman AS, Lee JD, Feinberg EH, Timmer JR, Anderson KV. 2005. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA 102:5913–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Otín AL, Guillou F. 2006. Mammalian genome targeting using site-specific recombinases. Front Biosci 11:1108–1136. [DOI] [PubMed] [Google Scholar]
- Gates H, Mallon AM, Brown SD EUMODIC Consortium . 2011. High-throughput mouse phenotyping. Methods 53:394–404. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetics Society of America (GSA) 2016. Genetics Society of America website: Announcing the Alliance of Genome Resources. Available online (http://genestogenomes.org/model-organism-databases-join-forces-announcing-the-alliance-of-genome-resources/), accessed on December 1, 2016.
- Gennequin B, Otte DM, Zimmer A. 2013. CRISPR/Cas-induced double-strand breaks boost the frequency of gene replacements for humanizing the mouse Cnr2 gene. Biochem Biophys Res Commun 441:815–819. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. 2013. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80:1368–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierut JJ, Jacks TE, Haigis KM. 2014. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb Protoc 2014:339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldowitz D, Frankel WN, Takahashi JS, Holtz-Vitaterna M, Bult C, Kibbe WA, Snoddy J, Li Y, Pretel S, Yates J, Swanson DJ. 2004. Large-scale mutagenesis of the mouse to understand the genetic bases of nervous system structure and function. Brain Res Mol Brain Res 132:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P the FlyBase Consortium . 2017. FlyBase at 25: Looking to the future. Nucleic Acids Res 45:D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. 2015. genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43:D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. 2014. WormBase 2014: New views of curated biology. Nucleic Acids Res 42:D789–D793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow JL, Steward CA, Frankish A, Gilbert JG, Gonzalez JM, Loveland JE, Mudge J, Sheppard D, Thomas M, Trevanion S, Wilming LG. 2014. The Vertebrate Genome Annotation browser 10 years on. Nucleic Acids Res 42:D771–D779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Baldock RA, Ringwald M. 2015. Mouse anatomy ontologies: Enhancements and tools for exploring and integrating biomedical data. Mamm Genome 26:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Wicks MN, Davidson DR, Burger A, Ringwald M, Baldock RA. 2013. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Semant 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC. 2016. Funding for model-organism databases in trouble. Nature; doi:10.1038/nature.2016.20134 Available online (http://www.nature.com/news/funding-for-model-organism-databases-in-trouble-1.20134), accessed on December 1, 2016. [Google Scholar]
- Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA. 2012. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 3:1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. 2002. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet 30:185–189. [DOI] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Eagle A, Fashena D, Frazer K, Kalita P, Mani P, Martin R, Moxon ST, Paddock H, Pich C, Ramachandran S, Ruzicka L, Schaper K, Shao X, Singer A, Toro S, Van Slyke C, Westerfield M. 2017. The Zebrafish Model Organism Database: New support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acids Res 45:D758–D768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabé de Angelis M, Balling R. 1998. Large scale ENU screens in the mouse: Genetics meets genomics. Mutat Res 400:25–32. [DOI] [PubMed] [Google Scholar]
- Hrabé de Angelis M, Nicholson G, Selloum M, White JK, Morgan H, Ramirez-Solis R, Sorg T, Wells S, Fuchs H, Fray M, Adams DJ, Adams NC, Adler T, Aguilar-Pimentel A, Ali-Hadji D, Amann G, André P, Atkins S, Auburtin A, Ayadi A, Becker J, Becker L, Bedu E, Bekeredjian R, Birling MC, Blake A, Bottomley J, Bowl MR, Brault V, Busch DH, Bussell JN, Calzada-Wack J, Cater H, Champy MF, Charles P, Chevalier C, Chiani F, Codner GF, Combe R, Cox R, Dalloneau E, Dierich A, Di Fenza A, Doe B, Duchon A, Eickelberg O, Esapa CT, Fertak LE, Feigel T, Emelyanova I, Estabel J, Favor J, Flenniken A, Gambadoro A, Garrett L, Gates H, Gerdin AK, Gkoutos G, Greenaway S, Glasl L, Goetz P, Da Cruz IG, Götz A, Graw J, Guimond A, Hans W, Hicks G, Hölter SM, Höfler H, Hancock JM, Hoehndorf R, Hough T, Houghton R, Hurt A, Ivandic B, Jacobs H, Jacquot S, Jones N, Karp NA, Katus HA, Kitchen S, Klein-Rodewald T, Klingenspor M, Klopstock T, Lalanne V, Leblanc S, Lengger C, le Marchand E, Ludwig T, Lux A, McKerlie C, Maier H, Mandel JL, Marschall S, Mark M, Melvin DG, Meziane H, Micklich K, Mittelhauser C, Monassier L, Moulaert D, Muller S, Naton B, Neff F, Nolan PM, Nutter LM, Ollert M, Pavlovic G, Pellegata NS, Peter E, Petit-Demoulière B, Pickard A, Podrini C, Potter P, Pouilly L, Puk O, Richardson D, Rousseau S, Quintanilla-Fend L, Quwailid MM, Racz I, Rathkolb B, Riet F, Rossant J, Roux M, Rozman J, Ryder E, Salisbury J, Santos L, Schäble KH, Schiller E, Schrewe A, Schulz H, Steinkamp R, Simon M, Stewart M, Stöger C, Stöger T, Sun M, Sunter D, Teboul L, Tilly I, Tocchini-Valentini GP, Tost M, Treise I, Vasseur L, Velot E, Vogt Weisenhorn D, Wagner C, Walling A, Wattenhofer-Donze M, Weber B, Wendling O, Westerberg H, Willershäuser M, Wolf E, Wolter A, Wood J, Wurst W, Yildirim AÖ, Zeh R, Zimmer A, Zimprich A EUMODIC Consortium. , Holmes C, Steel KP, Herault Y, Gailus-Durner V, Mallon AM, Brown SD. 2015. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet 47:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabé de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25:444–447. [DOI] [PubMed] [Google Scholar]
- Human-Mouse: Disease Connection (HMDC) 2016. The HMDC homepage and search form. Available online (www.diseasemodel.org), accessed on December 1, 2016.
- Hutton JJ, Coleman DL. 1969. Linkage analyses using biochemical variants in mice. II. Levulinate dehydratase and autosomal glucose 6-phosphate dehydrogenase. Biochem Genet 3:517–523. [DOI] [PubMed] [Google Scholar]
- Hutton JJ, Roderick TH. 1970. Linkage analyses using biochemical variants in mice. III. Linkage relationships of eleven biochemical markers. Biochem Genet 4:339–350. [DOI] [PubMed] [Google Scholar]
- International Knockout Mouse Consortium (IKMC) 2016. IKMC about page. Available online (https://www.mousephenotype.org/about-ikmc), accessed on December 1, 2016.
- International Mouse Knockout Consortium Collins FS, Rossant J, Wurst W. 2007. A mouse for all reasons. Cell 128:9–13. [DOI] [PubMed] [Google Scholar]
- International Mouse Phenotyping Consortium (IMPC) 2016. IMPC homepage. Available online (www.impc.org), accessed on December 1, 2016.
- International Mouse Strain Resource (IMSR) 2016. IMSR homepage. Available online (www.findmice.org), accessed on December 1, 2016.
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. 1999. Mouse ENU mutagenesis. Hum Mol Genet 8:1955–1963. [DOI] [PubMed] [Google Scholar]
- Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, Hu F, Smith R, Stěpán R, Sullivan J, Micklem G. 2014. InterMine: Extensive web services for modern biology. Nucleic Acids Res 42:W468–W472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. 2016. Funding for key data resources in jeopardy. Science 351:14 Available online (http://science.sciencemag.org/content/351/6268/14.full), accessed on December 1, 2016. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM. 2015. Disease Ontology 2015 update: An expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 43:D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, Fitzpatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jähn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Vooren SV, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, Leeuw ND, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. 2014. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res 42:D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, Baynam G, Bello SM, Boerkoel CF, Boycott KM, Brudno M, Buske OJ, Chinnery PF, Cipriani V, Connell LE, Dawkins HJ, DeMare LE, Devereau AD, de Vries BB, Firth HV, Freson K, Greene D, Hamosh A, Helbig I, Hum C, Jähn JA, James R, Krause R, F Laulederkind SJ, Lochmüller H, Lyon GJ, Ogishima S, Olry A, Ouwehand WH, Pontikos N, Rath A, Schaefer F, Scott RH, Segal M, Sergouniotis PI, Sever R, Smith CL, Straub V, Thompson R, Turner C, Turro E, Veltman MW, Vulliamy T, Yu J, von Ziegenweidt J, Zankl A, Züchner S, Zemojtel T, Jacobsen JO, Groza T, Smedley D, Mungall CJ, Haendel M, Robinson PN. 2017. The Human Phenotype Ontology in 2017. Nucleic Acids Res 45:D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen CK, Easty R, Di Fenza A, Fiegel T, Grifiths M, Horne A, Karp NA, Kurbatova N, Mason JC, Matthews P, Oakley DJ, Qazi A, Regnart J, Retha A, Santos LA, Sneddon DJ, Warren J, Westerberg H, Wilson RJ, Melvin DG, Smedley D, Brown SD, Flicek P, Skarnes WC, Mallon AM, Parkinson H. 2014. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data Nucleic Acids Res 42:D802–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Lloyd KC. 2014. Conditional targeting of Ispd using paired Cas9 nickase and a single DNA template in mice. FEBS Open Bio 4:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard C, Pendola JK, Hartford SA, Schimenti JC, Handel MA, Eppig JJ. 2004. New mouse genetic models for human contraceptive development. Cytogenet Genome Res 105:222–227. [DOI] [PubMed] [Google Scholar]
- Lewis WR, Malarkey EB, Tritschler D, Bower R, Pasek RC, Porath JD, Birket SE, Saunier S, Antignac C, Knowles MR, Leigh MW, Zariwala MA, Challa AK, Kesterson RA, Rowe SM, Drummond IA, Parant JM, Hildebrandt F, Porter ME, Yoder BK, Berbari NF. 2016. Mutation of Growth Arrest Specific 8 reveals a role in motile cilia function and human disease. PLoS Genet 12:e1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31:681–683. [DOI] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. 2015. Nature 521:520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. 2008. EuroPhenome and EMPReSS: Online mouse phenotyping resource. Nucleic Acids Res 36:D715–D718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks C. 2009. Models of Human Cancers Consortium (MMHCC) from the NCI. Dis Model Mech 2:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya H, Nakai Y, Motegi H, Niinaya N, Kida Y, Kaneko Y, Aritake H, Suzuki N, Ishii J, Koorikawa K, Suzuki T, Inoue M, Kobayashi K, Toki H, Wada Y, Kaneda H, Ishijima J, Takahashi KR, Minowa O, Noda T, Wakana S, Gondo Y, Shiroishi T. 2004. Development and implementation of a database system to manage a large-scale mouse ENU-mutagenesis program. Mamm Genome 15:404–411. [DOI] [PubMed] [Google Scholar]
- Menke DB. 2013. Engineering subtle targeted mutations into the mouse genome. Genesis 51:605–618. [DOI] [PubMed] [Google Scholar]
- Meyer M, de Angelis MH, Würst W, Kühn R. 2010. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA 107:15022–15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mianné J, Chessum L, Kumar S, Aguilar C, Codner G, Hutchison M, Parker A, Mallon AM, Wells S, Simon MM, Teboul L, Brown SD, Bowl MR. 2016. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Zhu QM, Lowenstein CJ. 2016. A CRISPR path to engineering new genetic mouse models for cardiovascular research. Arterioscler Thromb Vasc Biol 36:1058–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock B, Skurla R, Huppi K, D’Hoostelaere L, Klinman D, Mushinski JF. 1987. A restriction fragment length polymorphism at the murine c-myb locus. Nucleic Acids Res 15:4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EMY, Li X, Beutler B. 2013. Going forward with genetics: Recent technological advances and forward genetics in mice. Am J Path 182:1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AP, Welsh CE. 2015. Informatics resources for the Collaborative Cross and related mouse populations. Mamm Genome 26:521–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, Meziane H, Richardson D, Wells S, White J, Wood J EUMODIC Consortium , de Angelis MH, Brown SD, Hancock JM, Mallon AM. 2010. EuroPhenome: A repository for high-throughput mouse phenotyping data. Nucleic Acids Res 38:D577–D585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motenko H, Neuhauser SB, O’Keefe M, Richardson JE. 2015. MouseMine: A new data warehouse for MGI. Mamm Genome 26:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Informatics (MGI) 2016. a. Allele query. Available online (http://www.informatics.jax.org/allele/), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. b. Mammalian phenotype browser. Available online (http://www.informatics.jax.org/searches/MP_form.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. c. MGI batch data and analysis tools. Available online (http://www.informatics.jax.org/batch_data.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. d. MGI batch query form. Available online (http://www.informatics.jax.org/searches/MP_form.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. e. Gene expression database topic page. Available online (http://www.informatics.jax.org/expression.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. f. Gene expression literature index. Available online (http://www.informatics.jax.org/gxdlit), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. g. How to use MGI. Available online (http://www.informatics.jax.org/mgihome/other/homepage_usingMGI.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. h. MGI gene function topic page. Available online (http://www.informatics.jax.org/function.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. i. MGI gene page for Ren2 Available online (http://www.informatics.jax.org/marker/MGI:97899), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. j. MGI gene query. Available online (http://www.informatics.jax.org/marker), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. k. MGI GO download report. Available online (http://www.informatics.jax.org/downloads/reports/index.html#go), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. l. MGI homepage. Available online (www.informatics.jax.org), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. m. MGI marker coordinates report. Available online (http://www.informatics.jax.org/downloads/reports/MGI_MRK_Coord.rpt), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. n. MGI online submission forms page. Available online (www.informatics.jax.org/submit.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. o. MGI statistics. Available online (http://www.informatics.jax.org/mgihome/homepages/stats/all_stats.shtml), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. p. Mouse nomenclature homepage. Available online (http://www.informatics.jax.org/nomen), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. q. Phenotypes topic page. Available online (http://www.informatics.jax.org/allele/), accessed on December 1, 2016.
- Mouse Genome Informatics (MGI) 2016. r. SNPs query. Available online (http://www.informatics.jax.org/snp), accessed on December 1, 2016.
- Mouse Genome Sequencing Consortium Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigó R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. [DOI] [PubMed] [Google Scholar]
- Mouse Models of Human Cancer: The Mouse Tumor Biology Database (MTB) 2016. MTB homepage. Available online (http://tumor.informatics.jax.org), accessed on December 1, 2016.
- Murray SA, Eppig JT, Smedley D, Simpson EM, Rosenthal N. 2012. Beyond knockouts: cre resources for conditional mutagenesis. Mamm Genome 23:587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Balling R, Barsh G, Beier D, Brown SD, Bucan M, Camper S, Carlson G, Copeland N, Eppig J, Fletcher C, Frankel WN, Ganten D, Goldowitz D, Goodnow C, Guenet JL, Hicks G, Hrabe de Angelis M, Jackson I, Jacob HJ, Jenkins N, Johnson D, Justice M, Kay S, Kingsley D, Lehrach H, Magnuson T, Meisler M, Poustka A, Rinchik EM, Rossant J, Russell LB, Schimenti J, Shiroishi T, Skarnes WC, Soriano P, Stanford W, Takahashi JS, Wurst W, Zimmer A International Mouse Mutagenesis Consortium . 2001. Sequence interpretation. Functional annotation of mouse genome sequences. Science 291:1251–1255. [DOI] [PubMed] [Google Scholar]
- Nagy A. 2000. Cre recombinase: The universal reagent for genome tailoring. Genesis 26:99–109. [PubMed] [Google Scholar]
- NCBI CCDS Database 2016. CCDS homepage. Available online (https://www.ncbi.nlm.nih.gov/projects/CCDS/CcdsBrowse.cgi), accessed on December 1, 2016.
- NCBI Resource Coordinators 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms KA, Goodnow CC. 2001. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 15:409–418. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskiy I, Conrad DF, Chun S, Fay JC, Cheverud JM, Lawson HA. 2015. Using whole-genome sequences of the LG/J and SM/J inbred mouse strains to prioritize quantitative trait genes and nucleotides. BMC Genomics 16:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J. 2000. a. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 25:440–443. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Vizor L, Strivens M, Washbourne R, Hough T, Wells C, Glenister P, Thornton C, Martin J, Fisher E, Rogers D, Hagan J, Reavill C, Gray I, Wood J, Spurr N, Browne M, Rastan S, Hunter J, Brown SD. 2000. b. Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm Genome 11:500–506. [DOI] [PubMed] [Google Scholar]
- Pack AI, Galante RJ, Maislin G, Cater J, Metaxas D, Lu S, Zhang L, Von Smith R, Kay T, Lian J, Svenson K, Peters LL. 2007. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics 28:232–238. [DOI] [PubMed] [Google Scholar]
- Panda SK, Wefers B, Ortiz O, Floss T, Schmid B, Haass C, Wurst W, Kühn R. 2013. Highly efficient targeted mutagenesis in mice using TALENs. Genetics 195:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Füllgrabe A, Fuentes AM, Jupp S, Koskinen S, Mannion O, Huerta L, Megy K, Snow C, Williams E, Barzine M, Hastings E, Weisser H, Wright J, Jaiswal P, Huber W, Choudhary J, Parkinson HE, Brazma A. 2016. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 44:D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini N, Knopf JL, Gross KW. 1982. A DNA polymorphism, consistent with gene duplication, correlates with high renin levels in the mouse submaxillary gland. Cell 30:205–213. [DOI] [PubMed] [Google Scholar]
- Portales-Casamar E, Swanson DJ, Liu L, de Leeuw CN, Banks KG, Ho Sui SJ, Fulton DL, Ali J, Amirabbasi M, Arenillas DJ, Babyak N, Black SF, Bonaguro RJ, Brauer E, Candido TR, Castellarin M, Chen J, Chen Y, Cheng JC, Chopra V, Docking TR, Dreolini L, D’Souza CA, Flynn EK, Glenn R, Hatakka K, Hearty TG, Imanian B, Jiang S, Khorasan-zadeh S, Komljenovic I, Laprise S, Liao NY, Lim JS, Lithwick S, Liu F, Liu J, Lu M, McConechy M, McLeod AJ, Milisavljevic M, Mis J, O’Connor K, Palma B, Palmquist DL, Schmouth JF, Swanson MI, Tam B, Ticoll A, Turner JL, Varhol R, Vermeulen J, Watkins RF, Wilson G, Wong BK, Wong SH, Wong TY, Yang GS, Ypsilanti AR, Jones SJ, Holt RA, Goldowitz D, Wasserman WW, Simpson EM. 2010. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci USA 107:16589–16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. 2009. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Liu M, Chen Z, Shao Y, Pan H, Wei G, Yu C, Zhang L, Li X, Wang P, Fan HY, Du B, Liu B, Liu M, Li D. 2013. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res 41:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RH, Crowley MR, Moseley WS, Seldin MF. 1991. Comparison of interspecific to intersubspecific backcrosses demonstrates species and sex differences in recombination frequency on mouse chromosome 16. Mamm Genome 1:158–164. [DOI] [PubMed] [Google Scholar]
- Rhodes M, Straw R, Fernando S, Evans A, Lacey T, Dearlove A, Greystrong J, Walker J, Watson P, Weston P, Kelly M, Taylor D, Gibson K, Mundy C, Bourgade F, Poirier C, Simon D, Brunialti AL, Montagutelli X, Guénet JL, Haynes A, Brown SD. 1998. A high-resolution microsatellite map of the mouse genome. Genome Res 8:531–542. [DOI] [PubMed] [Google Scholar]
- Richardson JE. 2006. fjoin: Simple and efficient computation of feature overlaps. J Comput Biol 13:1457–1464. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Eppig JT, Nadeau JH. 1995. Building an integrated mouse genome database: A view from the front line. IEEE Eng Med Biol 14:718–724. [Google Scholar]
- Ring N, Meehan TF, Blake A, Brown J, Chen CK, Conte N, DiFenza A, Fiegel T, Horner N, Jacobsen JO, Karp N, Lawson T, Mason JC, Matthews P, Morgan H, Relac M, Santos L, Smedley D, Sneddon D, Pengelly A, Tudose I, Warren JW, Westerberg H, Yaikhom G, Parkinson H, Mallon A. 2015. A mouse informatics platform for phenotypic and translational discovery. Mamm Genome 26:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Eppig JT. 2011. Mouse mutants and phenotypes: Accessing information for the study of mammalian gene function. Methods 53:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L. 1989. Plan for genome centers sparks a controversy. Science 246:204–205. [DOI] [PubMed] [Google Scholar]
- Roderick TH, Davisson MT. 1985. The Mouse Linkage Map. Mouse News Letter.
- Roscoe B Jackson Memorial Laboratory Staff 1945. A chromosome map of the mouse. J Hered 36:271–274. [Google Scholar]
- Rosen B, Schick J, Wurst W. 2015. Beyond knockouts: The International Knockout Mouse Consortium delivers modular and evolving tools for investigating mammalian genes. Mamm Genome 26:456–466. [DOI] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MS, Thurston SJ, Birkenmeier EH. 1994. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome 5:253–274. [DOI] [PubMed] [Google Scholar]
- Royer N. 2015. The history of fancy mice. American Fancy Rat & Mouse Association. Available online (http://www.afrma.org/historymse.htm), accessed on September 20, 2016.
- Russell WL, Hunsicker PR, Carpenter DA, Cornett CV, Guinn GM. 1982. a. Effect of dose fractionation on the ethylnitrosourea induction of specific-locus mutations in mouse spermatogonia. Proc Natl Acad Sci USA 79:3592–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL, Hunsicker PR, Raymer GD, Steele MH, Stelzner KF, Thompson HM. 1982. b. Dose—response curve for ethylnitrosourea-induced specific-locus mutations in mouse spermatogonia. Proc Natl Acad Sci USA 79:3589–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Iulianella A, Melton KR, Lynn M, Walker M, Inman KE, Bhatt S, Leroux-Berger M, Crawford M, Jones NC, Dennis JF, Trainor PA. 2011. A phenotype-driven ENU mutagenesis screen identifies novel alleles with functional roles in early mouse craniofacial development. Genesis 49:342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85:5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. 2003. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21:562–565. [DOI] [PubMed] [Google Scholar]
- Schriml LM, Mitraka E. 2015. The Disease Ontology: Fostering interoperability between biological and clinical human disease-related data. Mamm Genome 26:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang SJ, Worthey E, Dwinell M, Jacob H. 2015. The Rat Genome Database 2015: Genomic, phenotypic and environmental variations and disease. Nucleic Acids Res 43:D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JD, Hilton DJ, Bahlo M, Kile BT. 2007. Probabilistic analysis of recessive mutagenesis screen strategies. Mamm Genome 18:5–22. [DOI] [PubMed] [Google Scholar]
- Simon MM, Moresco EM, Bull KR, Kumar S, Mallon AM, Beutler B, Potter PK. 2015. Current strategies for mutation detection in phenotype-driven screens utilizing next generation sequencing. Mamm Genome 9–10:486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. 2015. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. 2009. JBrowse: A next-generation genome browser. Genome Res 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Salimova E, Rosenthal N. 2011. Cre recombinase resources for conditional mouse mutagenesis. Methods 53:411–416. [DOI] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. 2012. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome 23:653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. 2015. Expanding the mammalian phenotype ontology to support automated exchange of high throughput mouse phenotyping data generated by large-scale mouse knockout screens. J Biomed Semant 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Eppig JT, Kadin JA, Richardson JE, Ringwald M. 2015. GXD: A community resource of mouse Gene Expression Data. Mamm Genome 7–8:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Lyne M, Lyne R, Kalderimis A, Rutherford K, Stepan R, Sullivan J, Wakeling M, Watkins X, Micklem G. 2012. InterMine: A flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 28:3163–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soewarto D, Fella C, Teubner A, Rathkolb B, Pargent W, Heffner S, Marschall S, Wolf E, Balling R, Hrabé de Angelis M. 2000. The large-scale Munich ENU-mouse-mutagenesis screen. Mamm Genome 11:507–510. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Bogue MA, Peters LL. 2003. Invited review: Identifying new mouse models of cardiovascular disease: a review of high-throughput screens of mutagenized and inbred strains. J Appl Physiol 94:1650–1659. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. 2012. High-resolution genetic mapping using the Mouse Diversity Outbred population. Genetics 190:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. 2012. Ten years of the collaborative cross. G3 2:153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ. 2016. Cre-Lox neurogenetics: 20 years of versatile applications in brain research and counting…. Front Genet 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J, Myer VE, Hsu PD. 2016. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell 63:355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium 2017. UniProt: The universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Deimling OH, Forejt J, Wienker TF. 1988. Allelic profile at 37 biochemical loci of two inbred strains of the house mouse derived from wild Mus musculus musculus. Lab Anim 22:61–66. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML, D’Eustachio P, Mock BA, Steinberg AD, Morse HC 3rd, Oakey RJ, Howard TA, Rochelle JM, Seldin MF. 1992. A linkage map of mouse chromosome 1 using an interspecific cross segregating for the gld autoimmunity mutation. Mamm Genome 2:158–171. [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC Sanger Institute Mouse Genetics Project , Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles MV, Qin W, Cheng AW, Wang H. 2015. CRISPR-Cas9-mediated genome editing and guide RNA design. Mamm Genome 26:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. 2013. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13:659–662. [DOI] [PubMed] [Google Scholar]
- Xie G, Clapcote SJ, Nieman BJ, Tallerico T, Huang Y, Vukobradovic I, Cordes SP, Osborne LR, Rossant J, Sled JG, Henderson JT, Roder JC. 2007. Forward genetic screen of mouse reveals dominant missense mutation in the P/Q-type voltage-dependent calcium channel, CACNA1A. Genes Brain Behav 6:717–727. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Bhomra A, Goodson M, Keane TM, Adams DJ, Flint J. 2012. The fine-scale architecture of structural variants in 17 mouse genomes. Genome Biol 13:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32:551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Shen Y, Chatterjee B, Siegfried BH, Leatherbury L, Rosenthal J, Lucas JF, Wessels A, Spurney CF, Wu YJ, Kirby ML, Svenson K, Lo CW. 2004. ENU induced mutations causing congenital cardiovascular anomalies. Development 131:6211–6223. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jia R, Palange NJ, Satheka AC, Togo J, An Y, Humphrey M, Ban L, Ji Y, Jin H, Feng X, Zheng Y. 2015. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One 10:e0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Chen Y, Li Y, Chen R, Mardon G. 2015. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep 5:8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, Leng Q, Huang X. 2014. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 46:49–55. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JE, Hale P, Baldarelli RM, Reed DJ, Recla JM, Sinclair R, Reddy TB, Bult CJ. 2015. A unified gene catalog for the laboratory mouse reference genome. Mamm Genome 26:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]